Abstract
The formation of translationally inactive 70S dimers (called 100S ribosomes) by hibernation-promoting factor is a widespread survival strategy among bacteria. Ribosome dimerization is thought to be reversible, with the dissociation of the 100S complexes enabling ribosome recycling for participation in new rounds of translation. The precise pathway of 100S ribosome recycling has been unclear. We previously found that the heat-shock GTPase HflX in the human pathogen Staphylococcus aureus is a minor disassembly factor. Cells lacking hflX do not accumulate 100S ribosomes unless they are subjected to heat exposure, suggesting the existence of an alternative pathway during nonstressed conditions. Here, we provide biochemical and genetic evidence that two essential translation factors, ribosome-recycling factor (RRF) and GTPase elongation factor G (EF-G), synergistically split 100S ribosomes in a GTP-dependent but tRNA translocation-independent manner. We found that although HflX and the RRF/EF-G pair are functionally interchangeable, HflX is expressed at low levels and is dispensable under normal growth conditions. The bacterial RRF/EF-G pair was previously known to target only the post-termination 70S complexes; our results reveal a new role in the reversal of ribosome hibernation that is intimately linked to bacterial pathogenesis, persister formation, stress responses, and ribosome integrity.
Keywords: ribosome, Staphylococcus aureus (S. aureus), GTPase, translation elongation factor, translation regulation, 100S ribosome, bacterial persistence, elongation factor G (EF-G), hibernation, ribosome-recycling factor (RRF)
Introduction
Bacterial ribosome hibernation has emerged as one of the pivotal cellular processes for entry into quiescence and subsequent resuscitation. Hibernation-promoting factor (HPF),2 either alone or in combination with ribosome modulation factor (RMF), induces the dimerization of 70S ribosomes to form the translational inactive 100S complexes in all bacterial phyla with the exception of Actinobacteria and hibernation factor YfiA (1, 2). The loss of HPF or RMF causes rapid ribosome degradation (3–5), translational derepression (4, 6, 7), reduced longevity (4, 8, 9), and antibiotic and stress susceptibility (10–12). Staaphylococcus aureus HPF is one of the predominant proteins induced upon host cell internalization and during infections (13, 14). For reviews of the topic, see Refs. 15–19.
Native 100S ribosomes from various bacteria are devoid of mRNA and tRNAs (20–24). In Firmicutes such as the human opportunistic pathogen S. aureus, an ∼200 amino acid-long HPF promotes ribosome dimerization via its self-dimerizing C-terminal domain, whereas its N-terminal domain occupies the tRNA- and mRNA-binding sites of the 30S subunits and thus sterically prevents translation (20–23). S. aureus 100S ribosomes (and HPF) are constitutively produced throughout the life cycle, as confirmed by time course immunoblotting and MS analyses of the HPF-bound 100S complexes (4, 25–27). A strong CodY-dependent promoter largely accounts for the high levels of HPF (28). The significance of 100S ribosomes during logarithmic growth is unclear, although they are thought to function as storage sites to preserve unused ribosomes (e.g. post-termination recycled ribosomes) from degradation (3–5, 29). In fact, 70S dimerization is strongly linked to the protection of ribosomes and the maintenance of active translation pools (3, 23, 30).
The hibernating 100S ribosomes serve as a reservoir to avoid futile translation and supply nutrient and translational machinery during bacterial regrowth from dormancy. To reactivate hibernating ribosomes for translation, 100S complexes need to be split into 70S monomers or 30S and 50S subunits concomitant with the removal of HPF. We previously showed that the evolutionarily conserved GTPase HflX is able to dissociate both 100S ribosomes and vacant 70S ribosomes in S. aureus but that GTP hydrolysis is required only for 100S complex splitting (31). Escherichia coli HflX rescues post-termination complex (PoTc)-like stalled 70S ribosomes from mRNA during thermal stress (32, 33). The expression levels of hflX in S. aureus and E. coli are undetectable during normal growth but are up-regulated by heat shock. The deletion of hflX only produces modest phenotypes (31, 32), implying that a more general housekeeping factor(s) is involved in the ribosome recycling of 100S complexes or stalled 70S complexes under nonstressed conditions. We posit that the alternative dissociation pathway of the 100S ribosomes involves a factor(s) that recognizes a PoTc-like substrate. The bacterial ribosome-recycling factor (RRF) and the GTPase elongation factor-G (EF-G) are known to break down the PoTc consisting of an mRNA and an uncharged P/E-site tRNA on a fully rotated 70S complex (34). The exact order of mRNA and tRNA release and 70S splitting remains controversial (35–39). In addition, PSRP1-induced hibernating ribosomes in chloroplasts, which are not dimerized and remain as 70S monomers, are bound with Chl-RRF at the intersubunit junction (40).
Here, we show that the 100S ribosome is a hitherto unknown target of RRF and EF-G in S. aureus. The splitting activity depends on GTP hydrolysis and not on the translocase activity of EF-G. Ectopic expression of HflX rescues the growth defect of an RRF-depleted strain, indicating functional redundancy of these factors. RRF/EF-G- and HflX-dependent pathways appear to functionally converge for the reactivation of hibernating ribosomes during stress adaptation.
Results
RRF and EF-G cooperatively dissociate the 100S ribosome in a GTP-dependent manner
We performed in vitro 100S ribosome dissociation assays using purified recombinant RRF (encoded by frr) and EF-G (encoded by fusA) proteins in the presence and absence of GTP analogs. Native 100S ribosomes were isolated to ≥70% homogeneity from methicillin-resistant S. aureus (MRSA) USA300 through a two-step density gradient fractionation and ultracentrifugation protocol. The purified RRF, EF-G, and HflX proteins or GTP alone failed to dissociate the 100S complexes (Fig. 1, panels i–v and ix). More than 50% of the 100S ribosomes were converted into 70S monomers in the presence of RRF, EF-G, and GTP (panel vii) or HflX and GTP (panel x). The 70S ribosomes were not further dissociated by RRF/EF-G into 50S and 30S subunits because the reactions did not contain the anti-association factor IF3 (41). Some dissociation of 100S complexes were observed in reactions containing GTP plus either RRF or EF-G (panels iii and v). This is likely due to the residual activity of the ribosome-associated RRF or EF-G because high-salt wash that otherwise removes ribosome-bound factor(s), was omitted from our 100S ribosome preparation to prevent the release of HPF. When GTP was substituted with nonhydrolyzable guanosine 5′-(β,γ-imido)triphosphate (GMPPNP), which is a higher-affinity binder of ribosome-bound EF-G (42), the RRF/EF-G pair was unsuccessful in splitting the 100S complexes (panel viii). In contrast, GMPPNP induced the HflX-mediated dissociation of both the 100S and 70S complexes (panel xi), consistent with previous observations that GMPPNP is a more effective ligand of HflX in splitting the empty 70S ribosome (31, 32).
Figure 1.
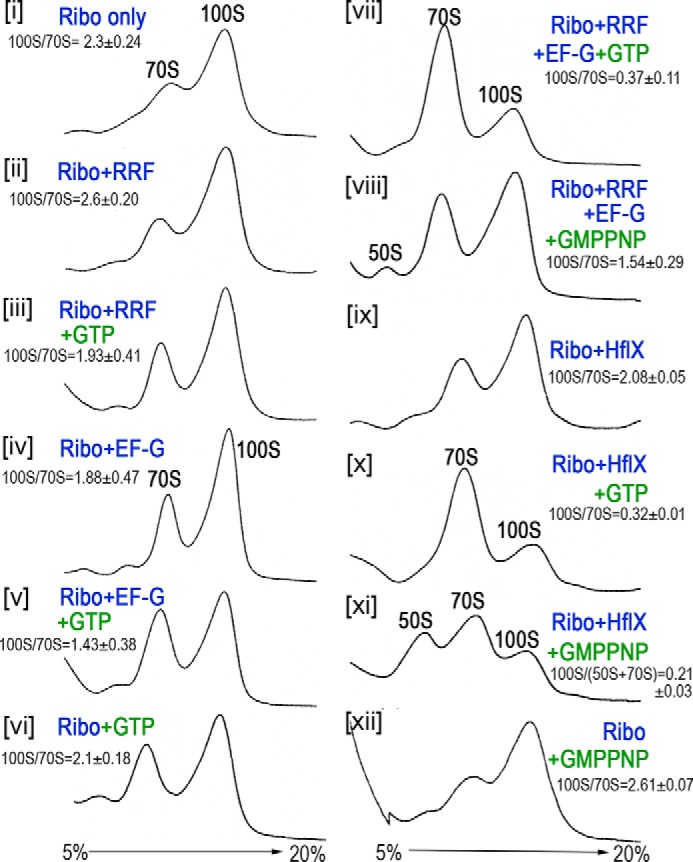
In vitro dissociation of the 100S ribosome by the RRF/EF-G pair and HflX in the presence and absence of guanosine analogs. Reactions were programmed with 0.2 μm ribosomes, 2 μm proteins, and 2 mm GTP analogs and incubated at 37 °C for 30 min. The samples were centrifuged in a 5–20% sucrose gradient, and ribosome profiles were monitored from the absorbance at 254 nm (y axis). Quantification of the 100S to 70S ratios were obtained from three technical replicates (of two independently prepared ribosomes and recombinant proteins); mean ± S.D.
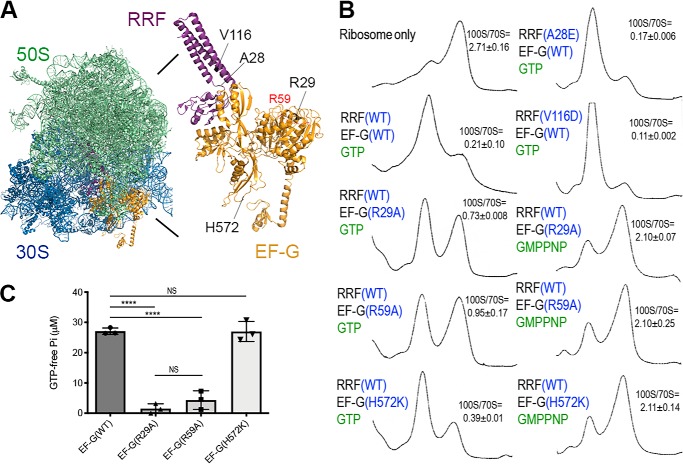
To validate these findings, we repeated the dissociation reactions using EF-G and RRF mutant proteins. Many loss-of-function mutations have been mapped to E. coli EF-G and RRF. For instance, a conserved Arg-29 is essential for GTP hydrolysis; a conserved Arg-59 affects tRNA translocation and ribosome binding but affects GTP hydrolysis only moderately; and H583K (His-572 in S. aureus numbering) conversion reduces tRNA translocation (43, 44). The substitution of Val-117 (V116 in S. aureus numbering) with an aspartate (frr14 allele) in RRF renders a temperature-sensitivity phenotype (45).
We introduced mutations at the equivalent positions of S. aureus EF-G and RRF, in addition to a benign A28E mutation in a flexible region of RRF (Fig. 2A). The R29A and R59A mutants of EF-G are impaired in 100S ribosome dissociation, whereas the H572K mutant retains its splitting activity at the wildtype (WT) level (Fig. 2B). The dissociation of the 100S ribosome by the EF-G(H572K) is abolished upon GMPPNP substitution. The impairments caused by R29A and R59A are due in part to a decrease in GTP hydrolysis (Fig. 2C) and not to reduced ribosome binding (Figs. S1, A and B). Species-specific activities of RRF and EF-G have been observed between several bacterial homologs, and noncognate RRF and EF-G pairing is nonfunctional (46, 47). These variations could account for the difference in GTPase activity observed for S. aureus EF-G(R59A) compared with that of E. coli (43). We conclude that the translocation function (His-572) of EF-G is not required for 100S ribosome disassembly but that GTP hydrolysis is indispensable.
Figure 2.
The effects of RRF and EF-G mutations in 100S ribosome disassembly. A, model of the RRF, EF-G and ribosome co-complex. A structural model was constructed from PDB accession numbers 4V54, 5OT7, and 4WPO. The mutations included in this study are indicated. R59 (red) was mapped to an unstructured region of EF-G in the crystal. B, the dissociation of 100S ribosomes is reduced in GTP-hydrolysis mutants (R29A and R59A) of EF-G but is unaffected by a mutation that compromises tRNA translocation (H572K). The dissociation reactions comprised 0.2 μm ribosomes, 2 μm proteins, and 2 mm GTP analogs and were incubated at 37 °C for 30 min. Ribosome profiles were analyzed via 5–20% sucrose density sedimentation, and the ribosomal species were monitored according to the absorbance at 254 nm. 100S to 70S ratios were obtained from three technical replicates (of two independently prepared ribosomes and recombinant proteins); mean ± S.D. C, malachite green GTPase assay showing the reduction of GTPase hydrolysis in the R29A and R59A mutants. The known translocation inactive H572K mutant does not present impaired GTPase activity. Error bars represent the S.E. obtained from three independent experiments using two different batches of purified proteins. p values were calculated by Student's unpaired t test, ****, p < 0.0001; NS, not significant.
Contrary to our prediction, RRF(V116D) showed an increase in 100S ribosome dissociation, whereas RRF(A28E) was fully functional, as expected (Fig. 2B). Limited proteolysis showed that RRF(V116) protein was more accessible to chymotrypsin digestion and may therefore adopt different conformations than the WT RRF (Fig. S2A). In addition, RRF(V116D) bound to the ribosome and recruited EF-G as efficiently as WT RRF (Figs. S1, C and D, and S2, B and C). Consistent with these findings, the V116D mutant allele fully complemented the growth defect of an RRF-depleted strain (Fig. 3D).
Figure 3.
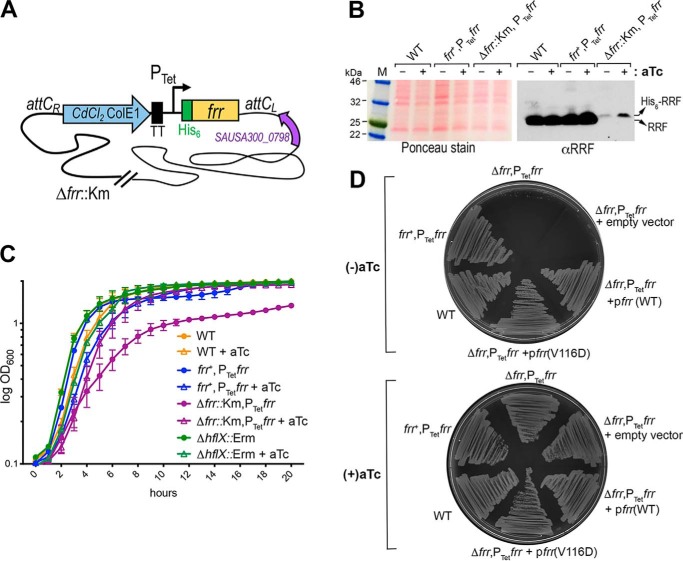
In vivo phenotypes of the RRF-depleted strain. A, construction of the RRF-depleted strain. His6-tagged frr was first placed under the control of a tetracycline (Tet)-inducible promoter on a cadmium chloride (CdCl2)-resistant integration plasmid. The plasmid was integrated into the chromosomal attC site, resulting in an S. aureus strain carrying an inducible frr (PTetfrr) and a native copy of frr (frr+). The WT copy of frr was deleted (Δfrr::Km, confers kanamycin resistance) from the frr+,PTetfrr strain by homologous recombination in the presence of 400 ng/ml of aTc. TT, transcriptional terminator. B, relative expression levels of native RRF and aTc-inducible His6-RRF. The expression of RRF was analyzed by immunoblotting with anti-RRF (1:4,000 dilutions). RRF was expressed from the native locus at a much higher level than the His6-RRF. His6-RRF migrated slightly more slowly than the native RRF. C, growth defects of RRF depletion in liquid media. S. aureus cells were grown in TSB at 37 °C with and without aTc. Real-time cell density was monitored on a TECAN plate reader according to the absorbance at 600 nm. The addition of aTc slightly suppressed bacterial growth in all tested strains, but it restored the growth of the RRF-depleted strain to levels comparable with those in other strains with aTc supplementation. Error bars are S.E. from three independent experiments, and each experiment included five replicates. D, the growth defects of RRF depletion were more significant on solid agar plates. Single colonies of different S. aureus strains were streaked on BactoTM agar plates containing TSB base, and bacterial growth was recorded after 16 h of incubation at 37 °C. The RRF-depleted strain (Δfrr,PTetfrr) and its derivative carrying the empty pLI50 plasmid only formed a few microcolonies in the absence of aTc. WT frr and frr(V116D) expressed in the pLI50 plasmid fully rescued the growth defects of Δfrr,PTetfrr.
HflX complements RRF deficiency
RRF binds to the A-site of PoTc and recruits EF-G to initiate 70S ribosome splitting. RRF null is deleterious. EF-G is also an essential factor for the translocation step during elongation (34). To avoid complications arising from EF-G knockdown, we focused on the effects of RRF depletion on ribosome recycling.
To discern between endogenous RRF and exogenously introduced RRF, we constructed an RRF-depleted strain by deleting the frr (locus SAUSA300_1152) and placing a hexahistidine-tagged frr under the control of an anhydrotetracycline (aTc)-inducible promoter. The His6-frr allele was then integrated into a neutral site of the S. aureus chromosome (Fig. 3A). The same chromosomal integration system (48) has been widely used to complement S. aureus knockouts. The His-tagging strategy does not impair splitting activity of S. aureus RRF (Figs. 1 and 2B) nor compromises the efficiency of a reconstituted cell-free translation system containing E. coli His6-RRF (49). We found that the aTc-inducible promoter was leaky, allowing basal expression of His6-RRF without aTc and an upshift in the production of His6-RRF in the presence of an inducer (Fig. 3B). In contrast to the hflX knockout mutant, which lacked a phenotype at 37 °C but exhibited better heat tolerance at 47 °C (31), the depletion of RRF significantly reduced growth in liquid media; this defect was rescuable by the addition of aTc (Fig. 3C), confirming the direct role of RRF deficiency. The relatively low amount of His6-RRF (compared with the WT strain) was sufficient for growth (Fig. 3B), implying that the WT concentration of RRF is greater than that required for normal growth.
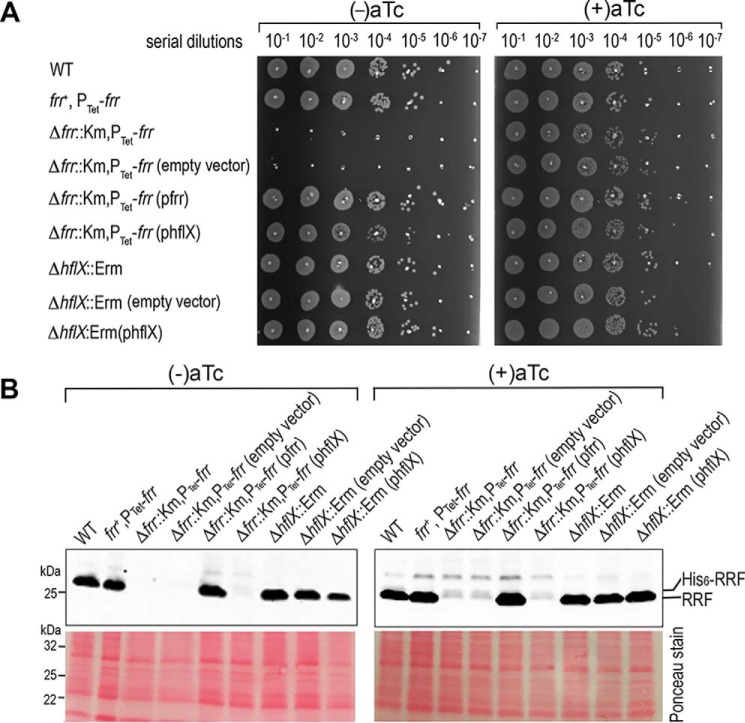
The growth defect of the RRF-depleted strain was accentuated on the solid agar media at all tested temperatures (30 °C-47 °C), in which only the supplementation of aTc supported growth (Fig. 3D). The severity of the growth defects observed on agar plates was only weakly associated with the type of gelling agents (Fig. S3). In the absence of aTc, RRF deficiency was fully complemented by expressing WT RRF or RRF(V116D) in a plasmid but not with an empty vector (Fig. 3D). Using the same complementation approach, we found that an hflX expressed in trans completely restored the growth defects of the RRF-depleted strain (Fig. 4A) and the expression of RRF in different genetic backgrounds was not affected by the introduction of plasmid (Fig. 4B). These results confirm that hflX functionally overlaps with RRF/EF-G in ribosome recycling. Furthermore, HflX is not expressed during growth at 37 °C and is exclusively up-regulated upon heat shock (Fig. S4) (31), explaining the dispensability of HflX under normal conditions.
Figure 4.
HflX complements the deficiency of RRF. A, spotting assay of the RRF-depleted strain and the hflX knockout harboring the indicated empty and complementation plasmids. Data are representative of three independent experiments. Cells were adjusted to an A600 of ∼0.2 and were serially diluted and spotted (3 μl/spot) on TSB-agar plates, then grown overnight at 37 °C to determine cell viability. The expression of hflX and frr in trans restored the growth of the RRF-depleted strain in the absence of anhydrotetracyline (aTc). B, expression of RRF in S. aureus TSB cultures with and without aTc. Western blots were performed using anti-RRF at 1:4,000 dilutions.
Consequences of inefficient ribosome recycling
We analyzed the effect of RRF depletion on ribosome pools by sucrose density gradient ultracentrifugation. Crude ribosomes were prepared from late log phase (an A600 of ∼1.4) cells grown without the aTc inducer. The cultures and sample preparation were free of any of the elongation inhibitors (e.g. chloramphenicol) that are normally used to preserve polysome species. Under these conditions, the WT cells accumulated 100S ribosomes but retained a very small fraction of polysomes (Fig. 5A, top left). In the RRF-depleted cells, PoTc could not be released from the mRNA templates, resulting in the queuing of the trailing ribosomes; these stalled polyribosomes were co-sedimented with the elongating polysomes (Fig. 5A, top middle). The lack of PoTc recycling reduced the level of ribosomal precursors available to form the 100S complexes, explaining the lower 100S peak in the RRF-depleted strain relative to the WT (Fig. 5B). The depletion of RRF was verified by immunoblotting (Fig. 5C). A very moderate effect on the 100S ribosome was observed in the hflX knockout (Fig. 5A, top right), consistent with previous findings (31, 32). We found that 5 μg/ml of RNase A was sufficient to fully collapse the S. aureus polysomes and 100S complexes but some polysomes in the RRF-depleted cells were resistant to ×4 access of RNase A and were preserved in a disome-like fraction (Fig. 5A, bottom), providing an additional clue to potential ribosome stacking at the termination codon.
Figure 5.
Effects of RRF depletion on ribosome pools and stop codon read-through. A, top panels, accumulation of unrecycled ribosomes and queuing of ribosomes in the RRF-depleted strain. Crude ribosomes were isolated from TSB cultures at A600∼ 1.4 (without the aTc inducer) grown at 37 °C and analyzed by 10–40% sucrose gradient sedimentation. No translational elongation inhibitor (e.g. chloramphenicol) was added throughout the experiments; thus, minimal polysomes were preserved in the WT strain. By contrast, unusually high levels of mixed polysomes and stalled ribosomes accumulated upon RRF depletion, which was also observed to a lesser extent in the hflX knockout mutant. Lower panels, conversion of polysomes into lower order ribosomal complexes by RNase A treatment. Excess RNase A (20 μg/ml) collapsed 100S ribosomes and the WT and ΔhflX polysomes, whereas a fraction of disome-like complexes in the RRF-depleted cells are resistant to RNase A. B, possible outcome of RRF depletion. RRF deficiency prevented the recycling of post-termination complexes, which in turn reduced the ribosome pools available for the formation of 100S ribosomes. C, analysis of RRF and His6-RRF production in the RRF-depleted strain and the hflX knockout mutant used in A. The deletion of hflX did not influence RRF expression. D, RRF deficiency reduced the translational capacity and promoted stop codon read-through. The fluorescence intensity of the GFP reporter plasmid (pDM4) was normalized according to the cell density and compared between the WT and RRF-depleted strains (Δfrr::Km,PTetfrr). RRF depletion reduced the expression of the WT GFP reporter. A stop codon was introduced at the Lys-54 position of the GFP to evaluate the degree of translational read-through. In the absence of the aTc inducer, ribosomes bypassed the Lys-54 stop codon, producing low levels of GFP. The addition of aTc to the Δfrr::Km,PTet-frr strain suppressed read-through. Error bars are S.D. from five biological replicates. p values were calculated by Student's unpaired t test (n = 5, ***, p < 0.001; ****, p < 0.0001; NS, not significant).
To investigate the impact of RRF depletion on stop codon read-through, we compared the synthesis of green fluorescent protein (GFP) between the WT and the RRF-depleted strain carrying a pDM4 reporter plasmid (50). RRF depletion (no aTc) reduced WT GFP fluorescence by 3-fold, suggesting that the perturbation of ribosome recycling prevents new rounds of translation (Fig. 5D). When a termination codon was introduced at the Lys-54 position of gfp, RRF depletion promoted a low level of translational read-through, resulting in an increase in fluorescence intensity relative to the WT. The read-through was reversed upon the addition of aTc (Fig. 5D). We conclude that PoTc recycling and the formation of the 100S ribosome are interdependent and that RRF depletion reduces the translational capacity.
Discussion
The recycling of PoTc, reactivation of the silent 100S ribosomes, and prevention of ribosome degradation are cost-saving activities because ribosome biogenesis is the most energy-consuming cellular process. A recent study offered the first clue that 100S dimers are disassembled and converted into active ribosomes in a reconstituted cell-free translation system consisting solely of purified components, including RRF and EF-G, but not the HflX (30).
In addition to the HflX-dependent disassembly pathway (31), we report here that the 100S ribosome is a previously unrecognized target of RRF and EF-G in S. aureus. RRF and EF-G split the 100S complexes via EF-G-mediated GTP hydrolysis, similar to the GTPase activity of HflX (Fig. 1). The two pathways appear to operate under different conditions, with RRF/EF-G acting as the primary dissociation factor for 100S dimers, whereas HflX serves as a secondary factor that is tightly regulated by heat stress. In fact, RRF and EF-G are highly abundant (∼20 μm) during active growth (46, 51), whereas HflX expression is almost undetectable at this stage (31). In addition, RRF/EF-G and HflX bind to the overlapping regions of the ribosome (32), further suggesting that the dissociation events mediated by these two pathways are mutually exclusive. When HflX is overexpressed in a plasmid, it functionally complements the loss of RRF that acts to recruit EF-G (Fig. 4). RRF/EF-G and HflX also recycle ribosomes from PoTc and thermally arrested ribosomes, respectively (Fig. 6) (32). It is conceivable that the existence of these distinct yet overlapping pathways is evolutionarily favorable for cells to cope with various cellular stresses.
Figure 6.
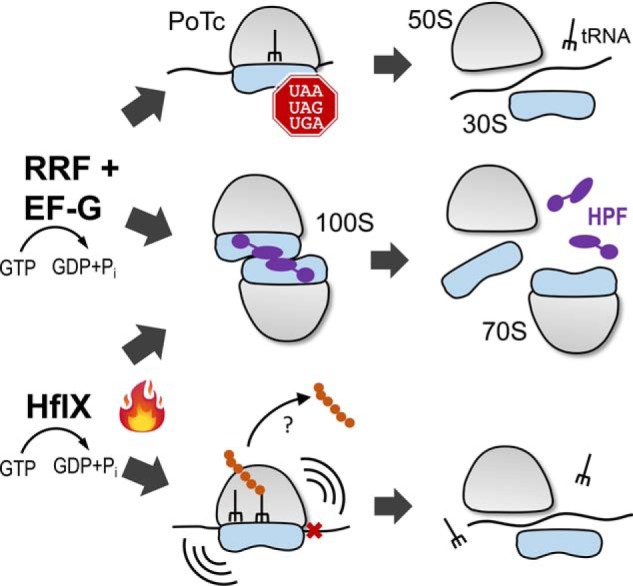
Disassembly pathways of S. aureus 100S ribosomes. RRF and EF-G act in concert to split the PoTc in a GTP hydrolysis-dependent manner. The hibernating 100S ribosome is the second substrate of RRF/EF-G. Thermal stress induces the expression of hflX. HflX recycles 100S ribosomes and rescues stalled 70S ribosomes. A question mark indicates an unknown factor responsible for the hydrolysis of peptidyl-tRNA. HPF is colored in purple.
Structural and biochemical studies revealed that RRF/EF-G and HflX bind to the 50S subunit and occupy the 30S–50S intersubunit space, presumably disrupting the intersubunit bridge B2a between helix 44 and helix 69 via GTP hydrolysis (32, 33, 38, 52). The sequence of events in 100S ribosome disassembly remains unclear. It is possible that the dimers are first split into 70S monomers, followed by dissociation into 30S and 50S subunits. The dimers may concurrently dissociate into 30S and 50S subunits in a single step. Alternatively, their dissociation may involve both single-step and two-step events. It has been shown that subunit dissociation is not essential for translational reinitiation. Instead, initiation could occur in a 70S-scanning mode (53, 54), and cells carrying a covalently linked 70S ribosomes are viable (55). Future kinetic studies are needed to dissect the precise order of 100S disassembly and the departure of HPF and the dissociation factors from the ribosomes. Furthermore, RRF depletion evokes ribosome queuing and translational read-through (Fig. 5). In principle, unrecycled ribosomes could allow reinitiation at the downstream cotranscribed ORF or reinitiation at the first AUG-like codon, provided that fMet-tRNAfMet is available. Alternatively, ribosome queuing could trigger one of the ribosome rescue systems (56, 57) that target stalled elongating ribosomes and the terminating ribosomes. Previous studies have employed heterologous reporter mRNAs to investigate 70S-scanning reinitiation (53, 54), the global impact of RRF insufficiency is not well-understood. Ribosome profiling and MS can be performed to address this outstanding question but are currently beyond the scope of this study.
The in vivo and in vitro activity of the temperature-sensitive E. coli RRF(V117D) mutant (equivalent to S. aureus V116D) has been extensively studied (45, 58, 59). At a nonpermissive temperature, E. coli RRF(V117D) cells are nonviable because the protein is misfolded and rapidly degraded at high temperature. Surprisingly, we found that the recombinant S. aureus RRF(V116D) protein is fully active, and perhaps more potent in dissociating 100S ribosomes in vitro (Fig. 2B). The overexpression of RRF(V116D) in a multicopy plasmid overcomes the growth defects of the RRF-depleted strain (Fig. 3). These observations are supported by findings that the RRF(V116D) protein is fully competent in ribosome binding and EF-G recruitment (Figs. S1 and S2) and that an excess amount of RRF(V116D) can compensate for the instability of the mutant protein in vivo. Consistent with this notion, overexpression of E. coli RRF(V117D) eliminates the temperature-sensitive phenotype (45).
The phenotype of the RRF-depleted strain is more severe on solid media than in the liquid media (Fig. 3, C and D). These striking differences could be due to the local depletion of nutrients and moisture or the accumulation of a toxic compound on the agar plates. For instance, specific solidifying agents may sequester essential micronutrients or contain toxic ingredients. Growth defects caused by the chelation of zinc on the agar plates have been observed in a zinc transporter mutant (60). To test this possibility, we compared the growth defects of the RRF-depleted strain on plates containing routinely used BactoTM agar, Gelrite that was free of phenolic compounds, agarose with a low ash content, and Eiken agar that retained more water. Although agarose and Eiken agar supported increased growth, the defects remained severe (Fig. S3), ruling out the contribution of a solidifying agent. It is possible that shaken liquid culture may provide additional aeration and spatial accessibility to nutrients, resulting in better growth.
Ribosome availability is directly linked to bacterial growth and revival, and ribosome homeostasis is essential for cellular function. The identification of the dual action of RRF/EF-G in maintaining a normal translational cycle and reactivating dormant ribosomes implies that RRF is an ideal antimicrobial target for inhibiting both the proliferation and regrowth of pathogens.
Experimental procedures
Bacterial strains, plasmids, and growth conditions
The strains and plasmids used in this study are listed in Table S1. The methicillin-resistant S. aureus (MRSA) USA300 strain JE2 (GenBank CP000255) was used throughout the study. The deletion mutant of hflX (SAUSA300_1198) has been described previously (31). Unless otherwise noted, S. aureus cells were grown at 37 °C in tryptic soy broth (TSB, Difco) at a 5:1 tube- or flask-to-medium ratio with a 1:100 dilution of an overnight seed culture. TSB agar plates were prepared using DifcoTM agar (Difco, BD 281230), Eiken agar (Eiken, E-MJ00), Gelrite (RPI, G35020), and agarose (GenMate, E-3120). When necessary, erythromycin, chloramphenicol, cadmium chloride, kanamycin, and anhydrotetracycline (all from Sigma-Aldrich) were used at 5 μg/ml, 10 μg/ml, 0.15 mm, 75 μg/ml, and 400 ng/ml, respectively. The restriction-deficient S. aureus RN4220 strain was used as a plasmid passage surrogate before the plasmid was transformed into the destination JE2 derivatives. E. coli cells harboring expression vectors were grown at 16 or 37 °C in LB (Difco). Antibiotics were used at 50 (kanamycin) or 100 μg/ml (ampicillin, Sigma-Aldrich). All GTP analogs were from Sigma-Aldrich. Primers were purchased from IDT DNA and are listed in Table S2.
Construction of conditional RRF knockdown strains MNY162 and MN165
An ∼0.7-kb DNA containing N terminally His6-tagged frr (SAUSA300_1152) was PCR amplified from S. aureus JE2 genomic DNA using the primers P1209 and P1210. The fragment was cloned into the KpnI and SacI sites of pRMC2 (61), generating pRMC2His6RRF under the control of the Tet promoter. The PTetHis6-frr region was released from pRMC2His6RRF by PstI and SacI digestion and cloned into the same sites of pJC1111 (48), generating pJC1111-PTetHis6-frr. This suicide plasmid was integrated into the chromosome of strain RN9011 following standard protocols (48). The PTetHis6-frr allele was subsequently transferred to the JE2 strain via Φ11 phage transduction, resulting in MNY147 bearing the native frr and an extra copy of PTetHis6-frr. The WT frr allele was deleted from MNY147 by homologous recombination using the plasmid pBT2Δfrr::Km. Two independent allelic exchanges were performed, generating the RRF-depleted strains MNY162 and MN165. To construct the pBT2Δfrr::Km plasmid, 1-kb flanking regions of frr were amplified with the primer pairs P1231/P1232 and P1233/P1234 using a two-step PCR protocol with JE2 genomic DNA as the template. The 2-kb SmaI-containing fragment was digested with SacI and SalI and cloned into the same sites of pBT2 (62) to produce pBT2Δfrr. A 1.4-kb kanamycin resistance cassette was released from pBTK (63), end repaired, and cloned into the SmaI site of pBT2Δfrr, generating pBT2Δfrr::Km.
Complementation plasmids
pLI50 (64) was used for genetic complementation. The construction of phflX was reported previously (31). To construct the frr complementation plasmid, a constitutively expressed CodY-dependent promoter (28) was attached to the frr via two-step PCR using the primer pairs P651/P1244 and P1245/P1121 with JE genomic DNA as the template. The PCR product was digested with BamHI and EcoRI and cloned into the same sites in pLI50, generating the pfrr plasmid. To construct pfrr(V116D), a site-directed QuikChange mutagenesis kit (Agilent Genomics) was employed to introduce a V116D substitution using the primers P1219/P1220.
Overexpression and reporter plasmids
The pET28a vector was used to overexpress HflX, RRF, and EF-G recombinant proteins. pET28-HflX has been described previously (31). To construct N terminally His6-tagged RRF, the frr coding region was PCR amplified with P1120/P1121 primers using JE2 genomic DNA as the template. The product was cloned into the NdeI and EcoRI sites of pET28a. The A28E and V116D alleles were introduced by QuikChange mutagenesis using primer pairs P1217/P118 and P1219/P1220, respectively. To construct C terminally His8-tagged EF-G, fusA (locus SAUSA300_0532) was PCR amplified with P1167/P1168, and the fragment was cloned into the NcoI and XhoI sites of pET28a. The R29A, R59A, and H572A mutations were introduced into EF-G by QuikChange mutagenesis using primer pairs P1121/P1122, P1213/P1214, and P1215/P1216, respectively. The K54stop mutation was introduced to the gfp reporter in pDM4 by QuikChange mutagenesis using primers P1255/P1256.
Native 100S ribosome isolation and dissociation
Crude ribosomes were isolated from late log-phase JE2 strains by cryo-milling methods (4, 25). Fifty absorbance units (A260) of ribosomes were layered on a 5–25% sucrose gradient that was prepared on a BioComp Gradient Master. The samples were centrifuged at 210,000 × g at 4 °C in a SW41 rotor in a Beckman Coulter Optima XPN-100 ultracentrifuge for 3 h. Fractionation was performed using a Brandel fractionation system equipped with a UA-6 UV detector. The 100S ribosome peaks were collected and centrifuged in a SW55 Ti rotor at 246,000 × g at 4 °C for 18 h. The ribosome pellet was resuspended in low-salt buffer (20 mm Hepes, pH 7.5, 14 mm Mg(OAc)2, 100 mm KOAc, 0.5 mm phenylmethylsulfonyl fluoride, 1 mm DTT) and quantified according to the A260 value (1 A260 = 23 pmol/ml of 70S).
In vitro 100S dissociation samples were assembled with a 0.2 μm 100S ribosomes, 2 μm proteins (RRF, EF-G, or HflX), and 2 mm GTP analogs in a 100-μl reaction. The reactions were incubated at 37 °C for 30 min before being spun through a 5–20% sucrose gradient. Ribosome profiles were recorded in real time traces probed at an absorbance of 254 nm. For quantification of 100S to 70S ratios, the baseline was first defined as the global minimum excluding the free nucleic acids and protein peak. The boundary of 100S–70S were manually selected from the trough between the peaks. The area under a peak was calculated by ImageJ and divided to obtain the ratio.
For ribosome queuing analysis, cells were grown in TSB at 37 °C until A600 ∼ 1.4 and harvested by rapid filtration and freezing in liquid nitrogen without the addition of chloramphenicol typically used to arrest translation in the ribosome profiling experiments (65). Frozen pellets were cryogenically pulverized on a Retsch Miller as described previously (4, 25). Four A260 units of crude ribosomes were centrifuged through a 10–40% sucrose gradient to separate the ribosome particles. To collapse polysomes, crude ribosomes were untreated or treated with increasing concentrations (5–20 μg/ml) of RNase A (Ambion AM2271) at 25 °C for 1 h prior to ultracentrifugation.
Purification of recombinant proteins and antibody production
The overexpression and purification of the His-tagged recombinant proteins have been described in detail previously (4, 31) and some minor modifications were made in protocol for bacterial growth for EF-G production. The overexpression of EF-G at 37 °C results in an insoluble protein. Cells carrying the EF-G plasmid were grown at 37 °C until an A600 of ∼0.4, after which isopropyl 1-thio-β-d-galactopyranoside was added at a final concentration of 0.5 mm, and induction was performed at 16 °C for 16 h before harvest. Polyclonal antibodies were raised in rabbits by Josman, LLC.
Western blotting
Total S. aureus total lysates were prepared in a bead beater as described previously (28). Immunoblotting was performed as reported (28, 66). Antibodies were used at the following dilution rates: anti-RRF (1:4,000), anti-HflX (1:1,000 dilutions after pre-absorption with lysates of ΔhflX knockout), and anti-EF-G (1:4,000).
GTPase activity assay
The PiColorLock Gold kit (Innova Biosciences) was used to determine the GTP hydrolysis of EF-G variants by measuring the amount of free Pi. Detailed protocols were reported previously (31).
GFP reporter assays
S. aureus cells harboring the pDM4 or pDM4(K54stop) plasmid were grown at 37 °C in TSB in the presence or absence of 400 ng/ml of aTc. Cells were harvested at an A600 of ∼0.3 by centrifugation. The pellets were resuspended and washed three times with sterile 1× PBS buffer to remove background fluorescence from TSB medium. The cell density was adjusted to an A600 of ∼0.2. Two-hundred microliters of the cell suspension was transferred to a black 96-well–flat-bottom plate (Corning Costar®) in triplicate, and GFP fluorescence was measured on a TECAN SPARK plate reader at excitation and emission wavelengths of 485 and 535 nm, respectively. The final fluorescence units were calculated by subtracting the fluorescence from the JE2 background and dividing the results by the final A600 to correct for cell density. The experiments were repeated five times. Statistical significance was evaluated with unpaired t test.
Data availability
All data are contained within the manuscript
Author contributions
A. B. data curation; A. B. and M.-N. Y. formal analysis; A. B. validation; A. B., K. E. S., and M.-N. Y. investigation; A. B. visualization; A. B., K. E. S., and M.-N. Y. methodology; A. B. and M.-N. Y. writing-original draft; M.-N. Y. conceptualization; M.-N. Y. resources; M.-N. Y. supervision; M.-N. Y. funding acquisition; M.-N. Y. project administration; M.-N. Y. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Victor Torres, Irnov Irnov, and Marat Sadykov for generously sharing the S. aureus chromosomal integration system and plasmids, and David Gohara for help with the structural modeling.
This work was supported by National Institutes of Health Grant GM121359 (to M.-N. Y.), Department of Defense Award W81XWH-18-1-0122 (to M.-N. Y.), and the Northwestern University Faculty Start-up fund. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4 and Tables S1 and S2.
- HPF
- hibernation-promoting factor
- RMF
- ribosome modulation factor
- RRF
- ribosome-recycling factor
- PoTc
- post-termination complex
- EF-G
- GTPase elongation factor-G
- aTc
- anhydrotetracyline
- GMPPNP
- guanosine 5′-(β,γ-imido)triphosphate
- TSB
- tryptic soy broth.
References
- 1. Li Y., Sharma M. R., Koripella R. K., Yang Y., Kaushal P. S., Lin Q., Wade J. T., Gray T. A., Derbyshire K. M., Agrawal R. K., and Ojha A. K. (2018) Zinc depletion induces ribosome hibernation in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 115, 8191–8196 10.1073/pnas.1804555115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Polikanov Y. S., Blaha G. M., and Steitz T. A. (2012) How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336, 915–918 10.1126/science.1218538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akiyama T., Williamson K. S., Schaefer R., Pratt S., Chang C. B., and Franklin M. J. (2017) Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. Proc. Natl. Acad. Sci. U.S.A. 114, 3204–3209 10.1073/pnas.1700695114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Basu A., and Yap M. N. (2016) Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic Acids Res. 44, 4881–4893 10.1093/nar/gkw180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shcherbakova K., Nakayama H., and Shimamoto N. (2015) Role of 100S ribosomes in bacterial decay period. Genes Cells 20, 789–801 10.1111/gtc.12273 [DOI] [PubMed] [Google Scholar]
- 6. Ueta M., Ohniwa R. L., Yoshida H., Maki Y., Wada C., and Wada A. (2008) Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J. Biochem. 143, 425–433 10.1093/jb/mvm243 [DOI] [PubMed] [Google Scholar]
- 7. Hood R. D., Higgins S. A., Flamholz A., Nichols R. J., and Savage D. F. (2016) The stringent response regulates adaptation to darkness in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A. 113, E4867–E4876 10.1073/pnas.1524915113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Akanuma G., Kazo Y., Tagami K., Hiraoka H., Yano K., Suzuki S., Hanai R., Nanamiya H., Kato-Yamada Y., and Kawamura F. (2016) Ribosome dimerization is essential for the efficient regrowth of Bacillus subtilis. Microbiology 162, 448–458 10.1099/mic.0.000234 [DOI] [PubMed] [Google Scholar]
- 9. Puri P., Eckhardt T. H., Franken L. E., Fusetti F., Stuart M. C., Boekema E. J., Kuipers O. P., Kok J., and Poolman B. (2014) Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol. Microbiol. 91, 394–407 10.1111/mmi.12468 [DOI] [PubMed] [Google Scholar]
- 10. Kline B. C., McKay S. L., Tang W. W., and Portnoy D. A. (2015) The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J. Bacteriol. 197, 581–591 10.1128/JB.02223-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McKay S. L., and Portnoy D. A. (2015) Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob. Agents Chemother. 59, 6992–6999 10.1128/AAC.01532-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Williamson K. S., Richards L. A., Perez-Osorio A. C., Pitts B., McInnerney K., Stewart P. S., and Franklin M. J. (2012) Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 194, 2062–2073 10.1128/JB.00022-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Michalik S., Depke M., Murr A., Gesell Salazar M., Kusebauch U., Sun Z., Meyer T. C., Surmann K., Pförtner H., Hildebrandt P., Weiss S., Palma Medina L. M., Gutjahr M., Hammer E., Becher D., et al. (2017) A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions. Sci. Rep 7, 9718 10.1038/s41598-017-10059-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palma Medina L. M., Becker A. K., Michalik S., Yedavally H., Raineri E. J. M., Hildebrandt P., Gesell Salazar M., Surmann K., Pförtner H., Mekonnen S. A., Salvati A., Kaderali L., van Dijl J. M., and Völker U. (2019) Metabolic cross-talk between human bronchial epithelial cells and internalized Staphylococcus aureus as a driver for infection. Mol. Cell. Proteomics 18, 892–908 10.1074/mcp.RA118.001138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gohara D. W., and Yap M. F. (2018) Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management. Curr. Genet. 64, 753–760 10.1007/s00294-017-0796-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matzov D., Bashan A., Yap M. F., and Yonath A. (2019) Stress response as implemented by hibernating ribosomes: a structural overview. FEBS J. 286, 3558–3565 10.1111/febs.14968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prossliner T., Skovbo Winther K., Sørensen M. A., and Gerdes K. (2018) Ribosome hibernation. Annu Rev. Genet 52, 321–348 10.1146/annurev-genet-120215-035130 [DOI] [PubMed] [Google Scholar]
- 18. Yoshida H., and Wada A. (2014) The 100S ribosome: ribosomal hibernation induced by stress. Wiley Interdiscip. Rev. RNA 5, 723–732 10.1002/wrna.1242 [DOI] [PubMed] [Google Scholar]
- 19. Trösch R., and Willmund F. (2018) The conserved theme of ribosome hibernation: from bacteria to chloroplasts of plants. Biol. Chem. 400, 879–893 [DOI] [PubMed] [Google Scholar]
- 20. Beckert B., Abdelshahid M., Schäfer H., Steinchen W., Arenz S., Berninghausen O., Beckmann R., Bange G., Turgay K., and Wilson D. N. (2017) Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J. 36, 2061–2072 10.15252/embj.201696189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Franken L. E., Oostergetel G. T., Pijning T., Puri P., Arkhipova V., Boekema E. J., Poolman B., and Guskov A. (2017) A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy. Nat. Commun. 8, 722 10.1038/s41467-017-00718-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khusainov I., Vicens Q., Ayupov R., Usachev K., Myasnikov A., Simonetti A., Validov S., Kieffer B., Yusupova G., Yusupov M., and Hashem Y. (2017) Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF. EMBO J. 36, 2073–2087 10.15252/embj.201696105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matzov D., Aibara S., Basu A., Zimmerman E., Bashan A., Yap M. N., Amunts A., and Yonath A. E. (2017) The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat. Commun. 8, 723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beckert B., Turk M., Czech A., Berninghausen O., Beckmann R., Ignatova Z., Plitzko J. M., and Wilson D. N. (2018) Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nat. Microbiol. 3, 1115–1121 10.1038/s41564-018-0237-0 [DOI] [PubMed] [Google Scholar]
- 25. Davis A. R., Gohara D. W., and Yap M. N. (2014) Sequence selectivity of macrolide-induced translational attenuation. Proc. Natl. Acad. Sci. U.S.A. 111, 15379–15384 10.1073/pnas.1410356111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ueta M., Wada C., Daifuku T., Sako Y., Bessho Y., Kitamura A., Ohniwa R. L., Morikawa K., Yoshida H., Kato T., Miyata T., Namba K., and Wada A. (2013) Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells 18, 554–574 10.1111/gtc.12057 [DOI] [PubMed] [Google Scholar]
- 27. Ueta M., Wada C., and Wada A. (2010) Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells 15, 43–58 10.1111/j.1365-2443.2009.01364.x [DOI] [PubMed] [Google Scholar]
- 28. Basu A., Shields K. E., Eickhoff C. S., Hoft D. F., and Yap M. N. (2018) Thermal and nutritional regulation of ribosome hibernation in Staphylococcus aureus. J. Bacteriol. 200, e00426–00418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Trauner A., Lougheed K. E., Bennett M. H., Hingley-Wilson S. M., and Williams H. D. (2012) The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J. Biol. Chem. 287, 24053–24063 10.1074/jbc.M112.364851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feaga H. A., Kopylov M., Kim J. K., Jovanovic M., and Dworkin J. (2020) Ribosome dimerization protects the small subunit. J. Bacteriol. 10.1128/JB.00009-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Basu A., and Yap M. N. (2017) Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase. Proc. Natl. Acad. Sci. U.S.A. 114, E8165–E8173 10.1073/pnas.1709588114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y., Mandava C. S., Cao W., Li X., Zhang D., Li N., Zhang Y., Zhang X., Qin Y., Mi K., Lei J., Sanyal S., and Gao N. (2015) HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat. Struct. Mol. Biol. 22, 906–913 10.1038/nsmb.3103 [DOI] [PubMed] [Google Scholar]
- 33. Dey S., Biswas C., and Sengupta J. (2018) The universally conserved GTPase HflX is an RNA helicase that restores heat-damaged Escherichia coli ribosomes. J. Cell Biol. 217, 2519–2529 10.1083/jcb.201711131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodnina M. V. (2018) Translation in prokaryotes. Cold Spring Harb. Perspect. Biol. 10, 10.1101/cshperspect.a032664 10.1101/cshperspect.a032664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen Y., Kaji A., Kaji H., and Cooperman B. S. (2017) The kinetic mechanism of bacterial ribosome recycling. Nucleic Acids Res. 45, 10168–10177 10.1093/nar/gkx694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iwakura N., Yokoyama T., Quaglia F., Mitsuoka K., Mio K., Shigematsu H., Shirouzu M., Kaji A., and Kaji H. (2017) Chemical and structural characterization of a model post-termination complex (PoTC) for the ribosome recycling reaction: evidence for the release of the mRNA by RRF and EF-G. PLoS ONE 12, e0177972 10.1371/journal.pone.0177972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prabhakar A., Capece M. C., Petrov A., Choi J., and Puglisi J. D. (2017) Post-termination ribosome intermediate acts as the gateway to ribosome recycling. Cell Rep. 20, 161–172 10.1016/j.celrep.2017.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fu Z., Kaledhonkar S., Borg A., Sun M., Chen B., Grassucci R. A., Ehrenberg M., and Frank J. (2016) Key intermediates in ribosome recycling visualized by time-resolved cryoelectron microscopy. Structure 24, 2092–2101 10.1016/j.str.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou D., Tanzawa T., Lin J., and Gagnon M. G. (2020) Structural basis for ribosome recycling by RRF and tRNA. Nat. Struct. Mol. Biol. 27, 25–32 10.1038/s41594-019-0350-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perez Boerema A., Aibara S., Paul B., Tobiasson V., Kimanius D., Forsberg B. O., Wallden K., Lindahl E., and Amunts A. (2018) Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat. Plants 4, 212–217 10.1038/s41477-018-0129-6 [DOI] [PubMed] [Google Scholar]
- 41. Hirokawa G., Iwakura N., Kaji A., and Kaji H. (2008) The role of GTP in transient splitting of 70S ribosomes by RRF (ribosome recycling factor) and EF-G (elongation factor G). Nucleic Acids Res. 36, 6676–6687 10.1093/nar/gkn647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zavialov A. V., Hauryliuk V. V., and Ehrenberg M. (2005) Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol. Cell 18, 675–686 10.1016/j.molcel.2005.05.016 [DOI] [PubMed] [Google Scholar]
- 43. Mohr D., Wintermeyer W., and Rodnina M. V. (2000) Arginines 29 and 59 of elongation factor G are important for GTP hydrolysis or translocation on the ribosome. EMBO J. 19, 3458–3464 10.1093/emboj/19.13.3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Savelsbergh A., Matassova N. B., Rodnina M. V., and Wintermeyer W. (2000) Role of domains 4 and 5 in elongation factor G functions on the ribosome. J. Mol. Biol. 300, 951–961 10.1006/jmbi.2000.3886 [DOI] [PubMed] [Google Scholar]
- 45. Janosi L., Mottagui-Tabar S., Isaksson L. A., Sekine Y., Ohtsubo E., Zhang S., Goon S., Nelken S., Shuda M., and Kaji A. (1998) Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 17, 1141–1151 10.1093/emboj/17.4.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Raj V. S., Kaji H., and Kaji A. (2005) Interaction of RRF and EF-G from E. coli and T. thermophilus with ribosomes from both origins: insight into the mechanism of the ribosome recycling step. RNA 11, 275–284 10.1261/rna.7201805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamami T., Ito K., Fujiwara T., and Nakamura Y. (2005) Heterologous expression of Aquifex aeolicus ribosome recycling factor in Escherichia coli is dominant lethal by forming a complex that lacks functional co-ordination for ribosome disassembly. Mol. Microbiol 55, 150–161 [DOI] [PubMed] [Google Scholar]
- 48. Chen J., Yoong P., Ram G., Torres V. J., and Novick R. P. (2014) Single-copy vectors for integration at the SaPI1 attachment site for Staphylococcus aureus. Plasmid 76, 1–7 10.1016/j.plasmid.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., and Ueda T. (2001) Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 10.1038/90802 [DOI] [PubMed] [Google Scholar]
- 50. Moormeier D. E., Endres J. L., Mann E. E., Sadykov M. R., Horswill A. R., Rice K. C., Fey P. D., and Bayles K. W. (2013) Use of microfluidic technology to analyze gene expression during Staphylococcus aureus biofilm formation reveals distinct physiological niches. Appl. Environ. Microbiol. 79, 3413–3424 10.1128/AEM.00395-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Caldas T., Laalami S., and Richarme G. (2000) Chaperone properties of bacterial elongation factor EF-G and initiation factor IF2. J. Biol. Chem. 275, 855–860 10.1074/jbc.275.2.855 [DOI] [PubMed] [Google Scholar]
- 52. Coatham M. L., Brandon H. E., Fischer J. J., Schümmer T., and Wieden H. J. (2016) The conserved GTPase HflX is a ribosome splitting factor that binds to the E-site of the bacterial ribosome. Nucleic Acids Res. 44, 1952–1961 10.1093/nar/gkv1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Qin B., Yamamoto H., Ueda T., Varshney U., and Nierhaus K. H. (2016) The termination phase in protein synthesis is not obligatorily followed by the RRF/EF-G-dependent recycling phase. J. Mol. Biol. 428, 3577–3587 10.1016/j.jmb.2016.05.019 [DOI] [PubMed] [Google Scholar]
- 54. Yamamoto H., Wittek D., Gupta R., Qin B., Ueda T., Krause R., Yamamoto K., Albrecht R., Pech M., and Nierhaus K. H. (2016) 70S-scanning initiation is a novel and frequent initiation mode of ribosomal translation in bacteria. Proc. Natl. Acad. Sci. U.S.A. 113, E1180–E1189 10.1073/pnas.1524554113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Orelle C., Carlson E. D., Szal T., Florin T., Jewett M. C., and Mankin A. S. (2015) Protein synthesis by ribosomes with tethered subunits. Nature 524, 119–124 10.1038/nature14862 [DOI] [PubMed] [Google Scholar]
- 56. Burroughs A. M., and Aravind L. (2019) The origin and evolution of release factors: implications for translation termination, ribosome rescue, and quality control pathways. Int. J. Mol. Sci. 20, e1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Keiler K. C. (2015) Mechanisms of ribosome rescue in bacteria. Nat. Rev. Microbiol. 13, 285–297 10.1038/nrmicro3438 [DOI] [PubMed] [Google Scholar]
- 58. Hirokawa G., Inokuchi H., Kaji H., Igarashi K., and Kaji A. (2004) In vivo effect of inactivation of ribosome recycling factor: fate of ribosomes after unscheduled translation downstream of open reading frame. Mol. Microbiol. 54, 1011–1021 10.1111/j.1365-2958.2004.04324.x [DOI] [PubMed] [Google Scholar]
- 59. Zhang L., Guo P., Zhang H., and Jing G. (2006) Cooperative unfolding of Escherichia coli ribosome recycling factor originating from its domain-domain interaction and its implication for function. Arch Biochem. Biophys. 450, 191–202 10.1016/j.abb.2006.03.029 [DOI] [PubMed] [Google Scholar]
- 60. Cerasi M., Liu J. Z., Ammendola S., Poe A. J., Petrarca P., Pesciaroli M., Pasquali P., Raffatellu M., and Battistoni A. (2014) The ZupT transporter plays an important role in zinc homeostasis and contributes to Salmonella enterica virulence. Metallomics 6, 845–853 10.1039/C3MT00352C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Corrigan R. M., and Foster T. J. (2009) An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmids 61, 126–129 10.1016/j.plasmid.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 62. Brückner R. (1992) A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122, 187–192 10.1016/0378-1119(92)90048-T [DOI] [PubMed] [Google Scholar]
- 63. Fuller J. R., Vitko N. P., Perkowski E. F., Scott E., Khatri D., Spontak J. S., Thurlow L. R., and Richardson A. R. (2011) Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front. Cell Infect. Microbiol. 1, 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee C. Y., Buranen S. L., and Ye Z. H. (1991) Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103, 101–105 10.1016/0378-1119(91)90399-V [DOI] [PubMed] [Google Scholar]
- 65. Mohammad F., Green R., and Buskirk A. R. (2019) A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. Elife 8, e42591 10.7554/eLife.42591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dzyubak E., and Yap M. N. (2016) The expression of antibiotic resistance methyltransferase correlates with mRNA stability independently of ribosome stalling. Antimicrob. Agents Chemother. 60, 7178–7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript