Abstract
Fatty acid esters of hydroxy fatty acids (FAHFAs) are a newly discovered class of signaling lipids with anti-inflammatory and anti-diabetic properties. However, the endogenous regulation of FAHFAs remains a pressing but unanswered question. Here, using MS-based FAHFA hydrolysis assays, LC-MS–based lipidomics analyses, and activity-based protein profiling, we found that androgen-induced gene 1 (AIG1) and androgen-dependent TFPI-regulating protein (ADTRP), two threonine hydrolases, control FAHFA levels in vivo in both genetic and pharmacologic mouse models. Tissues from mice lacking ADTRP (Adtrp-KO), or both AIG1 and ADTRP (DKO) had higher concentrations of FAHFAs particularly isomers with the ester bond at the 9th carbon due to decreased FAHFA hydrolysis activity. The levels of other lipid classes were unaltered indicating that AIG1 and ADTRP specifically hydrolyze FAHFAs. Complementing these genetic studies, we also identified a dual AIG1/ADTRP inhibitor, ABD-110207, which is active in vivo. Acute treatment of WT mice with ABD-110207 resulted in elevated FAHFA levels, further supporting the notion that AIG1 and ADTRP activity control endogenous FAHFA levels. However, loss of AIG1/ADTRP did not mimic the changes associated with pharmacologically administered FAHFAs on extent of upregulation of FAHFA levels, glucose tolerance, or insulin sensitivity in mice, indicating that therapeutic strategies should weigh more on FAHFA administration. Together, these findings identify AIG1 and ADTRP as the first endogenous FAHFA hydrolases identified and provide critical genetic and chemical tools for further characterization of these enzymes and endogenous FAHFAs to unravel their physiological functions and roles in health and disease.
Keywords: metabolism, enzyme inhibitor, diabetes, lipid signaling, glucose metabolism, androgen induced gene 1 (AIG1), androgen-dependent TFPI-regulating protein (ADTRP), fatty acid ester of hydroxy fatty acid (FAHFA), lipid ester, threonine hydrolase
Introduction
Fatty acid esters of hydroxy fatty acids (FAHFAs)2 were first identified as a new lipid class up-regulated in mice overexpressing the glucose transporter type 4 (GLUT4) in adipose tissue (AG4OX) (1). Paradoxically, AG4OX mice are obese but more glucose tolerant (2, 3), suggesting that FAHFAs might confer protection observed in AG4OX mice from the metabolic deterioration associated with obesity (3). There are multiple FAHFA families that are distinguished by their fatty acid composition and ester regioisomers that vary by the position of the ester linkage between the two acyl chains (1) (e.g. palmitic acid ester of 9-hydroxystearic acid, 9-PAHSA; oleic acid ester of 12-hydroxystearic acid, 12-OAHSA) (Fig. S1A). In humans, PAHSA levels correlate positively with insulin sensitivity, whereas insulin-resistant individuals have lower levels in subcutaneous white adipose tissue and serum (1). The association observed in mice and humans between endogenous FAHFA levels and regulation of systemic glucose homeostasis prompted analysis of the effects of FAHFAs in the context of metabolic disorders.
Pharmacological studies with two FAHFA isomers have shown that these lipids possess anti-diabetic and anti-inflammatory properties. Oral administration of synthetic 5- and 9-PAHSA improved glucose tolerance in chow- and HFD-fed mice. The effect in chow-fed mice was ascribed to the ability of these FAHFAs to induce glucagon-like peptide 1 (GLP-1) release and to directly stimulate insulin secretion (1, 4), whereas in HFD-fed mice, the effect resulted from increased insulin sensitivity (5). PAHSA treatment also decreased expression of inflammatory cytokines in adipose tissue macrophages that accompanies obesity (1). 5- and 9-PAHSA administration was also effective in decreasing islet inflammation in nonobese diabetic mice, a type 1 diabetes mouse model (6). Furthermore, co-administration of 5- and 9-PAHSA ameliorated disease severity in a mouse model of colitis (7), indicating that the anti-inflammatory activity of these PAHSAs extends beyond metabolic models. Additionally, cell-based assays have shown that 9-PAHSA treatment can decrease lipopolysaccharide-induced secretion of pro-inflammatory cytokines in cultured dendritic cells and monocytes (1, 8), supporting the hypothesis that the anti-inflammatory effects observed in FAHFA treatments likely result from their direct influence on immune cells. Since the initial discovery of FAHFAs, hundreds of additional members of this lipid class have been identified in various organisms (8–11), and two additional FAHFAs, 13-docosahexaenoic acid hydroxy linoleic acid (13-DHAHLA) and 13-linoleic acid hydroxy linoleic acid (13-LAHLA), have been shown to exert anti-inflammatory effects in cells (8, 12). Together, these studies have revealed FAHFAs as a structurally novel class of endogenous lipids with metabolically beneficial and anti-inflammatory properties. They have also suggested that pharmacological inhibition of the enzymes that degrade FAHFAs may constitute an attractive approach to treat metabolic and inflammatory disorders. Implementation of this strategy, however, must await description of the enzymes that synthesize and degrade FAHFAs in vivo.
Activity-based protein profiling (ABPP) provided the first clue that two poorly characterized genes, androgen-induced gene 1 (Aig1) and androgen-dependent TFPI-regulating protein (Adtrp), encoded enzymes that could be involved in FAHFA degradation (13) (Fig. S1B). ABPP measures the activity of enzymes in complex proteomes using activity-based chemical probes that covalently react with the active but not inactive or inhibited forms of enzymes, subsequently labeling them with reporter tags such as biotin, fluorophores, or reactive functional groups (e.g. azides and alkynes) (14–16). One class of activity-based probes feature fluorophosphonate (FP) warheads that covalently label the majority of serine hydrolases (15), a large and diverse enzyme family that use an active-site serine residue to hydrolyze a variety of substrates ranging from lipids to proteins (17). Enzymatic function had not been ascribed to ADTRP or AIG1 prior to recent studies, which demonstrated that these enzymes react with FP probes (13). This observation suggested that AIG1 and ADTRP possessed a functional nucleophilic residue similar to that of serine hydrolase enzymes. Interestingly, AIG1 and ADTRP lack catalytic serines; instead, their activity depends on a threonine nucleophile (18).
ADTRP was initially identified as a regulator of tissue factor pathway inhibitor (TFPI) (19). TFPI is an inhibitor of blood coagulation linked to bleeding and clotting disorders (20). Depletion of ADTRP in endothelial cells reduced TFPI levels, an observation that was attributed to the ability of these two proteins to interact in the plasma membrane (19). More recently, deletion of ADTRP in mice was reported to cause vascular dysfunction, although the mechanism by which this occurs is unclear (21). AIG1 is homologous to ADTRP and was discovered in human dermal papilla cells as an androgen-regulated gene (22). It is a transmembrane protein (13, 23) that has been implicated in regulation of calcium mobilization and sensitivity to oxidative stress-associated cell death (23).
Using a panel of putative lipid substrates, we recently described that both AIG1 and ADTRP can hydrolyze FAHFAs in vitro (13). These substrate assays were carried out by overexpressing, knocking down, or chemically inhibiting AIG1/ADTRP in the presence of excess lipid substrates (13). These assays, however, do not account for unknown substrates, cofactors, regulation by compartmentalization, or protein modifications that might be present and critical for the ability of AIG1/ADTRP to function as FAHFA hydrolases in vivo (24). Conclusive demonstration of the role of AIG1/ADTRP as FAHFA hydrolases requires perturbation of function in vivo followed by lipid substrate measurements. Hence, in this study, we generated and characterized AIG1-deficient (Aig1-KO), ADTRP-deficient (Adtrp-KO), and AIG1/ADTRP double knockout (DKO) mice. We have also used a complementary pharmacological approach to inhibit AIG1 and ADTRP in wildtype (WT) mice. Both genetic and chemical blockade of these enzymes led to increased 9-FAHFA and to a lesser extent 12/13-FAHFA levels in tissues. Furthermore, no significant changes in other metabolites by lipidomics were observed in the deficiency mouse models establishing that ADTRP and AIG1 are indeed endogenous FAHFA-degrading enzymes. The degree of up-regulation of FAHFAs observed in knockout mice, however, was not sufficient to confer protection from metabolic deterioration induced by high fat diet.
Results
Generation of AIG1- and ADTRP-deficient mice
Global Aig1- and Adtrp-KO mice were generated using CRISPR/Cas9 technology (25) to create an insertion/deletion (indel) mutation at corresponding genomic loci. Guide RNAs (gRNAs) were designed to introduce an indel before the catalytic threonine of each enzyme (Fig. S2, A–D). In parallel, we generated antibodies against AIG1 and ADTRP to assess protein expression in WT and mutant mice. AIG1 was detected in the brain of WT mice as a ∼21 kDa protein that was absent in Aig1-KO brain (Fig. S2C, right panel). Similarly, ADTRP in brown adipose tissue (BAT) was detected as a ∼21 kDa protein absent in Adtrp-KO BAT (Fig. S2D, right panel). Aig1- and Adtrp-KO mice were healthy in appearance and did not exhibit developmental defects (Fig. S2E, and data not shown). Pups from heterozygous crosses were born at the expected Mendelian ratios (Fig. S2F). Aig1-KO and Adtrp-KO mutants were intercrossed to generate AIG1/ADTRP DKO mice, which also lacked an overt phenotype (data not shown). Given the absence of confounding developmental abnormalities, we proceeded to biochemically characterize Aig1-KO, Adtrp-KO, and DKO mutants and their respective WT controls.
Analysis of serine hydrolase activity in AIG1- and ADTRP-deficient tissues
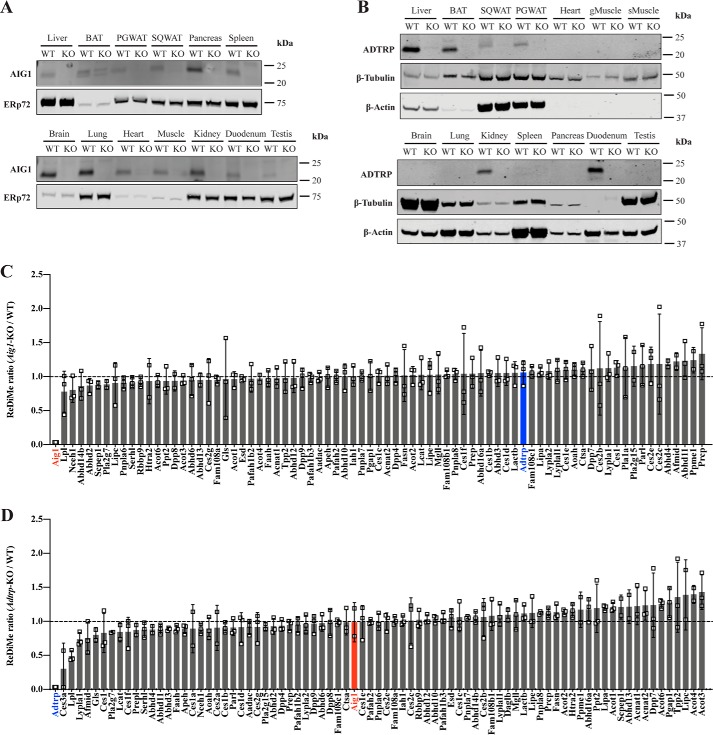
To discern the sites of greatest AIG1/ADTRP expression, we determined the distribution of AIG1/ADTRP protein across tissues using our in-house generated antibodies and samples from KO mice as negative control. AIG1 was detected in every tissue examined with the exception of BAT, where a confounding signal in Aig1-KO BAT precluded conclusive assignment (Fig. 1A). In contrast, ADTRP showed a restricted expression profile, with detectable levels found in liver, kidney, BAT, perigonadal white adipose tissue (PGWAT), subcutaneous (SQ) WAT, and duodenum (Fig. 1B).
Figure 1.
Tissue distribution of AIG1 and ADTRP and serine hydrolase activity analysis. A, tissue expression profile of AIG1 using microsomal fractions from tissues. ERp72 was used as a loading control for microsomal preparations. B, tissue expression profile of ADTRP using total lysates from tissues. β-Tubulin and β-actin were used as loading controls. gMuscle, gastrocnemius muscle; sMuscle, soleus muscle. ABPP-ReDiMe analysis of (C) WT versus Aig1-KO and (D) WT versus Adtrp-KO kidneys. Error bars represent S.D. (n = 3). Enzymes that were identified at least twice out of triplicates have been included. Tabulated peptide quantification data can be found in Table S1
To evaluate if deletion of AIG1 led to compensatory up-regulation of ADTRP activity and vice versa, or to the up-regulation of another hydrolase, we analyzed tissues from mutant mice via ABPP coupled with quantitative proteomics using duplex reductive demethylation methods (ReDiMe) where WT and KO proteomes were labeled with light and heavy formaldehyde, respectively (26). For this purpose, we used membrane fractions of kidney lysates because both AIG1 and ADTRP are expressed in this tissue. Deletion of AIG1 did not enhance ADTRP activity and no significant compensatory changes were seen in the activity across ∼80 other hydrolases detected (Fig. 1C and Table S1). Similarly, Adtrp-KO kidney lysates did not reveal compensatory up-regulation of AIG1 or other hydrolases detected (Fig. 1D and Table S1). These results suggest that AIG1 and ADTRP have specific expression patterns and do not compensate for each other.
9-PAHSA hydrolysis in AIG1- and ADTRP-deficient tissue lysates
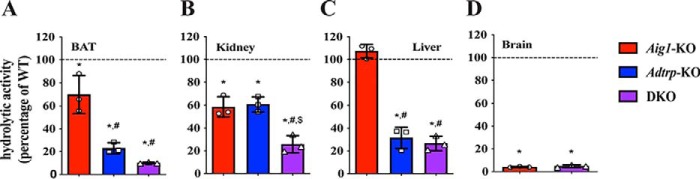
To evaluate the contribution of AIG1 and ADTRP to endogenous FAHFA hydrolytic activity, we selected a panel of tissues that express AIG1, and/or ADTRP such as BAT, liver, kidney, and brain and tested the respective membrane lysates in 9-PAHSA hydrolysis assays. Quantitative or liquid chromatography mass spectrometry (LC-MS) analysis of 9-hydroxystearic acid (9-HSA), one of the end products of 9-PAHSA hydrolysis (Fig. S1B), revealed that loss of AIG1, ADTRP, or both significantly decreased 9-PAHSA hydrolysis (Fig. 2). Dual deficiency of AIG1 and ADTRP resulted in an additive reduction of 9-PAHSA hydrolysis in BAT and kidney (Fig. 2, A and B). Decreased 9-PAHSA hydrolysis in Aig1-KO and DKO BAT lysates indicated that AIG1 is indeed expressed in BAT, which could not be definitively established by Western blot analysis alone (Fig. 1A). ADTRP was the primary FAHFA hydrolase in liver given Adtrp-KO and DKO liver hydrolytic activity was similar, whereas the activity in Aig1-KO remained unchanged compared with WT controls (Fig. 2C). Moreover, DKO liver and kidney had significant residual hydrolytic activity (Fig. S3), hinting at the existence of additional FAHFA hydrolases in these tissues. In brain lysates, where only AIG1 is expressed, 9-PAHSA hydrolytic activity was diminished in Aig1-KO lysates and was similar to DKO lysates demonstrating that AIG1 is the primary FAHFA hydrolase in brain (Fig. 2D). These data suggest that both AIG1 and ADTRP are major contributors to endogenous FAHFA hydrolytic activity.
Figure 2.
Tissue hydrolysis assays. 9-PAHSA hydrolysis assay on membrane lysates from (A) BAT, (B) kidney, (C) liver, and (D) brain from Aig1-KO, Adtrp-KO, and DKO mice represented as percent hydrolytic activity of their corresponding WT controls. Error bars represent S.D. (n = 3 per group; *, p < 0.05 versus WT control, t test; #, p < 0.05 versus Aig1-KO, t test; $, p < 0.05 versus ADTRP-KO, t test.
AIG1 and ADTRP regulate endogenous FAHFA levels
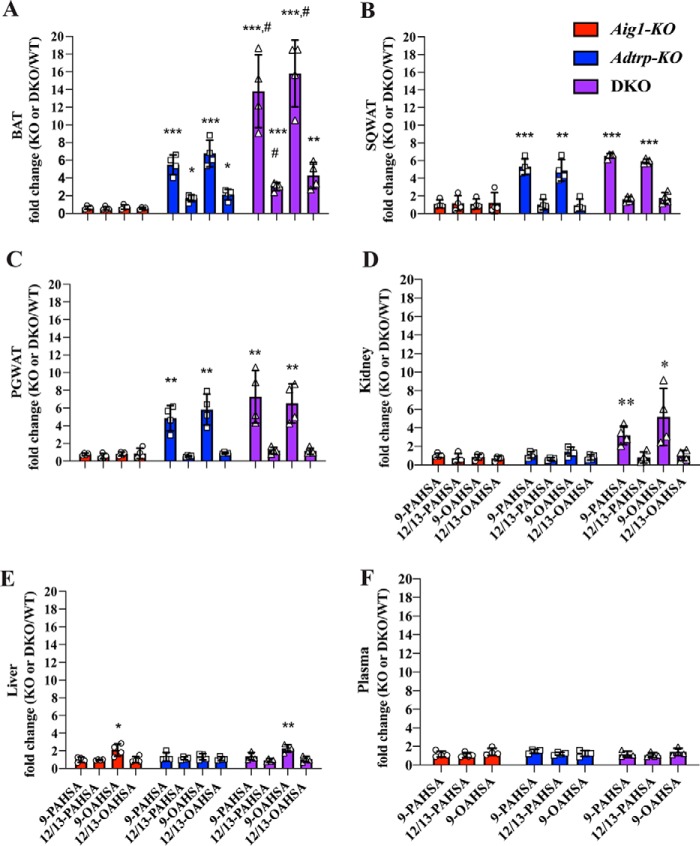
To examine the extent to which AIG1 and/or ADTRP regulate FAHFA degradation in vivo, we measured levels of endogenous 9- and 12/13-PAHSAs and OAHSAs in BAT, SQWAT, PGWAT, kidney, and liver using targeted LC-MS (27). Aig1-KO tissues showed little difference in FAHFA levels relative to WT controls. In contrast, 9-FAHFA levels were robustly increased in adipose tissue (WAT and BAT) harvested from Adtrp-KO mice, with no significant differences seen in liver or kidney (Fig. 3, and Table S2). Interestingly, the changes in FAHFA levels were isomer specific: 9-PAHSA and 9-OAHSA were increased ∼5-fold in all adipose depots from Adtrp-KO mice, whereas 12/13-PAHSA and 12/13-OAHSA levels increased only a modest ∼2-fold in BAT and were unchanged in SQWAT or PGWAT (Fig. 3, A–C). These data provide evidence of endogenous isomer-specific FAHFA degradation and imply the existence of multiple pathways that regulate FAHFA isomer metabolism.
Figure 3.
AIG1 and ADTRP regulate nonesterified FAHFA levels in vivo in a tissue-specific manner. Nonesterified 9- and 12/13-PAHSA and OAHSA measurements in (A) BAT, (B) SQWAT, (C) PGWAT, (D) kidney, (E) liver, and (F) plasma of Aig1-KO, Adtrp-KO, and DKO mice relative to the corresponding WT controls. Error bars represent S.D. (n = 4 per group. *, p < 0.05; **, p < 0.01; ***, p < 0.001, compared with WT. t test; #, p < 0.05; DKO compared with Adtrp-KO, t test).
Levels of 9-FAHFAs detected in white adipose depots of Adtrp-KO and DKO mice were similar, indicating that AIG1 has little or no FAHFA hydrolase activity in WAT (Fig. 3, B and C, and Table S2). However, loss of AIG1 further increased 9-FAHFA levels in DKO BAT relative to Adtrp-KO BAT (Fig. 3A and Table S2), showing that AIG1 contributes to degradation of 9-FAHFAs in BAT but only in the absence of ADTRP under the experimental conditions tested. Kidneys from DKO animals also showed increased 9-FAHFA levels, whereas these changes were not observed in Adtrp-KO mice or Aig1-KO mice, revealing that neither enzyme is the dominant FAHFA hydrolase in the kidney (Fig. 3D and Table S2). We observed minor differences in liver 9-OAHSA levels with very low changes in absolute levels (Fig. 3E and Table S2). Plasma levels of FAHFAs were not altered in any of the genetic models (Fig. 3F and Table S2). These findings designate AIG1 and ADTRP as the first bona fide endogenous FAHFA hydrolases identified, with ADTRP serving as the dominant FAHFA hydrolase in white and brown adipose tissue.
AIG1 and ADTRP regulate endogenous levels of TG-esterified FAHFAs in BAT
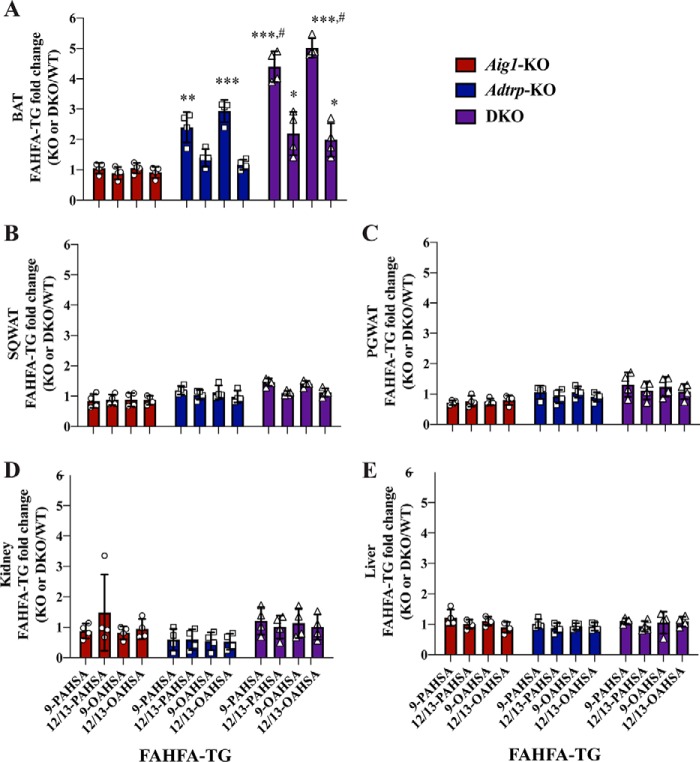
We recently described a novel pathway for FAHFA metabolism in which FAHFAs are esterified into triglycerides (TGs) to generate FAHFA-containing TGs (FAHFA-TG or TG-esterified FAHFAs) (28). FAHFA-TGs are potentially a storage form of FAHFAs; they are >100-fold more abundant in adipose tissues than nonesterified FAHFAs. TG-esterified FAHFAs are not expected to be direct substrates of AIG1 or ADTRP, but we hypothesized that increased FAHFA levels in tissues from Adtrp-KO and DKO mice might lead to greater abundance of TG-esterified FAHFAs. Indeed, we noted an increase in TG-esterified FAHFA levels in BAT of mutant mice (Fig. 4A and Table S3) that correlated with higher levels of nonesterified FAHFAs in this tissue (Fig. 3A). There was no change in the TG-esterified FAHFA pool in kidney or liver (Fig. 4, D and E, and Table S3), a finding consistent with minor changes in the nonesterified FAHFA pool in these tissues from mutant mice (Fig. 3, D and E). Surprisingly, in contrast to the elevation in nonesterified FAHFAs we noted in WAT from Adtrp-KO and DKO mice (Fig. 3, B and C), TG-esterified FAHFAs were not increased in WAT depots from these mice (Fig. 4, B and C, and Table S3). These results suggest that TG-esterified FAHFA pools are dependent on AIG1 and ADTRP function in a tissue-dependent manner.
Figure 4.
AIG1 and ADTRP regulate TG-esterified FAHFA levels in vivo in a tissue-specific manner. TG-esterified 9- and 12/13-PAHSA and OAHSA measurements in (A) BAT, (B) SQWAT, (C) PGWAT, (D) kidney, and (E) liver of Aig1-KO, Adtrp-KO, and DKO mice relative to the corresponding WT controls. Error bars represent S.D. (n = 4 per group. *, p < 0.05, **, p < 0.01, ***, p < 0.001, compared with WT, t test; #, p < 0.05, DKO compared with Adtrp-KO, t test).
ADTRP and AIG1 do not regulate other major lipid classes
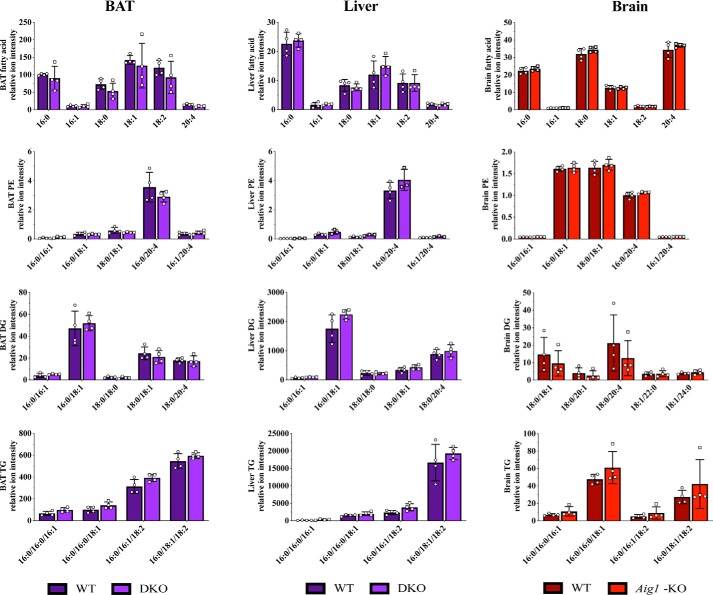
To determine whether AIG1 and ADTRP have additional substrates (24), we analyzed the lipidome of BAT and liver from DKO mice, and brain from Aig1-KO mice (Fig. 5). No significant differences in abundance of any lipid classes detected were found in any tissue examined. For instance, quantitation of fatty acid, phosphatidylethanolamine, diacylglycerol, and triacylglycerol species showed no alterations in BAT, liver, or brain of mutant mice (Fig. 5). The absence of substantial differences in other lipid species suggests that, in vivo, ADTRP and AIG1 function primarily as FAHFA-specific hydrolases.
Figure 5.
Lipidomics analysis of BAT and liver from DKO, and brain from Aig1-KO samples. Lipidomics analysis of various tissues did not identify any significant non-FAHFA changes in samples from DKO (BAT and liver) or Aig1-KO (brain) mice compared with corresponding WT controls. Fatty acids, phosphatidylethanolamine (PE), diglyceride (DG), and TG lipids are shown as representative lipid classes. Error bars represent S.D. (n = 4 per group).
Development of a dual AIG1/ADTRP inhibitor active in vivo
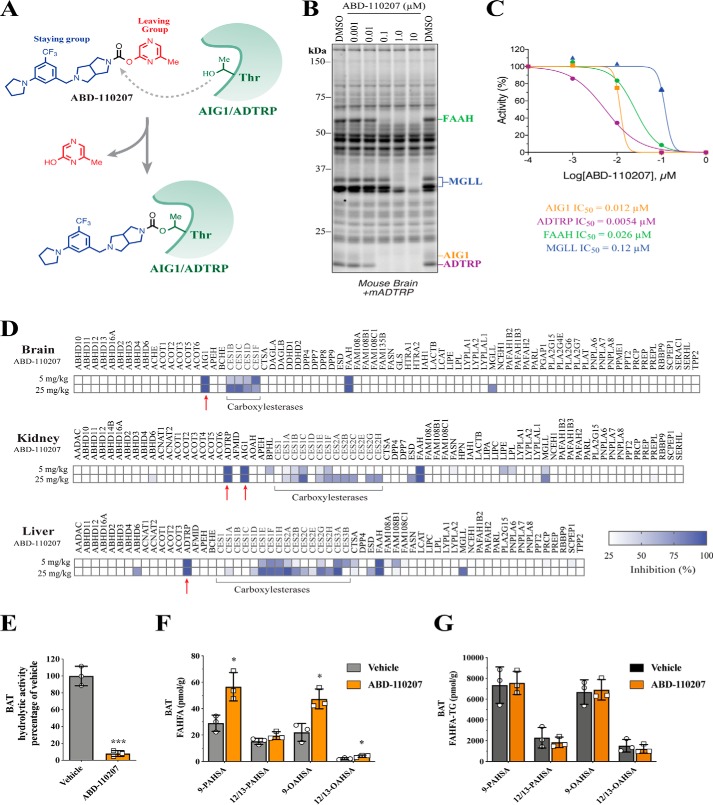
To facilitate further study of the biochemical and physiological roles of AIG1 and ADTRP, we set out to develop an in vivo active, chemical inhibitor of these enzymes. Our efforts were aided by the fact that AIG1 and ADTRP are amenable to ABPP with FP probes, and the finding that AIG1 is sensitive to electrophilic chemotypes, such as carbamates and lactones, commonly featured in serine hydrolase inhibitors (13). Thus, to identify dual AIG1/ADTRP inhibitors, a set of potent AIG1 hits identified from a serine hydrolase-directed library using ABPP-based methods in mouse brain were screened against recombinant mouse ADTRP. Compounds with potent inhibitory activity on both AIG1 and ADTRP were further triaged based on their selectivity against serine hydrolases. Among candidate AIG1/ADTRP tool compounds, ABD-110207 (Fig. 6A) showed an outstanding profile (AIG1, IC50 = 12 nm and ADTRP, IC50 = 5.4 nm) with little cross-reactivity to more than 20 serine hydrolases detectable in mouse brain (Fig. 6, B and C). We believe ABD-110207 inhibits AIG1/ADTRP through carbamylation of the active-site threonine residue (Fig. 6A) analogous to other covalent inhibitors that target fatty acid amide hydrolase (FAAH) and monoacylglycerol lipase (MGLL) and carbamylate the active-site serine of these enzymes (29, 30).
Figure 6.
Discovery and characterization of an in vivo active, dual AIG1/ADTRP inhibitor. A, structure of ABD-110207 and proposed mechanism of AIG1/ADTRP inhibition through carbamylation of active-site threonine. B, competitive gel-based ABPP profiles of ABD-110207 (0.001–10 μm) used to determine IC50 values of each serine/threonine hydrolase target. C, IC50 curves for ABD-110207 against AIG1, ADTRP, FAAH, and MGLL as determined by competitive ABPP with FP-Rh in mouse brain membrane proteomes spiked with mADTRP-transfected HEK293T cell proteomes. D, MS-based ABPP profiles in brain, kidney, and liver proteomes derived from mice treated with either vehicle or ABD-110207 (5 or 25 mg/kg, i.p.) for 4 h. Red arrows highlight the intended targets of ABD-110207, AIG1, and ADTRP. Tabulated peptide quantification data can be found in Table S4. E, 9-PAHSA hydrolytic activity in membrane lysates of BAT from mice treated with vehicle or ABD-110207 (n = 3 per group). F, nonesterified and G, TG-esterified FAHFA levels in BAT from mice treated with vehicle or ABD-110207 (n = 3 per group). Error bars represent S.D., *, p < 0.05; ***, p < 0.001, t test.
To test if ABD-110207 was suitable for use in vivo, we administered ABD-110207 to WT mice at 5 and 25 mg/kg by intraperitoneal (i.p.) injection and, 4 h later, collected brain, kidney, and liver to assess target engagement and selectivity across serine/threonine hydrolases. For this analysis, we used triplex ABPP-ReDiMe via labeling three groups differentially with light, medium, and heavy formaldehyde because it offers superior coverage, sensitivity, and resolution over gel-based ABPP (Fig. 6B). ABPP-ReDiMe profiles showed that, whereas both doses of ABD-110207 resulted in maximal (97%) inhibition of AIG1 in brain and kidney and ADTRP in kidney and liver, the 5 mg/kg dose elicited fewer off-targets (Fig. 6D). At this dose, the only off-target enzyme maximally inhibited by ABD-110207 was FAAH, whereas MGLL and several carboxylesterases showed submaximal inhibition only in a subset of tissues profiled (also see Table S4). AIG1 was not detected in the liver likely due to low expression and limited contribution to FAHFA hydrolysis observed in in vitro assays (Fig. 2C). These data support use of ABD-110207 as a first generation, in vivo active tool compound to study AIG1 and ADTRP function.
Acute inhibition of AIG1 and ADTRP increases FAHFA levels
Mouse models of whole-body deficiency can display secondary effects due to compensation mechanisms activated in response to chronic lack of a protein of interest. To rule out the possibility that elevated FAHFA levels in our AIG1/ADTRP genetic models were due to secondary changes, we tested the extent to which acute inhibition of AIG1 and ADTRP activity could result in accumulation of FAHFAs. WT mice were treated with a single dose of ABD-110207 (5 mg/kg, i.p.) and 4 h later tissues were collected to evaluate FAHFA hydrolase activity and FAHFA levels. Due to relatively low expression of AIG1 and ADTRP in BAT, ReDiMe-based assessment of enzyme inhibition was challenging. Hence 9-PAHSA hydrolytic activity was assessed in BAT membrane lysates where it was inhibited in ABD-110207–treated mice (Fig. 6E). Recapitulating what we observed in AIG1/ADTRP mutant mice (Fig. 3), acute inhibition of AIG1 and ADTRP consequentially elevated FAHFA levels in BAT (Fig. 6F). In contrast to the DKO mice, single-dose inhibition of AIG1 and ADTRP was not sufficient to increase the TG-esterified FAHFA pool in BAT (Fig. 6G), indicating that changes in this pool likely require chronic AIG1/ADTRP blockade. Free FAHFA levels in liver, and plasma of mice treated with the inhibitor were not altered (Fig. S4). These findings show that ABD-110207 is an effective in vivo active small-molecule AIG1/ADTRP inhibitor that blocks FAHFA hydrolase activity and elevates endogenous FAHFA levels in BAT, corroborating that these enzymes function as endogenous FAHFA hydrolases.
AIG1 and ADTRP deficiency are not sufficient to improve glucose metabolism in mice
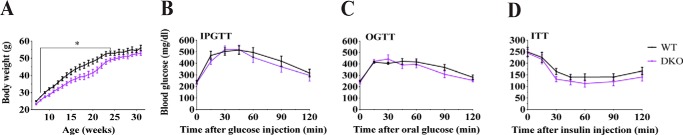
Pharmacological administration of 9- and/or 5-PAHSA improves glucose tolerance and insulin sensitivity in mice fed chow or HFD (1, 4, 5). This observation prompted us to test whether the increases we detected in endogenous 9-FAHFA levels in adipose tissue of Adtrp-KO and DKO mice are sufficient to influence glucose metabolism. Metabolic phenotyping of HFD-fed Aig1- and Adtrp-KO mice showed that deficiency of a single FAHFA hydrolase had no significant impact on body weight or glucose homeostasis via intraperitoneal glucose tolerance test (IPGTT), oral glucose tolerance tests (OGTTs), or insulin tolerance tests (ITTs) (Fig. S5). Thus, to assess if the greater elevation in endogenous FAHFAs seen in DKO tissues would affect systemic glucose metabolism, we fed DKO mice HFD and evaluated their response. Initially, DKO mice gained somewhat less weight, but this effect was transient and by the end of the study their body weight was similar to that of controls (Fig. 7A). More importantly, we found no difference in glucose tolerance or insulin sensitivity in AIG1/ADTRP-null mice relative to their WT littermates (Fig. 7, B–D). We conclude that the physiological increase in adipose tissue FAHFAs seen in AIG1/ADTRP mutant mice is unable to alter glucose homeostasis. Critically, circulating FAHFA levels are important because when FAHFAs are administered orally or by minipump, circulating FAHFA levels are increased 2–6–fold (1, 4). Hence, no change in plasma FAHFAs observed in AIG1/ADTRP mutant mice could contribute to the lack of phenotype in the mice.
Figure 7.
HFD-fed DKO mice do not have improved glucose metabolism compared with WT controls. A, body weight of WT and DKO male mice upon HFD feeding over time. Blood glucose levels during (B) OGTT after 2 g/kg of glucose administration (C) and IPGTT after 1 g/kg of glucose administration (D) of ITT after 0.8 units/kg of insulin administration in WT and DKO mice fed HFD. Error bars represent S.E., *, p < 0.05 (n = 6–8 per group).
Discussion
FAHFAs are emerging bioactive lipids that have anti-inflammatory and glucoregulatory properties, but, as newly discovered lipids, little is known about how their levels in tissues are controlled (1, 31). Using ABPP, we previously identified AIG1 and ADTRP as hydrolytic enzymes; hydrolysis assays revealed that AIG1 and ADTRP hydrolyzed FAHFAs in vitro (13). In this study, we generated genetic and chemical tools to assess the extent to which AIG1 and ADTRP function as FAHFA hydrolases in vivo. Our data show that these enzymes degrade endogenous FAHFAs, principally in adipose tissue, and appear to have no additional substrates.
Mice lacking AIG1, ADTRP, or both were developmentally similar to their WT littermates, and heterozygous crosses yielded pups in the expected Mendelian ratios (Fig. S2F). This finding contrasts with a recent report that described an independently-generated ADTRP-deficient mouse strain that exhibited moderately decreased viability and non-Mendelian offspring ratios (21). We suspect that differences between our knockout strategy, which removed 13-bases from the locus, and that of the reported approach, which deleted ∼38 kb of the genomic region between exons 3 and 5, might underlie the variance. Further studies will be required to understand the basis of the disparity between these two mouse strains.
Tissue distribution of ADTRP was restricted to a few tissues compared with the ubiquitous expression pattern observed for AIG1 (Fig. 1, A and B). This difference might suggest an evolutionary basis for tissue-specific function that has not been discovered yet. Whether FAHFA hydrolytic activity observed in vitro and in vivo for these enzymes has any biological relevance to tissue distribution remains to be studied.
Our results designate AIG1 and ADTRP as FAHFA-specific hydrolases that regulate FAHFA levels primarily in adipose tissue. Unexpectedly, not all mutant tissues in which we measured a decrease in total 9-PAHSA hydrolytic activity showed a corresponding increase in endogenous FAHFA levels (Figs. 2 and 3). For instance, endogenous FAHFA levels were unchanged in liver even though mutant lysates from this tissue hydrolyzed exogenously added 9-PAHSA at a reduced rate. This finding points to the existence of residual FAHFA hydrolytic activity in liver that compensates for the loss of AIG1 and ADTRP. It is also possible that the liver does not synthesize or store sufficient amounts of FAHFAs in the basal state to show significant accumulation upon loss of AIG1/ADTRP activity. Indeed, absolute levels of FAHFAs in liver are much lower than in adipose tissues (Table S2). Plasma levels of FAHFAs remained unchanged in AIG1 and ADTRP deficiency suggesting the up-regulation of FAHFAs observed in adipose depots were not reflected in circulating FAHFAs under the experimental conditions tested. Furthermore, most of the FAHFA up-regulation was attributed to ADTRP deficiency in vivo, whereas AIG1 deficiency had a milder effect only in the absence of ADTRP (Figs. 3 and 4). In the brain, where only AIG1 is expressed, very low amounts of FAHFAs were observed (data not shown), making it difficult to assess AIG1-dependent FAHFA hydrolysis in vivo. It is possible that AIG1-dependent FAHFA regulation occurs under certain conditions or in particular cell types or subcellular locations that escape detection in the whole tissue analysis methods used in this study.
Another unexpected finding was the specific regulation of 9-FAHFA regioisomers by AIG1 and ADTRP (Fig. 3), a phenomenon that was not observed in prior in vitro assays (13). The specificity might be due to the lower abundance of 12/13-FAHFAs relative to that of 9-FAHFAs, or to a greater biosynthetic flux, producing 9-FAHFAs over 12/13-FAHFAs. Irrespective of mechanism, this observation highlights the value of tissue measurements to identify and validate endogenous substrates, because multiple factors can influence the regulation of a substrate in vivo, in this case FAHFA regioisomers. These factors include its biosynthesis (32), colocalization with an enzyme (33), its concentrations (34), and the presence of other competitive substrates (35).
AIG1 and ADTRP deficiency also modulated TG-esterified FAHFA levels indirectly, in BAT, but not in WAT (Fig. 4). A possible explanation for this tissue specificity is that the FAHFA exchange rate between the TG-esterified and nonesterified pools might be different in BAT and WAT where there is a higher rate of incorporation of nonesterified FAHFAs into TGs in BAT versus WAT. Indeed, the absolute levels of TG-esterified FAHFAs are higher in BAT compared with WAT depots (Table S3). TG-esterified FAHFAs are depots for release of FAHFAs (28). TG-esterified FAHFAs are unlikely to be direct substrates of AIG1 and ADTRP because they are markedly different substrates than nonesterified FAHAs. Instead, the regulation of FAHFAs by AIG1 and ADTRP likely requires the release of FAHFAs from the TGs. Adipose triglyceride lipase has been identified as a hydrolase that can release FAHFAs from the TG pool (28, 36). Hence the activity of adipose triglyceride lipase, and AIG1 and ADTRP might coordinate FAHFA hydrolysis and signaling.
To complement our genetic loss-of-function studies (chronic inhibition of AIG1/ADTRP), we developed a first generation dual AIG1/ADTRP small-molecule inhibitor, named ABD-110207, that demonstrated potent activity in mice. MS-based ABPP analysis of multiple tissues from ABD-110207–treated mice revealed that this compound preferentially targets AIG1 and ADTRP across greater than 90 serine hydrolases, with only FAAH and, to a lesser extent, MGLL and several carboxylesterases as off-targets. Notably, the primary off-target enzymes detected (FAAH and MGLL) have well-defined biochemical pathways and would not be expected to interfere with the analysis herein. Acute inhibition of AIG1 and ADTRP using ABD-110207 led to a reduction in 9-PAHSA hydrolytic activity and rapid accumulation of FAHFAs in BAT (Fig. 6, E and F). The increase in FAHFA levels between vehicle- and inhibitor-treated samples was smaller than the difference seen between WT and KO models, which could be explained by the short duration of inhibitor treatment (4 h). Absence of FAHFA elevation in liver, and plasma (Fig. S4) is consistent with our observations in genetic models (Fig. 3). Furthermore, since ABD-110207 targets other serine hydrolases in liver and kidney (Fig. 6D and Table S4), these other enzymes do not appear to be involved in FAHFA hydrolysis under experimental conditions tested. Repeated dosing might lead to greater buildup of FAHFAs in BAT, and perhaps other tissues.
Pharmacological administration of 5- and/or 9-PAHSA has been shown to improve glucose metabolism and decrease inflammatory responses (1, 4, 6, 7, 37). Furthermore, the AG4OX mouse, which has dramatically elevated levels of endogenous FAHFAs, although obese, is nevertheless metabolically healthy (1). Study of the glucose metabolism of Aig1-KO, Adtrp-KO, and DKO mice in response to HFD failed to reveal protection from metabolic deterioration (Fig. S5 and Fig. 7), indicating that the extent of the increase in endogenous FAHFA levels in adipose tissue of Adtrp-KO and DKO mice, considerably lower than what is seen in multiple tissues of AG4OX mice, is not sufficient to affect systemic glucose homeostasis. Additionally, exogenous administration of FAHFAs can elevate FAHFA levels in multiple locations of metabolic relevance, such as liver and plasma. However, no increase in hepatic or circulating FAHFAs was detected in our mutant mice or with the inhibitor, which is a striking difference between systems and can explain the lack of a metabolic phenotype in the Aig1-KO, Adtrp-KO, and DKO mice. Furthermore, these results suggest that, at least for now, therapeutic approaches for targeting the FAHFA pathway are more likely to proceed by pharmacological intervention with FAHFAs rather than inhibition of these metabolic enzymes. Discordance between physiological and pharmacological function has been observed before. For instance, pharmacological treatment of fibroblast growth factor 21 (FGF21) improves several aspects of metabolic function in rodents, whereas its physiological role points to regulation of starvation response, torpor, and thirst (38, 39). Similarly, FAHFAs might have physiological functions that are distinct from those observed during pharmacological administration of these bioactive lipids, and FAHFA hydrolase-deficient mice and enzyme inhibitors will be valuable tools in dissecting such functions.
In summary, our genetic and chemical tools have revealed that 9-FAHFAs are the predominant metabolites regulated in vivo by AIG1 and ADTRP. Our detailed characterization of these unique tools demonstrates that they can be used to study the functional significance of modulating endogenous FAHFA levels in adipose tissue by limiting their degradation. Using pharmacological studies with FAHFAs as a guide, it will be of great interest to examine the effects of AIG1/ADTRP inhibition in the context of metabolism, immune responses, and vascular function. More generally, our results establish AIG1 and ADTRP as part of the druggable proteome and their role as endogenous FAHFA hydrolases.
Experimental procedures
Generation and maintenance of AIG1- and/or ADTRP-deficient mice
Aig1- and Adtrp-KO mice were generated using CRISPR/Cas9-mediated indel mutations. The sgRNAs were designed using an online tool (http://crispr.mit.edu/).3 DNA oligos corresponding to the variable sequence of sgRNAs (Table S5) were annealed and cloned into PX330 plasmid (Addgene plasmid number 42230) (40), which was used as the template to generate DNA strands that contain T7 promoter. HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs, E2040S) was used to synthesize sgRNA and the overall reaction was then purified using Mega Clear Kit (Ambion). The Transgenics core at the Salk Institute injected Cas9 mRNA and sgRNA into C57BL/6 embryos and transferred them into recipient mice to generate founder pups. Littermates were used in experiments including single deficiency mice. DKO mice were generated by crossing Aig1 and Adtrp-KO mice. WT controls were generated using common ancestor parents for DKO mice, and age-matched WT mice were used in experiments involving DKO mice. Aig1 mice were genotyped using allele-specific primers for PCR (Fig. S2C and Table S5). Adtrp mice were genotyped by PCR amplification (Fig. S2D and Table S5) and subsequent EcoRI digestion. Samples were run on 2% agarose gels for visualization. For mouse genotyping, a commercial laboratory was also utilized (Transnetyx Inc., Memphis, TN). Mice were housed in 12-h light/dark cycle and fed chow diet (PicoLab, 5053) or HFD (Open Source Diets, D12492). All animal procedures were approved by Institutional Animal Care and Use Committees of the Salk Institute, The Scripps Research Institute, and Explora Biolabs.
Western blotting
Total lysates of tissues were prepared using RIPA buffer (PierceTM, PI89901) and a homogenizer. The samples were centrifuged at 13,000 rpm for 15–30 min to clear debris. Supernatant was subsequently collected. Microsomal fractions were prepared by Dounce homogenization of tissues in PBS and serial centrifugation steps. Homogenates were centrifuged at 800 × g for 15 min at 4 °C. Supernatant was transferred and centrifuged at 6,000 × g for 15 min at 4 °C. Final supernatant was ultracentrifuged at 100,000 × g for 45 min at 4 °C. The resulting pellet was resuspended in PBS. 30 μg of protein was run on SDS-PAGE gels and stained for ADTRP (in-house antibody), AIG1 (in-house antibody), β-actin (Cell Signaling, 4970), or β-tubulin (Sigma-Aldrich, T8328) (1:1000 dilution for each primary antibody). Membranes were imaged using Licor Odyssey CLx Near-IR Fluorescence Imaging System.
ABPP-ReDiMe sample preparation
For ABPP-ReDiMe samples of WT and KO mouse tissues (three biological replicates per group), proteomes (1 mg/ml in 1 ml of PBS) were labeled with FP-biotin (5 μm) for 1 h at room temperature while rotating. For brain, liver, and kidney samples from vehicle or ABD-110207–treated mice, higher protein (3 mg/ml in 1 ml) and FP-biotin (10 μm) amounts were used to facilitate the detection of ADTRP. After labeling, the proteomes were denatured and precipitated using 4:1 MeOH/CHCl3, resuspended in 0.5 ml of 6 m urea in PBS, reduced using tris(2-carboxyethyl)phosphine (10 mm) for 30 min at 37 °C, and then alkylated using iodoacetamide (40 mm) for 30 min at room temperature in the dark. The biotinylated proteins were enriched with PBS-washed avidin-agarose beads (100 μl; Sigma-Aldrich) by rotating at room temperature for 1.5 h in PBS with 0.2% SDS (6 ml). The beads were then washed sequentially with 5 ml of 0.2% SDS in PBS (3×) and 5 ml of distilled H2O (3×). On-bead digestion was performed using sequencing-grade trypsin (2 μg; Promega) in 2 m urea in 100 mm triethylammonium bicarbonate buffer with 2 mm CaCl2 for 12–14 h at 37 °C (200 μl). Duplex or triplex reductive dimethylation was performed as previously described (26). Briefly, for duplex ReDiMe of WT versus KO tissues, either 13CD2O (heavy) or CH2O (light) was added to each sample (0.15%) followed by addition of NaBH3CN (22.2 mm). Tissue samples from vehicle- or inhibitor-treated mice (in two doses of single mice) were labeled using a triplex ReDiMe protocol where control samples (i.e. vehicle-treated) were labeled with CH2O (light) and inhibitor-treated samples were labeled with either CD2O (medium) or 13CD2O (heavy). Light and medium samples were then treated with NaBH3CN, whereas heavy samples were treated with NaBD3CN. Following a 1-h incubation period at room temperature, the reaction was quenched by addition of NH4OH (0.23%) and formic acid (0.5%). The samples were then combined and analyzed by LC/MS.
ABPP-ReDiMe MS and data analysis
MS was performed using an LTQ-Orbitrap or an Orbitrap Velos following previously described protocols (41). Peptides were eluted using a five-step multidimensional LC/MS protocol in which increasing concentrations of ammonium acetate are injected followed by a gradient of increasing acetonitrile, as previously described (42). For all samples, data were collected in data-dependent acquisition mode over a range from 400 to 1,800 m/z. Each full scan was followed by up to 7 or 30 fragmentation events for experiments using the LTQ and Orbitrap or Orbitrap Velos instruments, respectively. Dynamic exclusion was enabled (repeat count of 1, exclusion duration of 20 s) for all experiments. Data files (Thermo RAW) were converted into mzXML format using RawConverter (1.1.0.23) (43) using the monoisotopic precursor selection. The data were searched using the ProLuCID algorithm (2.1.5) (44) against an in-house reverse-concatenated mouse FASTA database (43,438 entries). This had been assembled from: 1) the UniProt Mouse Reference proteome (accessed 11/09/2012), cross-referenced with Ensembl, giving preference to long, reviewed entries, and filtered to 98% protein sequence identity using CD-HIT (21,602 forward entries) (45) and 2) common contaminant proteins (117 forward entries) from the Global Proteome Machine cRAP protein sequences (SCR_018187). ProLuCID searches specified static modification of cysteine residues (+57.021464; iodoacetamide alkylation), allowed differential modification of methionine residues (+15.994915; oxidation), and required peptides to contain at least one tryptic terminus. Each data set was independently searched with light, medium (if applicable), and heavy parameter files. For duplex ReDiMe, the search included static modifications on lysine and N termini (+28.031300 m/z (light) or + 34.063117 m/z (heavy)). For triplex ReDiMe, the search included static modifications on lysine and N termini (+28.031300 m/z (light), +32.056407 m/z (medium), or + 36.075670 m/z (heavy)). Mass tolerances were set to 50 and 600 ppm for precursor and fragment ions, respectively. The resulting peptide spectral matches were filtered using DTASelect (2.1.5) (46) with the maximum peptide false discovery rate set to the default (<5%) and enabling the following flags: use cleavage status when calculating probabilities (−trypstat), use separate models for modified peptides (−modstat), use δ mass for statistics (−dm), show all spectra for each sequence (−t 0), and remove proteins (−l-keratin) with descriptions matching “keratin.” ReDiMe ratios were quantified using in-house CIMAGE software (47). Briefly, spectral matches were used to localize a 10-min retention time window for ion chromatogram extraction using ±10 ppm mass accuracy. Extracted ion chromatograms of light and heavy peptides were required to have a coelution R2 (correlation score) of greater than or equal to 0.8 to be used for further analysis. Peptides detected as “singletons,” where extracted signal was present exclusively for either the m/z of the light or the heavy peptide, were given a ratio of 32, which is the maximum ReDiMe ratio reported here. Further quantitative analysis was restricted to FP-reactive serine and threonine hydrolases (see tables). Peptide ratios were log2 transformed and the median was used to represent the ReDiMe ratio of a protein. A per sample normalization was applied, correcting protein ratios by the median ReDiMe ratio of all hydrolases. The mean ± S.E. of ReDiMe ratios was calculated when combining replicates. We required proteins to have at least 2 unique quantified peptides and to be detected in at least 2 replicate experiments (where available). Protein ratios were calculated as the median ratio of unique peptides and transformed into percent inhibition values using the following equation.
| (Eq. 1) |
FAHFA hydrolysis assays and sample analysis using MS
Tissues were Dounce homogenized in PBS and sonicated. Crude membrane fractions were prepared using sequential centrifugation. Debris was cleared via centrifugation at 600 × g for 10 min. Supernatant was ultracentrifuged at 100,000 × g with a SW41 rotor for 45 min at 4 °C. The pellet was washed with PBS and ultracentrifuged again at 100,000 × g at 4 °C and the final pellet was resuspended in PBS. Denatured background samples were prepared by boiling lysates at 95 °C for 5 min and incubating on ice 4 times for 5 min. Separately 20–40 μg of the membrane preparation was incubated with 20 nmol of 9-PAHSA for 20 min at 37 °C. The reaction was stopped by adding 400 μl of 2:1 chloroform:methanol that contains 20 pmol of 9-hydroxyheptadecanoic acid (9-HHDA) as internal standard for quantification. Samples were vortexed and spun at 2200 × g for 5 min. The lower organic layer was transferred into a new vial and dried. Samples were resuspended in 100 μl of methanol and 5–10 μl of sample was injected to UPLC BEH C18 Column (Waters Acquity, 186002350). Species were resolved via a 15-min gradient with water with 5 mm ammonium acetate and 0.01% ammonium hydroxide (buffer A) and acetonitrile with 0.01% ammonium hydroxide (buffer B). HSA species were measured via pseudo-Multiple Reaction monitoring in negative ionization mode using Thermo Fisher TSQ Quantiva mass spectrometer (9-HSA, m/z 299.3 → m/z 299.3 (CE = 9 V); 9-HHDA, m/z 285.3 → m/z 285.3 (CE = 9 V)). 9-HSA was quantified via normalizing with internal standard 9-HHDA and subtracting the background signal obtained from the denatured samples. Hydrolysis activity was calculated per amount of protein per reaction time.
FAHFA measurements and data analysis
Tissues from ad lib fed female mice in C57BL/6J background at the age of 12–13 weeks were used for FAHFA analyses. FAHFA measurements and data analysis were performed as explained previously (28). 25–150 mg of tissue was homogenized in 1.5:1.5:3 ml of PBS:methanol:chloroform. 13C4–9-PAHSA and 13C16-PAHSA/16:0/16:0-TG (Sn1) were added as internal standards for quantification of nonesterified and TG-esterified FAHFAs, respectively. After homogenization, samples were vortexed for 15 s and centrifuged at 2500 × g for 6 min. The lower phase was transferred into a new glass vial and dried. Dried lipids were fractionated using SPE columns (28). First, columns were equilibrated with 6 ml of hexanes, the samples were then resuspended in 200 μl of chloroform and loaded on the column. Neutral lipid fraction containing TGs were eluted with 6 ml of 95:5% hexanes:ethyl acetate, and polar lipid fraction containing nonesterified FAHFAs were eluted using 4 ml of ethyl acetate. Nonesterified FAHFA fractions were dried and stored at −80 °C until analysis. TG fraction was resuspended in 200 μl of EtOH, and incubated with 0.1 m LiOH at room temperature for 24 h for saponification. The reaction was stopped with addition of 0.2 m HCl and lipids were extracted as above with addition of d31-9-PAHSA as internal standard for quantification of chemically hydrolyzed FAHFAs. Samples were passed through a second SPE and eluent containing liberated FAHFAs was collected with ethyl acetate. Samples were dried and resuspended in 60–200 μl of methanol and analyzed using LC-MS method as previously described (48). 2–10 μl of sample was injected to UPLC BEH C18 Column (Waters Acquity, 186002350). FAHFA measurements were performed using Thermo TSQ mass spectrometer via Multiple Reaction Monitoring using the transitions previously reported (1). Peak areas of the samples were normalized to the peak areas of the internal standards and the tissue weight for quantification of absolute levels.
Global lipidomics and data analysis
Lipids were extracted using a modified version of the Bligh-Dyer method (49). 25 mg of tissue was homogenized in 1:1:2 ml of PBS:methanol:chloroform with internal standards (13C16-palmitic acid, and d7-cholesterol). After homogenization, samples were vortexed for 15 s and centrifuged at 2500 × g for 5 min. Organic (bottom) phase was transferred into a new glass vial and dried under a gentle stream of nitrogen, and reconstituted in 2:1 chloroform:methanol for LC/MS analysis.
Lipidomic analysis was performed on a Vanquish HPLC online with a Q-Exactive quadrupole-orbitrap mass spectrometer equipped with an electrospray ion source (Thermo). Data were acquired in positive and negative ionization modes. Solvent A consisted of 95:5 water:methanol, solvent B was 60:35:5 isopropyl alcohol:methanol:water. For positive mode, solvents A and B contained 5 mm ammonium formate with 0.1% formic acid; for negative mode, solvents contained 0.028% ammonium hydroxide. A Bio-Bond (Dikma) C4 column (5 μm, 4.6 × 50 mm) was used. The gradient was held at 0% B between 0 and 5 min, raised to 20% B at 5.1 min, increased linearly from 20 to 100% B between 5.1 and 55 min, held at 100% B between 55 and 63 min, returned to 0% B at 63.1 min, and held at 0% B until 70 min. Flow rate was 0.1 ml/min from 0 to 5 min, 0.4 ml/min between 5.1 and 55 min, and 0.5 ml/min between 55 and 70 min. Spray voltage was 3.5 and 2.5 kV for positive and negative ionization modes, respectively. Sheath, auxiliary, and sweep gases were 53, 14, and 3, respectively. Capillary temperature was 275 °C. Data were collected in full MS/dd-MS2 (top 5). Full MS was acquired from 100 to 1500 m/z with resolution of 70,000, AGC target of 1 × 106, and a maximum injection time of 100 ms. MS2 was acquired with resolution of 17,500, a fixed first mass of 50 m/z, AGC target of 1 × 105 and a maximum injection time of 200 ms. Stepped normalized collision energies were 20, 30, and 40%.
Lipid identification was performed with LipidSearch (Thermo). Data alignment, peak integration, and comparison between sample conditions were performed with Dilu (github.com/bathyg/dilu). Mass accuracy, chromatography, and peak integration of all LipidSearch-identified lipids were verified with Skyline (50). Skyline and Dilu-generated peak areas were used in data reporting, data were normalized using internal standards.
Gel-based activity-based protein profiling
Inhibitor potency against AIG1, ADTRP, FAAH, and MGLL was determined using competitive gel-based ABPP using FP-Rh competition against ABD-110207. Full-length mouse ADTRP (Dharmacon) was recombinantly expressed in HEK293T cells. Cells were harvested 48 h following transfection and lysed by sonication in PBS. Brain tissue lysates were prepared from naive mouse sagitally-sectioned brain hemispheres by homogenizing in PBS using a TissueLyser II and sonication. Cell and tissue homogenates were fractionated by ultra-centrifugation (100,000 × g for 45 min at 4 °C) and membranes were resuspended in PBS. Mouse brain membrane proteomes (1 mg/ml) were mixed with mADTRP-transfected HEK293T proteomes (0.05 mg/ml) to allow simultaneous detection of AIG1 and ADTRP in a single assay. The mixed proteomes (50 μg) were treated with inhibitor or DMSO for 30 min at 37 °C and subsequently treated with FP-Rh (1.0 μm) for an additional 30 min at room temperature. Reactions were quenched with 4× SDS-PAGE loading buffer and FP-Rh–labeled enzymes were resolved by SDS-PAGE (10% acrylamide). In-gel fluorescence was visualized using a Bio-Rad ChemiDocTM XRS imager. Fluorescence is shown in gray scale. Quantification of enzyme activities was performed by densitometric analysis using ImageJ software (NIH). Integrated peak intensities were generated for bands corresponding to AIG1, ADTRP, FAAH, and MGLL. IC50 values were calculated through curve fitting semi-log–transformed data (x axis) by nonlinear regression with a four-parameter, sigmoidal dose response function (variable slope) in Prism software (GraphPad).
Inhibitor treatments
For measurement of 9-PAHSA hydrolytic activity and FAHFA levels in BAT upon inhibitor treatment, 2–3–month-old female C57BL/6 mice were used. For triplex ReDiMe experiments in liver, kidney, and brain 6–8–week-old male ICR mice were used. Mice were injected intraperitoneally with vehicle (18:1:1, PBS:EtOH:Emulphor) or inhibitor (5 or 25 mg/kg) in the morning. 4 h post-injection, mice were sacrificed, and tissues were collected for hydrolytic activity, FAHFA, and ABPP-ReDiMe analyses.
Metabolic phenotyping
Mice were weighed weekly for monitoring the body weight in HFD-fed mice.
Insulin tolerance test
Food was withdrawn 5 h prior to ITTs unless specified otherwise. Mice were injected intraperitoneally with the indicated amount of insulin, and blood glucose was measured over time using a glucometer (Bayer, Breeze2) via sampling from tail vein.
Intraperitoneal glucose tolerance test
Food was withdrawn 5 h prior to IPTTs unless specified otherwise. Mice were injected intraperitoneally with the indicated amount of glucose and blood glucose was measured over time using a glucometer (Bayer, Breeze2) via sampling from tail vein.
Oral glucose tolerance test
Food was withdrawn 5 h prior to OGTTs unless specified otherwise. Mice were gavaged orally with indicated amount of glucose and blood glucose was measured over time using a glucometer (Bayer, Breeze2) via sampling from tail vein.
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (51) partner repository with the dataset identifier PXD017539. Direct all other data inquiries to A. Saghatelian.
Author contributions
M. E. E., B. P. K., B. F. C., E. S., and A. S. conceptualization; M. E. E., W. H. P., D. T., C. J. D., A. F. M. P., N. N., K. M. L., C. L. H., A. R. C., M. J. N., and A. S. data curation; M. E. E., B. P. K., W. H. P., J. G. W., D. T., A. F. M. P., J. M. V., N. N., K. M. L., C. L. H., A. R. C., M. J. N., B. F. C., and E. S. formal analysis; M. E. E., B. P. K., W. H. P., J. G. W., D. T., M. J. N., and A. S. investigation; M. E. E. and A. S. writing-original draft; M. E. E., B. P. K., W. H. P., J. G. W., D. T., C. J. D., A. F. M. P., J. M. V., N. N., K. M. L., C. L. H., A. R. C., M. J. N., E. S., and A. S. writing-review and editing; C. J. D., J. M. V., N. N., K. M. L., C. L. H., A. R. C., and M. J. N. methodology; A. F. M. P. resources; B. F. C., E. S., and A. S. funding acquisition; A. S. project administration.
Supplementary Material
Acknowledgments
We thank members of Saghatelian, Saez, and Cravatt laboratories, and Dr. Barbara Kahn for discussions and experimental advice. The Mass Spectrometry Core of the Salk Institute was supported by National Institutes of Health, National Cancer Institute CCSG Grant P30 014195, National Institutes of Health Grant 1S10OD021815-01 and the Helmsley Center for Genomic Medicine, and the Transgenic Core Facility of the Salk Institute was supported by National Institutes of Health NCI CCSG P30 014195.
This work was supported by the National Institutes of Health Grants DK106210, DK114785, and DA033760, The Leona M. and Harry B. Helmsley Charitable Trust Grant 2012-PG-MED002 (to A. S.), National Cancer Institute Cancer Center Support Grant P30 CA014195 MASS core (to A. S.), a grant from the Dr. Frederick Paulsen Chair/Ferring Pharmaceuticals (to A. S.), National Institutes of Health F32 postdoctoral fellowship DK111159 (to M. E. E.), and a Hewitt Foundation for Medical Research Fellowship (to W. H. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S5 and Figs. S1–S5.
JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- FAHFA
- fatty acid esters of hydroxy fatty acids
- AIG1
- androgen-induced gene 1
- ADTRP
- androgen-dependent TFPI-regulating protein
- HFD
- high-fat diet
- GLUT4
- glucose transporter type 4
- AG4OX
- adipose GLUT4 overexpressing
- PAHSA
- palmitic acid hydroxy stearic acid
- ABPP
- activity-based protein profiling
- FP
- fluorophosphonate
- ReDiMe
- reductive demethylation methods
- DKO
- double knockout
- BAT
- brown adipose tissue
- SQWAT
- subcutaneous white adipose tissue
- PGWAT
- perigonadal white adipose tissue
- HSA
- hydroxy stearic acid
- OAHSA
- oleic acid hydroxy stearic acid
- TG
- triglyceride
- FAAH
- fatty acid amide hydrolase
- GTT
- glucose tolerance test
- MGLL
- monoacylglycerol lipase
- IPGTT
- intraperitoneal glucose tolerance test
- OGTT
- oral glucose tolerance test
- ITT
- insulin tolerance test
- 9-HHDA
- 9-hydroxyheptadecanoic acid
- GLP-1
- glucagon-like peptide 1
- TFPI
- tissue factor pathway inhibitor
- sgRNA
- single guide RNA
- i.p.
- intraperitoneal.
References
- 1. Yore M. M., Syed I., Moraes-Vieira P. M., Zhang T., Herman M. A., Homan E. A., Patel R. T., Lee J., Chen S., Peroni O. D., Dhaneshwar A. S., Hammarstedt A., Smith U., McGraw T. E., Saghatelian A., and Kahn B. B. (2014) Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 159, 318–332 10.1016/j.cell.2014.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shepherd P. R., Gnudi L., Tozzo E., Yang H., Leach F., and Kahn B. (1993) Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J. Biol. Chem. 268, 22243–22246 [PubMed] [Google Scholar]
- 3. Carvalho E., Kotani K., Peroni O. D., and Kahn B. B. (2005) Adipose-specific overexpression of GLUT4 reverses insulin resistance and diabetes in mice lacking GLUT4 selectively in muscle. Am. J. Physiol. Endocrinol. Metab. 289, E551–E561 10.1152/ajpendo.00116.2005 [DOI] [PubMed] [Google Scholar]
- 4. Syed I., Lee J., Moraes-Vieira P. M., Donaldson C. J., Sontheimer A., Aryal P., Wellenstein K., Kolar M. J., Nelson A. T., Siegel D., Mokrosinski J., Farooqi I. S., Zhao J. J., Yore M. M., Peroni O. D., Saghatelian A., and Kahn B. B. (2018) Palmitic acid hydroxystearic acids activate GPR40, which is involved in their beneficial effects on glucose homeostasis. Cell Metab. 27, 419–427.e4 10.1016/j.cmet.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou P., Santoro A., Peroni O. D., Nelson A. T., Saghatelian A., Siegel D., and Kahn B. B. (2019) PAHSAs enhance hepatic and systemic insulin sensitivity through direct and indirect mechanisms. J. Clin. Invest. 129, 4138–4150 10.1172/JCI127092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Syed I., Rubin de Celis M. F., Mohan J. F., Moraes-Vieira P. M., Vijayakumar A., Nelson A. T., Siegel D., Saghatelian A., Mathis D., and Kahn B. B. (2019) PAHSAs attenuate immune responses and promote beta cell survival in autoimmune diabetic mice. J. Clin. Invest. 129, 3717–3731 10.1172/JCI122445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee J., Moraes-Vieira P. M., Castoldi A., Aryal P., Yee E. U., Vickers C., Parnas O., Donaldson C. J., Saghatelian A., and Kahn B. B. (2016) Branched fatty acid esters of hydroxy fatty acids (FAHFAs) protect against colitis by regulating gut innate and adaptive immune responses. J. Biol. Chem. 291, 22207–22217 10.1074/jbc.M115.703835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuda O., Brezinova M., Rombaldova M., Slavikova B., Posta M., Beier P., Janovska P., Veleba J., Kopecky J. Jr., Kudova E., Pelikanova T., and Kopecky J. (2016) Docosahexaenoic acid–derived fatty acid esters of hydroxy fatty acids (FAHFAs) with anti-inflammatory properties. Diabetes 65, 2580–2590 10.2337/db16-0385 [DOI] [PubMed] [Google Scholar]
- 9. Hu T., Lin M., Zhang D., Li M., and Zhang J. (2018) A UPLC/MS/MS method for comprehensive profiling and quantification of fatty acid esters of hydroxy fatty acids in white adipose tissue. Anal. Bioanal. Chem. 410, 7415–7428 10.1007/s00216-018-1350-x [DOI] [PubMed] [Google Scholar]
- 10. Ma Y., Kind T., Vaniya A., Gennity I., Fahrmann J. F., and Fiehn O. (2015) An in silico MS/MS library for automatic annotation of novel FAHFA lipids. J. Cheminform. 7, 53 10.1186/s13321-015-0104-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu Q.-F., Yan J.-W., Zhang T.-Y., Xiao H.-M., and Feng Y.-Q. (2018) Comprehensive screening and identification of fatty acid esters of hydroxy fatty acids in plant tissues by chemical isotope labeling-assisted liquid chromatography-mass spectrometry. Anal. Chem. 90, 10056–10063 10.1021/acs.analchem.8b02839 [DOI] [PubMed] [Google Scholar]
- 12. Kolar M. J., Konduri S., Chang T., Wang H., McNerlin C., Ohlsson L., Härröd M., Siegel D., and Saghatelian A. (2019) Linoleic acid esters of hydroxy linoleic acids are anti-inflammatory lipids found in plants and mammals. J. Biol. Chem. 294, 10698–10707 10.1074/jbc.RA118.006956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parsons W. H., Kolar M. J., Kamat S. S., Cognetta A. B. 3rd, Hulce J. J., Saez E., Kahn B. B., Saghatelian A., and Cravatt B. F. (2016) AIG1 and ADTRP are atypical integral membrane hydrolases that degrade bioactive FAHFAs. Nat. Chem. Biol. 12, 367–372 10.1038/nchembio.2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cravatt B. F., Wright A. T., and Kozarich J. W. (2008) Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 77, 383–414 10.1146/annurev.biochem.75.101304.124125 [DOI] [PubMed] [Google Scholar]
- 15. Liu Y., Patricelli M. P., and Cravatt B. F. (1999) Activity-based protein profiling: the serine hydrolases. Proc. Natl. Acad. Sci. U.S.A. 96, 14694–14699 10.1073/pnas.96.26.14694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berger A. B., Vitorino P. M., and Bogyo M. (2004) Activity-based protein profiling: applications to biomarker discovery, in vivo imaging and drug discovery. Am J. Pharmacogenomics 4, 371–381 10.2165/00129785-200404060-00004 [DOI] [PubMed] [Google Scholar]
- 17. Bachovchin D. A., Ji T., Li W., Simon G. M., Blankman J. L., Adibekian A., Hoover H., Niessen S., and Cravatt B. F. (2010) Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc. Natl. Acad. Sci. U.S.A. 107, 20941–20946 10.1073/pnas.1011663107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kisselev A. F., Songyang Z., and of Chemistry G.-A. L. (2000) Why does threonine, and not serine, function as the active site nucleophile in proteasomes? J. Biol. Chem. 275, 14831–14837 10.1074/jbc.275.20.14831 [DOI] [PubMed] [Google Scholar]
- 19. Lupu C., Zhu H., Popescu N. I., Wren J. D., and Lupu F. (2011) Novel protein ADTRP regulates TFPI expression and function in human endothelial cells in normal conditions and in response to androgen. Blood 118, 4463–4471 10.1182/blood-2011-05-355370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wood J. P., Ellery P. E., Maroney S. A., and Mast A. E. (2014) Biology of tissue factor pathway inhibitor. Blood 123, 2934–2943 10.1182/blood-2013-11-512764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patel M. M., Behar A. R., Silasi R., Regmi G., Sansam C. L., Keshari R. S., Lupu F., and Lupu C. (2018) Role of ADTRP (androgen-dependent tissue factor pathway inhibitor regulating protein) in vascular development and function. J. Am. Heart. Assoc. 7, e010690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seo J., Kim J., and Kim M. (2001) Cloning of androgen-inducible gene 1 (AIG1) from human dermal papilla cells. Mol. Cells 11, 35–40 [PubMed] [Google Scholar]
- 23. Nickel N., Cleven A., Enders V., Lisak D., Schneider L., and Methner A. (2016) Androgen-inducible gene 1 increases the ER Ca2+ content and cell death susceptibility against oxidative stress. Gene 586, 62–68 10.1016/j.gene.2016.03.055 [DOI] [PubMed] [Google Scholar]
- 24. Saghatelian A., Trauger S. A., Want E. J., Hawkins E. G., Siuzdak G., and Cravatt B. F. (2004) Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry 43, 14332–14339 10.1021/bi0480335 [DOI] [PubMed] [Google Scholar]
- 25. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protocols 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inloes J. M., Hsu K. L., Dix M. M., Viader A., Masuda K., Takei T., Wood M. R., and Cravatt B. F. (2014) The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc. Natl. Acad. Sci. U.S.A. 111, 14924–14929 10.1073/pnas.1413706111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang T., Chen S., Syed I., Ståhlman M., Kolar M. J., Homan E. A., Chu Q., Smith U., Borén J., Kahn B. B., and Saghatelian A. (2016) A LC-MS-based workflow for measurement of branched fatty acid esters of hydroxy fatty acids. Nat. Protoc. 11, 747–763 10.1038/nprot.2016.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tan D., Ertunc M. E., Konduri S., Zhang J., Pinto A. M., Chu Q., Kahn B. B., Siegel D., and Saghatelian A. (2019) Discovery of FAHFA-containing triacylglycerols and their metabolic regulation. J. Am. Chem. Soc. 141, 8798–8806 10.1021/jacs.9b00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alexander J. P., and Cravatt B. F. (2006) The putative endocannabinoid transport blocker LY2183240 is a potent inhibitor of FAAH and several other brain serine hydrolases. J. Am Chem. Soc. 128, 9699–9704 10.1021/ja062999h [DOI] [PubMed] [Google Scholar]
- 30. Zvonok N., Pandarinathan L., Williams J., Johnston M., Karageorgos I., Janero D. R., Krishnan S. C., and Makriyannis A. (2008) Covalent inhibitors of human monoacylglycerol lipase: ligand-assisted characterization of the catalytic site by mass spectrometry and mutational analysis. Chem. Biol. 15, 854–862 10.1016/j.chembiol.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuda O., Brezinova M., Silhavy J., Landa V., Zidek V., Dodia C., Kreuchwig F., Vrbacky M., Balas L., Durand T., Hübner N., Fisher A. B., Kopecky J., and Pravenec M. (2018) Nrf2-mediated antioxidant defense and peroxiredoxin 6 are linked to biosynthesis of palmitic acid ester of 9-hydroxystearic acid. Diabetes 67, 1190–1199 10.2337/db17-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horton J. D., Goldstein J. L., and Brown M. S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 10.1172/JCI0215593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilfling F., Wang H., Haas J. T., Krahmer N., Gould T. J., Uchida A., Cheng J.-X., Graham M., Christiano R., Fröhlich F., Liu X., Buhman K. K., Coleman R. A., Bewersdorf J., Farese R. V. Jr., and Walther T. C. (2013) Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 24, 384–399 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tu W. C., Cook-Johnson R. J., James M. J., Mühlhäusler B. S., and Gibson R. A. (2010) Omega-3 long chain fatty acid synthesis is regulated more by substrate levels than gene expression. Prostaglandins Leukot. Essent. Fatty Acids 83, 61–68 10.1016/j.plefa.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 35. Portolesi R., Powell B. C., and Gibson R. A. (2007) Competition between 24:5n-3 and ALA for Δ6 desaturase may limit the accumulation of DHA in HepG2 cell membranes. J. Lipid Res. 48, 1592–1598 10.1194/jlr.M700081-JLR200 [DOI] [PubMed] [Google Scholar]
- 36. Paluchova V., Oseeva M., Brezinova M., Cajka T., Bardova K., Adamcova K., Zacek P., Brejchova K., Balas L., Chodounska H., Kudova E., Schreiber R., Zechner R., Durand T., Rossmeisl M., et al. (2020) Lipokine 5-PAHSA is regulated by adipose triglyceride lipase and primes adipocytes for de novo lipogenesis in mice. Diabetes 69, 300–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y. M., Liu H. X., and Fang N. Y. (2018) 9-PAHSA promotes browning of white fat via activating G-protein-coupled receptor 120 and inhibiting lipopolysaccharide/NF-κB pathway. Biochem. Biophys. Res. Commun. 506, 153–160 10.1016/j.bbrc.2018.09.050 [DOI] [PubMed] [Google Scholar]
- 38. Kliewer S. A., and Mangelsdorf D. J. (2010) Fibroblast growth factor 21: from pharmacology to physiology. Am. J. Clin. Nutr. 91, 254S–257S 10.3945/ajcn.2009.28449B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Song P., Zechner C., Hernandez G., Cánovas J., Xie Y., Sondhi V., Wagner M., Stadlbauer V., Horvath A., Leber B., Hu M. C., Moe O. W., Mangelsdorf D. J., and Kliewer S. A. (2018) The hormone FGF21 stimulates water drinking in response to ketogenic diet and alcohol. Cell Metab. 27, 1338–1347.e4 10.1016/j.cmet.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsu K. L., Tsuboi K., Adibekian A., Pugh H., Masuda K., and Cravatt B. F. (2012) DAGLβ inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat. Chem. Biol. 8, 999–1007 10.1038/nchembio.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Washburn M. P., Wolters D., and Yates J. R. 3rd. (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 10.1038/85686 [DOI] [PubMed] [Google Scholar]
- 43. He L., Diedrich J., Chu Y. Y., and Yates J. R. 3rd. (2015) Extracting accurate precursor information for tandem mass spectra by RawConverter. Anal. Chem. 87, 11361–11367 10.1021/acs.analchem.5b02721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu T., Park S. K., Venable J. D., Wohlschlegel J. A., Diedrich J. K., Cociorva D., Lu B., Liao L., Hewel J., Han X., Wong C. C. L., Fonslow B., Delahunty C., Gao Y., Shah H., and Yates J. R. 3rd. (2015) ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics 129, 16–24 10.1016/j.jprot.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fu L., Niu B., Zhu Z., Wu S., and Li W. (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tabb D. L., McDonald W. H., and Yates J. R. 3rd. (2002) DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res 1, 21–26 10.1021/pr015504q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weerapana E., Wang C., Simon G. M., Richter F., Khare S., Dillon M. B., Bachovchin D. A., Mowen K., Baker D., and Cravatt B. F. (2010) Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 10.1038/nature09472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kolar M. J., Nelson A. T., Chang T., Ertunc M. E., Christy M. P., Ohlsson L., Härröd M., Kahn B. B., Siegel D., and Saghatelian A. (2018) Faster protocol for endogenous fatty acid esters of hydroxy fatty acid (FAHFA) measurements. Anal. Chem. 90, 5358–5365 10.1021/acs.analchem.8b00503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 50. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 10.1093/bioinformatics/btq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (51) partner repository with the dataset identifier PXD017539. Direct all other data inquiries to A. Saghatelian.