Abstract
Heat shock factor 1 (HSF1) regulates cellular adaptation to challenges such as heat shock and oxidative and proteotoxic stresses. We have recently reported a previously unappreciated role for HSF1 in the regulation of energy metabolism in fat tissues; however, whether HSF1 is differentially expressed in adipose depots and how its levels are regulated in fat tissues remain unclear. Here, we show that HSF1 levels are higher in brown and subcutaneous fat tissues than in those in the visceral depot and that HSF1 is more abundant in differentiated, thermogenic adipocytes. Gene expression experiments indicated that HSF1 is transcriptionally regulated in fat by agents that modulate cAMP levels, by cold exposure, and by pharmacological stimulation of β-adrenergic signaling. An in silico promoter analysis helped identify a putative response element for activating transcription factor 3 (ATF3) at −258 to −250 base pairs from the HSF1 transcriptional start site, and electrophoretic mobility shift and ChIP assays confirmed ATF3 binding to this sequence. Furthermore, functional assays disclosed that ATF3 is necessary and sufficient for HSF1 regulation. Detailed gene expression analysis revealed that ATF3 is one of the most highly induced ATFs in thermogenic tissues of mice exposed to cold temperatures or treated with the β-adrenergic receptor agonist CL316,243 and that its expression is induced by modulators of cAMP levels in isolated adipocytes. To the best of our knowledge, our results show for the first time that HSF1 is transcriptionally controlled by ATF3 in response to classic stimuli that promote heat generation in thermogenic tissues.
Keywords: Heat shock factor protein 1 (HSF1), gene regulation, obesity, lipid metabolism, uncoupling protein, uncoupling protein 1 (UCP1), activating transcription factor 3 (ATF3), beta-adrenergic stimuli, brown fat/beige fat, thermogenesis
Introduction
Obesity is the result of an energy imbalance that causes excessive fat accumulation, ultimately leading to the development of the metabolic syndrome (1, 2). Several types of adipose depots exist, located in various anatomical locations, constituted by three main types of adipocytes: white, brown, and beige, each shown to possess characteristic gene expression profiles (3, 4). Specifically, it has been shown that classic epididymal white fat tissue (eWAT)3 present in the intra-abdominal area is involved in energy storage, whereas brown fat tissue (BAT), located in the interscapular region, controls energy dissipation via the process of heat generation called thermogenesis (5). In addition to brown fat cells, beige adipocytes, which are present in subcutaneous white adipose depots (scWAT), have also been shown to regulate the thermogenic processes and overall contribute to energy expenditure (3–7).
Activating transcription factor 3 (ATF3) belongs to the cAMP transcription factors (ATF/CREB) family, which currently includes eight members: CREB and ATF1 to ATF7 (8). It has been shown that ATF3 is rapidly induced by a wide range of challenges such as DNA damage, oxidative, and endoplasmic reticulum stress (9, 10), leading to the activation of signaling pathways required for the maintenance of cellular homeostasis (8, 11). ATF3 is known to activate and repress transcription through its binding to the canonical cis-regulatory element 5′-TGACGTCA-3′ via its basic region–leucine zipper domain as a homodimer or heterodimer, in combination with other ATFs or AP-1 transcription factors (12). Despite the evidence linking ATF3 to cellular stressors, there are currently no data on whether ATF3 levels are regulated by stimuli such as cold or β-adrenergic signaling in adipose depots.
We have previously demonstrated that heat shock factor 1 (HSF1) controls the levels of expression of the metabolic coactivator peroxisome proliferator-activated receptor γ coactivator 1-α (PGC1α) and induces thermogenic programs in vivo (13). In the present study we analyzed the expression profile of HSF1 in adipose tissues and the mechanisms of its transcriptional regulation in response to thermogenic cues. Here we provide novel evidence that HSF1 is highly expressed in mature BAT and scWAT and induced by agents that increase cAMP levels in vitro. Furthermore, we show that in vivo, HSF1 mRNA levels are induced by cold exposure and via pharmacological activation of the β-adrenergic receptor signaling pathway. Our detailed analysis of the molecular mechanisms that control HSF1 expression in thermogenic fat cells revealed that HSF1 is regulated by the activating transcription factor 3 (ATF3), which, in turn, is induced in response to thermogenic stimuli. Collectively, our studies provide novel evidence that HSF1 mRNA levels are rapidly up-regulated in response to thermogenic cues and identify for the first time the stress response transcription factor ATF3 as a critical new regulator of HSF1 levels in response to thermogenic signals.
Results
HSF1 is expressed selectively in thermogenic fat depots
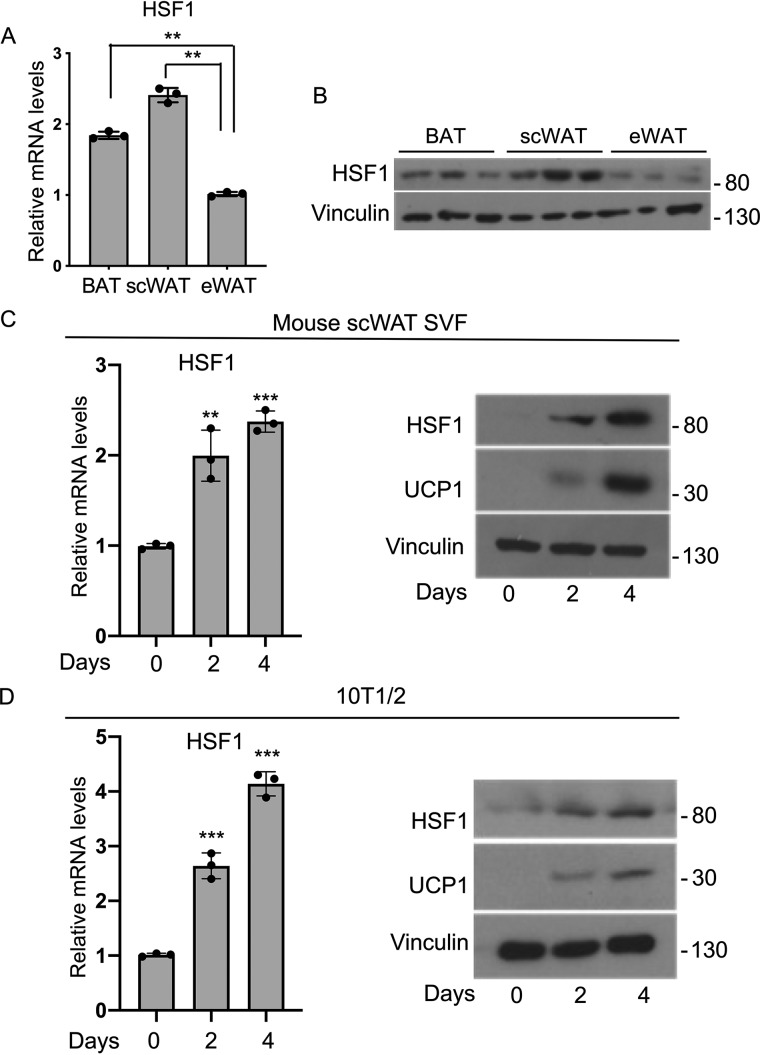
We have previously shown that HSF1 can directly control the expression of PGC1α in differentiated brown fat cells (13), but the relative levels of HSF1 in different adipose depots have not been previously investigated. To specifically determine the pattern of HSF1 expression in fat tissues in vivo, we analyzed HSF1 mRNA and protein levels in BAT, scWAT, and eWAT isolated from 9-week-old male mice. As shown in Fig. 1A, HSF1 mRNA levels were higher in BAT and scWAT than in eWAT. Western blotting analysis provided further confirmation that HSF1 is present at higher levels in BAT and scWAT than in eWAT (Fig. 1B). Next, we assessed HSF1 expression levels during a time course of brown fat differentiation of stromal vascular fraction (SVF) cells derived from mouse scWAT, as well as in the pluripotent mesenchymal cell line 10T1/2. As shown in Fig. 1 (C and D), HSF1 levels are elevated only in differentiated cells. We measured in parallel UCP1 and demonstrated that its levels are induced concomitantly to those of HSF1, confirming that the cells underwent brown-like differentiation (Fig. 1, C and D). These results demonstrate that HSF1 mRNA is selectively elevated in mature thermogenic tissues in vivo and in differentiated brown-like adipocytes in vitro.
Figure 1.
HSF1 is highly expressed in thermogenic adipocytes. A and B, HSF1 mRNA (A) and protein (B) levels in BAT, scWAT, and eWAT of male mice fed a normal chow diet (n = 3). C, HSF1 mRNA and HSF1 and UCP1 protein levels during a time course of differentiation of mouse SVF cells isolated from scWAT. D, HSF1 RNA levels and HSF1 and UCP1 protein levels during a time course of differentiation of 10T1/2 cells. Vinculin was used as a loading control for Western blots. The results are expressed as means ± S.E. from three independent experiments. **, p value < 0.005; ***, p value < 0.001.
HSF1 is induced by compounds that modulate cAMP levels
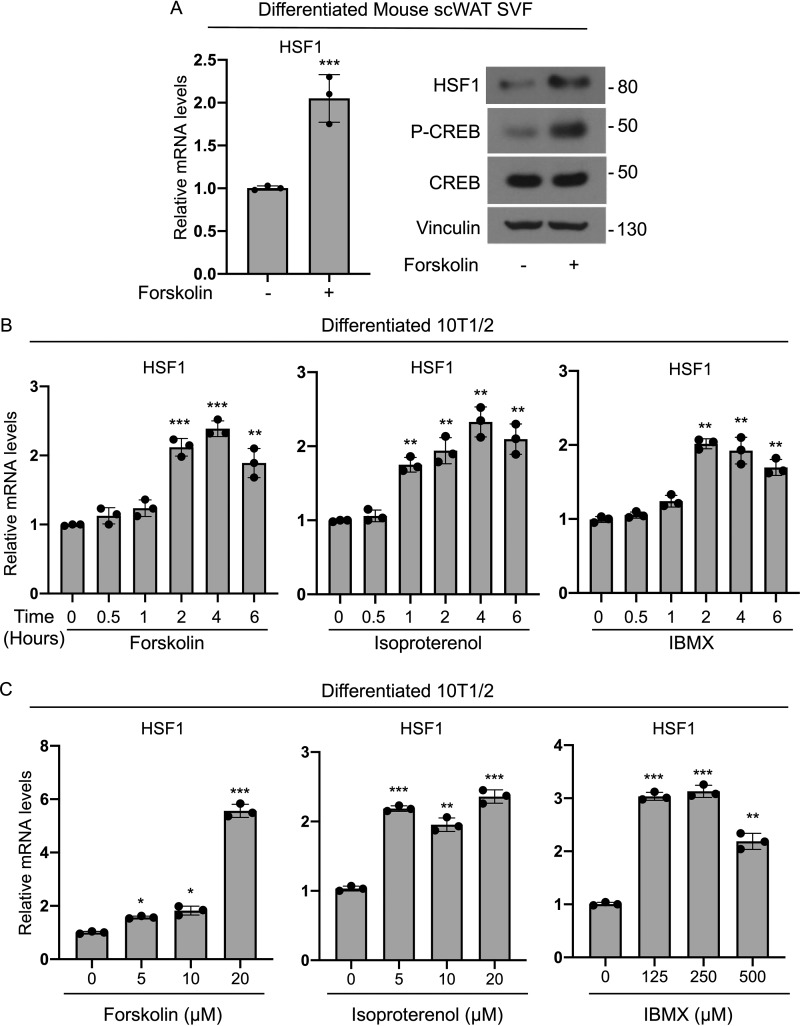
Given that HSF1 is highly expressed in differentiated brown-like adipocytes (Fig. 1), we assessed whether HSF1 levels are regulated via classic thermogenic signals known to be involved in the control of brown and beige cells functionality (14, 15). To test whether modulators of cAMP levels were able to induce HSF1, we treated brown differentiated SVF cells derived from scWAT with forskolin and observed an increase in HSF1 mRNA and protein levels compared with vehicle-treated cells (Fig. 2A). Increased phospho-CREB levels measured in parallel in the same cells confirmed the effectiveness of forskolin in inducing cAMP-dependent downstream signaling (Fig. 2A). To further investigate the dynamic of HSF1 induction by forskolin and the effects of other cAMP modulators on HSF1 levels, we performed detailed time-course and dose-response analyses in brown-like differentiated 10T1/2 cells and demonstrated that HSF1 mRNA levels are induced by these agents in differentiated cells (Fig. 2, B and C). These results suggest that HSF1 mRNA is regulated in response to modulators of cAMP levels in thermogenic adipocytes.
Figure 2.
cAMP activators up-regulate HSF1 mRNA levels in thermogenic adipocytes. A, HSF1 mRNA and protein levels of HSF1, phospho-CREB, and CREB in mouse SVF cells isolated from scWAT, differentiated into brown-like cells, and treated for 4 h with 10 μm forskolin. Vinculin was used as a loading control in Western blotting. B, time course of HSF1 mRNA levels in brown differentiated 10T1/2 cells treated either with 10 μm forskolin, 10 μm isoproterenol, or 500 μm IBMX for the indicated times. C, dose response of HSF1 mRNA levels in brown-like differentiated 10T1/2 cells treated for 2 h with the different concentrations indicated of either forskolin, isoproterenol or IBMX. The results are expressed as means ± S.E. from three independent experiments. *, p value < 0.01; **, p value < 0.005; ***, p value <0.001.
HSF1 is induced by low temperatures and by pharmacological stimulation of β-adrenergic signaling
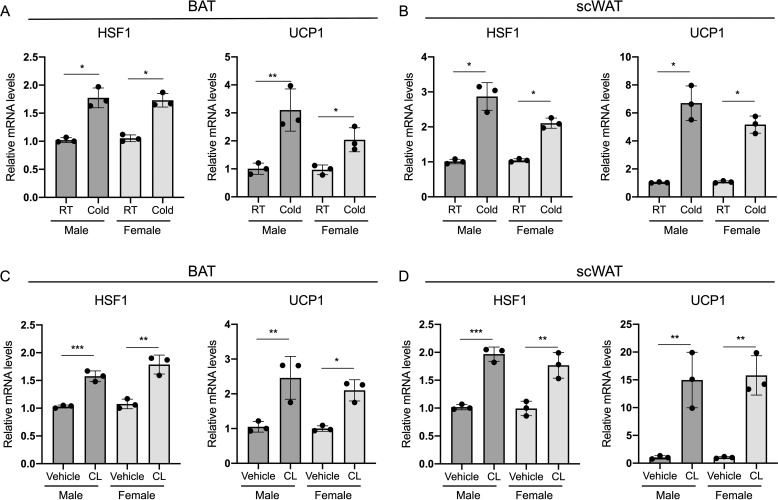
To determine whether HSF1 is regulated in response to classic thermogenic stimuli also in vivo, we analyzed the levels of HSF1 in BAT and scWAT obtained from male and female mice kept either at room temperature or exposed to cold. As shown in Fig. 3 (A and B) HSF1 mRNA levels are up-regulated when compared with those detected in control mice. Analysis of the expression levels of UCP1, a classic marker of brown fat functionality, demonstrated the efficacy of the cold exposure treatment in eliciting thermogenic gene expression (Fig. 3, A and B). To assess the responsiveness of HSF1 to pharmacological activation of β-adrenergic signaling in vivo, we treated male and female mice with the agonist CL316,243 (CL) and showed increased levels of HSF1 in both BAT and scWAT, compared with control treated mice, which occurred in parallel to the expected elevation in UCP1 levels (Fig. 3, C and D). Taken together, these results suggest that HSF1 mRNA expression is regulated in response to thermogenic stimuli both in cells, in vitro, and in vivo in fat tissues of both males and females.
Figure 3.
HSF1 is induced by cold exposure and pharmacological activation of β-adrenergic signaling in thermogenic tissues. A and B, HSF1 and UCP1 mRNA levels in BAT (A) and scWAT (B) of 9-week-old male and female mice kept at room temperature (RT) or exposed to cold for 6 h (n = 3). C and D, HSF1 and UCP1 mRNA levels in BAT (C) and scWAT (D) of 9-week-old male and female mice treated with either saline or CL316,243 (1 mg/kg of body weight) resuspended in saline for 3 h (n = 3). The results are expressed as means ± S.E. *, p value < 0.01; **, p value < 0.005; ***, p value <0.001.
ATF3 binds to the proximal region of the HSF1 promoter
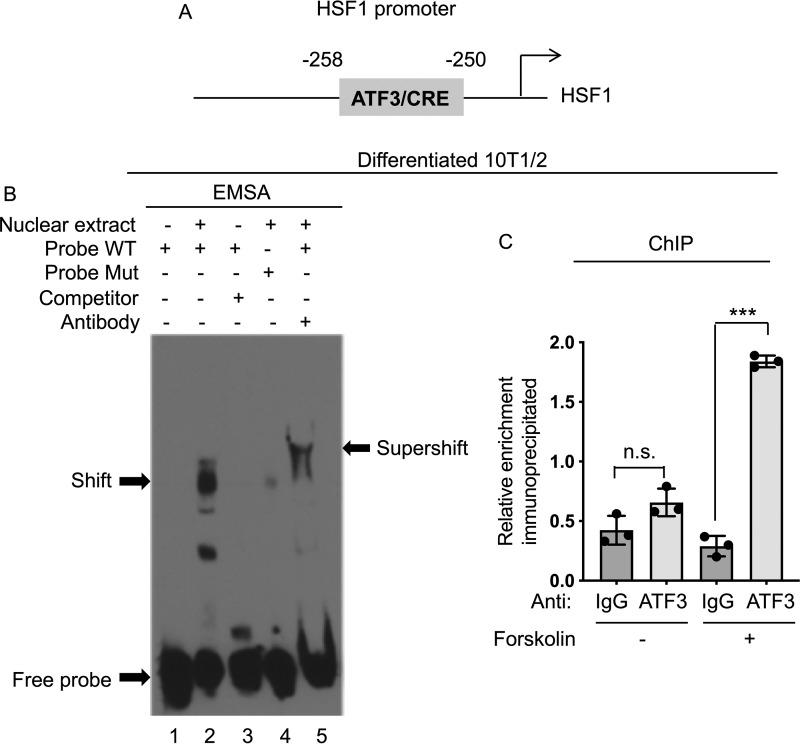
Given that ATF/CREB family members, such as ATF2 and CREB, have been shown to be involved in the regulation of the expression of genes involved in thermogenesis (16, 17), we hypothesized that an ATF family member may also control HSF1 levels in response to cAMP elevation and therefore screened in silico a 2-kb sequence upstream of the HSF1 transcription start site for putative ATF-binding motifs. Among the 13 ATF3 putative responsive elements identified by Genomatix, we chose to analyze the most proximal element located in position −258 to −250 (Fig. 4A) from the transcriptional start site. To assess whether ATF3 indeed binds to the HSF1 promoter at this site, we first performed electrophoretic mobility shift assays (EMSAs) and demonstrated that nuclear extracts obtained from differentiated 10T1/2 cells treated with forskolin are able to generate a shift when incubated with a labeled probe corresponding to the putative ATF3-binding element identified in silico in position −258 to −250, and a supershift, when an anti-ATF3 antibody was added. To test the specificity of ATF3 binding to this putative site, we tested whether the shift could be annulled if we pretreated nuclear extracts with excess unlabeled oligonucleotides and demonstrated that the binding was competed off. Further analysis using oligonucleotides containing a mutation in the putative ATF3-responsive element demonstrated the inability of these mutated oligonucleotides to generate a shift when incubated with nuclear extracts obtained from differentiated 10T1/2 treated with forskolin (Fig. 4B). To further confirm in vivo the binding of endogenous ATF3 present in brown-like adipocytes treated with forskolin to the HSF1 promoter, we performed ChIP assays. Our results demonstrated that ATF3 occupies the putative response element at position −258 to −250 (Fig. 4C). These data indicate that the ATF3 putative binding motif identified in the proximal region of the HSF1 promoter can recruit ATF3 in forskolin-stimulated 10T1/2 differentiated cells.
Figure 4.
ATF3 binds to the HSF1 promoter. A, in silico analyses using the Genomatix and Promo software programs identified a putative ATF3/CREB-binding motif located in the proximal promoter region of HSF1 at −258 to −250 bp. B, EMSA assay using nuclear extracts from brown-like differentiated 10T1/2 cells treated with 10 μm forskolin for 4 h. Nuclear extracts incubated with biotin-labeled DNA probes corresponding to the putative ATF3-binding sequences generated a specific shifted band (lane 2). The shift was detected only in the presence of a probe containing the WT ATF3-binding sequence (probe WT), but not with a mutated one (probe Mut). Supershift was achieved by adding an anti-ATF3 antibody (lane 5) to the nuclear extract–probe mixture. For competition assays, a 100-fold excess of unlabeled WT probe was used. Specific complexes are indicated with arrows. C, ChIP assay was performed in brown-like differentiated cells using either anti-IgG or an anti-ATF3 antibody to assess occupancy at the putative ATF3 site in the HSF1 promoter treated with vehicle or forskolin at the concentration of 10 μm for 4 h. The results are expressed as means ± S.E. from three independent experiments. ***, p value < 0.001. n.s., not significant.
ATF3 is induced in adipocytes by thermogenic stimuli
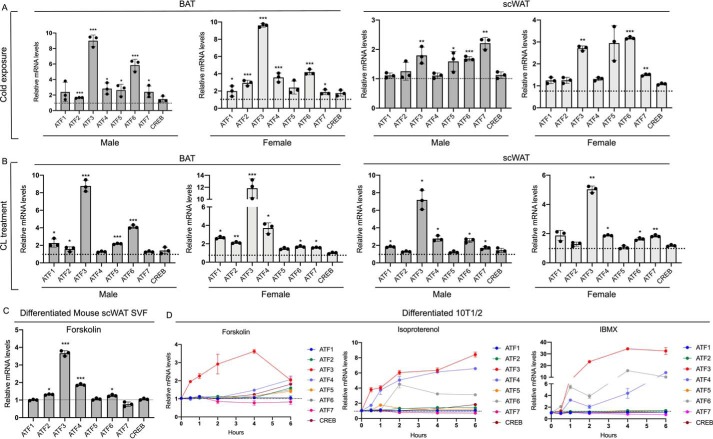
To further test whether ATF3 is regulated in vivo in response to thermogenic stimuli and to assess the relative induction of ATF3 compared with that of other ATF family members, we assessed the levels of all ATFs factors in fat depots in response to cold, compared with room temperature. Our results demonstrated that ATF3 is the most highly induced in BAT in response to these stimuli (Fig. 5A and Fig. S1) and among the factors with the highest induction in scWAT. Further analysis of fat tissue samples obtained from vehicle- and CL-treated mice revealed that ATF3 is the most highly induced ATF factor in both BAT and scWAT in response to this β-adrenergic agonist, compared with control treated mice (Fig. 5B). Although ATF3 appears to be similarly induced in males and females in response to cold and pharmacological stimuli in BAT and scWAT, other ATF family members, such as ATF4, 5, 6, and 7 showed differential inducibility between males and females, which appeared to be depot- and stimulus-dependent. These data suggest distinct mechanisms in males and females underlying the regulation of the expression of these genes. Comparison of the basal, unstimulated levels of all the ATFs demonstrated that ATF3 mRNAs are higher, or comparable, to those of ATF4, 5, 6, and 7 and CREB but lower than ATF1 and 2, in both BAT and scWAT, with the exception of ATF4, which appears to be higher than ATF3 selectively in BAT (Fig. S2). Given that ATFs family members are known to bind to DNA either as homodimers or heterodimers (18) and because ATF6 is the second most highly induced ATF factor after ATF3 in BAT in males (Fig. 5, A and B), we sought to investigate whether ATF6 may participate in HSF1 regulation via heterodimerization with ATF3 and via binding to the HSF1 promoter. As shown in Fig. S3, ATF3 antibodies did not immunoprecipitate ATF6 in differentiated adipocytes, and no occupancy of ATF6 at the HSF1 promoter at the −258 to −250 site was observed in these cells by ChIP assays.
Figure 5.
ATF3 is induced by cold exposure and pharmacological activation of β-adrenergic signaling in thermogenic tissues in vivo and by cAMP activators in vitro. A, mRNA levels of ATF/CREB family members in BAT and scWAT of 9-week-old male and female mice exposed to room temperature or cold for 6 h (n = 3). B, mRNA levels of ATF/CREB family members in BAT and scWAT of 9-week-old male and female mice treated for 3 h with saline or CL316,243 (1 mg/kg of body weight) resuspended in saline (n = 3). The dotted lines correspond to the levels of the control, set as 1. C, mRNA levels of ATF/CREB family in differentiated mouse SVF cells isolated from scWAT treated for 4 h with 10 μm forskolin. D, time course of the induction of mRNA levels of ATF/CREB family in brown-like differentiated 10T1/2 cells treated either with 10 μm forskolin, 10 μm isoproterenol, or 500 μm IBMX for the indicated time. The results are expressed as means ± S.E. from three independent experiments. *, p value < 0.01; **, p value < 0.005; ***, p value <0.001.
Given the paucity of data on the regulation of ATF3 levels in adipocytes, we assessed whether ATF3 is expressed in differentiated cells and whether its levels may be controlled by compounds that modulate cAMP. As shown in Fig. 5 (C and D), modulators of cAMP levels increased ATF3 mRNAs in differentiated SVF cells isolated from scWAT and in 10T1/2 cells (Fig. 5, C and D). ATF3 appeared to be the most highly induced among all ATF family members also in response to modulators of cAMP levels in vitro, similar to what observed in vivo (Fig. 5, C and D). Taken together these results demonstrate for the first time that ATF3 expression is highly regulated at the transcriptional level by thermogenic stimuli, both in vitro and in vivo.
ATF3 directly controls HSF1 mRNA levels
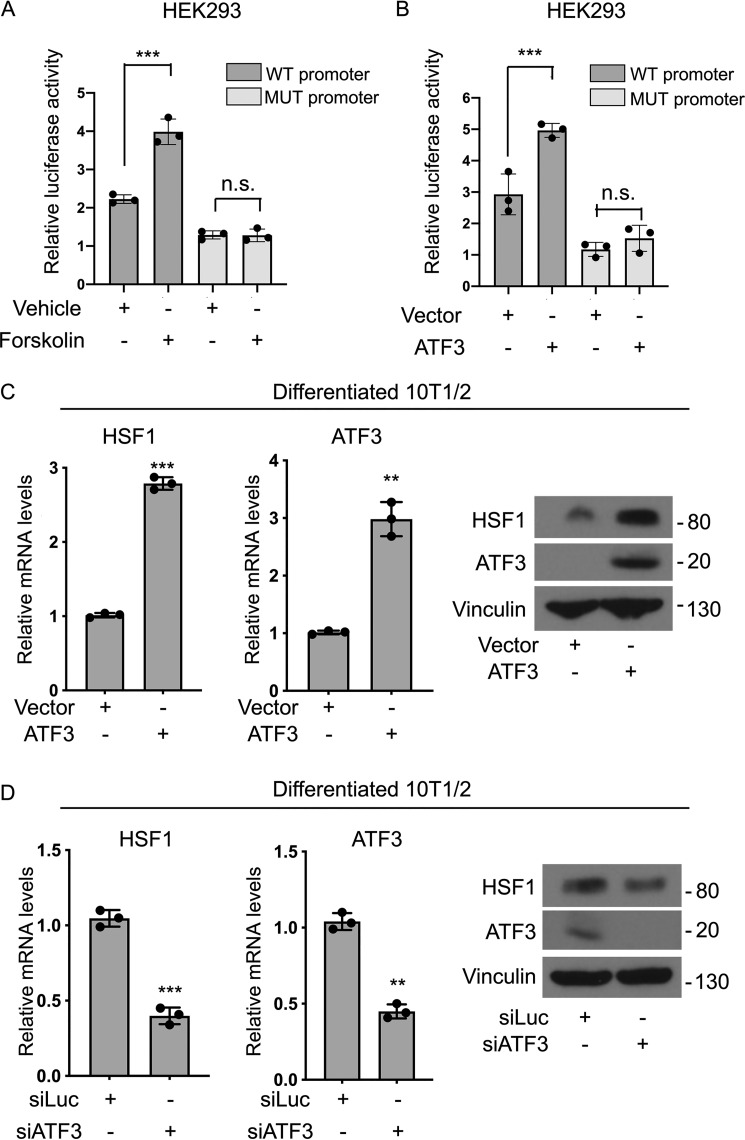
To confirm that ATF3 directly modulates HSF1 levels in response to thermogenic signals, we performed luciferase assays and gain- and loss-of-function studies. We first generated a HSF1 promoter reporter gene linked to luciferase containing 2 kb of the sequence upstream of the HSF1 transcription start site, which includes the putative ATF3-binding motif we newly identified. In addition to this WT-HSF1 promoter luciferase construct, we generated also a reporter containing a mutated putative ATF3-responsive element to assess its requirement for HSF1 regulation. As shown in Fig. 6A, luciferase assays revealed that the ATF3-binding site is required for the induction of the reporter gene in response to forskolin, confirming the involvement of the ATF3-binding motif located in the proximal promoter region in the regulation of HSF1 gene expression. Co-expression of ATF3 with the reporter constructs demonstrated that luciferase activity is induced only when ATF3 is co-transfected with the WT-HSF1 promoter but not in the presence of the mutated reporter (Fig. 6B). These results demonstrate that ATF3 is required for the induction of the HSF1 promoter luciferase reporter. To further establish the sufficiency and necessity of ATF3 for the regulation of the expression of endogenous HSF1 in cells, we performed gain- and loss-of-function experiments. As shown in Fig. 6C, overexpression of ATF3 in differentiated 10T1/2 cells led to the induction of HSF1, whereas ATF3 knockdown decreased HSF1 levels (Fig. 6D). All together, these data point for the first time to ATF3 as a novel regulator of HSF1 levels in brown and beige fat in response to classic extrinsic signals that control thermogenesis.
Figure 6.
ATF3 is required for the induction of HSF1 in response to elevation of cAMP levels. A, luciferase assays in HEK 293 cells expressing either a WT-HSF1-luc reporter vector containing the putative proximal ATF3 motif (WT promoter) or a HSF1 mutated luciferase reporter vector containing a mutated ATF3-binding site (MUT promoter) treated with vehicle or 10 μm forskolin for 4 h before harvesting. B, luciferase assay in HEK 293 cells co-transfected with either vector or ATF3 expression plasmid with either a WT (WT promoter) or a mutated HSF1-luc reporter promoter (MUT promoter) construct. C and D, RNA and protein levels of HSF1 and ATF3 in differentiated 10T1/2 brown-like cells expressing either vector, an ATF3 expression plasmid (C), a control siRNA (siLuc), or a siATF3 (D), treated with 10 μm forskolin for 2 h before harvesting. Vinculin was used as a loading control for Western blots. The results are expressed as means ± S.E. from three independent experiments. **, p value < 0.005; **, p value < 0.001. n.s., not significant.
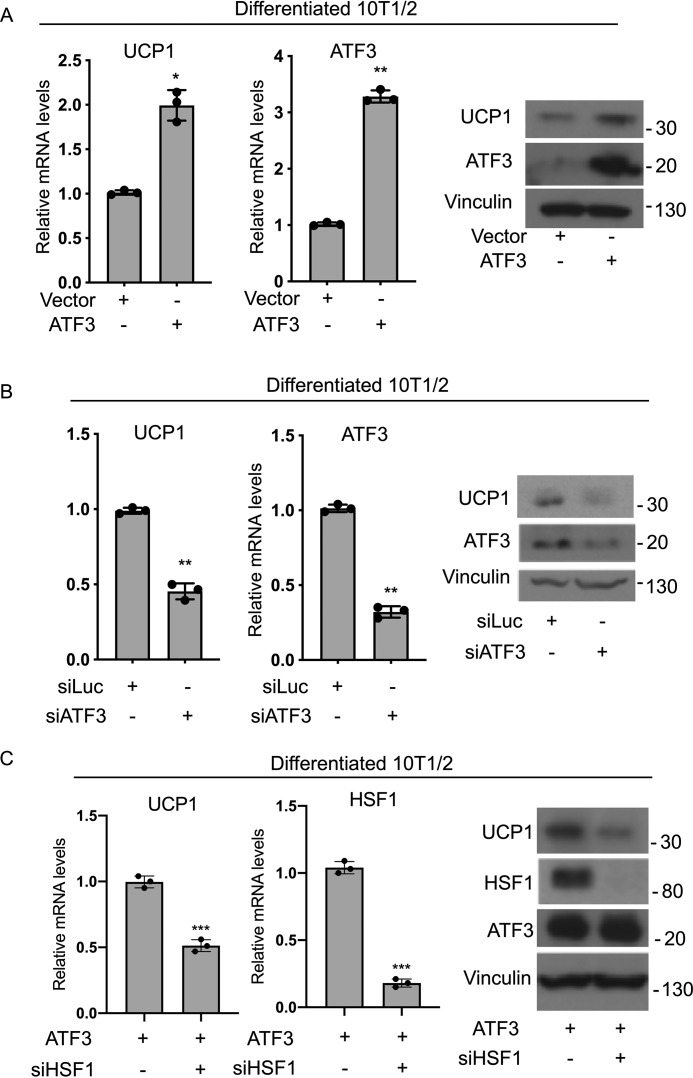
To assess whether ATF3 expression is required for the induction of UCP1 in brown-like adipocytes in vitro, we performed gain- and loss-of-ATF3-function in brown-like differentiated cells. As shown in Fig. 7 (A and B), ATF3 is sufficient and necessary to induce UCP1 in brown adipocytes. To test whether ATF3 requires the induction of HSF1 to regulate UCP1 in brown-like adipocytes, we assessed the effects of ATF3 overexpression on the levels of UCP1 in the presence or absence of HSF1. As shown in Fig. 7C, ATF3-mediated induction of UCP1 is reduced when HSF1 is knocked down. These results suggest that HSF1 is required, at least in part, for ATF3-mediated regulation of the brown fat functional marker UCP1.
Figure 7.
Requirement of the ATF3/HSF1 axis in the regulation of the brown fat marker UCP1. A and B, mRNA and protein levels of UCP1 in brown differentiated 10T1/2 cells after overexpression of ATF3 (A) or knockdown of ATF3 using siRNA (B). C, mRNA levels of UCP1 and HSF1 and protein levels of UCP1, HSF1, and ATF3 levels in differentiated 10T1/2 cells overexpressing ATF3 in the presence of either siRNA control (siLuc) or siHSF1. The results are expressed as means ± S.E. from three independent experiments. *, p value < 0.01; **, p value < 0.005; ***, p value <0.001.
Discussion
We have previously demonstrated that the stress-induced heat shock factor HSF1 is a novel regulator of energy metabolism, involved in the induction of the expression of the metabolic coactivator PGC1α in thermogenic cells (13). However, the questions of whether HSF1 is depot-selectively expressed in mature fat tissues in vivo and whether its levels are transcriptionally controlled in these fat tissues by classic stimuli that regulate the functionality of thermogenic adipose tissues have remained unanswered. The results presented here provide for the first time evidence that HSF1 is expressed at higher levels in thermogenic adipocytes and brown fat tissues than in classic white adipocytes, that HSF1 is induced in response to thermogenic stimuli in vitro and in vivo, and that it is transcriptionally controlled by the stress-induced transcription factor ATF3.
It has been previously shown that cold and β-adrenergic stimulation can rapidly induce the expression of factors involved in brown fat thermogenesis and functionality in fat tissues (17, 19, 20). Our data showing that cold exposure and elevation of cAMP levels lead to an increase in HSF1 mRNA levels provide the first evidence demonstrating that HSF1 is transcriptionally controlled by thermogenic stimuli in adipocytes and brown/beige fat tissues. These results pointing to a rapid induction of HSF1 levels is interesting in light of the fact that HSF1 has been previously shown to be predominantly regulated at the post-translational level in other tissues (21).
Our data link for the first time the stress-inducible transcription factor ATF3 to the transcriptional control of HSF1 by demonstrating that ATF3 binds to an upstream regulatory sequence present in the proximal region of the HSF1 promoter and that it is required for HSF1 induction. In addition, our in-depth analysis of the pattern of expression of ATF3 in fat tissues revealed that ATF3 is turned on at the transcriptional level in brown and beige tissues by thermogenic stimuli. These data suggest a possible role for ATF3 in the positive regulation of thermogenic responses via transactivation of HSF1, in addition to ATF3 recently described repressive effects on carbohydrate-responsive element-binding protein and stearoyl-CoA desaturase-1 (22). Further molecular studies focusing on the promoter of ATF3 will determine the specific mechanisms through which cAMP elevation leads to ATF3 transcriptional regulation in adipocytes.
Our survey of the levels of ATF family members in response to cold exposure and CL treatment has revealed for the first time that ATF3 is among the ATFs most highly induced at the transcriptional level by these stimuli in adipose tissue, supporting a critical role for ATF3 in the regulation of transcriptional responses to thermogenic stimuli leading to control of brown/beige fat tissue functionality. Although it has been shown that mice with total ablation of ATF3 develop obesity and insulin resistance (23) and have decreased browning (22), the genetic requirements for this transcription factor specifically in adipose tissues have not yet been investigated. Future studies involving the generation of new animal models with conditional knockout of ATF3 in adipose tissues will reveal the extent of ATF3 contribution to thermogenic functionality in fat tissues. Overall our data demonstrating that HSF1 is selectively expressed in brown/beige fat, that both HSF1 and ATF3 are turned on in response to thermogenic cues, and that ATF3 controls HSF1 levels provide novel insights into the specific molecular mechanisms controlling brown and beige fat functionality in response to thermogenic signals.
Experimental procedures
Cell culture and treatments
We used 10T1/2 and HEK-293 cells purchased from the American Type Culture Collection. The cells were maintained at 37 °C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% of fetal bovine serum (FBS; Thermo Fisher Scientific, catalog no. NC0959573) and with 1% penicillin/streptomycin (pen/strep; Thermo Fisher Scientific, catalog no. 15070063). SVF cells were obtained through isolation of cells from scWAT, according to a procedure previously described (24). Briefly, scWAT fat tissue pads were dissected from 9-week-old male mice, washed in 1× PBS buffer (Corning, catalog no. 21-040-CV), supplemented with 1% pen/strep, minced into small pieces with a sterile scissor, and digested with 1 mg/ml collagenase type IV (Roche, catalog no. 10269638001) for 1 h at 37 °C while shaking. The cell suspension was filtered using a 70-μm cell strainer (BD Falcon, catalog no. 352350) to remove any clump and debris and subsequently spun at 200 × g for 10 min. The pellet containing the SVF cells was resuspended in DMEM (Corning, catalog no. 10-013-CV), supplemented with 10% FBS and 1% pen/strep, plated in 12-well tissue culture plates, and maintained in DMEM containing 10% FBS and 1% pen/strep at 37 °C in conditions of 5% CO2 (13).
For brown-like differentiation, the cells were grown to confluence and induced to differentiate with a medium containing 10% FBS and 1% pen/strep supplemented with 20 nm insulin (Sigma, catalog no. I1507), 1 nm T3 (Sigma, catalog no. T2877), 125 μm indomethacin (Sigma, catalog no. I7378), 1 μm dexamethasone (Sigma, catalog no. D4902), 1 μm rosiglitazone (Sigma, catalog no. 557366-M), and 0.5 μm isobutylmethylxanthine (IBMX; Sigma, catalog no. I5879). 2 days after induction for 10T1/2 cells, or 4 days for scWAT SVF cells, the medium was replaced with maintenance medium containing DMEM supplemented with 10% FBS, 1% pen/strep, 1 nm T3, and 20 nm insulin and changed every 48 h. Differentiated brown-like adipocytes were stimulated with forskolin (Sigma, catalog no. F6886), isoproterenol (Sigma, catalog no. I6504), or IBMX for the time and the concentrations indicated in the legends, prior to harvesting RNA and/or protein.
Mice
8-week-old C57BL/6J male and female mice were purchased from Jackson Laboratories (catalog no. 000664) and exposed to a 12-h light/12-h dark cycle with free access to food and water. The mice were housed in the animal facility at 24 °C for 1 week to acclimate before the initiation of the studies. For cold exposure experiments, 9-week-old mice were kept at room temperature or placed at 4 °C for 6 h. The mice were then euthanized, and fat depots were harvested for subsequent analysis. To study the effects of pharmacological stimulation of the β-adrenergic pathway, 9-week-old mice were injected i.p. with saline or CL316,243 (Sigma, catalog no. C5976) dissolved in saline at 1 mg/kg/body weight. 3 hours after injection, the mice were euthanized, and adipose depots were harvested for molecular analyses. All animal procedures used in this study were approved by the Institutional Animal Care and Use Committee of New York University Langone Medical Center.
Plasmids and site-directed mutagenesis
The ATF3 expression plasmid was obtained from Origene (catalog no. MR201634); the pGL3 luciferase reporter vector (catalog no. E1751) and the pRL Renilla luciferase control reporter vector (catalog no. E2231) were purchased from Promega. The 2-kb sequence upstream of the transcriptional start site of the HSF1 gene was generated based on the reference sequence number ENSMUST 00000 226872.1_0 and cloned into the 5′ KpnI and 3′ XhoI sites of the pGL3vector (Genewiz). Point mutations were introduced in the WT HSF1 promoter luciferase reporter upstream sequence spanning from −258 to −250, by site-directed mutagenesis according to the manufacturer's protocol (New England Biolabs, catalog no. E0554S). The following oligonucleotides were used: HSF1_Fwd, GATCATGTGGGGGCTTACCACACGC; and HSF1_Rev, CCGTCCTCGCTGCCGGG GAAACGA.
Transient transfections
Differentiated 10T1/2 cells were transfected using DharmaFECT (Dharmacon, catalog no. T-2002-03), following the manufacturer's instructions. For ATF3 gain-of-function experiments, vector control- or ATF3-expressing brown-like differentiated 10T1/2 cells were harvested 2 days after transfection. For ATF3 knockdown experiments, brown-like differentiated 10T1/2 cells were transfected with 35 nm of either siRNA ATF3 (Dharmacon, catalog no. J-058604-13-0002) or siRNA luciferase (Dharmacon, catalog no. D-002050-01-20), used as a control, and harvested 3 days later. For assessing the effects of ATF3 overexpression in the presence or absence of HSF1, differentiated 10T1/2 cells were first transfected with 35 nm of either siRNA luciferase or siRNA HSF1 (Dharmacon, catalog no. L-040660-01-0005). 48 h later, the cells were transfected with the ATF3 expression plasmid and harvested 3 days later.
RNA isolation and RT-PCR analysis
Total RNA from cells was extracted with RNeasy (Qiagen, catalog no. 75144) and from tissues with TRIzol (Thermo Fisher Scientific, catalog no. 15596018). cDNA was reverse-transcribed using 1 μg of total RNA using the iScriptTM cDNA synthesis kit (Bio-Rad, catalog no. 1708890). RT-PCR analysis was performed in triplicate, using 25 ng of cDNA, 300 nm of primers, and iQTM SYBR® Green Supermix (Bio-Rad, catalog no. 1708880), according to manufacturer's instructions. Relative mRNA quantification was calculated using the ΔΔCt method, by normalizing each sample to the average change in cycle threshold value of the 36B4 gene used as control. For quantitative PCR analysis, the following primers were used: HSF1 forward, AGGCAGGAGCATAGATGAGA; HSF1 reverse, AGGATGGAGTCAATGAAGGC; ATF3 forward, GCCAAGTGTCGAAACAAGAAAAAG; ATF3 reverse, TCCTCGATCTGGGCCTTCAG; Ucp1 forward, GGCCCTTGTAAACAACAAAATAC; Ucp1 reverse, GGCAACAAGAGCTGACAGTAAAT; CREB forward, AGCAGCTCATGCAACATCATC; CREB reverse, AGTCCTTACAGGAG GA CTGAACT;ATF1 forward, AGACCTACCAGATCGTACCA; ATF1 reverse, GTCATCACCACGGTCTGC; ATF2 forward, CAGTGGATTGGTTGAGACTCAGTC; ATF2 reverse, GAGGAGTTGTGTGAGCTGGAGAAG; ATF4 forward, TCGATGCTCTGTTTCGAATG; ATF4 reverse, AGAATGTAAAGGGGGCAACC; ATF5 forward, GGCTGGCTCGTAGACTATGG; ATF5 reverse, CCAGAGGAAGGAGAGCTGTG; ATF6 forward, AATTCTCAGCTGATGGCTGT; ATF6 reverse, TGGAGGATCCTGGTGTCCAT; ATF7 forward, GGGTGTCCTCCCTGGAAAAG; ATF7 reverse, TCAGCTGAGCCACCTCATTGGGCAACAAGAGCTGACAGTAAAT; 36B4 forward, GCTT CATTGTGGGAGCAGAC; and 36B4 reverse, ATGGTGTTCTTGCCCATCAG.
Western blotting analysis and antibodies
To obtain protein extracts, adherent cells were washed with cold PBS, and snap-frozen tissues were homogenized with a tissue grinder (Powergen 700, Thermo Fisher Scientific, catalog no. GLH 115) and lysed with radioimmune precipitation assay buffer containing 20 mm Tris, 150 mm NaCl, 1% Triton X-100, supplemented with a mixture of protease inhibitors (Thermo Fisher Scientific, catalog no. PIA32953). 20 μg of protein lysates were run on 8% SDS-polyacrylamide gels and transferred onto 0.45-μm polyvinylidene difluoride membranes (Millipore, catalog no. IPVH00010). The blots were incubated with 5% nonfat dry milk (w/v) resuspended in 0.1% TBST buffer containing 50 mm Tris-HCl, 150 mm NaCl, pH 7.4, and 0.1% Tween 20 for 1 h at room temperature and subsequently incubated with primary antibodies overnight at 4 °C in 0.1% TBST buffer solution containing 1% BSA (Thermo Fisher Scientific; BP9703–100). The following antibodies were used at the indicated dilutions: anti-HSF1 (Cell Signaling Technology, catalog no. 4356) at 1:1000, anti-UCP1 (Abcam, catalog no. ab23841) at 1:1000, anti-ATF3 (Abcam, catalog no. ab207434) at 1:1000, anti-phospho-CREB Ser-133 (Cell Signaling, catalog no. 9198S) at 1:1000, anti-CREB (Cell Signaling, catalog no. 4820S) at 1:1000, anti-ATF1 (Proteintech, catalog no. J-057626-09-0002) at 1:1000, anti-ATF2 (Cell Signaling Technology, catalog no. 35031) at 1:1000, anti-ATF4 (Proteintech, catalog no. 10835–1-AP) at 1:1000, anti-ATF5 (Abcam, catalog no. ab184923) at 1:1000, anti-ATF6 (Abcam, catalog no. ab37149), anti-ATF7 (Abcam, catalog no. ab183507) at 1:1000, anti-β-actin (Cell Signaling Technology, catalog no. 3700S) at 1:1000, and anti-vinculin (Proteintech, catalog no. 66305-1-Ig) at 1:2000. After primary antibody incubation, the membranes were washed in TBST 0.1% (v/v) and incubated for 1 h at room temperature with an anti-rabbit (Bio-Rad, catalog no. 1706515) or anti-mouse (Bio-Rad, catalog no. 1706516) IgG horseradish peroxidase-=conjugated antibody in 0.1% TBST containing 2% nonfat dry milk (w/v) at a 1:20,000 dilution. After four additional washes in TBST 0.1% (v/v), the blots were developed using enhanced chemiluminescence (GE Healthcare, catalog no. RPN2108) using Hyblot CL autoradiography films (Thomas Scientific, catalog no. E3012) with an X-ray film developer (Konica Minolta, catalog no. SRX-101A).
Genomic data analysis
To identify the occurrence and frequency of ATF transcription factor–binding motifs in the HSF1 promoter, we used Genomatix software and the PROMO tool.
Electromobility shift assays
To perform EMSAs, we used nuclear extracts obtained from 10T1/2 cells differentiated into brown-like adipocytes and treated for 4 h with 10 μm forskolin (NE-PER kit, Thermo Fisher Scientific, catalog no. 78833). The extracts were incubated with biotin-labeled WT and mutated DNA probes corresponding to the sequence of the ATF3 putative binding element located in position −258 to −250 from the HSF1 transcriptional start site. The mutated oligonucleotide containing the sequence GTAGCACGT, instead of GTGACATGT, in positions −258 to −250, was used, and 100-fold excess of unlabeled WT probe was employed in competition assays. The protein–DNA complexes were resolved on a 4% TBE precast gel (Bio-Rad, catalog no. 4565013) at 4 °C, transferred onto a positively charged nitrocellulose membrane (GE HealthCare, catalog no. RPN3038), and UV cross-linked (Stratalinker, catalog no. 2400). Detection was performed following the manufacturer's instructions (Thermo Fisher Scientific, catalog no. 20148). The following oligonucleotides were used for EMSA assays: WT-promoter oligonucleotide, GCGATGCCGGTGACATGTGGGGCTTA; WT-RC oligonucleotide, TAAGCCCCACATGTCACCGGCATCGC; Mut-promoter oligonucleotide, GCGATGCCGGTTATATATGGGGCTTA; and Mut-RC oligonucleotide, TAAGCCCCATATATAACCGGCATCGC.
ChIP assays
ChIP experiments were performed using a ChIP assay kit (EMD Millipore, catalog no. 17-295), as previously described (13). Briefly, differentiated 10T1/2 cells vehicle or forskolin treated for 4 h were cross-linked by adding formaldehyde into the cell culture medium to a final concentration of 1%, subsequently washed with cold 1× PBS buffer supplemented with protease inhibitors (Millipore, catalog no. 20-283) and harvested according to the manufacturer's specifications. Sonication was performed on ice using Diagenode Bioruptor® for 30 min. The complexes between protein and DNA were immunoprecipitated overnight at 4 °C using either an anti-ATF3 antibody (Cell Signaling, catalog no. 33593), an anti-ATF6 antibody (Abcam, catalog no. ab37149), or an IgG antibody (Millipore, catalog no. 17-295) used as control. The complexes were then eluted at room temperature for 15 min, using elution reagent C (Millipore, catalog no. 20-294), after incubation with protein A–agarose beads (Millipore, catalog no. 16-201C). DNA fragments were purified using low- and high-salt immune buffer, as per vendor's protocol instructions and amplified by RT-PCR using primers designed to recognize the specific sequence spanning the putative ATF3-binding motif identified in position −258 to −250: PromoHSF1 forward, CTGATCAGGTTCCAAACGTC; and PromoHSF1 reverse, GAAGCGGCAGTAGATTCGGTAT
Co-immunoprecipitation assays
Co-immunoprecipitation assays were performed using PierceTM co-immunoprecipitation kit (Thermo Fisher Scientific, catalog no. 26149), according to the manufacturer's protocols. Briefly, lysates from differentiated 10T1/2 cells treated with vehicle or forskolin for 4 h were incubated with 5 μg of ATF3 antibody in the presence of 20 μl of the amine reactive coupling resin slurry (provided in the kit) overnight at 4 °C. Immunocomplexes were washed three times with lysis buffer and eluted at room temperature for 5 min, loaded on a 8% SDS-polyacrylamide gel, transferred onto 0.45-μm polyvinylidene difluoride membranes (Millipore, catalog no. IPVH00010) and incubated with either anti-ATF3 (Cell Signaling, catalog no. 33593), anti-ATF6 (Abcam, catalog no. ab37149), or anti-vinculin (Proteintech, catalog no. 66305-1-Ig) antibodies overnight at 4 °C and at room temperature for 1 h with secondary antibodies. The proteins were visualized using enhanced chemiluminescence (GE Healthcare, catalog no. RPN2108) using Hyblot CL autoradiography films (Thomas Scientific catalog no. E3012) with an X-ray film developer (Konica Minolta, catalog no. SRX-101A).
Luciferase reporter assays
pGL3 plasmids expressing the firefly luciferase gene driven by 2 kb of the upstream region of the HSF1 promoter containing either the intact WT putative ATF3-binding motif (WT promoter) or a mutated sequence at position −258 to −250 (MUT promoter) from the transcription start site were used for transient transfection studies. Briefly, HEK-293 cells were plated at the density of 20,000 cells/well in 12-well tissue culture plates. 24 h later, HEK293 cells were co-transfected with 0.5 μg of a reporter luciferase construct containing the HSF1 WT promoter (WT promoter) or with the HSF1 mutated promoter (MT promoter) at the ATF3-binding motif and 0.5 μg of control vector plasmid or 0.5 μg of a plasmid expressing ATF3 and 50 ng of Renilla reporter plasmid using Lipofectamine 2000 (Thermo Fisher Scientific, catalog no. 11668019). 48 h after transfection, the cells were harvested for luciferase analysis or treated with vehicle or forskolin (10 μm) for 4 h before collection, if indicated in the legend. The luciferase reporter activity was measured using the Dual-Luciferase reporter assay system kit (Promega, catalog no. E1910) and detected using the SpectraMax 96-well plate reader (Molecular Devices), normalized to Renilla activity.
Statistical analysis
The results were expressed as mean ± S.E., unless otherwise noted. Student's t test or one-way analysis of variance was used for comparison between groups. p values <0.05 were considered statistically significant. Statistical analyses were performed using the Prism 7 software (GraphPad Software).
Author contributions
N. V., L. P., and E. M. data curation; N. V., L. P., and E. M. software; N. V., L. P., and E. M. formal analysis; N. V., L. P., and E. M. validation; N. V., L. P., and E. M. investigation; N. V., L. P., and E. M. visualization; N. V., L. P., and E. M. methodology; N. V., L. P., and E. M. writing-original draft; E. M. conceptualization; E. M. resources; E. M. supervision; E. M. funding acquisition; E. M. project administration.
Supplementary Material
Acknowledgment
We are very thankful to Dr. Paola Di Marzio for support in the drafting of the manuscript.
This work was supported by start-up funds from New York University (to E. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
- eWAT
- epididymal white fat tissue
- HSF
- heat shock factor
- ATF
- activating transcription factor
- CREB
- cAMP response element–binding protein
- UCP
- uncoupling protein
- BAT
- brown fat tissue
- EMSA
- electrophoretic mobility shift assay
- sc
- subcutaneous
- PGC
- peroxisome proliferator-activated receptor γ coactivator
- SVF
- stromal vascular fraction
- CL
- CL316,243
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- pen/strep
- penicillin/streptomycin
- IBMX
- isobutylmethylxanthine.
References
- 1. Flegal K. M., Kit B. K., Orpana H., and Graubard B. I. (2013) Association of all-cause mortality with overweight and obesity using standard body mass index categories: a systematic review and meta-analysis. JAMA 309, 71–82 10.1001/jama.2012.113905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saydah S., Bullard K. M., Cheng Y., Ali M. K., Gregg E. W., Geiss L., and Imperatore G. (2014) Trends in cardiovascular disease risk factors by obesity level in adults in the United States, NHANES 1999–2010. Obesity (Silver Spring) 22, 1888–1895 10.1002/oby.20761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mueller E. (2016) Browning and graying: novel transcriptional regulators of brown and beige fat tissues and aging. Front. Endocrinol. 7, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giralt M., and Villarroya F. (2013) White, brown, beige/brite: different adipose cells for different functions? Endocrinology 154, 2992–3000 10.1210/en.2013-1403 [DOI] [PubMed] [Google Scholar]
- 5. Kajimura S., Spiegelman B. M., and Seale P. (2015) Brown and beige fat: physiological roles beyond heat generation. Cell Metab. 22, 546–559 10.1016/j.cmet.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harms M., and Seale P. (2013) Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263 10.1038/nm.3361 [DOI] [PubMed] [Google Scholar]
- 7. Sidossis L., and Kajimura S. (2015) Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis. J. Clin. Invest. 125, 478–486 10.1172/JCI78362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hai T., and Hartman M. G. (2001) The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 273, 1–11 10.1016/S0378-1119(01)00551-0 [DOI] [PubMed] [Google Scholar]
- 9. Zhao J., Li X., Guo M., Yu J., and Yan C. (2016) The common stress responsive transcription factor ATF3 binds genomic sites enriched with p300 and H3K27ac for transcriptional regulation. BMC Genomics 17, 335 10.1186/s12864-016-2664-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okamoto A., Iwamoto Y., and Maru Y. (2006) Oxidative stress-responsive transcription factor ATF3 potentially mediates diabetic angiopathy. Mol. Cell Biol. 26, 1087–1097 10.1128/MCB.26.3.1087-1097.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Allen-Jennings A. E., Hartman M. G., Kociba G. J., and Hai T. (2001) The roles of ATF3 in glucose homeostasis: a transgenic mouse model with liver dysfunction and defects in endocrine pancreas. J. Biol. Chem. 276, 29507–29514 10.1074/jbc.M100986200 [DOI] [PubMed] [Google Scholar]
- 12. Jadhav K., and Zhang Y. (2017) Activating transcription factor 3 in immune response and metabolic regulation. Liver Res. 1, 96–102 10.1016/j.livres.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma X., Xu L., Alberobello A. T., Gavrilova O., Bagattin A., Skarulis M., Liu J., Finkel T., and Mueller E. (2015) Celastrol protects against obesity and metabolic dysfunction through activation of a HSF1–PGC1α transcriptional axis. Cell Metab. 22, 695–708 10.1016/j.cmet.2015.08.005 [DOI] [PubMed] [Google Scholar]
- 14. Gauthier M. S., Miyoshi H., Souza S. C., Cacicedo J. M., Saha A. K., Greenberg A. S., and Ruderman N. B. (2008) AMP-activated protein kinase is activated as a consequence of lipolysis in the adipocyte: potential mechanism and physiological relevance. J. Biol. Chem. 283, 16514–16524 10.1074/jbc.M708177200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mottillo E. P., and Granneman J. G. (2011) Intracellular fatty acids suppress β-adrenergic induction of PKA-targeted gene expression in white adipocytes. Am. J. Physiol. Endocrinol. Metab. 301, E122–E131 10.1152/ajpendo.00039.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim H. B., Kim W. H., Han K. L., Park J. H., Lee J., Yeo J., and Jung M. H. (2010) cAMP-response element binding protein (CREB) positively regulates mouse adiponectin gene expression in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 391, 634–639 10.1016/j.bbrc.2009.11.111 [DOI] [PubMed] [Google Scholar]
- 17. Cao W., Daniel K. W., Robidoux J., Puigserver P., Medvedev A. V., Bai X., Floering L. M., Spiegelman B. M., and Collins S. (2004) p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell Biol. 24, 3057–3067 10.1128/MCB.24.7.3057-3067.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hai T., Wolfgang C. D., Marsee D. K., Allen A. E., and Sivaprasad U. (1999) ATF3 and stress responses. Gene Expr. 7, 321–335 [PMC free article] [PubMed] [Google Scholar]
- 19. Dempersmier J., Sambeat A., Gulyaeva O., Paul S. M., Hudak C. S., Raposo H. F., Kwan H. Y., Kang C., Wong R. H., and Sul H. S. (2015) Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol. Cell 57, 235–246 10.1016/j.molcel.2014.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen H. Y., Liu Q., Salter A. M., and Lomax M. A. (2013) Synergism between cAMP and PPARγ signalling in the initiation of UCP1 gene expression in HIB1B brown adipocytes. PPAR Res. 2013, 476049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vihervaara A., and Sistonen L. (2014) HSF1 at a glance. J. Cell Sci. 127, 261–266 10.1242/jcs.132605 [DOI] [PubMed] [Google Scholar]
- 22. Cheng C. F., Ku H. C., Cheng J. J., Chao S. W., Li H. F., Lai P. F., Chang C. C., Don M. J., Chen H. H., and Lin H. (2019) Adipocyte browning and resistance to obesity in mice is induced by expression of ATF3. Commun. Biol. 2, 389 10.1038/s42003-019-0624-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim S., Song N. J., Bahn G., Chang S. H., Yun U. J., Ku J. M., Jo D. G., and Park K. W. (2018) Atf3 induction is a therapeutic target for obesity and metabolic diseases. Biochem. Biophys. Res. Commun. 504, 903–908 10.1016/j.bbrc.2018.09.048 [DOI] [PubMed] [Google Scholar]
- 24. Perie L., Verma N., Xu L., Ma X., and Mueller E. (2019) Transcriptional regulation of ZNF638 in thermogenic cells by the cAMP response element binding protein in male mice. J. Endocr. Soc. 3, 2326–2340 10.1210/js.2019-00238 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.