Significance
Cryo-electron microscopy now shows what many massive protein molecular machines look like. What is needed is a methodology that can probe into the machinery under working conditions. This work demonstrates the ability of hydrogen exchange mass spectrometry to provide detailed structural, biophysical, and functional information on the hexameric 0.6-MDa AAA+ Hsp104 protein in its various static nucleotide-bound states and during functional cycling at near amino acid resolution.
Keywords: Hsp104, HX MS, HDX-MS, AAA+, molecular machine
Abstract
Hsp104 provides a valuable model for the many essential proteostatic functions performed by the AAA+ superfamily of protein molecular machines. We developed and used a powerful hydrogen exchange mass spectrometry (HX MS) analysis that can provide positionally resolved information on structure, dynamics, and energetics of the Hsp104 molecular machinery, even during functional cycling. HX MS reveals that the ATPase cycle is rate-limited by ADP release from nucleotide-binding domain 1 (NBD1). The middle domain (MD) serves to regulate Hsp104 activity by slowing ADP release. Mutational potentiation accelerates ADP release, thereby increasing ATPase activity. It reduces time in the open state, thereby decreasing substrate protein loss. During active cycling, Hsp104 transits repeatedly between whole hexamer closed and open states. Under diverse conditions, the shift of open/closed balance can lead to premature substrate loss, normal processing, or the generation of a strong pulling force. HX MS exposes the mechanisms of these functions at near-residue resolution.
The homohexameric Hsp104 disaggregase is a highly studied member of the AAA+ superfamily of molecular machines, which perform myriad biological functions (1–5). Hsp104 transduces the favorable energy of ATP binding and hydrolysis through local and global structure changes (6, 7) to drive the forceful energy-requiring disaggregation of substrate proteins by drawing them into and sometimes through its narrow axial pore (8–11). In Saccharomyces cerevisiae, it functions to reactivate proteins that aggregate due to environmental stress including heat and chemical shock (12–17). Perversely, WT-type Hsp104 can appear unable to unfold even weakly structured amorphous aggregates (in the absence of cochaperones Hsp70 and Hsp40), but it can both foster and disrupt prion amyloid (10, 18–22) and, when potentiated, it is able to dismantle even ultrastable aggregates such as TDP-43 fibrils and α-synuclein amyloid (19, 23–25). These properties suggest its possible use as a therapeutic agent (24). However, the structural and dynamical mechanisms that produce and relieve self-inhibition and regulate Hsp104 function more broadly remain poorly understood.
The structure of Hsp104 in its different functional forms has been solved at high resolution by cryo-electron microscopy (cryo-EM) and X-ray crystallography (6, 26–28). Each of the six Hsp104 protomers consists of an N-terminal domain (NTD), two conserved AAA+ nucleotide-binding domains (NBD1 and NBD2) with NBD1 split by an intervening middle domain (MD), and a C-terminal domain (CTD). Functionally important elements include a nucleotide-binding Walker A motif in both NBDs and loops responsible for binding substrate proteins and translocating them through a central axial pore (27, 29, 30). Cryo-EM results for Hsp104 obtained as snapshots during active cycling also define alternative global-scale hexamer conformations: the open, closed, and extended forms (6, 27, 31). The open state has a large central pore (30 Å diameter), unstructured pore loops, binds substrate protein weakly, and accounts for a dominant fraction of the cycling population. The same conformation is adopted by the static protein in the presence of bound ADP. The closed conformation has a narrow central pore (10 Å diameter), and tightly structured pore loops, which bind substrate strongly. It populates in the presence of the slowly hydrolyzing ATP analog ATPγS (6) and in a mutant that is able to bind but not hydrolyze ATP (31). An extended conformation seen by cryo-EM (6, 31) is much like the closed conformation, but is distinguished by a single offset protomer interface suggested to drive substrate translocation by an alternation (ratcheting) between the closed and extended states. Similar translocation models have been suggested for other AAA+ proteins (32).
We previously developed and used hydrogen exchange mass spectrometry (HX MS) (33–40) to study the structure of WT Hsp104 in physiological solution (7). HX MS was able to distinguish the large-scale open and closed/extended states under static and actively cycling conditions but not the closed and extended states, which we will refer to as the closed state. At a finer level HX MS proved able to characterize the various working and nonworking elements throughout Hsp104 including flexible elements that escape imaging methods. This paper reports the exploitation of these capabilities to study Hsp104 function and its regulation during active functional cycling.
A pivotal result is that the ATPase cycle is normally rate-limited by release of ADP from the NBD1 active site in the open state, which therefore becomes a principal regulatory control point. The slow rate-limiting ADP off rate prolongs occupation of the ADP-bound open state. This favors the loss of weakly bound substrate, counteracting the processing of even unstable amorphous aggregates. The autoinhibitory mechanism that down-regulates the potentially cytotoxic capability of WT Hsp104 is vested in an MD to MD cross-protomer bridge that forms only in the ADP-bound open state and suppresses the dynamics that allow ADP release. The three potentiated Hsp104 mutants studied here act by speeding ADP release through mechanisms that differ in detail, which we define. Faster ADP release from NBD1 potentiates activity in two ways. Accelerating the ATPase cycle increases the power output of the Hsp104 machine. Diminishing the cycle time spent in the ADP-bound open state counters substrate protein loss (6, 41, 42). Different conditions can cause these two determinants, ATPase activity and substrate loss, to concur or conflict, which helps to explain some otherwise puzzling behavior. A second formative result is that Hsp104 cycles repeatedly through the open state to allow ADP to ATP exchange. Transition to the open state promotes substrate loss, but it may also be coordinated to generate a strong translocation force by allowing the combined action of several ATPase events.
Results
Potentiation Enhances ATPase Activity.
The mutational potentiation of Hsp104 promotes faster ATP turnover and the ability to solubilize neurodegenerative amyloids and mitigate their toxicity (23, 24, 43). Many known potentiation sites are located in the Hsp104 MD, in the MD-NBD1 interface, or close to the nucleotide-binding site in NBD1 (24, 43). We selected three mutations (A503S, E360R, I187F) as representative of these three classes (43). Fig. 1 illustrates their position relative to the Hsp104 NBD1 and NBD2 nucleotide-binding sites.
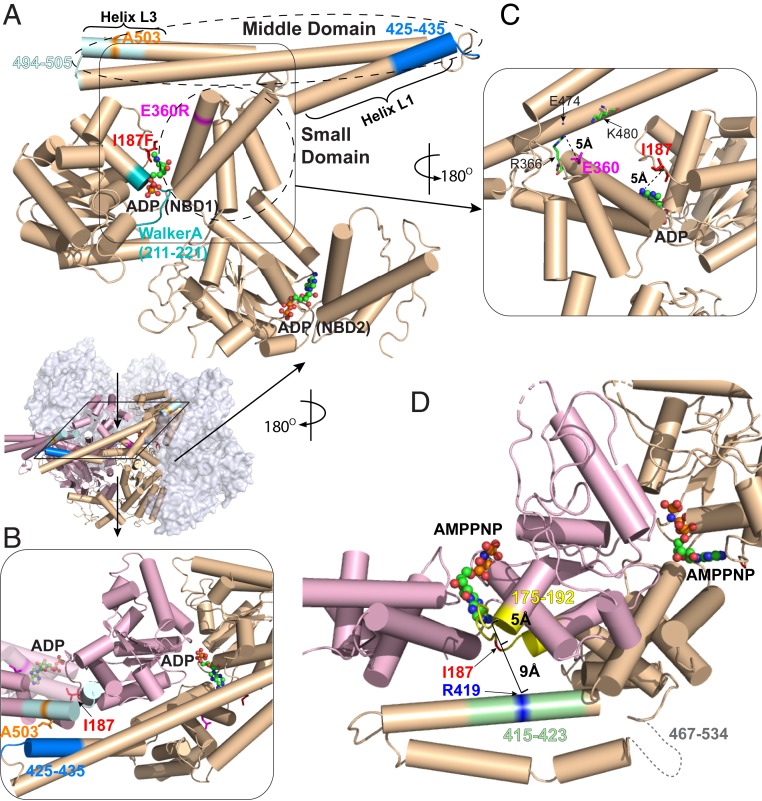
Fig. 1.
Location of potentiating Hsp104 mutations. (A) The three mutations studied here (A503S, E360R, I187F) are highlighted in stick representation along with other pertinent elements on a protomer (P3) in the ADP-bound open hexamer conformation (PDB ID code 5VY8) (6). Peptide fragments measured by HX MS are 494–505 covering helix L3 (light cyan), 425–435 covering a part of helix L1 (blue), and 211–221 covering Walker A (cyan). ADP is in ball-and-stick representation. The boundaries of the MD and the small domain of NBD1 are outlined. (B) Top view. A503 in MD helix L3 (light blue) interacts with L1 (dark blue) across an interprotomer interface (P3 in pink to P4 in wheat). Relevant structural elements are in the same colors as in A. (C) The NBD1 small domain. E360 forms a salt bridge with R366 (∼5 Å) and is close to other charged residues (e.g., K480 and E474) on the MD of the same protomer. (D) The main chain atoms of I187 are within 5 Å of the adenosine moiety of the bound nucleotide to one side and 9 Å from R419 (blue) across an interprotomer interface to the other side (P3 to P4). Segment 175–192 (interhelix B2-B3 loop) is in yellow and 415–423 in helix L1 is in light green. Bound AMPPNP is in ball-and-stick representation. The AMPPNP-bound cryo-EM structure (PDB ID code 5KNE) is used here to represent the ATP-bound state because the same MD configuration has been observed in a subclass of the ATPγS-bound protein (6, 27).
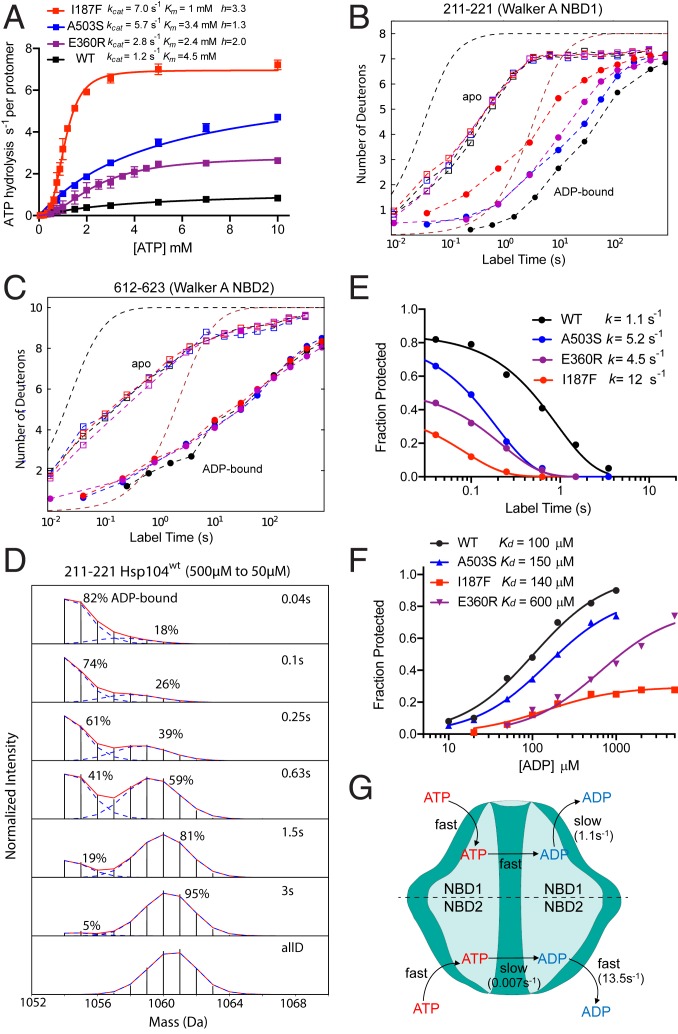
We measured ATPase activity over the range of ATP concentrations using a coupled enzyme assay (Fig. 2A and SI Appendix, Fig. S1). The WT parameters, kcat = 1.2 s−1 per protomer and Km = 4.5 mM, are in good agreement with previous reports given the different assay conditions applied (44, 45) (Methods). The accelerated ATPase activity of the potentiating mutants is due mainly to a kcat effect, ∼2-fold faster than WT for E360R and ∼5-fold for A503S and I187F. Accelerated ATPase is also associated with increased sigmoidicity, indicating increased cooperativity between protomers within the hexamer. The I187F variant with highest kcat shows the largest Hill coefficient of 3.3. The other mutants are less cooperative with smaller Hill coefficients of 2 for E360R and 1.3 for A503S. These parameters are listed in Table 1.
Fig. 2.
Potentiating mutations accelerate ADP off rate from NBD1 and increase steady-state ATPase turnover. (A) Steady-state ATP hydrolysis measured by a coupled enzyme assay (SI Appendix, Fig. S1). Hsp104 concentration was 2 μM. kcat is calculated by dividing maximum ATPase turnover rate by Hsp104 protomer concentration. Data points average three replicates. (B and C) HX MS centroid plots for time-dependent H to D exchange of Walker A segments in NBD1 and NBD2, with and without 5 mM ADP. Dashed curves reference HX protection factors (Pf) of 1- and 100-fold, i.e., a slowing of HX rate by 1-fold and 100-fold, relative to the known HX rates of unprotected amides (39, 46). (D) HX MS spectra measuring the time course of ADP dissociation from NBD1 Walker A of WT Hsp104, starting from 500 μM initial ADP concentration and jumping to 50 μM. (NBD2 is in SI Appendix, Fig. S5.) The two isotopic MS envelopes represent the still ADP-bound HX-protected population (lighter) and the increasing fraction that has experienced ADP dissociation and D-labeling (heavier) in the H to D exposure time indicated. (E) The time course of ADP dissociation. Fraction Protected refers to the fraction of Walker A sites still protected from HX due to bound ADP, measured as in D. The data are fit to a single exponential decay. The initial value reflects the equilibrium that exists between ADP-bound and unoccupied Hsp104 protomers at the beginning of the measurement (500 μM initial ADP). All protomers eventually become D-labeled as a result of ADP dissociation and irreversible H to D exchange. (F) Equilibrium ADP binding by NBD1 Walker A against varying initial free ADP concentration measured at short HX labeling time (40 ms) as in D and E. (G) The Hsp104 ATPase cycle with WT parameters. ATP NBD2 hydrolysis rate is from ref. 45. ADP release from NBD1 is accelerated by the potentiating mutations. In NBD2, slow ATP hydrolysis is rate-limiting and insensitive to the potentiating mutations studied.
Table 1.
Parameters for Hsp104 nucleotide binding
| ADP Kd, μM | ADP off rate, s−1 | ATPase kcat, s−1 per protomer* | Hill coefficient | ||
| NBD1 | WT | 100 ± 20 | 1.1 ± 0.1 | 1.2 ± 0.1 | 1 ± 0.1 |
| A503S | 150 ± 10 | 5.2 ± 0.2 | 5.7 ± 0.1 | 1.3 ± 0.1 | |
| E360R | 600 ± 100 | 4.5 ± 0.2 | 2.8 ± 0.2 | 2.0 ± 0.2 | |
| I187F | 140 ± 30 | 12 ± 1 | 7 ± 0.1 | 3.3 ± 0.2 | |
| NBD2 | WT | 12 ± 2 | 14 ± 7 | 0.007* | ND |
ADP dissociation rate from NBD1 measured for the static ADP-bound open state varies between 1 and 12 s−1 with equilibrium Kd in the 100–600 μM range (pDread 8.35, 25 °C). The I187F mutant has heterogeneous ADP binding; two protomers display the parameters listed and the other four have lower affinity with Kd ∼2 mM (SI Appendix, Fig. S6A). Its cooperative ATPase activity (high Hill coefficient) is due to its promotion of the ATP-bound state by formation of an interaction specific for this state (see text and Fig. 1D). In active ATPase cycling, the mutational variants hydrolyze between 1 and 7 ATP molecules per protomer per second (pH 8.35, 25 °C). NBD2 has much slower ATP hydrolysis, higher ADP affinity, faster ADP on and off kinetics, and it is not affected by the potentiating mutants.
NBD1 is thought to account for >99% of the hydrolysis (45). The errors of the first two columns are from fitting the single exponential data in Fig. 2 E and F. The errors of the last two columns are SD calculated for three replicates.
HX MS.
To seek the mechanistic determinants of Hsp104 interactions, we further developed and used a nonperturbing HX MS analysis (33, 36, 40, 47). The method obtains positional resolution of HX behavior by quenching timed H to D exchanged samples to a condition where HX is largely halted (pH 2.5, 0 °C), proteolyzing the D-labeled protein into many small fragments by brief exposure to pepsin, then roughly separating the fragments by fast liquid chromatography, and analyzing them for their D content by mass spectrometry. Each peptide fragment reports on the protein segment that it represents in the native protein. We reproducibly find ∼250 identifiable peptides that report on all segments throughout the entire Hsp104 protein in all datasets with good signal/noise. Peptides were monitored over their entire HX time scale, which provides a complete picture of the entire Hsp104 protein, avoiding a lamentable imperfection in many HX studies. A complete collection of time-dependent D-uptake centroid plots for WT Hsp104 and the three mutants studied in the apo, ADP-bound, and ATP-cycling states is in SI Appendix.
HX MS Can Measure Fast Rate-Limiting ADP Dissociation.
Release of the hydrolysis product ADP has been reported to be the rate-limiting step of several ATPases (48–52). If this is true for Hsp104, it would spend most of the ATPase cycle time in the ADP-bound open state. Consistently, cryo-EM snapshots find actively cycling Hsp104 predominantly (∼80%) in the open conformation, both with and without substrate (see figure 6A of ref. 6). Potentiating mutations might then increase ATPase activity by accelerating ADP dissociation.
Measuring ADP release from Hsp104 is challenging and has not been reported. Steady-state ATP turnover time is in the range of 1 s, making commonly applied methods difficult to implement. Also, the two NBD active sites of Hsp104, with different rates, can make measurement ambiguous. The ability of HX MS to distinguish HX throughout the protein makes it possible to resolve the fast kinetics of ADP release separately at the two NBD sites in the different potentiated variants. Fig. 2 B and C show H to D exchange results for two peptides that contain the Walker A segments responsible for nucleotide binding in NBD1 and NBD2 (7). In the absence of bound nucleotide, the half time for HX labeling of both Walker A segments is ∼200 ms corresponding to a low HX protection (protection factor ∼10, 22 °C, pD 7.85, Fig. 2). With ADP bound, HX rates of the Walker A segments in the different mutational variants are slowed by up to 100-fold (Fig. 2 B and C). This situation allows fast ADP release to be measured.
Scheme 1 formalizes the kinetic relationships. When ADP-bound Hsp104 is diluted into D2O with low free ADP concentration, the dissociation of bound ADP initiates a kinetic competition between the H to D exchange reaction (kex) and ADP rebinding (kon[ADP]), as in Scheme 1. If kex >> kon[ADP], H to D labeling will occur when ADP dissociates. Scheme 1 is then reduced to Scheme 2, in which case ADP dissociation from Walker A will be followed by irreversible D-labeling, measuring the rate of the ADP off reaction.
(Scheme 1)
(Scheme 2)
ADP Dissociation from NBD1.
For NBD1, the condition for Scheme 2 was ensured by using low free ADP concentration where rebinding is slow and the elevated pD of 8.35 where HX is fast. Hsp104 was preincubated with 500 μM ADP and then diluted by 10-fold into D2O to initiate ADP dissociation and H to D labeling. Timed samples were then submitted to HX MS analysis. Fig. 2D tracks the rate of ADP dissociation from the NBD1 Walker A nucleotide-binding segment 211–221 in WT Hsp104. When ADP dissociates, the segment is immediately D-labeled. The decreasing amplitude of the not-yet D-labeled mass spec envelope (lighter mass) measures the decreasing population fraction that has not yet experienced ADP dissociation and D-labeling during the H to D exposure time.
Fig. 2E plots the ADP dissociation rate. Analogous results for the potentiated proteins are in SI Appendix, Fig. S2. For WT Hsp104, ADP koff rate from NBD1 is 1.1 s−1, closely matching the steady state ATPase kcat of 1.2 s−1 (Fig. 2A and Table 1). NBD1 accounts for >99% of Hsp104 ATPase activity (45). This equivalence suggests that ADP dissociation is the rate-limiting step in the ATPase cycle. This view is supported by the analogous rates measured for the potentiated mutants. A second indication is that the functional ATPase cycle of Hsp104 carries it through the open state, where ADP dissociation occurs, as well as through the closed and extended states. This view is supported by other results described below (mechanisms of potentiation, Tyr loop HX, substrate loss during turnover).
Equilibrium ADP Binding at NBD1.
A similar experiment leads to the apparent equilibrium Kd for ADP dissociation and, therefore, also to the apparent ADP rebinding rate. We vary the initial free ADP concentration and find the fraction of the population of Walker A binding segments that is protected from HX initially, i.e., at short H to D exposure time, within 40 ms after dilution, where the protein still retains almost all of its initially bound ADP. The results for ADP binding by WT Hsp104 in NBD1 (Fig. 2F) measure a Kd ∼100 μM and, therefore, an apparent association rate constant of 104 M−1s−1. The potentiated Hsp104 variants exhibit accelerated ADP off rates and higher Kd for ADP binding to NBD1 (Fig. 2F and Table 1).
ADP Binding at NBD2.
The NBD2 site is not structurally blocked by the MD and, perhaps for that reason, it loses and rebinds ADP much more rapidly than NBD1. Even at low free ADP concentration (50 μM), the ADP rebinding rate is comparable to that for H to D labeling. This situation produces a major overlap between the MS envelope that represents the unprotected population and the still ADP-bound and slow exchanging MS envelope (SI Appendix, Fig. S3), which complicates the calculation of ADP on and off rates. We modeled site-resolved HX according to Scheme 1 by using MS isotopic envelope shape information (Methods). Fitting results are shown in SI Appendix, Fig. S5. The ADP off rate for NBD2, 14 s−1, is 10-fold greater than for NBD1. The fitted ADP on rate constant for WT NBD2, 1.1 × 106 M−1s−1, is 100-fold greater than for NBD1, leading to a 10-fold greater ADP affinity with Kd of 12 μM. In agreement, the equilibrium NBD2 Kd value was previously found to be 9 μM by fluorescence of two different Trp variants (8, 45), and Kd averaged between the two NBDs was 50 μM (53). These events are illustrated in Fig. 2G and parameters are listed in Table 1.
Potentiation Mechanisms.
All three potentiating mutations studied here enhance Hsp104 function by speeding ADP dissociation from NBD1. HX MS observations reveal their detailed mechanisms.
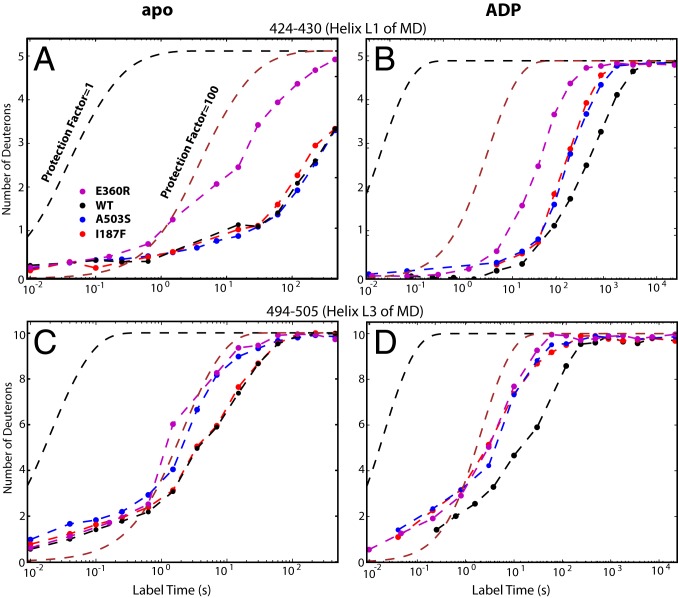
The potentiating A503S mutation placed in MD helix L3 (Fig. 1B) has only a very local structural effect. In the apo state the mutation destabilizes only its local helix L3 host (Fig. 3 A and C). The ADP-bound state brings L3 into a mutually stabilizing contact with L1 in the neighboring protomer to form a bridge across the interprotomer nucleotide-binding pocket of NBD1 (6, 7) (Fig. 1B). The A503S mutation diminishes the added interhelix stabilization and makes the whole MD less stable than in WT (Fig. 3 B and D and SI Appendix). These results indicate that the stability of the MD L3 to neighboring MD L1 crossbridge modulates the interprotomer structural dynamics, as noted in previous cross-linking experiments (28). The crossbridge acts as an external inhibitory gate that equally suppresses the rate of ADP escape from and rebinding to the interprotomer nucleotide-binding site, although the MD does not interact with bound ADP. The MD crossbridge controls the rate-limiting ADP release step in NBD1 and, thus, ATPase activity and functional power (Fig. 2 D and E and SI Appendix).
Fig. 3.
HX of the MD L1 and L3 helices. (A and C) In apo Hsp104. (B and D) In the ADP-bound state. The cross-protomer L3 to L1 bridge forms in the ADP state and down-regulates the rate of ADP dissociation. The potentiating mutations destabilize both crossbridging MD helices in the ADP-bound state (faster HX relative to WT) but cause only more local effects (except for E360R) in the apo state where the crossbridge is not formed.
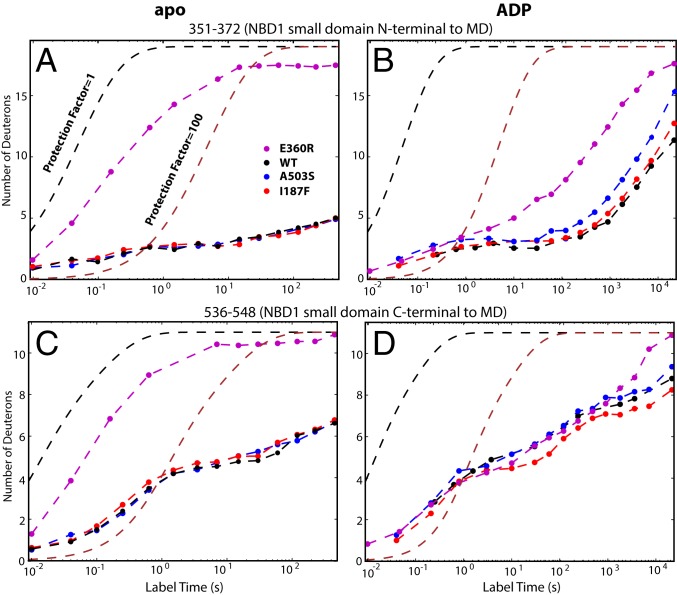
The E360R mutation places the positively charged R360 near both R366 (5 Å), N-terminal to the MD, and K480 in MD helix L2 (Fig. 1 A and C). In the apo state, these charge-charge repulsions greatly destabilize the entire small subdomain (Fig. 4 A and C and SI Appendix). In the ADP state, stability of the small subdomain is partially but not completely restored (Fig. 4 B and D) by the formation of contacts with the neighboring protomer and some MD residues (6). Unlike A503S, E360R speeds the ADP off rate but not its rebinding rate (Table 1). These observations implicate especially the destabilized small subdomain itself, which partly occludes the NBD1 nucleotide-binding site (Fig. 1C), in promoting ADP release, and possibly also the MD.
Fig. 4.
HX of some small domain peptides in apo (A and C) and ADP-bound (B and D) Hsp104, placed before and after the MD. E360R placed in the small domain greatly destabilizes the small subdomain, promotes its dynamics, and indirectly destabilizes the neighboring M domain (Fig. 3), which facilitates ADP escape. The other mutants have much smaller effects on this region.
The potentiating mutation I187F, placed in the interhelix B2-B3 loop, is brought close to the adenosine moiety of the bound nucleotide (<5 Å, Fig. 1C) in the ADP-bound open state. Among other tested substitutions at this position, only Tyr and Trp impose both a large potentiation and a cytotoxic effect at elevated temperature comparable to I187F (43). Ala and Ser with smaller side chains can also potentiate but less so, and they are not cytotoxic (43). These results imply that the bulky side chain promotes ADP release by direct steric interference with the bound nucleotide.
In addition, the I187F mutation makes the binding of ADP to NBD1 Walker A heterogeneous, inducing only ∼30%, or two of six, of the protomers to adopt the slow Walker A HX characteristic of other ADP-bound states. The other four protomers exhibit much lower ADP affinity (Fig. 2F). The heterogeneous population response to ADP binding, measured directly in Fig. 2F, is matched by the HX MS detection of two Walker A populations with different ADP binding. This is shown by HX protection due to ADP binding by two protomers at low ADP concentration (with Kd = 140 μM, Table 1) and the onset of binding in the other four protomers only at high ADP concentration (SI Appendix, Fig. S6A, Kd estimated to be ∼2 mM by the ADP-dependent centroid shift of this population). This binding heterogeneity seems to be caused by interference of the bulky I187F side chain with the protein’s response to ADP binding.
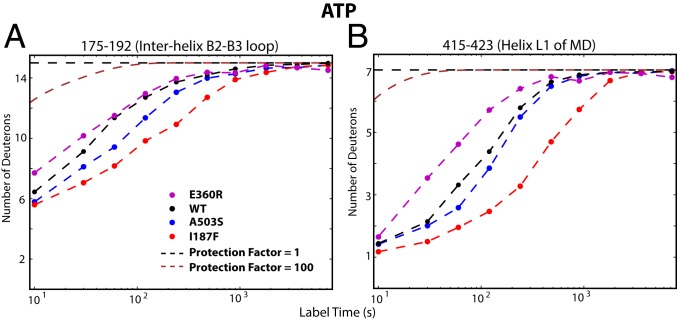
The I187F substitution also alters the response to ATP turnover. The sigmoidal ATPase activity profile and decreased Km indicates enhanced interprotomer cooperativity (Fig. 2A) implying that, in addition to speeding ADP dissociation, the mutation actively stabilizes the ATP-bound state. HX measured during ATP turnover points to the structural cause. During ATP turnover but not in the apo nor ADP-bound states, the B2-B3 interhelix loop and MD helix L1 are both made more stable (compare Fig. 5 with Fig. 3 A and B and with SI Appendix, Fig. S6 B and C). In the ATP state (Fig. 1D), these two segments are brought close to each other across the interprotomer interface in an MD-NBD1 configuration (6, 27). It appears that the phenylalanine side chain no longer clashes with the bound nucleotide but is displaced and binds to R419 in the L1 helix B2-B3 loop of the neighboring MD (Fig. 1D), perhaps by an interprotomer π-cation or π−π interaction (Fig. 1D). This interaction stabilizes both segments and, therefore, the ATP state.
Fig. 5.
I187F actively stabilizes the ATP-bound state. Placed in the interhelix B2-B3 loop, I187F stabilizes it (175-192; A) and MD helix L1 (415-423; B), apparently by forming a link across the protomer interface in the ATP state. This interaction during ATP turnover suggestively explains its role in enhancing ATP affinity and ATPase cooperativity (Fig. 2A). It is absent in the apo state (Fig. 3A and SI Appendix, Fig. S6B) and actively destabilizes helix L1 in the ADP state (Fig. 3B).
In summary, the ATPase activity of Hsp104 is rate-limited by ADP release from NBD1 in the open state. In the A503S variant, the ADP off rate and the steady-state ATP kcat are equally accelerated (4.7-fold). In the other two variants, the increase in ADP release is a bit greater than for ATP turnover, namely 4-fold versus 2.3-fold for E360R and 11-fold versus 6-fold for I187F. For these variants, ADP release has become sufficiently fast that other kinetic steps in the ATPase cycle (∼10 s−1) become partially rate-limiting. Another conclusion is that the MD bridge serves as an autoinhibitory element by acting as a kinetic gate to control ADP release. Notably, the solvent-exposed position of the MD places Hsp104 activity in easy reach of cellular control, such as Hsp70 and Hsp40. Also note that the potentiating mechanisms found here are specific for the ADP-bound open state and, therefore, indicate that, in its functional cycle, Hsp104 cycles through the open state.
Open-State Occupation during Turnover.
Speeding ADP release may also promote Hsp104 function by minimizing time in the open state and, thus, reduce substrate protein loss during the ATPase cycle (6, 41, 42). Substrate is held poorly in the open state and tends to be lost during processing, presumably due to the increased size of the central pore, from 10 Å in the closed/extended states to 30 Å in the open state (6, 27), and relaxation of the substrate-binding pore loops (7). To examine this possibility, we wish to discern the occupation of open and closed states during ATP cycling and its effect on substrate retention during active cycling. Previous work showed that the static open state can be recognized by the HX difference between the canonical and noncanonical (“lockwasher”) interprotomer interfaces described in cryo-EM studies (6, 27). However, during cycling this difference is time-averaged and obscured by fast protomer interchange.
Alternatively, the open/closed balance might be gauged by HX of the Tyr pore loops. In the static open state (apo, ADP) the Tyr loops of both NBDs display fast HX at close to the free peptide rate. In the closed state driven by bound ATPγS, HX rates are 100-fold slower (SI Appendix, Fig. S7). Consistently, cryo-EM observation finds that the pore loops are flexible in the static open state but they interact and become more structured when the channel contracts in the closed and extended states (6, 27, 31). In the case of fast open/closed interchange during active cycling, the degree of HX slowing relative to the static open state registers the time-averaged fractional open state occupation. Thus, Tyr loop HX can sensitively measure occupation of the closed and open states during cycling.
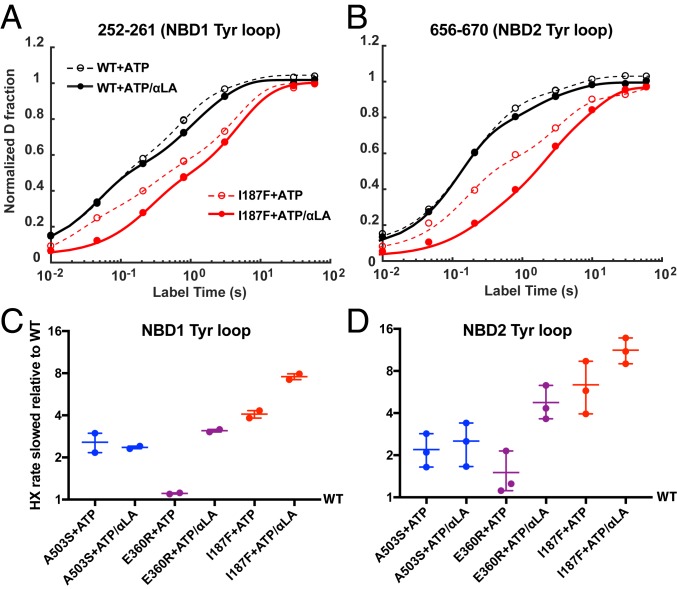
For WT Hsp104, Tyr loop HX in both NBDs measured during cycling is close to the fast open-state rate (SI Appendix, Fig. S7). This indicates that occupation of the open state is greatly favored, as shown by cryo-EM (6). Tyr loop HX is sensitive to the potentiating mutations. As an example, Fig. 6 A and B compare Tyr loop HX of the I187F mutant to WT Hsp104 during ATP turnover. HX is slowed in the mutant because it spends less time in the open state. Substrate protein further favors the closed state. Fig. 6 C and D compare the effect of the other mutations. The potentiating mutations slow Tyr loop HX in both NBDs in all protomers via contraction of the central pore, indicating a whole hexamer effect. The effect seen for each mutant form, with HX slowing of the Tyr loops between 3-fold and 10-fold, matches the reduction of cycle time in the open state due to increased ADP off rate, in the 4-fold to 11-fold range (Table 1). The one exception, E360R, reduces open-state occupation during steady-state turnover only with the added support of substrate protein (Fig. 6 C and D), perhaps because the mutation alone is so disruptive (Fig. 3). [The same behavior may account for its robust potentiation ability with substrate at normal growth temperature and low cytotoxicity at elevated growth temperature (43) where its reduced stability may restrain its activity.]
Fig. 6.
Tyr loop HX during ATP cycling. Potentiating mutations and substrate protein slow Tyr loop HX due to reduced time in the fast exchanging open state. (A and B) Two peptides in the I187F protein that cover the Tyr loop of NBD1 and NBD2 are shown as examples. The nonexponential nature of HX curves is due to residue-by-residue EX2 exchange, rather than concerted whole-segment EX1 exchange, and the fact that the peptides measured overlap into neighboring segments. These data points were overfitted with a sum of four exponential terms (dashed and continuous lines) to extract the midpoint for each curve. (C and D) Degree of Tyr loop HX slowing for the potentiated variants relative to WT. This is calculated by comparing the midpoint for each of the potentiated mutants with WT. Each point represents one peptide, namely 252–261 and 251–261 for NBD1 and 656–670, 656–674, 663–674 for NBD2. An ATP-regenerating system was used as described in Methods.
These results document and quantify the repeated cycling and major occupation of the Hsp104 open state during active turnover. The cycling protein visits the open state, presumably after several hydrolysis events (per hexamer), releasing a number of ADPs equal to the number of preceding ATPase reactions in the closed state.
Substrate Retention and Loss.
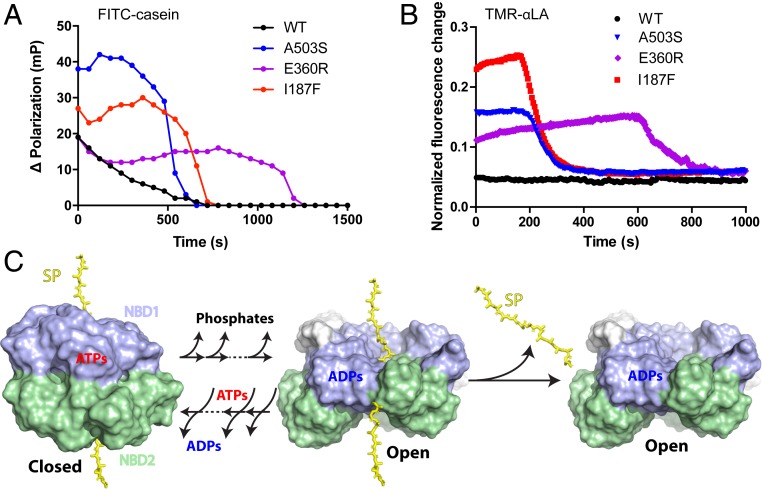
Does shortened time in the open state decrease substrate protein loss during cycling? To more definitively assess substrate protein interaction with Hsp104 during ATP cycling, we measured Hsp104 interaction with two different substrates. We labeled casein, an often-used model substrate protein, with fluorescein isothiocyanate (FITC-casein) and measured change of fluorescence polarization (Δanisotropy). These signals indicate slower tumbling upon binding to Hsp104. We also developed a probe, the 123 residue α-lactalbumin (αLA) protein to serve as a simpler homogeneous monomolecular substrate (53, 54). We unfolded αLA by disulfide reduction and carboxymethylation (55) and double labeled it near both termini with tetramethylrhodamine (TMR) (56–58) (SI Appendix, Fig. S8A). In free solution, unfolded αLA adopts a collapsed molten globular state (55) and the two terminal labels stack yielding a characteristic absorption spectrum and quenched low fluorescence (SI Appendix, Fig. S8 A–C). When αLA is drawn into the Hsp104 channel, the TMR labels unstack, marked by the expected absorbance change and fluorescence intensity enhancement.
WT Hsp104 or the potentiated variants were mixed with each labeled substrate in the presence of an ATP-regenerating system intended to keep inhibitory ADP concentration low, and fluorescence was measured during active turnover (Fig. 7 A and B). WT Hsp104 produces background signal for αLA and casein, consistent with the view that it spends most of its cycle time in the open state, therefore retains substrate poorly and experiences very low retention (Fig. 7 A and B). The potentiated variants show increased signal reflecting increased substrate retention due to less cycling time in the open state. In control experiments with a functionally inactive double pore-loop mutant (DPLA, Y257A/Y662A) (8, 9, 25) that specifically prohibits through-pore translocation (SI Appendix, Fig. S8D), only background-level fluorescence is seen, confirming that these observations reflect through-pore behavior. Further, the processing time before final substrate loss due to ATP exhaustion and ADP accumulation is quantitatively matched by the known ATPase rate of each variant (Table 1) and the available ATP (SI Appendix, Fig. S8E), confirming processing as expected.
Fig. 7.
The potentiating mutations promote substrate retention during active turnover. (A) Increased ΔPolarization signal due to retention of FITC-casein substrate during active ATP turnover. (B) Increased fluorescence due to retention of TMR-labeled αLA substrate during active ATP turnover. Low signal for WT Hsp104 (black) reflects the low steady-state level of bound-substrate due to substrate loss from the open state during ATP cycling. Increased steady state signal for the potentiated mutants reflects improved substrate retention due to less cycle time in the open state. Control experiments with Tyr pore loop mutants indicate that these signals reflect active substrate protein processing through the axial channel (SI Appendix, Fig. S8D) although the polarization signal probably also includes external protein. Final signal decay is due to ATP exhaustion, ADP accumulation, and substrate loss. Time in the steady-state substrate-bound condition before decay matches the expected exhaustion of the ATP regenerating capacity, dependent on the known ATP utilization rate (Table 1). The fluorescence traces (three replicates) are normalized to the high value obtained in the presence of ATPγS (SI Appendix, Fig. S8D). Pertinent background signals are subtracted. (4 μM Hsp104 in protomer; 10 mM phophoenolpyruvate for experiments shown in A and 5 mM for B; initial free ATP 10 mM; see also SI Appendix, Materials and Methods). (C) The kinetic competition during ATP/ADP cycling between loss of substrate protein from the ADP open state and ATP rebinding to induce the closed state. The transition between the closed and open states may involve multiple ATP hydrolysis or binding steps. The model omits possible multiple intermediate states bound with mixed ADP/ATP for simplicity. ADP release from the open state determines the fraction of time Hsp104 spend in this state. Slower ADP release leads to more time in the open state and, in turn, more substrate protein (SP) lost.
These results further support the conclusion that Hsp104 spends a large fraction of its cycling time in the open state, and they demonstrate that speeding the ADP off rate from the open state, thereby reducing open state occupation, reduces substrate loss during functional cycling.
Discussion
Biological evolution has elaborated hundreds of AAA+ protein machines that use the energy of ATP binding and hydrolysis to remodel macromolecular structures. It seems likely that these related proteins are based on similar principles and mechanisms with particular adaptations for the function at hand (3). Recent advances in cryo-EM technology now reveal what many of these large protein machines look like and allow inferences about how they might function (59). Testing and extending these inferences require studies under physiological conditions. We previously found that HX MS methods are able to separately resolve all of the functionally active and inactive elements of WT Hsp104 in physiological solution and measure their structural and biophysical properties (7). The work described here demonstrates the ability of HX MS to delve into the inner workings and uncover mechanisms and regulation of Hsp104 activity during active turnover.
Structural Basis for Hsp104 Potentiation.
The special ability of HX MS to separately measure ADP off rates from the two Walker A binding sites produce the pivotal finding that ATP turnover is rate-limited by dissociation of the reaction product ADP from NBD1. The three potentiating mutations we studied act by speeding the rate-limiting ADP dissociation from NBD1. The A503S mutation destabilizes and makes more dynamic the outer shell MD to MD helix L3 to L1 crossbridge and shows no other effect. This result makes clear the autoinhibitory function of the MD crossbridge and its mode of action. It is formed specifically in the ADP-bound state, well positioned to suppress the interprotomer dynamic motions needed to allow the rate-limiting ADP dissociation, and it is well placed for external cellular control. E360R, one layer deeper, destabilizes the NBD1 small subdomain, which forms part of the interprotomer interface and the nucleotide-binding pocket, and thus promotes the interprotomer dynamics needed for ADP release. An innermost mutation, I187F, promotes ADP dissociation by direct steric clash with the adenosine moiety of the bound ADP itself. The 187F aromatic ring may further enhance activity by reorienting to interact favorably with R419 and stabilize the closed state. Another class of potentiating mutation (N566I), not studied here, has recently been discovered in the NBD2 interprotomer interface (43). It seems likely that this mutation potentiates Hsp104 via some different mechanism.
Kinetic Basis for Hsp104 Regulation.
We find that the rate of ADP dissociation from Hsp104 NBD1 governs functional capability in two ways. It rate-limits the ATPase cycle and, therefore, controls the power output. It determines the fraction of cycle time in the open state and, thus, controls substrate protein loss during processing. Fig. 7C pictures the role of substrate retention during the cycle in terms of a kinetic competition between the loss of substrate protein from the ADP-bound open state and ATP rebinding with continued processing in the closed state. A prior study has similarly observed the influence of substrate dissociation on Hsp104 function during processing (41).
These results help to rationalize some puzzling observations. Ambient conditions can cause the functional impact of ATPase activity and substrate retention to cooperate, as for the mutations studied here, or to compete (25, 60). For example, despite its powerful unfoldase capability, Hsp104 can appear to be inactive for the disaggregation and unfolding of weak substrates (60). Present results suggest this occurs because of premature substrate loss from the dominant ADP-bound open state during the ATPase cycle (Fig. 7C). Upon dissociation, spontaneous refolding can reverse incomplete protein processing. Counterintuitively, unfoldase activity can increase rather than decline when ATP is diluted with slowly hydrolyzable ATPγS (60). This situation occurs when Hsp104 function is limited by substrate protein loss and not by processing power. Although the addition of ATPγS reduces peak processing power, it also promotes steady-state occupation of the closed state, therefore reduces occupation of the open state and counters substrate loss. Activity declines again at higher ATPγS where the power output due to remaining ATP becomes limiting. We note that the open/closed balance during cycling might similarly be influenced by conditions including ATP binding with mutationally inhibited hydrolysis (60, 61), which is like ATPγS binding, by hybridization with suitably modified protomers (25), and by interaction with collaborating chaperones Hsp70 and Hsp40 (14). With some substrates, early substrate release can support rather than inhibit function when only partial processing is sufficient. In some cases, it may even be required, as for initiating amyloidogenesis by assisting preamyloid oligomer formation (60, 62).
The Open State and Substrate Translocation.
A major implication of this work is that Hsp104 experiences repetitive large-scale transitions through its open and closed states during the ATP/ADP cycle and not only the closed and extended forms. The evidence for this view is as follows. For WT Hsp104, the steady-state ATPase rate during cycling matches the rate of ADP dissociation from the static open state (Table 1). The potentiating mutations speed the ATPase rate during cycling much as they speed the rate for ADP dissociation from the static open state (Table 1). The detailed mechanisms by which the potentiating mutations act are largely specific for the open state (Figs. 3 and 4). Tyr loop HX with and without substrate shows that the potentiating mutations decrease the time spent in the open state during cycling, with the size of the effects about as predicted by the accelerated ADP dissociation from the open state (Fig. 6). Decreased substrate loss during cycling tracks the shortened time in the open state due to the accelerated ADP dissociation (Fig. 7). In agreement, cryo-EM snapshots during steady-state Hsp104 cycling observe a dominant fraction of Hsp104 in the open state (see figure 6 in ref. 6), although this has gone largely unremarked.
In the closed/open transition, protomers experience a large relative vertical displacement (6). Protein substrate might be translocated accordingly, linked to either transition. A previously suggested translocation mechanism (6) for Hsp104 pictures stepwise substrate translocation, two amino acids in size, driven by a ratcheting between neighboring protomers. Results for other AAA+ proteins have similarly been taken to suggest small protomer-scale translocation steps, perhaps in a sequential hand over hand through the spirally positioned protomers and pore loops (32). The present HX MS results do not detect, but do not rule out, this kind of small-scale transition. Both large and small pulling modes may well occur. However, for Hsp104, the present results argue against substrate translocation that is driven by any mechanism without intervening visits to the whole hexamer open state for dissociation of the accumulated ADPs. Whether the large-scale transition itself contributes to the translocation process, either actively or passively, in Hsp104 or other AAA+ proteins, remains to be seen. If translocation must await the large-scale transition, this would allow a large and forceful pull that accumulates the energies of multiple prior ATP events for Hsp104 to disassemble highly resistant substrates (7, 25). This strategy appears able to explain the functional spring-loaded stroke in the NSF protein, another member of the AAA+ family, to unwind thermodynamically stable SNARE complexes (51, 63). It is also consistent with the observation of the large 14 amino acid stroke step measured for homologous ClpB (64).
Methods
Hsp104 mutational variants were generated, expressed, and purified as described (43, 65). HX MS experiments were done and analyzed as described in refs. 35, 40. More extensive details are in SI Appendix.
Data Availability.
Time-dependent mass centroid HX MS plots for all four versions of the Hsp104 protein under three different conditions (apo, ADP, and ATP) are in Dataset S1.
Supplementary Material
Acknowledgments
This work was supported by NIH Research Grants R01GM031847 (to S.W.E.), and R01GM099836 (to J.S.), National Science Foundation Grant MCB1020649 (to S.W.E.), the Mathers Charitable Foundation (S.W.E.), and an Alzheimer’s Association Research Fellowship (to J.L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921968117/-/DCSupplemental.
References
- 1.Duran E. C., Weaver C. L., Lucius A. L., Comparative analysis of the structure and function of AAA+ motors ClpA, ClpB, and Hsp104: Common threads and disparate functions. Front. Mol. Biosci. 4, 54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erzberger J. P., Berger J. M., Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Shorter J., Southworth D. R., Spiraling in control: Structures and mechanisms of the Hsp104 disaggregase. Cold Spring Harb. Perspect. Biol. 11, a034033 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olivares A. O., Baker T. A., Sauer R. T., Mechanical protein unfolding and degradation. Annu. Rev. Physiol. 80, 413–429 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Olivares A. O., Baker T. A., Sauer R. T., Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat. Rev. Microbiol. 14, 33–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gates S. N., et al. , Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357, 273–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye X., Lin J., Mayne L., Shorter J., Englander S. W., Hydrogen exchange reveals Hsp104 architecture, structural dynamics, and energetics in physiological solution. Proc. Natl. Acad. Sci. U.S.A. 116, 7333–7342 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lum R., Tkach J. M., Vierling E., Glover J. R., Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 279, 29139–29146 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Lum R., Niggemann M., Glover J. R., Peptide and protein binding in the axial channel of Hsp104. Insights into the mechanism of protein unfolding. J. Biol. Chem. 283, 30139–30150 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sweeny E. A., et al. , The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol. Cell 57, 836–849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castellano L. M., Bart S. M., Holmes V. M., Weissman D., Shorter J., Repurposing Hsp104 to antagonize seminal amyloid and counter HIV infection. Chem. Biol. 22, 1074–1086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez Y., Lindquist S. L., HSP104 required for induced thermotolerance. Science 248, 1112–1115 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Sanchez Y., Taulien J., Borkovich K. A., Lindquist S., Hsp104 is required for tolerance to many forms of stress. EMBO J. 11, 2357–2364 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glover J. R., Lindquist S., Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Wallace E. W., et al. , Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsell D. A., Kowal A. S., Singer M. A., Lindquist S., Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372, 475–478 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Parsell D. A., Sanchez Y., Stitzel J. D., Lindquist S., Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature 353, 270–273 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Shorter J., Lindquist S., Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304, 1793–1797 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Sweeny E. A., Shorter J., Mechanistic and structural insights into the prion-disaggregase activity of Hsp104. J. Mol. Biol. 428, 1870–1885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alberti S., Halfmann R., King O., Kapila A., Lindquist S., A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halfmann R., et al. , Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482, 363–368 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes D. L., Lancaster A. K., Lindquist S., Halfmann R., Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell 153, 153–165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackrel M. E., Yee K., Tariq A., Chen A. I., Shorter J., Disparate mutations confer therapeutic gain of Hsp104 function. ACS Chem. Biol. 10, 2672–2679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackrel M. E., et al. , Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell 156, 170–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeSantis M. E., et al. , Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell 151, 778–793 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michalska K., Zhang K., March Z. M., Hatzos-Skintges C. et al. , Structure of calcarisporiella thermophila Hsp104 disaggregase that antagonizes diverse proteotoxic misfolding events. Structure 27, 449–463.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokom A. L., et al. , Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation. Nat. Struct. Mol. Biol. 23, 830–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heuck A., et al. , Structural basis for the disaggregase activity and regulation of Hsp104. eLife 5, e21516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desantis M. E., Shorter J., The elusive middle domain of Hsp104 and ClpB: Location and function. Biochim. Biophys. Acta 1823, 29–39 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S., et al. , The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell 115, 229–240 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Lee S., Roh S. H., Lee J., Sung N. et al. , Cryo-EM structures of the Hsp104 protein disaggregase captured in the ATP conformation. Cell Rep. 26, 29–36.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puchades C., Sandate C. R., Lander G. C., The molecular principles governing the activity and functional diversity of AAA+ proteins. Nat. Rev. Mol. Cell Biol. 21, 43–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Englander J. J., Rogero J. R., Englander S. W., Protein hydrogen exchange studied by the fragment separation method. Anal. Biochem. 147, 234–244 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Englander S. W., Englander J. J., Structure and energy change in hemoglobin by hydrogen exchange labeling. Methods Enzymol. 232, 26–42 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Kan Z. Y., Mayne L., Chetty P. S., Englander S. W., ExMS: Data analysis for HX-MS experiments. J. Am. Soc. Mass Spectrom. 22, 1906–1915 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayne L., et al. , Many overlapping peptides for protein hydrogen exchange experiments by the fragment separation-mass spectrometry method. J. Am. Soc. Mass Spectrom. 22, 1898–1905 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walters B. T., Ricciuti A., Mayne L., Englander S. W., Minimizing back exchange in the hydrogen exchange-mass spectrometry experiment. J. Am. Soc. Mass Spectrom. 23, 2132–2139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kan Z. Y., Walters B. T., Mayne L., Englander S. W., Protein hydrogen exchange at residue resolution by proteolytic fragmentation mass spectrometry analysis. Proc. Natl. Acad. Sci. U.S.A. 110, 16438–16443 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen D., Mayne L., Phillips M., Englander S., Reference parameters for protein hydrogen exchange rates. J. Am. Soc. Mass Spectrom. 29, 1936–1939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kan Z. Y., Ye X., Skinner J. J., Mayne L., Englander S. W., ExMS2: An integrated solution for hydrogen-deuterium exchange mass spectrometry data analysis. Anal. Chem. 91, 7474–7481 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Durie C. L., et al. , Hsp104 and potentiated variants can operate as distinct nonprocessive translocases. Biophys. J. 116, 1856–1872 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weaver C. L., et al. , Avidity for polypeptide binding by nucleotide-bound Hsp104 structures. Biochemistry 56, 2071–2075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tariq A., Lin J., Jackrel M. E., Hesketh C. D. et al. , Mining disaggregase sequence space to safely counter TDP-43, FUS, and alpha-synuclein proteotoxicity. Cell Rep. 28, 2080–2095.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schirmer E. C., Queitsch C., Kowal A. S., Parsell D. A., Lindquist S., The ATPase activity of Hsp104, effects of environmental conditions and mutations. J. Biol. Chem. 273, 15546–15552 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Hattendorf D. A., Lindquist S. L., Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 21, 12–21 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai Y., Milne J. S., Mayne L., Englander S. W., Primary structure effects on peptide group hydrogen exchange. Proteins 17, 75–86 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallagher E. S., Hudgens J. W., Mapping protein-ligand interactions with proteolytic-fragmentation, hydrogen/deuterium exchange-mass spectrometry. Methods Enzymol. 566, 357–404 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Linder P., Jankowsky E., From unwinding to clamping–The DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Mayer M. P., Bukau B., Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hackney D. D., Kinesin ATPase: Rate-limiting ADP release. Proc. Natl. Acad. Sci. U.S.A. 85, 6314–6318 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryu J. K., et al. , Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover. Science 347, 1485–1489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye X., Lorimer G. H., Substrate protein switches GroE chaperonins from asymmetric to symmetric cycling by catalyzing nucleotide exchange. Proc. Natl. Acad. Sci. U.S.A. 110, E4289–E4297 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bösl B., Grimminger V., Walter S., Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J. Biol. Chem. 280, 38170–38176 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Cashikar A. G., et al. , Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell 9, 751–760 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Kuwajima K., Ikeguchi M., Sugawara T., Hiraoka Y., Sugai S., Kinetics of disulfide bond reduction in alpha-lactalbumin by dithiothreitol and molecular basis of superreactivity of the Cys6-Cys120 disulfide bond. Biochemistry 29, 8240–8249 (1990). [DOI] [PubMed] [Google Scholar]
- 56.Hamman B. D., et al. , Tetramethylrhodamine dimer formation as a spectroscopic probe of the conformation of Escherichia coli ribosomal protein L7/L12 dimers. J. Biol. Chem. 271, 7568–7573 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Okoh M. P., Hunter J. L., Corrie J. E., Webb M. R., A biosensor for inorganic phosphate using a rhodamine-labeled phosphate binding protein. Biochemistry 45, 14764–14771 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Blackman M. J., et al. , Structural and biochemical characterization of a fluorogenic rhodamine-labeled malarial protease substrate. Biochemistry 41, 12244–12252 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Bai X. C., McMullan G., Scheres S. H., How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 40, 49–57 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Doyle S. M., et al. , Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat. Struct. Mol. Biol. 14, 114–122 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Torrente M. P., et al. , Mechanistic insights into Hsp104 potentiation. J. Biol. Chem. 291, 5101–5115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shorter J., Lindquist S., Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell 23, 425–438 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao M., et al. , Mechanistic insights into the recycling machine of the SNARE complex. Nature 518, 61–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avellaneda M. J., et al. , Processive extrusion of polypeptide loops by a hsp100 disaggregase. Nature 578, 317–320 (2020). Erratum in Nature 578, E23 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Sweeny E. A., DeSantis M. E., Shorter J., Purification of hsp104, a protein disaggregase. J. Vis. Exp., 3190 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Time-dependent mass centroid HX MS plots for all four versions of the Hsp104 protein under three different conditions (apo, ADP, and ATP) are in Dataset S1.