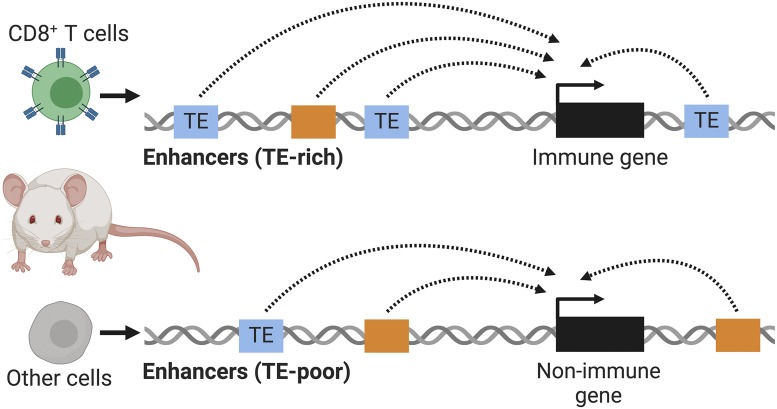
Mammalian genomes are replete with transposable elements (TEs): parasitic genetic sequences that can replicate to high copy numbers within host genomes (1). TEs are widely recognized as a potent source of cell type- and context-specific regulatory elements (2, 3). In PNAS, Ye et al. (4) analyze chromatin profiling data from mouse CD8+ T lymphocytes and find that multiple TE families contribute to predicted regulatory sequences. Compared to other cells, immune cells show the highest enrichment of TE-derived enhancers (Fig. 1), suggesting that TE cooption may preferentially influence immune regulatory networks.
Fig. 1.
Ye et al. (4) observe higher occurrences of TEs near immune genes. They hypothesize that the abundance of TEs in immune-associated regions of the mouse genome has enabled their regulatory cooption as immune enhancers, potentially facilitating the rapid regulatory evolution needed for adaptive immunity.
Transcriptional regulatory networks underlie the gene expression programs that determine cellular identity, function, and response to stimuli. In the genome, regulatory elements such as promoters and enhancers act as “wires” to connect genes into regulatory networks and control nearby gene expression. Changes to regulatory networks are recognized as an important mechanism for organismal evolution (5), but mechanisms driving the emergence of new regulatory elements are still poorly understood.
Owing to their ability to replicate throughout the host genome, TEs have long been hypothesized to play a role in the evolution of regulatory networks (6, 7). Although most TEs no longer encode functional proteins, many retain transcription factor binding sites and can thus alter the expression of nearby genes. Over the past decade, there have been numerous studies characterizing important roles for TEs in host gene regulation (reviewed in ref. 2). These findings point to the cooption of TEs as a general mechanism shaping the evolution of mammalian gene regulatory networks.
As studies in different species and biological systems uncover more examples of TE cooption, one key question that remains is whether the evolutionary trajectories of all regulatory networks are similarly affected by TEs. Nearly all mammalian cell types and tissues exhibit some degree of TE-derived regulatory activity, but the relative contribution of TEs to the genome-wide regulatory landscape of each tissue varies dramatically (3, 8). Increasing evidence suggests that certain cell types are particularly prone to TE-mediated gene regulation, including embryonic stem cells (9, 10), placental cells (11), and immune cells (12, 13). While our current knowledge of TE-derived regulatory elements is based only on a rather limited number of studies, it is intriguing to consider why these regulatory networks are prone to coopting TEs.
The high levels of TE-derived regulatory activity in reproductive and germline tissues can be explained by the fact that TEs are under strong evolutionary pressure to be transcriptionally active in those tissues (10, 14). From the selfish perspective of the TE, new insertions must occur in early embryonic or germline cells to be inherited. The abundance of TEs showing transcriptional activity in these cells may lead to an increased probability for some to be directly coopted for cellular gene regulation. Indeed, a large body of evidence indicates that many essential processes in embryonic and placenta development are regulated by TE-derived proteins, promoters, noncoding RNAs, regulatory elements, and topologically associated domain boundary elements (reviewed in ref. 15). These findings have been described in both humans and mice, which have largely distinct TE profiles, implying that TEs may have an evolutionarily widespread role in shaping regulatory networks underlying early development.
Do any other cell types show a propensity for TE-mediated gene regulation? A growing body of evidence suggests that TEs are also frequently coopted to regulate genes involved in immune processes. Studies have reported the use of TEs as promoters (16), interferon-inducible enhancers (12), and insulator elements in immune cells (13). Perhaps regulatory networks involved in immunity evolve under unique pressures that favor TE cooption. Protein-coding immune genes are among the most rapidly evolving genes in the genome, reflecting the constant need to adapt against new and evolving pathogens (17). Given that active or recently active TEs are a major source of genetic polymorphism, they may facilitate rapid adaptive evolution of immune responses at the gene regulatory level. However, these ideas remain speculative, as few studies to date have systematically examined TEs in the context of immune regulatory landscapes.
In PNAS, Ye et al. (4) assess the contribution of TEs to immune regulatory networks by analyzing genome-wide chromatin profiling data from mouse CD8+ T cells. The authors survey TE content within predicted enhancers using assay for transposase-accessible chromatin coupled with high-throughput sequencing (ATAC-seq) to generate genome-wide profiles of open chromatin, and chromatin immunoprecipitation sequencing (ChIP-seq) for histone modifications H3K27ac, H3K4me1, and H3K4me3 to identify potential regulatory elements in both naïve T cells and in vitro differentiated cells.
Ye et al. (4) first examine how TEs contribute to putative T cell enhancers. They partition each predicted enhancer region into three distinct domains: an accessible core (defined by the ATAC peak), proximal flanks, and distal flanks. They find that TEs contribute to all three domain components of predicted T cell enhancers and that different TE families colocalize to different domains. TEs come in a wide variety of different classes, families, and subfamilies, including endogenous retroviruses (ERVs) and long or short interspersed elements (LINEs and SINEs). The authors find that the accessible cores of lymphocyte enhancers are enriched for TEs belonging to the ERV and mammalian interspersed repeat SINE families, as well as L2 LINEs. In contrast, enhancer flanks are depleted of these families but enriched for other SINEs, particularly the rodent-specific B1 elements. The authors propose that different TEs may be predisposed to contribute distinct regulatory functions. For example, ERVs enriched at enhancer cores may provide transcription factor binding sites, while B1 SINEs enriched in enhancer flanks are more likely to facilitate chromatin organization. They note that primate genomes, which lack B1 SINEs, instead utilize the sequence-similar Alu SINEs to influence nucleosome positioning (18). These observations reveal a complex and multifaceted contribution by TEs to the functional sequences underlying T cell enhancers.
The authors next ask whether TE-derived enhancers regulate functionally distinct types of genes. They computationally predict enhancer gene targets based on coregulation of ATAC-seq and RNA-seq (over T cell differentiation) as well as Hi-C chromatin conformation data. Ye et al. (4) note that TE-derived enhancers are often linked to the same gene targets as TE-poor enhancers. Likewise, the majority of enhancers seem to contribute to the same overarching biological processes (e.g., lymphocyte migration and differentiation), regardless of whether they are TE-derived or not. This suggests that coopted TEs may have been recruited to act as secondary or backup enhancers, buffering against the potentially deleterious phenotypic consequences of enhancer loss.
The authors go on to compare enhancers between immune- and nonimmune-related cells, in order to distinguish elements that may be particularly important for immune functions. They use published datasets from the Enhancer Atlas database to obtain all available immune tissues and 14 other nonimmune tissues. Importantly, they find that TEs are enriched within enhancers that are specific to immune cells, in contrast to other cell types. In other words, a higher proportion of immune enhancers are TE-derived, consistent with the idea that immune tissues are more prone to coopting TEs as regulatory elements.
In PNAS, Ye et al. assess the contribution of TEs to immune regulatory networks by analyzing genome-wide chromatin profiling data from mouse CD8+ T cells.
Ye et al. (4) add to a growing number of analyses that implicate an elevated role for TEs in immune regulatory networks (8, 19). The authors propose several explanations for these observations. First, there may be pathogen-driven selection for TEs to be coopted and drive adaptive evolution of immunity. New TE insertions may confer novel gene expression patterns, or robustness to existing immune-related expression patterns, and become fixed in the species genome as a beneficial mutation. Alternatively, immune genes may simply be less essential, and TEs accumulate around them because there is reduced negative selection against TEs and other mutations, or strong selection for specific alleles of immune genes may have resulted in adjacent TEs also being preserved as genomic “hitchhikers” during a selective sweep. While the mechanisms driving TE accumulations near immune genes remain unclear, it is likely that their elevated abundance has facilitated regulatory cooption for immunity.
An outstanding question is how many of the TE-derived regulatory elements identified in this study (4) are truly biologically significant. Genome-wide analyses relying on chromatin profiling assays such as ATAC-seq or ChIP-seq implicitly assume that elements marked by a particular biochemical property (e.g., chromatin accessibility) have a gene regulatory function, which is often not the case. Recent studies using CRISPR-mediated silencing or deletion have confirmed important regulatory functions for a number of TE-derived enhancers (9, 12), but they have also revealed that many TEs bearing enhancer signatures do not affect the expression of any genes (20). Thus, while TEs may be a major source of predicted regulatory elements in immune cells, it remains possible that the majority of these elements have minimal or no beneficial function for immune cell biology.
Finally, the association between TE-derived enhancers and immune genes may have significant implications for understanding how genes are dysregulated in immune-related diseases. Most TEs in any given cell type are epigenetically silenced, and pathological TE dysregulation has been implicated in numerous diseases including autoimmunity (21) and cancer (22). This study (4) suggests that epigenetic dysregulation of TE-derived enhancers may result in the inappropriate activation of immune genes. The cooption of TEs for host immune evolution is therefore akin to a “double-edged sword”: TEs provide regulatory elements for immune genes but are potentially more prone to reactivation during pathogenesis.
It is now clear that TE cooption is an important and recurrent mechanism influencing the evolution of mammalian gene regulatory networks. Studies like Ye et al. (4) are beginning to reveal the “rules of cooption” by which TEs contribute as regulatory elements in both health and disease.
Footnotes
The authors declare no competing interest.
See companion article, “Specific subfamilies of transposable elements contribute to different domains of T lymphocyte enhancers,” 10.1073/pnas.1912008117.
References
- 1.Platt R. N. 2nd, Vandewege M. W., Ray D. A., Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 26, 25–43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chuong E. B., Elde N. C., Feschotte C., Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 18, 71–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundaram V., et al. , Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 24, 1963–1976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye M., et al. , Specific subfamilies of transposable elements contribute to different domains of T lymphocyte enhancers. Proc. Natl. Acad. Sci. U.S.A. 117, 7905–7916(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long H. K., Prescott S. L., Wysocka J., Ever-changing landscapes: Transcriptional enhancers in development and evolution. Cell 167, 1170–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britten R. J., Davidson E. H., Gene regulation for higher cells: A theory. Science 165, 349–357 (1969). [DOI] [PubMed] [Google Scholar]
- 7.McClintock B., The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. U.S.A. 36, 344–355 (1950). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pehrsson E. C., Choudhary M. N. K., Sundaram V., Wang T., The epigenomic landscape of transposable elements across normal human development and anatomy. Nat. Commun. 10, 5640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuentes D. R., Swigut T., Wysocka J., Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. eLife 7, e35989 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Römer C., Singh M., Hurst L. D., Izsvák Z., How to tame an endogenous retrovirus: HERVH and the evolution of human pluripotency. Curr. Opin. Virol. 25, 49–58 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Dunn-Fletcher C. E., et al. , Anthropoid primate-specific retroviral element THE1B controls expression of CRH in placenta and alters gestation length. PLoS Biol. 16, e2006337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuong E. B., Elde N. C., Feschotte C., Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., et al. , MIR retrotransposon sequences provide insulators to the human genome. Proc. Natl. Acad. Sci. U.S.A. 112, E4428–E4437 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haig D., Retroviruses and the placenta. Curr. Biol. 22, R609–R613 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Percharde M., Sultana T., Ramalho-Santos M., What doesn’t kill you makes you stronger: Transposons as dual players in chromatin regulation and genomic variation. BioEssays 42, e1900232 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Romanish M. T., Lock W. M., van de Lagemaat L. N., Dunn C. A., Mager D. L., Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 3, e10 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daugherty M. D., Malik H. S., Rules of engagement: Molecular insights from host-virus arms races. Annu. Rev. Genet. 46, 677–700 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Tanaka Y., Yamashita R., Suzuki Y., Nakai K., Effects of Alu elements on global nucleosome positioning in the human genome. BMC Genomics 11, 309 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simonti C. N., Pavlicev M., Capra J. A., Transposable element exaptation into regulatory regions is rare, influenced by evolutionary age, and subject to pleiotropic constraints. Mol. Biol. Evol. 34, 2856–2869 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Todd C. D., Deniz Ö., Taylor D., Branco M. R., Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. eLife 8, e44344 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokuyama M., et al. , ERVmap analysis reveals genome-wide transcription of human endogenous retroviruses. Proc. Natl. Acad. Sci. U.S.A. 115, 12565–12572 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang H. S., et al. , Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet. 51, 611–617 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]