Abstract
tRNAs universally carry a CCA nucleotide triplet at their 3′-ends. In eukaryotes, the CCA is added post-transcriptionally by the CCA-adding enzyme (CAE). The mitochondrion of the parasitic protozoan Trypanosoma brucei lacks tRNA genes and therefore imports all of its tRNAs from the cytosol. This has generated interest in the tRNA modifications and their distribution in this organism, including how CCA is added to tRNAs. Here, using a BLAST search for genes encoding putative CAE proteins in T. brucei, we identified a single ORF, Tb927.9.8780, as a potential candidate. Knockdown of this putative protein, termed TbCAE, resulted in the accumulation of truncated tRNAs, abolished translation, and inhibited both total and mitochondrial CCA-adding activities, indicating that TbCAE is located both in the cytosol and mitochondrion. However, mitochondrially localized tRNAs were much less affected by the TbCAE ablation than the other tRNAs. Complementation assays revealed that the N-terminal 10 amino acids of TbCAE are dispensable for its activity and mitochondrial localization and that deletion of 10 further amino acids abolishes both. A growth arrest caused by the TbCAE knockdown was rescued by the expression of the cytosolic isoform of yeast CAE, even though it was not imported into mitochondria. This finding indicated that the yeast enzyme complements the essential function of TbCAE by adding CCA to the primary tRNA transcripts. Of note, ablation of the mitochondrial TbCAE activity, which likely has a repair function, only marginally affected growth.
Keywords: Trypanosoma brucei, transfer RNA (tRNA), mitochondria, RNA processing, RNA interference (RNAi), mitochondrial tRNA import
Introduction
Unlike in most other eukaryotes the mitochondrion of the parasitic protozoan Trypanosoma brucei and its relatives does not encode any tRNAs (1, 2). However, organellar tRNAs are required for mitochondrial translation, which is essential in both the insect-stage and the bloodstream form of the parasite (3, 4). Thus, all mitochondrial tRNAs must be imported from the cytosol. Mitochondrial tRNA import is not restricted to trypanosomes but has been found in many organisms including some fungi, plants, and protozoans (2, 5–7). But in essentially all other systems, at least a subset of tRNAs is still encoded in the mitochondrial genome. tRNA import in trypanosomes has been studied in quite some detail and whereas there are still many unanswered questions regarding the process, some factors mediating tRNA import have recently been characterized (8–10). As in other systems a connection to mitochondrial protein import has emerged, even though in contrast to what has been proposed in yeast, trypanosomal tRNAs are not co-imported with proteins (9). Moreover, tRNAs appear to be imported as fully processed molecules (11, 12). The exclusive reliance on mitochondrial tRNA import has initiated a more general interest in trypanosomal tRNA biology (1). Aminoacyl-tRNA synthetases (aaRSs)2 have been studied extensively and it was shown that, likely due to mitochondrial tRNA import, most but interestingly not all aaRSs are dually targeted to the cytosol and the mitochondrion (3, 13–15). Furthermore, a mitochondrion-specific editing of the anticodon of the tRNATrp has been shown, which allows it to decode different codons in the cytosol and the mitochondrion, explaining how T. brucei accommodates the re-assignment of the stop codon UGA to Trp in mitochondria (16). This has fostered extensive work on tRNA modifications in general (17–19). Trypanosomes were also instrumental in deciphering the eukaryotic pathway for selenocysteinyl-tRNASec formation (20). Studies on that subject were aided by the fact that selenoproteins are neither essential for procyclic nor bloodstream forms of the parasite when cultured in vitro (21). Furthermore, unlike in other systems and in contrast to all other trypanosomal tRNAs, the tRNASec of T. brucei is transcribed by RNA polymerase II as it is embedded in a polycistron (22).
Eukaryotic tRNAs are transcribed by RNA polymerase III as precursors carrying 5′- and 3′-extensions and may carry introns. Correct processing of the precursor is essential for aminoacylation and thus for tRNA function (23, 24). 5′-Processing of tRNAs is generally catalyzed by RNase P, which in eukaryotes consists of a variable number of protein factors and of an RNA subunit, which carries the enzymatic activity. In trypanosomes this is different, a recent study has shown that T. brucei has an unusual protein-only RNase P and a similar situation has also been observed in plants (25, 26). Furthermore, a mitochondrial version of a protein-only RNase P enzyme has also been characterized in T. brucei, even though it is not clear what the in vivo substrate of that enzyme would be (26, 27). 3′-Processing of tRNAs involves cleavage of the 3′-extensions by RNases including RNase Z (24, 28). Subsequently CCA is added to the processed 3′-end by the nontemplated CCA-adding enzyme (CAE) creating the universal 3′ CCA end that is found on all tRNAs (whereas in many bacteria the CCA is already encoded in the tRNA gene this not the case in eukaryotes) (29). It is the 3′-terminal A that gets aminoacylated by the aaRSs. 3-Processing of tRNAs is one of the few aspects of tRNA biology that has so far not been studied in trypanosomes. This was the reason why we decided to characterize the trypanosomal CAE. In the present study, we show that the enzyme is essential for normal growth, because its ablation abolishes translation. We, furthermore, show that the protein is dually localized in mitochondria and the rest of the cell.
Results
TbCAE is essential for cytosolic translation
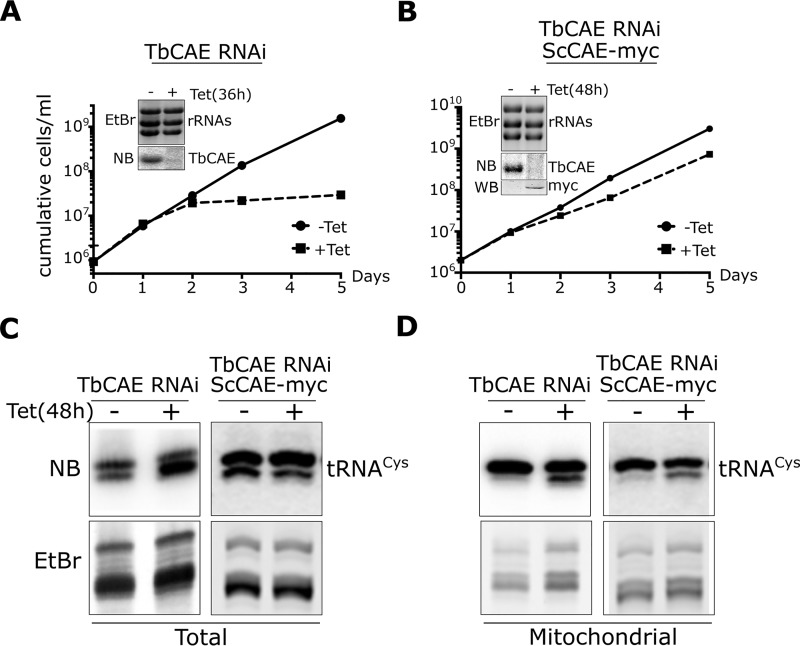
Doing BLAST searches using the sequences of the yeast or human CAEs as templates we identified a single ORF, Tb927.9.8780, that appears to encode the trypanosomal CAE, which we termed TbCAE. To functionally study the enzyme, we produced an inducible RNAi cell line in procyclic T. brucei that targets the ORF of the mRNA. Fig. 1A shows that addition of tetracycline results in the efficient ablation the TbCAE mRNA. Moreover, growth slows down after 1 day and stops after 2 days RNAi induction, which is typical for an essential gene.
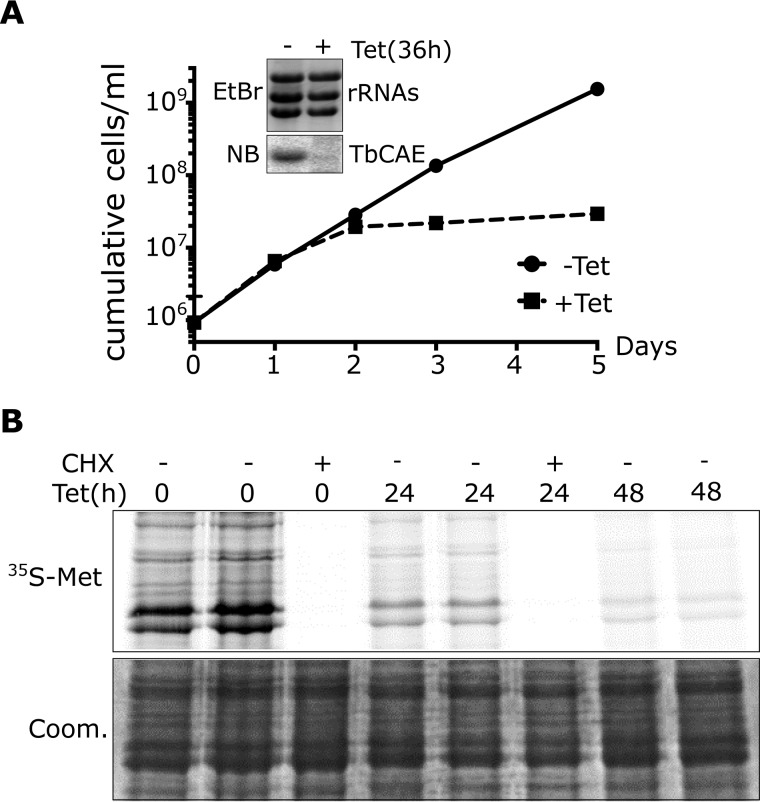
Figure 1.
TbCAE is essential for cytosolic translation. A, growth curve of uninduced (−tet) and induced (+tet) TbCAE RNAi cell line. The experiment has been performed in triplicate; the very small standard deviations are indicated. Insets show Northern blots (NB) of total RNA from uninduced or induced cells, which were probed for TbCAE mRNA. Ethidium bromide-stained rRNAs serve as loading controls. B, [35S]methionine labeling of the TbCAE RNAi cell line after 0, 24, and 48 h of RNAi induction. Duplicate samples are shown. The corresponding Coomassie (Coom)-stained gel serves as loading control. Cycloheximide (CHX) (300 μg/ml) is added to the negative control sample.
Ablation of the CAE should ultimately abolish translation as it would lead to accumulation of nonchargeable tRNAs with immature 3′-ends. To test this, we labeled cell culture with [35S]methionine, which gets incorporated into proteins during translation (Fig. 2B, left two lanes). When the experiment was done with the induced TbCAE-RNAi cell line we observed a great reduction of [35S]methionine incorporation after 24 h of induction when compared with uninduced cells. Moreover, after 48 h translation was essentially abolished. Virtually the same was seen if cells were treated with the translation inhibitor cycloheximide. Thus, ablation of TbCAE, as might be expected, rapidly abolishes translation.
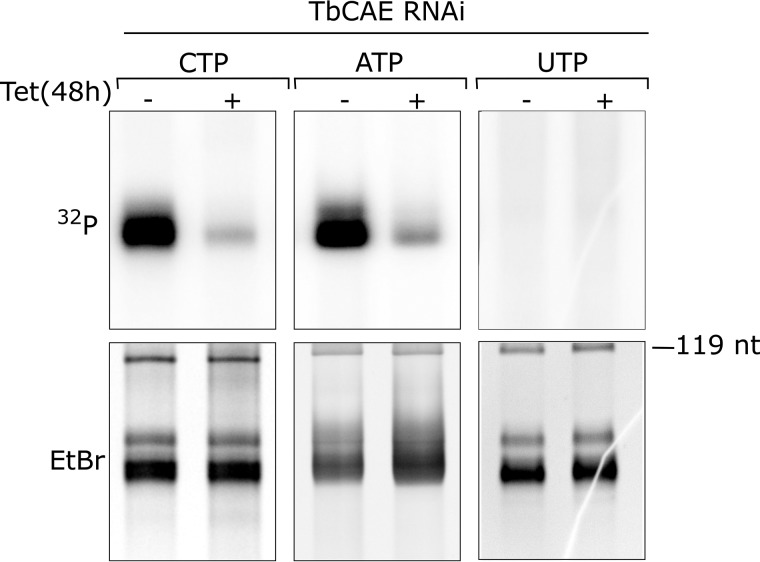
Figure 2.
Ablation of TbCAE inhibits its activity. Incorporation of [32P]CTP, [32P]ATP, and [32P]UTP into tRNAs was assayed in whole cell extracts prepared from the uninduced (−tet) and induced (+tet) TbCAE RNAi cell line. Only the tRNA regions of the gel are shown. The EtBr-stained gel serves as a loading control. nt, nucleotide.
Ablation of TbCAE inhibits its activity
To directly measure TbCAE activity we prepared total cellular extracts of the uninduced and 2-day induced TbCAE-RNAi cell line. The extracts were incubated in the presence of radioactive [α-32P]CTP and isolated total RNA (30). Subsequently, total RNA was re-isolated and separated by 8 m urea, 15% PAGE. The gel was stained by ethidium bromide, dried, and exposed on a PhosphorImager. The result in the upper left panel of Fig. 2 shows a highly efficient labeling of the tRNA region that was greatly reduced after ablation of TbCAE. The same result was obtained when [α-32P]ATP but not when [α-32P]UTP was added to the reaction. We concluded from these results that C and/or A were added to the small fraction of the newly synthesized and 3′-cleaved tRNAs present in the assay mixture. Thus, our results strongly suggest that TbCAE adds both C and A to the 3′-ends of newly transcribed and processed trypanosomal tRNAs. The absence of labeling in the presence [α-32P]UTP indicates that it is not caused by transcription.
Ablation of TbCAE differentially affects different tRNAs
To investigate how ablation of TbCAE affects different tRNA isoacceptor groups in vivo we isolated total RNA from uninduced and 2-day–induced cells, separated it by 8 m urea, 15% PAGE, and stained the gel by ethidium bromide (Fig. 3, lower panels). The tRNA regions of total RNA isolated from uninduced and induced cells look very similar. To look at individual tRNAs we analyzed the steady-state levels of all tRNA isoacceptor groups by Northern blotting using specific oligonucleotide probes (Fig. 3, upper panels). The results allow the separation of the tRNAs into four groups regarding how they react to the ablation of TbCAE.
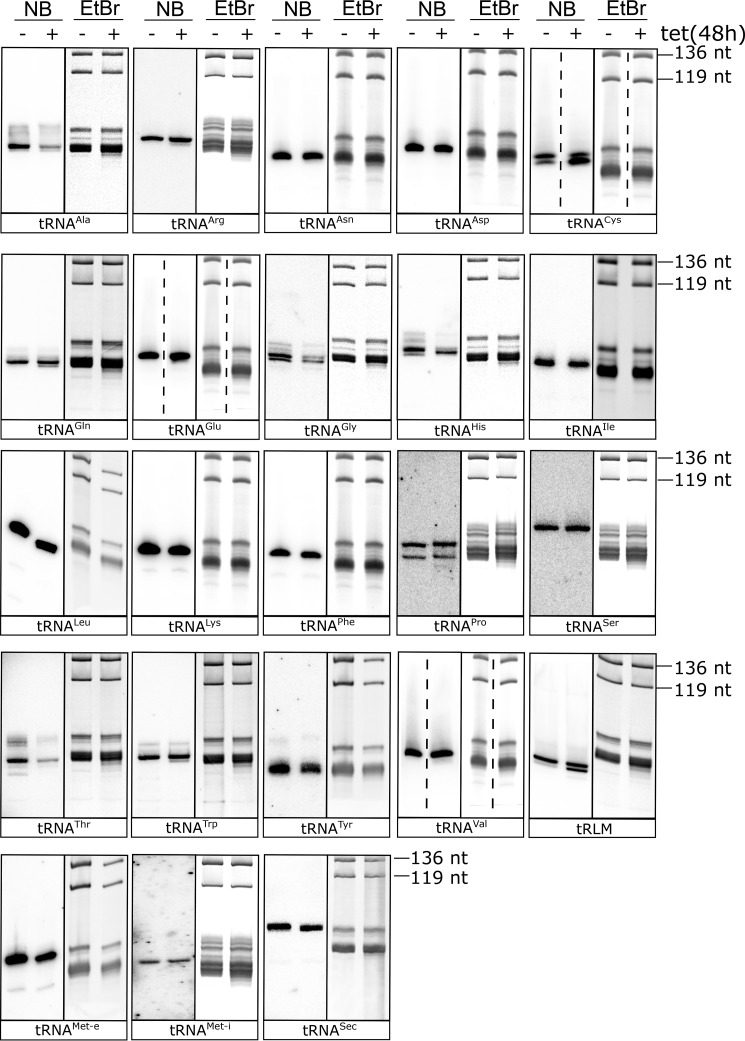
Figure 3.
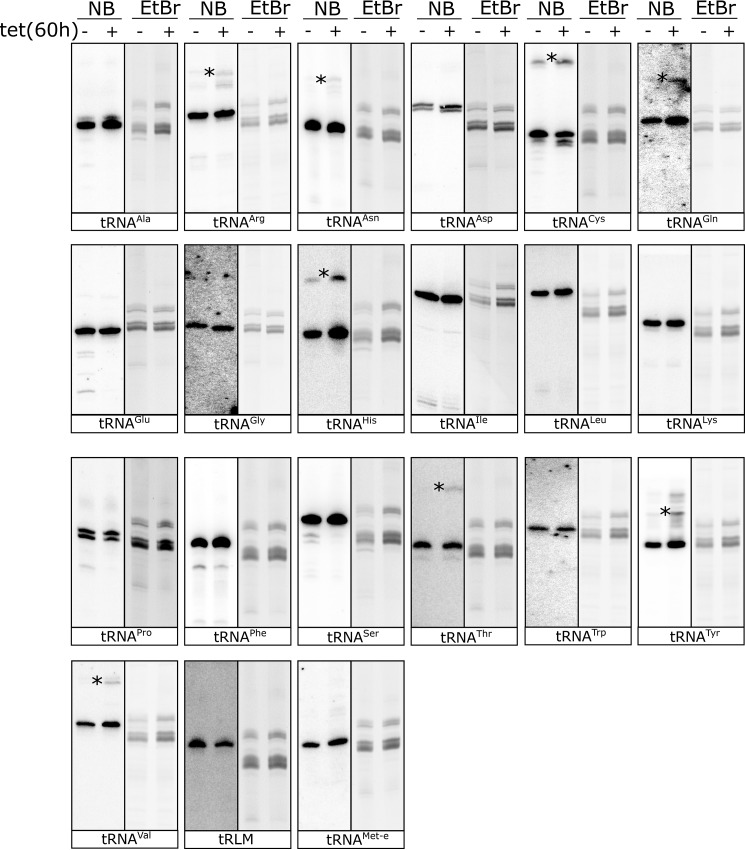
Ablation of TbCAE differentially affects different tRNAs. Northern blots (NB) of total cellular RNA isolated from the uninduced (−) and induced (+) TbCAE RNAi cell line probed by specific oligonucleotide hybridization, for all tRNA isoacceptor groups and the tRNA-like molecule (tRLM). Only the tRNA regions of the blots and EtBr-stained gels are shown. One blot has been reprobed. Thus, the loading controls (EtBr-stained gels) for tRNACys and tRNAGlu are identical. For oligonucleotides used for hybridization, see Table S1.
The strongest phenotypes were seen for tRNACys, tRNAHis, and the tRNA-like molecule that is part of the trypanosomal signal recognition particle (31). In all three cases we see efficient accumulation of a lower molecular weight band, suggesting that tRNAs lacking a part or the whole CCA-tail accumulate. In the case of tRNACys and tRNAHis the truncated molecules are the main species observed. For tRNAArg, tRNAGln, and tRNATrp, we see the same as above except that only very small amounts of truncated tRNAs are found.
For tRNAAla, tRNAGly, and tRNAThr, ablation of TbCAE causes a significant reduction in the steady-state levels when compared with the total tRNAs seen in the ethidium bromide-stained gel, which serves as a loading control. However, no specific accumulation of lower molecular weight forms is seen. Finally, and perhaps most surprisingly we find that 14 tRNA acceptor groups are not affected by the knockdown of the TbCAE.
For some tRNAs (Fig. 3, tRNAPro) we see additional low molecular weight bands. These might be other tRNA isoacceptors that cross-react with the oligonucleotide that was used as a probe. Alternatively they may represent hypo-modified tRNAs in the cases were the modification as added after the CCA addition.
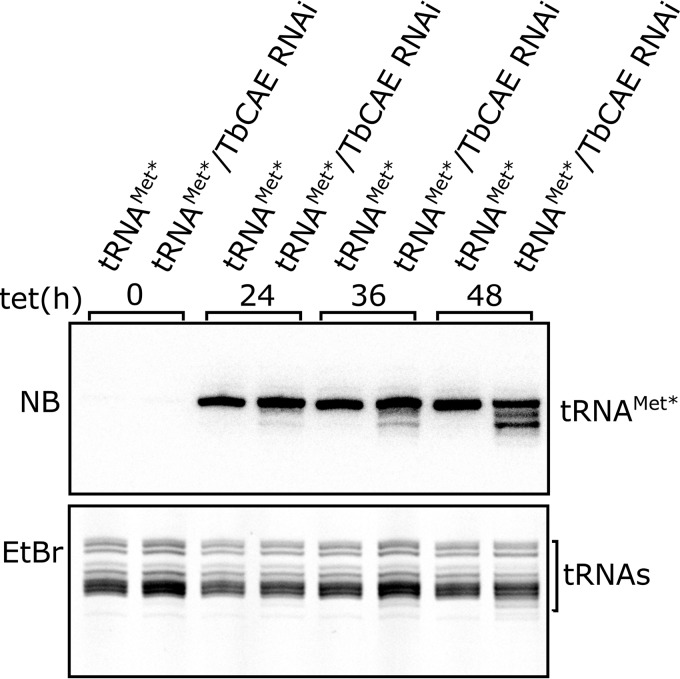
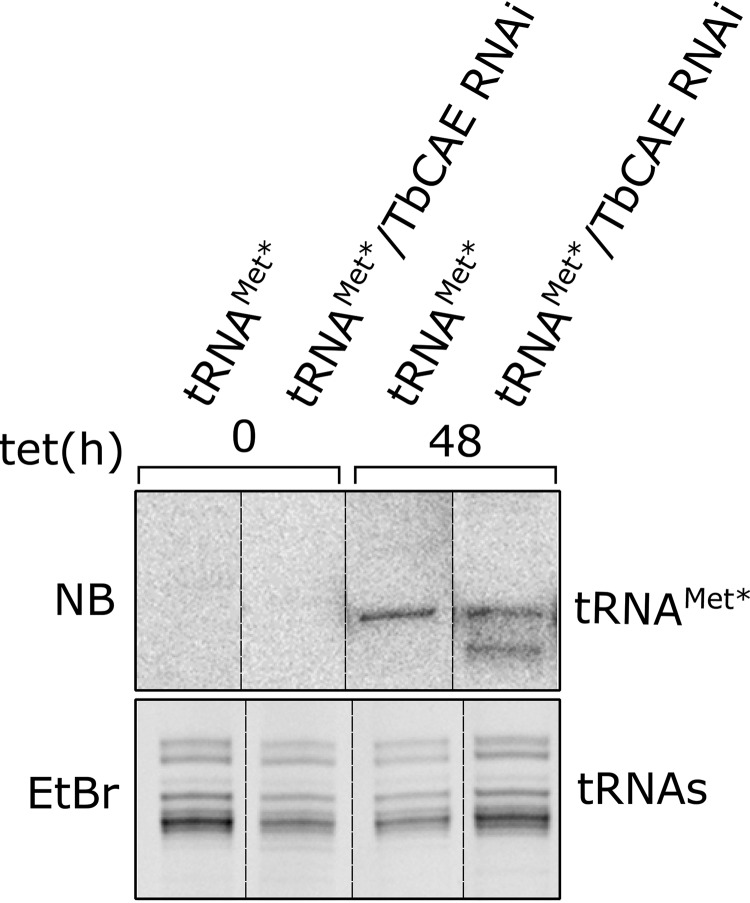
tRNA are known to be quite stable molecules. Thus, a caveat of our analysis is that the tRNA population, which was synthesized before the RNAi takes effect, might not be affected. To increase the sensitivity of the analysis we used a cell line in which upon addition of tetracycline not only TbCAE-RNAi is induced but also expression of a mutationally tagged tRNAMet occurs (11). (This is possible because its gene contains a 5′-flanking tetracycline-operator sequence.) With this system, we can selectively monitor the fate of the newly synthesized variant of tRNAMet (tRNAMet*) during induction of TbCAE RNAi. Interestingly, a significant accumulation of truncated tRNAMet* species was seen in such an experiment (Fig. 4), even though no phenotype was observed for either of the two endogenous tRNAsMet (tRNAMet-i and tRNAMet-e) when their steady-state levels were analyzed (Fig. 3).
Figure 4.
TbCAE RNAi causes accumulation of newly synthesized truncated tRNAMet*. Total RNA isolated from cell lines allowing either tetracycline-inducible expression of variant tRNAMet (tRNAMet*) only, or inducible expression of tRNAMet* in combination with RNAi against TbCAE were analyzed by Northern blots (NB) at the indicated time points. The newly synthesized tRNAMet* was detected by specific oligonucleotide hybridization. Only the tRNA region of the gel is shown. The EtBr-stained gel serves as a loading control.
In summary, these results show that ablation of TbCAE leads to accumulation of truncated tRNAs as would be expected. However, surprisingly there were large differences in how much truncated tRNAs accumulated, from 72.02% for tRNACys (S.D. ± 2.92%) to a very low percentage for tRNAMet, which could only be detected when newly synthesized tRNA was analyzed. RNAi inherently does not completely remove its target mRNA, indicating that a small amount of TbCAE is still present in induced cells. Assuming a similar half-life for all tRNA isoacceptor groups, the differential effects we see might be explained by different affinities of TbCAE to different tRNA isoacceptor groups. Thus, the tRNAs that are only marginally affected may have higher affinities for the enzyme and therefore the CCA-tail might still be added to these substrates even when only very little enzyme is present in induced RNAi cells. The simplest way to explain the reduced levels of tRNAAla, tRNAGly, and tRNAThr observed in induced RNAi cells is that the truncated tRNA lacking the CCA-tail becomes rapidly degraded.
A fraction of TbCAE is imported into mitochondria
To determine the intracellular localization of TbCAE, we produced a transgenic cell line expressing a version of TbCAE whose C terminus is tagged with the c-myc epitope. Fig. 5A shows that the TbCAE-myc is expressed. Moreover, when the cells are extracted with low concentration of digitonin the protein is essentially recovered in the soluble supernatant fraction. In the pellet that contains intact mitochondria approximately the same percentage of total tagged TbCAE is detected as is for the cytosolic markers elongation factor 1a (EF1a) and TbVCP. In line with this, TbCAE-myc is localized all over the cell when analyzed by immunofluorescence (Fig. 5B). In summary these results suggest that the large majority of TbCAE is cytosolically and to a lesser extent nuclearly localized. In other eukaryotic systems a fraction of CAEs is generally imported into mitochondria (32–35). The experiments shown in Fig. 5AB cannot confirm this for T. brucei. Moreover, the N terminus of TbCAE is not predicted to form a mitochondrial targeting sequence. However, not all mitochondrial proteins have typical N-terminal targeting sequences and import of a small fraction of TbCAE might be beyond the detection limit in these experiments. In fact, in a recent study we have experimentally defined the mitochondrial proteome of T. brucei using two criteria (15): to be scored as a component of the mitochondrial proteome a protein needs to be (i) present in a highly enriched gradient-purified mitochondrial fraction and (ii) its levels have to be reduced in mitochondria ablated for ATOM40, the import pore of the main translocase of the outer membrane (36, 37). TbCAE fulfilled both criteria in this study and therefore is a bona fide component of the mitochondrial proteome (Fig. 5C) (15).
Figure 5.
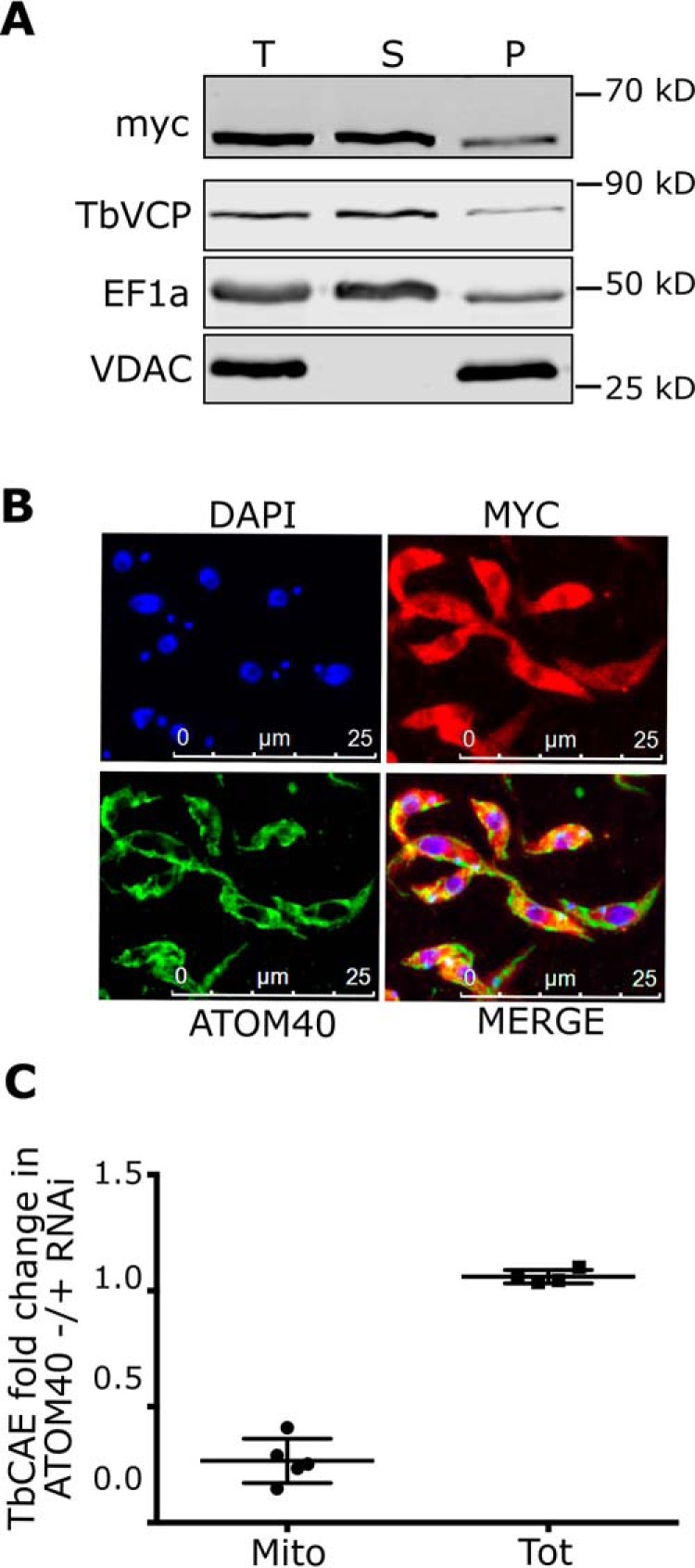
TbCAE is dually localized in cytosol/nucleus and inside mitochondria. A, digitonin-extracted total cells expressing triple c-myc–tagged TbCAE (T) to separate a mitochondria-enriched fraction (P) from the cytosol-containing supernatant (S). EF1a and TbVCP are used as a cytosolic and VDAC as a mitochondrial marker, respectively. B, immunofluorescence analysis of whole cells expressing triple c-myc–tagged TbCAE probed with anti-myc (red) and ATOM40 (green) antibodies. ATOM40 serves as a mitochondrial marker, nuclear and kinetoplast DNA is stained with DAPI (blue). C, fold-change of TbCAE amounts in isolated mitochondria (Mito) and total cells (Tot) from uninduced and induced ATOM40 RNAi cell lines, respectively, depicted as a scatter dot plot. Data are from a previously published SILAC-based proteomic analysis (15).
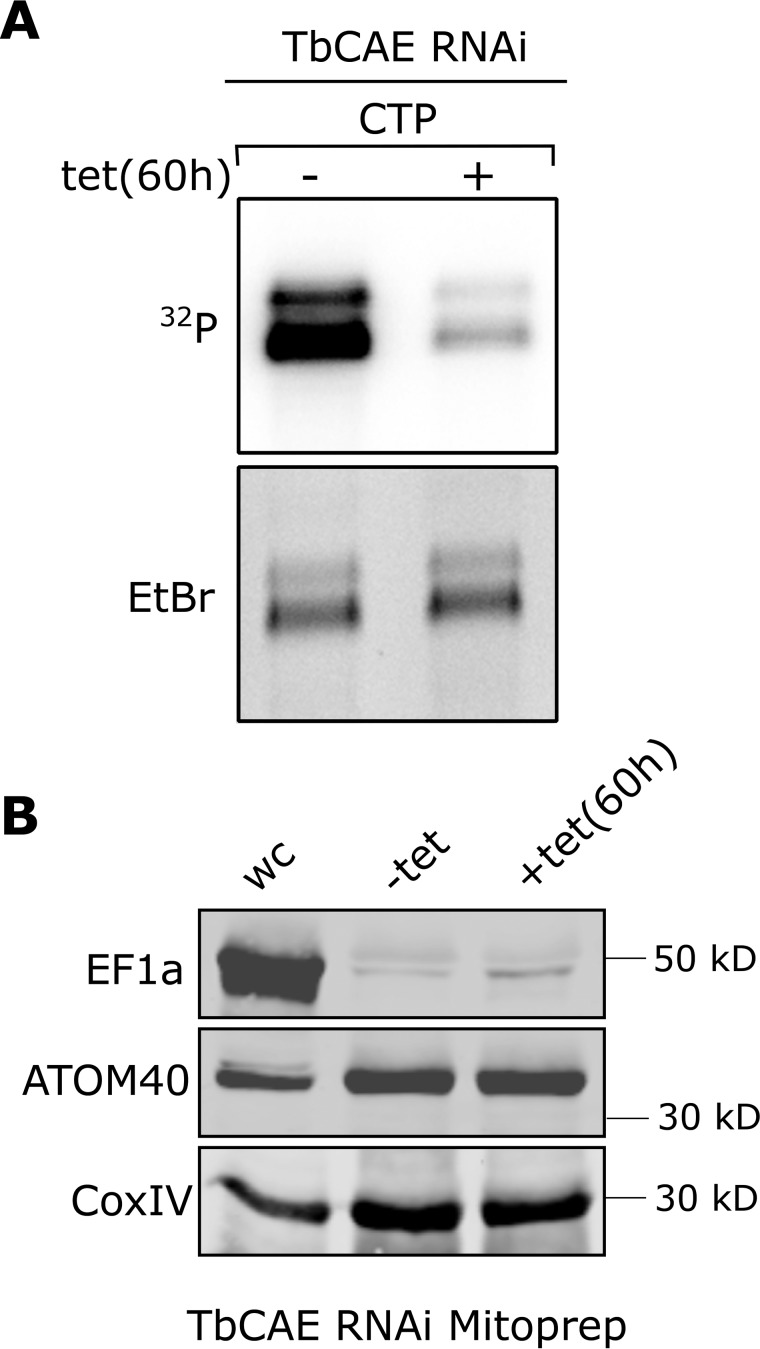
Should a small fraction of TbCAE be mitochondrially localized we would expect that its activity can be detected in mitochondrial extracts using the same assay that was used for total RNA (Fig. 2). Fig. 6 shows that this is the case. Mitochondrial tRNAs are strongly labeled with [α-32P]CTP in an organellar extract, however, after depletion of TbCAE by RNAi the labeled fraction is dramatically reduced. The immunoblots in Fig. 6B confirm that the mitochondrial extracts used in activity assays were not contaminated by cytosolic proteins. These results confirm that TbCAE is indeed a component of the mitochondrial proteome as has recently been determined (15), even though standard biochemical and cell biological methods are not sensitive enough to detect this (Fig. 5, A and B).
Figure 6.
TbCAE RNAi inhibits mitochondrial CAE activity. A, incorporation of [32P]CTP into tRNAs was assayed in mitochondrial extracts prepared from the uninduced (−tet) and induced (+tet) TbCAE RNAi cell line. Only the tRNA region of the gels is shown. The EtBr-stained gel served as a loading control. B, the total cellular extract (WC) and mitochondrial (Mito) extracts used in A were analyzed by immunoblots using a cytosolic (EF1a) and mitochondrial markers (ATOM40 and CoxIV), respectively.
Ablation of TbCAE only moderately affects mitochondrial tRNAs
As was done for total RNA (Fig. 3) we analyzed the effect ablation of the mitochondrial TbCAE activity has on the steady-state level of the different tRNA isoacceptor groups that were imported into mitochondria. To that end organellar RNA was isolated from digitonin-extracted uninduced and induced TbCAE RNAi cells that were treated with RNase to digest the contaminating cytosolic RNAs. Fig. S2 shows that the resulting mitochondrial RNA fractions, in contrast to total cellular RNA, is free of the cytosol-specific initiator tRNAMet and tRNASec.
In contrast to when tRNAs from total cellular extracts (of which less than 5% correspond to mitochondrial tRNA (38)) were analyzed, mitochondrially imported tRNAs do not seem to be affected by ablation of TbCAE (Fig. 7). The one exception is tRNACys for which accumulation of a small fraction of a lower molecular weight form is observed. Interestingly, tRNACys also shows the strongest phenotype when total tRNAs were analyzed (Fig. 3). For tRNAGln and tRNAHis we detected higher steady-state levels after ablation of TbCAE.
Figure 7.
TbCAE RNAi only moderately affects mitochondrial tRNAs. Northern blots (NB) of mitochondrial RNA isolated from the uninduced (−) and induced (+) TbCAE RNAi cell line probed by specific oligonucleotide hybridization for all tRNA isoacceptor groups and the tRNA-like molecule (tRLM). Only the tRNA regions of the blots and the gels are shown. The EtBr-stained gel serves as loading controls. Some blots have been reprobed. In these cases the corresponding loading controls (EtBr-stained gels) are identical. This applies for: tRNAAsp and tRNAGly; tRNAGlu and tRNAGln; tRNACys and tRNAThr; tRNALeu, tRNALys, and tRNATyr; and tRNAVal and tRNATrp. For oligonucleotides used for hybridization see Table S1. *, circular derivatives of tRNAs caused by the action of tRNA ligase present in the mitochondria.
To increase the sensitivity of our assay we again used the cell line in which upon addition of tetracycline both ablation of TbCAE and expression of the mutationally tagged tRNAMet are induced. However, instead of analyzing total tRNA (Fig. 4) we analyzed the mitochondrial tRNAs that were free of cytosolic tRNA (Fig. S3). The results show that ablation of TbCAE causes an ∼50% accumulation of a truncated newly synthesized mitochondrially-localized tRNAMet variant (Fig. 8).
Figure 8.
TbCAE RNAi causes accumulation of newly synthesized truncated tRNAMet variant in mitochondria. Mitochondrial RNAs isolated from cell lines allowing either tetracycline-inducible expression of variant tRNAMet (tRNAMet*) only, or inducible expression of the tRNAMet* in combination with TbCAE RNAi were analyzed by Northern blots (NB) at the indicated time points. The newly synthesized tRNAMet* was detected by specific nucleotide hybridization. Only the tRNA region of the gel is shown. The EtBr-stained gel serves as a loading control.
In summary, these results indicate that ablation of TbCAE affects at least a subset of mitochondrially imported tRNAs. However, the extent to which truncated forms accumulate is much lower than for total tRNA.
Complementation experiments using truncated TbCAE variants
To perform in vivo complementation experiments we used a synthetic gene that encodes a C terminally myc-tagged version of the TbCAE that is resistant to RNAi due to multiple synonymous codon replacements. Expression of this synthetic gene in the induced TbCAE-RNAi cell line resulted in essentially WT growth (Fig. 9A). This confirms that (i) the phenotypes observed in the TbCAE-RNAi cell line described above are not caused by off target effects and that (ii) the C-terminal myc-tag does not interfere with the function of the enzyme. Next, we transfected the TbCAE-RNAi cell line with constructs allowing inducible expression of three N-terminal–truncated variants of TbCAE lacking 10, 20, or 30 N-terminal amino acids, termed ΔN1–10TbCAE-myc, ΔN1–20TbCAE-myc, and ΔN1–30TbCAE-myc, respectively (Fig. 9, B–D). All complemented cell lines were tested for ablation of the endogenous TbCAE mRNA, growth, expression, and localization of the myc-tagged TbCAE variants as well as for TbCAE activity. The results show that in all cell lines the endogenous TbCAE mRNA was down-regulated and that the transgenic TbCAE variants were expressed (Fig. 9, insets, in growth curve panels). Moreover, whereas the cell line expressing ΔN1–10TbCAE-myc grew normally (Fig. 9B), like the cell line complemented with the full-length CAE (Fig. 9A), expression of ΔN1–20TbCAE-myc and ΔN1–30TbCAE-myc variants did not restore growth (Fig. 9CD). Finally, as might be expected, the lack of in vitro CAE activity observed for ΔN1–20TbCAE-myc and ΔN1–30TbCAE-myc variants (Fig. 9, C and D, right panels) correlates with their inability to complement the growth arrest (Fig. 8, middle panels).
Figure 9.
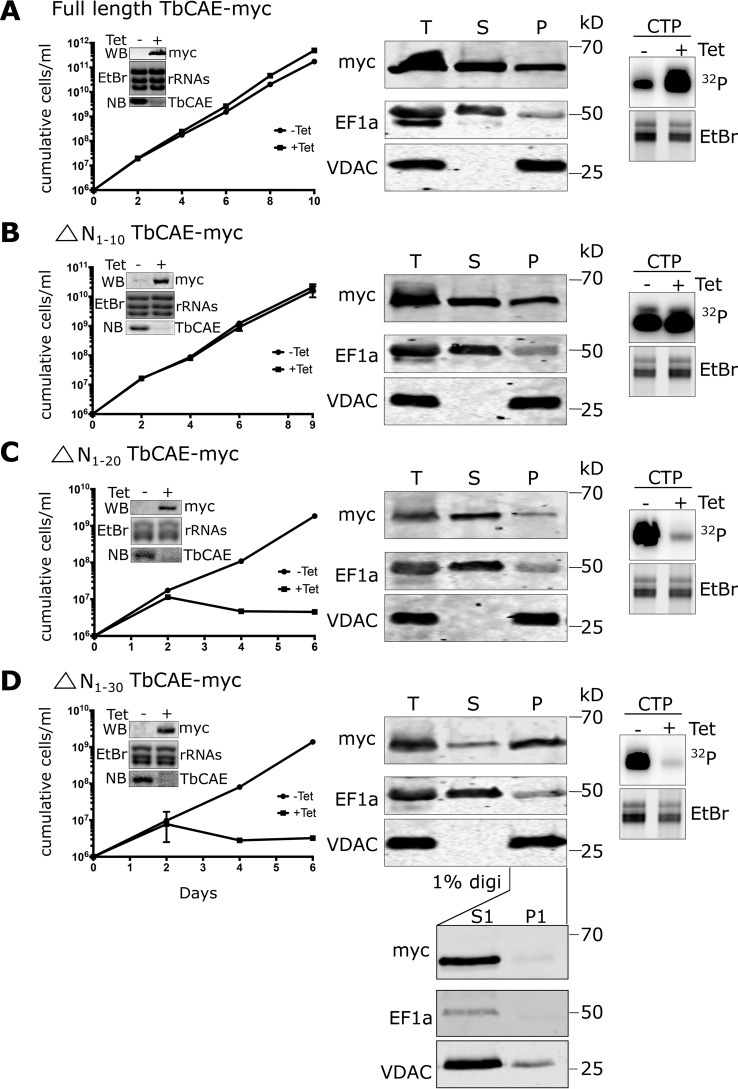
Analysis of function and localization of TbCAE variants by complementation experiments. The TbCAE RNAi cell line was complemented by ectopic expression of C terminally myc-tagged full-lengthTbCAE derived from a synthetic gene (A), or derivatives thereof lacking the N-terminal 10 (B), 20 (C) or 30 amino acids (D). The constructs were termed ΔN1-10TbCAE-myc, ΔN1-20TbCAE-myc, and ΔN1–30TbCAE-myc, respectively. Left panels, growth curves of the indicated uninduced (−tet) and induced (+tet) complemented cell lines. Insets in left panel, immunoblots of the myc-tagged TbCAE variant; loading control showing EtBr-stained tRNAs; Northern blots of total RNA from uninduced (−) and induced (+) cells probed for TbCAE mRNA after 48 h of tetracycline induction. Middle panels, digitonin extractions (0.025%) of the indicated induced complemented cell line. Ectopically expressed TbCAE was detected in total (T), mitochondria-enriched (P), and cytosol-containing supernatant (S) using anti-myc antibody. EF1a is used as a cytosolic and VDAC as a mitochondrial marker, respectively. Right panels, incorporation of [32P]CTP into tRNAs was assayed in total cell extracts prepared from the indicated uninduced (−tet) and induced (+tet) complemented cell lines. Only the tRNA region of the gels is shown. The EtBr-stained gels serves as a loading control. The bottom panel in D shows that ΔN1–30TbCAE-myc that co-fractionates with the mitochondria-enriched pellet (P) is fully soluble in 1% digitonin and is retained completely in the soluble mitochondrial fraction (S1) instead of the insoluble mitochondrial fraction (P1).
To determine the localization of the tagged TbCAE variants we used digitonin extraction as in Fig. 5A. The results in the left panel of Fig. 9A show that the full-length protein is recovered in both the pellet and supernatant fractions, which is consistent with the protein being dually localized to the cytosol and possibly the nucleus (both recovered in the supernatant fraction) as well as to the mitochondrion (recovered in the pellet). Interestingly, in contrast to the experiment shown Fig. 5A, we found a significant fraction of the protein in the pellet. However, it is important to consider that in the experiments shown in Fig. 9, in contrast to the one shown in Fig. 5A, the endogenous TbCAE is replaced with the TbCAE that is expressed from the recoded synthetic gene. Fig. S1 shows that the latter is significantly overexpressed, which could explain why it can now be detected in mitochondria. Complementation with the ΔN1-10TbCAE-myc version of the protein (Fig. 9B) shows the same results than the full-length protein indicating that the N-terminal 10 amino acids are neither required for mitochondrial localization nor for enzymatic activity of TbCAE. However, the ΔN1-20TbCAE-myc and ΔN1–30TbCAE-myc variants, both did not restore growth and were enzymatically inactive (Fig. 9, C and D). Interestingly, ΔN1–20TbCAE-myc was exclusively cytosolically localized (Fig. 9C), whereas the ΔN1–30TbCAE-myc variant was more efficiently imported than even the full-length protein (Fig. 9D). The lower panel in Fig. 9D shows that the pellet fraction of ΔN1–30TbCAE-myc is fully soluble in 1% of digitonin indicating it is indeed imported and does not reflect large protein aggregates. In summary, these results indicate that amino acids 11–20 are required for mitochondrial localization of TbCAE as well as for the CCA adding activity. Deleting a further 10 amino acids (21–30) then reveals a cryptic import signal that normally is buried within the protein (39) leading to an artifactual import of the truncated protein.
In summary, these results show that the N-terminal 10 amino acids are neither required for TbCAE activity nor for the mitochondrial localization of the protein. Further deletions, however, abolish both the mitochondrial localization as well as the enzymatic activity.
Complementation using yeast cytosolic CAE isoform
To determine whether the mitochondrial fraction of TbCAE is essential for growth, the TbCAE RNAi cell line was complemented by expressing the cytosolic/nuclear isoform of the CAE of Saccharomyces cerevisiae (ScCAE). Fig. 10 shows that expression of the yeast enzyme restores growth to nearly WT levels (Fig. 10, A and B). Northern blot analysis of total RNA isolated from the TbCAE RNAi cell lines reveals the expected (Fig. 3) accumulation of the slower migrating form of the tRNACys in the induced sample, which could be complemented by expression of ScCAE (Fig. 10C). In contrast, when mitochondrial RNAs were analyzed the accumulation of the slower migrating form of the tRNACys remained unaffected irrespectively of whether ScCAE was expressed or not (Fig. 10D). These results indicate that expression of the cytosolic/nuclear isoform of ScCAE in T. brucei selectively complements for the lack of the cytosolic fraction of TbCAE. However, because the complemented cell line is able to grow at near WT levels this demonstrates that the mitochondrial localized TbCAE is dispensable for growth.
Figure 10.
Complementation of the TbCAE RNAi cell line by yeast cytosolic/nuclear ScCAE. A, growth curve of the uninduced (−tet) and induced (+tet) TbCAE RNAi cell lines (same curve and Northern blotting as in Fig. 1A). B, growth curve of the TbCAE RNAi cell line complemented by expression of the triple c-myc–tagged cytosolic/nuclear isoform of yeast ScCAE. Insets, loading control showing EtBr-stained rRNA region; Northern blots (NB) of total RNA from uninduced and induced cells probed for TbCAE mRNA, and in the right panel, immunoblots demonstrating inducible expression of ScCAE-myc. C, left panel, NB of total cellular RNA isolated from uninduced (−tet) and induced (+tet) TbCAE RNAi cell line probed for tRNACys using specific oligonucleotide hybridization. D, right panel: as C, but isolated mitochondrial RNA of the TbCAE RNAi cell line complemented with ScCAE-myc was analyzed.
Discussion
We have identified TbCAE as the single protein responsible for both nuclear/cytosolic as well as mitochondrial CCA adding activity. Experiments using an inducible TbCAE-RNAi cell line strongly suggest that TbCAE catalyzes the addition of CCA to 3′-cleaved tRNA precursors. Because only tRNAs carrying an intact CCA 3′-end can be aminoacylated, TbCAE is expected to be essential. In line with this we observe that its ablation abolishes protein synthesis and cell growth. Moreover, it leads to accumulation of a subset of truncated tRNAs, although some tRNA isoacceptor groups are not affected, indicating that TbCAE may have different affinities for different tRNA isoacceptor groups. Interestingly, even though ablation of TbCAE inhibits the activity measured in total cellular as well as in mitochondrial extracts to essentially the same extent, imported mitochondrial tRNAs are much less affected than total tRNAs. In fact, accumulation of truncated mitochondrial forms at steady-state could only be observed for the tRNACys. How can this be explained?
In trypanosomes, in contrast to most other systems, all mitochondrial tRNAs are imported from the cytosol. It has been demonstrated that binding to cytosolic EF1a is a prerequisite for a tRNA to be imported into mitochondria. However, EF1a itself is not imported into mitochondria, but mediates the essential targeting step (11). Because elongation factors only recognize aminoacylated tRNAs (40) this implies that only charged tRNAs can be imported. This was independently confirmed in a recent publication showing that, whereas a mutant Escherichia coli tRNATyr expressed in T. brucei could not be imported into mitochondria, because it was not recognized by the trypanosomal tyrosyl-tRNA synthetase, its import could be induced by co-expression of the E. coli tyrosyl-tRNA synthetase (12). These two examples illustrate that in T. brucei only aminoacylated tRNAs are imported into mitochondria in vivo. Because only tRNAs with a mature CCA ends can be aminoacylated, it follows that only fully processed tRNAs carrying an intact CCA are imported into mitochondria.
This indicates that the mitochondrial TbCAE may have a different function than its cytosolic/nuclear counterpart. It does not need to add the CCA-tail to immature cleaved tRNAs because they are not present in mitochondria. Instead, it might be involved in repair of damaged 3′-ends. Such damage can occur by spontaneous or enzymatic hydrolysis, e.g. by one of the numerous ribonucleases that were detected in the T. brucei mitochondrion. The accumulation of truncated newly synthesized variant of tRNAMet (tRNAMet*) after TbCAE knockdown demonstrates that the CCA ends of imported tRNAs are indeed turned over.
Expression of the yeast CAE isoform in the induced TbCAE RNAi cell line allows to selectively complement the extra-mitochondrial TbCAE activity. In the resulting cell line, which is devoid of mitochondrial CAE activity, growth is restored to nearly WT levels illustrating that T. brucei can grow in the absence of a TbCAE-based mitochondrial tRNA repair mechanism. However, the fact that the complemented cell line grows a little bit slower suggest that even when kept in culture mitochondrial TbCAE provides a slight fitness advantage to the cells. Trypanosomes have a complex life cycle and mitochondrial translation is known to be essential in both the procyclic and the bloodstream forms (4). Mitochondrial TbCAE activity might therefore well be essential under certain conditions explaining why trypanosomes have maintained mitochondrial CAE activity, even though the organelle imports mature tRNAs from the cytosol.
A similar situation has been described for E. coli in which deletion of the CAE resulted in a slow growth phenotype (41, 42). Because all E. coli tRNA genes encode the CCA-tail this is likely caused by a deficient tRNA repair mechanism.
All eukaryotic CAEs investigated so far are targeted to multiple compartments. The yeast enzyme is the best studied one. Its mRNA has three in-frame AUG codons upstream of the ORF for the CAE. Moreover, transcription creates mRNAs with heterogenous 5′-ends. The enzyme variant synthesized from the most upstream AUG contains an N-terminal mitochondrial targeting signal and is imported into mitochondria, whereas the two shorter variants provide for the nuclear/cytosolic activities of the enzyme (32, 43). In plants the problem is even more complex and the single CAE is targeted to the nucleus/cytosol as well as to mitochondria and plastids. Similar to yeast multiple in-frame upstream start codons influence the localization of the enzyme. However, the stability of the mature part of the enzyme is also important for the intracellular distribution of the enzyme (34, 35). We did not find any in-frame AUG codons upstream of the TbCAE ORF, suggesting that the intracellular localization of the enzyme is not regulated by different translation start sites. Although differential transsplicing was shown to be responsible for dual targeting of trypanosomal isoleucyl-tRNA synthetase (44) there is no evidence for upstream alternative splice sites in the case of TbCAE. This makes differential trans-splicing an unlikely mechanism for differential localization. Our complementation experiments showed that amino acids 10–20 are required for mitochondrial targeting as well as for enzymatic activity. However, unlike a typical presequence, this segment cannot be cleaved off because this would inactivate the enzyme. Thus, it remains unclear how the multiple localization of TbCAE is regulated. One possibility is that, as has been shown for fumarase in yeast, the protein folding versus translocation rate determines how much of the protein is imported into mitochondria (45).
Finally, it has been shown that the single imported tRNALys in yeast mitochondria is co-imported with the precursor of the mitochondrial lysyl-tRNA synthetase (46). Because in trypanosomes a fraction of all but two tRNAs are imported into mitochondria, the partially imported TbCAE, which recognizes all tRNAs, appears to be an excellent candidate to piggyback tRNAs into the mitochondrion. However, the fact that ablation of TbCAE did not diminish the levels of any imported tRNAs and the same was the case for the fully viable cell line complemented with the cytosolic/nuclear isoform ScCAE demonstrates that TbCAE is not involved in tRNA import. The same has been shown previously for mitochondrial elongation factor Tu and various aaRSs. These results are therefore in line with a recent study showing that, in contrast to yeast, mitochondrial tRNAs in trypanosomes, whereas relying on some subunits of the protein import machinery, are not co-imported with proteins (9).
Materials and methods
Transgenic cell lines
Transgenic T. brucei cell lines are based on the procyclic strain 29-13 (47) and grown at 27 °C in SDM-79 containing 10% (v/v) fetal calf serum (Sigma). All the growth curves were performed in triplicates. Tetracycline (1 μg/ml; Sigma, catalog number. T7660) was added on day 0 in the indicated +Tet cells.
The inducible TbCAE RNAi cell line was produced using a pLew100-derived plasmid whose resistance cassette was changed from phleomycin to blasticidine. The RNAi targets nucleotides 300–856 of the ORF of TbCAE (Tb927.9.8780). The stem-loop DNA fragment was produced by cloning the 300–856 TbCAE gene fragment in opposing directions separated by a 460-bp stuffer DNA.
To produce the construct expressing the C-terminal triple c-myc–tagged version of the TbCAE, the respective ORF with the desired flanking restriction sites was amplified by PCR. The resulting PCR product was then ligated into a modified pLew100-based vector carrying a puromycin resistance and allowing the addition of the triple c-myc tag sequence at the C terminus.
For complementation experiments, a synthetic gene of TbCAE was used (Biomatik). The codons between positions 300 and 856 of the TbCAE gene were changed such that its transcript would be RNAi resistant but still translate into the same amino acid sequence found in the endogenous protein. The sequence of the synthetic TbCAE gene is shown in Fig. S4. Subsequently the synthetic gene was cloned into a pLew100-based vector allowing the addition of the triple c-myc tag at the C terminus and transfected into the TbCAE RNAi cell line. To produce the constructs allowing expression of the N terminally-truncated TbCAE variants that were used for complementation experiments, the corresponding DNA fragments were amplified from the synthetic gene.
For complementation with the yeast CCA adding enzyme, the ORF of the S. cerevisiae cca1 gene from the yeast strain BY4742 was amplified starting from the 2nd in-frame ATG codon (at 28 bp) because this translates into the cytosolic/nuclear-localized version of the protein (43). The resulting PCR product was cloned into the pLew100-based vector for C-terminal triple c-myc tagging.
Transfection of plasmids was done as described earlier (48). Expression was induced by adding tetracycline to a concentration of 1 μg/ml to the culture medium.
Antibodies
Commercially available monoclonal antibodies used and their working dilutions for immunofluorescence (IF) and immunoblotting (IB) are as follows: mouse c-Myc (Invitrogen, number 132500; IB 1:2000, IF 1:50), and mouse EF1a (Merck Millipore, number 05-235; IB 1:10,000). Anti-VDAC, anti-ATOM40, and anti-CoxIV polyclonal antibodies were produced in rabbits and described previously (49, 50). Working dilutions for IB and IF were: VDAC (IB 1:1000) and ATOM40 (IB 1:10,000; IF 1:1000). Mouse anti-TbVCP antibody was a generous gift from James D. Bangs and has been described previously (51).
Commercially available secondary antibodies for IB were IRDye 680LT goat anti-mouse and IRDye 800CW goat anti-rabbit (LI-COR Biosciences, 1:20,000). Secondary antibodies used for IF are goat anti-mouse Alexa Fluor 596 and goat anti-rabbit Alexa Fluor 488 (ThermoFisher Scientific, 1:1000).
Labeling of proteins with [35S]methionine
5 × 106 cells from an exponentially growing culture were washed with PBS and resuspended in 400 μl of SDM-80 medium (52) lacking l-methionine. Subsequently, the suspension was supplemented with 2 μl of [35S]methionine (10 μCi/μl, specific activity 1000 Ci/mmol, Hartmann Analytic). The cells were incubated at 27 °C for 20 min after which they were washed with PBS and resuspended in SDS-PAGE sample buffer. In the negative control, cycloheximide (300 μg/ml final concentration) was added to the culture 1 min before the addition of [35S]methionine. Proteins were denatured at 95 °C for 5 min, subjected to SDS-PAGE and subsequently stained with Coomassie. The gel was dried, and radioactive signal was analyzed using a PhosphorImager.
CCA adding enzyme assay
2 × 107 cells from exponentially growing cultures of the uninduced and induced TbCAE RNAi cell line were washed with PBS and re-suspended in 50 μl in CCA buffer (30 mm HEPES/KOH, pH 7.6, 30 mm KCl, 6 mm MgCl2) containing 0.1% Triton X-100 and incubated at 4 °C for 15 min. Subsequently, the samples were centrifuged at 16,000 × g in a table top centrifuge and the supernatants corresponding to the total cellular extract were collected. Next 5 μl each of the extracts and 2 μg of purified total RNA isolated from WT T. brucei cells were mixed with CCA buffer containing 10 μm DTT, subsequently unlabeled ATP (or CTP) (0.1 mm final) and [α-32P]CTP (or [α-32P]ATP or [α-32P]UTP) (10μCi, specific activity 3000 Ci/mmol; Hartmann Analytic) were added and the reaction was adjusted to 20 μl.
The reactions were incubated at 27 °C for 1 h and RNA was extracted using the acid guanidinium method (53). The extracted RNA samples were subjected to 8 m urea, 15% PAGE, the gel was stained with ethidium bromide, dried, and analyzed using a PhosphorImager.
To determine the mitochondrial TbCAE activity mitochondria were isolated from a culture of the uninduced and the 2.5-day– induced TbCAE RNAi cell line as described (54). The same protein equivalents of isolated mitochondria of each culture were washed with PBS and lysed in CCA buffer containing 0.5% of Triton X-100 and incubated for 15 min at 4 °C. To 50 μl of extracts unlabeled ATP (0.1 mm final) and [α-32P]CTP (10 μCi, specific activity 3000 Ci/mmol; Hartmann Analytic) were added, incubated for 1 h at 27 °C before the RNA was extracted using the acid guanidium method (53). The extracted RNA was subjected to 8 m urea, 15% PAGE, stained with ethidium bromide, and processed as described above.
RNA extraction and Northern blot analysis
For analysis of total tRNAs, uninduced and induced TbCAE RNAi cells were harvested and washed with PBS before the total RNA was extracted using the acid guanidium method (53). For isolation of the corresponding mitochondrial RNAs, the cells were permeabilized using low digitonin concentration (0.025%) and the contaminating cytosolic RNA was degraded using RNase A as described (38). Subsequently RNA was isolated from the resulting crude mitochondrial fractions by the acid guanidium method (53). 5 μg of total RNA or 2.5 μg of mitochondrial RNA each were separated on 8 m urea, 15% PAGE and visualized using ethidium bromide staining. Subsequently gels were electroblotted onto a charged nylon membrane using a semi-dry blotting apparatus (300 mAmps, 1 h, Bio-Rad) and probed with the corresponding labeled oligonucleotide (Table S1). The blots were hybridized in 6× SSPE (60 mm NaH2PO4, 0.9 m NaCl, 6 mm EDTA) containing 5× Denhardt's and 0.5% (w/v) SDS at 55 °C for 14–18 h using 10 pmol of the corresponding 5′-labeled oligonucleotide. Labeling of the oligonucleotides was done with [γ-32P]ATP (50 μCi specific activity 3000 Ci/mmol; Hartmann Analytic) using the polynucleotide kinase forward reaction. Northern blots were analyzed using a PhosphorImager.
For mRNA Northern blots, 7 μg of total RNA was separated on a 1% agarose gel in 20 mm MOPS buffer, pH 7.0, containing 0.5% formaldehyde. The probe used for the Northern blot analysis corresponds to the fragment (nucleotides 300–856) of the TbCAE ORF, which was gel-purified from a PCR product and radioactively labeled using the Prime-a-Gene labeling kit (Promega).
Digitonin extraction
1 × 108 of exponentially growing cells were incubated for 10 min on ice in 0.025% (w/v) digitonin made up in 1× SoTE (20 mm Tris-HCl, pH 7.5, 0.6 m sorbitol, 2 mm EDTA). These conditions selectively lyse the cell membrane but leave the mitochondrial membranes intact. After centrifugation in a table top centrifuge at 6,800 × g, at 4 °C, the resulting crude mitochondrial pellet was separated from the supernatant, corresponding to a crude cytosolic fraction. Total cells, crude cytosolic and mitochondrial fractions were mixed with SDS sample buffer and 2.5 × 106 cell equivalents of each fraction were subjected to 12% SDS-PAGE and immunoblotting.
Immunofluorescence
TbCAE-myc expression was induced for 24 h using tetracycline (1 μg/ml). Cells were fixed with 4% paraformaldehyde in 1× PBS, washed with 1× PBS, and allowed to adhere on a slide. Subsequently the cells were permeabilized on the slide with PBS containing 1% of Triton-X100 for 2 min after which primary antibody diluted in PBS was added for 1 h. The slides were washed three times with PBS and incubated with the corresponding secondary antibodies. After three more washes with PBS, the slides were mounted using VectaShield containing 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, P/N H-1200). Images were taken with a DFC360 FX monochrome camera (Leica Microsystems) mounted on a DMI6000B microscope (Leica Microsystems). Image analysis was done using LAS X software (Leica Microsystems).
Data availability
All data are contained within the manuscript.
Author contributions
S. S. and A. S. conceptualization; S. S. formal analysis; S. S. validation; S. S. investigation; S. S. visualization; S. S. methodology; S. S. and A. S. writing-review and editing; A. S. supervision; A. S. funding acquisition; A. S. writing-original draft; A. S. project administration.
Supplementary Material
Acknowledgment
We thank Dr. Moritz Niemann for help with the mitoprep.
This work was supported by NCCR “RNA & Disease” and Grant 175563 from the Swiss National Science Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S4.
- aaRSs
- aminoacyl-tRNA synthetases
- ATOM40
- atypical translocase of the outer membrane 40
- BLAST
- basic local alignment search tool
- CAE
- CCA-adding enzyme
- DAPI
- 4,6-diamidino-2-phenylindole
- EF1a
- eukaryotic elongation factor 1a
- SDM-79
- semi-defined medium-79
- TbCAE
- T. brucei CCA-adding enzyme
- TbVCP
- T. brucei valosin containing protein
- VDAC
- voltage-dependent anion channel
- IF
- immunofluorescence
- IB
- immunoblotting
- EtBr
- ethidium bromide
- NB
- Northern blot.
References
- 1. Shikha S., Brogli R., Schneider A., and Polacek N. (2019) tRNA biology in Trypanosomes. Chimia 73, 395–405 10.2533/chimia.2019.395 [DOI] [PubMed] [Google Scholar]
- 2. Alfonzo J. D., and Söll D. (2009) Mitochondrial tRNA import: the challenge to understand has just begun. Biol. Chem. 390, 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Niemann M., Schneider A., and Cristodero M. (2011) Mitochondrial translation in trypanosomatids: a novel target for chemotherapy? Trends Parasitol. 27, 429–433 10.1016/j.pt.2011.03.011 [DOI] [PubMed] [Google Scholar]
- 4. Cristodero M., Seebeck T., and Schneider A. (2010) Mitochondrial translation is essential in bloodstream forms of Trypanosoma brucei. Mol. Microbiol. 78, 757–769 10.1111/j.1365-2958.2010.07368.x [DOI] [PubMed] [Google Scholar]
- 5. Schneider A. (2011) Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu. Rev. Biochem. 80, 1033–1053 10.1146/annurev-biochem-060109-092838 [DOI] [PubMed] [Google Scholar]
- 6. Sieber F., Duchêne A. M., and Maréchal-Drouard L. (2011) Mitochondrial RNA import: from diversity of natural mechanisms to potential applications. Int. Rev. Cell Mol. Biol. 287, 145–190 10.1016/B978-0-12-386043-9.00004-9 [DOI] [PubMed] [Google Scholar]
- 7. Salinas T., Duchêne A. M., and Maréchal-Drouard L. (2008) Recent advances in tRNA mitochondrial import. Trends Biochem. Sci. 33, 320–329 10.1016/j.tibs.2008.04.010 [DOI] [PubMed] [Google Scholar]
- 8. Tschopp F., Charrière F., and Schneider A. (2011) In vivo study in Trypanosoma brucei links mitochondrial transfer RNA import to mitochondrial protein import. EMBO Rep. 12, 825–832 10.1038/embor.2011.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Niemann M., Harsman A., Mani J., Peikert C. D., Oeljeklaus S., Warscheid B., Wagner R., and Schneider A. (2017) tRNAs and proteins use the same import channel for translocation across the mitochondrial outer membrane of trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 114, E7679–E7687 10.1073/pnas.1711430114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seidman D., Johnson D., Gerbasi V., Golden D., Orlando R., and Hajduk S. (2012) Mitochondrial membrane complex that contains proteins necessary for tRNA import in Trypanosoma brucei. J. Biol. Chem. 287, 8892–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bouzaidi-Tiali N., Aeby E., Charrière F., Pusnik M., and Schneider A. (2007) Elongation factor 1a mediates the specificity of mitochondrial tRNA import in T. brucei. EMBO J. 26, 4302–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huot J. L., Shikha S., and Schneider A. (2019) Inducible orthogonal aminoacylation demonstrates that charging is required for mitochondrial tRNA import in Trypanosoma brucei. Sci. Rep. 9, 10836 10.1038/s41598-019-47268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Charrière F., O'Donoghue P., Helgadóttir S., Maréchal-Drouard L., Cristodero M., Horn E. K., Söll D., and Schneider A. (2009) Dual targeting of a tRNAAsp requires two different aspartyl-tRNA synthetases in Trypanosoma brucei. J. Biol. Chem. 284, 16210–16217 10.1074/jbc.M109.005348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Charrière F., Helgadóttir S., Horn E. K., Söll D., and Schneider A. (2006) Dual targeting of a single tRNATrp requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei. Proc. Natl. Acad. Sci. U.S.A. 103, 6847–6852 10.1073/pnas.0602362103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peikert C. D., Mani J., Morgenstern M., Käser S., Knapp B., Wenger C., Harsman A., Oeljeklaus S., Schneider A., and Warscheid B. (2017) Charting organellar importomes by quantitative mass spectrometry. Nat. Commun. 8, 15272 10.1038/ncomms15272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alfonzo J. D., Blanc V., Estevez A. M., Rubio M. A. T., and Simpson L. (1999) C to U editing of anticodon of imported mitochondrial tRNATrp allows decoding of UGA stop codon in Leishmania. EMBO J. 18, 7056–7062 10.1093/emboj/18.24.7056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paris Z., and Alfonzo J. D. (2018) How the intracellular partitioning of tRNA and tRNA modification enzymes affects mitochondrial function. IUBMB Life 70, 1207–1213 10.1002/iub.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wohlgamuth-Benedum J. M., Rubio M. A., Paris Z., Long S., Poliak P., Lukes J., and Alfonzo J. D. (2009) Thiolation controls cytoplasmic tRNA stability and acts as a negative determinant for tRNA editing in mitochondria. J. Biol. Chem. 284, 23947–23953 10.1074/jbc.M109.029421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crain P. F., Alfonzo J. D., Rozenski J., Kapushoc S. T., McCloskey J. A., and Simpson L. (2002) Modification of the universally unmodified uridine-33 in a mitochondria-imported edited tRNA and the role of the anticodon arm structure on editing efficiency. RNA 8, 752–761 10.1017/S1355838202022045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aeby E., Palioura S., Pusnik M., Marazzi J., Lieberman A., Ullu E., Söll D., and Schneider A. (2009) The canonical pathway for selenocysteine insertion is dispensable in Trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 106, 5088–5092 10.1073/pnas.0901575106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aeby E., Seidel V., and Schneider A. (2009) The selenoproteome is dispensable in bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 168, 191–193 10.1016/j.molbiopara.2009.08.007 [DOI] [PubMed] [Google Scholar]
- 22. Aeby E., Ullu E., Yepiskoposyan H., Schimanski B., Roditi I., Mühlemann O., and Schneider A. (2010) tRNASec is transcribed by RNA polymerase II in Trypanosoma brucei but not in humans. Nucleic Acids Res. 38, 5833–5843 10.1093/nar/gkq345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hartmann R. K., Gössringer M., Späth B., Fischer S., and Marchfelder A. (2009) The making of tRNAs and more: RNase P and tRNase Z. Prog. Mol. Biol. Transl. Sci. 85, 319–368 10.1016/S0079-6603(08)00808-8 [DOI] [PubMed] [Google Scholar]
- 24. Maraia R. J., and Lamichhane T. N. (2011) 3′ processing of eukaryotic precursor tRNAs. Wiley Interdiscip. Rev. RNA 2, 362–375 10.1002/wrna.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rossmanith W. (2012) Of P and Z: mitochondrial tRNA processing enzymes. Biochim. Biophys. Acta 1819, 1017–1026 10.1016/j.bbagrm.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taschner A., Weber C., Buzet A., Hartmann R. K., Hartig A., and Rossmanith W. (2012) Nuclear RNase P of Trypanosoma brucei: a single protein in place of the multicomponent RNA-protein complex. Cell Rep. 2, 19–25 10.1016/j.celrep.2012.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salavati R., Panigrahi A. K., and Stuart K. D. (2001) Mitochondrial ribonuclease P activity of Trypanosoma brucei. Mol. Biochem. Parasitol. 115, 109–117 10.1016/S0166-6851(01)00273-0 [DOI] [PubMed] [Google Scholar]
- 28. Mörl M., and Marchfelder A. (2001) The final cut: the importance of tRNA 3′-processing. EMBO J. 2, 17–20 10.1093/embo-reports/kve006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Betat H., and Mörl M. (2015) The CCA-adding enzyme: a central scrutinizer in tRNA quality control. Bioessays 37, 975–982 10.1002/bies.201500043 [DOI] [PubMed] [Google Scholar]
- 30. Hancock K., LeBlanc A. J., Donze D., and Hajduk S. L. (1992) Identification of nuclear encoded precursor tRNAs within the mitochondrion of Trypanosoma brucei. J. Biol. Chem. 267, 23963–23971 [PubMed] [Google Scholar]
- 31. Béja O., Ullu E., and Michaeli S. (1993) Identification of a tRNA-like molecule that copurifies with the 7SL RNA of Trypanosoma brucei. Mol. Biochem. Parasitol. 57, 223–230 10.1016/0166-6851(93)90198-7 [DOI] [PubMed] [Google Scholar]
- 32. Wolfe C. L., Lou Y. C., Hopper A. K., and Martin N. C. (1994) Interplay of heterogeneous transcriptional start sites and translational selection of AUGs dictate the production of mitochondrial and cytosolic/nuclear tRNA nucleotidyltransferase from the same gene in yeast. J. Biol. Chem. 269, 13361–13366 [PubMed] [Google Scholar]
- 33. Chen J. Y., Joyce P. B., Wolfe C. L., Steffen M. C., and Martin N. C. (1992) Cytoplasmic and mitochondrial tRNA nucleotidyltransferase activities are derived from the same gene in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 267, 14879–14883 [PubMed] [Google Scholar]
- 34. von Braun S. S., Sabetti A., Hanic-Joyce P. J., Gu J., Schleiff E., and Joyce P. B. (2007) Dual targeting of the tRNA nucleotidyltransferase in plants: not just the signal. J. Exp. Bot. 58, 4083–4093 10.1093/jxb/erm267 [DOI] [PubMed] [Google Scholar]
- 35. Leibovitch M., Bublak D., Hanic-Joyce P. J., Tillmann B., Flinner N., Amsel D., Scharf K. D., Mirus O., Joyce P. B., and Schleiff E. (2013) The folding capacity of the mature domain of the dual-targeted plant tRNA nucleotidyltransferase influences organelle selection. Biochem. J. 453, 401–412 10.1042/BJ20121577 [DOI] [PubMed] [Google Scholar]
- 36. Pusnik M., Schmidt O., Perry A. J., Oeljeklaus S., Niemann M., Warscheid B., Lithgow T., Meisinger C., and Schneider A. (2011) Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr. Biol. 21, 1738–1743 10.1016/j.cub.2011.08.060 [DOI] [PubMed] [Google Scholar]
- 37. Harsman A., Niemann M., Pusnik M., Schmidt O., Burmann B. M., Hiller S., Meisinger C., Schneider A., and Wagner R. (2012) Bacterial origin of a mitochondrial outer membrane protein translocase: new perspectives from comparative single channel electrophysiology. J. Biol. Chem. 287, 31437–31445 10.1074/jbc.M112.392118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tan T. H., Pach R., Crausaz A., Ivens A., and Schneider A. (2002) tRNAs in Trypanosoma brucei: genomic organization, expression and mitochondrial import. Mol. Cell Biol. 22, 3707–3717 10.1128/MCB.22.11.3707-3716.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hurt E. C., and Schatz G. (1987) A cytosolic protein contains a cryptic mitochondrial targeting signal. Nature 325, 499–503 10.1038/325499a0 [DOI] [PubMed] [Google Scholar]
- 40. Ribeiro S., Nock S., and Sprinzl M. (1995) Purification of aminoacyl-tRNA by affinity chromatography on immobilized Thermus thermophilus EF-Tu.GTP. Anal. Biochem. 228, 330–335 10.1006/abio.1995.1359 [DOI] [PubMed] [Google Scholar]
- 41. Zhu L., and Deutscher M. P. (1987) tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J. 6, 2473–2477 10.1002/j.1460-2075.1987.tb02528.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reuven N. B., Zhou Z., and Deutscher M. P. (1997) Functional overlap of tRNA nucleotidyltransferase, poly(A) polymerase I, and polynucleotide phosphorylase. J. Biol. Chem. 272, 33255–33259 10.1074/jbc.272.52.33255 [DOI] [PubMed] [Google Scholar]
- 43. Wolfe C. L., Hopper A. K., and Martin N. C. (1996) Mechanisms leading to and the consequences of altering the normal distribution of ATP(CTP):tRNA nucleotidyltransferase in yeast. J. Biol. Chem. 271, 4679–4686 10.1074/jbc.271.9.4679 [DOI] [PubMed] [Google Scholar]
- 44. Rettig J., Wang Y., Schneider A., and Ochsenreiter T. (2012) Dual targeting of isoleucyl-tRNA synthetase in Trypanosoma brucei is mediated through alternative trans-splicing. Nucleic Acids Res. 40, 1299–1306 10.1093/nar/gkr794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Regev-Rudzki N., Yogev O., and Pines O. (2008) The mitochondrial targeting sequence tilts the balance between mitochondrial and cytosolic dual localization. J. Cell Sci. 121, 2423–2431 10.1242/jcs.029207 [DOI] [PubMed] [Google Scholar]
- 46. Tarassov I., Entelis N., and Martin R. P. (1995) Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 14, 3461–3471 10.1002/j.1460-2075.1995.tb07352.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wirtz E., Leal S., Ochatt C., and Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 10.1016/S0166-6851(99)00002-X [DOI] [PubMed] [Google Scholar]
- 48. McCulloch R., Vassella E., Burton P., Boshart M., and Barry J. D. (2004) Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol. Biol. 262, 53–86 [DOI] [PubMed] [Google Scholar]
- 49. Niemann M., Wiese S., Mani J., Chanfon A., Jackson C., Meisinger C., Warscheid B., and Schneider A. (2013) Mitochondrial outer membrane proteome of Trypanosoma brucei reveals novel factors required to maintain mitochondrial morphology. Mol. Cell Proteomics 12, 515–528 10.1074/mcp.M112.023093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mani J., Desy S., Niemann M., Chanfon A., Oeljeklaus S., Pusnik M., Schmidt O., Gerbeth C., Meisinger C., Warscheid B., and Schneider A. (2015) Mitochondrial protein import receptors in kinetoplastids reveal convergent evolution over large phylogenetic distances. Nat. Commun. 6, 6646 10.1038/ncomms7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roggy J. L., and Bangs J. D. (1999) Molecular cloning and biochemical characterization of a VCP homolog in African trypanosomes. Mol. Biochem. Parasitol. 98, 1–15 10.1016/S0166-6851(98)00114-5 [DOI] [PubMed] [Google Scholar]
- 52. Lamour N., Rivière L., Coustou V., Coombs G. H., Barrett M. P., and Bringaud F. (2005) Proline metabolism in procyclic Trypanosoma brucei is down-regulated in the presence of glucose. J. Biol. Chem. 280, 11902–11910 10.1074/jbc.M414274200 [DOI] [PubMed] [Google Scholar]
- 53. Chomczynski P., and Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 10.1016/0003-2697(87)90021-2, 10.1006/abio.1987.9999 [DOI] [PubMed] [Google Scholar]
- 54. Schneider A., Charrière F., Pusnik M., and Horn E. K. (2007) Isolation of mitochondria from procyclic Trypanosoma brucei. Methods Mol. Biol. 372, 67–80 10.1007/978-1-59745-365-3_5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.