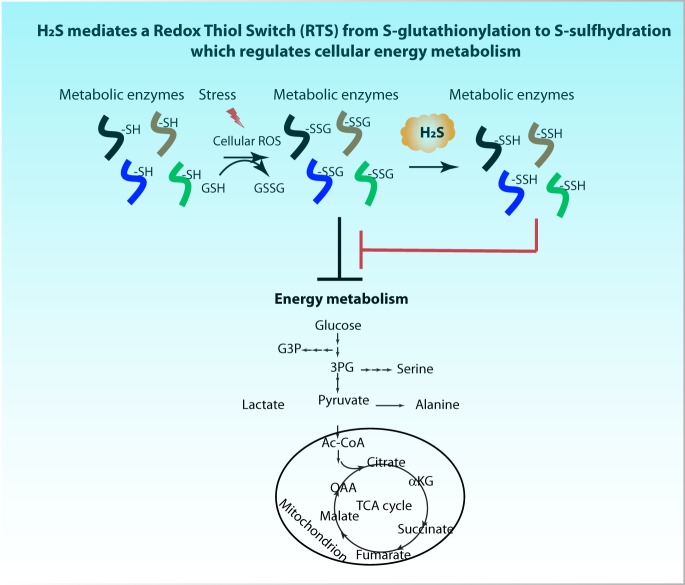
A mechanism for the cellular effects of the gas H2S, is the oxidative modification of protein cysteine residues. We developed a quantitative proteomics tool to profile protein S-persulfidation in cellular proteomes. We discovered a Redox Thiol Switch of S-glutathioinylation to S- persulfidation of proteins, among them enzymes in cellular energy metabolism. This work allows identification of redox regulation of cysteine residues of proteins in physiological and disease states and can assist design of therapeutics for diseases such cancer and diabetes.
Keywords: Post-translational modifications, thiol redox chemistry, energy metabolism, tandem mass spectrometry, quantification, chemoproteomics, diabetes, cysteine modifications, glutathionylation, H2S, persulfidation, protein sulfhydrome
Graphical Abstract
Highlights
Develop a TMT-based proteomics tool to profile cysteine persulfides in the cellular proteomes.
Discover a Redox Thiol Switch from protein S-glutathioinylation to S-persulfidation (RTSGS) with implications in the regulation of cellular energy metabolism under oxidative stress.
Abstract
The redox-based modifications of cysteine residues in proteins regulate their function in many biological processes. The gas molecule H2S has been shown to persulfidate redox sensitive cysteine residues resulting in an H2S-modified proteome known as the sulfhydrome. Tandem Mass Tags (TMT) multiplexing strategies for large-scale proteomic analyses have become increasingly prevalent in detecting cysteine modifications. Here we developed a TMT-based proteomics approach for selectively trapping and tagging cysteine persulfides in the cellular proteomes. We revealed the natural protein sulfhydrome of two human cell lines, and identified insulin as a novel substrate in pancreatic beta cells. Moreover, we showed that under oxidative stress conditions, increased H2S can target enzymes involved in energy metabolism by switching specific cysteine modifications to persulfides. Specifically, we discovered a Redox Thiol Switch, from protein S-glutathioinylation to S-persulfidation (RTSGS). We propose that the RTSGS from S-glutathioinylation to S-persulfidation is a potential mechanism to fine tune cellular energy metabolism in response to different levels of oxidative stress.
Hydrogen sulfide (H2S) is a gas molecule that can be produced endogenously in many organisms from bacteria to mammals (1, 2). H2S production at physiological concentrations is cytoprotective, as it reduces blood pressure (3), prevents neurodegeneration and extends lifespan (2, 4, 5, 6). H2S reacts with and modifies protein cysteine thiol groups to form persulfide bonds, known as protein S-persulfidation (also referred as S-sulfhydration). Because this gas is unstable, regulation of its synthesis can transiently alter protein cysteine modifications with an impact on cellular metabolism (7, 8).
It has been recognized as a great challenge to detect and quantify protein S-persulfidation in vivo, because of the high reactivity and instability of the cysteine persulfide bond in proteins (2, 9). Earlier, we developed a proteomics approach (8) exploiting the biotin thiol assay (BTA) to distinguish other types of cysteine-based PTMs from persulfidated proteins. We used this approach to show that the increase of H2S synthesis during endoplasmic reticulum (ER) stress in pancreatic beta cells promotes selective S-persulfidation of redox sensitive cysteine residues of proteins engaged in specific metabolic pathways (8).
However, the BTA approach can only be used to compare in parallel two different biological samples. Because of differences in the ionization efficiency and/or detectability of the many peptides in each sample, the changes of the resulting data between two samples cannot accurately reflect relative differences in their persulfidation levels. To cope with the challenges of large-scale proteomics analyses, we combined the BTA assay with the iodoacetyl isobaric tandem mass tag system (iodoTMT)1, thus enabling quantitative identification of persulfidated proteins from multiple samples. TMT is one of the most popular multiplexing methods, which uses isobaric tags for simultaneous labeling of peptides for identification and relative quantification by mass spectroscopy (10).
In the current study, we developed the TMT-BTA approach. With this approach, we show distinct patterns of the natural protein sulfhydromes in human pancreatic beta cells and hepatocytes, with hepatocytes exhibiting higher levels of persulfidated proteins compared with pancreatic beta cells. However, human pancreatic beta cells exhibited a unique landscape of persulfidated proteins, with the most enriched pathways being in intermediary metabolism. Furthermore, we identified insulin, the beta cell specific protein, as a S-persulfidation target.
The presence and extent of the conversion of S-persulfidation to other cysteine-based PTMs such as S-glutathionylation and vice versa in the proteome, is largely unknown. However, examples of proteins being glutathionylated and persulfidated on the same cysteine residue have been reported (7, 11, 12). One such example is the protein glyceraldehyde 3-phosphate dehydrogenase (GAPDH) which has been shown to have mixed disulfides at the redox sensitive cysteine residue150 (13). Glutathionylation of GAPDH at cysteine150 destabilizes the tertiary structure of the protein and decreases its enzymatic activity (13). In contrast, persulfidation of cysteine150, increases the catalytic activity of the protein (7). It is not known if there is a switch from one modification to another on the same cysteine residue, neither is known the physiological consequence for such a Redox Thiol Switch (RTS). Using the TMT-BTA approach and an in vitro assay, we show the existence of an RTS, which involves H2S-mediated reversal of S-glutathionylation to S-persulfidation (RTSGS) of specific cysteine residues in a subset of glutathionylated proteins. Finally, in order to understand the physiological significance of this RTSGS in cellular energy metabolism, we induced protein S-glutathionylation in pancreatic beta cells treated with diamide and measured metabolic flux of glucose. It is well known that protein S-glutathionylation can inhibit the activities of metabolic enzymes (14), and cause a decrease in the metabolic flux of glucose (15). In agreement with the RTSGS mechanism contributing to regulation of energy metabolism, exposure of cells to H2S, rescued the inhibited glucose flux in cells treated with diamide. We conclude that RTSGS, the redox thiol switch from S-glutathionylation to S-persulfidation is a potential mechanism to fine tune cellular energy metabolism in response to oxidative stress.
EXPERIMENTAL PROCEDURES
Antibodies
CBS: Abnova, H00000875–001p
CTH: Sigma, HPA023300
ATF4: Santa Cruz Biotechnology, SC-200
GAPDH: Santa Cruz Biotechnology, SC-32233 (6C5)
GSH: Virogen, 101-A (D8 clone)
MANF: Icosagen AS, 310–100
3MST: Santa Cruz Biotechnologies, SC-376168
PCK2: Cell Signaling, #6294
Cell Line and Cell Culture
Human pancreatic beta cells (EndoC-BH3) were purchased from the Univercell Biosolutions (Paris, France). The EndoC-BH3 cells were cultured in DMEM containing 5.6 mm glucose, 2% BSA fraction V, 50 μm 2-mercaptoethanol, 10 mm nicotinamide, 5.5 μg/ml transferrin, 6.7 ng/ml sodium selenite, penicillin (100 units/ml) and streptomycin (100 μg/ml). Ten μg/ml of puromycin (selective antibiotic) were added in the complete medium. The cells were seeded onto matrigel- and fibronectin-coated culture plates at 4 × 106 cells/plate.
MIN6 and INS1 cells were cultured as described previously (8)
Glucose-Stimulated Insulin Release (GSIS)
Glucose-stimulated insulin release was assayed INS1 cells as described previously (16)
Sample Preparation for Biotin Thiol Assay
Sample preparation and analysis were based on (8). In brief, proteins were extracted from cells with RIPA buffer (150 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, 0.5% deoxycholic acid, and 100 mm Tris pH 7.5) containing protease and phosphatase inhibitors. 2 mg of protein was incubated with 100 μm NM-biotin (Pierce) for 30 min, then mixed with Streptavidin-agarose resin (Thermo Scientific) and kept rotating overnight at 4 °C. The beads were washed and eluted with DTT or TCEP (10 mm). Eluted proteins were concentrated to a final volume of 25–40 μl with using of Amicon Ultracel 10K (Millipore), and used for gel electrophoresis followed by Western blot analysis.
Sample Preparation for TMT-BTA Assay
Proteins (2 mg) were extracted and biotinylated as described above. Biotinylated proteins were precipitated with ice cold acetone, resuspended in denaturation buffer (8 m urea, 1 mm MgSO4 and 30 mm Tris-HCl, pH 7.5) diluted with 10 volumes of buffer containing 1 mm CaCl2, 100 mm NaCl and 30 mm HEPES-NaOH pH 8.0, then incubated with MS-grade trypsin (Thermo, 90058) with occasional mixing for 18 h at 37 °C. The ratio of the enzyme to substrate was 1:40 (w/w). After digestion, trypsin was inactivated by incubation at 95 °C for 10 min, then reactions were mixed with streptavidin-agarose beads (0.5–2 ml) and incubated at 4 °C for 18 h following extensive washes in the presence of 0.1% SDS. Peptides were eluted with the buffer containing 10 mm TEAB with 10 mm TCEP. TCEP was removed with using a C18 column (Thermo). Peptides were eluted from the desalting column with 80% methanol, dried under vacuum, and suspended in buffer (50 mm TEAB buffer pH 8.0, Sigma). Free SH groups were alkylated by iodoTMT reagents at final volume of 150 μl. After 1h of TMT labeling under dark at 37 °C, addition of DTT (20 mm) terminated the alkylation reaction. The alkylated peptides were mixed and combined. After desalting through C18 columns, the mixed peptides were eluted with 70% methanol for LC-MS analysis.
TMT-BTA Peptide Identification and Quantification by LC-MS Analysis
This iodoTMT 6plex labeled peptide sample was reconstituted into 30 μl 1% acetic acid and was ready for MS analysis. The LC-MS system was a Thermo Ultimate 3000 UHPLC interfaced with a ThermoFisher Scientific Fusion Lumos tribrid mass spectrometer system. The HPLC column was a Dionex 15 cm × 75 μm id Acclaim Pepmap C18, 2 μm, 100 Å reversed-phase capillary chromatography column. Five μl volumes of the extract were injected, and the peptides eluted from the column by an acetonitrile/0.1% formic acid gradient at a flow rate of 0.3 μl/min were introduced into the source of the mass spectrometer on-line. The nanospray ion source was operated at 1.9 kV.
The digest was analyzed using both TMT-MS2 method and TMT-MS3 method. The TMT-MS2 is a data dependent acquisition method using HCD fragmentation for MS/MS scans of the precursor ions selected from MS1 full scan. The MS2 spectra were used for peptide identifications and the low mass region (100–140) of the same spectra was used simultaneously for quantifications of these peptides.
The TMT-MS3 method uses a technique called synchronized precursor selection (SPS) that can only be achieved on Thermo Fusion series MS. The method is also a data dependent acquisition using CID fragmentation for the MS2 scan of precursors selected from MS1. Then consecutive MS3 HCD scans at 100–500 m/z were performed on a combination of several of the most abundant ions (set at 10 in this study) selected from the MS2 scans. The CID MS2 spectra were used for peptide identifications and the HCD MS3 spectra were used for peptide quantifications.
Database Search Parameters and Acceptance Criteria for Identifications
Data from iodoTMT 6plex samples were searched against UniprotKB protein sequence database of proper species using Sequest program in the Thermo Proteome Discoverer V2.1 software package. Trypsin was used as the protease, and the maximum number of missed cleavage was set to 2. Peptide precursor mass tolerance was set to 10 ppm, and fragment mass tolerance was set to either 0.02 Th if Orbitrap was used as the detector or 0.6 Th if ion trap was used as the detector. Oxidation of Methionine and acetylation of protein N-terminus were set as Dynamic Modifications, and iodoTMT6plex of Cysteine was set as static modification. The false discovery rate (FDR) of peptide identification was set to 1%.
Posttranslational Modifications
Post translational modifications were identified using Sequest program in the Thermo Proteome Discoverer V2.1 software. For the detection of sulfide modification, protease and mass tolerance settings were the same as described in Database Search Parameters section, and in modification settings, Oxidation of Methionine, iodoTMT6plex of Cysteine, Sulfidation of Cysteine, and acetylation of protein N-terminus were set as Dynamic Modifications. The false discovery rate (FDR) of peptide identification was set to 1%.
Reporter Ion Quantification
Reporter abundance was quantified using the Thermo Proteome Discoverer V2.1 based on intensity and average reporter S/N threshold was set to 10. Normalization mode was set to total peptide amount. The settings on reporter ions quantifier node were: integration tolerance was set to 20 ppm, integration method was set to most confident centroid.
Sample Preparation for BioGEE Assay
Sample preparation and analysis were based on (17). Briefly, pancreatic beta cells were seeded in 102-cm dishes and cultured overnight. Cells were washed once with PBS and then incubated for 2.5 h at 37 °C in culture medium containing 0.03 mm BioGEE for 2.5 h. The cells were then washed twice, and the cells were lysed on ice with lysis buffer containing 10 mm NEM and protease inhibitor. 2 mg of protein was mixed with Streptavidin-agarose resin (Thermo Scientific) and kept rotating overnight at 4 °C. The beads were washed and eluted with TCEP (10 mm). Eluted proteins were concentrated to a final volume of 25–40 μl with using of Amicon Ultracel 10K (Millipore), and used for gel electrophoresis followed by Western blot analysis.
Bioinformatics Analysis of S-persulfidated Peptides
For pathway annotation: same as described previously (8). The pathways were scored based on persulfidated peptides using DAVID (www.david.ncifcrf.gov) program. Statistical significant of pathways are calculated, and pathways are ranked by the p values based on those tests.
For redox cysteine annotation: persulfidated peptide identified by MS, all exact matches in any of the RedoxDB database (https://biocomputer.bio.cuhk.edu.hk/RedoxDB/) on any oxidative modification cysteine sequences were collected.
Label-free MS Quantification of Full Proteome
The gel contains two samples derived from IHH and EndoC-BH3 cells. Each gel lane was cut into eight areas, these gel bands were washed/destained in 50% ethanol, 5% acetic acid and then dehydrated in acetonitrile. The bands were then reduced with DTT and alkylated with iodoacetamide prior to the in-gel digestion. All bands were digested in-gel using trypsin, by adding 5 μl 10 ng/μl chymotrypsin in 50 mm ammonium bicarbonate and incubating overnight digestion at room temperature to achieve complete digestion. The peptides that were formed were extracted from the polyacrylamide in two aliquots of 30 μl 50% acetonitrile with 5% formic acid. These extracts were combined and evaporated to <10 μl in Speedvac and then resuspended in 1% acetic acid to make up a finalvolume of ∼30 μl for LC-MS analysis.
The LC-MS system was a ThermoScientific Fusion Lumos mass spectrometry system. The HPLC column was a Dionex 15 cm × 75 μm id Acclaim Pepmap C18, 2 μm, 100 Å reversed-phase capillary chromatography column. Five μl volumes of the extract were injected, and the peptides eluted from the column by an acetonitrile/0.1% formic acid gradient at a flow rate of 0.3 μl/min were introduced into the source of the mass spectrometer on-line. The microelectrospray ion source is operated at 2.5 kV. The digest was analyzed using the data dependent multitask capability of the instrument acquiring full scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequence in successive instrument scans. These digests were analyzed utilizing LC gradient from 2 to 70% acetonitrile in 120 min on a capillary column and a data dependent acquisition with MS2-CID tandem MS.
The data were analyzed by using all CID spectra collected in the experiment to search the human UniProtKB database along with serum contaminants with the search programs MaxQuant. Protein quantitative value LFQ intensity was generated by a precursor intensity-based label-free quantification algorithm used by MaxQuant. LFQ intensities are the output of the Max-LFQ algorithm (18). These are based on the raw intensities that are normalized on multiple levels to ensure that profiles of LFQ intensities across samples accurately reflect the relative amounts of the proteins. Two different filters were used to identify proteins that have a different abundance in these samples, including a 2-fold difference in LFQ values, and a minimum of at least 5 peptides.
LC-MS Identification of Human Recombinant Insulin S-persulfidation
22.2 μm human recombinant insulin was incubated with 25 μg CTH in the presence of 4 mm l-cysteine and 1 mm PLP in buffer (100 mm HEPES-NaOH, pH7.4) for 1 h. The reaction mixture was dried and submitted for a LC-MS analysis.
The LC-MS system was a Thermo LTQ-Obitrap Elite hybrid mass spectrometer system. The HPLC column was a Dionex 15 cm × 75 μm id Acclaim Pepmap C18, 2 μm, 100 Å reversed-phase capillary chromatography column. Five μl volumes of the extract were injected, and the peptides eluted from the column by an acetonitrile/0.1% formic acid gradient at a flow rate of 0.3 μl/min were introduced into the source of the mass spectrometer on-line. The microelectrospray ion source is operated at 1.9 kV. The digest was analyzed using the data dependent multitask capability of the instrument acquiring full scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequence in successive instrument scans. The data was manually interpreted using the amino acid sequence of recombinant human insulin.
Generation of CRISPR-mediated knockout CTH INS1 cell lines
CTH null INS1 cells were generated using the protocol described in (19). In brief, sgRNAs targeting rat CTH were designed, amplified and cloned into transient lentiCRISPR v2 (Addgene plasmid # 52961) as a gift from Feng Zhang. Mammalian lentiviral particles harboring sgRNA-CTH plasmids were generated as described in (19). After 3 days of infection of INS1 cells, puromycin was added to enrich Cas9-expressing cells. Following 10 days of the puromycin selection, individual clone were isolated by flow cytometry and analyzed for decreased expression of CTH when overexpressed ATF4. sgRNA sequences for rat CTH gene.
sgCTH1: 5′-GGAGGGCGTCCTTCTGCATG-3′
sgCTH2: 5′-GAGGCGTCCTTCTGCATGCT-3′
Mismatch Detection Assay
Genomic DNA was extracted from individual clone INS1 cells using a DNA extraction kit (Zeymo). Target regions were PCR-amplified with pfu DNA polymerase (phusion, NEB, M0530S). PCR products were denatured at 95 °C for 10 min and re-annealed at −2 °C/second temperature ramp to 85 °C, followed by a −1 °C/second ramp to 25 °C. The heterocomplexed PCR products (5 μl) were incubated with 5 U T7E1 enzyme (New England Bio Labs) at 37 °C for 20 min. Products from mismatch assay were measured by electrophoresis on 2% agarose gel. Two different PCR primers (set 1 and set 2) were used to amplify the regions flanking the CRISPR targeting sites.
Primer 1 (F): 5′-GCTTCCTCATCCTGCAGACA-3′
Primer 1 (R): 5′-CCTGCTTACAGCCTGATCTTCT-3′
Primer 2 (F): 5′-CTCACCGGCTTCCTCATCCT-3′
Primer 2 (R): 5-GCCTGATCTTCTGGAGGCATTT-3′
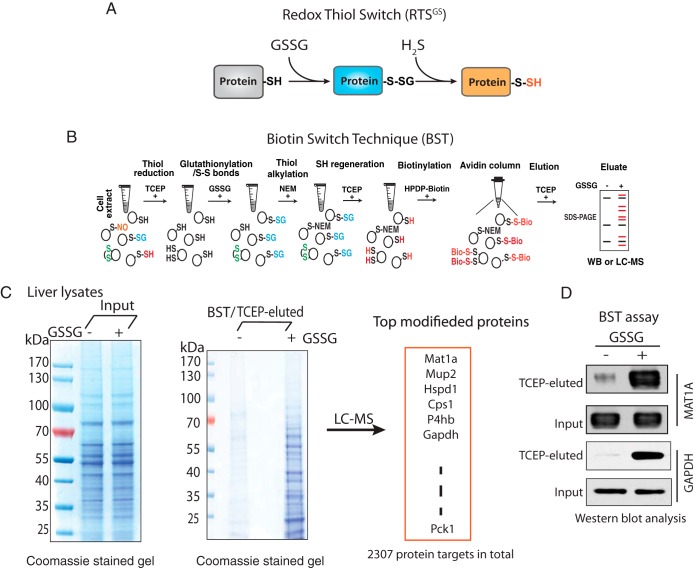
Identification of Protein S-glutathionylation by the Biotin Switch Technique (BST)
Similar to the BST used for the detection of nitrosylated proteins, detection of oxidized proteins from protein extracts was performed using the BST following the procedure described in (8) with minor modifications. Briefly, protein extraction was performed as described above. After alkylation with NEM, samples were precipitated with three volumes of cold acetone at −20 °C for 30 min to remove excess alkylating reagents, centrifuged (15,000 × g, 10 min, 4 °C), and resuspended at a concentration of 1 mg/ml in solubilization buffer (30 mm Tris-HCl pH 7.9, 1 mm EDTA and 100 mm NaCl) supplemented with 0.5% SDS. Reduction of oxidized proteins was achieved by adding 10 mm TCEP, then followed by cold acetone precipitation to remove excess TCEP. TCEP-treated protein pellets were dissolved in the solubilization buffer, then mixed with 0.25 mm HPDP-Biotin to label free thiols. After 60 min incubation at room temperature, the excess of HPDP-biotin was removed by acetone precipitation, and precipitated proteins were then resuspended at 1 mg/ml in solubilization buffer. The biotinylated sample was incubated with streptavidin beads (Thermo-20357) rotating at 4 °C for overnight. The beads were then washed 6 times with wash buffer (600 mm NaCl, 1 mm EDTA and 50 mm HEPES-NaOH, pH7.5) and eluted with TCEP. Protein eluate was separated by non-reducing SDS-PAGE following Western blotting with the indicated antibodies.
Metabolic Labeling of cells with [35S] Met
Sample preparation and analysis were based on (20). Briefly, INS1 cells were exposed to diamide at the indiciated concentrations for 1h, then washed with warm PBS. The untreated and treated INS1 cells incubated for 10 min in Met/Cys-free RPMI supplemented with 10% heat-inactive FBS in the presence of the treatment reagents. [35S]Met was added (30 μCi/ml) for an additional 30 min. INS1 cells were rinsed twice with cold PBS, and total proteins were precipitated three times with 5% TCA and 1 mm Met for 10 min on ice. Precipitates were dissolved in 200 μl of 1 N NaOH and 0.5% sodium deoxycholate for 1 h. Radioactivity was determined by liquid scintillation counting. Total cellular proteins were quantified by the DCTM Protein Assay (Bio-Rad) following the manufacturer's instruction. Incorporation of [35S] Met/Cys into total cellular proteins was calculated and normalized.
Metabolite Analysis by Targeted LC-MS: Determination of Glucose Flux
Treatment and assay of media [U-13C6] glucose enrichment. INS1 cells were seeded and cultured in the cell growth medium (RPMI, 1640 Thermo) containing 11.1 mm glucose, 10% heat-inactive FBS, 1 mm sodium pyruvate, 2 mm glutamine, 10 mm HEPES, 100 U/ml penicillin and 100 μg/ml streptomycin. Media were refreshed after 48h. INS1 cells were washed with warm PBS for one time, then incubated with glucose-free KRB buffer in the presence or absence of 0.1 mm diamide for 90 min. The untreated and treated cells were then exposed to the KRB buffer (a mixture of 8.4 mm glucose plus 8.4 mm of [U-13C6] glucose, and the indicated concentrations of H2S donor). After 90 min incubation, the cells were scraped, and pellets were stored at −80 °C until extraction of metabolites.
Metabolite extraction: Metabolites were extracted from cell pellets as described (8).
GC-MS conditions: Analyses were carried out on an Agilent 5973 mass spectrometer equipped with 6890 Gas Chromatograph. The details of GC-MS were described (8).
Experimental Design and Statistical Rationale
see Table I.
Table I.
| Figure # | F1B | F1E | F2A | F3A | F4B | F4H | F5B | F7A | F7C | F7I |
|---|---|---|---|---|---|---|---|---|---|---|
| Sample size | 6 | 5 | 6 | 2 | 6 | 6 | 1 | 12 | 6 | 6 |
| Control size | 3 | 1 | 3 | N/A | 3 | 3 | N/A | 4 | 2 | 2 |
| Biological replicate # | 3 | 1 | 3 | N/A | 3 | 3 | 1 | 4 | 1 | 2 |
| Statistical methods | t. test | N/A | t. test | N/A | t. test | t. test | N/A | t. test | N/A | N/A |
Sample size: The total number of samples performed and presented for each figure.
Control size: the numbers of controls employed.
Biological replicate: the number of biological replicates performed.
Statistical methods: the type of the statistical tests used for these analyses.
N/A: data unavailable
RESULTS
TMT-BTA proteomics to profile protein sulfhydromes
We have used the BTA assay (8) to develop a quantitative proteomics approach for profiling the changes in the protein sulfhydrome in different cellular contexts. The BTA assay involves the following processing steps: (i) biotinylation of thiolate groups with NM-Biotin, (ii) digestion of the biotinylated proteome with trypsin before capture in streptavidin beads, (iii) elution of the retained peptides that contain a persulfide bridge using a reducing agent, and (iv) LC-MS analysis of the eluted peptides.
Multiplexing proteomics strategies have been developed to relatively quantify multiple samples in parallel, through isotopically labeled peptides (21). TMT and iTRAQ are the most widely employed multiplexing methods, which use isobaric tags for simultaneous peptide identification and quantification (53). In particular, TMT has been successfully applied for the relative quantification of proteins and/or peptides (10, 22). We therefore included the process of iodoTMT multiplex labeling of the cysteine-containing peptides in the final step of the BTA assay (Fig. 1A). The TMT-labeled persulfidated peptides can be subsequently combined for LC-MS identification and quantification.
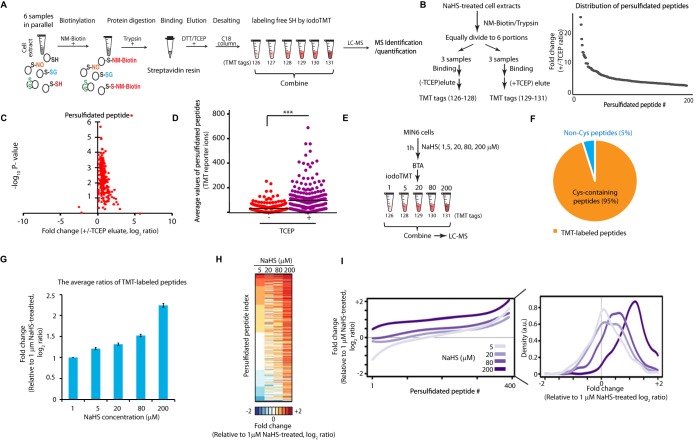
Fig. 1.
Quantitative proteomics approach to profile the protein sulfhydrome in vivo. A, Graphic representation of the Biotin Thiol Assay (BTA) (8) combined with alkylation of free SH groups with the tandem mass tags sixplex system (iodoTMT). Persulfidated cysteine residues in proteins from 6 samples were biotinylated with maleimide-PEG2-biotin (NM-Biotin). Subsequently, samples were processed with trypsin, passed through an avidin column and eluted with DTT or TCEP, which reduced the persulfide bonds and produced eluates containing the peptides with free SH groups. After desalting, free SH groups were alkylated with iodoTMT tags and the TMT-labeled samples were combined for LC-MS analysis. B, Schematic of workflow to determine the specificity of the TMT-BTA assay (left panel). Protein extracts from mouse pancreatic beta cells (MIN6) pretreated with an H2S donor (NaHS, 0.2 mm) were lysed, then subjected to the TMT-BTA assay. After trypsin digestion of the biotinylated proteome, the peptide sample was divided into six equal fractions. Each fraction was incubated with the same amount of avidin beads, then eluted without (−) or with (+) TCEP followed by the TMT labeling. The TMT-labeled samples were combined and subjected to LC-MS. Curve plot showing the distribution of relative changes of ratios (±TCEP) of the persulfidated peptides against the number (#) of identified peptides (right panel). Triplicates of each condition were used (C) Volcano plot showing the p value distribution against the relative changes of ratios (±TCEP) of the persulfidated peptides identified from the experimental data in (B). D, Dot plot showing the changes of average values of TMT report ion intensities of persulfidated peptides identified in (C). E, Schematic workflow to evaluate the TMT-BTA assay for the quantification of persulfidated proteins in vivo. MIN6 cells were treated with the H2S donor, NaHS, at the indicated concentrations for 1 h, then subjected to the TMT-BTA assay. LC-MS analysis was used to identify and quantify persulfidated peptides. F, Pie chart illustrating that 95% of the identified peptides in (D), were containing cysteine residues. G, Bar plot illustrating the dose-dependent increases of the average values of relative ratios (log2) of persulfidated peptides identified in (D). H, Heat map showing the relative persulfidated peptide enrichment ratios in MIN6 cells treated with the H2S donor at the indicated concentrations, as shown in (D). I, (Left panel) Curve plot showing the distribution of relative persulfidated peptide enrichment ratios against the number of identified peptides from the experimental data in (D), (Right panel) a histogram chart for the left panel.
In order to maximize TMT-labeled peptides in the eluate, we first optimized the buffer system and the concentration of reducing agents (DTT or TCEP) in the elution buffer of the BTA assay. We treated mouse pancreatic beta cells (MIN6) with a commonly used H2S donor (NaHS, sodium hydrosulfide), which promotes protein S-persulfidation both in cell extracts and in cells (2). The NaHS-treated MIN6 cells were lysed and subjected to the BTA assay. The persulfidated peptides were eluted in different buffer systems (supplemental Table S1) followed by the iodoTMT labeling and LC-MS analysis. The elution buffer containing 50 mm TEAB (triethylammonium bicarbonate) and 10 mm TCEP (tris-carboxyethyl phosphine hydrochloride) provided the highest labeling efficiency of persulfidated peptides (∼64%, supplemental Table S1). The poor TMT labeling efficiency of free thiol groups in persulfidated peptides could be because of residual TCEP, a thiol-free reducing reagent, which can interfere with labeling of proteins with the thiol alkylation reagent (23). We therefore introduced a desalting step to remove TCEP from cysteine-containing peptides before applying the labeling step. The TCEP-eluted peptides were passed through a C18 column, then eluted with 70% methanol containing 0.05% TFA (trifluoroacetic acid). The presence of TFA in the elution buffer can acidify and protonate the nascent free thiol groups to prevent auto-oxidation prior to thiol alkylation with iodoTMT tags. After desalting, cysteine-containing peptides were lyophilized, and each peptide sample was dissolved into 100 μl of 50 mm TEAB buffer, then alkylated with iodoTMT labeling reagents with constant concentration 1.33 μg/μl for 1 h in the dark. After combining all labeled peptide samples, the labeling reaction was quenched with 20 mm DTT, then applied to a C18 column for desalting. The TMT-labeled peptides were dried for MS identification and quantification. About 96% of total identified peptides were alkylated by iodoTMT 6plex reagents (data not shown). This labeling protocol was used for the subsequent studies.
Cysteine residues with either low signal intensity on report ions with under the detection threshold, or not fully labeled were discarded to minimize distortions of the proteomics data. However, across our studies, we found that less than 5% of total identified peptides were classified as unlabeled (did not contain cysteine residues) and were excluded from the subsequent analysis.
We previously reported that the BTA methodology identified specifically protein cysteine persulfides and not unmodified cysteine residues (with free SH groups) in the cellular proteome (8). If a peptide containing cysteine persulfide was biotinylated, then this peptide would be eluted with TCEP and be identified as a target for persulfidation. To establish this experimental system, we treated MIN6 cell lysates with NaHS and then applied the optimized TMT-BTA assay (Fig. 1B, left panel). Triplicate experiments were carried over for each condition (supplemental Table S2). Elution was performed either with a buffer containing TCEP, or NaCl (-TCEP) as a negative control. Eluates were labeled with iodoTMT reagents, followed by LC-MS analysis (Fig. 1B, left panel). About 200 persulfidated peptides were identified and quantified (Fig. 1B, right panel). Compared with the NaCl control (median intensity of TMT report ions, 227), TCEP-eluted samples exhibited significantly higher average intensities of report ions (657) on persulfidated peptides (Fig. 1C–1D). The TMT proteomics method demonstrates that the BTA assay is a highly selective method to identify the protein sulfhydromes in cell extracts.
We next determined the changes in the cellular sulfhydrome in MIN6 cells exposed to increasing H2S levels. The intracellular levels of H2S are on the range of nanomolar to micromolar, depending on cell types and cellular contexts (24). Under the highly reducing intracellular environments, protein S-persulfidation is unlikely the predominant modification over most protein free thiol groups in the proteome. Therefore, to determine if the TMT-BTA assay can quantify stoichiometric changes in the protein sulfhydrome in cells, we incubated MIN6 cells with increasing concentrations of NaHS (1–200 μM) followed by the TMT-BTA of MIN6 cell extracts (Fig. 1E, supplemental Table S3). We identified a total of 934 peptides, with the vast majority (890) containing cysteine residues, confirming that this method is highly selective in the identification of cysteine persulfides (Fig. 1F). As expected, the increased protein S-persulfidation was dependent on the NaHS concentrations (Fig. 1G–1I), although we did not observe a linear relationship between the increasing NaHS concentrations and the persulfidated peptides in the TMT proteomics approach (Fig. 1G). This may suggest that the H2S generated by NaHS may have differential accessibility and/or reactivity to redox sensitive cysteine residues as its concentration increases with increasing NaHS treatments. Finally, we found that all MS-identified persulfidated peptides were labeled by the TMT tags. Taken together, these data show that the TMT-BTA assay is a quantitative tool for the study of relative differences in the S-persulfidated proteome in cells and cell extracts and can be used to determine changes in the cellular sulfhydromes under different redox manipulations.
Quantitative Profiling of Protein Sulfhydromes Between Human Hepatocytes and Pancreatic Beta Cells by the TMT-BTA Approach
Next, we evaluated if the TMT-BTA approach can identify and quantify the relative differences of sulfhydromes between different cell types. It has been reported that protein S-persulfidation is one of the most prevalent redox post-translational modifications (PTMs) in liver tissue because of the high levels of expression of H2S-generating enzymes (7). The high abundance of protein S-persulfidation in liver cells made this cell type ideal for the purpose of comparing sulfhydromes between hepatocytes and other cell types. To this aim, we used the TMT-BTA assay to analyze the protein sulfhydrome in immortalized human hepatocytes, IHH, (25) and in human pancreatic beta cells, EndoC-BH3 (26) in biological triplicates (Fig. 2A, supplemental Table S4). About 447 peptides from 307 persulfidated proteins were identified in both human cell lines (Fig. 2B). To determine relative differences of persulfidated peptides between IHH and EndoC-BH3 cells, we performed a log2-transformation of TMT ratios (fold change) and used log2 TMT ratios as readout (Fig. 2B). As expected, human hepatocytes (IHH) exhibited significantly higher median levels of persulfidated peptides as compared with EndoC-BH3 cells (Fig. 2B–2D). Among all identified proteins, 33 proteins were persulfidated in EndoC-BH3 cells with ratios greater than 1.5-fold (Fig. 2C–2D). Bioinformatics clustering with a pathway annotation program (DAVID) of the identified shared sulfhydrome between hepatocytes and EndoC-BH3 cells, revealed an enrichment of those proteins involved in ribosome and metabolic pathways including glycolysis/gluconeogenesis (Fig. 2E, supplemental Table S5). Furthermore, proteins involved in mRNA translation and metabolic pathways are highly susceptible to S-persulfidation, in agreement with our previous studies showing that stress-induced inhibition of glycolytic flux was reversed by increased H2S biosynthesis in the presence of stress (8). The higher levels of protein S-persulfidation in hepatocytes as compared with EndoC-BH3 cells was further supported by the higher expression levels of H2S-generating enzymes in these cells (Fig. 2F). De novo synthesis of H2S is mediated by three enzymes: CTH (cystathionine γ lyase), CBS (cystathionine β synthase) and 3-MST (3-mercaptopyruvate sulfurtransferase) (27). Therefore, the levels of persulfidated proteins are in good correlation with the levels of the H2S-generating enzymes in these cells (Fig. 2F).
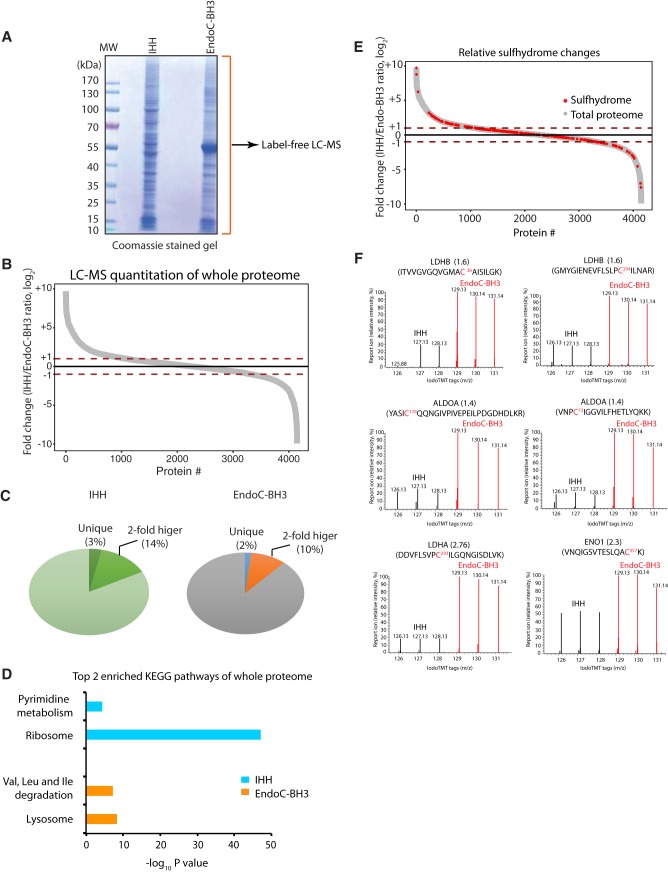
Fig. 2.
Quantitative profiling of the natural protein sulfhydrome in human hepatocytes and pancreatic beta cells by the TMT-BTA assay. A, Schematic of workflow for profiling the sulfhydrome in the two cell lines. Protein extracts from human hepatocytes (IHH) and pancreatic beta cells (EndoC-BH3) were subjected to the TMT-BTA assay, and the TMT-labeled peptides were mixed and analyzed by LC-MS. Triplicates of each cell line were used. B, (Left panel) Curve plot showing the distribution of relative changes (log2 IHH/EndoC-BH3 ratios) of the persulfidated peptides in IHH (light purple) and EndoC-BH3 cells (deep purple). (Right panel) Boxplot showing comparisons of the average levels of relative ratios (log2) of persulfidated peptides between the two cell lines. Triplicates of each cell line were used. C, Heat map of the relative ratios of the persulfidated peptides identified in experimental data in (B), illustrating the differences in the patterns of the natural protein sulfhydrome between IHH and EndoC-BH3 cells. D, Volcano plot showing the p value distribution against the fold change of ratios (log2 IHH/EndoC-BH3 ratios) of the persulfidated peptides identified from the experimental data in (B). E, Gene ontology biological pathways for all persulfidated proteins. PPP: pentose phosphate pathway. F, Evaluation of levels of H2S-generating proteins (CTH, CBS and 3-MST) in IHH and EndoC-BH3 by Western blot analysis.
Distinct Patterns of Natural Protein Sulfhydromes in Human Hepatocytes and Pancreatic Beta Cells
We hypothesized that the shared hepatic and beta cell sulfhydrome overlaid on their proteomes should show differential pathway enrichments. We first performed a label-free MS quantification (LFQ) experiment to determine the relative protein abundance in the different cellular proteomes (Fig. 3A, supplemental Table S6). IHH and EndoC-BH3 cell lysates were subjected to an SDS gel. Gel bands were cut in each lane, then reduced with DTT and alkylated with iodoacetamide prior to the in-gel digestion with trypsin. Those digests were analyzed using a ∼2h LC-MS/MS run. We ruled two peptides with one unique peptide required for protein identification. We normalized protein quantities expressed as labeled-free quantification (LFQ) intensities by MaxQuant. With this analysis, we identified and quantified almost 4500 proteins between the two cell lines (supplemental Table S6). About 2–3% of these proteins were solely identified either in the EndoC-BH3 cells, or in the hepatocytes (Fig. 3C); 10–13% of the proteins exhibited 2-fold differences between the two cell types (Fig. 3C), including CBS (0.3, EndoC-BH3/IHH ratio) and CTH (0.16, EndoC-BH3/IHH ratio), which were in agreement with the Western blotting analyses shown in Fig. 2F. To further validate the label-free MS data, we used Western blotting assays to test the expression of two proteins, MANF (mesencephalic astrocyte-derived neurotrophic factor) and PCK2 (mitochondrial phosphoenolpyruvate carboxykinase), which exhibited differential protein abundance in the two cell lines. In agreement with the MS data, EndoC-BH3 cells had higher expression of the MANF protein (5.77, EndoC-BH3/IHH ratio) (supplemental Fig. S1). In contrast, no significant change was found in the levels of metabolic enzymes GAPDH and PCK2 between hepatocytes and EndoC-BH3 cells, in agreement with the MS data (0.82 for GAPDH and 1.02 for PCK2, EndoC-BH3/IHH ratios) (Fig. 3B). Pathway analysis of the proteins with relative abundance greater than 2-fold between the two cell types (Fig. 3B), revealed that the branched-chain amino acid degradation and ribosome pathways were enriched in EndoC-BH3 cells and hepatocytes, respectively (Fig. 3D).
Fig. 3.
Protein S-persulfidation does not correlate with relative protein abundance between IHH and EndoC-BH3 cells. A, Identification and quantification of the full proteome from human hepatocytes (IHH) and EndoC-BH3 cells by LC-MS analysis. Cell extracts from both cell lines were resolved by reducing SDS-PAGE and stained with Coomassie blue. Gel lane was cut into fractions for in-gel digestions with trypsin. The peptides from each fraction were submitted for LC-MS identification and quantification. A total of 4664 proteins from hepatocytes and 4050 proteins from EndoC-BH3 cells were identified and quantified by LC-MS. B, Comparison of protein abundance between human hepatocytes and EndoC-BH3 cells in a curve plot showing the cell-type specific differences. Red dotted lines marks the persulfidation ratio (IHH/EndoC-BH3, log2) greater or less than 1. C, Pie charts of the data described in A and B. D, Gene ontology biological pathways for the relative most abundant proteins (> 1 in Fig. 3B) identified from IHH (blue bars) and EndoC-BH3 cells (orange bars). E, Comparison of the natural protein sulfhydrome in Fig. 2B with the full proteome reveals that a large fraction of persulfidated proteins have a moderate abundance in the cells. F, The TMT-BTA profiling of proteins containing cysteine residues with different reactivity to H2S from IHH and EndoC-BH3 cells. LC-MS-MS profiles for cysteine-containing peptides from LDHB, ALDOA, LDHA and ENO1 proteins are shown.
Next, we compared the relative changes in sulfhydromes between the two cell types, over their proteome abundance. To this end, we overlaid the protein abundance map with the natural sulfhydrome datasets (Fig. 3E). This analysis showed that 63 persulfidated proteins had a relative high abundance (log2 ratios greater than 1) in hepatocytes and 32 persulfidated proteins had relative high abundance (log2 ratios less than −1) in EndoC-BH3 cells. Among the 63 proteins, the enriched pathways were the ribosome and gluconeogenesis/glycolysis, whereas no pathway enrichment was noted in the 32 proteins from the EnoC-BH3 cells (supplemental Table S7). Furthermore, the remaining persulfidated proteins identified in both cell types showed the proteasome as an enriched pathway. Together, these data suggest that cellular sulfhydromes exhibit differential pathway enrichments as compared with their proteomes. A few metabolic proteins, such as lactate dehydrogenase (LDHA and LDHB), fructose-bisphosphate aldolase (ALDOA) and enolase 1 (ENO1) (Fig. 3F), were in relatively lower abundance, but were more persulfidated in the EndoC-BH3 cells than the hepatocytes. Overall, the evaluation of the relative abundance of sulfhydromes described here, also can be a very useful tool for identifying changes in the redox-mediated cellular responses by comparing two human cell types under different intracellular and extracellular cues.
ATF4-mediated CTH Expression Regulates Protein S-persulfidation in Rat Pancreatic Beta Cells
Next, we used the TMT-BTA assay to determine the regulation of the sulfhydrome in a single cell line under genetic manipulation of de novo H2S synthesis (Fig. 4A). We chose INS1 cells because the levels of the H2S-producing enzyme CTH, was undetectable in the absence of ER stress and thus made this cell line a good candidate to determine the effects of CTH expression on the sulfhydrome. We have previously shown that during chronic ER stress in MIN6 cells, increased protein S-persulfidation was in good correlation with increased expression of the transcription factor ATF4 and its transcription target, the CTH gene (27). In order to get more insights into the regulation of the sulfhydromes by the ATF4/CTH axis-mediated de novo H2S synthesis (Fig. 4A), we overexpressed the transcription factor ATF4 or GFP (green fluorescent protein as a negative control) in INS1 cells and applied the TMT-BTA assay to cell lysates. The TMT-BTA assay identified and quantified over 2000 persulfidated peptides (Fig. 4B, supplemental Table S8). Cysteine S-persulfidation levels were significantly increased by forced ATF4 expression but not by GFP expression (Fig. 4B–4C), in agreement to our previous study (8). Interestingly, few persulfidated peptides exhibited significantly decreased S-persulfidation in response to ATF4 overexpression. ATF4 has been identified as a master transcriptional regulator of stress-induced gene expression, including glutathione biosynthesis to protect cells against oxidative stress (28, 29). It is conceivable that the reversible cysteine persulfides in some proteins are being reduced very efficiently by the increased glutathione (GSH) in response to ATF4 overexpression.
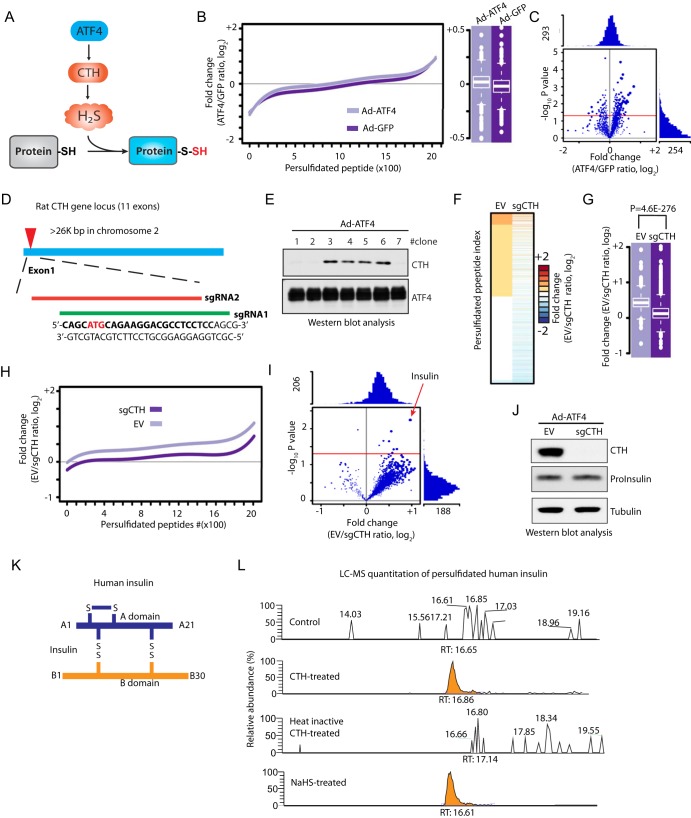
Fig. 4.
The TMT-BTA assay reveals insulin as a CTH-mediated persulfidated protein, in INS1 pancreatic beta cells. A, Schematic representation of the ATF4-induced CTH gene expression leading to increased intracellular H2S synthesis and S-persulfidation of proteins. B, Relative distribution of peptides containing persulfidated cysteine residues as determined by the TMT-BTA assay of cell extracts isolated from INS1 cells overexpressing ATF4 or GFP for 36 h. The relative changes (log2 value) of TMT ratios are plotted against the number of identified peptides. C, Volcano plot showing the relative change of the persulfidation ratios (ATF4/GFP, log2) of modified peptides plotted against the -log10 p value (n = 3). Red line indicates the p value ≤ 0.05. D, Schematic representation of CRISPR-cas9 mediated CTH knockout (KO) in INS1 cells. Two sgRNA sequences were designed for targeting exon 1 of the rat CTH gene. sgRNA targeting sequences are in bold. The translation initiation codon is shown in red. E, Western blot analysis of the effect of ATF4-overexpression in individual CRISPR-mediated CTH knockout INS1 cells. Clones 1, 2 and 7 did not express CTH protein. F, Heat map showing the relative enrichment values (log2 fold change) of persulfidated peptides identified in CTH KO (sgCTH) and control INS1 cells (empty vector, EV). CTH KO and EV INS1 cells were transfected to express ATF4, then subjected to the TMT-BTA assay, and followed by LC-MS identification and quantification. INS1 cells expressing cas9 protein without sgRNA were used as control. Triplicates of each condition were used. G, Boxplot showing comparisons of the average values of the log2 ratio of persulfidated peptides from CTH KO and EV INS1 cells expressing ATF4. H, Relative distributions of peptides containing persulfidated cysteine residues to their relative log2 values determined by the TMT-BTA assay. The relative changes (log2 value) of TMT ratios are plotted against the number of identified peptides from the experimental data in (F). I, Volcano plot showing the log2 fold change (sgCTH versus EV) of persulfidated proteins against the -log10 p value. Insulin is a major persulfidated protein regulated by the CTH protein levels. J, Western blot analysis of intracellular proinsulin levels showing no significant change of the levels of the protein in sgCTH INS1 cells as compared with the control cells. K, Schematic representation of insulin inter/intra-molecular disulfides. L, LC-MS analysis of CTH-mediated insulin persulfidation or NaHS-treated insulin in vitro. Human recombinant insulin was incubated with active or heat-inactivated recombinant CTH in the presence of PLP and l-cysteine for 1 h. The reaction mixture was subjected to LC-MS for the identification of persulfidated insulin. PLP: pyridoxal phosphate-a cofactor for the CTH enzyme. RT: retention time.
ATF4 can induce expression of many genes that can potentially increase the levels of the sulfhydrome (8, 14). To identify the subgroup of CTH-mediated persulfidated proteins (Fig. 4A), we looked for proteins with significant decreases in S-persulfidation in response to the depletion of CTH in ATF4-expressing INS1 cells. To this end, we employed the CRISPR-Cas9 technique to knock out the CTH-encoding gene in INS1 cells. We designed two single guide RNA sequences to target the translation start codon of the rat CTH gene (Fig. 4D, supplemental Fig. S2A), and then screened for individual INS1 clones to identify the ones with deficiency in CTH expression induced by ATF4 (Fig. 4E, supplemental Fig. S2B–S2D and supplemental Fig. S3). INS1 cells deficient for CTH, were infected with adenovirus that expressed ATF4, followed by the TMT-BTA assay. As expected, CTH depletion resulted in significant decreases of the protein sulfhydrome in INS1 cells expressing ATF4 (Fig. 4F–4I, supplemental Table S9) confirming that regulation of protein S-persulfidation by ATF4, is mediated in part via the induction of CTH. Among about 2000 persulfidated peptides, 157 peptides (corresponding to 130 proteins) exhibited more than 1.5-fold decrease in S-persulfidation with the loss of CTH induction. Notably, insulin exhibited a significant CTH-dependent decrease in S-persulfidation, suggesting that insulin may be a specific substrate of CTH in INS1 cells expressing ATF4. We identified a persulfidated insulin peptide in the B chain of insulin containing cysteines B7 and B19. Because of the close proximity of the two cysteine residues we were unable in this experiment to identify the exact position of persulfidation. Western blot analysis using antibodies for proinsulin revealed that the protein expression levels were not affected by CRISPR-mediated CTH knockout in INS1 cells expressing ATF4 (Fig. 4J), further supporting that the decreased insulin S-persulfidation is not because of changes in intracellular insulin content.
There are three disulfide bonds that are essential for mature insulin to maintain the correct structure (Fig. 4K). The CTH enzyme uses cysteine as the major source for H2S generation in mammals (30). To verify whether the enzymatic activity of CTH can regulate insulin S-persulfidation, we incubated recombinant human insulin with recombinant CTH protein (either active, or heat-inactivated) in the presence of cysteine and PLP as cofactor (pyridoxal phosphate), followed by LC-MS analysis. The samples were analyzed using a data dependent acquisition method. A persulfide modification on a cysteine gives a mass shift of +32 Th. Consistent with the data in INS1 cells, human insulin was persulfidated by H2S generated from the human recombinant CTH, but this modification was abolished by the inactivation of the CTH enzyme (Fig. 4L). Treatment of recombinant insulin with NaHS gave similar results to active CTH treatment (Fig. 4L). The combined data suggest that insulin S-persulfidation is the result of an enzymatic reaction that requires the presence of active CTH to catalyze protein cysteine persulfidation.
In Vitro Identification of Protein Disulfide Formations by the Biotin Switch Assay
We next tested the hypothesis that S-persulfidation may be used as a regulatory mechanism of the enzymatic activities of metabolic enzymes under oxidative stress conditions. It is known that redox sensitive cysteine glutathionylation inhibits the activities of metabolic enzymes (14). We have also previously shown that S-glutathionylation of GAPDH at the catalytic cysteine residue inhibits its enzymatic activity, and it can be restored by H2S treatment in vitro (8), suggesting that the S-glutathionylation to S-persulfidation RTS switch (Fig. 5A) could be a regulatory mechanism of energy metabolism.
Fig. 5.
A proteomics approach to detect the RTSGS switch in vitro. A, Schematic representation of the Redox Thiol Switch (RTSGS) from S-glutathionylation to S-persulfidation. This RTS is based on the idea that a reactive cysteine in a protein can be glutathionylated by oxidized glutathione (GSSG) via a thiol/disulfide exchange mechanism. The glutathione-modified cysteine residue can be interchanged to S-persulfidation by hydrogen sulfide (H2S). B, Schematic workflow of the Biotin Switch Technique (BST) to identify glutathionylated protein targets. The proteome in cell extracts was pretreated with TCEP to remove any cysteine-based reversible protein modifications. The reduced proteome was incubated with GSSG (5 mm) to introduce protein S-glutathionylation. After desalting, the GSSG-treated proteome was alkylated, reduced and biotinylated followed by an enrichment with avidin beads. The biotinylated proteins were eluted with TCEP. The proteins being modified by GSSG in the BST eluates can be further analyzed by Western blotting assays, or LC-MS. C, Determination of protein S-glutathionylation from mouse liver lysate treated with GSSG. The levels of modified proteins were evaluated in a SDS gel stained with Coomassie blue. The gel lane was excised and digested with trypsin for LC-MS identification. An excess of 2000 protein targets were identified by LC-MS analysis from the liver lysate treated with GSSG. D, Western blot analysis of the BST eluate in C, for two protein targets (MAT1A and GAPDH) being modified by GSSG in mouse liver lysates. The untreated and GSSG-treated liver lysates were subjected to the BST assay. Eluates were analyzed by Western blotting for the corresponding proteins.
Because there is a lack of methods that can measure specifically the changes of cysteine modifications between S-glutathionylation and S-persulfidation in cells, we decided to test our hypotheses in vitro. We first evaluated whether the PTM switch from S-glutathionylation to S-persulfidation could occur in the cellular proteome. It has been reported that protein S-glutathionylation can be promoted by treatment with oxidized glutathione (GSSG) in vitro (14, 31). We therefore incubated protein lysates isolated from mouse livers with the reducing agent TCEP in order to remove reversible cysteine modifications in the proteome. Subsequently, we treated lysates with GSSG to introduce S-glutathionylation. Of note, GSSG-treatment also may result in disulfide bond formation via the initial S-glutathionylation (14). The GSSG-treated lysates were subsequently subjected to an assay (32) referred as the Biotin Switch Technique (BST) to identify the proteins targeted by S-glutathionylation (Fig. 5B). We found that the content of glutathionylated proteins increased by the GSSG treatment (Fig. 5C, supplemental Fig. S4), consistent with previous reports (31). MS analysis identified more than 2300 protein targets for S-glutathionylation, including GAPDH (Fig. 5C, supplemental Table S10), which supports our previous observation of S-glutathionylation of GAPDH by GSSG treatment (8). Western blot analysis confirmed GAPDH and as an additional control, MAT1A (methionine adenosyltransferase 1A) as protein targets being modified by GSSG in vitro (Fig. 5D).
Determination of a Protein Cysteine Modification Switch from Disulfides to Persulfides In Vitro
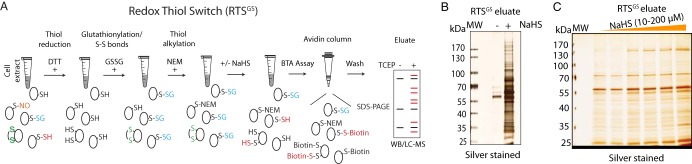
Does exposure of cell extracts to H2S convert proteome-wide S-glutathionylation to S-persulfidation? What proteins respond to this PTM thiol switch? To address this question, lysates isolated from mouse livers were exposed to TCEP followed by GSSG treatment. After desalting to remove excess GSSG, proteins with free cysteine thiol groups in the lysates were alkylated with NEM (N-ethylmaleimide) to block remaining free thiol groups. The lysates were incubated with NaHS, then subjected to the BTA assay which was further analyzed by SDS-PAGE (Fig. 6A). As shown in Fig. 6B, NaHS treatment increased the levels of S-persulfidated proteins from the GSSG-treated proteome compared with the untreated control; a dose-dependent increase in the levels of persulfidated proteins by H2S was also observed (Fig. 6C). These data provide strong support for the notion that the PTM switch from S-glutathionylation to S-persulfidation occurs on a proteome-wide scale. We named this protein cysteine PTM inter-conversion as Redox Thiol Switch S-Glutathionylation to S-persulfidation (RTSGS).
Fig. 6.
Identification of RTSGS targets by the BTA assay in vitro. A, Schematic of workflow to identify cysteine residues within the proteome, candidates for regulation by the RTSGS. Cell lysates were pretreated with TCEP to remove any reversible cysteine-based protein modifications, then incubated with GSSG to incorporate S-glutathionylation and disulfides to proteomes. The GSSG-treated protein extracts were treated with N-ethylmaleimide (NEM) to block any free thiol groups. After desalting, the lysates were exposed to H2S (NaHS) followed by the BTA assay. The RTS targets were analyzed in a SDS gel or by LC-MS. B, The BTA assay was performed to detect RTS targets in extracts from mouse liver. GSSG-treated mouse liver lysates were divided into two equal fractions, then treated without or with an H2S donor (NaHS, 2 mm) for 1h followed by the BTA assay. The eluates were analyzed by SDS-PAGE electrophoresis and silver staining of proteins. C, Mouse liver lysates (equal amount/treatment) were treated and analyzed as in B but with increasing concentrations of the H2S donor-NaHS (10, 20, 40, 80, 100, and 200 μm).
H2S Restores Oxidative Stress-Decreased Metabolic Flux in Glycolysis and the Mitochondrial TCA Cycle
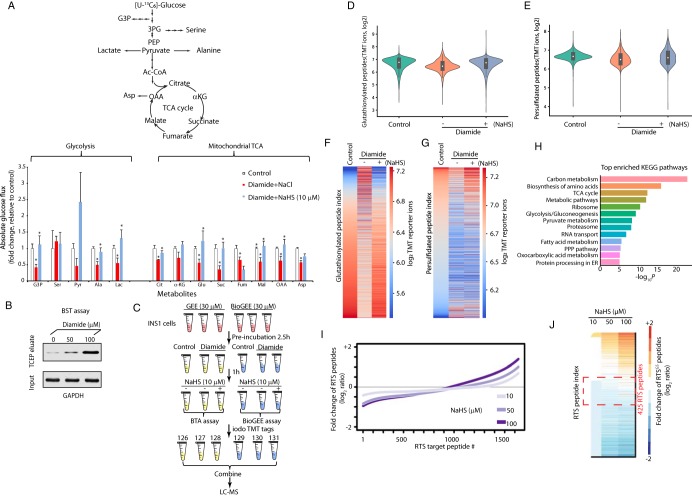
Pancreatic beta cells are highly sensitive to oxidative stress and the redox status of intracellular glutathione (GSH/GSSG) is important for their metabolism and glucose-stimulated insulin secretion (GSIS) (33). We have previously shown that H2S-treatment of MIN6 cells pre-exposed to oxidative stress can reverse stress-induced inhibitory effects on glucose metabolism (8). Therefore, pancreatic beta cells appeared to be a good experimental system to test the existence of the RTSGS in living cells and determine its significance on the cellular metabolism. We hypothesized that small changes in intracellular H2S levels can be sensed by metabolic enzymes via the RTSGS mechanism and thus adjust their glucose flux under oxidative stress (Fig. 7A, top panel). We tested this concept by exposing INS1 cells to a protein S-glutathionylation-promoting oxidant for a short time and measured the effects of H2S on the oxidant-induced changes of glucose flux. Diamide is a commonly used oxidant, whose short time addition to culture medium can cause oxidation of glutathione and increase protein S-glutathionylation and disulfides (34, 35). Treatment of INS1 cells with micromolar doses of diamide for 1 h increased the GSSG levels without any global changes in protein synthesis rates (supplemental Fig. S5A–S5B). Diamide-treated INS1 cells were washed to remove diamide, then exposed to NaHS (10 μm). Flux was measured in both conditions in the presence of glucose [U-13C6] (Fig. 7A, bottom panel). We previously reported that treatment of S-glutathionylated GAPDH with NaHS at concentrations 10–50 μM reversed the inhibition of GAPDH activity (8). Therefore we considered the concentration of 10 μM of NaHS within the physiological range for the regulation of metabolic activities of enzymes in cells. As expected, diamide-treatment alone significantly reduced glycolytic and mitochondrial TCA flux as shown by the low 13C label incorporation in glyceraldehyde 3-phosphate (G3P), alanine, lactate, citrate, glutamate, succinate, fumarate, malate, oxaloacetic acid (OAA) and aspartic acid (Fig. 7A, bottom panel, supplemental Table S11). This inhibition of the metabolic flux was correlated with increased amounts of oxidized GAPDH (Fig. 7B). When the diamide-treated cells were exposed to NaHS, the glycolytic and mitochondrial TCA flux were restored, as shown by the increase in 13C-labeling of G3P, alanine, lactate, citrate, glutamate, succinate, malate, OAA. These data suggest that the H2S-mediated RTSGS mechanism may sustain glycolytic and mitochondrial metabolism upon an oxidative challenge.
Fig. 7.
H2S restores oxidative stress-decreased metabolic flux in glycolysis and the mitochondrial TCA cycle. A, Measurement of [U-13C]-glucose flux in metabolites, expressed as the fold change of absolute glucose flux (ratio of control/treated). INS1 cells were treated with diamide (0.1 mm) in glucose-free growth medium for 90 min, then the cells were switched to 16.8 mm glucose in KRB buffer containing NaHS (0, 10 μm) for 90 min. [U-13C]-glucose replaced glucose in the KRB media for the last 90 min of treatments. 4 replicates of each condition were used. B, Evaluation of GAPDH S-glutathionylation in INS1 cells treated with diamide (0, 50 and 100 μM) at the indicated concentrations by the BST assay. INS1 cells were incubated without or with diamide for 90 min, then subjected to the BST assay to determine the levels of glutathionylated GAPDH protein using Western blot analysis (the details of experimental approach for the BST assay was described in Fig. 5B). C, Schematic workflow to identify and quantify glutathionylated and persulfidated proteins in vivo using the iodoTMT quantification proteomics approaches. INS1 cells were pre-incubated with either GEE or BioGEE (0.03 mm) for 2.5 h, then treated under the indicated conditions. After the treatments, all the GEE-treated samples were subjected to either the BTA assay for the detection of persulfidated proteins, and the samples labeled with BioGEE were subjected for the detection of glutathionylated proteins. After protein digestion and elution, all cysteine-containing peptides were labeled with the iodoTMT tags followed by LC-MS analysis. D, Violin plot showing the relative changes of S-glutathionylated peptides identified by the TMT-BioGEE assay. E, Violin plot showing the relative changes of S-persulfidated peptides identified by the TMT-BTA assay. F, Heat map showing the relative changes of S-glutathionylated peptides identified by the TMT-BioGEE assay. G, Heat map showing the relative changes of S-persulfidated peptides identified by the TMT-BTA assay. H, Gene ontology analysis of the persulfidated and glutathionylated targets in (C) showing that metabolic enzymes are highly enriched among all the identified proteins. I, Curve plot showing the dose-dependent increase of S-persulfidation levels on RTSGS peptides in the proteome treated with the H2S donor in vitro from the experimental data (supplemental Table S17). J, Heat map showing the relative changes of S-persulfidated peptides identified by the TMT-BTA assay. The RTSGS targets within the red dotted line frame showing that among the highly sensitive targets for RTSGS, are metabolic enzymes containing a redox domain with a P-loop motif.
In order to evaluate the changes in protein S-persulfidation and S-glutathionylation in culture cells upon exposure to diamide followed by H2S treatment, we pre-incubated INS1 cells with a biotinylated cell membrane-permeable derivative of glutathione (BioGEE), as well as the control reagent glutathione ethyl ester (GEE) (Fig. 7C). Protein S-glutathionylation induced by an oxidant such as diamide was evaluated using the BioGEE assay (supplemental Fig. S6A–S6C). We first treated those cells with diamide (0.1 mm), followed by 10 μM NaHS. Then, we used the BioGEE and BTA assays to simultaneously evaluate the changes of protein S-glutathionylation and S-persulfidation (Fig. 7C). Cysteine GSH-mixed disulfides and persulfides present in the INS1 proteome after these treatments were selectively reduced and labeled with iodoTMT 6plex reagents (Fig. 7C). After the TMT labeling, all cysteine-containing peptide samples were then combined, desalted and analyzed using LC-MS. MS analysis allowed the identification and quantification of 1501 TMT-labeled cysteine-containing peptides from about 740 proteins that underwent both modifications (Fig. 7D–7E, supplemental Table S12).
Diamide treatment induces protein S-glutathionylation and disulfide formation (14, 34). In agreement with this, we found that about 250 cysteine-containing peptides exhibited increases in S-glutathionylation upon diamide treatment (Fig. 7D, 7F). Western blot analysis confirmed GAPDH as a protein target glutathionylated by diamide in cells (supplemental Fig. S6). However, the average intensity of TMT-labeled glutathionylated peptides was decreased with diamide in INS1 cells (Fig. 7D), suggesting that in most cases, cysteines that form S-glutathionylation also may have formed other types of cysteine-based modifications. Crucially, these glutathionylated cysteines are not stable under oxidative stress conditions, and so other modifications occurred with higher frequency, leading to these modifications being dominant in terms of target occupancy, in agreement to a previous study (35). After subsequent exposure of diamide-treated INS1 cells to NaHS, the detection of the same group of peptides (Fig. 7F), which displayed increases in S-glutathionylation with diamide treatment, was largely reduced by the NaHS treatment. These data suggest that H2S reduced those glutathionylated cysteine GSH-mixed disulfide bonds, in agreement with the RTSGS concept. Surprisingly, the average levels of the total glutathionylated peptides were increased after the NaHS treatment (Fig. 7D, 7F), suggesting that the H2S signaling via glutathione persulfides can mediate protein S-glutathionylation under oxidative stress, for a subset of proteins. Together, these data highlight that diamide treatment induces S-glutathionylation of certain protein cysteine residues, that can be further modified by H2S.
In agreement with a previous report showing a dramatic reduction of total protein persulfide pools after cells being exposed to diamide (36), our MS analysis showed that the cellular sulfhydrome was largely reduced with diamide treatment, suggesting that the majority of protein persulfide pools were either being peroxidized with diamide or converted into other types of cysteine modifications (Fig. 7E). After NaHS treatment, the levels of persulfidated proteins were globally restored (Fig. 7E, 7G). This suggests that diamide-induced oxidative stress served as a catalyst to promote protein S-persulfidation. Taken together, we found that H2S signaling increases the levels of protein S-persulfidation of the group of proteins that show an increase of S-glutathionylation under oxidative stress. Bioinformatics analysis showed that cellular energy metabolism is the most enriched KEGG pathway among all the TMT-labeled proteins (Fig. 7H, supplemental Table S14). This further supports the notion that the RTSGS mechanism may regulate cellular metabolic flux under oxidative stress.
As previously mentioned, the identification of the RTSGS target proteins depends on the sequential conversion of the proteome from S-glutathionylation to S-persulfidation. We hypothesized that the increasing concentrations of NaHS would affect the physiological RTSGS protein sulfhydrome in a manner dependent on the sensitivity of the redox cysteine residues to the RTSGS PTM. We tested this hypothesis by evaluating the changes of protein S-glutathionylation. Lysates from MIN6 cells were exposed to TCEP, then to GSSG. An equal amount of GSSG-treated MIN6 cell extracts were incubated with NaHS at different concentrations 10, 50 and 100 μm in duplicates, then subjected to the TMT-BTA assay for LC-MS analysis (supplemental Fig. S7, Fig. 7I–7J, supplemental Table S13). We performed log2 transformation of TMT ratios and used log2 TMT ratios as readout.
Quantification of the sulfhydromes between the 10 and 100 μM NaHS treatment, revealed 990 RTSGS protein targets corresponding to 1535 cysteine-containing peptides. About 18% of these peptides exhibited substantial increases (1.5-fold, log2 ratio) in S-persulfidation (Fig. 7J), validating the results observed by SDS-PAGE (Fig. 6B–6C). Interestingly, we observed that 8% (124 peptides) of the total identified RTSGS cysteine peptides exhibited substantially decreases (1.5-fold, log2 ratio) in S-persulfidation, and at least 21 peptides (∼17%) contained two cysteine residues in their amino acid sequences. By contrast, only 5 dithiol peptides (∼1.8%) were found in the group of RTSGS targets (277 peptides) with increased S-persulfidation. These data suggest that the group of peptides with decreased S-persulfidation contain a higher percentage of two cysteine-containing peptides, as compared with the increased S-persulfidation group. Why did we observe this enrichment in the decreased S-persulfidation group? Our explanation is that two oxidized cysteine residues in a single peptide may have different sensitivity and reactivity to H2S. It is therefore possible that at higher concentrations of H2S, one cysteine in a glutathionylated protein is switched to a persulfide by H2S, but the second cysteine residue may be reduced to a free thiol, resulting in the retention of the peptide on the avidin column and therefore a decrease of the peptide in the eluate (supplemental Fig. S8). This can explain the enrichment of dithiol peptides in the decreased S-persulfidation group. Taken together, we conclude that the concentration of H2S in cells under oxidative stress, might be a determinant for the function of the RTSGS mechanism in the regulation of energy metabolism.
Among the 1535 RTSGS cysteine-containing peptides corresponding to 1021 proteins, 374 of those proteins did not change in S-persulfidation levels (log2 ratio between 0 and 0.6) with increasing concentration of NaHS (Fig. 7J, within the red dotted line frame). This finding suggests that a large number of the RTSGS target proteins have redox cysteine residues that exhibit greater sensitivity to H2S for the RTSGS. Low levels of H2S lead to the complete conversion from S-glutathionylation to S-persulfidation (supplemental Fig. S8). We speculate that highly sensitive RTSGS proteins (374 protein targets) may serve as H2S sensors that can quickly switch their oxidized cysteine residues to persulfides under low intracellular H2S levels in response to oxidative stress. Among these protein targets, were proteins with an NAD(P)H binding motif which are prevalent in the glycolytic and mitochondrial TCA (tricarboxylic acid) cycle pathways (supplemental Fig. S9, supplemental Table S15). Overall, the presented data may implicate the mechanism of the RTSGS as a modulator of energy metabolism under oxidative stress conditions.
DISCUSSION
The gas hydrogen sulfide (H2S) induces S-persulfidation of redox sensitive cysteine residues in the proteome and thus regulates protein function. The development of quantitative tools that can profile proteome-wide S-persulfidation is crucial to identify major protein targets and determine their relationship with the biological effects of this gas. Here, we introduced the TMT-BTA assay, which allowed the quantitative assessment of changes in protein-cysteine persulfidation in cells and in vitro under various cellular contexts. With this new quantitative proteomics approach, we have demonstrated a good correlation of the S-persulfidation status of intracellular proteins with the expression of H2S-generating enzymes. This PTM occurs only on redox sensitive cysteine residues of specific proteins. Our findings suggest the protein S-persulfidation mechanism as a potential dial switch in the regulation of cellular metabolism in pathophysiological states. Glycolytic proteins are the most highly persulfidated proteins in EndoC-BH3 cells, including fructose-bisphosphate aldolase (ALDOA), an enzyme that converts fructose-1, 6-bisphosphate (FBP) into dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3 phosphate (G3P). The regulation of FBP levels via synthesis and degradation is critical to control GSIS (37), the most important function of beta cells. ALDOA exhibits conserved amino acid residues across species, including the two cysteine residues (Cys73 and Cys178) which were extensively persulfidated in human pancreatic beta cells (Fig. 3F). A previous study showed that plant cytosolic ALDOA is subject to both S-glutathionylation and S-nitrosylation at the same cysteine residues, both resulting in inhibition of its enzymatic activity (38). Given this correlation, we speculate that S-persulfidation of Cys73 and or Cys178 might regulate ALDOA enzymatic activity, change glycolytic flux, and affect insulin secretion in pancreatic beta cells.
There are three H2S-generating enzymes, CTH, CBS and MST, in mammalian cells (1). Unlike the other two, CTH protein expression is transcriptionally induced by the stress-induced master transcription factor-ATF4, thus linking H2S synthesis to diverse physiological and pathological stress conditions (2, 27). The ATF4/CTH-mediated changes in the sulfhydrome are shown here for the first time. Pancreatic beta cells require the presence of an active stress response mechanism for their function, attributed to the easily disturbed proteostasis in the ER during fluctuations of proinsulin synthesis (39). Biochemical studies with recombinant insulin showed that the insulin B chain was a target for the ATF4/CTH-mediated S-persulfidation, thus prompting the question on the physiological significance of this regulation. Pancreatic beta cells synthesize proinsulin on ER-associated ribosomes. Proinsulin is then folded and processed to produce insulin which is stored in secretory granules and is secreted in response to nutrients, such as glucose (39). Increased H2S levels repressed GSIS in INS1 cells treated with H2S donors (supplemental Fig. S10) (40). The mechanism via which H2S decreases GSIS is not well understood. It has been shown that S-persulfidation of the KATP channel increased the activity of the ion channels, resulting in an inhibition of GSIS in pancreatic beta cells (41). We have shown here that in addition to this mechanism, S-persulfidation of insulin (specifically the B chain) may also contribute to inhibition of GSIS. Insulin B chain cysteine persulfidation is expected to break the disulfide bonds and unfold its structure (Fig. 4K), reducing the folded insulin in the secretory granules. This may be a novel physiological mechanism worth studying it further in the future for the fine-tuning of GSIS in pancreatic beta cells. Impairment of GSIS in pancreatic beta cells is associated with the development of diabetes in humans. A recent study showed that the insulin B chain fragment triggered an activation of immune T cells by enhancing the pathogenic B chain peptide loading to antigen-presenting cells, which contributed to the development of type 1 diabetes (42). Furthermore, it was recently shown that the B chain cysteine, CysB19 (also identified in the persulfidated peptide of this study, Fig. 4), is involved in aberrant disulfide-linked proinsulin complexes in the early development of diabetes (43). In addition, animal and cell culture models have showed that elevated H2S production is associated with the development of diabetes (44). Indeed, whether S-persulfidation of the insulin B chain in islets is an onset to trigger T cell activation leading to beta cell destruction remains to be further investigated.
In this report, we discovered that many protein targets (∼990 targets) were subjected to the RTSGS PTM switch from S-glutathionylation to S-persulfidation mediated by hydrogen sulfide. A key feature of this PTM switch is that both modifications occur on the same cysteine residue. The RedoxDB database, indicated about 9% of the identified RTSGS peptides are known to be modified by cysteine-based PTMs including disulfides, S-nitrosylation, S-glutathionylation and modification by sulfenic. This is in agreement to our functional annotation analysis that RTSGS proteins contain redox sensitive cysteine residues which are highly susceptible to oxidative stress associated PTMs. Pathway analyses also showed that RTSGS targets with the highest sensitivity to H2S were involved in energy metabolism. We can therefore speculate that the RTSGS mechanism and other related thiol-switch mechanisms, may regulate energy metabolism under chronic oxidative stress. Five RTSGS targets involved in glycolysis and the mitochondrial TCA cycle were found: Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), pyruvate kinase 2 (PKM2), isocitrate dehydrogenase (IDH), oxoglutarate dehydrogenase (OGDH) and pyruvate dehydrogenase (PDH). The enzymatic activities of those enzymes are inhibited by redox modifications under increased ROS levels (45). Thus, H2S-mediated RTSGS of these metabolic enzymes might be part of the mechanism to reverse the ROS-induced inhibition of glycolysis and the TCA cycle and restore metabolic flux under oxidative stress conditions.
The H2S-mediated protective effect against oxidative stress-mediated-deregulation of biological activities, is an evolutionary conserved process (24). For example, endogenous H2S production in multiple bacterial species offers resistance to oxidative stress and various classes of antibiotics (46). H2S reverses the age-associated impairment of the formation of new blood vessels in muscle (47), and the increase in H2S production in endothelial cells induced by sulfur amino acid restriction promotes glucose uptake and ATP production via glycolysis (48). However, the protective effects of H2S on cellular metabolism, may also enhance tumor growth, which is not a desired outcome for cancer therapeutics. Our studies may stimulate a revision of our current thinking of cancer cell metabolism. Tumors exhibit increased ATP production through glycolysis (49) and high H2S levels in tumors are associated with tumor growth (50). In addition, unique splicing isoforms of key metabolic enzymes contribute to increased glycolytic flux in tumors (51). For example, tumors express an alternatively spliced form of pyruvate kinase, PKM2, which enhances aerobic glycolysis and provides tumor growth advantage (32). PKM2 is a redox sensitive protein with a cysteine oxidation being inhibitory of its activity. Given that we have identified PKM2 as an RTSGS target, we suggest the idea that the H2S-mediated PTM switch on metabolic enzymes such as PKM2, is a potential mechanism for tumor growth.
In the current study, we have developed an experimental tool, named the TMT-BTA assay, for selectively trapping and tagging cysteine persulfides in the proteome of cells. Combined with tag-switch assay and quantitative MS proteomics, the TMT-BTA assay revealed the basal protein sulfhydrome of two human cell lines, and identified insulin as a novel persulfidation substrate in pancreatic beta cells. With this approach, we have identified biological pathways being targets for RTSGS, a specific reversible thiol modification in cysteine residues under oxidative stress conditions. Although the TMT-BTA assay can be used to identify S-persulfidated cysteine residues in proteins, we believe that some hyperreactive cysteines including cysteine persulfides in proteins are likely to be oxidized in the absence of thiol-blocking reagents under aerobic conditions during protein extraction. Those artificially oxidized cysteine thiols can't be labeled by biotin-conjugated NEM, nor they can be identified as S-persulfidated targets. If we lyse cells with RIPA buffer containing thiol-alkylating reagents such as IAM and NEM, free thiol groups from cysteine persulfides are alkylated during the sample preparation, and clearly the BTA would never work because Biotin-NEM tag can't attach to those “blocked” persulfide bonds in the first step of the BTA involved biotinylation of S-persulfidated cysteine thiols. Given this known potential problem, a future modification can be the addition of trichloroacetic acid (TCA) to quench protein thiol oxidation before performing the BTA assay (52). NM-Biotin selectively targets probe-accessible cysteines in native proteins. In this way, a specific fraction of cysteine persulfides buried within the body of a protein cannot react and be labeled by the probe. We envision that NM-Biotin probes can be created to profile the reactivity of different S- persulfides in the proteome, even the ones hidden in highly structured proteins. We therefore believe that other cysteine-reactive electrophilic probes such as IAM-Biotin may prove more suitable for such cysteine residues
The findings suggest that the amounts of H2S-producing enzymes control the sulfhydromes of cells, thus modulating energy metabolism. The identified pool of protein targets subjected to the RTSGS can serve as a valuable resource to study the effects of H2S on redox homeostasis and metabolism in physiological and pathological states. Although in physiological states, protein S-persulfidation may protect normal cells from ROS-induced metabolic deregulation, in cancer cells, it may be an unwanted mechanism. Cancer cells are known to reroute signaling pathways to gain a metabolic advantage to prevent oxidative damage under stress conditions, which is a signature phenotype associated with most tumors (49). We therefore envision that the H2S-mediated RTSGS may be a major contributing mechanism to reprogram cancer cell metabolism for their proliferation and survival. In this case, it would provide a rationale for cancer therapeutics to modulate the activity of the H2S-producing enzymes in tumor cells.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the PRIDE PRoteomics IDEntifications (PRIDE) database (https://www.ebi.ac.uk/pride/archive/) with the dataset identified PXD015307.
Supplementary Material
Acknowledgments
We thank Dr. Ruma Banerjee for providing recombinant CTH used in this study. We thank Dr. Danny Manor for providing IHH cells used in this study. We thank Ken Farabough for providing editorial assistance. We acknowledge the Univercell Biosolutions (Paris, France) providing the EndoC-BH3 cells used in this study.
Footnotes
* This work was supported by NIH grants R37-DK060596, R01-DK053307 (to M.H.), DK48280 (P.A.), American Diabetes Association Postdoctoral Fellowship 1-16-PDF-018 (to X.-H.G.) and National Science Centre (Poland) 2018/30/E/NZ1/00605 to D.K. The Orbitrap Elite and Fusion Lumos LC-MS instruments were purchased via an NIH shared instrument grants 1S10RR031537-01, 1S10OD023436-01 (to B.W.). The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- iodoTMT
- iodoacetyl isobaric tandem mass tags
- 3-MST
- 3-mercaptopyruvate sulfurtransferase
- Ala
- Alaine
- ALDOA
- Fructose-bisphosphate aldolase
- Asp
- Aspartic acid
- ATF4
- Activating transcription factor 4
- BioGEE
- Biotinylated glutathione ethyl ester
- BTA-TMT
- Biotin thiol assay conjugated with tandem mass tag system
- CBS
- cystathionine β synthase
- Cit
- Citric acid
- CRISPR
- clustered regularly interspaced short palindromic repeats
- CTH
- cystathionine γ lyase
- DAVID
- Bioinformatics clustering with a pathway annotation program
- DHAP
- Dihydroxyacetone phosphate
- ENO1
- Enolase 1
- FBP
- Fructose-1,6-bisphosphate
- FDR
- False discovery rate
- Fum
- Fumaric acid
- G3P
- Glyceraldehyde 3-phosphate
- GAPDH
- Glyceraldehyde 3-phosphate dehydrogenase
- GFP
- Green fluorescent protein
- Glu
- Glutamic acid
- GSH
- Reduced glutathione
- GSIS
- Glucose-stimulated insulin secretion
- GSSG
- Oxidized glutathione
- HPDP-Biotin
- N-[6-(Biotinamido)hexyl]-3 ′-(2 ′-pyridyldithio) propionamide
- IAM
- Iodoacetamide
- IDH
- Isocitrate dehydrogenase
- IHH
- Human immortalized human hepatocytes
- INS1
- rat pancreatic beta cells
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- KRB
- Krebs-Ringer Modified Buffer
- Lac
- Lactic acid
- LC
- Liquid chromatography
- LDHA and LDHB
- lactate dehydrogenase
- LFQ
- Label-free MS quantification
- Mal
- Maleic acid
- MANF
- Mesencephalic astrocyte-derived neurotrophic factor
- MAT1A
- Methionine adenosyltransferase 1A
- MIN6
- mouse pancreatic beta cells
- MS
- Mass spectrometry
- NaHS
- Sodium hydrosulfide
- NEM
- N-Ethylmaleimide
- NM-Biotin
- Maleimide-PEG2-Biotin
- OAA
- Oxaloacetic acid
- OGDH
- Oxoglutarate dehydrogenase
- PCK2
- Mitochondrial phosphoenolpyruvate carboxykinase
- PKM2
- Pyruvate kinase 2
- PLP
- Pyridoxal phosphate
- PPP
- Pentose phosphate pathway
- Pyr
- Pyruvate
- S-SG
- S-glutathionylated cysteine
- S-SH
- S-persulfidated cysteine
- Ser
- Serine
- sgRNA
- Single guide RNA
- Suc
- Succinic acid
- TCA
- Citric acid cycle
- TCEP
- Tris-carboxyethyl phosphine hydrochloride
- TEAB
- Triethylammonium bicarbonate
- TFA
- Trifluoroacetic acid
- α-KG
- α-Ketoglutaric acid.
REFERENCES
- 1. Paul B. D., and Snyder S. H. (2012) H(2)S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 13, 499–507 [DOI] [PubMed] [Google Scholar]
- 2. Paul B. D., and Snyder S. H. (2015) H2S: A Novel Gasotransmitter that Signals by Sulfhydration. Trends Biochem. Sci. 40, 687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang G., Wu L., Jiang B., Yang W., Qi J., Cao K., Meng Q., Mustafa A. K., Mu W., Zhang S., Snyder S. H., and Wang R. (2008) H2S as a physiologic vasorelaxant: hypertension in mice with deletion of cystathionine gamma-lyase. Science 322, 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hine C., Harputlugil E., Zhang Y., Ruckenstuhl C., Lee B. C., Brace L., Longchamp A., Trevino-Villarreal J. H., Mejia P., Ozaki C. K., Wang R., Gladyshev V. N., Madeo F., Mair W. B., and Mitchell J. R. (2015) Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell 160, 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kabil O., and Banerjee R. (2010) Redox biochemistry of hydrogen sulfide. J. Biol. Chem. 285, 21903–21907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miller D. L., and Roth M. B. (2007) Hydrogen sulfide increases thermotolerance and lifespan in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 104, 20618–20622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mustafa A. K., Gadalla M. M., Sen N., Kim S., Mu W., Gazi S. K., Barrow R. K., Yang G., Wang R., and Snyder S. H. (2009) H2S signals through protein S-sulfhydration. Sci. Signaling 2, ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao X. H., Krokowski D., Guan B. J., Bederman I., Majumder M., Parisien M., Diatchenko L., Kabil O., Willard B., Banerjee R., Wang B., Bebek G., Evans C. R., Fox P. L., Gerson S. L., Hoppel C. L., Liu M., Arvan P., and Hatzoglou M. (2015) Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. eLife 4, e10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mishanina T. V., Libiad M., and Banerjee R. (2015) Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat. Chem. Biol. 11, 457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McAlister G. C., Huttlin E. L., Haas W., Ting L., Jedrychowski M. P., Rogers J. C., Kuhn K., Pike I., Grothe R. A., Blethrow J. D., and Gygi S. P. (2012) Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 84, 7469–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yun J., Mullarky E., Lu C., Bosch K. N., Kavalier A., Rivera K., Roper J., Chio I. I., Giannopoulou E. G., Rago C., Muley A., Asara J. M., Paik J., Elemento O., Chen Z., Pappin D. J., Dow L. E., Papadopoulos N., Gross S. S., and Cantley L. C. (2015) Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science 350, 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shenton D., and Grant C. M. (2003) Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 374, 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hwang N. R., Yim S. H., Kim Y. M., Jeong J., Song E. J., Lee Y., Lee J. H., Choi S., and Lee K. J. (2009) Oxidative modifications of glyceraldehyde-3-phosphate dehydrogenase play a key role in its multiple cellular functions. Biochem. J. 423, 253–264 [DOI] [PubMed] [Google Scholar]
- 14. Mieyal J. J., Gallogly M. M., Qanungo S., Sabens E. A., and Shelton M. D. (2008) Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxidants Redox Signaling 10, 1941–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hiranruengchok R., and Harris C. (1995) Diamide-induced alterations of intracellular thiol status and the regulation of glucose metabolism in the developing rat conceptus in vitro. Teratology 52, 205–214 [DOI] [PubMed] [Google Scholar]
- 16. Haataja L., Snapp E., Wright J., Liu M., Hardy A. B., Wheeler M. B., Markwardt M. L., Rizzo M., and Arvan P. (2013) Proinsulin intermolecular interactions during secretory trafficking in pancreatic beta cells. J. Biol. Chem. 288, 1896–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao X. H., Bedhomme M., Veyel D., Zaffagnini M., and Lemaire S. D. (2009) Methods for analysis of protein glutathionylation and their application to photosynthetic organisms. Mol. Plant 2, 218–235 [DOI] [PubMed] [Google Scholar]
- 18. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanjana N. E., Shalem O., and Zhang F. (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guan B. J., Krokowski D., Majumder M., Schmotzer C. L., Kimball S. R., Merrick W. C., Koromilas A. E., and Hatzoglou M. (2014) Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2alpha. J. Biol. Chem. 289, 12593–12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Altelaar A. F., Munoz J., and Heck A. J. (2013) Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genetics 14, 35–48 [DOI] [PubMed] [Google Scholar]
- 22. Chung H. S., Murray C. I., Venkatraman V., Crowgey E. L., Rainer P. P., Cole R. N., Bomgarden R. D., Rogers J. C., Balkan W., Hare J. M., Kass D. A., and Van Eyk J. E. (2015) Dual labeling biotin switch assay to reduce bias derived from different cysteine subpopulations: a method to maximize S-nitrosylation detection. Circulation Res. 117, 846–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Henkel M., Rockendorf N., and Frey A. (2016) Selective and efficient cysteine conjugation by maleimides in the presence of phosphine reductants. Bioconjugate Chem. 27, 2260–2265 [DOI] [PubMed] [Google Scholar]
- 24. Filipovic M. R., Zivanovic J., Alvarez B., and Banerjee R. (2018) Chemical biology of H2S signaling through persulfidation. Chem. Rev. 118, 1253–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ulatowski L., Dreussi C., Noy N., Barnholtz-Sloan J., Klein E., and Manor D. (2012) Expression of the alpha-tocopherol transfer protein gene is regulated by oxidative stress and common single-nucleotide polymorphisms. Free Radical Biol. Med. 53, 2318–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benazra M., Lecomte M. J., Colace C., Muller A., Machado C., Pechberty S., Bricout-Neveu E., Grenier-Godard M., Solimena M., Scharfmann R., Czernichow P., and Ravassard P. (2015) A human beta cell line with drug inducible excision of immortalizing transgenes. Mol. Metabolism 4, 916–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sbodio J. I., Snyder S. H., and Paul B. D. (2018) Regulators of the transsulfuration pathway. Br. J. Pharmacol. 176, 583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., and Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 29. Han J., Back S. H., Hur J., Lin Y. H., Gildersleeve R., Shan J., Yuan C. L., Krokowski D., Wang S., Hatzoglou M., Kilberg M. S., Sartor M. A., and Kaufman R. J. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chiku T., Padovani D., Zhu W., Singh S., Vitvitsky V., and Banerjee R. (2009) H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and homolanthionine and is responsive to the grade of hyperhomocysteinemia. J. Biol Chem 284, 11601–11612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zaffagnini M., Bedhomme M., Groni H., Marchand C. H., Puppo C., Gontero B., Cassier-Chauvat C., Decottignies P., and Lemaire S. D. (2012) Glutathionylation in the photosynthetic model organism Chlamydomonas reinhardtii: a proteomic survey. Mol. Cell. Proteomics 11, M111.014142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anastasiou D., Poulogiannis G., Asara J. M., Boxer M. B., Jiang J. K., Shen M., Bellinger G., Sasaki A. T., Locasale J. W., Auld D. S., Thomas C. J., Vander Heiden M. G., and Cantley L. C. (2011) Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334, 1278–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gerber P. A., and Rutter G. A. (2017) The Role of Oxidative Stress and Hypoxia in Pancreatic Beta-Cell Dysfunction in Diabetes Mellitus. Antioxidants Redox Signaling 26, 501–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fratelli M., Demol H., Puype M., Casagrande S., Eberini I., Salmona M., Bonetto V., Mengozzi M., Duffieux F., Miclet E., Bachi A., Vandekerckhove J., Gianazza E., and Ghezzi P. (2002) Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 99, 3505–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hansen R. E., Roth D., and Winther J. R. (2009) Quantifying the global cellular thiol-disulfide status. Proc. Natl. Acad. Sci. U.S.A. 106, 422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wedmann R., Onderka C., Wei S., Szijarto I. A., Miljkovic J. L., Mitrovic A., Lange M., Savitsky S., Yadav P. K., Torregrossa R., Harrer E. G., Harrer T., Ishii I., Gollasch M., Wood M. E., Galardon E., Xian M., Whiteman M., Banerjee R., and Filipovic M. R. (2016) Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem. Sci. 7, 3414–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kebede M., Favaloro J., Gunton J. E., Laybutt D. R., Shaw M., Wong N., Fam B. C., Aston-Mourney K., Rantzau C., Zulli A., Proietto J., and Andrikopoulos S. (2008) Fructose-1,6-bisphosphatase overexpression in pancreatic beta-cells results in reduced insulin secretion: a new mechanism for fat-induced impairment of beta-cell function. Diabetes 57, 1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van der Linde K., Gutsche N., Leffers H. M., Lindermayr C., Muller B., Holtgrefe S., and Scheibe R. (2011) Regulation of plant cytosolic aldolase functions by redox-modifications. Plant Physiol. Biochem. 49, 946–957 [DOI] [PubMed] [Google Scholar]
- 39. Liu M., Hodish I., Haataja L., Lara-Lemus R., Rajpal G., Wright J., and Arvan P. (2010) Proinsulin misfolding and diabetes: mutant INS gene-induced diabetes of youth. Trends Endocrinol. Metabolism 21, 652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang G., Zhang L., Yang G., Wu L., and Wang R. (2013) Hydrogen sulfide-induced inhibition of L-type Ca2+ channels and insulin secretion in mouse pancreatic beta cells. Diabetologia 56, 533–541 [DOI] [PubMed] [Google Scholar]
- 41. Yang W., Yang G., Jia X., Wu L., and Wang R. (2005) Activation of KATP channels by H2S in rat insulin-secreting cells and the underlying mechanisms. J. Physiol. 569, 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wan X., Zinselmeyer B. H., Zakharov P. N., Vomund A. N., Taniguchi R., Santambrogio L., Anderson M. S., Lichti C. F., and Unanue E. R. (2018) Pancreatic islets communicate with lymphoid tissues via exocytosis of insulin peptides. Nature 560, 107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arunagiri A., Haataja L., Pottekat A., Pamenan F., Kim S., Zeltser L. M., Paton A. W., Paton J. C., Tsai B., Itkin-Ansari P., Kaufman R. J., Liu M., and Arvan P. (2019) Proinsulin misfolding is an early event in the progression to type 2 diabetes. eLife 8, pii: e44532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Szabo C. (2012) Roles of hydrogen sulfide in the pathogenesis of diabetes mellitus and its complications. Antioxidants Redox Signaling 17, 68–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mailloux R. J., Jin X., and Willmore W. G. (2014) Redox regulation of mitochondrial function with emphasis on cysteine oxidation reactions. Redox Biol. 2, 123–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mironov A., Seregina T., Nagornykh M., Luhachack L. G., Korolkova N., Lopes L. E., Kotova V., Zavilgelsky G., Shakulov R., Shatalin K., and Nudler E. (2017) Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 114, 6022–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Das A., Huang G. X., Bonkowski M. S., Longchamp A., Li C., Schultz M. B., Kim L. J., Osborne B., Joshi S., Lu Y., Trevino-Villarreal J. H., Kang M. J., Hung T. T., Lee B., Williams E. O., Igarashi M., Mitchell J. R., Wu L. E., Turner N., Arany Z., Guarente L., and Sinclair D. A. (2018) Impairment of an endothelial NAD(+)-H2S signaling network is a reversible cause of vascular aging. Cell 173, 74–89.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Longchamp A., Mirabella T., Arduini A., MacArthur M. R., Das A., Trevino-Villarreal J. H., Hine C., Ben-Sahra I., Knudsen N. H., Brace L. E., Reynolds J., Mejia P., Tao M., Sharma G., Wang R., Corpataux J. M., Haefliger J. A., Ahn K. H., Lee C. H., Manning B. D., Sinclair D. A., Chen C. S., Ozaki C. K., and Mitchell J. R. (2018) Amino acid restriction triggers angiogenesis via GCN2/ATF4 regulation of VEGF and H2S production. Cell 173, 117–129.e114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Szabo C., Coletta C., Chao C., Modis K., Szczesny B., Papapetropoulos A., and Hellmich M. R. (2013) Tumor-derived hydrogen sulfide, produced by cystathionine-beta-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 12474–12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Luo W., and Semenza G. L. (2012) Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol. Metabolism 23, 560–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Longen S., Richter F., Köhler Y., Wittig I., Beck K. F., and Pfeilschifter J. (2016) Quantitative persulfide site identification (qPerS-SID) reveals protein targets of H2S releasing donors in mammalian cells. Sci Rep. 6, 29808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pappireddi N., Martin L., and Wühr M. (2019) a review on quantitative multiplexed proteomics. Chembiochem 20, 1210–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the PRIDE PRoteomics IDEntifications (PRIDE) database (https://www.ebi.ac.uk/pride/archive/) with the dataset identified PXD015307.