Abstract
Bacterial biofilms are cellular communities that produce an adherent matrix. Exopolysaccharides are key structural components of this matrix and are required for the assembly and architecture of biofilms produced by a wide variety of microorganisms. The human bacterial pathogens Escherichia coli and Salmonella enterica produce a biofilm matrix composed primarily of the exopolysaccharide phosphoethanolamine (pEtN) cellulose. Once thought to be composed of only underivatized cellulose, the pEtN modification present in these matrices has been implicated in the overall architecture and integrity of the biofilm. However, an understanding of the mechanism underlying pEtN derivatization of the cellulose exopolysaccharide remains elusive. The bacterial cellulose synthase subunit G (BcsG) is a predicted inner membrane–localized metalloenzyme that has been proposed to catalyze the transfer of the pEtN group from membrane phospholipids to cellulose. Here we present evidence that the C-terminal domain of BcsG from E. coli (EcBcsGΔN) functions as a phosphoethanolamine transferase in vitro with substrate preference for cellulosic materials. Structural characterization of EcBcsGΔN revealed that it belongs to the alkaline phosphatase superfamily, contains a Zn2+ ion at its active center, and is structurally similar to characterized enzymes that confer colistin resistance in Gram-negative bacteria. Informed by our structural studies, we present a functional complementation experiment in E. coli AR3110, indicating that the activity of the BcsG C-terminal domain is essential for integrity of the pellicular biofilm. Furthermore, our results established a similar but distinct active-site architecture and catalytic mechanism shared between BcsG and the colistin resistance enzymes.
Keywords: biofilm, enzyme mechanism, glycobiology, cellulose, structural biology, virulence factor, bacterial cellulose synthase subunit G (BcsG), phosphoethanolamine, phosphoryl transfer, polysaccharide, surface adhesion
Introduction
A common survival strategy for many bacteria is biosynthesis and export of polysaccharides to form a protective biofilm. A biofilm consists of a cluster of microorganisms fixed within a synthesized matrix of exopolymers and allows bacteria to thrive in a range of environments because of the adherence and protection provided during colonization (1). Horizontal gene transfer between organisms and expression of genes conferring antimicrobial resistance are also promoted in biofilm communities (2). In addition, persistent colonization by the widely distributed pathogens Escherichia coli and Salmonella spp. has been linked to production of biofilms composed of the exopolysaccharide cellulose and amyloid protein fibers, termed curli (3). Cellulose-containing biofilms have also shown importance for Acetobacter, Sarcina, Rhizobium, Agrobacterium, Cronobacter, and Pseudomonas species to form symbiotic and pathogenic interactions (4–7). Secretion of bacterial cellulose produced by a broad host of different bacteria is accompanied by modification of the linear β-(1–4)-glucan polymer into a diverse range of complex biomaterials. At present, at least three distinct operon classes have been described that possess different accessory complexes for enzymatic modification of the cellulose polymer (4, 8–11). These glycans possess unique properties to suit the needs of the organism(s) producing them as part of their extracellular matrix. Furthermore, these modifications enhance bacterial persistence (12), a phenomenon only beginning to unfold in the literature. In addition to persistence, the cellulose polymer has also been proven to be important for establishment of infection and may serve as a virulence factor under some circumstances (13).
To date, all characterized cellulose biosynthesis operons encode an essential protein complex that polymerizes and exports cellulose. This complex is referred to as the bacterial cellulose synthesis (Bcs)4 complex (Fig. 1) and typically consists of the bcsABZC operon (14). The proposed function of the Bcs protein complex was originally based on its resemblance to other polysaccharide excretion systems, like the better-characterized alginate system from Pseudomonas spp. (15, 16). However, structural and functional studies have provided evidence of a role of specific gene products. Biosynthesis of cellulose chains occurs on the cytoplasmic face of the inner membrane, where BcsA acts in a processive fashion to add glucose subunits from UDP-glucose substrates onto the growing chain (17–19). Polymerization occurs concomitantly with transport across the inner membrane through a pore in BcsA that leads to emergence of the growing chain into the periplasm, where it interacts with BcsB, a protein containing a carbohydrate-binding domain (18). Although the precise molecular mechanism of cellulose export across the periplasm and outer membrane of bacteria is still unknown, in the analogous alginate system, this process occurs through the coordinated action of a tetratricopeptide repeat–containing protein (AlgK) and an outer membrane β-barrel–containing protein (AlgE), with release of the nascent chain occurring through the action of alginate lyase (AlgL) (15, 16, 20, 21). BcsC in the cellulose system has been proposed to carry out the functions of AlgK and AlgE because it has homologous regions for the tetratricopeptide repeat and β-barrel domains (15, 21). Studies with recombinant BcsZ have provided further evidence of this model, as functional and structural results demonstrated that BcsZ hydrolyzes cellulose polymers and may reside in the periplasm, thereby positioning it to act in a similar manner as AlgL (20, 22).
Figure 1.
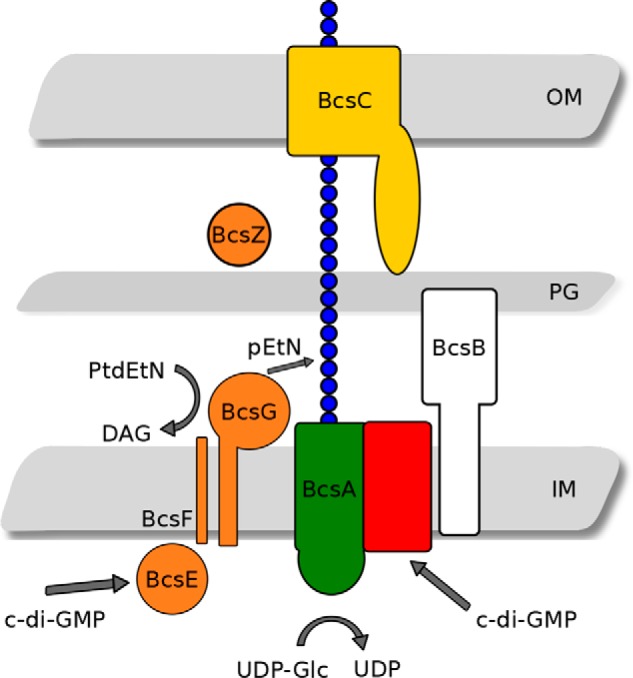
The enterobacterial cellulose synthase. The role of bcsEFG is proposed to be pEtN modification of the cellulose polymer in the periplasmic space (model adapted from Thongsomboon et al. (26). OM, outer membrane; PG, peptidoglycan; IM, inner membrane layers of the bacterial cell wall.
The role of accessory genes in these operons, previously of unknown function, is beginning to be understood. In type II cellulose operons, the function of the bcsEFG operon adjacent to the core bcsABCZ operon remained unknown and was simply thought to be necessary for maximal cellulose production (23–25). BcsE has been shown to encode a novel c-di-GMP receptor resembling the GGDEF motif inhibitory site, termed a GGDEF I site–like domain, that is necessary for optimal cellulose biosynthesis (24). More recently, BcsG has been demonstrated to aid in modification of the cellulose polymer with a phosphoethanolamine (pEtN) group on every other glucosyl residue and participate in a direct interaction with BcsA (26). The specific role of BcsG has been proposed to be as a pEtN transferase directly modifying the cellulose polymer, although its sufficiency to do so has not been demonstrated before. Sun et al. (27) have shown that BcsG exhibits phospholipase activity, fulfilling at least part of its proposed role in pEtN cellulose biosynthesis (27). Furthermore, the structure of the C-terminal catalytic domain from Salmonella enterica serovar Typhimurium has also been reported by Sun et al. (27) (PDB code 5OLT), and site-directed mutagenesis studies of a limited set of residues reflect particular phenotypes, pointing to the importance of bcsG to the pEtN cellulose biofilm. In conjunction, the role of the BcsG N-terminal domain in assembly of the mature bacterial cellulose synthase was confirmed (27). However, the consequences of pEtN-unmodified cellulose regarding the properties of the biofilm and the complete function and mechanism of BcsG as a cellulose pEtN transferase have not yet been demonstrated.
Here, we report the structure of the C-terminal catalytic domain of BcsG from E. coli (EcBcsGΔN) and outline the essential catalytic motifs required for pEtN modification of cellulose in E. coli AR3110 through a clearly defined functional complementation experiment specific for pellicle biofilm phenotypes. We also demonstrate for the first time that EcBcsGΔN acts as a phosphoethanolamine transferase capable of acting alone (i.e. in the absence of other Bcs proteins) to directly decorate cello-oligosaccharides in vitro. Finally, we present evidence that EcBcsG proceeds through a Ping Pong Bi Bi reaction pathway via a pEtN-Ser278 covalent intermediate, a novel mechanism for the pEtN transferase family.
Results and discussion
BcsGΔN acts as a phosphoethanolamine transferase in vitro
The C-terminal catalytic domain of EcBcsG was subcloned using the expression construct composed of residues Ala164 to Gln559 from UniProt entry P37659 and included an N-terminal hexahistidine tag conferred by the pET28a expression vector. Protein from this expression construct was purified to apparent homogeneity via nickel affinity chromatography and size-exclusion chromatography. To assess the ability of EcBcsGΔN to catalyze the transfer of pEtN to cellulose, we developed an in vitro activity assay. We provided recombinant EcBcsGΔN with the commercially available substrate analogues O-phosphorylethanolamine (Sigma) and the lysophospholipid 1-myristoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine (14:0 LysoPE, Avanti), but we were unable to detect an enzymatic product with these substrates using TLC or MS. Instead, we synthesized the chromogenic substrate analogue para-nitrophenyl phosphorylethanolamine (p-NPPE) as reported previously (28) and assessed the ability of EcBcsGΔN to cleave the pEtN group. EcBcsGΔN was able to cleave the pEtN group, liberating free para-nitrophenolate, with a specific activity of 3.1 ± 0.2 nmol min−1 mg of protein−1. We then further investigated whether EcBcsGΔN was capable of transferring this pEtN group to cellulose by addition of a 100-fold excess of d-glucose (Sigma) or cello-oligosaccharides as substrate analogs for cellulose, with lengths ranging from n = 2 to 6 (Megazyme). Due to the limited solubility of cellohexaose, it was tested at 3 mm, the maximum our assay allowed. We observed a significant increase in the rate of formation of para-nitrophenolate in the presence of the tetrasaccharide, pentasaccharide, and hexasaccharide (16.8 ± 0.5, 31.6 ± 1.5, and 33.1 ± 1.1 nmol min−1 mg of protein−1, respectively; ANOVA, F(6,20) = 1283, p < 0.0001; Fig. 2). To test the substrate specificity of EcBcsGΔN, we repeated this experiment using chito-oligosaccharides (n = 2 to 6) as acceptors (Fig. S1). EcBcsGΔN displayed significantly different activity on chito-oligosaccharides compared with cello-oligosacchairdes (two-way ANOVA, F(1,24) = 124.7, p < 0.0001). A post hoc Sidak's test showed that there was no significant difference in activity for chito-oligosaccharides we tested compared with an acceptor-free control (p > 0.2 for all cases). These results suggest that chitin, a linear polysaccharide of the amino sugar GlcNAc with β-1,4 linkage, is a poor substrate for EcBcsGΔN.
Figure 2.
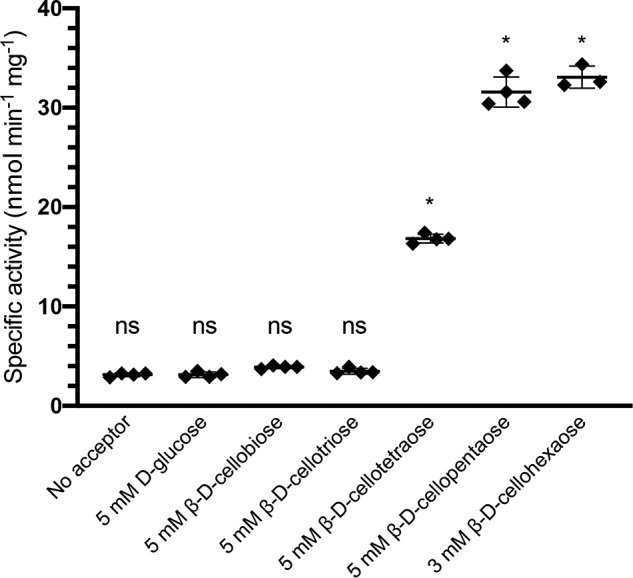
The measured rate of p-NPPE cleavage by EcBcsGΔN in the presence and absence of cello-oligosaccharides. The presence of cello-oligosaccharides with a length of >3 significantly increased the rate of p-NPPE cleavage. Asterisks denote significant differences from the acceptor-free control (ANOVA, F(6,20) = 1283; *, p < 0.0001; n = 4 for all groups except hexasaccharide, where n = 3). ns, not significant. Error bars depict S.D.
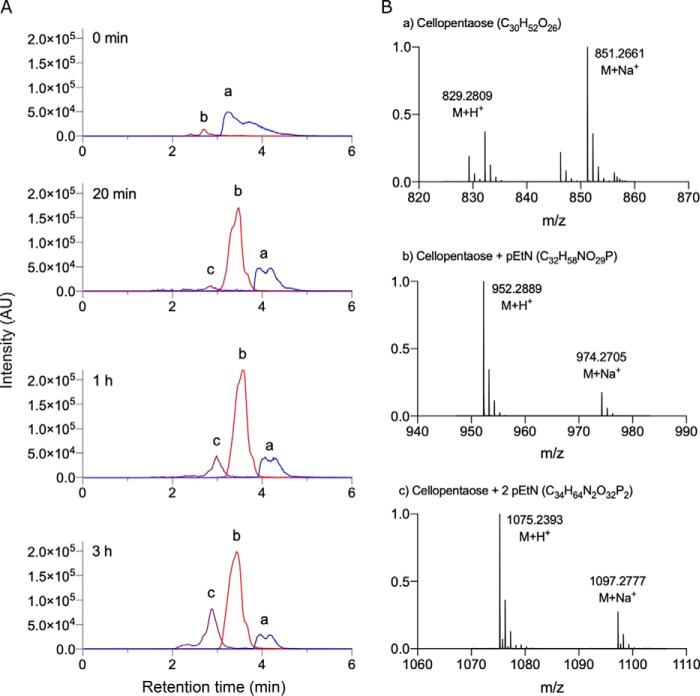
To confirm that the increase in the rate of turnover of p-NPPE by EcBcsGΔN was the result of transferase activity, we separated and analyzed the reaction products over time from the sample containing cellopentaose by LC-MS. In addition to unreacted p-NPPE ([C8H12NO6P]+H+, m/z 263.16) and cellopentaose ([C30H52O26]+H+, m/z 829.28), we observed accumulation of two related ions over time with reduced retention times and m/z values corresponding to the expected molecular mass of the mono- and disubstituted pentasaccharide ([C32H58NO29P]+H+, m/z 952.29; [C34H64N2O32P2]+H+, m/z 1075.24; Fig. 3). We did not observe these species in an enzyme-free control, which instead contained only the m/z species corresponding to the starting materials. Interestingly, we never observed ions that corresponded to modification with more than two pEtN groups. Acquired MS/MS spectra indicated that the two pEtN groups were linked to the two terminal sugars of the pentasaccharide, which represent the least sterically hindered sites for addition. Although no reliable cross-ring cleavage products were identified in MS/MS scans that would suggest linkage of the pEtN groups, these modifications are presumably present on the C6 hydroxyl group, as demonstrated by Thongsomboon et al. (26).
Figure 3.
Identification of the EcBcsGΔN enzymatic products by LC-MS. A, extracted ion chromatograms for cellopentaose (a), the mono-pEtN–substituted cellopentaose ion (b), and the di-pEtN–substituted cellopentaose ion (c), sampled from enzyme reactions over time, show the accumulation of products in the reaction mixture. B, representative mass spectra of ions a, b, and c from A, used to identify the enzyme products and unreacted starting material.
Our results support the model proposed by Thongsomboon et al. (26), which identified BcsG as the likely pEtN transferase based on bcsG deletion phenotypes. For the first time, we show at the protein level that the C-terminal domain of BcsG is sufficient for pEtN transfer in vitro. Furthermore, Sun et al. (27) demonstrated that the C-terminal domain of BcsG from S. enterica serovar Typhimurium is capable of cleaving pEtN phospholipids, but evidence of transfer to a cellulose acceptor was not demonstrated. Our results further expand these data to include direct transfer of the pEtN group to cello-oligosaccharides by the BcsG C-terminal domain in the absence of Bcs proteins, which was only predicted from the model originally proposed (26).
The overall structure of EcBcsGΔN
Using the same construct of EcBcsGΔN as described above, the de novo crystal structure was determined using the single-wavelength anomalous dispersion method with anomalous scattering on crystals prepared with uniformly l-selenomethionine–labeled EcBcsGΔN (hereafter called EcBcsGΔN-Se). The high-resolution diffraction data were used to determine a P 21 21 21 space group with two copies of EcBcsGΔN-Se in the asymmetric unit, although we observed that EcBcsG purified by gel filtration as a monomer (data not shown). Following model building, the structure was refined to an Rwork and Rfree of 0.215 and 0.225, respectively (see Table S1 for data and model statistics). The EcBcsGΔN-Se model presented excellent stereochemical parameters and covered the apparent entirety of the catalytic domain. The structure (numbered according to the amino acid position of the full-length protein, 164–559) is complete at the C terminus (i.e. to amino acid 559) and truncates in an unstructured loop immediately adjacent to the His6 tag at the N-terminal region of the soluble domain. Ramachandran analysis of the structure was satisfactory, and the model had only two (0.32%) side-chain rotamer outliers that were present only in the B chain. The atomic coordinates and relevant data for EcBcsGΔN-Se were deposited to the Protein Data Bank and are available under the accession number 6PCZ.
The completed structure of EcBcsGΔN-Se was used as a molecular replacement search model to solve the structure of a native EcBcsGΔN crystal (see Table S1 for data and model statistics). A high-quality dataset, in full agreement with the structure of EcBcsGΔN-Se, with a P 1 21 1 space group was resolved to 1.75 Å. This model also contained two copies in the asymmetric unit, but packing of the two protein molecules was observed along an alternate face compared to EcBcsGΔN-Se. Refinement of the native structure resulted in Rwork and Rfree of 0.165 and 0.198, respectively. This model also covers the entire C-terminal catalytic domain with truncation at the N terminus near the His6 tag. Similar stereochemical parameters are observed in the native structure compared with EcBcsGΔN-Se, and three Ramachandran outliers are present across the A and B chains near the disordered termini of the polypeptide. The structural data for the native model of EcBcsGΔN were also deposited into the Protein Data Bank under accession number 6PD0.
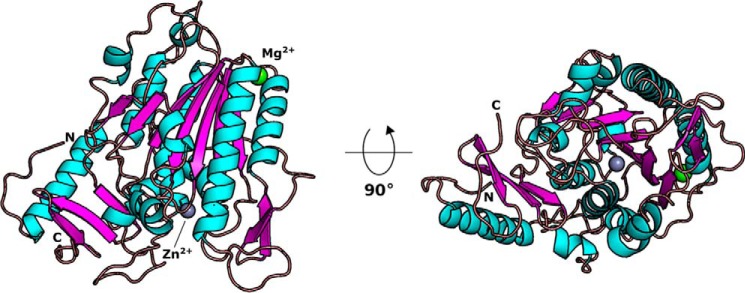
The crystal structure of EcBcsGΔN reveals a three-dimensional fold bearing 15 α-helical and 13 β-sheet secondary structure elements (Fig. 4). A seven-strand parallel β-sheet is present at the core of the protein, flanked by five α-helices. A large α-helical component and a small four-stranded antiparallel β-sheet are present at the C-terminal region of the fold, packed against the core of the protein. The structure truncates in a loop, presumably the linker between the catalytic domain and the N-terminal transmembrane domain.
Figure 4.
The molecular structure of EcBcsGΔN. Shown is a composite ribbon model of EcBcsGΔN containing Mg2+ and Zn2+ ions. The EcBcsGΔN C-terminal domains folds in an overall α/β fold, typical of the alkaline phosphatase superfamily.
We observed electron density in our calculated Fourier Fo-Fc maps that corresponded to two unique metal ligands in our structures of EcBcsGΔN (Fig. S2, A and B). Although virtually all known pEtN transferases contain one or more Zn2+ ions in their respective active sites, we were hesitant to model Zn2+ into these sites without experimental evidence to support their identity because of the low sequence similarity of EcBcsG to other pEtN transferases. In addition, Zn2+ was not a component introduced in any step of EcBcsGΔN purification or crystallization. Instead, we modeled Mg2+ into both sites, as we observed that Mg2+ had a stabilizing effect on our recombinant EcBcsGΔN and so was included in all steps of protein purification and crystallization. Metal validation by the CheckMyMetal server (29) revealed that one of these sites was a good fit for Mg2+, although the second site was more typical for Co2+, Cu2+, or Zn2+. To unambiguously demonstrate the metal content of our recombinant EcBcsGΔN, we digested the protein in trace metal–grade nitric acid and analyzed these samples by inductively coupled plasma-atomic emission spectroscopy (ICP-AES) (Fig. S1C). The resultant spectra from analysis demonstrated that the metal ligands we observed in EcBcsGΔN were one equivalent each of Mg2+ and Zn2+, and so these metals were modeled into the final structure of EcBcsGΔN.
BcsG is a member of the pEtN transferase family
In good agreement with the structure of BcsG from S. enterica serovar Typhimurium, our structure shows that the soluble periplasmic domain of EcBcsGΔN folds in a globular structure that is typical of the alkaline phosphatase superfamily, with global alignment resulting in an average root mean square deviation of 0.531 Å across 2278 atoms.
A structural homology search on the DALI server further suggested that EcBcsGΔN belongs to the pEtN transferase family, as among the top scores are the representative family members EptC from Campylobacter jejuni (CjEptC, PDB code 4TN0 (30)), EptA from Neisseria meningitidis (NmEptA, PDB code 4KAY (31)), and the mobile colistin resistance factor 1 (MCR-1, PDB code 5K4P) with z-scores of 20.4, 20.2, and 20.4, respectively. Structural superposition of EcBcsGΔN with these three structures resulted in an average root mean square deviation of 3.0 Å across 146 equivalent Cα atoms contained within the conserved central β/α fold (Fig. S3).
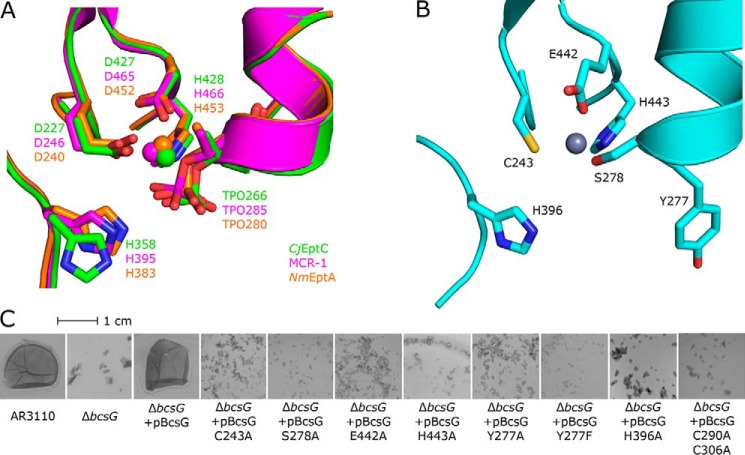
A subsequent amino acid sequence alignment of EcBcsGΔN, MCR-1, CjEptC, and NmEptA revealed several blocks of conserved residues among these structures. However, these sequences aligned poorly with EcBcsG (Fig. S4) because of large alignment gaps, and the catalytic features of EcBcsG needed to be aligned with these pEtN transferases manually (Figs. S3 and S4). A review of the literature on the pEtN transferase family revealed that these blocks of conservation are primarily contained within a highly conserved active site fold, typical for the family (32). This conserved active site is shaped by a solvent-accessible pocket at the C terminus of the central β-sheet fold and shows binding of a conserved catalytic Zn2+ ion (Fig. 5B). This catalytic Zn2+ is coordinated by an aspartic acid/glutamic acid/histidine triad strictly conserved in the family (Fig. S3, blocks 4 and 1, and Fig. 5B). Additionally, a conserved threonine residue (Fig. S3, block 2, and Fig. 5B) serves as the catalytic nucleophile, as evidenced by the observation of this residue as phosphothreonine in crystallo in almost all structures of pEtN transferase family members available to date (30, 31, 33).
Figure 5.
The active-site architecture of the lipid A pEtN transferases is partially conserved in EcBcsGΔN. A, structural superposition of the conserved active-site pocket of CjEptC (green), MCR-1 (magenta), and NmEptA (orange). B, the active-site pocket of EcBcsGΔN. C, biofilm phenotypes of a nonpolar ΔbcsG deletion in E. coli AR3110 functionally complemented by pBcsG or single alanine variants thereof. Replacement of the Zn2+-binding triad Cys243, Glu442, and His443; the catalytic nucleophile Ser278; His396; the Cys pair Cys290-Cys306; or the surface-exposed Tyr277 with alanine resulted in failure to rescue the fragile pellicle phenotype, demonstrating the importance of these residues for transferase activity in vivo.
In EcBcsGΔN, the conserved histidine is present as His443, and residues equivalent to the Asp and Glu are present as Glu442 and Cys243, respectively (Fig. 5A). In place of the catalytic threonine, Ser278 is equivalently positioned in a plausible orientation for nucleophilic attack and accommodation of a pEtN group. However, we did not observe density in our structure attributable to a covalently bonded phosphate group, which is in contrast to typical crystal structures of this family. In an attempt to resolve such a substrate-bound state of EcBcsGΔN, we collected high-resolution datasets on intact crystals grown and/or harvested in solutions containing pEtN and related phosphorylated substrates. Unfortunately, no clear electron density attributable to pEtN or phosphate was observed, and so we could not confirm the identity of Ser278 as the catalytic nucleophile from the structure of the holoenzyme alone.
Furthermore, in the structural model of EcBcsGΔN, we observed a strictly conserved histidine required for activity of other family members as His396 (30, 33). We also observed Tyr277 lining the active-site pocket at a position occupied by Ser or Ala in other family members (30, 31, 33). We hypothesized that the presence of this tyrosine in EcBcsG may facilitate accommodation of cellulose in close proximity to the catalytic serine and the Zn2+ ion at the active center, as the Tyr277 hydroxyl is oriented outward toward bulk solvent and not inward to the face of the Zn2+ at the catalytic center.
The transfer of pEtN to substrates other than lipid A or the lipopolysaccharide core layer has been demonstrated for other enzymes in the pEtN transferase family. For example, in addition to its cellular role as an intrinsic colistin resistance factor, CjEptC has been shown to modify periplasmic N-linked glycan of various endogenous proteins in an analogous fashion (34, 35) and is essential for flagellar assembly and motility in C. jejuni (30, 35). Additionally, OpgE in E. coli has been shown to transfer pEtN to osmoregulated periplasmic glucan, a substrate similar to cellulose (36). To test whether EcBcsG might also serve a secondary role as an intrinsic colistin resistance factor, we performed a minimum inhibitory concentration assay for colistin under biofilm-permissive conditions in the cellulose+ E. coli K12 strain AR3110 and nonpolar chromosomal deletion of bcsG in this background (AR3110 ΔbcsG). We observed a measured minimum inhibitory concentration value of 2 μg ml−1 in both backgrounds (Fig. S5), indicating susceptibility to colistin and demonstrating that EcBcsG does not confer intrinsic colistin resistance at endogenous levels.
BcsG shares a conserved active site with the pEtN transferase family
To map the active site of BcsG, we performed a functional complementation experiment in the E. coli AR3110 ΔbcsG background, acquired from Thongsomboon et al. (26). We supplemented, in trans, bcsG and site-directed mutants of bcsG that contained single alanine substitutions of residues Cys243, Tyr277, Ser278, His396, Glu442, and His443 and assessed the biofilm phenotype of these cells (Fig. 5C). In agreement with the findings of Thongsomboon et al. (26), the E. coli AR3110 background produced a rigid pellicle phenotype when cultured under biofilm-permissive conditions. Nonpolar deletions of bcsG in this background (E. coli AR3110 ΔbcsG) instead grew as a fragile pellicle phenotype that was highly susceptible to shearing under mechanical stress. In a similar fashion as Sun et al. (27), we then constructed a library of BcsG variants and assessed their ability to functionally complement a bcsG deletion background, although we used a distinct phenotype experiment in which we directly assessed the integrity of the biofilm. We observed that the fragile pellicle phenotype of E. coli AR3110 ΔbcsG was rescued by supplementation of WT bcsG in trans. Furthermore, complementation with single alanine substitutions of the metal-binding triad Cys243, Glu442, and His443 failed to rescue the fragile pellicle phenotype and demonstrated that Zn2+ binding is indispensable for catalytic activity of BcsG and that these residues truly are the functional equivalents of the catalytic residues of polymyxin resistance factors. Furthermore, complementation with alanine substitutions of Ser278, His396, and Tyr277 also failed to rescue the fragile pellicle phenotype, verifying their role in the catalytic cycle, as proposed from examination of the structure. Our EcBcsG biofilm phenotype assay confirmed similar trends as for the putative orthologue BcsG from S. enterica serovar Typhimurium (StBcsG) while also expanding the active site to include Tyr277.
We hypothesized that involvement of Tyr277 in catalysis would likely be via accommodation of cellulose glycans because of its solvent-facing orientation at the surface of our structure. To test this hypothesis, we further constructed a Y277F variant, which would be expected to retain this interaction and functionally complement the bcsG deletion. Surprisingly, the Y277F variant also failed to rescue the fragile pellicle phenotype, demonstrating direct involvement of the tyrosine hydroxyl in catalysis.
Stogios et al. (37) reported the only structure to date of a pEtN transferase family member bound to pEtN, Moraxella catarrhalis ICR (ICRMc). This structure demonstrated that a tyrosine residue in close proximity to the catalytic nucleophile threonine was involved in polar contact with the electronegative phosphate group of pEtN (37). However, ICRMc was reported to function as a homodimer, and the Tyr338 residue forming the polar contact was buried at the dimer interface and belonged to the second molecule of the dimer and not to the same polypeptide chain coordinating the Zn2+ to which the pEtN was bound (37). Instead, the monomeric structure of BcsG and the positioning of Tyr277 in our structure suggest that this residue may form polar contacts with glucan hydroxyl groups during the latter step of catalysis, in which EcBcsG transfers pEtN from Ser278 to the C6 hydroxyl of cellulose. The role of polar contacts between a Tyr hydroxyl and glucan chains in substrate binding to carbohydrate-active enzymes is well-established, particularly in glycosyl hydrolases. This is also true of the BcsZ endo-β-1,4-glucanase, encoded on a shared operon with the bcsA–bcsB synthase complex and adjacent to the bcsEFG operon (22).
The conserved pEtN transferase disulfide bond
Although the number of total disulfide bonds identified varies from three (CjEptC and MCR-1) to five (NmEptA) in the pEtN transferase structures examined here, only a single conserved disulfide bond is shared by all three of these structures (30, 31, 33). This disulfide bond is shaped by the conserved cysteine pairs Cys262-Cys272 in CjEptC, Cys281-Cys291 in MCR-1, and Cys276-Cys286 in NmEptA (Fig. 6) (28–30). The conserved disulfide bond directly fixes the helical secondary structural elements of NmEptA, CjEptC, and MCR-1, which contribute the respective catalytic threonine nucleophiles to the loops immediately N-terminal of each helix (30, 31, 33). These conserved disulfides thus orient the catalytic nucleophiles in close proximity to the Zn2+ ion present at the respective catalytic centers. Recently, the structure of EptC from E. coli (EcEptC, PDB code 6A83) has been reported to have a similar overall fold and share structural similarity with pEtN transferase family 3 (38). However, EcEptC did not confer polymyxin resistance in vivo despite sharing the conserved active-site architecture and catalytic Zn2+ binding discussed here (38). Instead, Zhao et al. (38) proposed that the absence of Cys pairs forming disulfides in EcEptC were responsible for the lack of observed activity and showed that disruption of Cys pairs in MCR-1 or CjEptC also abrogated polymyxin resistance when expressed in E. coli C43 to levels observed with expression of EcEptC.
Figure 6.
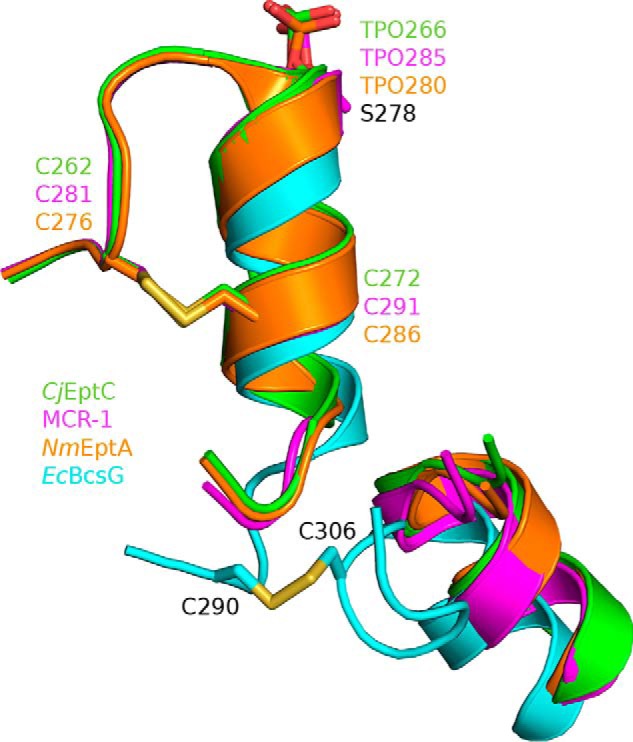
The pEtN transferase disulfide bond. A conserved, disulfide-bonded cysteine pair in CjEptC (green), MCR-1 (magenta), and NmEptA (orange) fixes the helix presenting the catalytic threonine adjacent to the catalytic Zn2+ at the active site and is important for activity. In EcBcsGΔN (cyan), a similar disulfide is observed, although not at the conserved location.
To understand the importance of disulfide bond presence in EcBcsG, we examined our structure for the presence of the conserved disulfide bond. Amino acids equivalent to the conserved cysteine pairs from CjEptC, NmEptA, and MCR-1 described above are observed as Ala274 and Arg284 in EcBcsGΔN. Instead, we observed an unambiguous disulfide bond in our native and Se structures of EcBcsGΔN between the cysteine pair Cys290-Cys306 (Fig. 6). This unusual disulfide bond in EcBcsG appears to serve the same function as the conserved pEtN transferase disulfide bond despite being structurally distinct. Instead of fixing the helix presenting the catalytic nucleophile to a loop directly N-terminal to it, as in the polymyxin resistance factors, the EcBcsG disulfide instead links the loop immediately C-terminal of the equivalent helix with a neighboring helical element packed against the central β-sheet fold. This novel disulfide retains the conserved function, orienting the catalytic EcBcsG nucleophile, Ser278, in proximity to the catalytic Zn2+ at the in EcBcsGΔN active site in a fashion analogous to CjEptC, NmEptA, and MCR-1.
To further investigate the hypothesis that this disulfide bond is important for catalytic activity of EcBcsG, we constructed a disulfide-deficient variant, EcBcsG C290A/C306A, and assessed its activity in vivo using our functional complementation experiment. We observed that the EcBcsG C290A/C306A did not functionally complement E. coli AR3110 ΔbcsG and failed to rescue the fragile pellicle phenotype (Fig. 5C). Our data suggest that the disulfide is indispensable for catalytic activity in vivo and that disruption of this disulfide results in a phenotype indistinguishable from EcBcsG variants with disruptions of the essential catalytic architecture, supporting the hypothesis that the EcBcsG disulfide bond is an important catalytic feature of the enzyme.
Identification of the covalent enzyme intermediate
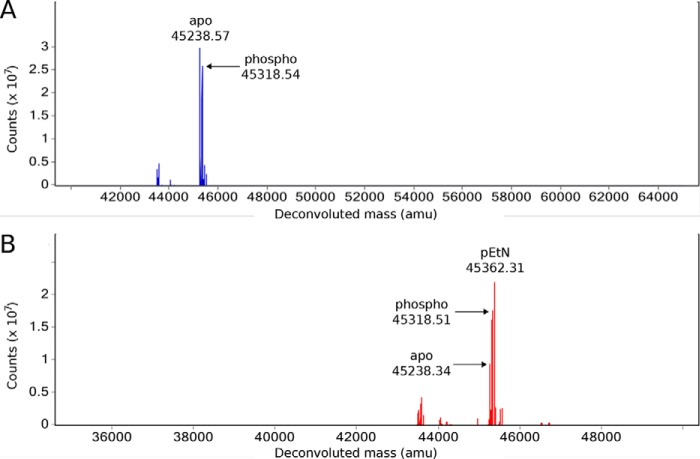
A nucleophilic attack and subsequent transfer of pEtN by EcBcsG or other pEtN transferase family members plausibly functions through a Ping Pong Bi Bi mechanism and so would require formation of a covalent pEtN–enzyme intermediate during the double-displacement catalytic cycle, a finding experimentally supported by Hinchliffe et al. (33) for MCR-1. However, given the rate of esterase activity observed using the substrate analog p-NPPE, we suspected that this intermediate would be particularly short-lived and solvent-labile with active EcBcsGΔN in aqueous solution. Accordingly, to test the hypothesis that EcBcsG might proceed through a covalent pEtN-Ser278 intermediate, we performed intact protein LC-MS of EcBcsGΔN alone and immediately following p-NPPE exposure (Fig. 7). The mass spectra for the EcBcsGΔN holoenzyme demonstrated a mixed population of m/z species of 45238.57 and 45318.54. The m/z 45238.57 species is in agreement with the predicted molecular mass of the recombinant EcBcsGΔN polypeptide less the most C-terminal residue Gln (45230.88 Da), which may be lost in the E. coli cytoplasm. The m/z 45318.54 species is in agreement with phosphorylated EcBcsGΔN (+79.97 Da). We then introduced p-NPPE to EcBcsGΔN immediately prior to LC-MS analysis and observed a new m/z species of 45362.31, corresponding to a covalent pEtN intermediate (+123.74 Da), accompanied by a decrease in relative signal intensity for the unmodified polypeptide at 45238.57. Interestingly, no apparent change in the relative abundance of the monophosphorylated EcBcsGΔN species (45318.54 m/z) was observed.
Figure 7.
The intact protein mass spectra of EcBcsGΔN. A, the mass spectrum of EcBcsGΔN prior to substrate exposure is composed of apparent m/z species corresponding to the expected EcBcsGΔN sequence and a phospho (+79.97 Da) EcBcsGΔN polypeptide. B, the appearance of a new m/z species in the mass spectrum of EcBcsGΔN following exposure to p-NPPE corresponds to addition of pEtN (+123.97 Da).
The existence of a phosphorylated form of EcBcsGΔN, site-specific to the catalytic nucleophile, would not be surprising as such a phosphoform has been commonly observed in the structures of other pEtN transferase family members (30, 31, 33, 37). However, it was surprising that this phosphorylation was not evident in our structure of EcBcsGΔN. We rationalized this disparity in our structural data as being due to important crystal contacts between the face of EcBcsGΔN containing the active site in the B chain with a neighboring molecule in the lattice. In this scenario, an altered electrostatic surface, or altered conformation of EcBcsGΔN because of a phosphorylated intermediate, may not have contributed productively to crystal packing. As phosphate was not a component of the buffers used in routine purification of EcBcsGΔN, phosphorylation must have occurred in the cytoplasm of the E. coli expression host. This observation of phospho-EcBcsGΔN long after host cell lysis indicates that the phosphoform of the enzyme was not solvent-labile in aqueous solution and was stable enough to persist in considerable quantity on the order of days following purification. Furthermore, we never observed monophosphorylated cellopentaose in the mass spectra of our in vitro transferase assays, indicating that EcBcsGΔN did not productively catalyze the transfer of this phosphate to cellulose.
Ser278 is the BcsG catalytic nucleophile
In an attempt to localize the covalent phosphate and pEtN modifications, we subsequently digested the intact and pEtN-linked EcBcsGΔN in sequencing-grade trypsin and analyzed the tryptic peptide library via LC–quadrupole-TOF MS/MS. Approximately 91% coverage was achieved, with total agreement with the EcBcsGΔN sequence. We detected nonspecifically phosphorylated and pEtNylated peptides containing surface-exposed residues, although these sites were matched to MS/MS data with low confidence and high mass error and did not fit our structural data. Instead, a single unique phosphopeptide containing the putative catalytic nucleophile Ser278 was observed in multiple scans. In addition, we observed a nonspecific cleavage fragment of this same peptide between residues Thr275 and Ser276. Each of these precursor ions could be reliably matched to the sequence and phosphorylation validated by MS/MS fragmentation (Table S2). However, we failed to detect the presence of similar peptides with a mass shift consistent with the addition of a pEtN group (+123.74 Da) to the same site. The absence of a covalent pEtN-Ser278 intermediate in aqueous solution was not surprising, given the appreciable rate of p-NPPE esterase activity by EcBcsGΔN that we observed under acceptor-free conditions (e.g. 3.1 ± 0.2 nmol min−1 mg of protein−1). This observation supported our hypothesis that such a species must be short-lived and likely would not persist through tryptic digestion. Subsequently, we performed MS/MS analysis of this unique Ser278-containing peptide. Analysis of these peptide MS/MS data unambiguously localized the 79.97-Da mass of the phosphate group to Ser278 with high confidence for all peptides we detected (Table S2 and Fig. S6), providing evidence of the role of Ser278 as the catalytic nucleophile.
Determination of the EcBcsGΔN reaction order
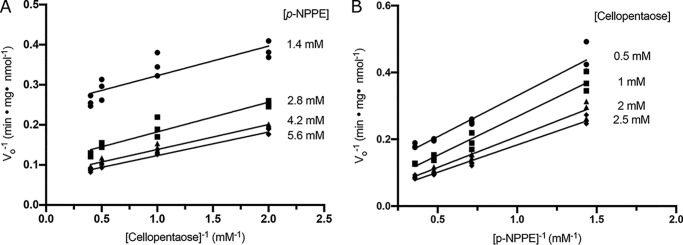
Cleland's rules (39) were applied to investigate the catalytic mechanism of EcBcsG. To investigate the reaction order and test the hypothesis that EcBcsGΔN must follow a Ping Pong Bi substrate, Bi product mechanism, the initial velocity was measured at cosubstrate concentrations near the Km for each (1.4, 2.8, 4.2, and 5.6 mm with Km = 2.4 ± 0.3 for p-NPPE and 0.5, 1, 2, and 2.5 mm with Km = 1.7 ± 0.2 for cellopentaose). Double reciprocal plots of initial velocity against p-NPPE concentration at a fixed cellopentaose concentration (1) and cellopentaose concentration at a fixed p-NPPE concentration (2) were prepared and fit to linear models (Fig. 8).
Figure 8.
The reaction order of EcBcsGΔN by kinetic analysis. A and B, Lineweaver–Burk plots of initial rates of reaction using 50 μm EcBcsGΔN at 37 °C (pH 7.5). Reactions were performed using fixed concentrations of p-NPPE with varied cellopentaose concentration (A) or vice versa (B).
The resulting linear model data were analyzed with a regular one-way ANOVA, determining that the slopes were not significantly different from each other (F(3,12) = 1.275, p = 0.3273 (1); F(3,12) = 1.086, p = 0.3661 (2)). A Tukey's test was conducted post-analysis for pairwise comparison of each slope, and none were found to be significantly different from any other (p > 0.3 for all cases). A subsequent analysis of covariance (ANCOVA) was performed to test the null hypothesis that the intercepts are equal, and the intercepts were found to be significantly different from each other (F(3,11) = 188.1, p < 0.0001).
The observation of double reciprocal initial velocity versus cosubstrate concentration plots bearing parallel slopes with unique intercepts suggests that each cosubstrate used in the EcBcsGΔN activity assay interacts with a different form of the enzyme. Hence, according to Cleland's rules, such a finding supports the hypothesis that EcBcsGΔN must proceed through a Ping Pong Bi Bi reaction mechanism (39).
In summary, the crystal structure of EcBcsGΔN reveals a protein fold typical of the pEtN transferase family and the presence of a conserved active site typical for the family, albeit with amino acid substitutions that may define it as a subfamily worthy of further delineation. Furthermore, our functional complementation results show that the enzyme is strictly dependent on a conserved Zn2+ binding triad, a conserved histidine, the catalytic nucleophile Ser278, a novel tyrosine residue lining the active site pocket, and a proximal disulfide bond for proper orientation of Ser278. Based on the results presented here, we propose that EcBcsG proceeds by a similar Zn2+-dependent catalytic mechanism, making it mechanistically indistinguishable but functionally unique to the biochemically characterized pEtN transferase family members. We also demonstrate, for the first time, that the C-terminal domain of EcBcsG is sufficient for transfer of pEtN onto cellulose acceptors, which is in contrast to prior work suggesting that other members of the cellulose synthetic complex were essential for activity (i.e. BcsA, BcsB, and/or BcsE) and that BcsG may be part of a pathway that modifies intermediates (e.g. BcsA, other periplasmic proteins, peptidoglycan, or osmoregulated periplasmic glucan) instead of cellulose directly (26, 27).
An understanding of BcsG-directed pEtN cellulose biosynthesis offers opportunities for structure-based drug discovery to inhibit the production of pEtN cellulose. The identification of pEtN cellulose as a critical virulence factor in establishment of uropathogenic E. coli (13), and likely other pathogens, highlights the clinical importance of BcsG as a therapeutic target. Furthermore, an understanding of the mechanism of bcsEFG-mediated pEtN cellulose biosynthesis would provide avenues for engineering new cellulosic materials or could confer the benefits of pEtN cellulose in new organisms. The structural and mechanistic insights into BcsG we report here, for the first time, broaden our understanding of phosphoethanolamine transferases beyond the realm of polymyxin resistance factors and provide a unifying theme with respect to bacterial persistence that is conferred by this enzyme family.
Experimental procedures
The bcsGΔN and bcsG genes were amplified from E. coli K12 chromosomal DNA and cloned into the pET28a expression vector (Table S3). Recombinant EcBcsGΔN was expressed in E. coli BL21(DE3) cells growing in SB medium (32 g tryptone, 20 g yeast extract, and 10 g of NaCl per litre of H2O) containing kanamycin (50 μg ml−1). EcBcsGΔN was expressed the same way, but in complete M9 minimal medium containing l-selenomethionine (20 μg ml−1). Cultures were incubated at 37 °C with shaking until they reached an optical density reading (600 nm wavelength) of 0.6–0.8, when expression was induced by addition of isopropyl 1-thio-β-d-galactopyranoside (1 mm), and growth was allowed to continue for 16–20 h at 23 °C. Cell pellets were collected by centrifugation and lysed using a Constant Systems cell disruptor operating at 17,000 psi (117,211 kPa). Recombinant EcBcsGΔN was purified to apparent homogeneity using nickel affinity chromatography and size-exclusion chromatography. EcBcsGΔN was crystallized using the hanging drop vapor diffusion method and solved using the single-wavelength anomalous diffraction method. The model of EcBcsGΔN-Se was used as a search model to solve the structure of the EcBcsGΔN crystal using molecular replacement. The structural homology search of EcBcsG was carried out using the DALI server (RRID: SCR_013433). The pEtN transferase sequences were aligned using ClustalOmega (RRID: SCR_001591) and visualized in JalView (RRID: SCR_006459). Synthesis of p-NPPE was performed as described previously (28), but using N-tert-butyloxycarbonyl ethanolamine. The pellicle phenotype was assessed by growth in culture tubes for 5 days at 30 °C without shaking using salt-free LB medium (10 g tryptone, 5 g yeast extract, and 10 g of NaCl per litre of H2O) containing 50 μg ml−1 kanamycin. The biofilms were detached from the tubes by inversion, washed in deionized water, and stained in a 0.01% (v/w) solution of Congo Red. The EcBcsGΔN activity assay was performed in 96-well microtiter plates by mixing purified EcBcsGΔN (50 μm) with p-NPPE (7 mm) in assay buffer (50 mm HEPES:NaOH (pH 7.5) and 50 mm NaCl) with addition of cello-oligosaccharides (3–5 mm), as specified, in final volumes of 100 μl. These reactions were monitored in a Cytation5 imaging microplate reader (BioTek) at 414 nm and 37 °C for 30 min. The reaction order experiment was performed in the same manner, but with cosubstrate concentrations as specified. Mass spectrometry was carried out using an Agilent UHD 6530 quadrupole-TOF mass spectrometer at the Mass Spectrometry Facility of the Advanced Analysis Centre, University of Guelph. Full experimental details, including NMR spectra of the p-NPPE synthetic intermediate and product, are provided in Figs. S7–S12.
Data availability
The atomic coordinates and reflection files for EcBcsGΔN-Se and EcBcsGΔN were deposited into the Protein Data Bank under accession codes 6PCZ and 6PD0, respectively. The raw LC-MS data for the tryptic EcBcsGΔN peptide library and the corresponding peptide search results have been deposited in Figshare (10.6084/m9.figshare.11538342 and 10.6084/m9.figshare.11918253, respectively). All remaining data are contained within the article.
Author contributions
A. C. A., K. E. M., and J. T. W. conceptualization; A. C. A. and J. T. W. resources; A. C. A., A. J. N. B., L. H., K. E. M., and J. T. W. data curation; A. C. A. and A. J. N. B. software; A. C. A., A. J. N. B., L. H., and J. T. W. formal analysis; A. C. A., K. E. M., and J. T. W. validation; A. C. A., A. J. N. B., L. H., and J. T. W. investigation; A. C. A. visualization; A. C. A., A. J. N. B., L. H., and J. T. W. methodology; A. C. A. and L. H. writing-original draft; A. J. N. B., K. E. M., and J. T. W. writing-review and editing; J. T. W. supervision; J. T. W. funding acquisition; J. T. W. project administration.
Supplementary Material
Acknowledgments
We thank Dyanne Brewer and Armen Charchoglyan of the Mass Spectrometry Facility (Advanced Analysis Centre, University of Guelph) for expert technical assistance and advice regarding MS experiments. We thank Michael Suits (Department of Chemistry and Biochemistry, Wilfrid Laurier University) for technical assistance with XRD experiments. The research described in this paper was performed using beamline 08ID-1 at the Canadian Light Source, which is supported by the Canada Foundation for Innovation, the Natural Sciences and Engineering Research Council of Canada, the University of Saskatchewan, the Government of Saskatchewan, Western Economic Diversification Canada, National Research Council Canada, and the Canadian Institutes of Health Research.
This work was supported by Natural Sciences and Engineering Research Council of Canada Grant 418310 (to J. T. W.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S12, Tables S1–S3, Materials and Methods, and References.
- Bcs
- bacterial cellulose synthase
- pEtN
- phosphoethanolamine
- p-NPPE
- para-nitrophenyl phosphorylethanolamine
- ANOVA
- analysis of variance.
References
- 1. Mah T. F., and O'Toole G. A. (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39 10.1016/S0966-842X(00)01913-2 [DOI] [PubMed] [Google Scholar]
- 2. del Pozo J. L., and Patel R. (2007) The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Ther. 82, 204–209 10.1038/sj.clpt.6100247 [DOI] [PubMed] [Google Scholar]
- 3. Saldaña Z., Xicohtencatl-Cortes J., Avelino F., Phillips A. D., Kaper J. B., Puente J. L., and Girón J. A. (2009) Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 11, 992–1006 10.1111/j.1462-2920.2008.01824.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spiers A. J., Bohannon J., Gehrig S. M., and Rainey P. B. (2003) Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50, 15–27 10.1046/j.1365-2958.2003.03670.x [DOI] [PubMed] [Google Scholar]
- 5. Ross P., Mayer R., and Benziman M. (1991) Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55, 35–58 10.1128/MMBR.55.1.35-58.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh N., Patil A., Prabhune A. A., Raghav M., and Goel G. (2017) Diverse profiles of N-acyl-homoserine lactones in biofilm forming strains of Cronobacter sakazakii. Virulence 8, 275–281 10.1080/21505594.2016.1226713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aly M. A., Reimhult E., Kneifel W., and Domig K. J. (2019) Characterization of biofilm formation by Cronobacter spp. isolates of different food origin under model conditions. J. Food Prot. 82, 65–77 [DOI] [PubMed] [Google Scholar]
- 8. Saxena I. M., and Brown R. M. (1995) Identification of a second cellulose synthase gene (acsAII) in Acetobacter xylinum. J. Bacteriol. 177, 5276–5283 10.1128/JB.177.18.5276-5283.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krasteva P. V., Bernal-Bayard J., Travier L., Martin F. A., Kaminski P. A., Karimova G., Fronzes R., and Ghigo J. M. (2017) Insights into the structure and assembly of a bacterial cellulose secretion system. Nat. Commun. 8, 2065 10.1038/s41467-017-01523-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Römling U., and Galperin M. Y. (2015) Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 23, 545–557 10.1016/j.tim.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maeda K., Tamura J., Okuda Y., Narikawa R., Midorikawa T., and Ikeuchi M. (2018) Genetic identification of factors for extracellular cellulose accumulation in the thermophilic cyanobacterium Thermosynechococcus vulcanus: proposal of a novel tripartite secretion system. Mol. Microbiol. 109, 121–134 10.1111/mmi.13977 [DOI] [PubMed] [Google Scholar]
- 12. Whitfield G. B., Marmont L. S., and Howell P. L. (2015) Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front. Microbiol. 6, 471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hollenbeck E. C., Antonoplis A., Chai C., Thongsomboon W., Fuller G. G., and Cegelski L. (2018) Phosphoethanolamine cellulose enhances curli-mediated adhesion of uropathogenic Escherichia coli to bladder epithelial cells. Proc. Natl. Acad. Sci. 115, 10106–10111 10.1073/pnas.1801564115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morgan J. L., McNamara J. T., and Zimmer J. (2014) Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat. Struct. Mol. Biol. 21, 489–496 10.1038/nsmb.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whitney J. C., and Howell P. L. (2013) Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 21, 63–72 10.1016/j.tim.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keiski C.-L., Harwich M., Jain S., Neculai A. M., Yip P., Robinson H., Whitney J. C., Riley L., Burrows L. L., Ohman D. E., and Howell P. L. (2010) AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure 18, 265–273 10.1016/j.str.2009.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morgan J. L., Strumillo J., and Zimmer J. (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493, 181–186 10.1038/nature11744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Omadjela O., Narahari A., Strumillo J., Mélida H., Mazur O., Bulone V., and Zimmer J. (2013) BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 110, 17856–17861 10.1073/pnas.1314063110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morgan J. L., McNamara J. T., Fischer M., Rich J., Chen H.-M., Withers S. G., and Zimmer J. (2016) Observing cellulose biosynthesis and membrane translocation in crystallo. Nature 531, 329–334 10.1038/nature16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolfram F., Arora K., Robinson H., Neculai A. M., Yip P., and Howell P. L. (2012) Expression, purification, crystallization and preliminary X-ray analysis of Pseudomonas aeruginosa AlgL. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 584–587 10.1107/S1744309112012808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan J., Rouse S. L., Li D., Pye V. E., Vogeley L., Brinth A. R., El Arnaout T., Whitney J. C., Howell P. L., Sansom M. S. P., and Caffrey M. (2014) A conformational landscape for alginate secretion across the outer membrane of Pseudomonas aeruginosa. Acta Crystallogr. Sect. D Biol. Crystallogr. 70, 2054–2068 10.1107/S1399004714001850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mazur O., and Zimmer J. (2011) Apo- and cellopentaose-bound structures of the bacterial cellulose synthase subunit BcsZ. J. Biol. Chem. 286, 17601–17606 10.1074/jbc.M111.227660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Solano C., García B., Valle J., Berasain C., Ghigo J. M., Gamazo C., and Lasa I. (2002) Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43, 793–808 10.1046/j.1365-2958.2002.02802.x [DOI] [PubMed] [Google Scholar]
- 24. Fang X., Ahmad I., Blanka A., Schottkowski M., Cimdins A., Galperin M. Y., Römling U., and Gomelsky M. (2014) GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol. Microbiol. 93, 439–452 10.1111/mmi.12672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adcox H. E., Vasicek E. M., Dwivedi V., Hoang K. V., Turner J., and Gunn J. S. (2016) Salmonella extracellular matrix components influence biofilm formation and gallbladder colonization. Infect. Immun. 84, 3243–3251 10.1128/IAI.00532-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thongsomboon W., Serra D. O., Possling A., Hadjineophytou C., Hengge R., and Cegelski L. (2018) Phosphoethanolamine cellulose: A naturally produced chemically modified cellulose. Science 359, 334–338 10.1126/science.aao4096 [DOI] [PubMed] [Google Scholar]
- 27. Sun L., Vella P., Schnell R., Polyakova A., Bourenkov G., Li F., Cimdins A., Schneider T. R., Lindqvist Y., Galperin M. Y., Schneider G., and Römling U. (2018) Structural and functional characterization of the BcsG subunit of the cellulose synthase in Salmonella typhimurium. J. Mol. Biol. 430, 3170–3189 10.1016/j.jmb.2018.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dimitrijević M., Grujić-Injac B., and Lajsić S. (1979) The synthesis of a new phospholipid from the Koilin glandular layer of chicken gizzard. Hoppe Seylers Z Physiol. Chem. 360, 477–480 10.1515/bchm2.1979.360.1.477 [DOI] [PubMed] [Google Scholar]
- 29. Zheng H., Chordia M. D., Cooper D. R., Chruszcz M., Müller P., Sgeldrick G. M., and Minor W. (2014) Validating metal binding sites in macromolecule structures using the CheckMyMetal web server. Nat. Protoc. 9, 156–170 10.1038/nprot.2013.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fage C. D., Brown D. B., Boll J. M., Keatinge-Clay A. T., and Trent M. S. (2014) Crystallographic study of the phosphoethanolamine transferase EptC required for polymyxin resistance and motility in Campylobacter jejuni. Acta Crystallogr. D Biol. Crystallogr. 70, 2730–2739 10.1107/S1399004714017623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wanty C., Anandan A., Piek S., Walshe J., Ganguly J., Carlson R. W., Stubbs K. A., Kahler C. M., and Vrielink A. (2013) The structure of the neisserial lipooligosaccharide phosphoethanolamine transferase A (LptA) required for resistance to polymyxin. J. Mol. Biol. 425, 3389–3402 10.1016/j.jmb.2013.06.029 [DOI] [PubMed] [Google Scholar]
- 32. Harper M., Wright A., St Michael F., Li J., Deveson Lucas D., Ford M., Adler B., Cox A. D., and Boyce J. D. (2017) Characterization of two novel lipopolysaccharide phosphoethanolamine transferases in Pasteurella multocida and their role in resistance to Cathelicidin-2. Infect. Immun. 85, e00557–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hinchliffe P., Yang Q. E., Portal E., Young T., Li H., Tooke C. L., Carvalho M. J., Paterson N. G., Brem J., Niumsup P. R., Tansawai U., Lei L., Li M., Shen Z., Wang Y., et al. (2017) Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci. Rep. 7 10.1038/srep39392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scott N. E., Nothaft H., Edwards A. V., Labbate M., Djordjevic S. P., Larsen M. R., Szymanski C. M., and Cordwell S. J. (2012) Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J. Biol. Chem. 287, 29384–29396 10.1074/jbc.M112.380212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cullen T. W., and Trent M. S. (2010) A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. U.S.A. 107, 5160–5165 10.1073/pnas.0913451107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bontemps-Gallo S., Cogez V., Robbe-Masselot C., Quintard K., Dondeyne J., Madec E., and Lacroix J.-M. (2013) Biosynthesis of osmoregulated periplasmic glucans in Escherichia coli: the phosphoethanolamine transferase is encoded by opgE. Biomed. Res. Int. 2013, 371429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stogios P. J., Cox G., Zubyk H. L., Evdokimova E., Wawrzak Z., Wright G. D., and Savchenko A. (2018) Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis. ACS Chem. Biol. 13, 1322–1332 10.1021/acschembio.8b00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao Y., Meng Q., Lai Y., Wang L., Zhou D., Dou C., Gu Y., Nie C., Wei Y., and Cheng W. (2019) Structural and mechanistic insights into polymyxin resistance mediated by EptC originating from Escherichia coli. FEBS J. 286, 750–764 [DOI] [PubMed] [Google Scholar]
- 39. Cleland W. W. (1963) The kinetics of enzyme-catalyzed reactions with two or more substrates or products: III: prediction of initial velocity and inhibition patterns by inspection. Biochim. Biophys. Acta 67, 188–196 10.1016/0926-6569(63)90227-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and reflection files for EcBcsGΔN-Se and EcBcsGΔN were deposited into the Protein Data Bank under accession codes 6PCZ and 6PD0, respectively. The raw LC-MS data for the tryptic EcBcsGΔN peptide library and the corresponding peptide search results have been deposited in Figshare (10.6084/m9.figshare.11538342 and 10.6084/m9.figshare.11918253, respectively). All remaining data are contained within the article.