Abstract
Background
Glioblastoma (GBM) is the most common primary malignant tumor in adult central nervous system and results in disappointing survival outcomes. Although the diagnosis and therapy approach have been developed recently, the prognosis of GBM remains poor. A novel, minimally invasive biomarker for GBM is necessary for early diagnosis or prognosis prediction.
Methods
All circRNAs were detected by qRT-PCR in GBM samples including training and validation sets. We used the risk score analysis to assume the diagnosis ability for GBM. The receiver operating characteristic curve was also employed.
Results
Among the 14 candidates, circRNA, circNT5E, circFOXO3, circ_0001946, circ_0029426, circ-SHPRH, and circMMP9 were detected with increased levels in the training set. Further investigation in the validation set indicated that circFOXO3, circ_0029426, and circ-SHPRH might be the fingerprints for GBM compared with controls. The risk score analysis revealed that the combination of three circRNAs could distinguish the GBM from healthy control with the area under curve value of 0.980 and 0.906, respectively.
Conclusion
The three circRNAs might be novel fingerprints for predicting the occurrence of GBM.
Keywords: glioma, circRNA, plasma, biomarker, serum
Introduction
Gliomas are defined as human malignant tumors derived from glial cells. Gliomas account for almost 80% of all malignant brain tumors.1 To date, gliomas have beendivided into four subgroups from I to IV according to the pathologic grading system.2 The prognosis was associated with the subgroup. For the gliomas, lower grade frequently indicated better prognosis; for the higher grade, a worse prognosis was predicted.3 Research has found that for the grade IV gliomas, also known as glioblastoma, survival time is about one year,4 and for grade II and III gliomas, the survival time is better.5 Thus, it is important to identify new biomarkers for the early diagnosis of glioblastomas (GBM).
Recently, the existence of circRNA in human body fluid has been identified, opening up a new field for the potential function of circRNAs as fingerprints in human disease, especially in human malignant cancers.6,7 For example, researchers have proven that circ-KIAA1244 could serve as a novel circulating biomarker for the detection of gastric cancer;8 the circRNAs circRNA_0001178 and circRNA_0000826 may serve as a potential diagnostic biomarker for liver metastases from colorectal cancer.9 However, to our knowledge, biomarker-based investigation focusing on circRNA in human GBM in the Chinese Han population has been poorly investigated.
In this study, we conducted our experiment based on the identified circRNAs that were identified in tissue samples as having important functions in GBM development according to previous reports. For example, circular RNA circ_0001946 could act as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1.10 Circ-0001801 contributes to cell proliferation, migration, invasion, and epithelial to mesenchymal transition (EMT) in glioblastoma by regulating the miR-628-5p/HMGB3 axis.6 Besides, overexpressed circ_0029426 in glioblastoma forecasts unfavorable prognosis and promotes cell progression by sponging miR-197,11 etc. None of these circRNAs has been investigated in circulating samples such as plasma or serum. We enrolled 14 candidate circRNAs for screening. We hypothesized that these GBM-related circRNAs might be utilized to detect and monitor GBM. We aimed to explore the potential circRNA acting as a fingerprint for the early identification of GBM.
Materials and Methods
Samples and Screening Phase
One hundred fresh GBM plasma samples were obtained from patients who received therapy between March 2009 and August 2015 in Department of Neurosurgery Huai’an First People’s Hospital. All control participants were healthy volunteers. We have confirmed that all volunteers were free of health problems. All patients enrolled in this study gave written informed consent. The GBM patients were diagnosed histopathologically. We obtained the blood samples before the operation. All plasma samples were extracted immediately and were stored at −80°C for further analysis. All experiments were performed in compliance with government policies and the Helsinki Declaration. The individuals were informed about the study and gave consent prior to the specimen collection, and the research has been approved by an ethics committee of the affiliated Huai’an No.1 People’s Hospital of Nanjing Medical University (Project identification code: IRB-KPJ2017-003-01). All patients’ information has been summarized in Table 1.
Table 1.
Clinicopathologicl Features Analysis of Glioblastoma (GBM) Patients and Cancer-Free Control Samples
| GBM | Control | P value | |
|---|---|---|---|
| N | 100 | 100 | |
| Age mean (SE) year | 52.38 (0.12) | 51.99 (0.24) | 0.21a |
| Sex (male/female) | 87/13 | 88/12 | 0.22b |
| Tumor Ssze (cm) | |||
| ≤ 5cm | 66 | ||
| > 5cm | 34 | ||
| Preoperative KPS | |||
| ≥ 80 | 59 | ||
| < 80 | 41 | ||
| Surgery Procedure | |||
| GTR | 32 | ||
| PR | 68 |
Notes: aStudent’s t-test. bChi-square test.
RNA Extraction, Microarray Detection, and Data Analysis Workflow
TIANamp Virus RNA Kitwith TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used for total RNAs isolation. The synthetic C. elegans miRNA (cel-miR-39, Applied Biosystems, Foster City, CA, USA) was added to each sample as external reference. The cDNA was generated by using the Super-Script First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). Five micrograms of DNase-treated total RNA was incubated with or without 7.5 units of RNase R (Lucigen, Catalog No. RNR07250) for 10 min at 37 °C. RNA was purified and concentrated as described previously before being subjected to RT-PCR or RT-qPCR analysis. The experiment was repeated an additional two times, and three biological replicates of the samples were analyzed. Meanwhile, the GAPDH was used for the internal normalization.
Risk Score Analysis
We performed the risk score to analyze the predicting ability of these candidate plasma circRNAs. In brief, the upper 95% reference interval (95% CI) of each circRNA value in the controls group was used as the cut-off for the expression level of the certain circRNA. If the expression of circRNA in this sample was higher than the 95% CI, we assigned it as 1, if lower, we assigned it as 0. The risk score function (RSF) to predict the GBM group was defined according to a linear combination of the expression level for each circRNA. For example, the RSF for sample i using information from three circRNAs was: rsfi = ∑3j – 1Wj.sij. In the above equation, sij is the risk score for circRNA j on sample i, and Wj is the weight of the risk score of circRNA j. The risk score of three circRNAs were calculated using the weight by the regression coefficient that was derived from the univariate logistic regression analysis of each circRNA. Samples were ranked according to their RSF and then divided into a high-risk group, representing the GBM patients, and a low-risk group, representing the predicted control individuals. Frequency tables and receiver operating characteristic (ROC) curves were then used to evaluate the diagnostic effects of the profiling and to find the appropriate cut-off point, and to validate the procedure and cut-offs in the next validation sample set.
Statistical Analysis
All statistical analyses were carried out by SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). Chi-square test and Fisher’s exact test were used to analyze the association between clinicopathologic parameters and expressions. All confidence intervals (CIs) were stated at the 95% confidence level. All statistical tests were two-sided and a P < 0.05 was considered as statistically significant.
Results
GBM-Associated circRNAs in the Training Set
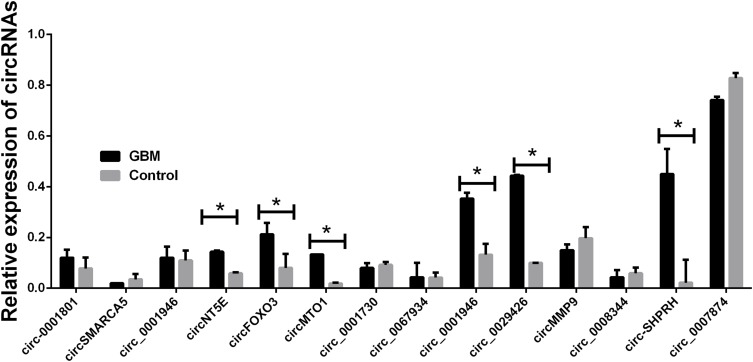
The 14 candidate circRNAs included circ-0001801, circSMARCA5, circ_0001946, circNT5E, circFOXO3, circMTO1, circ_0001730, circ_0067934, circ_0001946, circ_0029426, circMMP9, circ_0008344, circ-SHPRH, and circ_0007874. Forty samples were first used to detect the candidates as a training set. We found six circRNAs (circNT5E, circFOXO3, circ_0001946, circ_0029426, circ-SHPRH, and circMMP9) were significantly different in GBM patients compared with either patients with healthy control (Figure 1).
Figure 1.
Relative expression of 14 candidate circRNAs. Total 40-paired plasma from GBM patients and 40 cancer-free controls were used in RT-qPCR analysis. Data were presented as mean ± SEM. Data were analyzed with Student’s t-test. * indicates P < 0.05.
Confirming the Candidate Biomarker in Validation Set
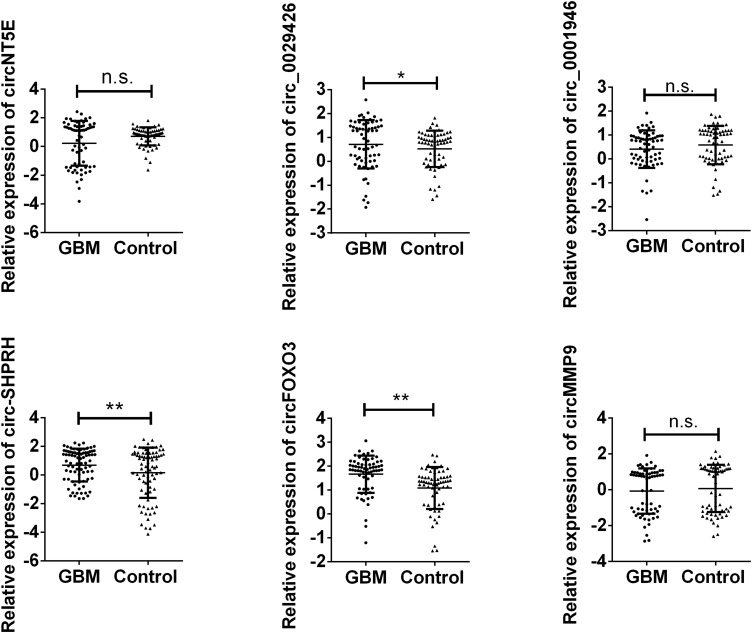
The following investigation was used to detect the expression of the six circRNAs in the validation set. As presented in Figure 2, circFOXO3, circ_0029426, and circ-SHPRH were finally selected in the validation set, indicating they might be potential biomarkers for predicting GBM.
Figure 2.
Validation of six different expressed circRNA in validation set. Plasma from 60 GBM patients and 80 cancer-free controls were enrolled. Data were presented as plot of the median and range of log-transformed relative expression level and was analyzed with Student’s t-test. *indicates P < 0.05 and **indicates P < 0.01. n.s. indicates no significance.
Diagnostic Potency Prediction by Risk Score Analysis
Researcher has identified that circRNAs might be potential biomarkers for other human malignant tumors; however, they only focused on the predictive value of these circRNAs in tissue samples, and no evidence was given for these circRNAs to predict the GBM from healthy controls in plasma samples. Next, to further examine the detailed accuracy and specificity of these three circRNAs as a GBM potential signature, the risk score formula was applied to assess the diagnostic value of the three circRNAs profiling system.
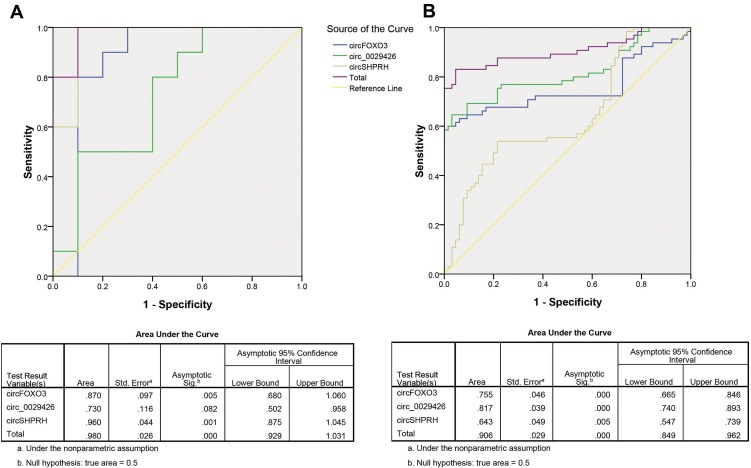
At first, the three circRNAs were analyzed to detect whether they could predict GBM from the controls. By using the risk score analysis, the training set was divided into a higher-risk group and a lower-risk group, which indicated the GBM group and the NC group. Next, the cut-off value, defined as the maximal value for the value of sensitivity + specificity, was used. We applied the value of 4.311 as the cut-off, and obtained the positive predictive value (PPV) and negative predictive value (NPV) of 90% and 80% in the training set, respectively. The same method was used in the validation set, where the PPV and NPV were 89% and 82%, respectively (Table 2). The predicting ability was also calculated by ROC curve. The areas under the ROC curves (AUC) of the three circRNAs in the training set were 0.870, 0.730, and 0.960, respectively while the combination of the three factors possessed a moderate ability for discrimination between GBM patients and controls with an AUC of 0.980 (Figure 3A). Besides, the AUCs in the validation set were 0.755, 0.817 and 0.643, respectively, while the combination was 0.906 (Figure 3B).
Table 2.
Risk Score Analysis of in Glioblastoma (GBM) and Cancer-Free Control Plasma Samples
| Score | 0–4.311 | 4.311–12.96 | PPVa | NPVb |
|---|---|---|---|---|
| Training set | 0.90 | 0.80 | ||
| GBM | 2 | 18 | ||
| Control | 16 | 4 | ||
| Validation set | 0.89 | 0.82 | ||
| GBM | 11 | 89 | ||
| Control | 82 | 18 |
Notes: aPPV, positive predictive value. bNPV, negative predictive value.
Figure 3.
Receiver operating characteristic (ROC) analysis of the three potential biomarkers for glioblastoma (GBM) using risk score analysis. (A) ROC analysis of the three potential biomarkers for GBM in the training set. (B) ROC analysis of the three potential biomarkers for GBM in the validation set.
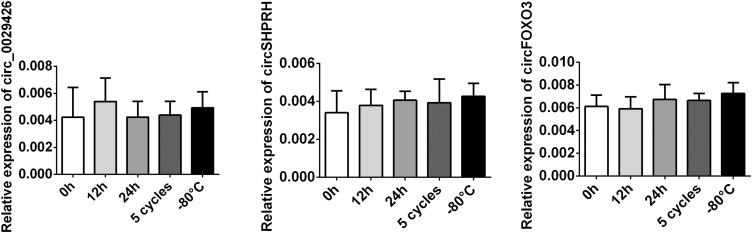
Endogenous Expression Stable of circRNA in Human Plasma
The endogenous expression of the three circRNAs was further detected in healthy controls. The PCR product band was further examined after different treatment for the three plasma samples obtained from controls by agarose electrophoresis. The expression stable ability for the circRNAs in plasma sample was confirmed under treatment with different time in room temperature and five cycles of freez–thawing. None of the expressions of the three circRNA expression was suppressed (Figure 4). We found that the expression level of the three circRNA was not alternated, indicating that circFOXO3, circ_0029426, and circ-SHPRH weres stably expressed and detectable in human plasma.
Figure 4.
The products of the amplification fragment were detected by agarose electrophoresis. Data were presented as mean ± SEM.
Discussion
Glioblastoma is the most common tumor with high malignancy in the human central nervous system.12 Although the diagnosis and treatment approach have been developed, the prognosis for the glioblastoma remains poor. Early diagnosis for the GBM is very necessary and the development of prognosis or diagnosis biomarker for GBM is urgent.
With the development of high-throughput sequencing technology, various human non-coding RNA were found and identified as important regulation factors in human malignant tumors.13 Increasing evidence has indicated that circRNAs play a crucial role including carcinogenesis, progression and clinical outcomes of various human cancers.14,15 For example, researchers have found that circEPSTI1 could promote cell proliferation and apoptosis through sponging miR-4753 and miR-6809 to regulate the BCL11A level in triple-negative breast cancer.16 The circNT5E could function as a sponge for the miR-422a, which is involved in cell proliferation, migration, and invasion in GBM.17 Based on the information above, the diagnostic and prognostic value of circRNAs is also very important in research hotspots.18,19 Glioblastoma is the most common primary cancer derived from the central nervous system. Little is known regarding the fingerprint for GBM of circRNA. In this study, we enrolled the identified 14 circRNAs as candidates. The 14 circRNAs have proven to be vital factors either for the occurrence and development of GBM and were differently expressed in human GBM tissue samples. Among the 14 candidates, we first screened six circRNAs in a 40-paired plasma sample with stable expression and increased in GBM plasma samples. The validation set was also employed with a larger sample size. Followed by risk score analysis, the prediction ability of three circRNAs was measured by ROC analysis. Here in this study, we presented that higher circulating circRNA expression in GBM is correlative with GBM. However, limited information is available on the underlying mechanism concerning the observations. Further studies on genetic and epigenetic mechanisms should be carried out to clarify the issue.
Conclusion
We identified three circRNAs, circFOXO3, circ_0029426, and circ-SHPRH, which have the potential marker for the tumorigenesis prediction of GBM in this study. This preliminary study is limited by the small sample size in this work. A deeper potential function of the three circRNAs in regulating the pathogenesis of GBM is necessary for us to explore in the future.
Acknowledgment
The authors would like to show gratitude to all members in the Department of Neurology of the Affiliated Huai’an No. 1 People’s Hospital of Nanjing Medical University.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205(12):613–621. doi: 10.1016/j.cancergen.2012.10.009 [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng YS, Lin C, Cheng YP, Yu YL, Tang CT, Hueng DY. Epithelial cell transformation sequence 2 is a potential biomarker of unfavorable survival in human gliomas. Neurol India. 2014;62(4):406–409. doi: 10.4103/0028-3886.141278 [DOI] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 5.Bell C, Dowson N, Fay M, et al. Hypoxia imaging in gliomas with 18F-fluoromisonidazole PET: toward clinical translation. Semin Nucl Med. 2015;45(2):136–150. doi: 10.1053/j.semnuclmed.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 6.Chen WL, Jiang L, Wang JS, Liao CX. Circ-0001801 contributes to cell proliferation, migration, invasion and epithelial to mesenchymal transition (EMT) in glioblastoma by regulating miR-628-5p/HMGB3 axis. Eur Rev Med Pharmacol Sci. 2019;23(24):10874–10885. doi: 10.26355/eurrev_201912_19791 [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, Liao K, Miao Z, et al. CircFOXO3 promotes glioblastoma progression by acting as a competing endogenous RNA for NFAT5. Neuro-Oncology. 2019;21(10):1284–1296. doi: 10.1093/neuonc/noz128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17(1):137. doi: 10.1186/s12943-018-0888-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, Wang C, Song H, Xu Y, Ji G. RNA-Seq profiling of circular RNAs in human colorectal cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18(1):8. doi: 10.1186/s12943-018-0932-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J Cell Physiol. 2019;234(8):13807–13819. doi: 10.1002/jcp.28061 [DOI] [PubMed] [Google Scholar]
- 11.Zhang G, Sun W, Zhu L, Feng Y, Wu L, Li T. Overexpressed circ_0029426 in glioblastoma forecasts unfavorable prognosis and promotes cell progression by sponging miR-197. J Cell Biochem. 2019;120(6):10295–10302. doi: 10.1002/jcb.28313 [DOI] [PubMed] [Google Scholar]
- 12.Lv X, Wang M, Qiang J, Guo S. Circular RNA circ-PITX1 promotes the progression of glioblastoma by acting as a competing endogenous RNA to regulate miR-379-5p/MAP3K2 axis. Eur J Pharmacol. 2019;863:172643. doi: 10.1016/j.ejphar.2019.172643 [DOI] [PubMed] [Google Scholar]
- 13.Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9(1):4475. doi: 10.1038/s41467-018-06862-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Song G, Yuan J, et al. Circular RNA circ_0003204 inhibits proliferation, migration and tube formation of endothelial cell in atherosclerosis via miR-370-3p/TGFbetaR2/phosph-SMAD3 axis. J Biomed Sci. 2020;27(1):11. doi: 10.1186/s12929-019-0595-9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Fan M, Wang Y, Gao S. Circular RNA circMTO1 acts as an antitumor factor in rectal cancer cell lines by downregulation of miR-19b-3p. J Cell Biochem. 2019. doi: 10.1002/jcb.29570 [DOI] [Google Scholar]
- 16.Xie J, Wang S, Li G, et al. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med. 2019;23(5):3597–3602. doi: 10.1111/jcmm.14260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R, Zhang S, Chen X, et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. 2018;78(17):4812–4825. doi: 10.1158/0008-5472.CAN-18-0532 [DOI] [PubMed] [Google Scholar]
- 18.Begum S, Yiu A, Stebbing J, Castellano L. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene. 2018;37(30):4055–4057. doi: 10.1038/s41388-018-0230-3 [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. doi: 10.1038/s41388-017-0019-9 [DOI] [PubMed] [Google Scholar]