The glycosylation profile for IgG and PBMC and their associations with AITD and one of its biomarkers, TPOAb, have been determined in three independent European cohorts by applying couple of different assays, including HILIC-UPLC, Lectin-probed Western blotting, tandem mass-spectrometer, RNA-Seq, GWAS. The results reported reveal a decreased level of IgG core-fucosylation and PBMC antennary α1,2 fucosylation with TPOAb level and AITD and a possible role of the glycosylation of TPOAb driven by tissue-specific FUT8 and IKZF1 expression in AITD patients.
Keywords: Immunology, glycosylation, antibodies, glycoproteins, patient cohorts, antibody-dependent cell-mediated cytotoxicity (ADCC), autoimmune thyroid disease, fucosylation, IgG, thyroid peroxidase antibodies (TPOAb)
Graphical Abstract
Highlights
Core fucosylation of IgG and antennary α1,2 fucosylation of peripheral blood mononuclear cells (PBMCs) is negatively associated with AITD and TPOAb positivity.
An association of the decrease in the core fucosylation with two enzymes: IKZF1 and FUT8.
No relevant gene mutation falling within the genes for those enzymes or general transcription deregulation in plasma of patients were observed.
Suggestion that alteration of gene expression happens in subgroup of cells, such as cells from thyroid germinal centers previously identified in patients with AITD.
Abstract
Autoimmune thyroid diseases (AITD) are the most common group of autoimmune diseases, associated with lymphocyte infiltration and the production of thyroid autoantibodies, like thyroid peroxidase antibodies (TPOAb), in the thyroid gland. Immunoglobulins and cell-surface receptors are glycoproteins with distinctive glycosylation patterns that play a structural role in maintaining and modulating their functions. We investigated associations of total circulating IgG and peripheral blood mononuclear cells glycosylation with AITD and the influence of genetic background in a case-control study with several independent cohorts and over 3,000 individuals in total. The study revealed an inverse association of IgG core fucosylation with TPOAb and AITD, as well as decreased peripheral blood mononuclear cells antennary α1,2 fucosylation in AITD, but no shared genetic variance between AITD and glycosylation. These data suggest that the decreased level of IgG core fucosylation is a risk factor for AITD that promotes antibody-dependent cell-mediated cytotoxicity previously associated with TPOAb levels.
Autoimmune thyroid diseases (AITD)1 are a class of chronic, organ-specific autoimmune disorders that disturb the function of the thyroid gland. They affect close to 5% of the European population (with a gender disparity) and so, represent the most common group of autoimmune diseases (1). AITD encompass a spectrum of conditions including Hashimoto's thyroiditis (HT) and Graves' disease (GD). One of the features of AITD is the production of autoantibodies against components of thyroid cells that are also detected in the bloodstream.
The autoantibodies are produced against the three core thyroid proteins: thyroid peroxidase (TPO), thyroglobulin (Tg), and the thyroid-stimulating hormone receptor (TSHR). Except for antibodies against TSH receptors (TSAb), which are known to stimulate the production of thyroid hormones by binding TSH receptors in GD (2), little is known about the role of two other thyroid autoantibodies, thyroid peroxidase antibodies (TPOAb) and thyroglobulin antibodies (TgAb). Circulating TPOAb is the most common and diagnostically useful marker of AITD, detectable in the serum of most HT (95%) and GD (85%) patients (3). In recent years, using TPOAb as a marker has been challenged because it appears in ∼10% of apparently healthy individuals (4). Even though autoantibodies are often a hallmark of autoimmune disorders, they can appear years before the first symptoms (5), which poses the question about their causative role. Some evidence exists that autoantibodies can trigger autoimmunity, and IgG isotype seems to be connected with the development of autoimmune diseases potentially through regulation of IgG effector functions by alternative glycosylation (5). However, it is yet to be determined if anti-thyroid antibodies cause AITD, or whether additional control mechanisms, such as post-translational modifications, are required to trigger the disease onset.
The most abundant and diverse form of post-translational modification is glycosylation, the attachment of sugar moieties to proteins, and various glycans are involved in virtually all physiological processes (6). Glycans attached to Immunoglobulin G (IgG) are indispensable for its effector function and control of inflammation (7–10). There are two glycosylation sites within the fragment crystallizable (Fc) of IgG that affect the molecule's 3D-conformation and affinity for binding to Fcγ-receptors (FcγRs) on a range of immune cells (6, 11, 12). Additional N-glycosylation sites are present in ∼20% of IgG fragment antigen binding (Fab) and play a role in immunity, such as the affinity of epitope-binding site (13–16). Previous analysis of IgG glycosylation with other autoimmune diseases showed a reduction of IgG galactosylation and sialylation, which trigger inflammatory response (17–20). In relation to AITD, two small studies (62 and 146 patients respectively) looked at the TgAb glycosylation and reported differences between AITD, papillary thyroid cancers, and controls (21, 22). First, TgAb IgG from HT patients showed higher levels of core fucose than the control group, as well as of terminal sialic acid and mannose (21). On the other hand, it was observed that among HT, GD, and papillary thyroid cancer groups, HT patients had significantly lower core fucose content on TgAb than the other two groups (22). Furthermore, because recent genome-wide association studies (GWAS) identified novel loci associated with IgG glycosylation, which were known to be strongly associated with autoimmune conditions (23, 24) and the heritability of AITD is estimated to 55–75% (25–27), the next logical step was to examine if there was any common genetic background between those features. No data is currently available on common genetic variants associated with IgG glycosylation traits and AITD, and no large study on associations of the glycosylation of total IgG with the level of thyroid autoantibodies or with AITD has been performed.
Our goal was to determine if there are any IgG or peripheral blood mononuclear cells glycan structures associated with the AITD or TPOAb positivity and examine if there are any common heritable factors between AITD and glycan structures. We investigated the association of total serum or plasma IgG glycome composition with TPOAb level in two cohorts: TwinsUK discovery cohort of 2297 individuals (988 controls, 1309 TPOAb positive) and Croatian replication cohort (73 controls, 90 TPOAb positive). We then focused our analysis of the IgG glycosylation on individuals with AITD in discovery cohort (988 controls, 203 AITD), and compared them to Polish replication cohort (114 control, 105 HT). In a fraction of the samples from Polish cohort we analyzed peripheral blood mononuclear cells glycosylation. Additionally, we looked for possible common genetic background between AITD and N-glycans, and at the gene expression of relevant genes. In total, we found an association between the decreased level of IgG core fucosylation and PBMCs antennary α1,2 fucosylation with TPOAb level as well as with AITD. We observed the association of significantly affected IgG N-glycan traits with FUT8 and IKZF1 genes (responsible for IgG fucosylation), but we could not identify SNPs or a general dysregulation of gene expression in whole blood; suggesting a restricted dysregulation of glycosylation in a subpopulation of B cells.
EXPERIMENTAL PROCEDURES
Study Sample for the TwinsUK Cohort - The Discovery Cohort
The discovery sample consisted of twins from the UK Adult Twin Registry (TwinsUK cohort). The TwinsUK cohort is comprised of virtually 12,000 monozygotic and dizygotic twins unselected for any disease or trait. The cohort is from Northern European/UK ancestry and has been shown to be representative of singleton populations and the UK population in general (29) (30). The project was approved by the local Ethics Committee, and informed consent was obtained from all participants.
A more detailed description of the subjects and analyses performed on the TwinsUK cohort can be found in the supplemental Table S1. TwinsUK glycomic, transcriptomic, genetic data and GWAS results are publicly available upon request on the department website (http://www.twinsuk.ac.uk/data-access/accessmanagement/).
The blood collected for the detection of different biological markers (TPOAb levels, plasma IgG N-glycan traits, transcriptomic data, and genetic variants) may have come from different time points for the same individuals between 1997 and 2013. As aging is an important modulator of IgG glycosylation (31) and evolution of AITD, we restricted our sample collection/selection protocol in such a way that the largest absolute time difference between different samples collected from the same individuals is five years. This was performed in order to have enough subjects, but with a weak, potential aging effect between two samples. The criteria are not used on genomic data as it is almost stable relative to aging. For the current study, only twins who have phenotype data collected within the same year as the collection of plasma used for the IgG glycosylation profiling were selected in order to account for the potential effects of aging. Consequently, only 2,297 twins have glycan data and TPOAb level measured by Roche immunoassay and 392 twins from Abbott immunoassay (Table I.1 in supplemental Table S1).
The blood to assess gene expression has been collected between 2009 and 2011. For the current analysis, only twins who have phenotype data collected within a 5-year range around the collection of blood used for the gene expression were selected in order to account for the potential effects of aging. Consequently, gene expression data was collected in only 180 twins that have gene expression data and AITD status, 199 twins with TPOAb level and 326 twins with IgG N-glycan traits (Table I.2 in supplemental Table S1).
Study Sample for the Polish and Croatian Cohorts
Two other cohorts were used to test the findings obtained in the TwinsUK cohort. Both cohorts are described in the Table I.1 in supplemental Table S1. The recruitment in the two replicated cohorts was performed to have age and gender-matched population between cases and controls.
Croatian replication cohort includes 73 control individuals and 90 case individuals from Croatia. Individuals were considered as a control if they have TPOAb and TgAb levels within reference ranges (0–4.11 IU/ml for TPOAb and 0–5.61 IU/ml for TgAb). On the other hand, subjects were considered as case individuals if they have a high level of TPOAb or TgAb (with the antibody level at least double the upper limit of its reference range) and with normal complete blood count. The blood was collected via venous puncture for the detection of different biological markers from March to June 2015. The study was approved by the Ethics Committee of Genos Ltd. Informed consent for participation in the study was obtained from all participants.
The second replication cohort consisted of patients from the Endocrinology Department of the University Hospital in Krakow and Jagiellonian University staff, Poland, and included 219 individuals (105 HT patients and 114 healthy donors)—a cohort of mainly females (103 HT and 106 in control group) with an average age of 36 (Table I.1 supplemental Table S1). All HT patients have been on levothyroxine sodium substitutive therapy and were diagnosed based on a high TSH level and the TPOAb and/or TgAb positivity. Moreover, patients do not have any other concomitant autoimmune diseases, other thyroid diseases and history of cancers. The control group consisted of 114 individuals with a negative history of HT, other autoimmune diseases, other thyroid diseases and cancers with negative/low antibody titers of TPOAb, TgAb and anti-TSHR and TSH level within the reference range. Having a small number of men in this cohort, we ran an analysis of IgG N-glycan traits with only data from women. After quality control of data obtained after IgG glycome analysis, 106 control subjects and 103 HT patients were analyzed concerning IgG glycosylation profiling and to keep the age and gender match. 24 blood samples from 219 collected from the Polish replication cohort were also used to analyze PBMC glycosylation: 11 patients with HT and 13 healthy donors. The groups were matched for sex and age. Informed consent for participation in the study was obtained from all participants. The blood was collected via venous puncture for the detection of different biological markers from May 2014 to July 2015. The study was approved by the Ethics Committee of the Jagiellonian University Medical College in Krakow.
Detection of TSH and TPOAb and Definition of AITD in TwinsUK
The study was performed using TPOAb level as a continuous variable and a clinical AITD definition as a binary trait. Not having a clinical diagnostic for all twins, TPOAb level, and TSH level were used to define individuals with AITD and healthy groups. Individuals were considered to have AITD if they had a positive TPOAb titer (3 times more than the threshold set by the manufacturer (18 IU/ml for Abbott assay and 100 IU/ml for Roche assay)) or had a TSH level more than 10 mIU/L. We considered subjects to have HT when they have a TSH level ≥10 mIU/L or a positive TPOAb titer as decribed previously with TSH level more than 4 mIU/L. We considered individuals as heathy controls if they had a TSH level in the normal range and a negative TPOAb titer, with no previous clinical diagnosis of thyroid disease and not treated with thyroid medications or steroids. Individuals with a history of thyroid cancer or thyroid surgery (because of benign conditions) were excluded.
The sera to assess TPOAb and TSH levels have been collected by a trained nurse or phlebotomist using venipuncture and a SafetyLokTM Blood Collection Kit (21G3/4 Needles) and plain 10 ml serum-separating tube vacutainer (no additives) during twin visits. After collection from the study subject, whole blood was held at 22 °C for 50 min at room temperature for a clot to form and serum separated within 60 min of collection. Processing of blood was performed using a refrigerated (4 °C) clinical centrifuge at 3000 × g for 10 min with the serum supernatant subsequently collected, transferred to a 2 ml screw capped Nunc Cryotubes and immediately frozen at −80 °C and kept frozen in 2 ml screw capped Nunc Cryotube at −80 °C until use. The quantitative determination of TSH and TPOAb (only IgG class) levels was done on the sera by a chemiluminescent microparticle immunoassay (ARCHITECT® Anti-TPO or TSH (ABBOTT Diagnostics Division, Wiesbaden, Germany, 2005)) (a TPOAb titer >6 IU/ml considered positive; normal range of TSH level is between 0.4 mIU/L and 4 mIU/L). For samples analyzed by an electrochemiluminescence immunoassay “ECLIA” (Elecsys and cobas e analyzers, (Roche Diagnostics, Indianapolis, lN, 2010)) (a TPOAb titer >34 IU/ml considered positive; normal range of TSH level is between 0.4 mIU/L and 4 mIU/L).
Detection of Thyroid Hormones, TSH, TgAb and TPOAb Levels for Croatian and Polish Cohorts
Sera samples for Croatian cohort were collected via venous puncture from March to June 2015 into 10 ml serum-separating vacutainer tube without additives, centrifuged at 3500 rpm for 10 min, after which the serum was transferred to 2 ml Eppendorf tubes and kept frozen at −20 °C until use. Quantitative determination of TSH, free T3, free T4, TPOAb and TgAb was performed by a chemiluminescent micro-particle immunoassay (ARCHITECT® TSH, free T3, free T4, Anti-TPO or Anti-Tg (ABBOTT Diagnostics Division, Wiesbaden, Germany, 2015)).
Sera and PBMCs for Polish cohort were collected via venous puncture for the detection of different biological markers from May 2014 to July 2015 using S-Monovette Serum Tubes 2.7 ml that contains Clotting Activator/Serum (Sarstedt, 05.1557.001) for collection. Blood samples were left at room temperature for five hours for blood coagulation and centrifuged at 1200 × g for 10 min at 4 °C. Serum samples were stored in deep freezing in Safe Lock Tubes 2 ml (Eppendorf, 0030123.344). The TSH level in sera samples were determined by immunoradiometric assay (IRMA) (DIAsource TSH-IRMA Kit (DIAsource ImmunoAssays S.A., Louvain-la-Neuve, Belgium)). Anti-TPO and anti-Tg levels were measured by radioimmunoassays (RIAs) (BRAHMS anti-TPOn RIA, BRAHMS anti-Tgn RIA (BRAHMS GmbH, Hennigsdorf, Germany)). The sera, as well as PBMC samples, were kept frozen at −70 °C until the analysis of glycosylation.
IgG Glycosylation Profiling
The IgG glycosylation profiling was applied on total IgG glycome from blood (combined Fc and Fab glycans and all IgG subclasses). It is noted that glycosylation patterns of total IgG reflect mostly IgG Fc glycans because of the small proportion of IgG Fab glycans in the total IgG N-glycan structures (16). It was performed in Genos Glycoscience Research Laboratory in Croatia using UPLC analysis of 2AB-labeled glycans. The IgG glycosylation of the discovery cohort was performed in four batches whereas the replications cohorts were performed in one batch per cohort. However, the protocol for replication cohorts followed the same protocol than the protocol used for the two last batches of the discovery cohort and described below. Effects of technical confounders (96-well plate and run-day batch effects) are minimized by careful experimental design (blocking on case-control status, sex and age, whereas the rest was randomized) (32). Additionally, batch-effects are controlled by introducing in-house plasma standards to each 96-well plate (32).
Isolation of Total IgG from Human Blood
The IgG was isolated using 96-well protein G monolithic plates (BIA Separations, Ljubljana, Slovenia) as described previously (33). Briefly, blood samples (70–100 μl) were diluted eight times with phosphate buffered saline (PBS), pH 7.4, and applied to protein G monolithic plates. Following this, IgGs were instantly washed with PBS, pH 7.4 and eluted with 1 ml of 0.1 m formic acid (Merck, Darmstadt, Germany), pH 2.5 and neutralized with 1 m ammonium bicarbonate (Merck) to pH 7.0. IgG concentration in eluates was measured using Nanodrop 8000 Spectrophotometer (Thermo Scientific, Waltham, MA, SAD), with coefficient variations for standard samples being below 15% within each cohort.
IgG N-glycan Structure Release and Labeling
Volume of 300 μl of isolated IgG eluate was dried and IgG samples were dissolved in 30 μl 1.33% SDS (w/v) (Invitrogen, Carlsbad, CA) and denatured by incubation at 65 °C for 10 min. After incubation, samples were left to cool down to room temperature for 30 min. Afterward, ten μl of 4% Igepal CA-630 (Sigma-Aldrich, St. Louis, MO) was added to the samples and incubated on a shaker for 15 min. Glycans were released by adding 1.25 mU of PNGase F (ProZyme, Hayward, CA) in 10 μl 5 × PBS to the samples, followed by overnight incubation at 37 °C. The released N-glycans were labeled with 2-aminobenzamide (2-AB) fluorescent dye. The labeling mixture was freshly prepared by dissolving 2-AB (Sigma-Aldrich, St. Louis, MO) in DMSO (Sigma-Aldrich, St. Louis, MO) and glacial acetic acid (Merck, Darmstadt, Germany) mixture (70:30, v/v) and by adding 2-picoline borane (Sigma-Aldrich, St. Louis, MO) to a final concentration of 19.2 mg/ml for 2-AB and 44.8 mg/ml for 2-picoline borane. A volume of 25 μl of labeling mixture was added to each N-glycan sample in the 96-well plate, and the plate was sealed using the adhesive seal. Mixing was achieved by shaking for 10 min, followed by 2h incubation at 65 °C.
Clean-up of 2-AB Labeled IgG N-glycan Structures
The two first batches of the TwinsUK cohort and batches of Croatian and Polish cohorts were cleaned up using the protocol described previously (31) whereas the two last batches of the TwinsUK cohort were prepared as described below.
Free fluorescent label, an excess of reagents and proteins were removed from the samples using (HILIC-SPE). After cooling down to room temperature for 30 min, 700 μl of acetonitrile (previously cooled down to 4 °C, Fluka) was added to each sample. Clean-up procedure was performed on hydrophilic 0.2 μm AcroPrep GHP filter plate. The solvent was removed by application of vacuum using a vacuum manifold at around 25 mm Hg. All wells were prewashed with 200 μl 70% ethanol, 200 μl ultrapure water and 200 μl 96% acetonitrile in water that was previously cooled down to 4 °C. The samples were loaded into the wells, and after short incubation subsequently washed 5 × 200 μl acetonitrile in water (ϕ = 96%, previously cooled down to 4 °C). Glycans were eluted with 2 × 90 μl of ultrapure water after 15 min shaking at room temperature, and combined eluates were stored at −20 °C until ultra-performance liquid chromatography (UPLC) analysis.
IgG N-glycan Structures Hydrophilic Interaction Chromatography (HILIC)-UPLC
Fluorescently labeled N-glycans were separated by HILIC on a Waters Acquity UPLC instrument (Waters, Milford, MA) as previously described (31) consisting of a quaternary solvent manager, sample manager and an FLR fluorescence detector set with excitation and emission wavelengths of 250 and 428 nm, respectively. The instrument was under the control of Empower 3 software, build 3471 (Waters, Milford, MA). Labeled N-glycans were separated on a Waters BEH Glycan chromatography column, 100 × 2.1 mm i.d., 1.7 μm BEH particles, with 100 mm ammonium formate, pH 4.4, as solvent A and acetonitrile as solvent B. Separation method used linear gradient of 75–62% acetonitrile (v/v) at flow rate of 0.4 ml/min in a 25 min analytical run. Samples were maintained at 10 °C before injection, and the separation temperature was 60 °C. The system was calibrated using an external standard of hydrolyzed and 2-AB labeled glucose oligomers from which the retention times for the individual glycans were converted to glucose units. Data processing was performed using an automatic processing method with a traditional integration algorithm after which each chromatogram was manually corrected to maintain the same intervals of integration for all the samples. The chromatograms were all separated in the same manner into 24 peaks, and the amount of glycans in each peak was expressed as a percentage of the total integrated area. As many of these structures share the same structural features (galactose, sialic acid, core-fucose, bisecting N-acetylglucosamine (GlcNAc)), 53 additional derived traits were calculated that average these features across multiple glycans (supplemental Table S2).
Data Pre-processing and Normalization of IgG N-glycan Structures
Before any transformation and standardization of data, we excluded the glycan called GP3 because of its co-elution with a contaminant that significantly affected its values in some samples. We combined two glycan peaks (GP) GP20 and GP21 (Zagreb code) into a single trait called GP2021 (Zagreb code) because of the difficulty to distinguish these two peaks well in some samples (33). Because of right-skewness of the distribution of IgG N-glycan structures, a global normalization (34) and natural logarithm transformation were applied on 22 directly measured glycan structures. We estimated 53 derived glycan levels from the 22 normalized and non-transformed directly measured glycans using R package called glycanr (35) and we then applied natural logarithm transformation on the 53 new derived glycan traits. Following this, we removed the technical confounders (batch and run-day effects) using ComBat (36) developed in R package sva (37). The 22 directly measured glycan structures and 53 derived glycan traits were centered and scaled to have a mean of 0 and standard deviation (S.D.) of 1. Samples being more than 6 S.D. from the mean were considered as outliers and excluded from the analysis. Additionally, batch-effects are controlled by introducing in-house plasma standards to each 96-well plate. Average coefficient variation of directly measured IgG N-glycans in standard samples of the largest cohort (Twins batch 4) was below 12%, with 10 most abundant glycans having average CV below 7%. For the two replicate cohorts, average coefficient variations were below 10%.
Adjustment of Covariates for IgG N-glycan Traits
For the discovery and replication cohorts, we adjusted IgG N-glycan traits for age, age squared, sex and interaction between sex and age, as they are known biological covariates, using a linear model. The residuals of IgG N-glycan traits were used for the analysis described here.
IgG and PBMC Glycosylation Profiling Using Lectin-probed Western Blotting
Lectin-probed Western blotting (LB) for AAL was performed to IgG glycosylation analysis in three batches whereas LB for AAL, GNA, MAL-II, PHA-L, PHA-E, SNA and UEA-I was used to assess the glycosylation features from PBMCs of participants described in supplemental Table S1 (one batch per lectin assay, only for UEA I two batches).
Isolation of PBMCs
PBMCs were isolated from heparinized blood sampled from the patients with HT and the healthy donors by density-gradient centrifugation at 1600 rcf for 15 min at 4 °C over Gradisol L. The collected cells were washed twice with PBS and lysed in RIPA buffer containing protease inhibitors for 45 min at 4 °C. The cell homogenates were centrifuged at 21,000 rcf for 10 min at 4 °C. The supernatants containing pure protein extracts were collected, and protein concentration was assayed using Lowry method modified by Peterson (38).
Lectin Blotting
The glycosylation of IgGs and PBMCs was analyzed using biotinylated lectins in LB previously described (39). LB is also considered for quantitative analysis of glycosylation as described in previous studies (40, 41). The lectin specificity was determined based on the glycoprotein reaction with lectins blocked by the inhibiting sugars or 200 mm acetic acid for 24 h at 4 °C.
Experimental Design and Statistical Rationale
Tandem mass spectrometry (MS/MS) (Orbitrap Velos Pro system, Thermo Scientific, San Jose, CA) was used to identify glycoproteins in the band marked the red frame in Fig. 2D in the manuscript, according to Ochwat et al. (42) with minor modifications. We selected three samples, one Hashimoto thyroiditis donor and two healthy donors, as representatives of our Polish cohort (biological replicates). We did not perform any technical replicate. Proteins extracted from PBMC isolated from the blood of Hashimoto thyroiditis and healthy donors were separated on 10% SDS-PAGE gel. Coomassie Brilliant Blue-stained bands were cut out based on the UEA-I reaction performed on PVDF membrane. Proteins in the excised pieces of gels were reduced with 10 mm dithiothreitol (DTT) for 30 min at 56 °C and then alkylated with iodoacetamide (IAA) in darkness for 45 min at room temperature. Then samples were subjected to the standard procedure of trypsin digestion by overnight incubation with trypsin in the concentration 10 ng/μl and analyzed by MS/MS in the Mass Spectrometry Laboratory (Institute of Biochemistry and Biophysics, Warsaw, Poland). The number of missed cleavages was set to 0 or 1. Raw data files were pre-processed with Mascot Distiller software (version 2.4.2.0, MatrixScience). The obtained peptide masses and fragmentation spectra were matched to the Sprot non-redundant database (version 2015_01, 547,357 sequences, 194,874,700 residues) with a Homo sapiens filter, reducing to 20,274 sequences, using the Mascot search engine (Mascot Daemon v. 2.4.0, Mascot Server v. 2.4.1, MatrixScience). Spectra were searched using a ± 0.1 Da mass tolerance for fragment ions mass and 30 ppm for mass tolerance of precursor ions. Oxidation of methionine and carboxymethylation of lysine was set as a variable modification. Protein identification was performed using the Mascot search engine (MatrixScience) with the probability-based algorithm. Protein scores are derived from ions scores as a non-probabilistic basis for ranking protein hits. Ions score is −10*Log(P), where P is the probability that the observed match is a random event. Data were searched with automatic decoy database and were filtered to obtain a false discovery rate below 1% (UEA1: p value< 0.05, Individual ions scores > 27; UEA2 p value <0.00075; Individual ions scores > 45; UEA3: p value< 0.00065, Individual ions scores > 46) (43). Because the aim of the MS/MS analysis was the identification of glycoprotein(s) responsible for the altered UEA-I reaction, only glycosylated proteins were selected among all identified proteins based on their descriptions in Uniprot database using protein ID. Based on the relative molecular weight of UEA-I-positive protein(s) determined in SDS-PAGE (120 kDa) only proteins with appropriate molecular mass were chosen.
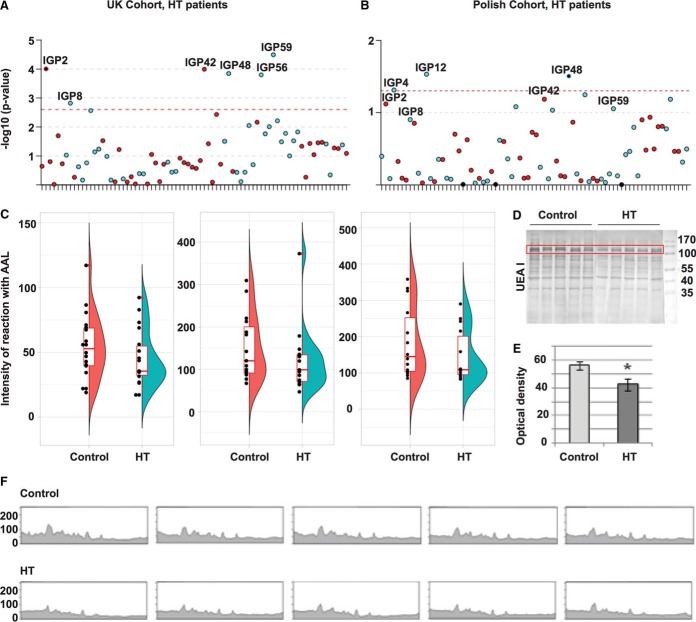
Fig. 2.
Glycosylation analysis of the human IgG and PBMC proteins from control and HT groups. A, Manhattan plot of 75 IgG N-glycan traits for the HT patients of the UK discovery cohort (supplemental Table S3 - Sheet 3). The Manhattan plot is drawn with colors corresponding to the direction of associations (blue = negative, red = positive). The red dashed line corresponds to the level of significance in the discovery cohort (p value≤2.5 × 10−3). Image created with R package called coMET. B, Manhattan plot of 75 IgG N-glycan traits for the HT Polish replication cohort (Sheet 5 of supplemental Table S3). The Manhattan plot is drawn with colors corresponding to the direction of association (blue = negative, red = positive). The red dashed line corresponds to the level of significance p value in the replication cohort (0.05). Image created with R package called coMET. C, Reduction (not significant), of the binding of AAL to IgG core fucose in HT patients compared with control healthy individuals from the Polish cohort in the three batches using lectin blotting assay (p value>0.05; the number of samples per group in each batch is 18, 15, 14 respectively); (supplemental Table S4). The plot combines a flat violin plot, box plot with whiskers and different data. A violin plot is a hybrid of box plot and kernel density plot. Box plot with whiskers represents five summary statistics (the median, the first and third quartiles for two hinges and 1.5 times interquantile range from the hinges for two whiskers). Outliers are labeled by the red dot adjacent to the black dot of the measurement. Image created with R package called ggplot2. D, PBMC protein extracts were resolved on 10% SDS-PAGE gels in reducing conditions. After electrophoresis, the proteins were electrotransferred to a PVDF membrane and stained with the UEA I lectin (ncase = 9 and ncontrol = 10; supplemental Table S5). The molecular markers are shown on the last lane in kilo Dalton (kDa) (PageRuler Prestained Protein Ladder, 26616, Thermo Scientific). E, Densitometric measurement of the UEA I-positive protein marked in the red frame of Fig. 2D. Duncan's test was used to compare mean values of optical density of the lectin binding. Statistically significant difference is marked with an asterisk (p value<0.05). f) The lectin staining of Fig. 2D measured densitometrically.
Statistical Analysis
The intensity of lectin staining was measured by densitometry analysis of all PBMC proteins in a sample reacted with the lectin using UVImap Image Quantification software (UVItec, Cambridge, UK) and was normalized to CBB staining. Because of the use of stain-free gels (Bio-Rad), the intensity of IgG reaction with lectin was normalized to IgG protein level. Data were plotted as means ± standard deviation. Mann-Whitney U test was used to compare mean values of optical density of the lectin reaction intensity between HT and healthy donor samples whereas Levene's test was used to compare variance. p value<0.05 values were recognized significant.
Detection of Gene Expression in Whole blood
The gene expression in whole blood was measured on 384 female twins from the TwinsUK Adult twin registry. In the present study, 180 twins have both RNA-Seq data and AITD status data collected within five years, 199 individuals for TPOAb level and 326 for IgG N-glycan structures. The protocol was described previously (44). Briefly, after preparing for sequencing with the Illumina TruSeq sample preparation kit according to the manufacturer's instructions, samples were sequenced on a HiSeq 2000 machine (Illumina, San Diego, CA). The 49-bp sequenced paired-end reads were mapped to the GRCh37 reference genome with the BurrowsWheeler Aligner (BWA) v0.5.9. GENCODE 10 annotation was used to define exons of genes identified as protein coding. All overlapping exons of a gene were merged into meta-exons.
Genotyping for the Discovery Cohort
Genotyping of the TwinsUK data set was done with a combination of 4 Illumina arrays (HumanHap300 (45, 46), HumanHap610Q, 1 m-Duo and 1.2MDuo 1 m; Illumina). After quality control steps, the merged data set consists of 5654 individuals (2040 from the HumanHap300, 3,461 from the HumanHap610Q and 153 from the HumanHap1M and 1.M arrays) and up to 874,733 SNPs depending on the data set (HumanHap300: 303,940, HumanHap610Q: 553,487, HumanHap1M and 1.M: 874,733). Then samples were imputed using the IMPUTE software package (v2) (47) using two reference panels, P0 (HapMap2, rel. 22, combined CEU+YRI+ASN panels) and P1 (610k+, including the combined HumanHap610k and 1 m reduced to 610k SNP content).
Statistical Methods
All statistical analyses were run using R version 3.2.3. Linear mixed effect models were performed in using R lme of package lme4 (48), and linear models were performed in using R function lm of package stat.
Determination of Effective Number of Independent Tests for Glycomic Data
Because of high and partial correlations between glycans, we decided to use the method five proposed by Li & Ji (2005) (49) to define an effective number (Meff) of independent tests. We then used this number to determine the effective Bonferroni p value threshold such as 0.05/Meff instead of 0.05/M. We then used the effective number to set the effective Bonferroni p value threshold. 20 independent tests were estimated for 76 glycans. Consequently, to account for multiple testing in the discovery cohort, we present results surpassing a conservative Bonferroni correction assuming 20 independent tests, thus giving a significant threshold of (p value <2.5 × 10−3 = 0.05/20). The association between TPOAb level or AITD status and IgG N-glycan trait were considered replicated if the glycan has the same direction as in the discovery cohort and has a nominal p value (>0.05) for one tail in at least one of two replication cohorts.
IgG Glycome-wide Association Scan of TPOAb Level or AITD Status in the Discovery and Replication Cohort
To examine whether IgG glycome composition was significantly associated with the TPOAb level or AITD status in the discovery cohort, we compared the fitted model in equation (2) with a model that did not include the residual of glycan in equation (1):
| (1) |
| (2) |
where Yi represents the TPOAb levels or AITD status for individual i and Gij is IgG N-glycan trait of type j among 75 N-glycans for the same individual i. A random intercept was added only in the discovery cohort in order to model the family-relatedness.
In the two replication cohorts, Wilcoxon test was performed on the residuals of IgG N-glycan traits
Heritability Analysis of IgG N-glycan Traits and AITD
Using the advantage of having twin data, ADCE models (additive genetics (A), dominance genetics (D), shared environment (C) and non-shared environment (E)) were employed in the estimate of heritability of glycosylation structures and AITD. An R package called mets that allows the analysis of monozygotic, dizygotic twins and unrelated individuals was used. The significance of variance components A, D, and C was assessed by dropping each component sequentially from the full model (ADCE) and comparing the sub-model fit to the full model. Sub-models were compared with full models by hierarchical χ2 tests. The difference between log-likelihood values between sub-model and full model is asymptotically distributed as χ2 with degrees of freedom (df) equal to the difference in df of the sub-model and the full model. A statistical indicator of goodness-of-fit, the Akaike information criterion (AIC), computed as χ2 - 2df; sub-models are accepted as the best-fitting model if there is no significant loss of fit when a latent variable (A, C, D, or E) is fixed to equal zero, was used to determine the most parsimonious model. When two sub-models have the same AIC compared with the full model, we decide to keep the model that was most likely (with additive genetic variance) or with the lowest p value for different components.
Genome-wide Association Analysis on IgG N-glycan Traits and AITD
To define the list of SNPs associated with glycosylation profiles regardless to any specific phenotypes in the TwinsUK cohort, we performed an analysis using the GenABEL software package (50), which is designed for GWAS analysis of family-based data by incorporating pairwise kinship matrix calculated using genotyping data in the polygenic model to correct relatedness and hidden population stratification. We selected SNPs for each glycan that had a p value under the GWAS significance threshold (p value < 5 × 10−8). Moreover, we added the list of SNPs previously defined and reported by Lauc, G. et al. (51).
To define the list of SNPs associated with AITD and thyroid functions, we selected the SNPs listed in the NHGRI GWAS catalogue (52) on 17/06/2015, after searching for the terms “thyroid” or “Graves” or “Hashimoto”.
Determination of Shared SNPs and Genes Between IgG N-glycan Traits and Thyroid Functions and Diseases
To examine whether IgG N-glycan traits and thyroid functions and diseases shared SNPs or genes, we compared the list of SNPs from GWASs performed on Croatian and TwinsUK data for IgG N-glycan traits and from the NHGRI GWAS catalogue for thyroid traits (cf. above). As SNPs detected by GWASs could be lead SNPs and not necessarily causal SNPs (53), we extended the list of SNPs to other variants in linkage disequilibrium (LD) with a r2 threshold of 0.8 from 1000G Phase 1 European population. Using HaploReg V4.1 (54), we extracted the closest annotated genes and transcript from GENCODE.
Enrichment of Genes Associated with IgG Glycosylation
To define if there was an enrichment of genes with SNPs associated with IgG N-glycan traits, we performed a one-sided Fisher's exact test on a list of the closest genes of published SNPs linked to IgG N-glycans traits (23). It was performed only on nine groups of genes that were associated with at least one of significant IgG N-glycan traits in the discovery cohort (ST6GAL1, IKZF1, ABCF2-SMARCD3, B4GALT1, FUT8, SMARCB1-DERL3, SYNGR1-TAB1-MGAT3-CACNA1l, BACH2, HLA-DQA2/HLA-DQB2).
Transcriptomic-wide Association Scan of TPOAb Level or AITD Status in the Discovery
To examine whether gene expression was significantly associated with the TPOAb level or AITD status in the discovery cohort, we performed linear mixed model with gene expression as outcome and TPOAb level or AITD as exposure. To correct for variation in sequencing depth between samples, all read count quantifications were normalized by the median number of well-mapped reads. Following this, the variation from technical covariates and age were removed in adding them as fixed effect in the mixed linear model where the family-relatedness was modeled as random effect.
RESULTS
TPOAb level and AITD are Associated with a Decreased Level of IgG Core Fucosylation
The presence of autoimmune antibodies, such as TPOAb and TgAb, is not a definite sign of the AITD. Indeed, the diagnosis of AITD is based on the combination of clinical signs and symptoms with biomarkers of thyroid function and immune responses. The TPOAb level is a continuous variable (contrary to AITD that is a binary variable) and is considered as the most sensitive test for both Graves's disease and HT because ∼95% of people with HT and 85% with Graves's disease show TPOAb detectable (3). However, 5–10% of healthy people with normal range of thyroid hormones (TSH, fT3, fT4) levels (called also euthyroid) test positively for TPOAb. So, we wanted to test whether IgG glycosylation status can play a role in active AITD and correlated with the TPOAb level or AITD. We investigated the associations between total plasma IgG glycome composition and peripheral blood TPOAb level and AITD status in 2,297 individuals and 1,191 individuals (988 controls and 203 AITD) respectively from the TwinsUK cohort (Discovery Cohort; supplemental Table S1) using hydrophilic interaction chromatography ultra-performance liquid chromatography (HILIC UPLC). HILIC-UPLC chromatograms were all separated into 24 glycan peaks (GP-Zagreb code, IGP-Edinburgh code, Fig 1A, supplemental Table S2), and the amount of glycans in each peak was expressed as a percentage of the total integrated area. Furthermore, we excluded glycan peak GP3 because of the co-elution with the contaminant and combined GP20 and GP21 to get 22 directly measured N-glycan traits (Fig. 1C, 1D; supplemental Table S2). As many of these structures share the same features (terminal galactose, terminal sialic acid, core-fucose, bisecting N-acetylglucosamine (GlcNAc)), we calculated 53 additional derived traits that average these features across multiple glycans (Fig. 1C, 1D; supplemental Table S2). Latter structural features were found to be more closely related to individual enzymatic activities in different cellular compartments (55) and underlying genetic polymorphisms (23) than the 24 original glycan peaks. As all these structural features partially correlated (Fig. 1B), we estimated only 20 independent glycan traits (23) in our data using the Equation (5) method proposed by Li & Ji (2005) (49). The p value for discovery step (0.05/20 = 2.5 × 10−3) was determined using Bonferroni correction for multiple testing with number of independent features (23). We found eleven directly measured and derived IgG N-glycan traits negatively associated with increased TPOAb level (IGP7, IGP8, IGP15, IGP33, IGP48, IGP56, IGP58, IGP59, IGP60, IGP62, and IGP63) and six directly measured and derived IgG N-glycan traits positively associated (IGP2, IGP21, IGP36, IGP42, IGP45, and IGP46) following Bonferroni correction for multiple testing (p value <2.5 × 10−3; circle symbol in Fig. 1C; Sheet 1 of supplemental Table S3). Only two derived IgG N-glycan traits from the TPOAb positive group (IGP48 and IGP59) stayed negatively associated with AITD status whereas three directly measured and derived IgG N-glycan traits (IGP2, IGP21, and IGP42) stayed positively associated with AITD status (triangle symbol in Fig. 1C; Sheet 2 of supplemental Table S3), but no new significant IgG N-glycan traits appeared in the AITD group when compared with the TPOAb positive group of the Twins UK cohort.
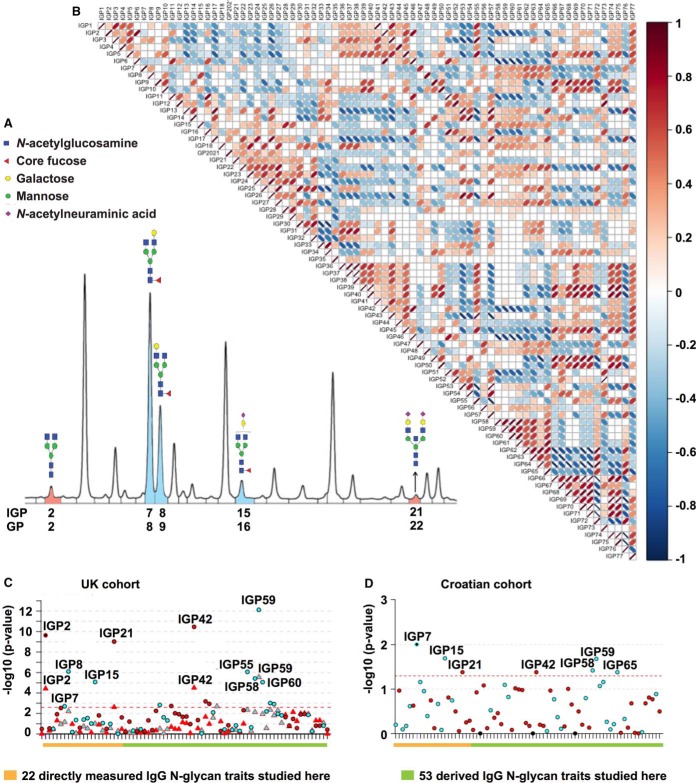
Fig. 1.
UPLC analysis of the IgG N-glycome and Manhattan plot of IgG N-glycan traits with AITD status and TPOAb level in two cohorts. A, Representative chromatogram showing separation of labeled IgG N-glycan structures into 24 chromatographic IgG glycan peaks (IGPs) is shown with indicated structures of glycans that significantly decreased (blue) or significantly increased (red) with TPOAb level and/or AITD status. B, Correlation matrix between 75 individual IgG N-glycan traits in a general population (the TwinsUK cohort). Image created with R package corrplot. C, Manhattan plot of 75 IgG N-glycan traits (directly measured and derived) in the discovery TwinsUK cohort (supplemental Table S3 - Sheet 1 and 2). The Manhattan plot is drawn with colors corresponding to the direction of associations (blue = negative, red = positive) and phenotypes are distinguished by the shapes of dots (circle = TPOAb level, triangle = AITD status). The red dashed line corresponds to the level of significance in the discovery cohort (p value ≤ 2.5 × 10−3). The orange line highlights 22 original glycan peaks detected by HILIC UPLC and analyzed in this study where a chromatogram is represented in Fig 1A whereas the green line highlights 53 additional IgG N-glycan derived traits. D, Manhattan plot of 75 IgG N-glycan traits for the Croatian replication cohort (TPOAb-positivity; Supplemental Table S3 - Sheet 4). The Manhattan plot is drawn with colors corresponding to the direction of associations (blue = negative, red = positive). The red dashed line corresponds to the level of significance p value in the replication cohort (0.05). The orange line highlights 22 original glycan peaks detected by HILIC UPLC and analyzed in this study where a chromatogram is represented in Fig 1A whereas the green line highlights 53 additional IgG N-glycan derived traits. Image created with R package coMET(28).
Next, we wanted to test if the same TPOAb-associated IgG N-glycan traits are conserved in a cohort of individuals with TPOAb-positivity from Croatia (unknown clinical status). We collected data from 73 control individuals (TPOAb 0.43 (0.53) IU/ml) and 90 case individuals (TPOAb 324.32 (408.46) IU/ml) from Croatia (replication Cohort; Croatian; supplemental Table S1). Four IgG N-glycan traits with negative association (IGP7, IGP15, IGP58 and IGP59) and two IgG N-glycan traits with positive association (IGP21 and IGP42) in the discovery cohort were significantly associated with TPOAb-positivity without established clinical diagnosis in the Croatian cohort (analogous to the ones observed in the TwinsUK discovery cohort) (Fig. 1D; Sheet 4 of supplemental Table S3– IgG N-glycan traits are bold and blue for negative association or red for positive association).
Investigating the structural composition of directly measured and derived IgG N-glycan traits associated with AITD and the TPOAb level (supplemental Table S2), we observed an increase in IgG N-glycan traits without core fucose (IGP2, IGP21, IGP42, IGP46) and structures with bisecting GlcNAc (IGP36, IGP45) whereas a decrease in IgG N-glycan traits containing core fucose and without bisecting GlcNAc (IGP7, IGP8, IGP15, IGP48, IGP62, IGP63) as well as structures with core fucose regardless of bisecting GlcNAc (IGP58, IGP59, IGP60). Additionally, a decrease in IGP33, which refers to the ratio of all fucosylated (regardless of bisecting GlcNAc) mono- and di-sialylated structures, was observed. Overall, we found 17 IgG N-glycan traits associated with the TPOAb level, which suggests a decrease in abundance of glycan structures containing core fucose, and five of those structures remain associated with AITD status (Fig. 1C, 1D).
HT is Associated with the Decreased Level of IgG Core Fucose and the PBMC Antennary α1,2 Fucose
Next, we focused our IgG N-glycan trait analysis on individuals with HT. In the TwinsUK Cohort (726 individuals - 675 controls and 51 HT), active HT was identified as TSH>10 mIU/L, or TSH>4 mIU/L accompanied by TPOAb > 100 IU/ml (Discovery cohort; HT; supplemental Table S1). To verify the finding, we had an additional cohort of a group of 103 HT patients and 106 control subjects from Poland (Replication cohort; Polish; supplemental Table S1) with clinical manifestation of HT but unknown TPOAb and TSH values at the time of the sampling. Six of the TPOAb associated IgG N-glycan traits remain significant in the UK HT cohort: two IgG N-glycan traits without core fucose (IGP2, IGP42) are increased, whereas IgG N-glycan traits containing core fucose and without bisecting GlcNAc (IGP8, IGP48), as well as traits representing fucosylation of agalactosylated glycans (IGP59) and percentage of monogalactosylated structures in total neutral IgG N-glycans structures (IGP56) remain decreased (Fig. 2A). In contrast, only one of the 17 IgG N-glycan traits that were significant in the discovery cohort (TPOAb level and AITD status; Sheet 3 of the supplemental Table S3III) was replicated with HT patients in the Polish cohort (IGP48; control versus patients with HT; Fig. 2B; Sheet 5, supplemental Table S3). However, other significant IgG N-glycan traits from the TwinsUK discovery cohort had the same direction for their effect sizes in the Polish replication cohort (Sheet 3 and Sheet 5 of supplemental Table S3; relevant IgG N-glycan traits are bold). The results from different discovery and replication cohorts suggest a decrease in abundance of glycan structures containing core fucose with the TPOAb level and a reduction of an IgG N-glycan structure with core fucose and galactose in HT.
N-Glycosylation of IgG extracted from 20 HT versus 20 healthy donors in Polish cohort (supplemental Table S1) was also examined in three batches by lectin blotting using Aleuria aurantia lectin (AAL), specific for α1,6-linked core fucose (56). We observed a consistently decreased level of IgG core fucosylation associated with HT status using AAL in the three batches, but none were significant with sample size tested (Fig. 2C, p value> 0.05; supplemental Table S4). We further tested whether the decreased level of core fucosylation in IgG extends to PBMCs and thus whether a specific suite of enzymes associated with a core fucosylation is generally altered in immune cells of AITD patients. N-Glycosylation of proteins extracted from PBMC homogenates from HT versus healthy donors (supplemental Table S1) was examined by lectin blotting using seven lectins with different sugar specificity: AAL, Galanthus nivalis lectin (GNL), Mackia amurensis lectin (MAL-II), Phaseolus vulgaris erythroagglutinin (PHA-E), Phaseolus vulgaris leucoagglutinin (PHA-L), Sambucus nigra agglutinin (SNA), and Ulex europaeus agglutinin (UEA I). Lectin blotting with UEA l identified a significant reduction of antennary α1,2 fucosein patients with HT (Fig. 2D) whereas blotting with AAL, which preferentially binds core fucose (α1,6), as well as all other lectin blotting assays, showed no difference in the content of particular glycan species between HT and control group (supplemental Fig. S6, supplemental Table S5). Coomassie Brilliant Blue staining control of total protein amount confirmed that there was no significant difference in protein profiles between control and HT samples (supplemental Fig. S7). Tandem mass spectrometry (MS/MS) was used to identify glycoproteins in the band marked the red frame in Fig. 2D in the manuscript according to Ochwat et al. (42) with minor modifications. Nine glycoproteins were identified in ca. 120 kDa-band and their functions based on the references were presented in supplemental Table S8. To summarize, we found six significant glycans in the UK HT discovery cohort that were also associated with the TPOAb level (one was replicated in the Polish cohort) and a decrease of PBMC antennary α1,2 fucose associated with HT. Moreover, a set of glycoproteins have been identified in 120 kDa band of PBMCs antennary α1,2 fucose, but further studies need to confirm them as biomarkers of AITD.
AITD and IgG N-glycan Traits are Highly Heritable, but No Shared Genetic Effect Between Them Could Be Determined
We and others identified several loci associated with IgG and plasma glycosylation and autoimmune conditions in the previous GWAS (24, 25, 57, 58), so we wanted to relate those findings to the genetic background of the subjects in this study to determine whether there is a shared heritability of AITD and IgG N-glycan traits. We estimated the proportions of genetic and environmental variance in the phenotypic variation of AITD status and different IgG N-glycan traits using the twin data from the TwinsUK cohort and the maximum likelihood variance component model fitting, also known as a structural equation-modeling or ADCE model (59). The best models of AITD status and TPOAb-positivity estimated were the DE model - with only a dominance genetic variance (D) and a unique environmental variance (E) - with about 57–63% of heritability (supplemental Table S9), as previously estimated in a Danish twin cohort (25–27). However, because the TPOAb does not follow a normal distribution, we could not predict with accuracy the best ADCE model. Regarding IgG N-glycan traits, most of IgG N-glycan traits have AE (model with only an additive genetic variance (A) and a unique environmental variance (E)) for the best model and the average heritability for all IgG N-glycan traits was of 55.15% (CI; min = 50.54; max = 59.74; supplemental Table S2). Given that the heritability for AITD and TPOAb-positivity predicts only a dominance genetic variance as a fraction of genetic variance, and the heritability of IgG N-glycan traits was estimated to be composed with mostly additive genetic variance, the portion of shared genetic variances between AITD and IgG N-glycan traits using a bivariate heritability analysis (60) could not be determined.
Lead SNPs Associated with IgG Glycan FA2G1S1 and TPOAb-positivity Fall in the Same High LD Nearby HCP5 Gene
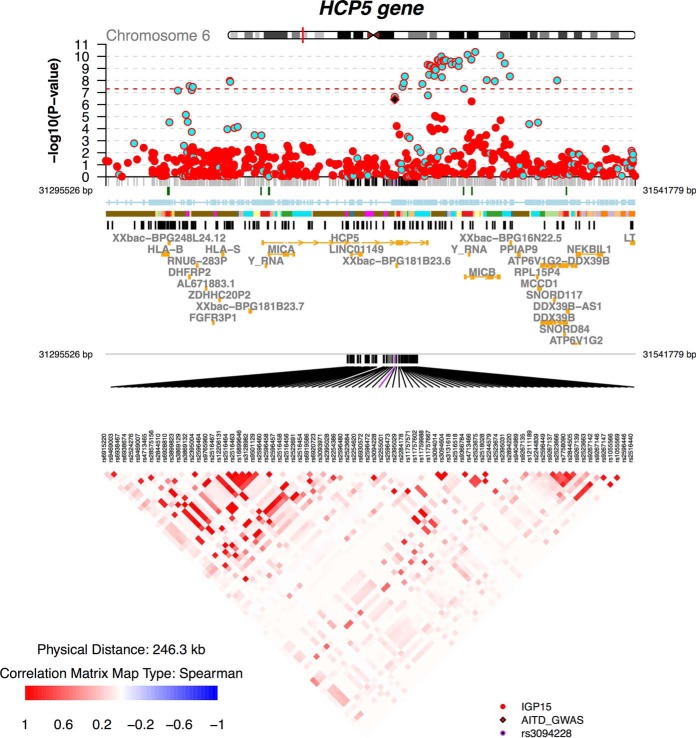
Next, we investigated whether specific genetic variants determined by previous GWASs could explain some genetic components potentially shared between AITD status and the specific IgG N-glycan traits identified here as associated with AITD and TPOAb level. We compared the results from internal GWASs of IgG N-glycan traits in the TwinsUK cohort (around 4500 individuals) (24) and previous GWASs of IgG N-glycan traits (23) with results from the GWAS catalogue for thyroid diseases and functions including GD, TPOAb-positivity, hypothyroidism, hyperthyroidism, levels of thyroid hormones (63) (supplemental Table S10). No lead SNPs could be found to be associated with at least one of thyroid phenotype and one of IgG N-glycan traits. By exploiting linkage disequilibrium (LD; r2>0.8) around the lead SNPs detected by these previous GWASs, we observed that the lead SNP of IGP15 (rs3094014) fell in the same high LD block around HCP5 (HLA Complex P5) gene on the chromosome 6 within the MHC class I region, as the lead SNP of TPOAb-positivity (rs3094228) (Fig. 3). Moreover, we found that the lead SNP of IGP14 and IGP54 (rs199442) clustered with hypothyroidism SNP (rs77819282) in the same high LD around the NSF (N-Ethylmaleimide Sensitive Factor, Vesicle Fusing ATPase) gene on the chromosome 17. Neither IGP14 nor IGP54 was associated with AITD and TPOAb-positivity. However, one of the significant IgG N-glycan traits, IGP15 also known as IgG glycan FA2G1S1, is correlated with the TPOAb level in our current study. Both SNPs relevant to both IGP15 and TPOAb-positivity (rs3094014 and rs3094228), as well as the other SNPs in this high LD (rs3099840, rs3131620, rs3128987), have a cell-type specific expression quantitative trait loci (eQTLs). For example, rs3094014 is an eQTL for C4A (thyroid, p value = 2.2 × 10−14; whole blood p value = 1.5 × 10−7), CYP21A1P (thyroid, p value = 4.7 × 10−12; whole blood p value = 2.0 × 10−9), HLA-C (thyroid, p value = 1.9 × 10–6), HCP5 (thyroid, p value = 1.1 × 10−9; whole blood, p = 1.2 × 10−9), and CYP21A2 (whole blood, p value = 7.1 × 10−8), whereas rs3094228 is associated with expression of C4A (thyroid, p value = 3.4 × 10−13; whole blood p value = 1.07 × 10−7), CYP21A1P (thyroid, p value = 1.1 × 10−9; whole blood p value = 3.79 × 10−8), HLA (PBMC, p value = 2.09 × 10−7 to 8.77 × 10−23), HCP5 (thyroid, p value = 1.3 × 10−9; whole blood, p value = 1.86 × 10−8), and CYP21A2 (whole blood, p value = 3.24 × 10−7) (64–66). This data indicates that SNPs associated with both IGP15 and TPOAb-positivity clustered around HCP5 gene alter the expression of genes in the thyroid cells and whole blood cells and fall in DNase sensitive site in T-cells and B-cells.
Fig. 3.
coMET plot for SNP association data on IGP15 in whole blood and TPOAb-positivity in the HCP5 gene region. Regional Manhattan plot from a GWAS of IGP15 performed on the TwinsUK cohort and TPOAb-positivity from meta-analysis GWAS (61). The regional Manhattan plot is drawn with colors corresponding to the direction of association (blue = negative, red = positive), the solid diamond symbol (black) represents the SNP rs3094228, which was associated with TPOAb-positivity (61). The red dashed line corresponds to the level of significance (p value = 5 × 10−8). Annotation tracks corresponding to CpG island (UCSC), Encode Dnase cluster (UCSC), GM12878 Chromatin State Segmentation by HMM from Encode/Broad (UCSC), SNPs from GWAS Catalogue (UCSC) and transcripts from ENSEMBL are displayed below the regional Manhattan plot. An LD matrix computed from genotypes of twins from the TwinsUK cohort (r2 using PLINK software (62)) is also provided. For Encode/Broad ChromHMM the colors correspond to: Red = 1. Active Promoter, Pink = 2. Weak Promoter, Purple = 3. Poised Promoter, Orange = 4 Strong Enhancer, Lilac = 5. Strong Enhancer, Yellow = 6. Weak Enhancer, Light Orange = 7. Weak Enhancer, Blue = 8.Insulator, Light Green = 9. Txn Transition, Green = 10. Txn Elongation, Cyan = 11. Weak Txn, Magenta = 12. Repressed, Brown = 13. Heterochrom/lo. Image created with R package called coMET (28).
Enrichment of IgG N-glycan Traits Associated With Two Enzymes, Fut8 and Ikzf1, Essential for the Core Fucosylation of IgGs in TPOAb Positive Subjects
We then focused on the nine groups of genes that were found to be associated with modifications in IgG glycosylation pattern from the previous GWASs (23), and we evaluated which groups of genes associated with IgG glycosylation patterns that are over-represented among IgG N-glycan traits associated with TPOAb positivity and AITD in the current study. Using the 17 significant IgG N-glycan traits in the discovery cohort and out of the nine groups of genes, we observed an enrichment of IgG N-glycan traits associated with FUT8 and IKZF1 genes (PFUT8 = 3.52 × 10−3; odd ratioFUT8 = 7.28 (IGP2, IGP42, IGP46, IGP58, IGP60, IGP63); PIKZF1 = 8.74 × 10−4; odd ratioIKZF1 = 9.19 (IGP2, IGP42, IGP46, IGP58, IGP59, IGP60, IGP62), Fig. 4). Both were associated with core fucosylation of IgGs regardless of bisecting GlcNAc (23). When we repeated the analysis with only the subset 7 replicated IgG N-glycan traits replicated in Croatian cohort, the enrichments became non-significant (PFUT8 = 7.41 × 10−2; odd ratioFUT8 = 4.85 (IGP42, IGP58); PIKZF1 = 9.21 × 10−2; odd ratioIKZF1 = 4.30 (IGP42, IGP58, IGP59) respectively). On the other hand, none of the 17 significant IgG N-glycan traits, and so of 7 replicated IgG N-glycan traits, was associated with the LAMB1 gene which was associated with core fucosylation of IgGs with the bisecting GlcNAc (PLAMB1 = 1; odd ratioLAMB1 = 0) in Lauc et al., 2013 (23). Overall, we observed an enrichment of IgG N-glycan traits associated with two genes, FUT8 and IKZF1, essential for the formation of IgG core fucose in TPOAb positive subjects.
Fig. 4.
Enrichment in AITD patients of IgG N-glycan traits associated with IKZF1 and FUT8 genes. Correlation network between 75 IgG N-glycan traits, highlighting 17 IgG N-glycan traits associated with AITD and TPOAb level and those associated with SNPs close to HLA, FUT8, IKZF1 genes. Nodes represent different IgG N-glycan traits (the color of nodes is related to the associations performed in discovery and replicated cohorts) and connections show the Pearson correlation between IgG N-glycan traits (only correlations more than 0.75 are visualized, red for negative correlation and blue for positive correlation, the strength of correlation is represented by the width of edge). The three clouds above of nodes cluster the different IgG N-glycan traits previously found to be associated with SNPs within/close to HLA gene (blue cloud), the FUT8 gene (yellow cloud) and IKZF1 gene (red cloud (23). Image created with R package qgraph.
The Decreased Level of IgG Core Fucose and PBMC Antennary α1,2 Fucose in AITD Is Not Associated with a Modification of Gene Expressions in Circulating Whole Blood Cells
A decrease in IgG core fucose, PBMC antennary α1,2 fucose, and no lead SNPs in the fucosylation genes associated with TPOAb positivity or AITD could all be explained by general dysregulation in gene expression belonging to fucosylation pathways. To explore this possibility, we performed transcriptome-wide association scan (TWAS) of the TPOAb level and AITD status on up to 199 individuals from the TwinsUK cohort (67). We further performed TWAS of 75 N-glycan traits on general TwinsUK cohort regardless of TPOAb level or AITD status (326 individuals) (sheet 2 of the supplemental Table S1). No significant modification of gene expressions (p value <5.82 × 10–7) was genome-wide associated with AITD and TPOAb at the exon level, even for subsets of genes such as those belonging to the pathways of fucosylation, including FUT1, FUT2, FUT8, LAMB1, and IKZF1. The decreased level of IgG and PBMC fucosylation in AITD could not be confirmed to be a consequence of a general depletion of fucosylation pathways in whole blood cells. However, in general population regardless for AITD status or TPOAb level (supplemental Table S11), IGP58 and IGP60 were associated with ENSG00000211949.2, the main exon of IGHV3–23, immunoglobulin heavy variable 3–23 present on chromosome 14 (beta = 0.25; p value<4.9 × 10−7 for both IgG) whereas IGP45 was associated with ENSG00000101986.7, the 7th exon of ABCD1 gene (beta = −0.35; p value = 2.43 × 10−7) and with ENSG00000072310.10, the 10th exon of SREBF1 gene (beta = −0.34; p value = 5.18 × 10−7). Decreased core fucosylation and PBMC antennary α1,2 fucose are not accompanied by the alteration of gene expression in PBMC.
DISCUSSION
The role of the autoantibodies in the development of AITD is still unknown. AITD are among the most frequent autoimmune disorders occurring in almost 5% of general population (1). If individuals harboring positive antibodies against thyroid proteins but without active disease are included, that adds up to 15% of the general population affected by thyroid autoimmunity (4). Three central thyroid autoantibodies (TPOAb, TgAb, TSAb) were identified in patients with AITD, and their role in AITD is still unclear. Because the presence of TPOAb does not necessarily indicate active AITD, but IgG glycosylation was previously shown to be an essential factor in regulating autoantibody function in autoimmune diseases (18, 19), we examined the IgG glycosylation status in AITD patients and TPOAb positive individuals. This study is the first that investigated the potential association of IgG and PBMC glycosylation with AITD and TPOAb level, genetic background between them and hypothesized about the causality of these relationships.
We observed a decrease in IgG core fucose level, which presented as altered levels of 17 IgG N-glycan traits in the whole blood of individuals with TPOAb, and depletion of PBMC antennary α1,2 fucose in patients with HT. Six out of seventeen significant IgG N-glycan traits for TPOAb positivity were replicated in a Croatian cohort. Only one out of these six IgG N-glycan traits was replicated in a Polish cohort, however, the other five significant structures from the UK cohort showed the same trend in the Polish cohort. Several healthy individuals in the Polish cohort have high thyroid autoantibodies (TgAb or TPOAb), but lower than the threshold set by the manufacturer for reporting of clinically elevated levels, whereas some HT patients have low thyroid autoantibodies. Additionally, all HT patients are under treatment with thyroid drugs that could affect their levels of thyroid autoantibodies (68, 69). Therefore, a possible reason why only one significant glycan feature was replicated in the Polish HT cohort might be the level of TPOAb. Unfortunately, there are not enough samples to stratify the Polish cohort according to TPOAb level or to classify the Croatian cohort according to clinical diagnosis to test the importance of TPOAb level in findings using UPLC data. However, when we reduced the case group from Croatia to 83 individuals with very high TPOAb (>100 IU/ml in Roche assay, the same criteria to define AITD status in the TwinsUK cohort), the associations for the six replicated IgG N-glycan traits became more significant whereas the beta values remained similar to the preceding analysis (Sheet 2 of supplemental Table S3). Using lectin blotting in the Polish cohort, we validated the reduction of core-fucosylated IgG in HT patients, but because of the lack of data on TPOAb levels for HT patients (i.e. analyzed in the same blood collected for IgG glycosylation analysis), we could not confirm the association with TPOAb level rather than AITD status. Overall, our results suggest that the decreased fraction of core-fucosylated IgG could be more related to the level of TPOAb than the status of AITD.
Several studies showed that 95% of IgG N-glycan structures in a healthy individual have core fucose and it acts as a “safety switch”, attenuating potentially harmful ADCC (70–74). Using FUT8 knockout Chinese hamsters, one previous study produced de-fucosylated CD20 antibodies and showed that afucosylated antibodies have a much higher affinity for FcγRIIIa (CD16a) - an immunoglobulin receptor distributed on natural killer (NK) cells, macrophages, and γδ T cells. They enhanced ADCC over 100-fold more than fucosylated CD20 antibodies (75). Production of antibodies without the core fucose has recently revolutionized antibody therapies, by providing substantially enhanced ADCC (76, 77). Thus, the deficiency of IgG core fucose observed in the people with AITD and TPOAb-positivity in our data and the activities of IgG core fucose on the immune system previously identified, suggest an increase of ADCC in TPOAb positive individuals.
Interestingly, ADCC was previously reported in AITD without restriction to subgroups of patients, however, more patients with HT than with GD presented ADCC activities (78, 79). Although no strong correlation of TPOAb level (total IgG or IgG subclasses) measured by enzyme-linked immunosorbent assay (ELISA) with ADCC activity could be found in previous studies, and other thyroid antigens besides TPO could be involved in ADCC, TPO was described as the primary antigen involved in the thyroid ADCC (78–81). Deglycosylation of TPOAb was shown to reduce the binding to FcγRs and thus inhibit the apoptosis of thyroid cancer cells via complement-dependent cytotoxicity (CDC) and cytotoxicity via ADCC (80). Although IgG glycosylation studied in the present paper is of total and not antigen-specific IgG, we found a relationship between TPOAb level and IgG core fucose, and we suggest that the depletion of core fucose observed in TPOAb circulating in blood could enhance their cytotoxicity activities on thyrocytes through ADCC (Fig. 5).
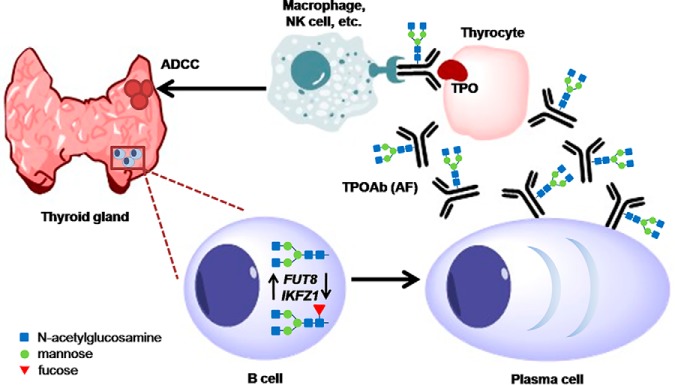
Fig. 5.
Hypothetic model of the possible role of the glycosylation of TPOAb driven by tissue-specific FUT8 and IKZF1 expression in AITD patients. Because of the lack of the significant deregulation of the expression of FUT8 and IKZF1 genes in whole blood of AITD patients, these genes could have an aberrant expression in a tissue-specific manner. It was previously suggested that TPOAb producing B-cells are generated in the germinal centers that are appearing in the thyroid gland of the AITD patients. Those cells could have aberrant expression of FUT8 and IKZF1 genes resulting in afucosylated (AF) TPOAb antibodies that are stimulating potentially harmful ADCC directed against the thyroid epithelial cells (thyrocytes), and therefore contributing to the AITD development.
Furthermore, taking advantage of the twin study design of the discovery cohort, we tested shared genetic and environmental effects between different significant glycan features found to be associated with TPOAb positivity and AITD in this study. Surprisingly, although previous GWASs on IgG N-glycan traits showed several SNPs also associated with autoimmune diseases (23, 82), no shared genetic variance or lead SNPs could be detected between IgG N-glycan traits with AITD status or the TPOAb level. In exploring in LD of different lead SNPs associated with thyroid diseases and IgG N-glycan traits, we found that a SNP associated with IGP14 and IGP54 (rs199442) clustered with hypothyroidism SNP (rs77819282) in the same high LD block around NSF, but because neither IGP14 or IGP54 were found to be associated with TPOAb and AITD, NSF could potentially have a pleiotropic effect on hypothyroidism that we could not address with current data. SNPs rs3094014 and rs3094228 appear to be associated with IGP15 and AITD respectively, are in high LD, are in the HCP5 gene region and indeed are eQTLs for HCP5 and other genes associated with immune response. These two lead SNPs and four others of the same LD block fall in a regulatory element, more precisely an enhancer and an open-chromatin in B-cell and thyroid tissue. Even though we identified two SNPs significantly associated with the IgG N-glycan structure IGP15, the cross-sectional nature of the cohorts and lack of independence of previous GWASs with the TwinsUK cohort do not allow conclusions to be drawn on the causal relationship between IgG core fucosylation and AITD.
Based on the findings of GWASs performed on the IgG N-glycan traits (23), we also showed an enrichment of the afucosylated IgG N-glycan traits that are associated with FUT8 and IKZF1 genes. Both genes were considered leading players in the core fucosylation of IgGs, although the mechanism of IKZF1 gene in the fucosylation is still unclear (23). The IKZF1 gene encodes a transcription factor belonging to the family of zinc-finger DNA-binding proteins associated with chromatin remodeling and regulating lymphocyte differentiation. Interestingly, several SNPs around the IKZF1 gene have been associated with autoimmune diseases based on the GWASs' findings; including systemic lupus erythematosus (83), Crohn's disease (84), inflammatory bowel disease (84), and other diseases such as acute lymphoblastic leukemia (85). On the other hand, the FUT8 gene encodes fucosyltransferase 8, a known enzyme catalyzing the addition of fucose in α1,6 linkage to the first GlcNAc residue (core fucose), but no SNPs around FUT8 genes have been associated with immune phenotypes other than IgG and plasma N-glycan structures from the previous GWASs (23, 57, 58, 63). However, even though these two fucosylation enzymes were found to be associated with the significant N-glycan structures from the present study, we could not find a direct association of the SNPs falling within or near their genomic regions with AITD or TPOAb at the current sample size. Because FUT8 and IKZF1 are essential players in the regulation of the core fucose expression and as we found no identifiable genomic mutations occurring in or near those genes in TPOAb positive individuals or AITD patients, we went on to examine the expression of those genes in whole blood.
The transcriptomic analysis in the whole blood cells performed in the TwinsUK cohort suggested that the general decrease of fucosylation associated with AITD and TPOAb level was not a consequence of dysregulated gene expression of known fucosylation genes in the circulating blood. Furthermore, with the current data, we were also not able to find a dysregulation in PBMC transcriptome associated with AITD that is linked to PBMC fucosylation. We are unable to hypothesize about the functionalities and pathways of production of PBMC antennary α1,2 fucose as up to now, PBMC glycosylation is not well characterized because of the difficulty in the isolation of a sufficient amounts of individual types of PBMC (e.g. B-cells and CD4+ T-cells) for glycan analysis. In comparison, IgG glycan structures are well studied as the IgG comprises ∼75% of total serum immunoglobulins. Considering we measured IgG glycosylation from whole blood and found the enrichment of afucosylated N-glycan traits, the statement that we found no general dysregulation of the fucosylation pathway in whole blood might seem confusing at first. However, although antibodies circulate and are detected in the blood, most plasma cells that produce circulating IgGs are in germinal centers of secondary lymphoid organs like the spleen and lymph nodes (86). In the case of AITD, the production of antibodies against thyroid components was suggested to happen directly in the thyroid gland where formations of germinal centers have occurred in patients with AITD (87–89). Consequently, in addition to the previous detection of germinal centers in the thyroid gland of patients with AITD, our finding from transcriptomic analysis suggests that the IgG glycosylation pattern observed in circulating blood of people with AITD status is not directly associated with immune cells in whole blood, but could be a dysregulation of specific immune cell-types such as thyroid-derived lymphocytes that produce thyroid autoantibodies (90) (Fig. 5).
In this study, we used fixed volumes of total IgG solution and not the same initial mass of IgG for the UPLC analysis of released glycans. Additionally, IgG glycosylation was analyzed on the level of total IgG, not on the level of a specific antibody. This approach was chosen because of the fixed amount of residual salts and the difficulty in obtaining enough material for analysis of IgG glycosylation of an antibody (e.g. absence or low concentration of thyroid autoantibodies in the total IgG in healthy individuals). Preparations of reference of TPOAb level were made from a pool of serum from patients with AITD and were prepared and lyophilized 35 years ago and use the international unit per milliliter (IU/ml) as the reference unit. Unfortunately, there is no simple way to convert TPOAb level from IU/ml provided by the assays in the present study to a concentration expressed as microgram per milliliter (μg/ml) and check if the concentration of total IgG is altered by the secretion of thyroid autoantibodies. The concentrations of TPOAb and TgAb were previously measured (up to 1.4 mg/ml for TPOAb and 0.7 mg/ml for TgAb) (91–93). Knowing that normal IgG concentration range is 5.1–15.8 mg/ml, in extreme cases, thyroid autoantibodies could represent up to 50% of total IgGs, and their presence in the blood can also increase the serum levels of total IgGs (94, 95). Thus, the secretion of thyroid antibodies can alter the concentration of total IgG in AITD.
If the modification of IgG glycosylation observed in the current study is the consequence of increased TPOAb concentration in total IgGs, the hypothesis that the depletion of IgG core fucose is a biomarker of TPOAb and its activity, and thereby a risk factor of harmful ADCC in the thyroid cells triggered by TPOAb (Fig. 5), becomes more likely. The cross-sectional nature of cohorts in this study and lack of independence of previous GWASs with the TwinsUK cohort do not allow conclusions on the causal relationship between IgG core fucosylation and AITD. Therefore, further analysis needs to be performed to describe genes and mechanisms playing in IgG and PBMC glycosylation in AITD in more specific tissues and cell-types to test this hypothesis. Studies looking at the expression of the known fucosylation genes in immune cells from thyroid tissue of healthy subjects, TPOAb positive individuals, and AITD patients might be especially valuable in dissecting the role of the fucosylation in the AITD disease genesis. However, to determine the causality between TPOAb level, fucosylation, ADCC and possibly other factors involved in the AITD genesis, prospective longitudinal studies that look at these traits before and after the diagnostic of AITD, as well as Mendelian randomization (96, 97) in large independent cohorts would be required.
This study identified for the first time an association of decreased level of IgG core-fucosylation and PBMC antennary α1,2 fucosylation with TPOAb level and AITD. This is also the first time that a decrease of fucosylation on IgG and PBMC is associated with one autoimmune disease and one of its biomarkers. Our findings could not be explained by common genetic background or general dysregulation of gene expression in the whole blood. However, drawing from the knowledge generated in many previous studies, we could speculate that this reduction of IgG core fucose could be a consequence of tissue-specific aberrant gene expression. Moreover, we hypothesized that the decreased IgG core fucosylation could be a novel risk factor for potentially harmful ADCC in the thyroid gland, associated with TPOAb and AITD. Further studies of the glycosylation of thyroid autoantibodies and their interactions with other immune features (e.g. immune cell-types, secreted proteins) and thyroid cells may be helpful to elucidate the potential role of autoantibodies and their glycosylation patterns in the pathogenesis and the treatment of thyroid diseases.
DATA AVAILABILITY
TwinsUK glycomic, phenotype, genetic data and GWAS results are publicly available upon request on the department website (http://www.twinsuk.ac.uk/data-access/accessmanagement/). RNA-seq data are available in the European Genome-phenome Archive (EGA) under accession EGAS00001000805.
The Polish tandem mass-spectrometry data (raw and annotation) are publicly available on the PRIDE archive (http://www.ebi.ac.uk/pride) under the identifier: PXD015684.
Supplementary Material
Acknowledgments
We would like to thank Massimo Mangino, Jonas Zierer, and Maxim Freydin, King's College London, UK, for discussion in the analysis of glycosylation data. The pre-print of this paper has been deposited on bioRxiv pre-print server (98). We acknowledged the different participants in the TwinsUK cohort, Polish cohort, and Crotian cohort. The glycomic and clinical data of the Polish cohort are publicly available upon request to Dr. Ewa Pocheć (ewa.pochec@uj.edu.pl). The glycomic and clinical data of the croatian cohort are publicly available upon request to Dr. Gordan Lauc (glauc@pharma.hr).
Footnotes
* The study was funded by TwinsUK cohort and the Wellcome Trust; European Community's Seventh Framework Programme (FP7/2007-2013). The study also receives support from the National Institute for Health Research (NIHR)- funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London and the Australian National Health and Medical Research Council (PG 1087407). Polish cohort: The study was supported by the grants from the Jagiellonian University (PBMC glycosylation, K/DSC/001749; IgG glycosylation, K/DSC/002341). IgG glycan analysis in the three cohorts was performed in Genos Glycoscience Research Laboratory and partly supported by the European Union's Horizon 2020 grants SYSCID (grant agreement No. 733100), IMforFUTURE (grant agreement No. 721815), GlySign (grant agreement No. 722095) and by the European Structural and Investment Funds IRI grant (#KK.01.2.1.01.0003) and Croatian National Centre of Research Excellence in Personalized Healthcare grant (#KK.01.1.1.01.0010). The authors have declared a conflict of interest. G.L. is the founder and CEO of Genos Ltd, a private research organization that specializes in high-throughput glycomic analysis and has several patents in this field. M.P., M.V., I.T.-A., and M.M.K. are employees of Genos Ltd. All authors declared no other conflicts of interest.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- AITD
- Autoimmune thyroid disease
- ADCC
- antibody-dependent cell-mediated cytotoxicity
- FcãRs
- Fcã-receptors
- FUT8
- Fucosyltransferase 8 (Alpha (1,6) Fucosyltransferase)
- GD
- Graves' disease
- GlcNAc
- N-acetylglucosamine
- GWAS
- genome-wide association studies
- HT
- Hashimoto's thyroiditis
- IgG
- Immunoglobulin G
- IKZF1
- IKAROS Family Zinc Finger 1
- LD
- linkage disequilibriumv
- NK
- Natural killer cell
- NSF
- N-Ethylmaleimide Sensitive Factor
- PBMC
- peripheral blood mononuclear cells
- Tg
- thyroglobulin
- TgAb
- thyroglobulin antibodies
- TPO
- thyroid peroxidase
- TPOAb
- Thyroid peroxidase antibodies
- TSAb
- antibodies against TSH receptors
- TSH
- thyroid-stimulating hormone
- TSHR
- TSH receptor
- TWAS
- transcriptome-wide association studies.
REFERENCES
- 1. Wang B., Shao X., Song R., Xu D., and Zhang J.-A. (2017) The emerging role of epigenetics in autoimmune thyroid diseases. Front. Immunol. 8, 174–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. De Groot L. (2015) Graves' disease and the manifestations of thyrotoxicosis. In Thyroid Disease Manager (Feingold K. R., Anawalt B., Boyce A., Chrousos G., Dungan K., Grossman A., Hershman J. M., Kaltsas G., Koch C., Kopp P., Korbonits M., McLachlan R., Morley J. E., New M., Perreault L., Purnell J., Rebar R., Singer F., Trence D. L., Vinik A., Wilson D. P., eds), SourceEndotext [Internet] MDText.com, Inc., South Dartmouth, MA [Google Scholar]
- 3. Mariotti S., Caturegli P., Piccolo P., Barbesino G., and Pinchera A. (1990) Antithyroid peroxidase autoantibodies in thyroid diseases. J. Clin. Endocrinol. Metabolism 71, 661–669 [DOI] [PubMed] [Google Scholar]
- 4. Hollowell J. G., Staehling N. W., Flanders W. D., Hannon W. H., Gunter E. W., Spencer C. A., and Braverman L. E. (2002) Serum TSH, T sub4/sub, and Thyroid Antibodies in the United States Population (1988 to 1994): National Health and Nutrition Examination Survey (NHANES III). J. Clin. Endocrinol. Metabolism 87, 489–499 [DOI] [PubMed] [Google Scholar]
- 5. Seeling M., Brückner C., and Nimmerjahn F. (2017) Differential antibody glycosylation in autoimmunity: sweet biomarker or modulator of disease activity? Nat. Rev. Rheumatol. 13, 621–630 [DOI] [PubMed] [Google Scholar]
- 6. Kobata A. (2008) The N-linked sugar chains of human immunoglobulin G: their unique pattern, and their functional roles. Biochim. Biophys. Acta 1780, 472–478 [DOI] [PubMed] [Google Scholar]
- 7. Shade K.-T., and Anthony R. (2013) Antibody glycosylation and inflammation. Antibodies 2, 392–414 [Google Scholar]
- 8. Novokmet M., Lukić E., Vučković F., Đurić Ž, Keser T., Rajšl K., Remondini D., Castellani G., Gašparović H., Gornik O., and Lauc G. (2014) Changes in IgG and total plasma protein glycomes in acute systemic inflammation. Sci. Reports 4, 4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marth J. D., and Grewal P. K. (2008) Mammalian glycosylation in immunity. Nat. Rev. Immunol. 8, 874–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maverakis E., Kim K., Shimoda M., Gershwin M. E., Patel F., Wilken R., Raychaudhuri S., Ruhaak L. R., and Lebrilla C. B. (2015) Glycans in the immune system and The Altered Glycan Theory of Autoimmunity: a critical review. J. Autoimmun. 57, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Subedi G. P., and Barb A. W. (2016) The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc γ receptor. MAbs 8, 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arnold J. N., Wormald M. R., Sim R. B., Rudd P. M., and Dwek R. A. (2007) The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 25, 21–50 [DOI] [PubMed] [Google Scholar]
- 13. Bondt A., Rombouts Y., Selman M. H. J., Hensbergen P. J., Reiding K. R., Hazes J. M. W., Dolhain R. J. E. M., and Wuhrer M. (2014) Immunoglobulin G (IgG) Fab glycosylation analysis using a new mass spectrometric high-throughput profiling method reveals pregnancy-associated changes. Mol. Cell. Proteomics 13, 3029–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spiegelberg H. L., Abel C. A., Grey H. M., and Fishkin B. G. (1970) Localization of the carbohydrate within the variable region of light and heavy chains of human γG myeloma proteins. Biochemistry 9, 4217–4223 [DOI] [PubMed] [Google Scholar]
- 15. Abel C. A., Spiegelberg H. L., and Grey H. M. (1968) Carbohydrate content of fragments and polypeptide chains of human .gamma.G-myeloma proteins of different heavy-chain subclasses. Biochemistry 7, 1271–1278 [DOI] [PubMed] [Google Scholar]
- 16. van de Bovenkamp F. S., Hafkenscheid L., Rispens T., and Rombouts Y. (2016) The emerging importance of IgG Fab glycosylation in immunity. J. Immunol. 196, 1435–1441 [DOI] [PubMed] [Google Scholar]
- 17. Jennewein M. F., and Alter G. (2017) The immunoregulatory roles of antibody glycosylation. Trends Immunol. 38, 358–372 [DOI] [PubMed] [Google Scholar]
- 18. Vučkovic F., Krištic J., Gudelj I., Teruel M., Keser T., Pezer M., Pučic-Bakovic M., Štambuk Trbojevic-Akmačic J. I., Barrios C., Pavić T., Menni C., Wang Y., Zhou Y., Cui L., Song H., Zeng Q., Guo X., Pons-Estel B. A., McKeigue P., Leslie Patrick A., Gornik O., Spector T. D., Harjaček M., Alarcon-Riquelme M., Molokhia M., Wang W., and Lauc G. (2015) Association of systemic lupus erythematosus with decreased immunosuppressive potential of the IgG glycome. Arthritis Rheumatol. 67, 2978–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trbojević-Akmačić I., Ventham N. T., Theodoratou E., Vučković F., Kennedy N. A., Krištić J., Nimmo E. R., Kalla R., Drummond H., Štambuk Dunlop J. M. G., Novokmet M., Aulchenko Y., Gornik O., Campbell H., Pučić Baković M., Satsangi J., Lauc G., and Consortium IBD-BIOM. (2015) Inflammatory bowel disease associates with proinflammatory potential of the immunoglobulin G glycome. Inflammatory Bowel Dis 21, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harre U., Lang S. C., Pfeifle R., Rombouts Y., Fruhbeisser S., Amara K., Bang H., Lux A., Koeleman C. A., Baum W., Dietel K., Grohn F., Malmstrom V., Klareskog L., Kronke G., Kocijan R., Nimmerjahn F., Toes R. E. M., Herrmann M., Scherer H. U., and Schett G. (2015) Glycosylation of immunoglobulin G determines osteoclast differentiation and bone loss. Nat. Commun. 6, 6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yuan S., Li Q., Zhang Y., Huang C., Wu H., Li Y., Liu Y., Yu N., Zhang H., Lu G., Gao Y., Gao Y., and Guo X. (2015) Changes in anti-thyroglobulin IgG glycosylation patterns in Hashimoto's thyroiditis patients. J. Clin. Endocrinol. Metabolism 100, 717–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao L., Liu M., Gao Y., Huang Y., Lu G., Gao Y., Guo X., and She B. (2013) Glycosylation of sera thyroglobulin antibody in patients with thyroid diseases. Eur. J. Endocrinol. 168, 585–592 [DOI] [PubMed] [Google Scholar]
- 23. Lauc G., Huffman J. E., Pučić M., Zgaga L., Adamczyk B., Mužinić A., Novokmet M., Polašek O., Gornik O., Krištić J., Keser T., Vitart V., Scheijen B., Uh H. W., Molokhia M., Patrick A. L., McKeigue P., Kolčić I., Lukić I. K., Swann O., van Leeuwen F. N., Ruhaak L. R., Houwing-Duistermaat J. J., Slagboom P. E., Beekman M., de Craen A. J., Deelder A. M., Zeng Q., Wang W., Hastie N. D., Gyllensten U., Wilson J. F., Wuhrer M., Wright A. F., Rudd P. M., Hayward C., Aulchenko Y., Campbell H., and Rudan I. (2013) Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet. 9, e1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shen X., Klarić L., Sharapov S., Mangino M., Ning Z., Di Wu Trbojević-Akmačić I., Pučić-Baković M., Rudan I., Polašek O., Hayward C., Spector T. D., Wilson J. F., Lauc G., and Aulchenko Y. S. (2017) Multivariate discovery and replication of five novel loci associated with Immunoglobulin G N-glycosylation. Nat. Communications 8, 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brix T. H., Kyvik K. O., and Hegedüs L. (2000) A population-based of chronic autoimmune hypothyroidism in Danish twins. JCEM 85, 536–539 [DOI] [PubMed] [Google Scholar]
- 26. Brix T. H., Kyvik K. O., Christensen K., and Hegedüs L. (2001) Evidence for a major role of heredity in Graves' disease: a population-based study of two Danish twin cohorts. J. Clin. Endocrinol. Metabolism 86, 930–934 [DOI] [PubMed] [Google Scholar]
- 27. Hansen P. S., Brix T. H., Bennedbæk F. N., Bonnema S. J., Iachine I., Kyvik K. O., and Hegedüs L. (2006) The relative importance of genetic and environmental factors in the aetiology of thyroid nodularity: A study of healthy Danish twins. Clin. Endocrinol. 62, 380–386 [DOI] [PubMed] [Google Scholar]
- 28. Martin T. C., Yet I., Tsai P.-C., and Bell J. T. (2015) coMET: visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinformatics 16, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spector T. D., and Williams F. M. (2006) The UK Adult Twin Registry (TwinsUK). Twin Res Hum Genet. 9, 899–906 [DOI] [PubMed] [Google Scholar]
- 30. Moayyeri A., Hammond C. J., Valdes A. M., and Spector T. D. (2013) Cohort Profile: TwinsUK and Healthy Ageing Twin Study. Int. J. Epidemiol. 42, 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krištić J., Vučković F., Menni C., Klarić L., Keser T., Beceheli I., Pučić-Baković M., Novokmet M., Mangino M., Thaqi K., Rudan P., Novokmet N., Sarac J., Missoni S., Kolčić I., Polašek O., Rudan I., Campbell H., Hayward C., Aulchenko Y., Valdes A., Wilson J. F., Gornik O., Primorac D., Zoldoš V., Spector T., and Lauc G. (2013) Glycans are a novel biomarker of chronological and biological ages. J. Gerontol. 69, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ugrina I., Campbell H., and Vučkovic F. (2017) Laboratory experimental design for a glycomic study. Methods Mol. Biol. 1503, 13–19 [DOI] [PubMed] [Google Scholar]
- 33. Pučić M., Knežević A., Vidič J., Adamczyk B., Novokmet M., Polašek O., Gornik O., Šupraha Goreta -S., Wormald M. R., Redžić I., Campbell H., Wright A., Hastie N. D., Wilson J. F., Rudan I., Wuhrer M., Rudd P. M., Josić D., and Lauc G. (2011) High throughput isolation and glycosylation analysis of IgG–variability and heritability of the IgG glycome in three isolated human populations. Mol. Cell. Proteomics 10, M111.010090-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dieterle F., Ross A., Schlotterbeck G., and Senn H. (2006) Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. application in 1H NMR metabonomics. Anal. Chem. 78, 4281–4290 [DOI] [PubMed] [Google Scholar]
- 35. Ugrina I., Klaric L., and Vučković F. (2015) Glycanr: Tools for analysing N-Glycan data. 1–2. Available at: https://github.com/iugrina/glycanr
- 36. Johnson W. E., Li C., and Rabinovic A. (2006) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 [DOI] [PubMed] [Google Scholar]
- 37. Leek J. T., Johnson W. E., Parker H. S., Jaffe A. E., and Storey J. D. (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peterson G. L. (1983) Determination of total protein. Methods Enzymol. 91, 95–121 [DOI] [PubMed] [Google Scholar]
- 39. Hoja-Łukowicz D, Link-Lenczowski P., Carpentieri A., Amoresano A., Pocheć E., Artemenko K. A., Bergquist J., and Lityńska A. (2012) L1CAM from human melanoma carries a novel type of N-glycan with Galβ1–4Galβ1- motif. Involvement of N-linked glycans in migratory and invasive behaviour of melanoma cells. Glycoconj J. 30, 205–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Demina E. P., Pierre W. C., Nguyen A. L. A., Londono I., Reiz B., Zou C., Chakraberty R., Cairo C. W., Pshezhetsky A. V., and Lodygensky G. A. (2018) Persistent reduction in sialylation of cerebral glycoproteins following postnatal inflammatory exposure. J. Neuroinflammation 15, 336–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okada M., Chikuma S., Kondo T., Hibino S., Machiyama H., Yokosuka T., Nakano M., and Yoshimura A. (2017) Blockage of core fucosylation reduces cell-surface expression of PD-1 and promotes anti-tumor immune responses of T cells. Cell Reports 20, 1017–1028 [DOI] [PubMed] [Google Scholar]
- 42. Ochwat D., Hoja-Łukowicz Lityńska D. A. (2004) N-glycoproteins bearing beta1–6 branched oligosaccharides from the A375 human melanoma cell line analysed by tandem mass spectrometry. Melanoma Res. 14, 479–485 [DOI] [PubMed] [Google Scholar]
- 43. Paziewska A., Polkowski M., Rubel T., Karczmarski J., Wiechowska-Kozlowska A., Dabrowska M., Mikula M., Dadlez M., and Ostrowski J. (2018) Mass spectrometry-based comprehensive analysis of pancreatic cyst fluids. Biomed. Res. Int. 2018, 7169595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Buil A., Brown A. A., Lappalainen T., Viñuela A., Davies M. N., Zheng H.-F., Richards J. B., Glass D., Small K. S., Durbin R., Spector T. D., and Dermitzakis E. T. (2014) Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat. Genet. 47, 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Richards J. B., Rivadeneira F., Inouye M., Pastinen T. M., Soranzo N., Wilson S. G., Andrew T., Falchi M., Gwilliam R., Ahmadi K. R., Valdes Arp A. M. P., Whittaker P., Verlaan D. J., Jhamai M., Kumanduri V., Moorhouse M., van Meurs J. B., Hofman A., Pols H. A., Hart D., Zhai G., Kato B. S., Mullin B. H., Zhang F., Deloukas P., Uitterlinden A. G., and Spector T. D. (2008) Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet 371, 1505–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Soranzo N., Rivadeneira F., Chinappen-Horsley U., Malkina I., Richards J. B., Hammond N., Stolk L., Nica A., Inouye M., Hofman A., Stephens J., Wheeler E., Arp P., Gwilliam R., Jhamai P. M., Potter S., Chaney A., Ghori M. J., Ravindrarajah R., Ermakov S., Estrada K., Pols H. A., Williams F. M., McArdle W. L., van Meurs J. B., Loos R. J., Dermitzakis E. T., Ahmadi K. R., Hart D. J., Ouwehand W. H., Wareham N. J., Barroso I., Sandhu M. S., Strachan D. P., Livshits G., Spector T. D., Uitterlinden A. G., and Deloukas P. (2009) Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genet. 5, e1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Howie B. N., Donnelly P., and Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bates D. (2016) lme4: linear mixed-effects models using ”Eigen“ and S4. 1–114. https://cran.r-project.org/web/packages/lme4/index.html
- 49. Li J., and Ji L. (2005) Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95, 221–227 [DOI] [PubMed] [Google Scholar]
- 50. Zhong K., Karssen L. C., Kayser M., and Liu F. (2016) CollapsABEL: an R library for detecting compound heterozygote alleles in genome-wide association studies. BMC Bioinformatics 17, 443–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lauc G., Huffman J. E., Pučić M., Zgaga L., Adamczyk B., Mužinić A., Novokmet M., Polašek O., Gornik O., Krištić J., Keser T., Vitart V., Scheijen B., Uh H. W., Molokhia M., Patrick A. L., McKeigue P., Kolčić I., Lukić I. K., Swann O., van Leeuwen F. N., Ruhaak L. R., Houwing-Duistermaat J. J., Slagboom P. E., Beekman M., de Craen A. J., Deelder A. M., Zeng Q., Wang W., Hastie N. D., Gyllensten U., Wilson J. F., Wuhrer M., Wright A. F., Rudd P. M., Hayward C., Aulchenko Y., Campbell H., and Rudan I. (2013) Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet. 9, e1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., and Parkinson H. (2013) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Farh K. K.-H., Marson A., Zhu J., Kleinewietfeld M., Housley W. J., Beik S., Shoresh N., Whitton H., Ryan R. J. H., Shishkin A. A., Hatan M., Carrasco-Alfonso M. J., Mayer D., Luckey C. J., Patsopoulos N. A., De Jager P. L., Kuchroo V. K., Epstein C. B., Daly M. J., Hafler D. A., and Bernstein B. E. (2014) Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ward L. D., and Kellis M. (2016) HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 44, D877–D881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Moremen K. W., Tiemeyer M., and Nairn A. V. (2012) Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Miyoshi E., Moriwaki K., and Nakagawa T. (2008) Biological function of fucosylation in cancer biology. J. Biochem. 143, 725–729 [DOI] [PubMed] [Google Scholar]
- 57. Huffman J. E., Kneþeviã A., Vitart V., Kattla J., Adamczyk B., Novokmet M., Igl W., Puèiã M., Zgaga L., Johannson Å, Redzic I., Gornik O., Zemunik T., Polasek O., Kolcic I., Pehlic M., Koeleman C. A., Campbell S., Wild S. H., Hastie N. D., Campbell H., Gyllensten U., Wuhrer M., Wilson J. F., Hayward C., Rudan I., Rudd P. M., Wright A. F., and Lauc G. (2011) Polymorphisms in B3GAT1, SLC9A9 and MGAT5 are associated with variation within the human plasma N-glycome of 3533 European adults. Human Mol. Gen. 20, 5000–5011 [DOI] [PubMed] [Google Scholar]
- 58. Lauc G., Essafi A., Huffman J. E., Hayward C., Knežević A., Kattla J. J., Polašek O., Gornik O., Vitart V., Abrahams J. L., Pučić M., Novokmet M., Redžić I., Campbell S., Wild S. H., Borovečki F., Wang W., Kolčić I., Zgaga L., Gyllensten U., Wilson J. F., Wright A. F., Hastie N. D., Campbell H., Rudd P. M., and Rudan I. (2010) Genomics meets glycomics-the first gwas study of human N-glycome identifies HNF1A as a master regulator of plasma protein fucosylation. PLoS Genet. 6, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rijsdijk V., and Sham P. C. (2002) Analytic approaches to twin data using structural equation models. Briefings Bioinformatics 3, 119–133 [DOI] [PubMed] [Google Scholar]
- 60. Visscher P. M. (2004) Power of the classical twin design revisited. Twin Res. 7, 505–512 [DOI] [PubMed] [Google Scholar]
- 61. Medici M., Porcu E., Pistis G., Teumer A., Brown S. J., Jensen R. A., Rawal R., Roef G. L., Plantinga T. S., Vermeulen S. H., Lahti J., Simmonds M. J., Husemoen L. L., Freathy R. M., Shields B. M., Pietzner D., Nagy R., Broer L., Chaker L., Korevaar T. I., Plia M. G., Sala C., Völker U., Richards J. B., Sweep F. C., Gieger C., Corre T., Kajantie E., Thuesen B., Taes Y. E., Visser W. E., Hattersley A. T., Kratzsch J., Hamilton A., Li W., Homuth G., Lobina M., Mariotti S., Soranzo N., Cocca M., Nauck M., Spielhagen C., Ross A., Arnold A., van de Bunt M., Liyanarachchi S., Heier M., Grabe H. J., Masciullo C., Galesloot T. E., Lim E. M., Reischl E., Leedman P. J., Lai S., Delitala A., Bremner A. P., Philips D. I., Beilby J. P., Mulas A., Vocale M., Abecasis G., Forsen T., James A., Widen E., Hui J., Prokisch H., Rietzschel E. E., Palotie A., Feddema P., Fletcher S. J., Schramm K., Rotter J. I., Kluttig A., Radke D., Traglia M., Surdulescu G. L., He H., Franklyn J. A., Tiller D., Vaidya B., de Meyer T., Jørgensen T., Eriksson J. G., O'Leary P. C., Wichmann E., Hermus A. R., Psaty B. M., Ittermann T., Hofman A., Bosi E., Schlessinger D., Wallaschofski H., Pirastu N., Aulchenko Y. S., de la Chapelle A., Netea-Maier R. T., Gough S. C., Meyer Zu Schwabedissen H., Frayling T. M., Kaufman J. M., Linneberg A., Räikkönen K., Smit J. W., Kiemeney L. A., Rivadeneira F., Uitterlinden A. G., Walsh J. P., Meisinger C., den Heijer M., Visser T. J., Spector T. D., Wilson S. G., Völzke H., Cappola A., Toniolo D., Sanna S., Naitza S., Peeters R. P. (2014) Identification of novel genetic Loci associated with thyroid peroxidase antibodies and clinical thyroid disease. PLoS Genet. 10, e1004123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., Bender D., Maller J., Sklar P., de Bakker P. I. W., Daly M. J., and Sham P. C. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., and Hindorff Parkinson L. H. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, 1001–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Westra H. J., Peters M. J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M. W., Fairfax B. P., Schramm K., Powell J. E., Zhernakova A., Zhernakova D. V., Veldink J. H., Van den Berg L. H., Karjalainen J., Withoff S., Uitterlinden A. G., Hofman A., Rivadeneira F., Hoen PAC' Reinmaa E., Fischer K., Nelis M., Milani L., Melzer D., Ferrucci L., Singleton A. B., Hernandez D. G., Nalls M. A., Homuth G., Nauck M., Radke D., Völker U., Perola M., Salomaa V., Brody J., Suchy-Dicey A., Gharib S. A., Enquobahrie D. A., Lumley T., Montgomery G. W., Makino S., Prokisch H., Herder C., Roden M., Grallert H., Meitinger T., Strauch K., Li Y., Jansen R. C., Visscher P. M., Knight J. C., Psaty B. M., Ripatti S., Teumer A., Frayling T. M., Metspalu A., van Meurs J. B. J., and Franke L. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. GTEx Consortium Human genomics. (2015) The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., Eleftheriadis M., Sinning C. R., Schnabel R. B., Lubos E., Mennerich D., Rust W., Perret C., Proust C., Nicaud V., Loscalzo J., Hübner N., Tregouet D., Münzel T., Ziegler A., Tiret L., Blankenberg S., and Cambien F. (2010) Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS ONE 5, e10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Buil A., Brown A. A., Lappalainen T., Viñuela A., Davies M. N., Zheng H.-F., Richards J. B., Glass D., Small K. S., Durbin R., Spector T. D., and Dermitzakis E. T. (2015) Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat. Genet. 47, 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Drutel A., Archambeaud F., and Caron P. (2013) Selenium and the thyroid gland: More good news for clinicians. Clin Endocrinol (Oxf). 78, 155–164 [DOI] [PubMed] [Google Scholar]
- 69. Schmidt M., Voell M., Rahlff I., Dietlein M., Kobe C., Faust M., and Schicha H. (2008) Long-term follow-up of antithyroid peroxidase antibodies in patients with chronic autoimmune thyroiditis (Hashimoto's Thyroiditis) treated with levothyroxine. Thyroid 18, 755–760 [DOI] [PubMed] [Google Scholar]
- 70. Kanda Y., Yamada T., Mori K., Okazaki A., Inoue M., Kitajima-Miyama K., Kuni-Kamochi R., Nakano R., Yano K., Kakita S., Shitara K., and Satoh M. (2007) Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: The high-mannose, hybrid, and complex types. Glycobiology 17, 104–118 [DOI] [PubMed] [Google Scholar]
- 71. Shields R. L., Lai J., Keck R., O'Connell L. Y., Hong K., Gloria Meng Y., Weikert S. H. A., and Presta L. G. (2002) Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J. Biol. Chem. 277, 26733–26740 [DOI] [PubMed] [Google Scholar]
- 72. Niwa R., Hatanaka S., Shoji-Hosaka E., Sakurada M., Kobayashi Y., Uehara A., Yokoi H., Nakamura K., and Shitara K. (2004) Enhancement of the antibody-dependent cellular cytotoxicity of low-fucose IgG1 Is independent of FcgammaRIIIa functional polymorphism. Clin. Cancer Res. 10, 6248–6255 [DOI] [PubMed] [Google Scholar]
- 73. Shinkawa T., Nakamura K., Yamane N., Shoji-Hosaka E., Kanda Y., Sakurada M., Uchida K., Anazawa H., Satoh M., Yamasaki M., Hanai N., and Shitara K. (2003) The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 278, 3466–3473 [DOI] [PubMed] [Google Scholar]
- 74. Niwa R., Sakurada M., Kobayashi Y., Uehara A., Matsushima K., Ueda R., Nakamura K., and Shitara K. (2005) Enhanced natural killer cell binding and activation by low-fucose IgG1 antibody results in potent antibody-dependent cellular cytotoxicity induction at lower antigen density. Clin. Cancer Res. 11, 2327–2336 [DOI] [PubMed] [Google Scholar]
- 75. Yamane-Ohnuki N., Kinoshita S., Inoue-Urakubo M., Kusunoki M., Iida S., Nakano R., Wakitani M., Niwa R., Sakurada M., Uchida K., Shitara K., and Satoh M. (2004) Establishment of FUT8 knockout Chinese hamster ovary cells: An ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol. Bioengineering 87, 614–622 [DOI] [PubMed] [Google Scholar]
- 76. Yamane-Ohnuki N., and Satoh M. (2009) Production of therapeutic antibodies with controlled fucosylation. MAbs 1, 230–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Okeley N. M., Alley S. C., Anderson M. E., Boursalian T. E., Burke P. J., Emmerton K. M., Jeffrey S. C., Klussman K., Law C. L., Sussman D., Toki B. E., Westendorf L., Zeng W., Zhang X., Benjamin D. R., and Senter P. D. (2013) Development of orally active inhibitors of protein and cellular fucosylation. Proc. Natl. Acad. Sci. U.S.A. 110, 5404–5409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rodien P., Madec A. M., Ruf J., Rajas F., Bornet H., Carayon P., and Orgiazzi J. (1996) Antibody-Dependent cell-mediated cytotoxicity in autoimmune thyroid disease: relationship to antithyroperoxidase antibodies. J. Clin. Endocrinol. Metabolism 81, 2595–2600 [DOI] [PubMed] [Google Scholar]
- 79. Metcalfe R. A., Oh Y. S., Stroud C., Arnold K., and Weetman A. P. (1997) Analysis of antibody-dependent cell-mediated cytotoxicity in autoimmune thyroid disease. Autoimmunity 25, 65–72 [DOI] [PubMed] [Google Scholar]
- 80. Rebuffat S. A., Morin M., Nguyen B., Castex F., Robert B., and Péraldi-Roux S. (2010) Human recombinant anti-thyroperoxidase autoantibodies: in vitro cytotoxic activity on papillary thyroid cancer expressing TPO. Br. J. Cancer 102, 852–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rebuffat S. A., Nguyen B., Robert B., Castex F., and Peraldi-Roux S. (2008) Antithyroperoxidase antibody-dependent cytotoxicity in autoimmune thyroid disease. J. Clin. Endocrinol. Metabolism 93, 929–934 [DOI] [PubMed] [Google Scholar]
- 82. Roederer M., Quaye L., Mangino M., Beddall M. H., Mahnke Y., Chattopadhyay P., Tosi I., Napolitano L., Terranova Barberio M., Menni C., Villanova F., Di Meglio P., Spector T. D., and Nestle F. O. (2015) The genetic architecture of the human immune system: A bioresource for autoimmunity and disease pathogenesis. Cell 161, 387–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Morris D. L., Sheng Y., Zhang Y., Wang Y.-F., Zhu Z., Tombleson P., Chen L., Cunninghame Graham D. S., Bentham J., Roberts A. L., Chen R., Zuo X., Wang T., Wen L., Yang C., Liu L., Yang L., Li F., Huang Y., Yin X., Yang S., Rönnblom L., Fürnrohr B. G., Voll R. E., Schett G., Costedoat-Chalumeau N., Gaffney P. M., Lau Y. L., Zhang X., Yang W., Cui Y., and Vyse T. J. (2016) Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat. Genet. 48, 940–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. de Lange K. M., Moutsianas L., Lee J. C., Lamb C. A., Luo Y., Kennedy N. A., Jostins L., Rice D. L., Gutierrez-Achury J., Ji S.-G., Heap G., Nimmo E. R., Edwards C., Henderson P., Mowat C., Sanderson J., Satsangi J., Simmons A., Wilson D. C., Tremelling M., Hart A., Mathew C. G., Newman W. G., Parkes M., Lees C. W., Uhlig H., Hawkey C., Prescott N. J., Ahmad T., Mansfield J. C., Anderson C. A., and Barrett J. C. (2017) Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49, 256–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Treviño L. R., Yang W., French D., Hunger S. P., Carroll W. L., Devidas M., Willman C., Neale G., Downing J., Raimondi S. C., Evans PUI CHWE, Relling M. V. (2009) Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 41, 1001–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nutt S. L., Hodgkin P. D., Tarlinton D. M., and Corcoran L. M. (2015) The generation of antibody-secreting plasma cells. Nat. Rev Immunol. 15, 160–171 [DOI] [PubMed] [Google Scholar]
- 87. Aust G., Krohn K., Morgenthaler N. G., Schröder S., Schütz A., Edelmann J., and Brylla E. (2006) Graves“ disease and Hashimoto”s thyroiditis in monozygotic twins: case study as well as transcriptomic and immunohistological analysis of thyroid tissues. Eur. J. Endocrinol. 154, 13–20 [DOI] [PubMed] [Google Scholar]
- 88. Caturegli P., De Remigis A., Chuang K., Dembele M., Iwama A., and Iwama S. (2013) Hashimoto's thyroiditis: celebrating the centennial through the lens of the Johns Hopkins Hospital surgical pathology records. Thyroid 23, 142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Armengol M. P., Juan M., Lucas-Martin A., Fernández-Figueras M. T., Jaraquemada D., Gallart T., and Pujol-Borrell R. (2001) Thyroid autoimmune disease: demonstration of thyroid antigen-specific B cells and recombination-activating gene expression in chemokine-containing active intrathyroidal germinal centers. Am. J. Pathol. 159, 861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Weetman A. P., McGregor A. M., Lazarus J. H., and Hall R. (1982) Thyroid antibodies are produced by thyroid-derived lymphocytes. Clin. Exp. Immunol. 48, 196–200 [PMC free article] [PubMed] [Google Scholar]
- 91. Rapoport B., and McLachlan S. M. (2001) Thyroid autoimmunity. J. Clin. Investig. 108, 1253–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Beever K., Bradbury J., Phillips D., McLachlan S. M., Pegg C., Goral A., Overbeck W., Feifel G., and Smith B. R. (1989) Highly sensitive assays of autoantibodies to thyroglobulin and to thyroid peroxidase. Clin. Chem. 35, 1949–1954 [PubMed] [Google Scholar]
- 93. Nakatake N., Sanders J., Richards T., Burne P., Barrett C., Pra C. D., Presotto F., Betterle C., Furmaniak J., and Smith B. R. (2006) Estimation of serum TSH receptor autoantibody concentration and affinity. Thyroid 16, 1077–1084 [DOI] [PubMed] [Google Scholar]
- 94. Yamauchi K., Yamada T., Sato A., Inazawa K., and Aizawa T. (2010) Elevation of serum immunoglobulin G in Hashimoto's thyroiditis and decrease after treatment with L-thyroxine in hypothyroid patients. Int. Med. 49, 267–271 [DOI] [PubMed] [Google Scholar]
- 95. Kawashima S. T., Tagami T., Nakao K., Nanba K., Tamanaha T., Usui T., Naruse M., Minamiguchi S., Mori Y., Tsuji J., Tanaka I., and Shimatsu A. (2014) Serum levels of IgG and IgG4 in Hashimoto thyroiditis. Endocrine 45, 236–243 [DOI] [PubMed] [Google Scholar]
- 96. Swerdlow D. I., Kuchenbaecker K. B., Shah S., Sofat R., Holmes M. V., White J., Mindell J. S., Kivimaki M., Brunner E. J., Whittaker J. C., Casas J. P., and Hingorani A. D. (2016) Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. Int. J. Epidemiol. 45, 1600–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Haycock P. C., Burgess S., Wade K. H., Bowden J., Relton C., and Smith G. D. (2016) Best (but oft-forgotten) practices: The design, analysis, and interpretation of Mendelian randomization studies. Am. J. Clin. Nutrition 103, 965–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Martin T. C., Simurina M., Zabczynska M., Martinic Kavur M., Rydlewska M., Pezer M., Kozlowska K., Burri A., Vilaj M., Turek-Jabrocka R., Krnjajić-Tadijanović M., Trofimiuk-Müldner M., Ugrina I., Lityńska A., Hubalewska-Dydejczyk A., Trbojevic-Akmacic I., Lin E. M., Walsh J. P., Pocheć E., Spector T. D., Wilson S. G., and Lauc G. (2018) Decreased IgG core fucosylation, a player in antibody-dependent cell-mediated cytotoxicity, is associated with autoimmune thyroid diseases. bioRxiv 1–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TwinsUK glycomic, phenotype, genetic data and GWAS results are publicly available upon request on the department website (http://www.twinsuk.ac.uk/data-access/accessmanagement/). RNA-seq data are available in the European Genome-phenome Archive (EGA) under accession EGAS00001000805.
The Polish tandem mass-spectrometry data (raw and annotation) are publicly available on the PRIDE archive (http://www.ebi.ac.uk/pride) under the identifier: PXD015684.