Abstract
The transfer of a phosphate from ATP to a protein substrate, a modification known as protein phosphorylation, is catalyzed by protein kinases. Protein kinases play a crucial role in virtually every cellular activity. Recent studies of atypical protein kinases have highlighted the structural similarity of the kinase superfamily despite notable differences in primary amino acid sequence. Here, using a bioinformatics screen, we searched for putative protein kinases in the intracellular bacterial pathogen Legionella pneumophila and identified the type 4 secretion system effector Lpg2603 as a remote member of the protein kinase superfamily. Employing an array of biochemical and structural biology approaches, including in vitro kinase assays and isothermal titration calorimetry, we show that Lpg2603 is an active protein kinase with several atypical structural features. Importantly, we found that the eukaryote-specific host signaling molecule inositol hexakisphosphate (IP6) is required for Lpg2603 kinase activity. Crystal structures of Lpg2603 in the apo-form and when bound to IP6 revealed an active-site rearrangement that allows for ATP binding and catalysis. Our results on the structure and activity of Lpg2603 reveal a unique mode of regulation of a protein kinase, provide the first example of a bacterial kinase that requires IP6 for its activation, and may aid future work on the function of this effector during Legionella pathogenesis.
Keywords: bacterial protein kinase, bacterial pathogenesis, inositol phosphate, enzyme structure, allosteric regulation, effector, inositol hexikisphosphate, Legionella
Introduction
Protein kinases are a class of enzymes that catalyze phosphorylation, a post-translational modification involving the transfer of the terminal phosphate from ATP to protein substrates (1). The human genome encodes over 500 protein kinases that play a crucial role in cellular function and are implicated in virtually every cellular activity (2). The protein kinase superfamily can be broadly divided into two groups: the eukaryotic protein kinases and the atypical protein kinases. Eukaryotic protein kinases consist of an N-lobe and a C-lobe that harbor conserved amino acid motifs necessary for catalytic function. In contrast, atypical protein kinases lack easily detectable sequence similarity to the eukaryotic protein kinases but still retain catalytic activity. Several atypical members of the kinase superfamily have emerged in recent years that share the structural kinase fold despite significant sequence divergence (3).
We have taken a bioinformatics approach to analyze and identify these atypical and uncharacterized members of the protein kinase superfamily. With this strategy, we identified the Golgi casein kinase, Fam20C, that phosphorylates serine residues within the Ser-X-Glu/pSer consensus motif, found in roughly 75% of human plasma and cerebrospinal fluid phosphoproteins (4, 5). While searching for proteins with sequence similarity to Fam20C, we identified the atypical protein kinase, CotH, present in many bacterial and eukaryotic spore forming organisms. CotH phosphorylates spore coat proteins for effective germination in Bacillus subtilis (6). Furthermore, we identified a distant member of the kinase superfamily, the SelO family, that is conserved from bacteria to humans and catalyzes protein AMPylation instead of phosphorylation (7).
A subset of proteins that our bioinformatics approach continues to bring to the forefront are bacterial effector proteins. Bacterial effector proteins serve as a diverse pool of proteins in which many atypical and novel biological mechanisms can be found (8). HopBF1, for example, is a family of bacterial effector proteins from Pseudomonas syringae that utilize a novel molecular “mimicry” to phosphorylate host cell Hsp90 to evade the host immune defense during infection (9). Likewise, Legionella pneumophila, a Gram-negative intracellular pathogen known to be the causative agent of Legionnaires disease, contains a plethora of effector proteins with exciting biology. L. pneumophila translocates over 300 effectors to alter host cellular processes to form a replicative niche and evade degradation (10, 11). One such effector protein, SidJ, also retains a protein kinase-like fold but catalyzes protein polyglutamylation that depends on the host cofactor calmodulin (12–15).
Interestingly, the L. pneumophila genome encodes for five eukaryotic-like protein kinases that manipulate host cell signaling (16, 17). LegK1–4 and LegK7 are serine/threonine kinases that target various host pathways including actin remodeling, protein synthesis, the immune response, the protein folding machinery and the Hippo pathway (18–23). In search of atypical protein kinases in L. pneumophila, we identified a kinase domain in the type 4 secretion system effector protein, Lpg2603 (lem28, sdmB), which has also been predicted to have a kinase fold by Burstein et al. (24). Previously, a conserved phosphatidylinositol-4-phophate (PI4P)–binding3 domain at the C terminus of Lpg2603 was identified and shown to localize the protein to the Legionella-containing vacuole during infection (25). This localization is shared by L. pneumophila type 4 secretion system effectors Lpg1101/Lem4 and DrrA/sidM, which harbor the conserved PI4P-binding domain at their C termini but differ in their N-terminal domains (25). DrrA is a multidomain effector with a nucleotidyltransferase domain and a guanine nucleotide exchange factor domain in addition to the PI4P-binding domain (26, 27). In contrast, Lpg1101 contains an N-terminal haloacid dehalogenase domain with tyrosine phosphatase activity (28). However, the structure and biochemical activity of Lpg2603 remained uncharacterized.
Here, we show that Lpg2603 is an active protein kinase with several unusual structural features not typical of canonical protein kinases. Importantly, we found that Lpg2603 requires binding of the host cofactor inositol hexakisphosphate (IP6) for activation through a unique mode of active site rearrangement. Our results not only provide insights into the mechanism by which Lpg2603 catalyzes phosphorylation but also highlight a unique mechanism of allosteric regulation of kinases by IP6.
Results
Lpg2603 has sequence similarity to protein kinases
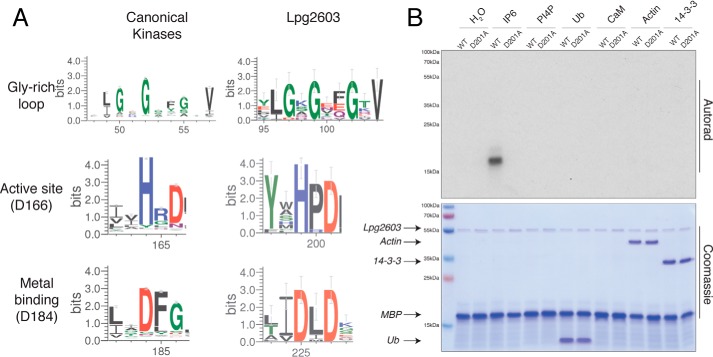
A bioinformatic screen for Legionella effectors bearing sequence similarity to protein kinases was performed using the FFAS algorithm (29) and yielded Lpg2603 as a likely protein kinase, with 10–18% sequence identity to known bacterial kinases including OspG, NleH, YopO, PpkA, and several human kinases. The FFAS alignments allowed unequivocal assignments of the typical features of protein kinases: the Gly-rich ATP-binding loop, as well as the predicted catalytic Asp201 and Mg2+ ion-binding Asp225 (Fig. 1A). In contrast, the ion pair Lys72 and Glu91 (protein kinase A (PKA) nomenclature) are absent in Lpg2603 (Fig. S1). Homologs of Lpg2603 are found only in a range of strains from the Legionella/Fluoribacter genus.
Figure 1.
Lpg2603 is an active kinase in the presence of IP6. A, sequence logo built using 74 homologs of Lpg2603 depicting conserved motifs present in kinases including the Gly-rich loop and active site residues (numbering in parentheses corresponds to PKA). The logo for canonical kinases was built using 3998 homologs (Pfam domain PF00069). B, autoradiograph depicting the incorporation of 32P from [γ-32P]ATP into MBP by recombinant Lpg2603 WT or the inactive mutant, D201A, in the presence of different cofactors: IP6, PI4P, ubiquitin (Ub), calmodulin (CaM), globular actin, or 14-3-3 protein. Note that CaM migrates at the same molecular weight as MBP. The reactions were terminated after 10 min by the addition of EDTA, and the products were resolved by SDS-PAGE and visualized by Coomassie Blue staining (lower panel) and autoradiography (upper panel). The results are representative of three independent experiments.
Lpg2603 is an active kinase
To determine whether Lpg2603 is an active kinase, we expressed L. pneumophila Lpg2603 in Escherichia coli as a His6–Sumo fusion protein and purified the protein by nickel–nitrilotriacetic acid affinity chromatography. We also purified recombinant Lpg2603 containing an alanine mutation in the predicted catalytic Asp201 (PKA nomenclature Asp166). Following removal of the His6–Sumo tag, we performed kinase assays in the presence of [γ-32P]ATP. Recombinant L. pneumophila Lpg2603, however, did not phosphorylate the generic protein kinase substrate, myelin basic protein (MBP) (Fig. 1B, first and second lanes). Bacterial effectors often utilize eukaryotic specific cofactors to regulate their activity within the host cell while remaining inactive in the bacterial cell (30). Therefore, we tested the activity of Lpg2603 in the presence of some common cofactors. Remarkably, WT Lpg2603, but not the predicted catalytically inactive D201A mutant, phosphorylated MBP in the presence of inositol hexakisphosphate (IP6) but none of the other cofactors that we tested (Fig. 1B).
Lpg2603 requires IP6 for optimal activity
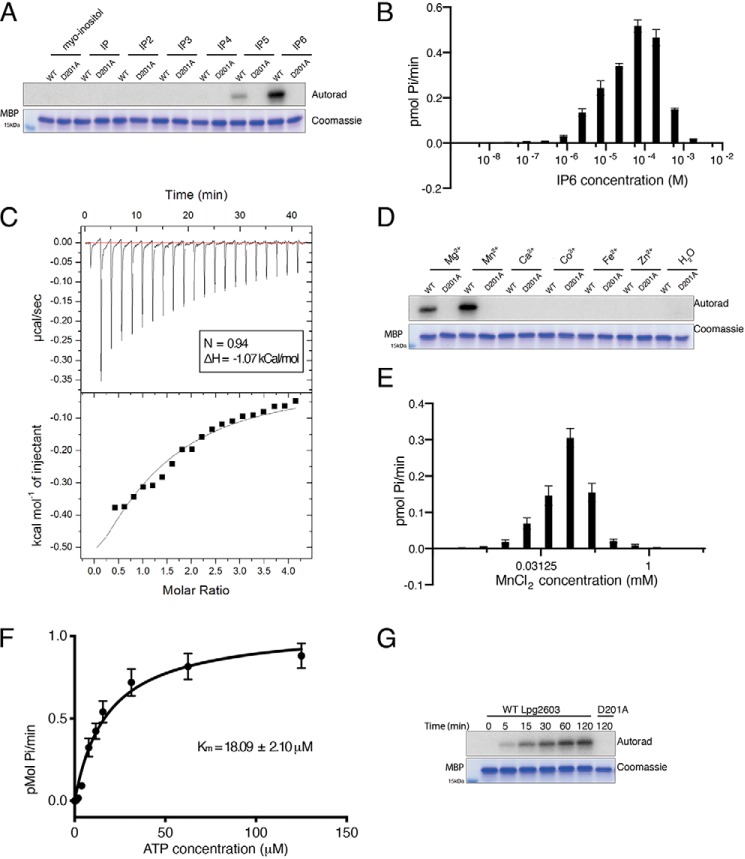
Given the structural similarity in inositol phosphates, we tested whether other inositol phosphates could activate Lpg2603. Lpg2603 displayed a strong preference for IP6 compared with myo-inositol, inositol phosphate, inositol diphosphate, inositol triphosphate, inositol tetraphosphate, and inositol pentaphosphate with optimal MBP phosphorylation observed at ∼65 μm IP6 (Fig. 2, A and B). To determine the binding affinity of IP6 to Lpg2603, we measured the dissociation constant (Kd) using isothermal titration calorimetry (ITC). IP6 bound to Lpg2603 with a Kd of ∼315 μm, which is in accordance with eukaryotic cellular IP6 concentration (Fig. 2C) (31).
Figure 2.
Lpg2603 activity requires IP6 and Mn2+. A, in vitro kinase assay showing incorporation of 32P from [γ-32P]ATP into MBP by Lpg2603 in the presence of myo-inositol, inositol phosphate (IP), inositol diphosphate (IP2), inositol triphosphate (IP3), inositol tetraphosphate (IP4), inositol pentaphosphate (IP5), and IP6. The reactions were terminated and analyzed as in Fig. 1B. The results are representative of three independent experiments. B, incorporation of γ-32P from [γ-32P]ATP into MBP by Lpg2603 with increasing concentrations of IP6. The reactions were terminated after 10 min by the addition of EDTA, the products were resolved by SDS-PAGE, and Coomassie-stained MBP bands were excised for scintillation counting. The results are representative of three independent experiments. The error bars represent standard deviation from three replicates from an individual experiment. C, representative isothermal calorimetry data for Lpg2603 binding to IP6. Lpg2603 is at 200 μm in the cell, and IP6 is at 8 mm in the titration syringe for a final molar ratio of 1:4. Best fit parameters were as follows: n = 0.94; ΔH = −1.07 kCal/mol. D, in vitro kinase assay showing incorporation of γ-32P from [γ-32P]ATP into MBP by Lpg2603 in the presence or absence of MgCl2, MnCl2, CaCl2, CoCl2, FeCl2, and ZnCl2. The reactions were terminated and analyzed as in Fig. 1B. The results are representative of three independent experiments. E, incorporation of γ-32P from [γ-32P]ATP into MBP by Lpg2603 with varying concentrations of MnCl2. The reactions were terminated and analyzed as in B. The results are representative of three independent experiments. The error bars represent standard deviation from three replicates from an individual experiment. F, kinetic analysis depicting the concentration dependence of Mn2+/ATP on the rate of MBP phosphorylation by Lpg2603. Km for Mn2+/ATP = 18.09 ± 2.10 μm; Vmax = 1.05 ± 0.04 pmol/min Reactions were terminated and analyzed as in B. The results are representative of three independent experiments. The error bars represent standard error of the mean from three replicates from an individual experiment. Kinetic measurements were fitted into the Michaelis–Menten equation using GraphPad Prism. G, time-dependent incorporation of γ-32P from [γ-32P]ATP into MBP by Lpg2603. The reactions were terminated and analyzed as in Fig. 1B. The results are representative of three independent experiments.
We next assayed the optimal divalent metal for kinase activity. Lpg2603, but not the inactive mutant, phosphorylated MBP in the presence of Mg2+and Mn2+ (Fig. 2D). Lpg2603 demonstrates a preference for Mn2+ which, interestingly, is inhibitory at high concentrations (Fig. 2E). Similar to canonical kinases, Lpg2603 has a Km for ATP of 18 μm (Fig. 2F). Furthermore, Lpg2603, but not the inactive mutant, phosphorylated MBP in a time-dependent manner (Fig. 2G). Collectively, our results demonstrate that Lpg2603 is a bacterial effector kinase that requires IP6 for catalytic activity.
Crystal structure of Lpg2603 reveals an IP6-binding pocket and mechanism of activation
To gain further insight into the mechanism of Lpg2603 activity, we solved the crystal structure of a fragment of Lpg2603, which lacks the PI4P-binding region at a resolution of 2.10 Å (Fig. 3A). The apo structure displayed a unique N-terminal extension that is rich in β-strands. There was no observable electron density for residues 76–117 of the N-lobe, indicating a flexible protein region. Electron density is absent for residues that are important for canonical kinase activity including the Gly-rich loop and ion pair Lys from the VAIK motif. The C-lobe consists of five α-helices including the catalytic Asp201 and metal-binding Asp225. The apo structure can be superimposed onto PKA with an RMSD of 4.8 Å over 178 C-α backbone atoms.
Figure 3.
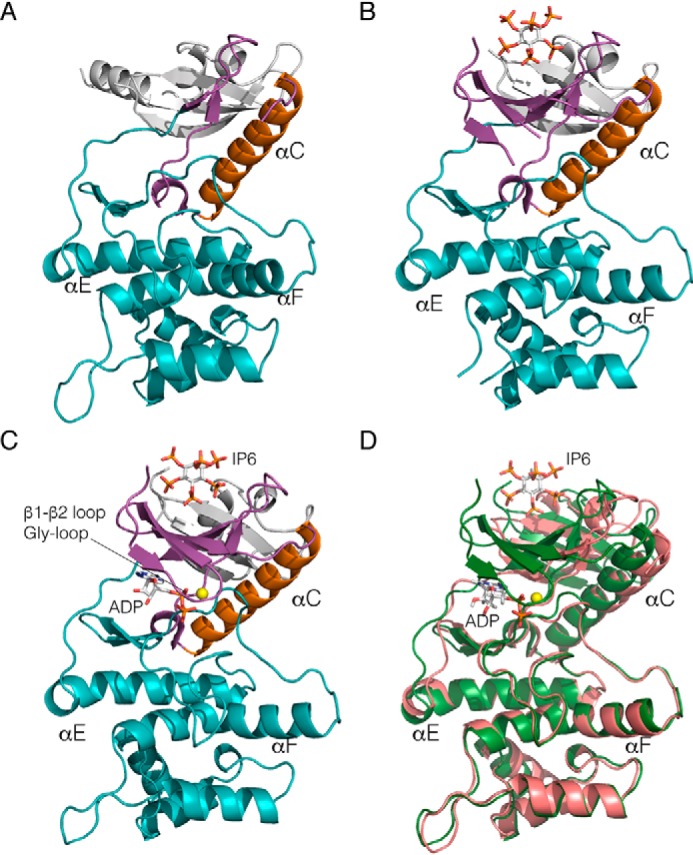
Crystal structure of Lpg2603 reveals a unique mode of kinase activation by IP6. A, ribbon representation of apo-Lpg2603. The N-lobe and C-lobe are depicted in magenta and teal, respectively. The αC-helix and the N-terminal extensions are shown in orange and white, respectively. B, ribbon representation of Lpg2603 bound to IP6. The ligand IP6 is shown in ball-and-stick form, colored according to atom. C, ribbon representation of Lpg2603 bound to IP6 and ADP. The nucleotide ADP and ligand IP6 are shown in ball-and-stick form, colored according to atom. The Mn2+ ion is shown as a yellow sphere. D, superposition of the apo-Lpg2603 (shown in pink) with the nucleotide and ligand-bound Lpg2603 (shown in green) depicting the disordered N-lobe in the apo structure.
Next, we sought to compare the apo structure to the IP6-bound structure to determine the effect of cofactor binding. Surprisingly, the crystal structure of Lpg2603 bound to IP6 revealed a kinase fold with a highly ordered N-lobe with clear electron density for residues 92–117, which form three of the five strands of the canonical N-lobe β-sheet (Fig. 3B). The IP6 is coordinated in a positively charged pocket composed of residues from the N-lobe β-sheet (corresponding to strands β1–β5 of classical kinases) and residues from the N-terminal extension that includes a three-stranded β-sheet. The IP6-binding site is very well-conserved in Lpg2603 homologs, suggesting a common mechanism of activation (Fig. S1). Although the C-lobes of the apo- and IP6-bound structures remain similar, conformational shifts are observed in the IP6-binding cradle, αC-helix, and metal-binding residues that favor ATP binding (Fig. 3, A and B). The apo structure can be superimposed onto the IP6-bound structure with an RMSD of 2.3 Å over 251 C-α backbone atoms. Thus, we hypothesize that binding of IP6 stabilizes the N-lobe of Lpg2603 to allow for ATP binding and kinase activity.
We next solved the crystal structure of Lpg2603 bound to IP6 and adenosine diphosphate (ADP) to identify the residues that facilitate nucleotide coordination upon IP6 binding (Fig. 3C). The overall conformation of the kinase in the presence and absence of nucleotide is very similar with an RMSD of 0.7 Å over 274 C-α backbone atoms (Fig. 3D). One notable difference upon ADP binding is the ordering of the Gly-rich loop that folds over the nucleotide in the active site (Fig. S2). Structural homology searches using DALI identified membrane-associated tyrosine-and threonine-specific cdc2-inhibitory kinase (Myt1), troponin I–interacting kinase (TNNI3K), and Bruton's tyrosine kinase (BTK) as the closest structural homologs of Lpg2603.
Structure guided mutagenesis highlight the residues important for nucleotide and IP6 coordination
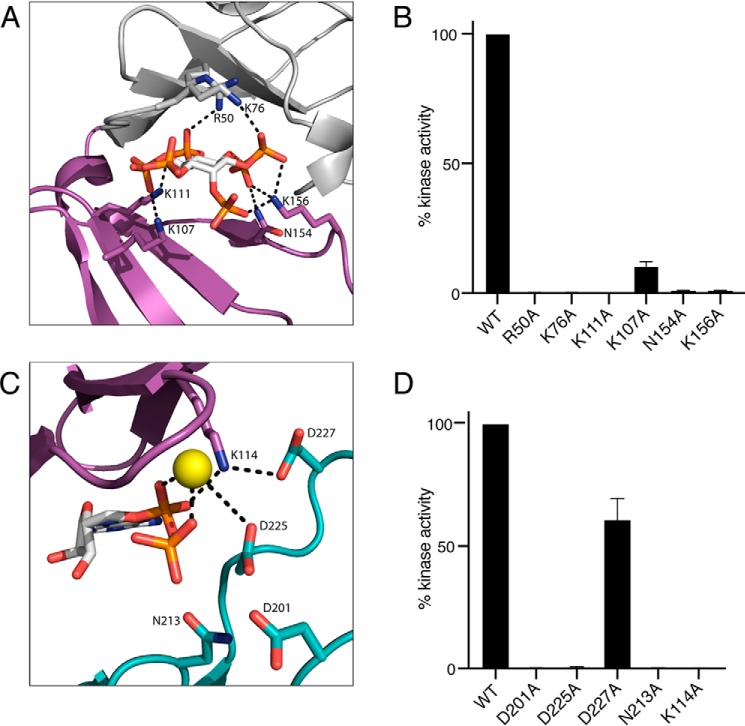
Using the Lpg2603 holoenzyme structure, we identified key residues that form the IP6-binding pocket (Fig. 4A). To identify IP6-binding residues that are important for kinase activity, we purified recombinant Lpg2603 proteins with alanine substitutions at Lys111, Lys156, Asn154, Lys107, Arg50, and Lys76. As expected, mutations in these residues to an Ala either weakened or completely abolished Lpg2603 activation by IP6 (Fig. 4B). These results support our hypothesis that proper coordination of IP6 is necessary for Lpg2603 activation.
Figure 4.
Mutational analysis highlights residues in Lpg2603 important for catalysis. A, enlarged image of IP6-binding pocket showing residues and interactions important for IP6 binding. IP6 is shown in ball-and-stick form, colored according to atom. B, activity of Lpg2603 or mutants were assayed as in Fig. 2B. Activity is expressed relative to the WT enzyme. The results are representative of three independent experiments. The error bars represent standard deviation. C, enlarged image of the nucleotide-binding pocket showing residues and interactions important for ADP binding and catalysis. The bound ADP is shown in ball-and-stick form, colored according to atom. Mn2+ ion is shown as a yellow sphere. Note that Lys114 lies within the VAIK motif common in protein kinases. D, Lpg2603 or mutants were assayed as in Fig. 2B. Activity is expressed relative to the WT enzyme. The results are representative of three independent experiments. The error bars represent standard deviation.
Next, we assayed recombinant Lpg2603 proteins with mutations in the nucleotide-binding pocket (Fig. 4C). The highly conserved catalytic loop in Lpg2603 consists of a noncanonical HPD motif with Asp201 likely acting as the catalytic base. Mutation of the catalytic Asp201 abolished kinase activity of Lpg2603 (Fig. 4D). A metal binding DXD motif is present in Lpg2603 where the Asp225 coordinates the Mn2+ ions in the active site. Mutation of the predicted metal-binding residues, Asp225 and Asn213 eliminated activity of the kinase. Lys114 from the β3 strand (PKA equivalent Lys72) coordinates the α and β phosphates of ATP (Fig. 4C). However, the glutamate corresponding to the canonical Glu91 (PKA) from the αC-helix that forms the conserved Lys72–Glu91 ion pair is positioned away from the active site, whereas Asp227 coordinates the ion pair Lys114. Interestingly, mutation of Lys114 completely abolished kinase activity, whereas mutation of Asp227, which forms an ion pair with Lys114, retained more than 50% of activity. This is reminiscent of the hypomorphic E74A ion pair mutant of the bacterial effector kinase HopBF1 (9). Collectively, our results highlight the residues important for IP6 binding and phosphotransfer.
IP6 binding facilitates nucleotide binding in Lpg2603
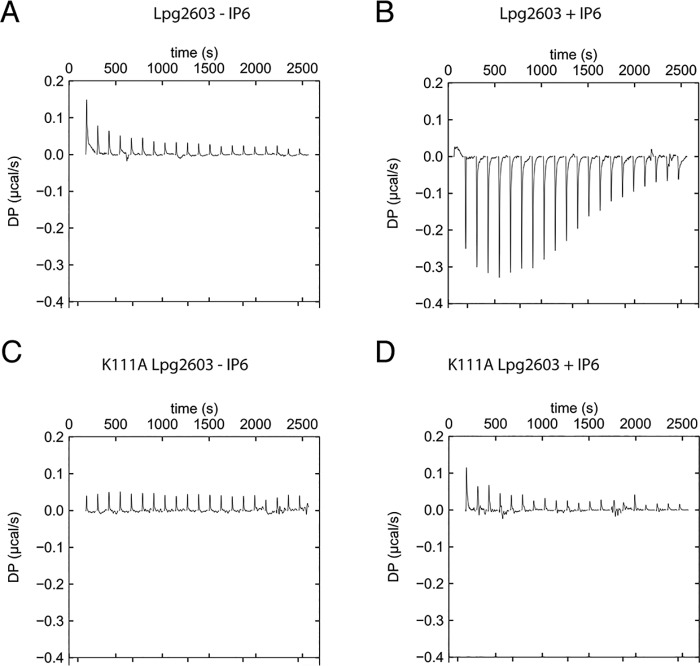
Given the structural reorganization observed with IP6, we investigated whether IP6 binding is required for nucleotide binding. To assay for ATP binding, we performed ITC with the weakly hydrolyzable ATP analog AMP-PNP. In the absence of IP6, Lpg2603 no longer binds to the nucleotide (Fig. 5A). Upon addition of IP6, Lpg2603 binds to AMP-PNP with a Kd of 3.6 μm (Fig. 5B). The IP6-bound Lpg2603 crystal structure and the in vitro kinase assays indicate Lys111 as a key residue for IP6-induced activation of kinase activity. Lpg2603 K111A does not bind to AMP-PNP in the presence or absence of IP6 (Fig. 5, C and D). These results provide further evidence that IP6 allosterically activates Lpg2603 kinase activity.
Figure 5.
Nucleotide binding to Lpg2603 depends on IP6 binding. A, representative isothermal titration calorimetry data for Lpg2603 binding to AMP-PNP in the absence of IP6. For all instances, Lpg2603 is at 100 μm in the cell, and AMP-PNP is present at 2 mm in the titration syringe to a final molar ratio of 1:4. B, representative isothermal titration calorimetry data for Lpg2603 binding to AMP-PNP in the presence of IP6. Best fit parameters were as follows: n = 1.54; ΔH = −5.79 kCal/mol. C, representative isothermal titration calorimetry data for Lpg2603 K111A binding to AMP-PNP. D, representative isothermal titration calorimetry data for Lpg2603 K111A binding to AMP-PNP in the presence of IP6. The results are representative of two independent experiments.
Discussion
Lpg2603 harbors a conserved Legionella-effector PI4P-binding region at the C terminus, which is required for anchoring the kinase to the Legionella containing vacuole during an infection (25). We have modeled this domain by using the homologous PI4P-binding domain from the Legionella effector DrrA and docked it against the kinase domain (Fig. S3). The full-length structure model displays the lipid-binding domain adjacent to the C-lobe of the kinase, suggesting that PI4P-binding and/or membrane localization may affect kinase function of Lpg2603. This also suggests that its substrates may be membrane-anchored or membrane-proximal. Despite our efforts to identify interacting proteins, we were unable to determine the host substrate of Lpg2603. Nevertheless, our results of the IP6-dependent kinase activation will pave the way to identify the in vivo substrate during Legionella infection.
We have identified a new mechanism of bacterial effector kinase regulation by a host signaling molecule, IP6. IP6 is a eukaryote-specific ligand and is therefore, absent in bacteria (32). Interestingly, it is also the most abundant inositol phosphate in eukaryotic cells where it regulates several processes including growth factor signaling, cell cycle progression, and vesicle trafficking (31). Cleverly, bacterial effectors have evolved to hijack host signaling molecules such as IP6 to spatially regulate activity (30). Less than a handful of other bacterial effectors from different pathogens are activated by IP6 including acetyl transferases (HopZ1, YopJ, and AvrA) and a cysteine protease (VPA1380) (30). Thus, the bacterial enzyme remains inactive within the bacterial cell until it is delivered to its destination within the eukaryotic host cell. Notably, this is the first example of a bacterial effector kinase that requires IP6 for activation. The IP6-binding site of Lpg2603 is spatially distinct from the kinase active site, suggesting allosteric regulation rather than IP6 acting as a cofactor for catalysis (Fig. 3C). Hence, we hypothesize that binding of IP6 at the N-terminal extension triggers conformational changes in multiple amino acids that functionally link the IP6-binding pocket to the kinase active site.
In addition to bacterial effectors, two eukaryotic protein kinases, BTK and protein kinase CK2, have also been shown to be activated by IP6 binding (33, 34). BTK is a nonreceptor tyrosine kinase that undergoes IP6-induced dimerization and activation (34). BTK is critical for proper B-cell function, and mutations are implicated in chronic lymphocytic leukemia, X-linked agammaglobulinemia, and several other autoimmune diseases (35). BTK is composed of multiple domains: pleckstrin homology (PH)–Tec homology domain, kinase domain, SH2 and SH3 domain (36). IP6 binds to the PH–Tec homology domain to induce dimerization and trans-autophosphorylation of the kinase domains, leading to activation (34). Notably, the mechanism of IP6 activation of BTK is distinct from Lpg2603 because we did not observe any dimerization of Lpg2603 upon incubation with IP6 (data not shown). In contrast to BTK, IP6 binds to the basic patch in the substrate recognition site of the C-lobe in CK2 (33). CK2 is a serine/threonine kinase that regulates cell proliferation (37); however, the physiological relevance of regulation of CK2 by IP6 is unclear.
Compared with the IP6-binding site of BTK (Protein Data Bank code 4Y94) and CK2 (Protein Data Bank code 3W8L), the IP6-binding site in Lpg2603 is more tightly coordinated by charged residues (Figs. 4A and 6). Moreover, in BTK and CK2, the sites are rather shallow and are built by residues from two different monomers (Fig. 6). The N-lobe of Lpg2603 and the N-terminal extension together form a β-sandwich that binds IP6 reminiscent of PH domains (Fig. S4) (38). However, connectivity of the strands in the two β-sheets is different in Lpg2603 from that of PH domains. This is likely an example of convergent evolution of a structural subdomain. The IP6-binding pocket in Lpg2603 consists of amino acids from the N-terminal extension and the N-lobe of the kinase that are highly conserved within homologs of Lpg2603. Furthermore, the apo structure revealed a fairly disordered N-lobe and active site in the absence of IP6, highlighting the importance of binding this ligand for activity. Hence, our crystallographic and biochemical analysis reveals a previously undocumented mode of allosteric regulation of the kinase superfamily.
Figure 6.
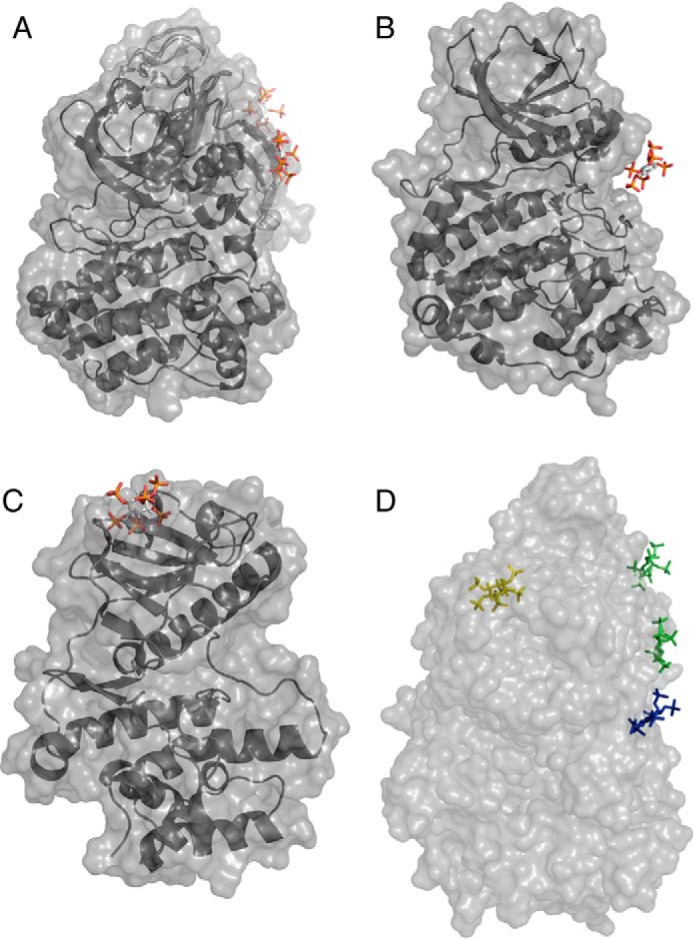
Structural comparison reveals unique mode of IP6 binding to Lpg2603. A, surface representation of BTK bound to IP6 (Protein Data Bank codes 4Y93 and 4Y94). B, surface representation of CK2 bound to IP6 (Protein Data Bank code 3W8L). C, surface representation of Lpg2603 bound to IP6. D, surface representation of the superposition of BTK, CK2, and Lpg2603. IP6 bound to BTK, CK2, and Lpg2603 is shown in green, blue, and yellow, respectively.
Experimental procedures
Reagents
Selenomethionine media was purchased from Molecular Dimensions (MD12-500). Inositol 1,3,4,5-tetraphosphate (Q-1345), inositol 1,4-bisphosphate (Q-0014), inositol 1,4,5 phosphate (Q-0145), and phosphatidylinositol 4-phosphate (P4008a) were purchased from Echelon Biosciences. d-myo-Inositol-1,3,4,5,6 pentaphosphate (10009851) was purchased from Cayman Chemicals. myo-Inositol 1-dihydrogen phosphate (S860360), myo-inositol (I7508), inositol hexakisphosphate (P8810), myelin basic protein (M1891), and AMP-PNP (A2647) were purchased from Sigma. Ubiquitin and calmodulin were expressed as His6 fusion proteins and purified from E. coli as described (12). Globular actin was a generous gift from Michael Rosen (39).
Generation of constructs
L. pneumophila Lpg2603 residues 10-C, 21-322, or 10-322 Lpg2603 were cloned into a modified pet28a bacterial expression vector (ppSumo), containing an N-terminal His6 tag followed by the yeast Sumo (smt3). The coding sequence for the yeast homolog of 14-3-3, BMH2, was amplified by PCR using Saccharomyces cerevisiae BY4741 gDNA as a template. The amplified open reading frames were cloned into pGEX4T containing an N-terminal GST tag.
Protein expression and purification
L. pneumophila Lpg2603 10-C, 21-322, or 10-322 ppSumo were transformed into E. coli Rosetta (DE3) competent cells. The cells were grown in LB medium at 37 °C until the A600 reached ∼0.5–0.7. Protein expression was induced by 0.4 mm isopropyl β-d-thiogalactoside overnight at room temperature. The cells were harvested by centrifugation and lysed in 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, and 0.1% β-mercaptoethanol) by sonication. Cell lysates were centrifuged at 25,000 × g for 25 min. The cleared lysate was incubated with nickel–nitrilotriacetic acid–agarose for approximately 1 h at 4 °C. The beads were passed over a column and washed with 20 column volumes of 50 mm Tris, pH 8, 300 mm NaCl, 10 mm imidazole. Protein was eluted off the beads with 50 mm Tris, pH 8, 300 mm NaCl, and 300 mm imidazole. The proteins were cut overnight at 4 °C with His6-tagged ULP Sumo protease and further purified using a Superdex 200 size-exclusion chromatography column attached to an AKTA Pure FPLC system (GE Healthcare).
For purification of the yeast homolog of 14-3-3, BMH2 pGEX4T was transformed into E. coli Rosetta (DE3) competent cells. The cells were grown in LB medium at 37 °C until the A600 reached ∼0.5–0.7. Protein expression was induced by 0.4 mm isopropyl β-d-thiogalactoside overnight at room temperature. The cells were harvested by centrifugation and lysed in 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, and 0.1% β-mercaptoethanol) by sonication. Cell lysates were centrifuged at 25,000 × g for 25 min. The cleared lysate was incubated with GSH–Sepharose for approximately 1 h at 4 °C. The beads were passed over a column and washed with 20 column volumes of 50 mm Tris, pH 8, 300 mm NaCl, 0.1% β-mercaptoethanol. Protein was eluted off the beads with 50 mm Tris, pH 8, 300 mm NaCl, 10 mm reduced GSH.
In vitro kinase assays
In vitro kinase assays were performed using untagged Lpg2603 10-C in a reaction mixture containing 50 mm Tris, pH 7.5, 100 μm MnCl2, 100 μm [γ-32P]ATP (SA = 1000 cpm/pmol), 1 mm DTT, 167 μg/ml MBP, 21 μg/ml Lpg2603, and 20 μm IP6. Reactions were incubated at 25 °C for 10 min and terminated by the addition of 0.5 mm EDTA. SDS loading buffer was added to the samples and boiled. The reaction products were separated by SDS-PAGE and visualized by Coomassie Blue staining.
For comparison of cofactors, reactions were performed as above with the following modifications: reactions contained 20 μm IP6, 188 μm PI4P, 40 μg/ml ubiquitin, 40 μg/ml calmodulin, or 40 μg/ml BMH2. For comparison of inositols, reactions were performed as above with the following modifications: reactions were incubated with 20 μm of the indicated inositols. For comparison of metals, reactions were performed as above with the following modifications: reactions contained 100 μm of the indicated metals. For comparison of Lpg2603 point mutants, reactions were performed as above with the following modifications: reactions contained 100 μm [γ-32P]ATP (SA = 5000 cpm/pmol). For the kinetic analysis, reactions were performed as above with the following modifications: reactions contained 300 μm MnCl2, 300 μm [γ-32P]ATP (SA = 5000 cpm/pmol). Reactions were incubated at 25 °C for 15 min.
Crystallization and structure determination
For apo-Lpg2603, recombinant Lpg2603 10-322 in 5 mm Tris-HCl, pH 8, 30 mm NaCl was concentrated to 10 mg/ml. The crystals were grown at 20 °C by the sitting-drop vapor-diffusion method using a 1:1 ratio of protein:reservoir solution containing 18% PEG3350 and 0.2 m LiCl and were flash-frozen in 20% PEG3350, 0.2 m LiCl, 30 mm NaCl, and 30% ethylene glycol. Apo-Lpg2603 crystals exhibited the symmetry of space group P3121 with cell dimensions of a = 62.96 Å, c = 176.41 Å, contained one apo-Lpg2603 per asymmetric unit, and diffracted to a minimum Bragg spacing (dmin) of 2.10 Å when exposed to synchrotron radiation. Selenomethionine labeled-protein was obtained by expressing Lpg2603 10-322 in B834 cells grown in SelenoMetTM media. SeMet Lpg2603 10-322 in 5 mm Tris-HCl, pH 8, 30 mm NaCl was concentrated to 20 mg/ml. The crystals were grown at 20 °C by the sitting-drop vapor-diffusion method using 1:1 ratio of protein:reservoir solution containing 13% PEG3350 and 0.2 m LiCl and were flash-frozen in 15.5% PEG3350, 0.2 m LiCl, 30 mm NaCl, and 35% ethylene glycol. SeMet apo-Lpg2603 crystals exhibited the symmetry of space group P3121 with cell dimensions of a = 63.11 Å and c = 175.51 Å, contained one SeMet apo-Lpg2603 per asymmetric unit, and diffracted to a minimum Bragg spacing (dmin) of 2.15 Å when exposed to synchrotron radiation.
For IP6-bound Lpg2603, Lpg2603 10-322 in 5 mm Tris-HCl, pH 8, 30 mm NaCl, and 1 mm IP6 was concentrated to 16 mg/ml. The crystals were grown at 20 °C by the sitting-drop vapor-diffusion method using a 1:1 ratio of protein:reservoir solution containing 0.1 m sodium acetate, pH 5.75, and were flash-frozen in 0.1 m sodium acetate, pH 5.75, 30 mm NaCl, 1 mm IP6, and 45% ethylene glycol. IP6-bound Lpg2603 crystals exhibited the symmetry of space group P212121 with cell dimensions of a = 52.93 Å, b = 73.21 Å, and c = 261.25 Å, contained three IP6-bound Lpg2603 molecules per asymmetric unit, and diffracted to a minimum Bragg spacing (dmin) of 2.65 Å when exposed to synchrotron radiation.
For IP6- and ADP-bound Lpg2603, SeMet Lpg2603 21-322 in 5 mm Tris-HCl, pH 8, 30 mm NaCl, 1 mm DTT, 0.5 mm IP6, 1 mm MnCl2, and 1 mm AMP-PNP was concentrated to 10 mg/ml. The crystals were grown at 20 °C by the sitting-drop vapor-diffusion method using a 1:1 ratio of protein:reservoir solution containing 0.1 m citric acid, pH 4.0, 6% (+/−)-2-methyl-2,4-pentanediol (MPD). The wells were allowed to equilibrate for ∼24 h, and crystal growth was initiated by microseeding. The crystals were flash-frozen in 0.1 m citric acid, pH 3.5, 7% (+/−)-2-methyl-2,4-pentanediol (MPD), 30 mm NaCl, 0.5 mm IP6, 1 mm MnCl2, 1 mm AMP-PNP, and 35% ethylene glycol. IP6- and ADP-bound Lpg2603 crystals exhibited the symmetry of space group P21212 with cell dimensions of a = 52.54 Å, b = 77.07 Å, c = 72.56 Å contained one IP6- and ADP-bound Lpg2603 per asymmetric unit and diffracted to a minimum Bragg spacing (dmin) of 1.77 Å when exposed to CuKα radiation from a home source. Although we added AMP-PNP to the protein, we observed ADP in our crystal structure. This may be due to contaminating ATP or weak hydrolysis of AMP-PNP.
Diffraction data were collected at 100 K at the Advanced Photon Source Beamline 19-ID for all data sets except the IP6- and ADP-bound Lpg2603, which was collected with CuKα radiation from a Rigaku Xtal MM003 source. Anomalous data for the SeMet apo-Lpg2603 were collected near the Se K-edge. The data were indexed, integrated, and scaled using the HKL-3000 program package (40). Data collection statistics are provided in Table S1.
Phase determination and structure refinement
Phases for SeMet apo-Lpg2603 were obtained from a single-wavelength anomalous dispersion experiment using a selenomethionyl-derivatized protein crystal with data collected at the selenium K-edge to a dmin of 2.15 Å. Seven selenium sites were located using the program SHELXD (41), and phases were refined with the program SHELXE (42), resulting in an over-all figure-of-merit of 0.66 for data between 46.39 and 2.15 Å. Phases were further improved by density modification in the program dm (43). An initial model containing 74% of all apo-Lpg2603 residues was automatically generated in the program ARP/wARP (44).
Because the selenomethionyl derivatized and native crystals were isomorphous, all further calculations for the native structure were performed versus the native data. Additional residues for apo-Lpg2603 were manually modeled in the program Coot (45). Positional and isotropic ADP, as well as TLS ADP refinement, was performed to a resolution of 2.10 Å using the program Phenix (46) with a random 5% of all data set aside for an Rfree calculation. The current model contains one apo-Lpg2603 monomer; included are residues 1–75, residues 118–332, and 196 water molecules. The Rwork is 0.176, and the Rfree is 0.211. A Ramachandran plot generated with MolProbity (47) indicated that 97.4% of all protein residues are in the most favored regions, and none are in the disallowed regions.
Phases for the IP6-bound Lpg2603 were obtained by the molecular replacement method in the program Phaser (48) using the coordinates for the apo Lpg2603 monomer with residues 1–15 removed. Model building and refinement were performed to a resolution of 2.65 Å using a similar protocol to the apo structure. Three IP6-bound Lpg2603 molecules were located in the asymmetric unit, and the electron density for chain C is substantially weaker than for chains A and B. The Rwork is 0.253, and the Rfree is 0.282; the presence of anisotropy in the data and the weak electron density for chain C is likely the cause of the higher than expected Rwork and Rfree for this model. A Ramachandran plot generated with MolProbity indicates that 95.4% of all protein residues are in the most favored regions and none in disallowed regions.
Phases for the IP6- and ADP-bound Lpg2603 were obtained by the molecular replacement method in the program Phaser using the coordinates for the IP6-bound Lpg2603 monomer. Model building and refinement were performed to a resolution of 1.77 Å using a similar protocol to the apo structure. One IP6- and ADP-bound Lpg2603 molecule was located in the asymmetric unit. The Rwork is 0.213, and the Rfree is 0.237. A Ramachandran plot generated with MolProbity indicates that 95.4% of all protein residues are in the most favored regions, and none are in the disallowed regions. Phasing and model refinement statistics for all structures are provided in Table S1.
Lpg2603 sequence logo
The similarity of Lpg2603 to kinases was identified by screening the set of L. pneumophila subsp. Philadelphia effectors using the FFAS server (29). Homologs of the Lpg2603 kinase domain were collected using BLAST and aligned by Mafft (50). Sequence logos were produced using the WebLogo 3.0 server (51).
Structural modeling of full-length Lpg2603
The structural model of the DrrA domain was built using the 4mxp structure as template for the I-Tasser server (52). The full-length structure model of Lpg2603 was built by docking together the kinase-like domain structure and the DrrA domain structure model using the AIDA server (53).
Structural comparisons
Structures were visualized and analyzed in PyMOL. Structure database searches were conducted using the Dali and Fatcat servers (49, 54).
Isothermal calorimetry
Recombinant Lpg2603 10-C WT or Lpg2603 10-C K111A was produced as described above under “Protein expression and purification.” For AMP-PNP binding assays, the final buffer consisted of 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm DTT, 1 mm MgCl2, and 1 mm IP6. For IP6 binding, MgCl2 and IP6 were excluded from the buffer.
For AMP-PNP binding assays, 2 mm AMP-PNP was titrated into Lpg2603 10-C at 100 μm cell concentration using the Malvern MicroCal iTC200 system at 20 °C. Initial injection was 0.5 μl, and the following 20 injections were 1.9 μl with stirring at 750 rpm. Initial injections were not included in the final analysis. Spacing between injections were 120 s to allow for proper baseline equilibration. Resulting thermograms were integrated using the NITPIC software, and SEDPHAT was used to fit the isotherms assuming a binary interaction model.
For IP6 binding assays, 8 mm IP6 was titrated into Lpg2603 10-C at 400 μm cell concentration as above. Initial injections were 0.5 μl, and the following 20 injections were 1.9 μl. Initial injections were, once again, not included in the final analysis. Spacing between injections were adjusted to 300 s to allow for proper baseline equilibration. The resulting thermograms were analyzed using the Origin software to obtain Kd and confidence intervals.
Data availability
All of the data are contained within the manuscript. The Lpg2603 structures have been deposited in the Protein Data Bank with accession codes as follows: apo-Lpg2603 under code 6VVC; Lpg2603 bound to IP6 under code 6VVD; and Lpg2603 bound to IP6, Mn2+, and ADP under code 6VVE.
Author contributions
A. S., K. P., and V. S. T. conceptualization; A. S., D. R. T., and V. S. T. data curation; A. S., C. N., B. C. P., D. R. T., and V. S. T. formal analysis; A. S., C. N., B. C. P., D. R. T., and V. S. T. investigation; A. S., D. R. T., and V. S. T. methodology; A. S. and V. S. T. writing-original draft; A. S., K. P., and V. S. T. writing-review and editing; K. P. and D. R. T. software; K. P. and D. R. T. validation; V. S. T. supervision; V. S. T. funding acquisition.
Supplementary Material
Acknowledgments
We thank Joshua Sheetz, Shaeri Mukherjee, Kim Orth, Samantha Yee, and members of the Tagliabracci lab for valuable input. We thank Chad Brautigam and Shih-Chia Tso for help with ITC. The results shown in this report are derived from work performed at the Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source.
This work was supported by National Institutes of Health Grants R00DK099254 and DP2GM137419 (to V. S. T.) and T32DK007257-37 (to A. S.), Welch Foundation Grants I-1911 (to V. S. T.), Polish National Agency for Scientific Exchange Scholarship PPN/BEK/2018/1/00431 (to K. P.), and Award S10OD025018 (D. R. T.) from the Office of the Director, National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S4.
- PI4P
- phosphatidylinositol-4-phophate
- IP6
- inositol hexakisphosphate
- PKA
- protein kinase A
- MBP
- myelin basic protein
- ITC
- isothermal titration calorimetry
- RMSD
- root-mean-square deviation
- BTK
- Bruton's tyrosine kinase
- AMP-PNP
- adenosine 5′-(β,γ-iminotriphosphate)
- CK2
- casein kinase 2
- PH
- pleckstrin homology
- SeMet
- selenomethionine
- ADP
- adenosine diphosphate.
References
- 1. Fischer E. H. (2013) Cellular regulation by protein phosphorylation. Biochem. Biophys. Res. Commun. 430, 865–867 10.1016/j.bbrc.2012.10.024 [DOI] [PubMed] [Google Scholar]
- 2. Manning G., Whyte D. B., Martinez R., Hunter T., and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 3. Kanev G. K., de Graaf C., de Esch I. J. P., Leurs R., Würdinger T., Westerman B. A., and Kooistra A. J. (2019) The landscape of atypical and eukaryotic protein kinases. Trends Pharmacol. Sci. 40, 818–832 10.1016/j.tips.2019.09.002 [DOI] [PubMed] [Google Scholar]
- 4. Tagliabracci V. S., Engel J. L., Wen J., Wiley S. E., Worby C. A., Kinch L. N., Xiao J., Grishin N. V., and Dixon J. E. (2012) Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science 336, 1150–1153 10.1126/science.1217817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tagliabracci V. S., Wiley S. E., Guo X., Kinch L. N., Durrant E., Wen J., Xiao J., Cui J., Nguyen K. B., Engel J. L., Coon J. J., Grishin N., Pinna L. A., Pagliarini D. J., and Dixon J. E. (2015) A single kinase generates the majority of the secreted phosphoproteome. Cell 161, 1619–1632 10.1016/j.cell.2015.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nguyen K. B., Sreelatha A., Durrant E. S., Lopez-Garrido J., Muszewska A., Dudkiewicz M., Grynberg M., Yee S., Pogliano K., Tomchick D. R., Pawlowski K., Dixon J. E., and Tagliabracci V. S. (2016) Phosphorylation of spore coat proteins by a family of atypical protein kinases. Proc. Natl. Acad. Sci. U.S.A. 113, E3482–E3491 10.1073/pnas.1605917113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sreelatha A., Yee S. S., Lopez V. A., Park B. C., Kinch L. N., Pilch S., Servage K. A., Zhang J., Jiou J., Karasiewicz-Urbańska M., Lobocka M., Grishin N. V., Orth K., Kucharczyk R., Pawlowski K., et al. (2018) Protein AMPylation by an evolutionarily conserved pseudokinase. Cell 175, 809–821.e19 10.1016/j.cell.2018.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salomon D., and Orth K. (2013) What pathogens have taught us about posttranslational modifications. Cell Host Microbe 14, 269–279 10.1016/j.chom.2013.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lopez V. A., Park B. C., Nowak D., Sreelatha A., Zembek P., Fernandez J., Servage K. A., Gradowski M., Hennig J., Tomchick D. R., Pawlowski K., Krzymowska M., and Tagliabracci V. S. (2019) A bacterial effector mimics a host HSP90 client to undermine immunity. Cell 179, 205–218.e21 10.1016/j.cell.2019.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hubber A., and Roy C. R. (2010) Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283 10.1146/annurev-cellbio-100109-104034 [DOI] [PubMed] [Google Scholar]
- 11. Liu Y., and Luo Z. Q. (2007) The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect. Immun. 75, 592–603 10.1128/IAI.01278-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Black M. H., Osinski A., Gradowski M., Servage K. A., Pawlowski K., Tomchick D. R., and Tagliabracci V. S. (2019) Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases. Science 364, 787–792 10.1126/science.aaw7446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bhogaraju S., Bonn F., Mukherjee R., Adams M., Pfleiderer M. M., Galej W. P., Matkovic V., Lopez-Mosqueda J., Kalayil S., Shin D., and Dikic I. (2019) Inhibition of bacterial ubiquitin ligases by SidJ-calmodulin catalysed glutamylation. Nature 572, 382–386 10.1038/s41586-019-1440-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sulpizio A., Minelli M. E., Wan M., Burrowes P. D., Wu X., Sanford E. J., Shin J. H., Williams B. C., Goldberg M. L., Smolka M. B., and Mao Y. (2019) Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ. eLife 8, e51162 10.7554/eLife.51162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gan N., Zhen X., Liu Y., Xu X., He C., Qiu J., Liu Y., Fujimoto G. M., Nakayasu E. S., Zhou B., Zhao L., Puvar K., Das C., Ouyang S., and Luo Z. Q. (2019) Regulation of phosphoribosyl ubiquitination by a calmodulin-dependent glutamylase. Nature 572, 387–391 10.1038/s41586-019-1439-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park B. C., Reese M., and Tagliabracci V. S. (2019) Thinking outside of the cell: secreted protein kinases in bacteria, parasites, and mammals. IUBMB Life 71, 749–759 10.1002/iub.2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haenssler E., and Isberg R. R. (2011) Control of host cell phosphorylation by Legionella pneumophila. Front. Microbiol. 2, 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ge J., Xu H., Li T., Zhou Y., Zhang Z., Li S., Liu L., and Shao F. (2009) A Legionella type IV effector activates the NF-κB pathway by phosphorylating the IκB family of inhibitors. Proc. Natl. Acad. Sci. U.S.A. 106, 13725–13730 10.1073/pnas.0907200106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Losick V. P., Haenssler E., Moy M. Y., and Isberg R. R. (2010) LnaB: a Legionella pneumophila activator of NF-κB. Cell Microbiol. 12, 1083–1097 10.1111/j.1462-5822.2010.01452.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hervet E., Charpentier X., Vianney A., Lazzaroni J. C., Gilbert C., Atlan D., and Doublet P. (2011) Protein kinase LegK2 is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila. Infect. Immun. 79, 1936–1950 10.1128/IAI.00805-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Michard C., Sperandio D., Baïlo N., Pizarro-Cerdá J., LeClaire L., Chadeau-Argaud E., Pombo-Grégoire I., Hervet E., Vianney A., Gilbert C., Faure M., Cossart P., and Doublet P. (2015) The Legionella kinase LegK2 targets the ARP2/3 complex to inhibit actin nucleation on phagosomes and allow bacterial evasion of the late endocytic pathway. mBio 6, e00354–00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee P. C., and Machner M. P. (2018) The Legionella effector kinase LegK7 hijacks the host Hippo pathway to promote infection. Cell Host Microbe 24, 429–438.e6 10.1016/j.chom.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moss S. M., Taylor I. R., Ruggero D., Gestwicki J. E., Shokat K. M., and Mukherjee S. (2019) A Legionella pneumophila kinase phosphorylates the Hsp70 chaperone family to inhibit eukaryotic protein synthesis. Cell Host Microbe 25, 454–462.e6 10.1016/j.chom.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burstein D., Amaro F., Zusman T., Lifshitz Z., Cohen O., Gilbert J. A., Pupko T., Shuman H. A., and Segal G. (2016) Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat. Genet. 48, 167–175 10.1038/ng.3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hubber A., Arasaki K., Nakatsu F., Hardiman C., Lambright D., De Camilli P., Nagai H., and Roy C. R. (2014) The machinery at endoplasmic reticulum-plasma membrane contact sites contributes to spatial regulation of multiple Legionella effector proteins. PLoS Pathog. 10, e1004222 10.1371/journal.ppat.1004222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Müller M. P., Peters H., Blümer J., Blankenfeldt W., Goody R. S., and Itzen A. (2010) The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949 10.1126/science.1192276 [DOI] [PubMed] [Google Scholar]
- 27. Murata T., Delprato A., Ingmundson A., Toomre D. K., Lambright D. G., and Roy C. R. (2006) The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8, 971–977 10.1038/ncb1463 [DOI] [PubMed] [Google Scholar]
- 28. Beyrakhova K., Li L., Xu C., Gagarinova A., and Cygler M. (2018) Legionella pneumophila effector Lem4 is a membrane-associated protein tyrosine phosphatase. J. Biol. Chem. 293, 13044–13058 10.1074/jbc.RA118.003845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu D., Jaroszewski L., Li Z., and Godzik A. (2014) FFAS-3D: improving fold recognition by including optimized structural features and template re-ranking. Bioinformatics 30, 660–667 10.1093/bioinformatics/btt578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson D. M., Feix J. B., and Frank D. W. (2015) Cross kingdom activators of five classes of bacterial effectors. PLoS Pathog. 11, e1004944 10.1371/journal.ppat.1004944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shamsuddin A. M. (1999) Metabolism and cellular functions of IP6: a review. Anticancer Res. 19, 3733–3736 [PubMed] [Google Scholar]
- 32. Abul Kalam S., and Sanchita B. (2012) IP6 (Inositol Hexaphosphate) as a signaling molecule. Curr. Signal. Transduction Ther. 7, 289–304 10.2174/157436212802481628 [DOI] [Google Scholar]
- 33. Lee W. K., Son S. H., Jin B. S., Na J. H., Kim S. Y., Kim K. H., Kim E. E., Yu Y. G., and Lee H. H. (2013) Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140. Proc. Natl. Acad. Sci. U.S.A. 110, 19360–19365 10.1073/pnas.1304670110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Q., Vogan E. M., Nocka L. M., Rosen C. E., Zorn J. A., Harrison S. C., and Kuriyan J. (2015) Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate. Elife 4, e06074 10.7554/eLife.06074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weber A. N. R., Bittner Z., Liu X., Dang T. M., Radsak M. P., and Brunner C. (2017) Bruton's tyrosine kinase: an emerging key player in innate immunity. Front. Immunol. 8, 1454 10.3389/fimmu.2017.01454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hyvönen M., and Saraste M. (1997) Structure of the PH domain and Btk motif from Bruton's tyrosine kinase: molecular explanations for X-linked agammaglobulinaemia. EMBO J. 16, 3396–3404 10.1093/emboj/16.12.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Litchfield D. W. (2003) Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 369, 1–15 10.1042/bj20021469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ferguson K. M., Kavran J. M., Sankaran V. G., Fournier E., Isakoff S. J., Skolnik E. Y., and Lemmon M. A. (2000) Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6, 373–384 10.1016/S1097-2765(00)00037-X [DOI] [PubMed] [Google Scholar]
- 39. Doolittle L. K., Rosen M. K., and Padrick S. B. (2013) Measurement and analysis of in vitro actin polymerization. Methods Mol. Biol. 1046, 273–293 10.1007/978-1-62703-538-5_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Minor W., Cymborowski M., Otwinowski Z., and Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution: from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 10.1107/S0907444906019949 [DOI] [PubMed] [Google Scholar]
- 41. Schneider T. R., and Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 10.1107/S0907444902011678 [DOI] [PubMed] [Google Scholar]
- 42. Sheldrick G. M. (2002) Macromolecular phasing with SHELXE. Zeitschrift für Kristallographie Crystalline Materials 217, 644–650 10.1524/zkri.217.12.644.20662 [DOI] [Google Scholar]
- 43. Cowtan K. (1999) Error estimation and bias correction in phase-improvement calculations. Acta Crystallogr. D Biol. Crystallogr. 55, 1555–1567 10.1107/S0907444999007416 [DOI] [PubMed] [Google Scholar]
- 44. Langer G., Cohen S. X., Lamzin V. S., and Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 10.1038/nprot.2008.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Afonine P. V., Mustyakimov M., Grosse-Kunstleve R. W., Moriarty N. W., Langan P., and Adams P. D. (2010) Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 66, 1153–1163 10.1107/S0907444910026582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ye Y., and Godzik A. (2004) FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 32, W582–W585 10.1093/nar/gkh430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Katoh K., Rozewicki J., and Yamada K. D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu D., Jaroszewski L., Li Z., and Godzik A. (2015) AIDA: ab initio domain assembly for automated multi-domain protein structure prediction and domain–domain interaction prediction. Bioinformatics 31, 2098–2105 10.1093/bioinformatics/btv092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Holm L., and Laakso L. M. (2016) Dali server update. Nucleic Acids Res. 44, W351–W355 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data are contained within the manuscript. The Lpg2603 structures have been deposited in the Protein Data Bank with accession codes as follows: apo-Lpg2603 under code 6VVC; Lpg2603 bound to IP6 under code 6VVD; and Lpg2603 bound to IP6, Mn2+, and ADP under code 6VVE.