Abstract
Prominins (proms) are transmembrane glycoproteins conserved throughout the animal kingdom. They are associated with plasma membrane protrusions, such as primary cilia, as well as extracellular vesicles derived thereof. Primary cilia host numerous signaling pathways affected in diseases known as ciliopathies. Human PROM1 (CD133) is detected in both somatic and cancer stem cells and is also expressed in terminally differentiated epithelial and photoreceptor cells. Genetic mutations in the PROM1 gene result in retinal degeneration by impairing the proper formation of the outer segment of photoreceptors, a modified cilium. Here, we investigated the impact of proms on two distinct examples of ciliogenesis. First, we demonstrate that the overexpression of a dominant-negative mutant variant of human PROM1 (i.e. mutation Y819F/Y828F) significantly decreases ciliary length in Madin–Darby canine kidney cells. These results contrast strongly to the previously observed enhancing effect of WT PROM1 on ciliary length. Mechanistically, the mutation impeded the interaction of PROM1 with ADP-ribosylation factor–like protein 13B, a key regulator of ciliary length. Second, we observed that in vivo knockdown of prom3 in zebrafish alters the number and length of monocilia in the Kupffer's vesicle, resulting in molecular and anatomical defects in the left-right asymmetry. These distinct loss-of-function approaches in two biological systems reveal that prom proteins are critical for the integrity and function of cilia. Our data provide new insights into ciliogenesis and might be of particular interest for investigations of the etiologies of ciliopathies.
Keywords: prominin-1 (PROM1), cilia, zebrafish, membrane protein, ADP-ribosylation factor (ARF), development, CD133, left-right asymmetry, MDCK, membrane polarity, ADP-ribosylation factor-like protein 13B (Arl13b), cilium, Kupffer's vesicle, ciliopathy, development
Introduction
Prominins (proms)6 are cholesterol-binding, pentaspan membrane glycoproteins conserved throughout the animal kingdom and expressed in numerous embryonic and adult epithelial cells (1–4). In mammals, two prom proteins were identified (2). The first member of the family, PROM1 (CD133), has become a widely used surface antigenic marker for the identification and isolation of a broad range of somatic stem cell populations, and potentially cancer-initiating cells (reviewed in Ref. 5). In the visual system, PROM1 gene mutations are associated with various forms of retinal degeneration (6, 7), which can be phenocopied in genetically modified mice carrying either a null allele or a mutated dominant form of human PROM1 (8, 9). Murine models deficient in prom1 show a severe disorganization of photoreceptor outer segments, which are specialized sensory cilia (reviewed in Ref. 10). In zebrafish, the corresponding mammalian PROM1 gene is duplicated, and the co-orthologues are referred to as prom1a and prom1b (2, 11–14). The knockdown of prom1b, but not prom1a, also led to photoreceptor degeneration (15). This observation was surprising, given that both prom1a/b are co-expressed in fish retina (13, 16, 17). Prom1 is found in rods and cones, where it is concentrated in the outer segments, particularly at the edges of open disks, irrespective of the species investigated (6, 13, 18) (reviewed in Refs. 7 and 19). These morphological and pathological data together with the presence of prom1 (or its paralogue prom2) in nonmotile and motile cilia found in various mammalian organ systems lay evidence of a conserved involvement of proms in the maintenance of ciliary structure and function (20–22). Besides cilia, prom1 and prom2 are associated with various types of cellular protrusions, such as microvilli, filopodia, and lamellipodia, indicating a strong preference for highly curved membrane domains (1, 2, 23, 24). The association of proms with small membrane vesicles that are budding from microvilli and primary cilia is in line with this preference (20, 21, 25).
Mutations in the ganglioside-binding site of PROM1 or variations in the level of membrane cholesterol influence its specific subcellular localization as well as the organization of membrane protrusions and/or the dynamics of the vesicles budding thereof (21, 26–28). The direct interaction of prom1 (or prom2) with cholesterol (21, 26, 29) and potentially other membrane lipids, such as monosialotetrahexosylganglioside (GM1) (30, 31), might account for these peculiarities. These prom/lipid complexes might stimulate the interaction of prom1 with various membrane and/or cytoplasmic protein partners and hence modulate the architecture of the given protrusion. For instance, the binding of prom1 to protocadherin 21 organizes the nascent precursor of photoreceptive membranes at the base of the outer segment (9). The interaction of prom1 with the actin-related protein 2/3 (Arp2/3) complex, which mediates branching of actin networks, favors the formation of clusters of microvilli in epithelial cells and filopodia in nonepithelial cells (28). The latter observation might be associated with the implication of prom1 in cancer cell migration (32). The selective binding of prom1 to ADP-ribosylation factor–like protein 13B (Arl13b) or histone deacetylase 6 (HDAC6) can orchestrate the functionality and dynamics of primary cilia and consequently the activation of stem cells (33). The interactions of Prom1 with Arl13b/HDAC6 appear to be dependent on cytoplasmic lysine 138 (numbered according to prom1 splice variant s2) (33, 34), whereas binding to Arp2/3 complex is stimulated by phosphorylation of a C-terminal tyrosine residue located at position 819/828 (s1/s2 variant) (28, 35). This post-translational modification also promotes the interaction of prom1 with the p85 subunit of phosphoinositide 3-kinase (PI3K), thereby activating the transformation of phosphatidylinositol 4,5-bisphosphate (PIP2) into phosphatidylinositol 3,4,5-trisphosphate (PIP3) at the inner leaflet of the plasma membrane (36), which plays a pivotal role in the organization of plasma membrane protrusions (28). For example, the expression of a mutant form of human PROM1 containing a single amino acid substitution in s1 and s2 splice variants (i.e. Tyr819/828 → Phe; Y819F or Y828F mutant, respectively) impaired the elongation of microvilli (28). Altogether, alterations in prom1, as observed in vivo in retinal pathologies or in vitro with various cell types, may influence the organization of its surrounding membranous microenvironment and its cross-talk with cytoskeleton regulators. As a consequence, the general structure of the plasmalemma and, hence, dynamic protrusions such as microvilli and cilia would be affected.
Here, we further dissect the role of proms in ciliary architecture by determining the impact of Y819/828F mutation in PROM1 splice variants s1 and s2 on the primary cilium of mammalian cells and the down-regulation of one of three prom paralogues in zebrafish (12, 13). Zebrafish prom3 has attracted our attention due to its expression in the Kupffer's vesicle, an embryonic organ implicated in the left-right (L-R) axis determination via a cilium-based nodal flow mechanism (37). Being composed of monociliated cells, Kupffer's vesicle represents an excellent model for disentangling cilium-related questions (38). Our data revealed a direct implication of mammalian and nonmammalian proms for two separate paradigms of ciliogenesis.
Results
Mutation of tyrosine 819/828 in human PROM1 reduces ciliary length
We recently demonstrated that the mutation Y819F or Y828F in human PROM1.s1 and PROM1.s2 splice variants, respectively, impairs tyrosine phosphorylation and the elongation of microvilli in epithelial Madin–Darby canine kidney (MDCK) cells (28, 39). The prom1 splice variants differ solely by the inclusion of a 9-amino-acid-encoding exon in the N-terminal domain of prom1.s2 (40, 41). By using scanning EM (SEM), we evaluated whether the same mutation would affect the primary cilium. We observed that cilia were indeed shorter in cells expressing the Y819/828F mutant proteins (Fig. 1A and Fig. S1), and some cells did not harbor a cilium (Fig. 1A, asterisk). In contrast, very long primary cilia with more than 10 μm in length were regularly observed on WT PROM1-transfected cells (Fig. 1B and Fig. S1). These observations prompted us to quantify ciliary length. WT cells and those expressing the mutant proteins were subjected to double immunofluorescence using anti-PROM1 and anti-acetylated α-tubulin (AcTub) antibodies. The latter visualizes the ciliary axoneme, indicating ciliary length. Samples were analyzed by confocal laser-scanning microscopy (CLSM). In agreement with SEM data, primary cilia were significantly shorter in cells expressing Y819/828F mutants compared with WT cells at 7 days post-confluence (dpc) (1.7 ± 0.81 μm (n = 203) and 1.7 ± 0.68 μm (n = 236) for PROM1.s1 Y819F and PROM1.s2 Y828F, respectively, versus 2.5 ± 1.17 μm (n = 224) in control cells; p < 0.001, Mann–Whitney test) (Fig. 1C, left). Similar data were obtained at 14 dpc (Fig. 1C, right). These data differ from those previously obtained with the expression of WT PROM1, where an increase in ciliary length was observed for s1 (3.1 ± 1.4 μm (n = 569)) and s2 (3.1 ± 1.4 μm (n = 953)) variants (Fig. 1C; 7 dpc; see also Ref. 33). The number of cells harboring no cilium was also increased upon expression of PROM1 Y819/828F mutants (Fig. 1D) (33).
Figure 1.
Mutating tyrosine 819/828 of human PROM1 shortens primary cilia. A–D, polarized WT cells (MDCK), those expressing human PROM1.s1 or PROM1.s2 splice variants, or corresponding Y819/828F mutants were grown for 7 dpc (A–C) or 14 dpc (C and D) and processed for SEM (A and B) and CLSM (C and D). SEM micrographs revealed long (green) and short (red) primary cilia in cells expressing WT and mutated PROM1, respectively (A). Note the presence of very long cilia (>10 μm) in WT PROM1-positive cells (B; see also Fig. S1). Arrow, primary cilium; dashed line, origin of cilium; *, cell without primary cilium; #, artifact of SEM preparation. For the immunofluorescence analysis (C and D), cells were double-immunolabeled for PROM1 and AcTub. The length of AcTub-labeled cilia was evaluated by CLSM and classified into three categories: <3, 3–5, and >5 μm (C). The number of analyzed primary cilia is displayed in parentheses. Data (purple) obtained from cells expressing PROM1.s2 were taken from our recent study (33) and used for comparison. The percentage of total (blue) or PROM1+ (black) cells with no cilium is indicated (D). The mean and S.D. (error bars) are presented. Scale bars, 500 nm (top panels) and 2 μm (bottom panels) (A) and 1 μm (B).
Because the assembly and disassembly of primary cilia are tightly coordinated with the cell cycle, we evaluated the number of cells after 7 and 14 dpc. Surprisingly, the number of cells expressing PROM1 Y819/828F mutants was significantly increased compared with control cells or cells expressing WT PROM1.s1 or PROM1.s2 variant, suggesting an extensive proliferation at an early stage of polarization (Fig. S2). At 14 dpc, cell numbers were, however, similar in all cell lines (Fig. S2) (data not shown), suggesting that cell division might not be the only cause accounting for the appearance of short cilia. These data suggest that mammalian prom1 positively influences the length of primary cilia.
Mutation of tyrosine 819/828 in human PROM1 reduces its interaction with Arl13b and impacts Arl13b subcellular localization
We recently reported that the K138Q mutation impedes the interaction of PROM1 with Arl13b, a member of the Arl (ADP-ribosylation factor (Arf)–related) subfamily of the Ras superfamily of small GTPases that regulates ciliary length (42). This mutation results in the formation of shorter cilia (33). To find out whether the interaction of PROM1 with Arl13b also mediates the negative impact of the Y819/828F mutation on ciliary length, we immunoisolated PROM1 (WT and mutants) after cell solubilization using paramagnetic bead–conjugated AC133 mAb and determined its interaction with Arl13b by immunoblotting. We observed that the Y819/828F mutation interfered with PROM1-Arl13b interaction for both s1 and s2 splice variants (Fig. 2, A and B). Similar data were obtained with the reciprocal experiment (i.e. co-immunoisolation of PROM1 using Arl13b as bait) (data not shown). These data suggest that the impact of the Y819/828F mutation on primary cilium length involves PROM1-Arl13b interaction. To further dissect this issue, we investigated the subcellular localization of Arl13b within the ciliary compartment. As expected, the CLSM analyses of MDCK cells expressing PROM1.s2 revealed the presence of Arl13b along the ciliary axoneme (Fig. 2C). A partial co-localization with PROM1 was observed. The latter was nonuniformly distributed along the cilium as reported previously (20, 33), which might indicate its association with specific membrane microdomains (30) (see “Discussion”). In contrast, the corresponding PROM1 mutant seemed to be uniformly expressed, like Arl13b, in short cilia (Fig. 2C). Unexpectedly, we observed an accumulation of Arl13b at the base of cilia (Fig. 2, C and D) together with a reduction at the level of axonemal microtubules in cells expressing PROM1 mutant (0.69 ± 0.29 (n = 6)) compared with those harboring WT PROM1 (1.55 ± 0.44 (n = 9); p < 0.01, Mann–Whitney test) as indicated by the Arl13b/AcTub immunoreactivity ratio (see also Fig. 2C, top right panels). Overall, these observations suggest that the mutation of the cytoplasmic tyrosine 819/828 in PROM1 causes some defects in the intraflagellar transport of Arl13b and its reduction in the ciliary axoneme and hence influences the ciliary length.
Figure 2.

Mutation of tyrosine 819/828 in human PROM1 reduces its interaction with Arl13b. A–D, polarized WT cells (MDCK)or those expressing human PROM1.s1 or PROM1.s2 splice variants or the corresponding Y819/828F mutants were subjected either to immunoisolation (IS) using paramagnetic beads conjugated to mAb against PROM1 (AC133) or anti-fluorescein isothiocyanate (FITC) antibody as negative control (A and B) or processed for indirect immunofluorescence followed by CLSM (C and D). For immunoisolation, the input (PROM1, 1:10; Arl13b, 1:40) and immunoisolated materials were probed for PROM1 and Arl13b by immunoblotting (A). The arrowhead and asterisks indicate plasma membrane and endoplasmic reticulum–associated species of PROM1, respectively, whereas arrows show Arl13b. Molecular mass markers (kDa) are indicated (left). The ratio of Arl13b/PROM1 (plasma membrane species only) immunoreactivities was quantified (B). 3–6 independent experiments were performed. Data are presented as mean and S.D. (error bars). Individual values are shown. **, p < 0.01; ***, p < 0.001 (two-tailed unpaired Student's t test). For immunofluorescence, cells were triple-immunolabeled for PROM1 (green), Arl13b (red), and AcTub (white). Nuclei (nu) were counterstained with DAPI (blue). The three-dimensional view (x-z orientation) was built from 38–45 x-y sections throughout the ciliary compartment (C, left panels) or x-y section (right top panels) and a composite of 5–10 x-y sections (right bottom panels) taken at the axoneme level (Ax, green bar) or at the base (red bar) of the primary cilium (C), respectively. The top view of three-dimensional reconstructions highlights Arl13 at the base of the cilium (D, dashed red line). Cells expressing either WT (left) or mutant (right) PROM1 are displayed. Green arrow, punctate PROM1 staining at the ciliary membrane of WT PROM1+ cells. Asterisk, PROM1 associated with microvillar membrane. Scale bars, 1 μm.
Expression and distribution of proms in zebrafish
Until now most of the experimental evidence linking proms with the structure and dynamics of cilia was derived from analysis of mammalian proteins. As prom paralogues are expressed redundantly in many tissues, very few models permit documenting their individual roles (2). Developing nonmammalian vertebrates offer effective in vivo functional assay systems for testing loss-of-function scenarios with well-defined readouts. We used zebrafish (Danio rerio) as an established vertebrate model to gain insights into the physiological function of a given nonredundantly expressed prom (2, 13). Indeed, besides prom1a and prom1b, the zebrafish genome contains a third prom gene, named prom3 (Gene ID: 556596; chromosome 13). As is the case for other proms, several splice forms of prom3 are transcribed. We observed at least two distinct splice variants that differ by the inclusion of a facultative exon (exon B) in the 5′-UTR (Table S1).
Distribution analysis of the three zebrafish proms by whole-mount in situ hybridization (ISH) revealed spatially and temporally restricted, but dynamically changing, expression patterns during embryogenesis. These differences are already obvious at very early stages of development as, for instance, maternal deposition of prom1a transcripts could be detected as described previously (12). A detailed description of zebrafish prom expression domains starting at early blastula stage through somitogenesis stages (6-, 8-, and 16-somite stages) as well as in pharyngulas (at 22 h post-fertilization (hpf)) is presented in the supporting Results (Figs. S3 and S4).
The unique expression of prom3 in the transient embryonic structure called the Kupffer's vesicle (Fig. S3) caught our attention owing to its implication in the L-R axis determination via a cilium-based nodal flow mechanism. The fact that knocking down zebrafish Arl13b elicited phenotypes that are related to ciliary structures within Kupffer's vesicle (44) further prompted us to concentrate on zebrafish prom3, while using prom1a as a negative control, given that both prom1 paralogues are absent from this anatomical structure.
Depletion of zebrafish prom3 leads to left-right asymmetry defects
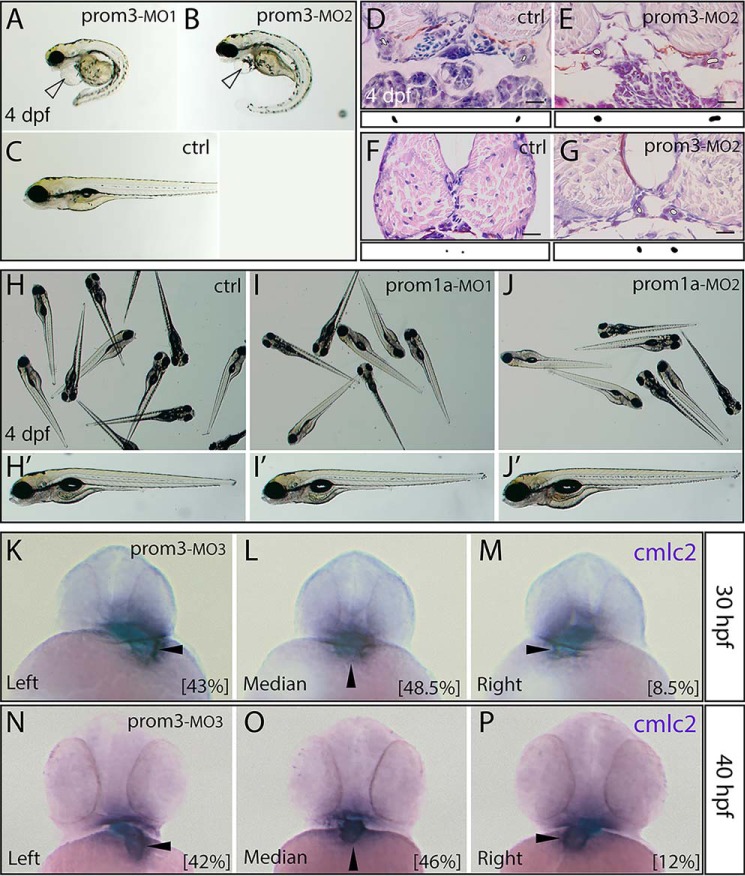
To address the biological role of prom3, an antisense depletion approach was undertaken using three independent morpholino-oligonucleotides (MOs). Two distinct strategies were employed. First, a translation blockage was utilized using two independent MOs (MO1 and MO2). To interfere with mRNA translation, the oligonucleotides were made complementary either to the translation initiation codon region of the message or the 5′-UTR (in a sequence stretch found in both splice variants; Fig. S5A). Second, the blockage of pre-mRNA splicing was applied using a third MO (MO3). It was designed to block pre-mRNA splicing allowing also for the direct evaluation of specificity by monitoring the impairment of the pre-mRNA processing (Fig. S5B). The impact of MO3 on mRNA splicing was verified by RT-PCR (Fig. S5, C and D). Following oligonucleotide injection, a gross morphological observation at 4 days post-fertilization (dpf), revealed severe impairments in morphants. Indeed, the prom3 morphants induced by either MO1 (5 ng injected) or MO2 (10 ng) suffered from pericardial edema, and their body axis was frequently curved ventrally downward with a kink in the tail (Fig. 3, A and B). The body size was reduced (Fig. 3, A and B versus C). Similar observations were made with MO3 (10 ng) (Fig. S6). In line with the localization of prom3 transcripts in the intermediate mesoderm and in its successor structure, the pronephric duct (Fig. S4C), the rostral and middle portion of the pronephric duct appeared to be dilated compared with the control (standard control MO, 10 ng) (Fig. 3, E and G), yet no typical glomerular cyst formation was observed. In contrast, neither the control nor the antisense knockdown of prom1a resulted in any obvious gross morphological defects (Fig. 3, H–J′).
Figure 3.
Morphological phenotypes upon prom3 knockdown. A–P, embryos were injected at the one-cell stage with distinct MO against either prom3 (A, MO1, 5 ng; B, E, and G, MO2, 10 ng; K–P, MO3, 10 ng) or prom1a (I and I′, MO1, 10 ng; J and J′, MO2, 10 ng) or with control MO (C, D, F, H, and H′, ctrl, 10 ng). At different developmental stages as indicated, specimens were either observed in a native, unfixed state (A–C and H–J), analyzed histologically with hematoxylin and eosin staining (D–G), or processed for whole-mount ISH using antisense DIG-labeled probe against cmlc2 (K–P). At early (K–M; 30 hpf) and late (N–P; 40 hpf) pharyngula periods, a randomized position (left, median, or right) of the heart tube highlighted by cmlc2 (arrowhead) is observed in prom3 morphants. Numbers in parentheses indicate the frequencies of phenotypes (30 hpf, n = 35; 40 hpf, n = 59). At 4 dpf, prom3 morphants, but neither the control nor prom1a MO-injected ones (C and H–J′), displayed severe morphological disturbances including swollen pericardium (A and B; hollow arrowhead), ventrally curved body (A and B), and an increased lumen size of the pronephric duct (E and G; outlined white surface). The surface areas of the lumen size are depicted as black blobs under the corresponding image. Orientation is as follows. A–C and H′–J′, lateral; H–J, random; K–P, anterior (dorsal toward top). Scale bars (D–G), 16 μm.
These observations prompted us to investigate the consequence of the depletion of prom3 at earlier stages of development (i.e. 30 and 40 hpf). The injection of MO3 resulted in an alteration of normal laterality (handedness). In contrast to control embryos where the normal leftward looping of the heart tube (situs solitus) was observed, the prom3 morphants had a randomized heart position as detected by ISH of cardiac myosin light chain 2 (cmlc2) (Fig. 3, K–P). They displayed either dextrocardia (situs inversus) (i.e. inversion of L-R asymmetry with respect to the median-sagittal plane as judged by left-right displacement of the developing heart tube) (Fig. 3, M and P; 8.5 and 12% of cases at 30 and 40 hpf, respectively; n = 35 and 59) or an absence of laterality resulting in a median heart tube position (Fig. 3, L and O; 48.5 and 46% of cases at 30 and 40 hpf, respectively). Interestingly, the simultaneous application of one-fourth of two independent MOs blocking translation (i.e. MO1 (1.25 ng) and MO2 (2.5 ng)) resulted in a synergistic effect on fish morphology at 58 hpf leading to a severe phenotype similar to what has been observed upon injection of MO2 alone at an amount of 10 ng (Fig. 4, A–D). The combined injection of one-eighth of the single effective dose of each individual MO (MO1 + MO2) elicited a laterality defect as observed with cmlc2 ISH (Fig. 4, E and F).
Figure 4.
Simultaneous injection of two nonoverlapping MOs against prom3 results in a synergistic effect. A–F, embryos were injected at the one-cell stage with control MO (A, ctrl, 10 ng), MO2 (D, 10 ng) or a combination of MO1 + MO2 against prom3 at the doses indicated (B, C and E, F). At 58 hpf, larvae were observed in a native, unfixed state (A–D) or processed for whole-mount ISH using antisense DIG-labeled probe against cmlc2 (E and F). At the effective dose (10 ng), MO2 elicited mild to severe phenotypes (D). The simultaneous injection of one-fourth of their single effective amounts of MO1 (5 ng) and MO2 (10 ng) results in body axis distortion and fluid backup (C) similar to the severe phenotype detected upon injection of MO2 alone at the effective dose (D). Application of one-eighth of effective amounts caused no changes in body axis development (B) but often elicited L-R defects of the heart tube (F) in contrast to the normal position (situs solitus) (E). Orientation is as follows. A–D, random; E and F, anterior (dorsal toward the top). A, atrium; V, ventricle.
Next, we examined whether the randomized anatomical laterality of the morphants was evidenced by altered molecular asymmetries earlier in development. The expression of lefty-2, a member of the transforming growth factor-β superfamily, was analyzed as a typical example of asymmetrically expressed genes during the segmentation period (45, 46). In contrast to control embryos, prom3 morphants displayed a randomized gene expression at 25 hpf (Fig. 5, A–D). Four configurations of lefty-2 localization were detected with different frequencies, depending on the nature and the amount of the MO used (Fig. 5E). Either right-sided or bilateral or even no expression was observed in addition to the normal left-sided appearance of this molecule. Thus, the antisense depletion of prom3 resulted in a L-R randomization of gene expression well before the manifestation of anatomical signs of handedness, suggesting that prom3 acts upstream of lefty-2 in the sequence of events leading to L-R determination in zebrafish.
Figure 5.
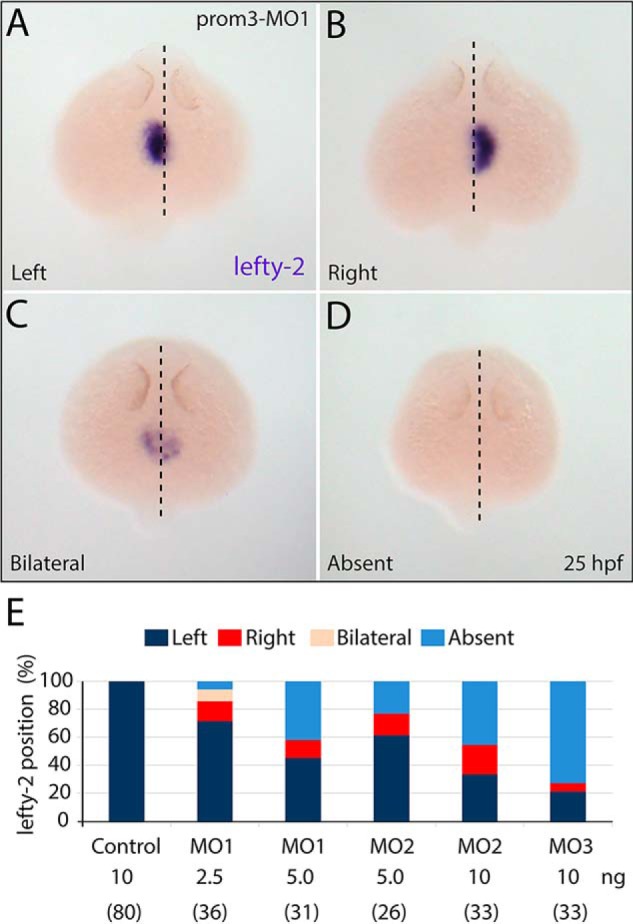
Prom3 influences molecular left-right asymmetry. A–E, embryos were injected at the one-cell stage with either one of the three MOs against prom3 (A–D, MO1 (2.5 ng); E, MO1 (2.5 and 5 ng), MO2 (5 and 10 ng), and MO3 (10 ng)) or control MO (E, 10 ng). At 25 hpf, specimens were processed for whole-mount ISH using antisense DIG-labeled lefty-2 probe (A–D). Note the randomized expression of lefty-2 (left, right, and bilateral) with regard to the median-sagittal plane (dashed line) or its absence. Shown are frequencies of the lefty-2 position (see color code) observed in prom3-morphants upon application of a given MO at various amounts (E). The number of embryos analyzed is indicated in parentheses. Orientation is as follows. A–D, dorsal (rostral toward the top).
Zebrafish prom3 regulates ciliary length in Kupffer's vesicle
What could explain the molecular and anatomical laterality defects of prom3 morphants? As indicated above, prom3 is, contrarily to prom1a and prom1b, selectively expressed in Kupffer's vesicle (Fig. S4, E–G). This transient embryonic structure of teleosts plays a pivotal role in the L-R axis determination. It is composed of around 50–60 monociliated cells (47). The cilia, 3–5 μm in length, have a major impact on the generation of a local microflow required for the establishment of correct laterality. Thus, Kupffer's vesicle serves as an integration focus between the early upstream epigenetic L-R information generated during cleavage stages by maternally deposited proton pumps and the well-conserved asymmetric Nodal/Lefty/Pitx2 signaling cascade in the left lateral plate mesoderm (45). To gain insight into the role of prom3 in this developing structure, we applied prom3 MO2 and MO3. The samples were analyzed at the 8-somite stage by double immunolabeling using anti-AcTub antibody and anti-atypical protein kinase C (aPKCζ) antibody. (Despite the fact that the generation of antisera against prom3 was attempted, no specific antibody could be raised that would work on immunohistochemical staining. This might be explained by a high level of N-glycosylation in prom3 (see below).) Although the formation of Kupffer's vesicle itself was not impaired, we noticed that AcTub-labeled ciliary structures were severely affected (Fig. 6, A and B). Both the number of cilia per Kupffer's vesicle (Fig. 6C) and their length (Fig. 6D) were significantly reduced upon prom3 MO application. The antisense depletion altered neither the polarity of epithelial cells in Kupffer's vesicle nor their nuclear distribution as observed by aPKCζ and 4′,6-diamidino-2-phenylindole (DAPI) staining, respectively (Fig. 6, A and B). This strongly argues for a direct implication of prom3 in ciliogenesis rather than an indirect effect on cell polarity and proliferation in general. Altogether, these data suggest that compromised ciliary morphology was responsible for the observed laterality defects. The exact mechanisms by which zebrafish prom3 impacts on cilia will need to be further clarified once stable loss-of-function mutants and specific antibody tools against this protein are available. However, its analysis, through the molecular and morphological/anatomical readouts of the effective bioassay system applied, delivers strong evidence for the involvement of proms in ciliary biology in metazoan organisms, irrespective of their phylogenetic position during evolution.
Figure 6.
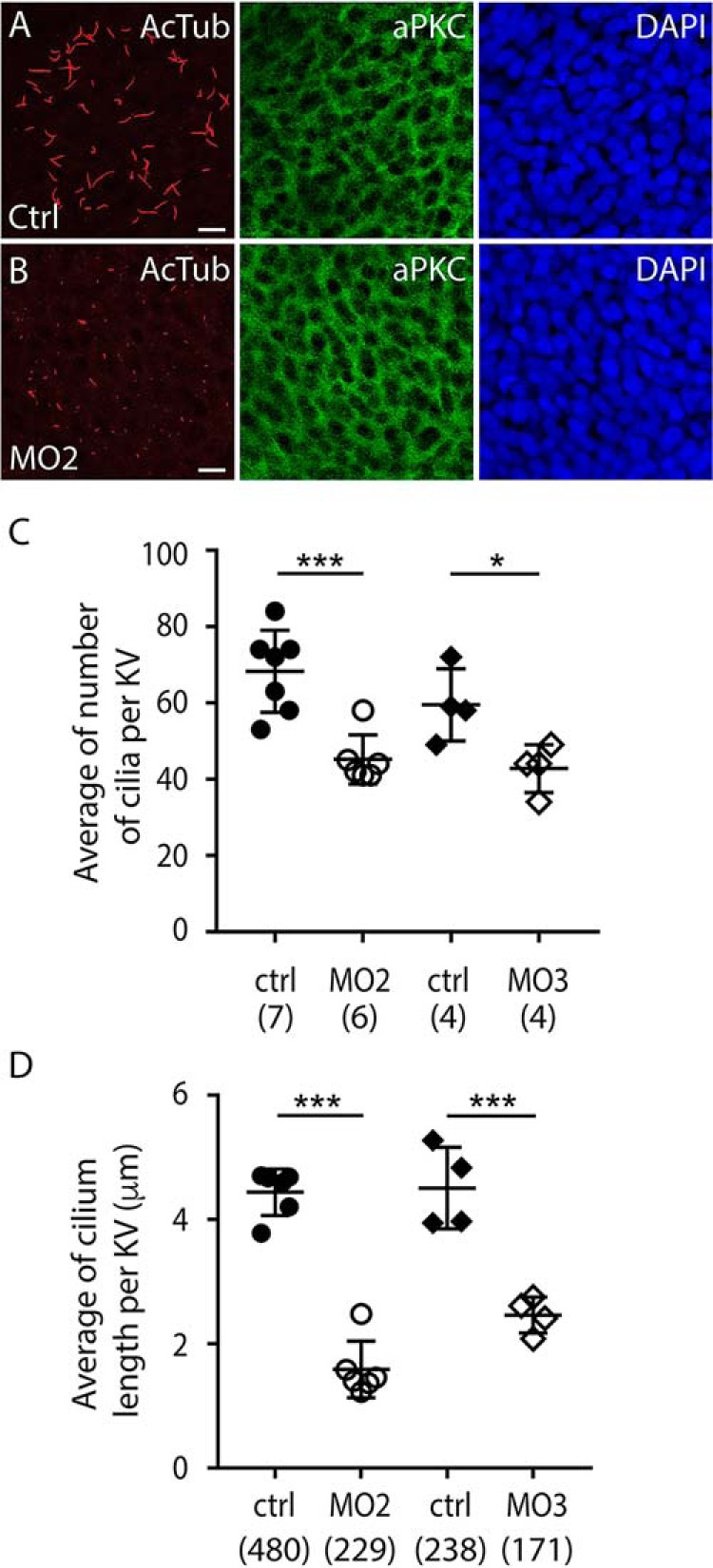
Prom3 regulates the number and length of cilia in the Kupffer's vesicle. A–D, embryos were injected at the one-cell stage with control (A, C, and D, ctrl, 10 ng) and MO against prom3 (B, C, and D, MO2; C and D, MO3, 10 ng) and processed at the 8-somite stage for double immunofluorescence using anti-AcTub (red) and anti-aPKCζ (green) antibodies, counterstained with DAPI (blue), and analyzed by CLSM. The number (C) and length (D) of cilia in the Kupffer's vesicle (KV) were quantified. The total numbers of Kupffer's vesicles and cilia analyzed are indicated in parentheses. The mean and S.D. (error bars) are presented. Individual values are shown. *, p < 0.05; ***, p < 0.001. Scale bars (A and B), 10 μm.
The localization of zebrafish prom3 mimics that of mammalian prom2 and co-localizes with Arl13b within the ciliary compartment
Mammalian prom1 is selectively associated with the apical membrane of polarized epithelial cells, whereas its paralogue prom2 is distributed in a nonpolarized fashion (21, 48). We therefore investigated the localization of zebrafish prom3 upon ectopic expression in MDCK cells. To that end, we generated expression constructs of zebrafish prom-3 either tagged with hemagglutinin (HA) epitope or as a fusion protein consisting of the enhanced GFP (EGFP) linked to the cytoplasmic C-terminal domain of prom3. In previous studies, we showed that GFP does not interfere with the proper intracellular trafficking and targeting of mammalian prom1 and prom2 to plasma membrane protrusions (20, 21). Upon high-speed centrifugation of transfected MDCK cell homogenates, prom3-HA/GFP was solely recovered in the membrane fraction, indicating that the fusion proteins were, as expected, tightly associated with the membrane (Fig. S7, A and B). Peptide-N-glycosidase F treatment led to the shift of their apparent molecular mass from ∼160 to 90 kDa in the case of prom3-HA or from ∼180 to 110 kDa for prom3-GFP, consistent with the presence of 20 potential N-glycosylation sites (Fig. S7, A and B) (data not shown).
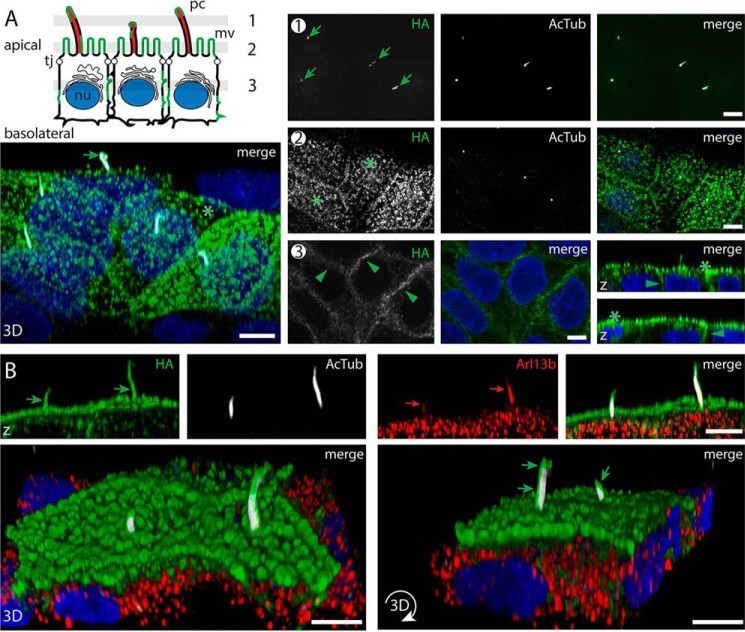
Interestingly, CLSM analysis of cells growing as a polarized cell monolayer revealed that prom3-HA was expressed at both apical and basolateral membranes (Fig. 7A). Similar data were observed with prom3-GFP (Fig. S7C). Within the apical domain, prom3-HA– and prom3-GFP–tagged versions were observed at the primary cilium highlighted by AcTub staining as well as in microvilli (Fig. 7A and Fig. S7C). The subcellular localization of prom3 is reminiscent of that of mammalian prom2 (21). We also observed a co-localization of zebrafish prom3-HA with Arl13b in the ciliary compartment (Fig. 7B, arrow) as demonstrated for mammalian prom1, suggesting that prom-Arl13b interaction may be conserved among prom molecules and across species. As with PROM1 (Fig. 2C), the staining of prom3-HA is often asymmetrically distributed in the ciliary membrane. Prom3-HA can be either concentrated on one side of the ciliary membrane throughout the entire axoneme (Fig. 7B and Video S1) or appear in a patched fashion, including its concentration at the ciliary tip (Video S2). The ciliary length (i.e. 3.1 ± 1.3 μm; n = 85) of cells expressing prom3-HA is similar to that of cells positive for WT PROM1 (see above).
Figure 7.
Zebrafish prom3 is distributed in a nonpolarized fashion in polarized epithelial cells and co-localized with Arl13b at the primary cilium. A and B, zebrafish prom3-HA–expressing MDCK cells growing as a polarized cell monolayer (7 dpc) were either double-immunolabeled (A) for prom3-HA using anti-HA antibody (green) and anti-AcTub antibody (white) or triple-immunolabeled (B) with the detection of Arl13b (red). Nuclei (nu) were counterstained with DAPI (blue) prior to CLSM analyses. Three to four single optical x-y section planes (0.4-μm slice each) throughout the cell monolayer as illustrated in the cartoon (A, sections 1–3) revealed the presence of prom3-HA in primary cilia (pc, sections 1 and 2, green arrow) highlighted with AcTub staining and microvilli (mv, section 2, asterisk) present at the apical membrane. Prom3-HA is also detected at lateral membranes (section 3, green arrowhead). Three-dimensional views (x-z orientation (z), top panels; top-side orientations (3D), bottom panels) were built from 42 x-y sections throughout cells. The immunolabeling revealed the co-localization of prom3-HA and Arl13b in the ciliary compartment (B, green and red arrows, respectively). Note the asymmetric distribution of prom3-HA at the ciliary membrane (B, bottom panels, green arrow). Tj, tight junction. Scale bars, 5 μm.
Although the information concerning the molecular trafficking of membrane proteins in and out the ciliary compartment is still incomplete and sometimes conflicting (49, 50), it has been shown that the proper targeting of some cilium-associated transmembrane proteins (e.g. seven-transmembrane receptor-1, rhodopsin, and smoothened) depends on two consecutive conserved hydrophobic and basic residues (51). Such residues (tyrosine and arginine) are present in the cytoplasmic C-terminal domain of mammalian prom1 and zebrafish prom3 in the vicinity of the last transmembrane segment (Fig. S7D). Mutation of these two residues to alanine (YR→AA) in prom3-HA, however, did not affect its proper targeting. Just like the WT version, the mutated prom3 was found associated with primary cilia (Fig. S7E). The ciliary targeting motif of proms thus remains to be discovered.
Discussion
PROM1 has received widespread attention due to its clinical translation relevance as a surface marker of stem cells (52) and cancer-initiating cells, thus representing a target for cancer eradication (5). Besides, various retinal degenerative diseases are associated with mutations in the PROM1 gene, qualifying it as a candidate for gene replacement therapy. Despite that, the function of prom1 and other members of the prom's family remained hypothetical for a long time (24). Recently, two studies have clarified the role of prom1 in the architecture of plasma membrane protrusions, such as microvilli, filopodia, and primary cilia (28, 33). Prom1 can modulate their structures and functions by interacting with regulators engaged in their biogenesis and/or dynamics.
The dual interaction of prom1 with Arl13b or HDAC6 could influence the dynamics of ciliary length either in a positive or negative manner, respectively (53–57). As demonstrated with dental epithelial stem cells, such interactions can regulate temporal and spatial activation of stem cells and perhaps cancer-initiating cells (33). The implication of prom1 in ciliogenesis might have consequences in numerous cilium-based diseases known as ciliopathies (58). In the retina, its ablation prevents the proper formation of photoreceptor outer segments that represent modified sensory cilia. The deletion of Arl13b in adult mice via tamoxifen-induced Cre/loxP recombination caused similar retinal phenotypes (59, 60). Some of the phenotypes of zebrafish prom3 morphants are strikingly reminiscent of those of a group of mutants with compromised pronephric function (61). Mutations in genes responsible for the assembly/motility of cilia frequently result in edematic phenotypes, occasionally with cystic distension of the glomerulus or the pronephric tubule/duct (44, 53, 62). The link between proms and Arl13b regarding the regulation of cilia is strengthened by the decreased length of monocilia associated with the Kupffer's vesicle in prom3 knockdown. As a result, molecular and anatomical L-R asymmetry defects, a phenotype similar to that obtained with both zebrafish Arl13b mutants and morphants (44), were observed. These data suggest that across species, prom-Arl13b interaction could play a key role in many cellular systems, which involve motile or nonmotile cilia, such as primary cilia. The latter host various signal transducers (e.g. sonic hedgehog and Wnt pathways) responsible for proper organ development (63). Our data may also explain the limited phenotypic and physiological defects after the ablation of a given prom paralogue in tissues expressing two of them simultaneously because a compensatory mechanism could occur (2, 8, 43).
Mechanistically, the mutation of the residue Tyr-819/828 located at the C terminus of PROM1 is particularly instructive because the reduced Arl13b binding suggests that tyrosine phosphorylation may be involved in regulating such interaction, similarly to what has been reported for PROM1 binding to PI3K-p85 subunit and Arp2/3 complex (28, 36). Whether this modification would interfere with HDAC6 interaction that is mediated, as for Arl13b, by a lysine residue located in the first small intracellular loop of prom1 remains to be evaluated (33, 34). Similarly, it remains to be determined whether the interaction of prom1 with the Arp2/3 complex, which is also dependent on its tyrosine 819/828 phosphorylation (28), could influence the ciliary structure (64). Investigating this interaction at the base of the cilium and/or its tip during the shedding of prom1+ membrane vesicles (20, 28, 33, 65) will be of particular interest because it has been shown that actin polymerization in these subcompartments depends on the Arp2/3 complex (66–68). In photoreceptors, the prom1-Arp2/3 complex interaction could stimulate the growth of membrane evaginations from the connecting cilium at the base of the outer segment, thus generating a new photoreceptor disc (69).
A further exciting question is how proms can influence microvilli and cilia, thus examining the connection of cholesterol-binding membrane proteins with actin- and tubulin-based cytoskeleton structures. Other constituents indirectly implicated in such interaction might be gangliosides, whose presence in the ciliary membrane was demonstrated (30). A recent study on the structural impact of mutations in the ganglioside-binding site of PROM1 revealed their influence on the interaction of prom1 with PI3K in a tyrosine phosphorylation–dependent manner (28). Therefore, the morphological disturbance of primary cilia observed upon overexpression of mutants Y819/828F may also reflect a disrupted interaction of proms with cholesterol- and/or ganglioside-rich membrane microdomains (21, 26, 30, 31), as suggested for the altered microvillar structure (28). The local activity of PI3K that converts PIP2 into PIP3 in the inner leaflet of the plasma membrane can regulate the interaction of the lipid bilayer with the underlying structural core of cellular protrusions and/or the transport of membrane components therein (28, 70–74). The accumulation of Arl13b at the ciliary base of cells expressing the PROM1 mutant may result from a defect in the aforementioned mechanism. It is known that Arl13b acts as a guanine-nucleotide-exchange factor for Arl3, and both proteins coordinate the intraflagellar transport (75–78). Thus, prom-containing membrane microdomains may act as building units that elaborate and/or maintain the proper composition of microvillar and ciliary membranes and thus influence interactions with cytoplasmic regulatory proteins. They could also affect membrane fluidity by creating distinct liquid phases and, hence, influence the diffusion of their constituents (74). The uneven distribution of PROM1 (or prom3) observed in cilia of WT PROM1 (or prom3-HA)+ cells compared with its uniform spread in mutated PROM1+ cells suggests the disruption of these membrane microdomains (Fig. 2C). A more in-depth study should dissect the impact of proms on each subdomain of the cilium, including the basal body and the transition zone as well as the molecular players involved in the intraflagellar transport (79). The interaction of proms with other membrane (e.g. protocadherin 21) or secreted (spacemaker) proteins can also organize the extracellular space between cellular protrusions (9, 80, 81). Altogether, these distinct interactions could explain the implication of proms in various cellular processes such as proliferation, differentiation, autophagy, migration, and carcinogenesis.
Interestingly, prom1 also plays a role in autophagy, where a phosphorylation-dependent interaction with γ-aminobutyric acid receptor–associated protein (GABARAP), an initiator of autophagy, leads to the suppression of GABARAP-mediated ULK1 kinase activation and the subsequent initiation of autophagy (82, 83). The prom1 phosphorylation could link ciliogenesis and autophagy (84, 85), which in turn may regulate the cell proliferation versus differentiation (86). This model is thrilling because cell differentiation has been linked to a reduction or loss of prom1 (52, 87, 88). A relationship between cell proliferation, cilia, and prom3 was suggested in zebrafish hair cells (89).
The apico-basolateral distribution of zebrafish prom3 is reminiscent of that of mammalian prom2 (21, 48), which contrasts with the exclusive apical localization of prom1 (90). However, the cytoplasmic C-terminal domain of prom3 is distinct from that of prom2, which is known to be almost perfectly conserved among mammals (2). Also, genomic analysis revealed that zebrafish prom3 is not a true orthologue of the mammalian prom2 gene. Although both genes display a higher degree of compactness compared with prom1 (Table S1), they do not show a conserved synteny (11). Still, prom3 might have evolved from the same ancestral locus as prom2 generated during the first round of vertebrate genome duplication (11, 12, 91).
In conclusion, our data revealed a participation of mammalian and nonmammalian prom paralogues in ciliary architecture and function beyond photoreceptors. With that, we contribute to the ongoing dissection of molecular and cellular networks underlying not only ciliopathies, but also developmental pathways linked to the integrity and dynamics of cilia.
Experimental procedures
Animals
Animal experiments were performed in strict accordance with German Animal Welfare legislation, and all procedures were performed in compliance with the UK Animals Act (Scientific Procedures, 1986) approved by the Institutional Animal Welfare Officer (Tierschutzbeauftragter). All efforts were made to minimize the suffering of animals, and those used for organ removal were sacrificed according to the Schedule 1 methods of humane killing. Experiments were carried out at the Max-Planck-Institute of Molecular Cell Biology and Genetics (MPI-CBG, Dresden, Germany), which holds licenses (Az.:74-9165.40-9-2000-1 and 74-9168.25-9-2001-1) for keeping and breeding of laboratory animals (Erteilung und Erlaubnis für das Züchten und Halten von Wirbeltieren zu wissenschaftlichen Zwecken) and collecting organs and tissues (Anzeigepflichtige Tierversuche), both issued by Regierungspräsidium Dresden, Saxony. No genetically modified animals were used or generated in the current study.
Fish embryos
Fish were raised and kept under standard conditions at 27 °C at the fish facility of the MPI-CBG, Dresden. Freshly laid fertilized eggs were harvested into E3 embryo medium (5 mm NaCl, 0.17 mm KCl, 0.33 mm CaCl2, 0.33 mm MgSO4, 0.00001% (w/v) methylene blue). Embryos were raised at 28.5 °C and staged according to Kimmel et al. (92).
Cloning of zebrafish prom3
By searching the fourth assembly of the zebrafish genome of the Wellcome Trust Sanger Institute using the murine prom1 amino acid sequence as a query, a genomic contig (Zv4_scaffold1778) encoding for a previously unannotated, partial prom molecule was identified. With the sequence information gained, a cDNA fragment of this prom homologue, designated as prom3, was amplified by PCR from a 72-hpf whole-larvae cDNA pool using the oligonucleotides 5′-GACTGATCCTGAGCTGTCTG-3′ (jj-499) and 5′-ACAATGACTTACTCATACACATCTG-3′ (jj-500) as forward and reverse primers, respectively. Total RNA was used to prepare a first strand cDNA template with Superscript II reverse transcriptase (Invitrogen). As a result, a 1.2-kb cDNA fragment was obtained, subcloned into pCRII-TOPO TA vector (Invitrogen), and sequenced. The missing extremities were amplified by 5′- and 3′-RACEs using a SMART RACE cDNA synthesis kit (catalog no. 634914, Clontech) according to the manufacturer's instructions. The gene-specific oligonucleotides used for the first and second round of amplification of 5′-RACE were 5′-GATGAAGTGTAGTCCACAAGGGTGCGTTCTG-3′ (jj-593) and 5′-ACTAACATGAAGATCCAGGAGATCAG-3′ (jj-594). Those for 3′-RACE were 5′-GACATCACATCTAACATAATGCCTTTGCTGCTG-3′ (jj-590) and 5′-GTGAGAGCTGAGAGCAAGGTGTTCGTAGACTG-3′ (jj-591). PCR products were extracted from the agarose gel (Qiagen GmBH, Helden, Germany) and subcloned into pCRII-TOPO TA vector. The identity of the inserts was determined by sequencing. Finally, the entire zebrafish prom3 coding sequence was amplified with Accuprime Taq High Fidelity DNA polymerase (Invitrogen) using the oligonucleotides 5′-CAGAGGGAGACATTGGAGCGGAC-3′ (jj-616) and 5′-GTGATTTCGGATACAGCCAAGTG-3′ (jj-621) as forward and reverse primers, respectively. PCR products were subcloned into pCRII-TOPO Dual Promoter vector (Invitrogen). Under these conditions, we have identified two distinct splice variants of zebrafish prom3 that differ in their 5′-UTR. Their sequences were deposited in the GenBankTM database (accession numbers DQ341400 and HQ698925). Sequencing reactions were performed using an Applied Biosystems 3730 Genetic Analyzer at the MPI-CBG sequencing facility.
The nucleotide and protein sequences were analyzed by the HUSAR compilation of sequence analysis tools accessible online at the German Cancer Research Center (Deutsches Krebsforschungszentrum, Heidelberg, Germany) or European Molecular Biology Laboratory–European Bioinformatics Institutes (EMBL-EBI). The genomic organization of the Prom3 gene is presented in Table S1.
Plasmid construction
Eukaryotic expression plasmids Zf PB2-1 and Zf PB1-3 containing full-length coding sequence of zebrafish prom3 (DQ341400) fused in-frame at its C terminus to either EGFP or HA epitope tag nucleotide sequence, respectively, were constructed by selective PCR amplification. The 5′ primer was 5′-GTCAGATCCGCTAGCAACATGAAGTCCAGAACGAAAGCAG-3′ (jj-685N), whereas the 3′ ones were 5′-GACTGCACTCGAGTTGTTTCGCATTTTCCACGAACTC-3′ (jj-687) and 5′-GACTGCACTCGAGTCAGGCGTAGTCGGGGACGTCGTAGGGGTATTGTTTCGCATTTTCCACGAACTC-3′ (jj-686) for the EGFP and HA tag, respectively. In the latter oligonucleotide, TGA stop codon (boldface type) and coding sequence of HA tag (27 nucleotides double underlined) are indicated. The 5′ primer contained an NheI restriction site, whereas the 3′ primers carried an XhoI recognition site (underlined), allowing for directional subcloning into pEGFP-N1 vector (BD Bioscience Clontech) digested with the corresponding restriction enzymes. In all cases, prom sequences within the expression plasmids were sequenced to confirm the absence of any reading mistakes introduced by the DNA polymerase and/or subcloning steps.
Expression plasmids (hprom1.s1 and hprom1.s2) containing full-length coding sequence of human PROM1 splice variant s1 (GenBankTM AF507034.1, cDNA clone MGC: 20041 IMAGE: 4644690) and s2 (AF027208.1; Ref. 9) into the enhanced yellow fluorescent protein (EYFP)-N1 vector (BD Bioscience Clontech) were recently described (28). Note that the EYFP was excised during the cloning procedure. Plasmids containing human PROM1 mutant Y819F or Y828F were also reported (28).
Site-directed mutagenesis
A two-amino acid motif (Tyr796-Arg797) located at the zebrafish prom3 C-terminal domain was mutated using the QuikChange® II site-directed mutagenesis kit (catalog no. 200523, Stratagene) according to the manufacturer's instructions. Zf PB1–3 plasmid (see above) was used as template. The oligonucleotides were 5′-CTGTTAAACTGGCCAAGTACGCTGCTAGGATGAAGTATTCAGATGTG-3′ (jj-689YR→AA) and 5′-CACATCTGAATACTTCATCCTAGCAGCGTACTTGGC-CAGTTTAACAG-3′ (jj-690YR→AA), as sense and antisense primers, respectively. They carry two nucleotide triplets (underlined) encoding for two alanine residues (AA) instead of the native amino acids (YR). The resulting plasmid (Zf PD2-3) was sequenced to confirm mutations.
Cell culture and transfection
MDCK cells (strain II, American Type Culture Collection, catalog no. ATCC CCL-34) were cultured as described previously (28). They were maintained in a humidified incubator at 37 °C under a 5% CO2 atmosphere. MDCK cells were stably transfected with either Zf PB2-1 (prom3-EGFP), Zf PB1–3 (prom3-HA), or Zf PD2-3 (prom3YR-AA-HA) plasmids using Amaxa® Nucleofector® technology (Amaxa, Cologne, Germany). Two days after transfection, cells were selected by introducing 500–600 μg/ml GeneticinTM (G418 Sulfate, Thermo Fisher Scientific) into the culture medium and further expanded. After selection, the concentration of the selective agent was reduced to 250 μg/ml. MDCK cells expressing human PROM1.s1 and PROM1.s2 and the corresponding Y819/828F mutants as well as those containing solely control vector were recently described (28). Note that under these conditions, Geneticin did not affect the ciliary length, as demonstrated with transfected cells containing expression vector alone (data not shown), ruling out a direct effect of the drug on polyphosphoinositides (93, 94). To enhance the expression of the recombinant protein, 10 mm sodium butyrate was added to the culture medium for 16 h before certain experiments as described (90). Cells were used 7 and 14 dpc.
Probes for in situ hybridization
Antisense complementary DIG-labeled RNA (cRNA) probes were generated using a DIG labeling mix (Roche Molecular Biochemicals). To synthesize cRNA probes, the entire cDNA inserts containing the coding sequence of either zebrafish prom1a (HQ386793) or prom3 (DQ341400) were used. For zebrafish prom1b (AF373869), a 1.365-kb cDNA fragment (nucleotides 234–1,599) was used. The average homology of the zebrafish prom1 co-paralogues along the stretch used for ISH is ∼60% (13). Combined with the stringent hybridization and post-hybridization conditions, this precludes any cross-hybridization. To generate templates for zebrafish lefty2 (BC097092) and cmlc2 (BC076232) probes, cDNA clones were obtained from the German Resource Center and Primary Database (German Science Center for Genome Research, RZPD, Berlin).
Microinjection of morpholino-oligonucleotides
Standard control and custom MOs were purchased from GeneTools. MOs made complementary to the AUG translational initiation site, the 5′-UTR, and the splice site of zebrafish prom3 were 5′-CTGCTTTGCTTCTGGACTTCATGTT-3′ (MO1), 5′-GAGCGTGTTCCAGTTAATTCAGGTC-3′ (MO2), and 5-TGGTGCTCACAGAATAAGTACAGTG-3′ (MO3), respectively. MOs directed against the AUG initiation coding site and 5′-UTR of zebrafish prom1a were 5′-AGCATGATTAGTTCTAGTGTTGAAG-3′ (MO1) and 5′-GTTCAGTAAGACACAATCACTGACA-3′ (MO2), respectively. Control MO was 5′-CCTCTTACCTCAGTTACAATTTATA-3′. MOs were dissolved in H2O at a concentration of 40 μg/μl (stock solution) and further diluted (5 ng/nl) for injection in Danieau buffer containing 0.2% phenol red as tracking dye. They were injected at the one-cell stage into the yolk using a PV830 pneumatic PicoPump (WPI) in a maximum volume of 2 nl. At a given period of the developmental stage, zebrafish larvae were either observed in a native, unfixed state or further processed as described below.
PCR detection of nuclear pre-mRNA processing
Control MO and prom3-MO3–injected embryos were manually dechorionated with fine-tip watchmaker's forcipes and snap-frozen at the 9-somite stage. After total RNA isolation and reverse transcription using Superscript II reverse transcriptase (Invitrogen), PCR amplification was performed using oligonucleotides 5′-GATCCTCTACATTGTCCTGATG-3′ (jj-719) and 5′-GACATCATCTAAGATGCCTG-3′ (jj-720) as forward and reverse primers, respectively. Cycling parameters (denaturation at 94 °C for 30 s, annealing at 50 °C for 45 s, extension at 72 °C for 1 min) were used for 40 cycles. To exclude amplification of genomic DNA that might be present in total RNA preparations, DNase digestion of the RNA sample was performed (10 min, room temperature) followed by a clean-up step (RNeasy kit, Qiagen) before cDNA synthesis.
Histological sections
Embryos were fixed in 4% paraformaldehyde (PFA), washed three times with PBS (pH 7.4), dehydrated, and embedded in paraffin. Serial sections were cut at 4 μm with a rotary microtome. Slides were deparaffinized, stained with hematoxylin-eosin, and coverslipped with xylene-based mounting medium.
Whole-mount in situ
Embryos were PFA-fixed at the required stage of development. After dechorionation, embryos were dehydrated in methanol and stored at −20 °C until use. Whole-mount ISH was performed as described (95). Stained embryos were post-fixed in 4% PFA and observed using either an Olympus SZX12 microscope or a Zeiss Axioscope. Images were captured without glycerol clearing.
Whole-mount immunohistochemistry
Embryos were fixed in 4% PFA at the required stage of development. After dechorionation, embryos were washed three times for 10 min in PBST (PBS + 0.1% Tween 20) and subsequently incubated with 0.005% SDS in 0.2% gelatin-PBS for 30 min. Embryos were then rinsed once with 0.2% gelatin-PBS followed by three periods of incubation in 0.2% saponin-PBS for 30 min each and one period of incubation in blocking buffer I (5% normal goat serum and 2% BSA diluted with 0.2% saponin-PBS) for 1 h at room temperature. Afterward, the samples were incubated simultaneously with anti-AcTub mouse mAb (1:800; clone 6-11B-1, catalog no. T6793, Sigma–Aldrich, Darmstadt, Germany) and anti-aPKCζ rabbit polyclonal antibody (1:200; catalog no. sc-216, Santa Cruz Biotechnology, Inc.) diluted in blocking buffer I overnight at 4 °C. Primary antibodies were detected using goat anti-mouse Alexa Fluor®546– or anti-rabbit Alexa Fluor®488–conjugated secondary antibodies, respectively (1:750; Thermo Fisher Scientific) diluted in blocking buffer I, respectively. Embryos were counterstained with DAPI (1 μg/ml; Thermo Fisher Scientific) to facilitate the identification of the Kupffer's vesicle according to its typical nuclear configuration. After washing with PBS, the caudal halves of the embryos were dissected and freed from yolk with fine tungsten needles. Samples were subsequently flat-mounted with Mowiol 4.88 (Calbiochem). Images were captured using a Zeiss 780 confocal laser-scanning microscope (Jena, Germany). Composite images were prepared from the digital data files using Fiji (96) and Adobe® Photoshop and Illustrator software (San Jose, CA).
Immunoisolation
WT MDCK cells or those expressing human PROM1 variants were lysed in solubilization buffer (0.2% Triton X-100, 150 mm NaCl, 20 mm Tris/HCl, pH 7.5) containing completeTM protease inhibitor mixture (Roche Diagnostics GmbH, Manheim, Germany), 2 mm sodium orthovanadate and 10 mm sodium fluoride (both from New England Biolabs Inc., Ipswich, MA), and 10 mm β-glycerophosphate (Sigma–Aldrich) on ice for 30 min. After centrifugation (10 min, 16,000 × g, 4 °C), an aliquot of detergent lysates was removed as an input sample, and remaining materials were divided into two fractions. They were incubated with either 65 μl of mAb AC133-MicroBeads, or 130 μl of anti-FITC- MicroBeads as negative control, for 1.5 h at 4 °C prior to magnetic separation using MS columns (all from Miltenyi Biotec, Bergisch Gladbach, Germany). Materials retained in columns were washed five times with 1 ml of ice-cold PBS containing 2 mm EDTA and 0.1% Triton X-100 and then eluted with preheated (95 °C) Laemmli sample buffer according to the manufacturer's instructions. Input and bound fractions were analyzed by immunoblotting for PROM1 and Arl13b (see below).
Alternatively, Arl13b was immunoisolated. After precleaning the detergent cell lysates with 100 μl of μMACS Protein A MicroBeads (Miltenyi Biotec) for 30 min at 4 °C, followed by the magnetic separation, lysates were divided into two fractions. One was incubated with 4.5 μl of rabbit antiserum against Arl13b (catalog no. 17711-1-AP, Proteintech, Manchester, UK), and the other was incubated without as a negative control, for 20 min at 4 °C. Afterward, 100 μl of μMACS Protein A MicroBeads was added for 1 h at 4 °C. Bound fractions were isolated with a magnetic column and processed as described above.
Membrane preparation
MDCK cells expressing either zebrafish prom3-HA, prom3YR-AA-HA, or prom3-EGFP were scraped with a rubber policeman in ice-cold PBS and collected by centrifugation at 1,200 × g for 5 min at 4 °C. Cell pellets were homogenized with a glass-Teflon pestle in sucrose buffer (300 mm sucrose, 5 mm EDTA, 10 mm Hepes-KOH, pH 7.5, containing Complete Protease Inhibitor (Roche Molecular Biochemicals) at 4 °C and subsequently incubated for 30 min on ice. Lysates were centrifuged at 1,000 × g for 10 min at 4 °C, and the resulting supernatants were subjected to ultracentrifugation at 60,000 × g for 60 min at 4 °C. The pellets containing membrane fractions were resuspended in 1% SDS, 20 mm sodium phosphate and boiled for 2 min at 95 °C, whereas cytosol fractions were recovered in supernatants. Membrane fractions were subjected to endoglycosidase treatment (see below) prior to their analysis by SDS-PAGE and immunoblotting.
Endoglycosidase digestion and immunoblotting
MDCK membrane extracts were incubated in the absence or presence of 1 unit of peptide-N-glycosidase F overnight at 37 °C according to the manufacturer's instructions (Roche Diagnostics GmbH).
All samples were subjected to SDS-PAGE (7.5%) and transferred to polyvinylidene difluoride membranes (pore size: 0.45 μm; Millipore Corp., Belford, MA) using a semi-dry transfer cell system (Cti, Idstein, Germany). After transfer, membranes were incubated overnight at 4 °C in blocking buffer II (PBS containing 0.1% Tween 20 and 5% milk powder) prior to a 1-h incubation at room temperature with either mouse monoclonal antibodies against human PROM1 (1:2,000, clone 80B258 (22), Fig. 2A; 1:200, clone W6B3C1 (Miltenyi Biotec); data not shown) or Arl13b antiserum (1:1,000; Proteintech) diluted in blocking buffer II. Recombinant epitope-tagged zebrafish prom3 proteins were detected using either mouse monoclonal GFP antibody (1:1,000; catalog no. sc-9996, Santa Cruz Biotechnology) or rat monoclonal HA tag antibody (1:1,000; clone 3F10, Roche Molecular Biochemicals). In all cases, antigen-antibody complexes were revealed using the appropriate horseradish peroxidase–conjugated secondary antibody (Dianova, Hamburg, Germany) followed by chemiluminescence detection (ECL System, GE Healthcare Life Sciences). Membranes were exposed to Amersham Biosciences Hyperfilm ECL (GE Healthcare Life Sciences). When necessary, Fiji software was used for the quantification of immunoblotted materials.
Immunofluorescence and confocal microscopy
MDCK cells were cultured on fibronectin (BD Biosciences)-coated glass coverslips in silicon 8-well chambers (flexiPerm (Greiner Bio-One GmbH) or μ-slide ibidi (ibidi GmbH, Gräfelfing, Germany)).
MDCK cells expressing human PROM1
Cells were rinsed with Ca/Mg buffer (PBS containing 1 mm CaCl2 and 0.5 mm MgCl2) and incubated in blocking buffer III (Ca/Mg buffer containing 0.2% gelatin) for 30 min at 4 °C. For cell surface staining of PROM1, cells were incubated with mouse mAb CD133/1 (1:50; Miltenyi Biotec) diluted in blocking buffer III for 1 h at 4 °C. After washing with PBS, cells were fixed in 4% PFA in PBS for 30 min at room temperature and then incubated for 10 min in PBS containing 50 mm NH4Cl. Fixed cells were permeabilized and blocked with 0.2% saponin in 0.2% gelatin/PBS for 30 min at room temperature. To visualize the primary cilium, cells were labeled with mouse monoclonal AcTub antibody (1:1000; clone 6-11B-1, Sigma–Aldrich) for 30 min in permeabilization buffer, followed by Alexa Fluor®546 (or 647)–conjugated goat anti-mouse IgG2b (1:600; Thermo Fisher Scientific), whereas Alexa Fluor®488-conjugated goat anti-mouse IgG1 antibody (1:500; Thermo Fisher Scientific) was used to detect anti-PROM1 antibody as described (33). In some experiments, Arl13b was detected with the rabbit antiserum against Arl13b and Alexa Fluor®555–conjugated donkey anti-rabbit IgG (H+L) (1:500; Thermo Fisher Scientific). Nuclei were visualized with DAPI (1 μg/ml). Finally, cells were washed three times in PBS containing 0.2% gelatin, three times in PBS, and twice in H2O and mounted in Mowiol 4.88. For the measurements of the ciliary length, cells were not mounted but were directly observed with a Zeiss LSM 700 or LSM 780 confocal laser-scanning microscope (Jena, Germany).
MDCK cells expressing zebrafish prom3
Cells were washed with PBS and then fixed and permeabilized as described above. To evaluate the distribution of the transgenes, zebrafish prom3-HA– or prom3YR-AA-HA–expressing cells were labeled with a rat monoclonal HA tag antibody (1 μg/ml) followed by the goat anti-rat IgG (H+L) conjugated either to Cy2TM (1:600; Jackson ImmunoResearch Inc.) or Alexa Fluor®488 (1:500; Thermo Fisher Scientific), whereas in prom3-EGFP–transfected cells, the green fluorescence was monitored. The detection of AcTub and Arl13b was performed as described above. After mounting, cells were observed with a Zeiss LSM 780 or 510 Meta confocal laser-scanning microscope. The microscope settings were such that multipliers were within their linear range. The images shown were prepared from data files of LSM 5 LIVE (Göttingen, Germany) using Volocity 3D Analysis software (PerkinElmer Life Sciences), Fiji, and Arivis Vision 4D software (Arivis AG) as well as Adobe® Photoshop and Illustrator software.
Measurements
Primary cilia of MDCK cells were quantified using Huygens Professional software (Scientific Volume Imaging, Hilversum, The Netherlands) as described (33). After deconvolution of Z-stack images, the advanced object analyzer was used to obtain ciliary length. Fluorescence intensities of Arl13b and AcTub in the axoneme were quantified with Fiji. The number and length of cilia in Kupffer's vesicles were evaluated using the segmented line tool of the Fiji software on maximum projection of Z-stack images captured from flat-mounted tissues with a Zeiss LSM 780 confocal laser-scanning microscope.
Scanning EM
MDCK cells were prepared for SEM analysis as described (21). Briefly, cells grown on fibronectin-coated coverslips were fixed in 2% glutaraldehyde overnight at 4 °C. After dehydration in an acetone gradient (25–100%), cells were critical point–dried in a CO2 system (Critical Point Dryer CPD 300, Leica). Samples were mounted on aluminum stubs, coated with gold in a sputter coater (SCD 050, BAL-TEC GmbH), and examined with a scanning electron microscope (JSM-7500F, JEOL).
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 6.00 for Mac (GraphPad Software, La Jolla, CA) or Microsoft Excel software. Two-tailed unpaired Student's t test was used for the analysis of the nuclei numbers observed in MDCK monolayer for a given surface area, Arl13b/PROM1 ratio, cilium number, and their length in Kupffer's vesicle, whereas a Mann–Whitney test was applied for the length of cilium in MDCK cells and Arl13b/AcTub ratio. Observed differences were regarded as significant if the calculated p values were ≤0.05. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Data availability
All data are contained within the article and supporting materials. The zebrafish prom3 sequences were deposited in the public GenBankTM database under accession numbers DQ341400 and HQ698925.
Acknowledgments
We express our gratitude to Sylvi Graupner for skilled assistance. We thank the Electron Microscopy/Histology (EMH) and Light Microscopy Facilities at the Technology Platform of the Center for Molecular and Cellular Bioengineering (Technische Universität Dresden) for instrument access and technical support, in particular Dr. Tomas Kurth, Dr. Hella Hartmann, and Ellen Geibelt. The European Regional Development Fund supports the Technology Platforms at the Center for Molecular and Cellular Bioengineering.
Author contributions
J. J., K. T., J. K., W. B. H., and D. C. conceptualization; J. J., K. T., P. J., W. B. H., and D. C. resources; J. J., K. T., J. K., P. J., and D. C. data curation; J. J., K. T., J. K., P. J., C. A. F., and D. C. formal analysis; J. J., K. T., P. J., and D. C. supervision; J. J., W. B. H., and D. C. funding acquisition; J. J., K. T., P. J., and D. C. validation; J. J., K. T., J. K., P. J., C. A. F., and D. C. investigation; J. J., K. T., J. K., and D. C. visualization; J. J., K. T., J. K., P. J., C. A. F., and D. C. methodology; J. J., K. T., C. A. F., and D. C. writing-original draft; J. J. and D. C. project administration; J. J., K. T., J. K., P. J., C. A. F., W. B. H., and D. C. writing-review and editing.
Supplementary Material
This work was supported by Intramural Funds of the Medical Faculty of TU Dresden (MeDDrive grant) and Deutsche Forschungsgemeinschaft (DFG) Grant CO298/5-1 (to J. J.), DFG Grant SFB655 A2 (to W. B. H.), and DFG Grants SFB655 B3 and CO298/5-1 (to D. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1, Figs. S1–S7, and Videos S1 and S2.
- prom
- prominin
- Arp
- actin-related protein
- Arl
- ADP-ribosylation factor–like protein
- HDAC6
- histone deacetylase 6
- PI3K
- phosphoinositide 3-kinase
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PIP3
- phosphatidylinositol 3,4,5-trisphosphate
- L-R
- left-right
- MDCK
- Madin–Darby canine kidney
- SEM
- scanning EM
- AcTub
- acetylated α-tubulin
- CLSM
- confocal laser-scanning microscopy
- dpc
- days post-confluence
- ISH
- in situ hybridization
- hpf
- hours post-fertilization
- MO
- morpholino-oligonucleotide
- dpf
- days post-fertilization
- aPKC
- atypical protein kinase C
- DAPI
- 4′,6-diamidino-2-phenylindole
- HA
- hemagglutinin
- EGFP and EYFP
- enhanced green and yellow fluorescent protein, respectively
- GABARAP
- γ-aminobutyric acid receptor–associated protein
- contig
- group of overlapping clones
- DIG
- digoxigenin
- cRNA
- complementary RNA
- PFA
- paraformaldehyde.
References
- 1. Weigmann A., Corbeil D., Hellwig A., and Huttner W. B. (1997) Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 94, 12425–12430 10.1073/pnas.94.23.12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fargeas C. A., Florek M., Huttner W. B., and Corbeil D. (2003) Characterization of prominin-2, a new member of the prominin family of pentaspan membrane glycoproteins. J. Biol. Chem. 278, 8586–8596 10.1074/jbc.M210640200 [DOI] [PubMed] [Google Scholar]
- 3. Kania G., Corbeil D., Fuchs J., Tarasov K. V., Blyszczuk P., Huttner W. B., Boheler K. R., and Wobus A. M. (2005) Somatic stem cell marker prominin-1/CD133 is expressed in embryonic stem cell-derived progenitors. Stem Cells 23, 791–804 10.1634/stemcells.2004-0232 [DOI] [PubMed] [Google Scholar]
- 4. Corbeil D., Karbanová J., Fargeas C. A., and Jászai J. (2013) Prominin-1 (CD133): Molecular and cellular features across species. Adv. Exp. Med. Biol. 777, 3–24 10.1007/978-1-4614-5894-4_1 [DOI] [PubMed] [Google Scholar]
- 5. Grosse-Gehling P., Fargeas C. A., Dittfeld C., Garbe Y., Alison M. R., Corbeil D., and Kunz-Schughart L. A. (2013) CD133 as a biomarker for putative cancer stem cells in solid tumours: limitations, problems and challenges. J. Pathol. 229, 355–378 10.1002/path.4086 [DOI] [PubMed] [Google Scholar]
- 6. Maw M. A., Corbeil D., Koch J., Hellwig A., Wilson-Wheeler J. C., Bridges R. J., Kumaramanickavel G., John S., Nancarrow D., Röper K., Weigmann A., Huttner W. B., and Denton M. J. (2000) A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum. Mol. Genet. 9, 27–34 10.1093/hmg/9.1.27 [DOI] [PubMed] [Google Scholar]
- 7. Gurudev N., Florek M., Corbeil D., and Knust E. (2013) Prominent role of prominin in the retina. Adv. Exp. Med. Biol. 777, 55–71 10.1007/978-1-4614-5894-4_4 [DOI] [PubMed] [Google Scholar]
- 8. Zacchigna S., Oh H., Wilsch-Bräuninger M., Missol-Kolka E., Jászai J., Jansen S., Tanimoto N., Tonagel F., Seeliger M., Huttner W. B., Corbeil D., Dewerchin M., Vinckier S., Moons L., and Carmeliet P. (2009) Loss of the cholesterol-binding protein prominin-1/CD133 causes disk dysmorphogenesis and photoreceptor degeneration. J. Neurosci. 29, 2297–2308 10.1523/JNEUROSCI.2034-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang Z., Chen Y., Lillo C., Chien J., Yu Z., Michaelides M., Klein M., Howes K. A., Li Y., Kaminoh Y., Chen H., Zhao C., Chen Y., Al-Sheikh Y. T., Karan G., et al. (2008) Mutant prominin 1 found in patients with macular degeneration disrupts photoreceptor disk morphogenesis in mice. J. Clin. Invest. 118, 2908–2916 10.1172/JCI35891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fargeas C. A., Büttner E., and Corbeil D. (2015) Commentary: “prom1 function in development, intestinal inflammation, and intestinal tumorigenesis”. Front. Oncol. 5, 91 10.3389/fonc.2015.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wotton K. R., Weierud F. K., Dietrich S., and Lewis K. E. (2008) Comparative genomics of Lbx loci reveals conservation of identical Lbx ohnologs in bony vertebrates. BMC Evol. Biol. 8, 171 10.1186/1471-2148-8-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McGrail M., Batz L., Noack K., Pandey S., Huang Y., Gu X., and Essner J. J. (2010) Expression of the zebrafish CD133/prominin1 genes in cellular proliferation zones in the embryonic central nervous system and sensory organs. Dev. Dyn. 239, 1849–1857 10.1002/dvdy.22307 [DOI] [PubMed] [Google Scholar]
- 13. Jászai J., Fargeas C. A., Graupner S., Tanaka E. M., Brand M., Huttner W. B., and Corbeil D. (2011) Distinct and conserved prominin-1/CD133-positive retinal cell populations identified across species. PLoS ONE 6, e17590 10.1371/journal.pone.0017590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jászai J., Graupner S., Tanaka E. M., Funk R. H., Huttner W. B., Brand M., and Corbeil D. (2013) Spatial distribution of prominin-1 (CD133)-positive cells within germinative zones of the vertebrate brain. PLoS ONE 8, e63457 10.1371/journal.pone.0063457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu Z., Hu X., Reilly J., Jia D., Liu F., Yu S., Liu X., Xie S., Qu Z., Qin Y., Huang Y., Lv Y., Li J., Gao P., Wong F., et al. (2019) Deletion of the transmembrane protein Prom1b in zebrafish disrupts outer-segment morphogenesis and causes photoreceptor degeneration. J. Biol. Chem. 294, 13953–13963 10.1074/jbc.RA119.008618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Corbeil D., Fargeas C. A., and Jászai J. (2019) Deciphering the roles of prominins in the visual system. J. Biol. Chem. 294, 17166 10.1074/jbc.L119.011198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu Z., Hu X., Reilly J., Jia D., Liu F., Yu S., Liu X., Xie S., Qu Z., Qin Y., Huang Y., Lv Y., Li J., Gao P., Wong F., Shu X., Tang Z., and Liu M. (2019) Reply to Corbeil et al.: deletion of the transmembrane protein Prom1b in zebrafish disrupts outer-segment morphogenesis and causes photoreceptor degeneration. J. Biol. Chem. 294, 17167 10.1074/jbc.RL119.011224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han Z., Anderson D. W., and Papermaster D. S. (2012) Prominin-1 localizes to the open rims of outer segment lamellae in Xenopus laevis rod and cone photoreceptors. Invest. Ophthalmol. Vis. Sci. 53, 361–373 10.1167/iovs.11-8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldberg A. F., Moritz O. L., and Williams D. S. (2016) Molecular basis for photoreceptor outer segment architecture. Prog. Retin. Eye Res. 55, 52–81 10.1016/j.preteyeres.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dubreuil V., Marzesco A. M., Corbeil D., Huttner W. B., and Wilsch-Bräuninger M. (2007) Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J. Cell Biol. 176, 483–495 10.1083/jcb.200608137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Florek M., Bauer N., Janich P., Wilsch-Braeuninger M., Fargeas C. A., Marzesco A. M., Ehninger G., Thiele C., Huttner W. B., and Corbeil D. (2007) Prominin-2 is a cholesterol-binding protein associated with apical and basolateral plasmalemmal protrusions in polarized epithelial cells and released into urine. Cell Tissue Res. 328, 31–47 10.1007/s00441-006-0324-z [DOI] [PubMed] [Google Scholar]
- 22. Karbanová J., Missol-Kolka E., Fonseca A. V., Lorra C., Janich P., Hollerová H., Jászai J., Ehrmann J., Kolár Z., Liebers C., Arl S., Subrtová D., Freund D., Mokry J., Huttner W. B., and Corbeil D. (2008) The stem cell marker CD133 (Prominin-1) is expressed in various human glandular epithelia. J. Histochem. Cytochem. 56, 977–993 10.1369/jhc.2008.951897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Freund D., Bauer N., Boxberger S., Feldmann S., Streller U., Ehninger G., Werner C., Bornhäuser M., Oswald J., and Corbeil D. (2006) Polarization of human hematopoietic progenitors during contact with multipotent mesenchymal stromal cells: effects on proliferation and clonogenicity. Stem Cells Dev. 15, 815–829 10.1089/scd.2006.15.815 [DOI] [PubMed] [Google Scholar]
- 24. Corbeil D., Röper K., Fargeas C. A., Joester A., and Huttner W. B. (2001) Prominin: a story of cholesterol, plasma membrane protrusions and human pathology. Traffic 2, 82–91 10.1034/j.1600-0854.2001.020202.x [DOI] [PubMed] [Google Scholar]
- 25. Marzesco A. M., Janich P., Wilsch-Bräuninger M., Dubreuil V., Langenfeld K., Corbeil D., and Huttner W. B. (2005) Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J. Cell Sci. 118, 2849–2858 10.1242/jcs.02439 [DOI] [PubMed] [Google Scholar]
- 26. Röper K., Corbeil D., and Huttner W. B. (2000) Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat. Cell Biol. 2, 582–592 10.1038/35023524 [DOI] [PubMed] [Google Scholar]
- 27. Marzesco A. M., Wilsch-Bräuninger M., Dubreuil V., Janich P., Langenfeld K., Thiele C., Huttner W. B., and Corbeil D. (2009) Release of extracellular membrane vesicles from microvilli of epithelial cells is enhanced by depleting membrane cholesterol. FEBS Lett. 583, 897–902 10.1016/j.febslet.2009.01.048 [DOI] [PubMed] [Google Scholar]
- 28. Thamm K., Šimaitė D., Karbanová J., Bermúdez V., Reichert D., Morgenstern A., Bornhäuser M., Huttner W. B., Wilsch-Bräuninger M., and Corbeil D. (2019) Prominin-1 (CD133) modulates the architecture and dynamics of microvilli. Traffic 20, 39–60 10.1111/tra.12618 [DOI] [PubMed] [Google Scholar]
- 29. Karbanová J., Lorico A., Bornhäuser M., Corbeil D., and Fargeas C. A. (2018) Prominin-1/CD133: lipid raft association, detergent resistance, and immunodetection. Stem Cells Transl. Med. 7, 155–160 10.1002/sctm.17-0223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janich P., and Corbeil D. (2007) GM1 and GM3 gangliosides highlight distinct lipid microdomains within the apical domain of epithelial cells. FEBS Lett. 581, 1783–1787 10.1016/j.febslet.2007.03.065 [DOI] [PubMed] [Google Scholar]
- 31. Taïeb N., Maresca M., Guo X. J., Garmy N., Fantini J., and Yahi N. (2009) The first extracellular domain of the tumour stem cell marker CD133 contains an antigenic ganglioside-binding motif. Cancer Lett. 278, 164–173 10.1016/j.canlet.2009.01.013 [DOI] [PubMed] [Google Scholar]
- 32. Liu C., Li Y., Xing Y., Cao B., Yang F., Yang T., Ai Z., Wei Y., and Jiang J. (2016) The interaction between cancer stem cell marker CD133 and Src protein promotes focal adhesion kinase (FAK) phosphorylation and cell migration. J. Biol. Chem. 291, 15540–15550 10.1074/jbc.M115.712976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singer D., Thamm K., Zhuang H., Karbanová J., Gao Y., Walker J. V., Jin H., Wu X., Coveney C. R., Marangoni P., Lu D., Grayson P. R. C., Gulsen T., Liu K. J., Ardu S., et al. (2019) Prominin-1 controls stem cell activation by orchestrating ciliary dynamics. EMBO J. 38, e99845 10.15252/embj.201899845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mak A. B., Nixon A. M., Kittanakom S., Stewart J. M., Chen G. I., Curak J., Gingras A. C., Mazitschek R., Neel B. G., Stagljar I., and Moffat J. (2012) Regulation of CD133 by HDAC6 promotes β-catenin signaling to suppress cancer cell differentiation. Cell Rep. 2, 951–963 10.1016/j.celrep.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boivin D., Labbé D., Fontaine N., Lamy S., Beaulieu E., Gingras D., and Béliveau R. (2009) The stem cell marker CD133 (prominin-1) is phosphorylated on cytoplasmic tyrosine-828 and tyrosine-852 by Src and Fyn tyrosine kinases. Biochemistry 48, 3998–4007 10.1021/bi900159d [DOI] [PubMed] [Google Scholar]
- 36. Wei Y., Jiang Y., Zou F., Liu Y., Wang S., Xu N., Xu W., Cui C., Xing Y., Liu Y., Cao B., Liu C., Wu G., Ao H., Zhang X., and Jiang J. (2013) Activation of PI3K/Akt pathway by CD133-p85 interaction promotes tumorigenic capacity of glioma stem cells. Proc. Natl. Acad. Sci. U.S.A. 110, 6829–6834 10.1073/pnas.1217002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Essner J. J., Amack J. D., Nyholm M. K., Harris E. B., and Yost H. J. (2005) Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development 132, 1247–1260 10.1242/dev.01663 [DOI] [PubMed] [Google Scholar]
- 38. Amack J. D. (2014) Salient features of the ciliated organ of asymmetry. Bioarchitecture 4, 6–15 10.4161/bioa.28014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thamm K., Graupner S., Werner C., Huttner W. B., and Corbeil D. (2016) Monoclonal antibodies 13A4 and AC133 do not recognize the canine ortholog of mouse and human stem cell antigen prominin-1 (CD133). PLoS ONE 11, e0164079 10.1371/journal.pone.0164079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu Y., Flint A., Dvorin E. L., and Bischoff J. (2002) AC133–2, a novel isoform of human AC133 stem cell antigen. J. Biol. Chem. 277, 20711–20716 10.1074/jbc.M202349200 [DOI] [PubMed] [Google Scholar]
- 41. Fargeas C. A., Huttner W. B., and Corbeil D. (2007) Nomenclature of prominin-1 (CD133) splice variants—an update. Tissue Antigens 69, 602–606 10.1111/j.1399-0039.2007.00825.x [DOI] [PubMed] [Google Scholar]
- 42. Larkins C. E., Aviles G. D., East M. P., Kahn R. A., and Caspary T. (2011) Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol. Biol. Cell 22, 4694–4703 10.1091/mbc.e10-12-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walker T. L., Wierick A., Sykes A. M., Waldau B., Corbeil D., Carmeliet P., and Kempermann G. (2013) Prominin-1 allows prospective isolation of neural stem cells from the adult murine hippocampus. J. Neurosci. 33, 3010–3024 10.1523/JNEUROSCI.3363-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Duldulao N. A., Lee S., and Sun Z. (2009) Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development 136, 4033–4042 10.1242/dev.036350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hirokawa N., Tanaka Y., Okada Y., and Takeda S. (2006) Nodal flow and the generation of left-right asymmetry. Cell 125, 33–45 10.1016/j.cell.2006.03.002 [DOI] [PubMed] [Google Scholar]
- 46. Bisgrove B. W., and Yost H. J. (2006) The roles of cilia in developmental disorders and disease. Development 133, 4131–4143 10.1242/dev.02595 [DOI] [PubMed] [Google Scholar]
- 47. Essner J. J., Vogan K. J., Wagner M. K., Tabin C. J., Yost H. J., and Brueckner M. (2002) Conserved function for embryonic nodal cilia. Nature 418, 37–38 10.1038/418037a [DOI] [PubMed] [Google Scholar]
- 48. Jászai J., Farkas L. M., Fargeas C. A., Janich P., Haase M., Huttner W. B., and Corbeil D. (2010) Prominin-2 is a novel marker of distal tubules and collecting ducts of the human and murine kidney. Histochem. Cell Biol. 133, 527–539 10.1007/s00418-010-0690-1 [DOI] [PubMed] [Google Scholar]
- 49. Hsiao Y. C., Tuz K., and Ferland R. J. (2012) Trafficking in and to the primary cilium. Cilia 1, 4 10.1186/2046-2530-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Malicki J., and Avidor-Reiss T. (2014) From the cytoplasm into the cilium: bon voyage. Organogenesis 10, 138–157 10.4161/org.29055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y., and Reiter J. F. (2005) Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–1021 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- 52. Fargeas C. A., Fonseca A.-V., Huttner W. B., and Corbeil D. (2006) Prominin-1 (CD133): from progenitor cells to human diseases. Future Lipidol. 1, 213–225 10.2217/17460875.1.2.213 [DOI] [Google Scholar]
- 53. Sun Z., Amsterdam A., Pazour G. J., Cole D. G., Miller M. S., and Hopkins N. (2004) A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131, 4085–4093 10.1242/dev.01240 [DOI] [PubMed] [Google Scholar]
- 54. Lu H., Toh M. T., Narasimhan V., Thamilselvam S. K., Choksi S. P., and Roy S. (2015) A function for the Joubert syndrome protein Arl13b in ciliary membrane extension and ciliary length regulation. Dev. Biol. 397, 225–236 10.1016/j.ydbio.2014.11.009 [DOI] [PubMed] [Google Scholar]
- 55. Caspary T., Larkins C. E., and Anderson K. V. (2007) The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell 12, 767–778 10.1016/j.devcel.2007.03.004 [DOI] [PubMed] [Google Scholar]
- 56. Pintado P., Sampaio P., Tavares B., Montenegro-Johnson T. D., Smith D. J., and Lopes S. S. (2017) Dynamics of cilia length in left-right development. R. Soc. Open Sci. 4, 161102 10.1098/rsos.161102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roy K., Jerman S., Jozsef L., McNamara T., Onyekaba G., Sun Z., and Marin E. P. (2017) Palmitoylation of the ciliary GTPase ARL13b is necessary for its stability and its role in cilia formation. J. Biol. Chem. 292, 17703–17717 10.1074/jbc.M117.792937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Badano J. L., Mitsuma N., Beales P. L., and Katsanis N. (2006) The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7, 125–148 10.1146/annurev.genom.7.080505.115610 [DOI] [PubMed] [Google Scholar]
- 59. Hanke-Gogokhia C., Wu Z., Sharif A., Yazigi H., Frederick J. M., and Baehr W. (2017) The guanine nucleotide exchange factor Arf-like protein 13b is essential for assembly of the mouse photoreceptor transition zone and outer segment. J. Biol. Chem. 292, 21442–21456 10.1074/jbc.RA117.000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dilan T. L., Moye A. R., Salido E. M., Saravanan T., Kolandaivelu S., Goldberg A. F. X., and Ramamurthy V. (2019) ARL13B, a Joubert syndrome–associated protein, is critical for retinogenesis and elaboration of mouse photoreceptor outer segments. J. Neurosci. 39, 1347–1364 10.1523/JNEUROSCI.1761-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Drummond I. A., Majumdar A., Hentschel H., Elger M., Solnica-Krezel L., Schier A. F., Neuhauss S. C., Stemple D. L., Zwartkruis F., Rangini Z., Driever W., and Fishman M. C. (1998) Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development 125, 4655–4667 [DOI] [PubMed] [Google Scholar]
- 62. Sullivan-Brown J., Schottenfeld J., Okabe N., Hostetter C. L., Serluca F. C., Thiberge S. Y., and Burdine R. D. (2008) Zebrafish mutations affecting cilia motility share similar cystic phenotypes and suggest a mechanism of cyst formation that differs from pkd2 morphants. Dev. Biol. 314, 261–275 10.1016/j.ydbio.2007.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. May-Simera H. L., and Kelley M. W. (2012) Cilia, Wnt signaling, and the cytoskeleton. Cilia 1, 7 10.1186/2046-2530-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Copeland J. (2019) Actin-based regulation of ciliogenesis: the long and the short of it. Semin. Cell Dev. Biol. 10.1016/j.semcdb.2019.12.005 [DOI] [PubMed] [Google Scholar]
- 65. Marzesco A. M. (2013) Prominin-1-containing membrane vesicles: origins, formation, and utility. Adv. Exp. Med. Biol. 777, 41–54 10.1007/978-1-4614-5894-4_3 [DOI] [PubMed] [Google Scholar]
- 66. Yeyati P. L., Schiller R., Mali G., Kasioulis I., Kawamura A., Adams I. R., Playfoot C., Gilbert N., van Heyningen V., Wills J., von Kriegsheim A., Finch A., Sakai J., Schofield C. J., Jackson I. J., and Mill P. (2017) KDM3A coordinates actin dynamics with intraflagellar transport to regulate cilia stability. J. Cell Biol. 216, 999–1013 10.1083/jcb.201607032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nager A. R., Goldstein J. S., Herranz-Pérez V., Portran D., Ye F., Garcia-Verdugo J. M., and Nachury M. V. (2017) An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell 168, 252–263.e14 10.1016/j.cell.2016.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Drummond M. L., Li M., Tarapore E., Nguyen T. T. L., Barouni B. J., Cruz S., Tan K. C., Oro A. E., and Atwood S. X. (2018) Actin polymerization controls cilia-mediated signaling. J. Cell Biol. 217, 3255–3266 10.1083/jcb.201703196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Spencer W. J., Lewis T. R., Phan S., Cady M. A., Serebrovskaya E. O., Schneider N. F., Kim K. Y., Cameron L. A., Skiba N. P., Ellisman M. H., and Arshavsky V. Y. (2019) Photoreceptor disc membranes are formed through an Arp2/3-dependent lamellipodium-like mechanism. Proc. Natl. Acad. Sci. U.S.A. 116, 27043–27052 10.1073/pnas.1913518117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sheetz M. P., Sable J. E., and Döbereiner H. G. (2006) Continuous membrane-cytoskeleton adhesion requires continuous accommodation to lipid and cytoskeleton dynamics. Annu. Rev. Biophys. Biomol. Struct. 35, 417–434 10.1146/annurev.biophys.35.040405.102017 [DOI] [PubMed] [Google Scholar]
- 71. Bae Y. K., Kim E., L'Hernault S. W., and Barr M. M. (2009) The CIL-1 PI 5-phosphatase localizes TRP polycystins to cilia and activates sperm in C. elegans. Curr. Biol. 19, 1599–1607 10.1016/j.cub.2009.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jensen V. L., Li C., Bowie R. V., Clarke L., Mohan S., Blacque O. E., and Leroux M. R. (2015) Formation of the transition zone by Mks5/Rpgrip1L establishes a ciliary zone of exclusion (CIZE) that compartmentalises ciliary signalling proteins and controls PIP2 ciliary abundance. EMBO J. 34, 2537–2556 10.15252/embj.201488044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Garcia-Gonzalo F. R., Phua S. C., Roberson E. C., Garcia G. 3rd, Abedin M., Schurmans S., Inoue T., and Reiter J. F. (2015) Phosphoinositides regulate ciliary protein trafficking to modulate Hedgehog signaling. Dev. Cell 34, 400–409 10.1016/j.devcel.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee S., Tan H. Y., Geneva I. I., Kruglov A., and Calvert P. D. (2018) Actin filaments partition primary cilia membranes into distinct fluid corrals. J. Cell Biol. 217, 2831–2849 10.1083/jcb.201711104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cevik S., Hori Y., Kaplan O. I., Kida K., Toivenon T., Foley-Fisher C., Cottell D., Katada T., Kontani K., and Blacque O. E. (2010) Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J. Cell Biol. 188, 953–969 10.1083/jcb.200908133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li Y., Wei Q., Zhang Y., Ling K., and Hu J. (2010) The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J. Cell Biol. 189, 1039–1051 10.1083/jcb.200912001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ivanova A. A., Caspary T., Seyfried N. T., Duong D. M., West A. B., Liu Z., and Kahn R. A. (2017) Biochemical characterization of purified mammalian ARL13B protein indicates that it is an atypical GTPase and ARL3 guanine nucleotide exchange factor (GEF). J. Biol. Chem. 292, 11091–11108 10.1074/jbc.M117.784025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nozaki S., Katoh Y., Terada M., Michisaka S., Funabashi T., Takahashi S., Kontani K., and Nakayama K. (2017) Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J. Cell Sci. 130, 563–576 10.1242/jcs.197004 [DOI] [PubMed] [Google Scholar]
- 79. Copeland S. J., McRae A., Guarguaglini G., Trinkle-Mulcahy L., and Copeland J. W. (2018) Actin-dependent regulation of cilia length by the inverted formin FHDC1. Mol. Biol. Cell 29, 1611–1627 10.1091/mbc.E18-02-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zelhof A. C., Hardy R. W., Becker A., and Zuker C. S. (2006) Transforming the architecture of compound eyes. Nature 443, 696–699 10.1038/nature05128 [DOI] [PubMed] [Google Scholar]
- 81. Bachor T. P., Karbanová J., Büttner E., Bermúdez V., Marquioni-Ramella M., Carmeliet P., Corbeil D., and Suburo A. M. (2017) Early ciliary and prominin-1 dysfunctions precede neurogenesis impairment in a mouse model of type 2 diabetes. Neurobiol. Dis. 108, 13–28 10.1016/j.nbd.2017.07.010 [DOI] [PubMed] [Google Scholar]
- 82. Bhattacharya S., Yin J., Winborn C. S., Zhang Q., Yue J., and Chaum E. (2017) Prominin-1 is a novel regulator of autophagy in the human retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 58, 2366–2387 10.1167/iovs.16-21162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Izumi H., Li Y., Shibaki M., Mori D., Yasunami M., Sato S., Matsunaga H., Mae T., Kodama K., Kamijo T., Kaneko Y., and Nakagawara A. (2019) Recycling endosomal CD133 functions as an inhibitor of autophagy at the pericentrosomal region. Sci. Rep. 9, 2236 10.1038/s41598-019-39229-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Orhon I., Dupont N., and Codogno P. (2016) Primary cilium and autophagy: the avengers of cell-size regulation. Autophagy 12, 2258–2259 10.1080/15548627.2016.1212790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Morleo M., and Franco B. (2019) The autophagy-cilia axis: an intricate relationship. Cells 8, E905 10.3390/cells8080905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mizushima N., and Levine B. (2010) Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12, 823–830 10.1038/ncb0910-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kosodo Y., Röper K., Haubensak W., Marzesco A. M., Corbeil D., and Huttner W. B. (2004) Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 23, 2314–2324 10.1038/sj.emboj.7600223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bauer N., Wilsch-Bräuninger M., Karbanová J., Fonseca A. V., Strauss D., Freund D., Thiele C., Huttner W. B., Bornhäuser M., and Corbeil D. (2011) Haematopoietic stem cell differentiation promotes the release of prominin-1/CD133-containing membrane vesicles—a role of the endocytic-exocytic pathway. EMBO Mol. Med. 3, 398–409 10.1002/emmm.201100147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Steiner A. B., Kim T., Cabot V., and Hudspeth A. J. (2014) Dynamic gene expression by putative hair-cell progenitors during regeneration in the zebrafish lateral line. Proc. Natl. Acad. Sci. U.S.A. 111, E1393–E1401 10.1073/pnas.1318692111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Corbeil D., Röper K., Hannah M. J., Hellwig A., and Huttner W. B. (1999) Selective localization of the polytopic membrane protein prominin in microvilli of epithelial cells—a combination of apical sorting and retention in plasma membrane protrusions. J. Cell Sci. 112, 1023–1033 [DOI] [PubMed] [Google Scholar]
- 91. Fargeas C. A. (2013) Prominin-2 and other relatives of CD133. Adv. Exp. Med. Biol. 777, 25–40 10.1007/978-1-4614-5894-4_2 [DOI] [PubMed] [Google Scholar]
- 92. Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., and Schilling T. F. (1995) Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- 93. Tüscher O., Lorra C., Bouma B., Wirtz K. W., and Huttner W. B. (1997) Cooperativity of phosphatidylinositol transfer protein and phospholipase D in secretory vesicle formation from the TGN–phosphoinositides as a common denominator? FEBS Lett. 419, 271–275 10.1016/S0014-5793(97)01471-3 [DOI] [PubMed] [Google Scholar]
- 94. Jones A. T., and Wessling-Resnick M. (1998) Inhibition of in vitro endosomal vesicle fusion activity by aminoglycoside antibiotics. J. Biol. Chem. 273, 25301–25309 10.1074/jbc.273.39.25301 [DOI] [PubMed] [Google Scholar]
- 95. Jászai J., Reifers F., Picker A., Langenberg T., and Brand M. (2003) Isthmus-to-midbrain transformation in the absence of midbrain-hindbrain organizer activity. Development 130, 6611–6623 10.1242/dev.00899 [DOI] [PubMed] [Google Scholar]
- 96. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and supporting materials. The zebrafish prom3 sequences were deposited in the public GenBankTM database under accession numbers DQ341400 and HQ698925.