Significance
DNA oxidative damage by reactive oxygen species represents a major challenge to genome integrity that contributes to the aging process as well as many human diseases. In particular, guanine is the most vulnerable of the nucleobases to oxidation and leads to transcription-blocking lesions: 5-guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp). The molecular mechanism of transcription stalling of these nonbulky but helix-distorting DNA lesions remains completely unknown. This work systematically investigates how these lesions affect transcription and provides structural snapshots of how Pol II is stalled by a nonbulky Gh lesion in a stepwise manner, revealing an unexpected mechanism of transcriptional stalling. Important insights are gained into understanding the mechanisms of lesion recognition, transcriptional stalling, transcriptional bypass, and mutagenesis of oxidative DNA lesions.
Keywords: oxidative damage, DNA lesions, transcription arrest, guanidinohydantoin, spiroiminodihydantoin
Abstract
Oxidation of guanine generates several types of DNA lesions, such as 8-oxoguanine (8OG), 5-guanidinohydantoin (Gh), and spiroiminodihydantoin (Sp). These guanine-derived oxidative DNA lesions interfere with both replication and transcription. However, the molecular mechanism of transcription processing of Gh and Sp remains unknown. In this study, by combining biochemical and structural analysis, we revealed distinct transcriptional processing of these chemically related oxidized lesions: 8OG allows both error-free and error-prone bypass, whereas Gh or Sp causes strong stalling and only allows slow error-prone incorporation of purines. Our structural studies provide snapshots of how polymerase II (Pol II) is stalled by a nonbulky Gh lesion in a stepwise manner, including the initial lesion encounter, ATP binding, ATP incorporation, jammed translocation, and arrested states. We show that while Gh can form hydrogen bonds with adenosine monophosphate (AMP) during incorporation, this base pair hydrogen bonding is not sufficient to hold an ATP substrate in the addition site and is not stable during Pol II translocation after the chemistry step. Intriguingly, we reveal a unique structural reconfiguration of the Gh lesion in which the hydantoin ring rotates ∼90° and is perpendicular to the upstream base pair planes. The perpendicular hydantoin ring of Gh is stabilized by noncanonical lone pair–π and CH–π interactions, as well as hydrogen bonds. As a result, the Gh lesion, as a functional mimic of a 1,2-intrastrand crosslink, occupies canonical −1 and +1 template positions and compromises the loading of the downstream template base. Furthermore, we suggest Gh and Sp lesions are potential targets of transcription-coupled repair.
DNA is under constant attack from reactive oxygen species, chemicals, spontaneous hydrolysis, etc. (1–4). Admist this, some DNA lesions not only interfere with replication but also cause severe pausing and arrest of transcribing RNA polymerase II (Pol II) elongation complex (EC) (5–7). Prolonged stalling of EC might lead to cell cycle arrest and apoptosis (2, 8). DNA damage accumulation, in particular oxidative damage, contributes to the aging process itself as well as human diseases such as cancer (9).
Guanine (G) is the most vulnerable of nucleobases to oxidation. Indeed, 8-oxo-7,8-dihydroguanine (8OG) is one of the most well-known and abundant oxidative DNA lesions (10). It is noteworthy that 8OG is orders of magnitude more prone to further oxidation, having a redox potential much lower than that of guanine (11, 12). Further oxidation from 8OG destroys the purine ring and results in several nonbulky hydantoin lesions, including 5-guanidinohydantoin (Gh) and two stereoisomeric spiroiminodihydantoin (R-Sp and S-Sp) lesions (Fig. 1A) (13, 14). Gh exists as a pair of interconverting diastereomers at neutral pH that cannot be individually studied, while Sp also has a stereocenter; its two stable diastereomers can be individually purified and studied (15, 16). The oxidation products Sp and Gh are detected in mouse tissues experiencing inflammation with two orders of magnitude lower frequency when compared to 8OG (17).
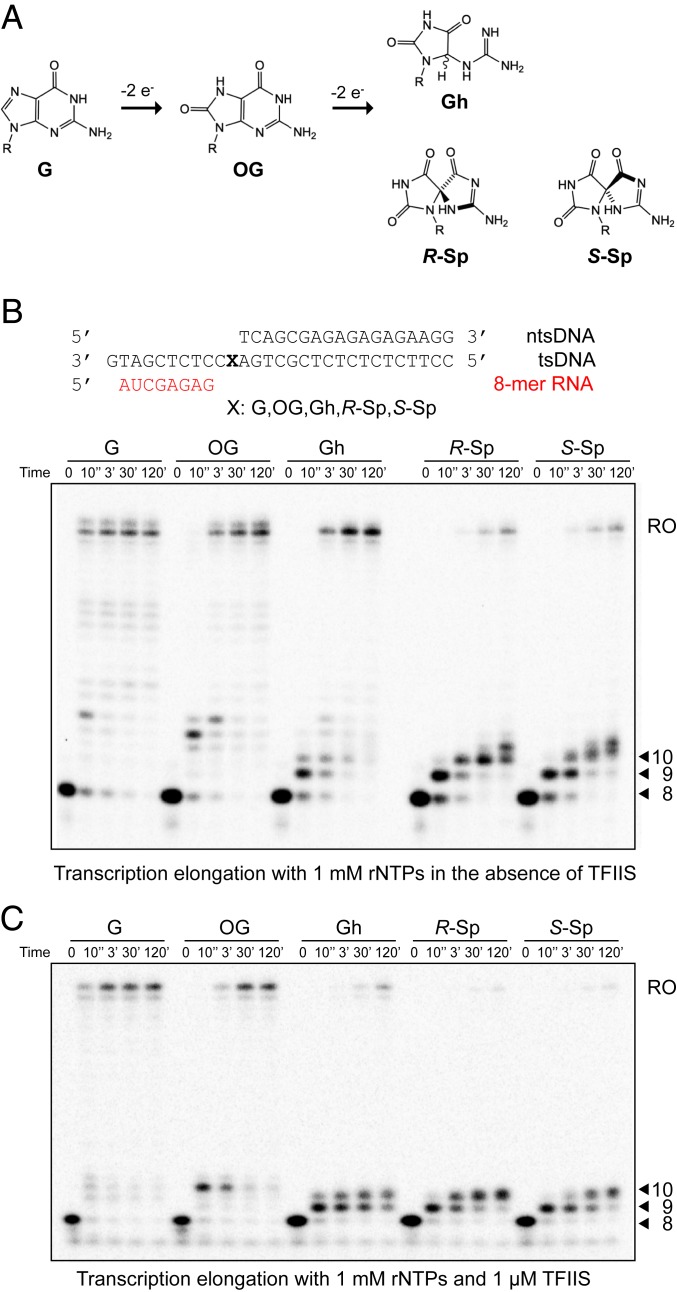
Fig. 1.
Impact of consecutive oxidation products of guanine (8OG, Gh, R-Sp, and S-Sp) on RNA polymerase II transcription. (A) Schematic representation of consecutive oxidation products of guanine resulting in 8OG, then Gh or Sp, respectively. (B) Transcription assay with 1 mM rNTPs in the absence of TFIIS. The sequences of transcription scaffolds contain site-specific lesions. X represents G, 8OG, Gh, R-Sp, or S-Sp, respectively. The template strand (tsDNA) and nontemplate strand DNA (ntsDNA) are shown in black. RNA primer (8mer) is shown in red. Transcription reaction was started by adding 1 mM of rNTPs to Pol II EC. Time points are 0, 10 s, 3 min, 30 min, and 2 h unless stated otherwise. The position of the run-off transcript is indicated as RO. (C) Transcription assay with 1 mM rNTPs in the presence of 1 μM of TFIIS. Pol II EC-TFIIS was preincubated for 10 min before adding 1 mM of rNTPs. Time points are 0, 10 s, 3 min, 30 min, and 2 h.
These chemically related guanine-derived oxidized lesions interfere with replication and transcription processes. Sp and Gh lesions are highly mutagenic for replication in Escherichia coli with nearly 100% mutation rate, thus these hydantoin DNA lesions negatively impact the genome at a similar frequency as the mildly mutagenic 8OG lesion (18). In addition, Sp and Gh lesions cause replication blocks by phage DNA polymerase and lead to slow misincorporation of A or G by reverse transcriptases (18–20). As for transcription, in sharp contrast to effective transcriptional bypass of 8OG (1, 21, 22), an early study using HeLa nuclear extract reported that the presence of Gh or Sp in template-strand DNA (tsDNA) causes severe transcriptional stalling (23). However, it is not clear how these nonbulky lesions (much smaller size in comparison with guanine and other bulky lesions) lead to strong stalling. It is also not clear how these lesions affect transcriptional kinetics and fidelity checkpoint steps (nucleotide incorporation, extension, proofreading) by RNA Pol II. Furthermore, the role of transcription elongation factors in modulating transcriptional stalling also remains elusive.
Here we systematically investigated the molecular basis of how these guanine-derived oxidized lesions affect RNA Pol II transcription using a combined biochemical and structural approach. We found that both Gh and Sp lesions cause two consecutive transcriptional stalling events. Transcription factors Rad26 (yeast Cockayne syndrome B protein [CSB] ortholog) and TFIIS failed to rescue transcriptional stalling. We further investigated how these lesions affect individual transcriptional fidelity checkpoint steps. Finally, we performed comprehensive structural analysis that captures structural snapshots of Pol II processing a site-specific Gh lesion in a stepwise manner. We reveal how Gh is accommodated when Pol II initially encounters the Gh lesion, how ATP binds and is incorporated opposite the Gh lesion, how the translocation of Gh-AMP pair is jammed via a unique reconfiguration of the Gh lesion, and how Pol II gets arrested and backtracked. Taken together, these results provide mechanistic understanding of how Pol II deals with guanine-derived oxidative damage during transcription.
Result
Nonbulky Gh and Sp Lesions, but Not 8OG, Lead to Strong Pol II Stalling.
To investigate the effect of these damaged bases on transcription, we performed in vitro transcription assays using purified 12-subunit yeast RNA polymerase II with either a miniscaffold (Fig. 1B) or full-bubble scaffold (Fig. 2) containing a site-specific damaged base in the tsDNA. We found very distinct Pol II behaviors that range from full bypass to almost complete stalling for these DNA lesions (Fig. 1B). We observed dominant run-off transcripts in undamaged dG or 8OG templates. In sharp contrast, we observed few run-off transcripts, but at least two strong consecutive stalling bands for R-Sp and S-Sp templates. These two consecutive stalling bands (9mer and 10mer) suggest that nucleotide incorporation opposite DNA lesions and subsequent extension steps are significantly retarded. For transcription from the Gh-containing template, we observed strong pausing bands at early time points as well as accumulative run-off transcripts with prolonged incubation.
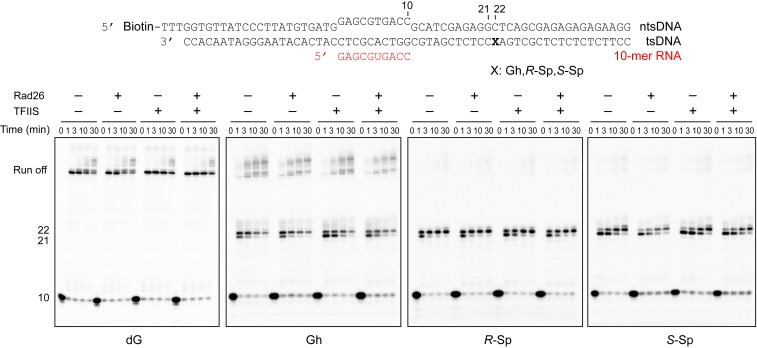
Fig. 2.
Transcription assay in the presence of Rad26 and TFIIS. Pol II ECs were prepared with a full-bubble scaffold, and transcription assay was performed in the presence or absence of 100 nM Rad26 and 300 nM TFIIS. Time points are 0 (control, without rNTPs), 1 min, 3 min, 10 min, and 30 min. Rad26 was added before the reaction started, while TFIIS was added together with 1 mM of rNTPs because preincubation with TFIIS and EC leads to partial degradation of RNA. Addition of Rad26 or TFIIS did not change the stalling patterns of the Gh or Sp lesion-containing scaffold.
TFIIS and Rad26 Cannot Rescue Transcription Stalling by Gh and Sp Lesions.
TFIIS is an elongation factor that stimulates cleavage of backtracked transcripts and reactivates Pol II elongation. As a result, TFIIS enhances overall bypass over some types of transcriptional barriers and DNA lesions (24, 25). To test whether TFIIS can rescue transcriptional stalling induced by these oxidative lesions, we performed the transcription assay in the presence of TFIIS. It is intriguing to note that the presence of TFIIS significantly reduced the run-off product levels from templates with Gh and Sp lesions in a TFIIS dose-dependent manner, while the impact on run-off transcripts from dG and 8OG templates is minimal (Fig. 1C and SI Appendix, Fig. S1). Similarly, TFIIS is unable to rescue transcriptional stalling induced by Gh or Sp lesions using the full-bubble scaffold (Fig. 2). These results indicate that RNA transcripts with Gh or Sp lesions are active targets of TFIIS-stimulated cleavage activity (see following section and Discussion).
Rad26/CSB, a key factor in transcription-coupled repair, is recruited to stalled Pol II and initiates transcription-coupled repair (26–28). Our recent structure revealed the molecular basis of how Rad26 helps to distinguish different types of transcriptional arrest during the DNA lesion recognition step: upon binding upstream of stalled Pol II, Rad26 can facilitate Pol II forward translocation and therefore rescue some types of transcriptional stalling imposed by noncovalent transcriptional barriers and small DNA lesions, whereas Pol II stalled by bulky lesions remains arrested in the presence of Rad26 and commits to recruitment of downstream repair factors (26–28). To investigate whether oxidative lesion-induced transcriptional stalling can be rescued by Rad26, we performed the transcription assay using the full-bubble scaffold shown in Fig. 2. The result shows that the transcription stalling induced by Gh or Sp lesions cannot be rescued by adding Rad26 and TFIIS. We speculate that these oxidative lesion-induced prolonged transcriptional stalling events provide a strong signal for transcription-coupled repair in vivo (29).
Error-Prone Incorporation of Purines opposite Gh and Sp Lesions.
To further understand how these DNA lesions affect nucleotide selectivity at the incorporation step, we performed a single-nucleotide incorporation assay by adding individual rNTPs to the 9mer RNA scaffold (Fig. 3 A and B and SI Appendix, Fig. S2). As shown in Fig. 3, Gh and Sp strongly prefer error-prone incorporation, whereas the dG template favors error-free (C) incorporation and the 8OG template supports both effective error-free (C) and error-prone (A) incorporation (21). The preference for nucleotide incorporation for Gh and Sp template is ranked as follows: A > G >> C ∼ U.
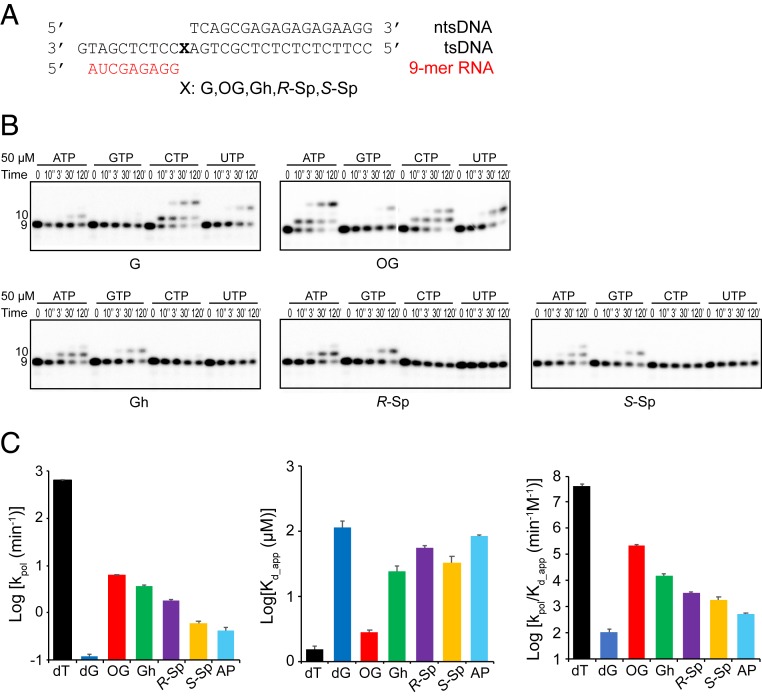
Fig. 3.
Impact of consecutive oxidation products of guanine on substrate selectivity and ATP incorporation kinetics. (A) The sequences of the transcription scaffold containing site-specific lesions. X represents G, 8OG, Gh, R-Sp, or S-Sp, respectively. The color code is the same as in Fig. 1B. (B) Single nucleotide incorporation assay. Pol II EC with Gh, R-Sp, or S-Sp prefers A and G incorporation (purines), whereas 8OG preferentially incorporates both A and C. A total of 50 μM of each rNTP was used as final concentration. The time points are 0, 10 s, 3 min, 30 min, and 2 h. (C) Kinetic parameters of ATP incorporation opposite each lesion. We compared rate constants (kpol), apparent substrate binding affinity (Kd.,app), and overall catalytic efficiency (Log kpol/Kd,app). The color codes for parameters are: black (dT), blue (dG), red (OG), green (Gh), purple (R-Sp), yellow (S-Sp), and cyan (abasic site [AP]). Raw gel data are shown in SI Appendix, Fig. S3. All data were processed by using Prism. Error bars indicate SE calculated from regression fit. Kinetic parameters of dT and abasic site are referred to from our previous paper (30). Numerical values are indicated in SI Appendix, Table S1.
To compare the relative efficiency of ATP misincorporation opposite these chemically related oxidative lesions, we measured kinetic parameters of ATP incorporation for the different bases (Fig. 3C and SI Appendix, Fig. S3 and Table S1) and compared them with those of abasic site and nondamaged dT from our previous reports (30). As shown in Fig. 3C and SI Appendix, Table S1, the efficiency of ATP misincorporation is extremely low for the undamaged dG template with a kpol/Kd,app value of 1.09 × 10−4 (min−1⋅μM−1). The presence of oxidative DNA lesions dramatically enhances the efficiency of ATP misincorporation. For the 8OG template, the efficiency of ATP misincorporation opposite 8OG is increased by 2,100-fold compared to that of nondamaged dG (compared by kpol/Kd,app), further supporting previous reports that 8OG can form a Hoogsteen base pair with A (21, 31). For the Gh template, there is a 141-fold increase of kpol/Kd,app for ATP misincorporation compared to that of the undamaged dG template. Interestingly, the kpol value of Gh is about two times lower than that of 8OG, while Kd,app is about eight times weaker. This result suggests that 8OG is a better base-pairing partner for ATP, compared to Gh. We also compared the two stereoisomers R-Sp and S-Sp. The efficiencies for ATP misincorporation are about 30- and 17-fold higher than that for undamaged dG, respectively. It is also intriguing to note that the efficiencies for ATP incorporation of all four oxidative lesions (8OG, Gh, R-Sp, and S-Sp) are higher than that for the abasic site, varying from 3-fold to over 400-fold. These results indicate distinct mechanisms among these oxidative lesions in facilitating ATP misincorporation other than the canonical nontemplated A-rule (30, 32).
Slow Error-Prone Bypass, but Not Error-Free Bypass, Is Allowed for the Gh Lesion.
To investigate the effect of these oxidative lesions on extension steps, we systematically performed extension assays using scaffolds containing 10mer RNA with either C (10C, extension from error-free bypass), A or G (10A or 10G, extension from error-prone bypass) at the 3′ end (Fig. 4A).
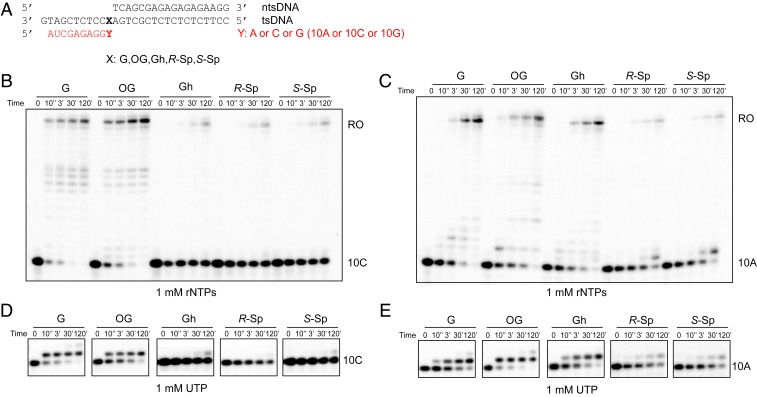
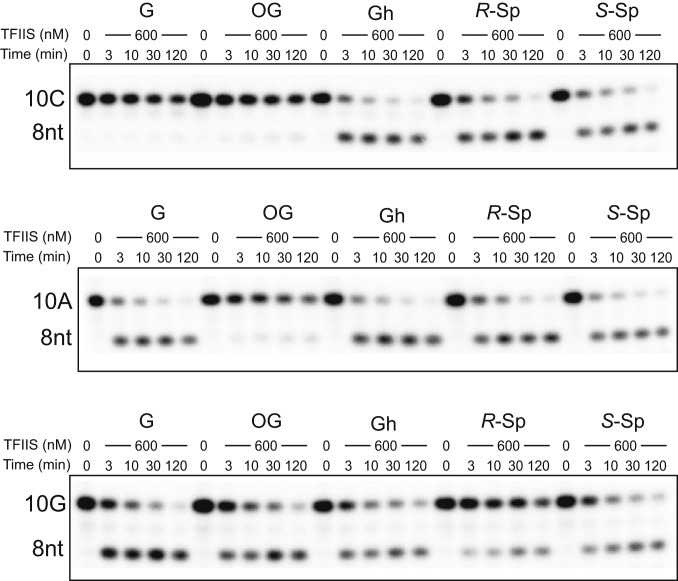
Fig. 4.
Transcriptional pausing or bypass of consecutive oxidative lesions. (A) Sequences of scaffold used in transcription extension assay. X represents G, 8OG, Gh, R-Sp, or S-Sp, respectively. Y represents A, C, or G, respectively. The color code is the same as Fig. 1B. (B–E) Extension (Upper) assay beyond the oxidative lesion using 10C (Fig. 4 B and C), 10A (Fig. 4 D and E), and 10G (SI Appendix, Fig. S4) primer, respectively. The reaction was incubated with either a mixture of four rNTPs (1 mM) (Fig. 4 B and D and SI Appendix, Fig. S4) or individual rNTPs as shown in the figure panels (Fig. 4 C and E and SI Appendix, Fig. S4). The time points are 0, 10 s, 3 min, 30 min, and 2 h.
For the extension after error-free nucleotide incorporation (Fig. 4A, 10C), we observed few run-off and extension products for Gh and Sp templates, which is in sharp contrast with results from nondamaged G and 8OG templates (Fig. 4B). These results suggest that error-free extension is greatly inhibited for Gh and Sp templates.
For the extension after ATP misincorporation (Fig. 4A, 10A), we observed run-off transcripts with prolonged incubation for G, 8OG, and Gh templates, suggesting slow error-prone bypass of these lesions after ATP misincorporation (Fig. 4C). In contrast, few run-off and extension products were observed for Sp templates. We also observed a similar trend for the extension after GTP misincorporation (SI Appendix, Fig. S4).
We further investigated whether the presence of the lesion affects the fidelity of the extension step (Fig. 4 D and E). In all lesions tested, UTP is preferred (if there is an extension product). It is noteworthy that no obvious rNTP incorporation was observed in the scaffolds with 10C containing Gh and Sp lesions (error-free scaffold), suggesting the error-free extension is greatly inhibited. In sharp contrast, slow UTP incorporations were observed for the scaffolds that mimic error-prone extension (10A and 10G) (Fig. 4 and SI Appendix, Fig. S4).
In summary, these chemically related lesions derived from guanine oxidation have distinct effects on transcription extension. The presence of 8OG in tsDNA supports both error-free (10C) and error-prone extension (10A), which is consistent with previous reports suggesting error-prone bypass of the 8OG lesion (21, 31). Contrastingly, the presence of Gh and Sp lesions strongly inhibits error-free incorporation and extension. Only slow error-prone extension is allowed for the Gh lesion.
Effect of Oxidative Lesions on TFIIS-Stimulated Proofreading Cleavage.
Transcription elongation factor TFIIS plays an important role in stimulating proofreading cleavage of mismatched RNA transcripts (24, 25). To test whether misincorporation opposite oxidative lesions can be recognized by TFIIS-stimulated cleavage, we incubated TFIIS with Pol II EC harboring different types of 10mer RNA and damage, in the absence of rNTPs (Fig. 5). As shown in Fig. 5, TFIIS preferentially cleaves mismatched pairs (10A:G or 10G:G pairs) but not a matched pair (10C:G) for the undamaged dG scaffold. Intriguingly, the presence of 8OG inhibits cleavage of both 8OG-10C and 8OG-10A. This result indicates that 8OG-C and 8OG-A base pairs are recognized as “matched” pairs and therefore misincorporation of ATP opposite 8OG escapes Pol II proofreading.
Fig. 5.
The presence of an oxidative lesion changes Pol II proofreading dramatically. TFIIS-stimulated cleavage analysis of 10mer RNA scaffolds with different lesions were performed by incubation with 600 nM of TFIIS. Time points are 0 (control, without TFIIS), 3 min, 10 min, 30 min, and 2 h. Pol II fails to distinguish 10A:OG mismatched pair and 10C:OG pair, as the TFIIS-stimulated cleavage activity of 10A:OG and 10C:OG are greatly abolished. In contrast, the 10G:OG is recognized as a mismatched pair with high cleavage activity. Intriguingly, Gh and both Sp lesions always lead to high cleavage activities regardless of the 3′ end primers (10A, 10C, and 10G).
In sharp contrast, almost all RNA transcripts opposite Gh or Sp lesions we tested are effectively cleaved by TFIIS, indicating that after addition of a nucleotide to the +1 site, these lesions are prone to backtracking and cleavage in the absence of rNTPs. We also found that the scaffold containing R-Sp and 10G was significantly more resistant to TFIIS-induced cleavage. It is noteworthy that the R-Sp lesion was previously shown to be more resistant to nuclease P1 digestion and NEIL1 cleavage compared to the S-Sp (33, 34).
Structural Insights into Gh Lesion-Induced Transcriptional Stalling.
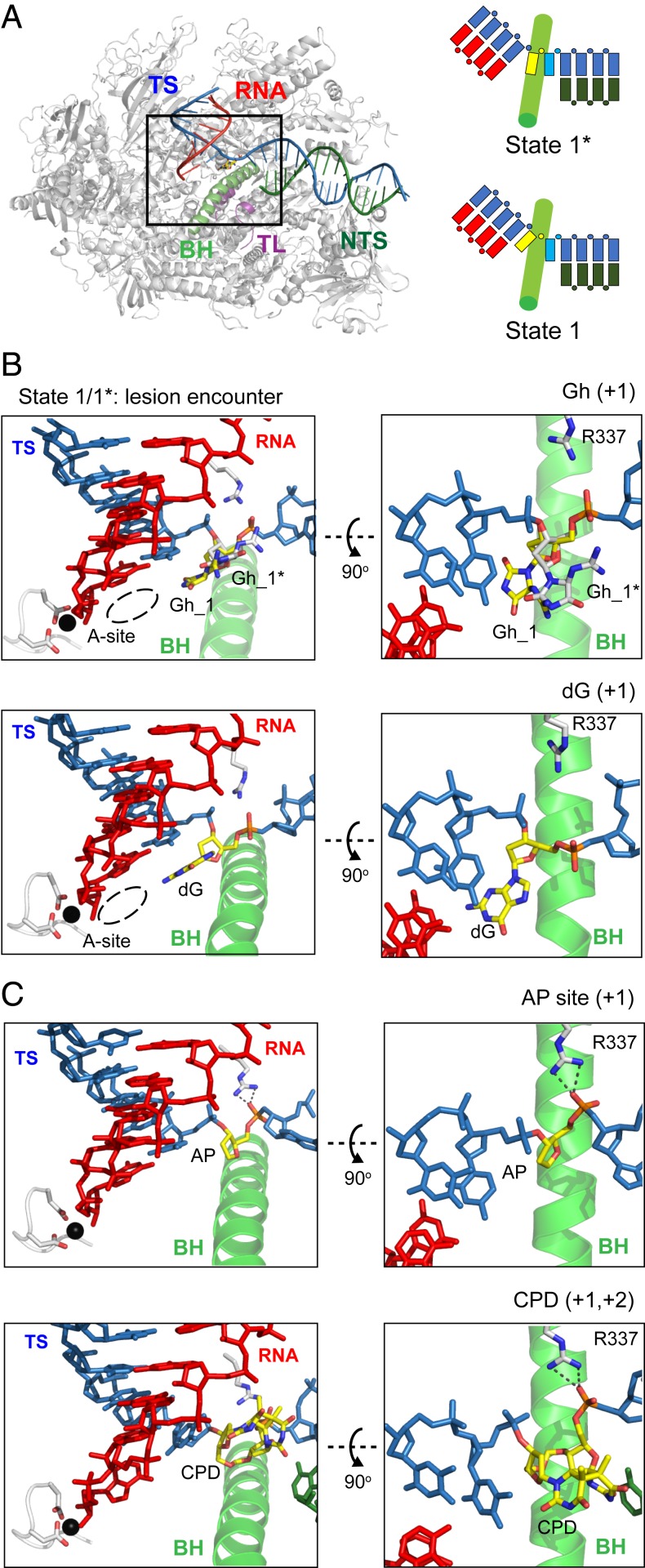
To understand the molecular basis of Gh-induced transcriptional stalling, we solved seven Pol II structures (SI Appendix, Table S2): six Gh lesion-containing structures and one structure containing the undamaged template (as a control). These structures capture snapshots of Pol II processing the Gh lesion in a stepwise manner: encounter (before ATP binding, states 1 and 1*), ATP binding (before chemistry, state 2E), nucleotide incorporation (postchemistry, state 3), translocation (state 4), and extension and backtracking (states 5 and 6), respectively.
To understand how Gh is loaded into the active site when Pol II encounters a Gh lesion, we solved the encounter structure at 3.4-Å resolution (before ATP binding, states 1 and 1*). Pol II EC is essentially in a posttranslocation state in which the active site is available for substrate binding (Fig. 6 A and B). Interestingly, we observed two conformers of the Gh lesion in a single crystal that fit the electron density: one is located in the +1 canonical template position (state 1) and the other is above the bridge helix (state 1*) (Fig. 6B and SI Appendix, Fig. S5A and Movie S1). The relative occupancy of states 1 and 1* is 0.45 and 0.55, respectively. The Gh lesion in state 1* represents a half-way translocation position (Fig. 6B). The nucleobase of Gh (hydantoin ring) is shifted about 3 Å compared with the corresponding undamaged dG at the canonical +1 template position (Movie S2). Furthermore, the distance between the guanidinium group of the Gh lesion and its own 5′ phosphate group is only 2.9 Å, suggesting potential hydrogen bonding between these two groups (Fig. 6B and SI Appendix, Fig. S5A). In addition, the phosphate backbone is also rotated toward R337 in Rpb1 about 3.4 Å and forms a hydrogen bond. This results in a sandwiched 5′ phosphate group of Gh by its guanidinium group and R337 in Rpb1. The state 1* (Gh) is similar to the half-way translocation positions observed in Pol II EC containing other DNA lesions (Fig. 6C) (30, 32, 35). The rmsd value for the structure of Pol II containing the Gh lesion was less than 0.7 with Pol II containing an abasic site (Protein Data Bank [PDB]: 6BLO), a cyclobutane pyrimidine dimer (CPD, PDB: 4A93), an 8,5′-cyclo-2′-deoxyadenosine (CydA, PDB: 4X6A) lesion in DNA (21, 30, 32, 35). The presence of Gh in the DNA template does not change the overall structure of Pol II EC beyond the active site.
Fig. 6.
Crystal structure of Pol II elongation complex encountering a Gh lesion. (A) Overall structure of Pol II EC harboring a Gh lesion is shown. For clarity, part of Rpb2 (residues from 20 to 770) is omitted. Color codes are as follows: tsDNA (TS, blue), ntsDNA (NTS, dark green), RNA (red), bridge helix (BH, green), trigger loop (TL, purple), Mg2+ ion (black), and other elements (gray). Schematic representation of miniscaffold with bridge helix is shown with the same color codes. The Gh lesion and downstream undamaged 5′ template nucleobase are shown in yellow and cyan, respectively. (B and C) Structural comparison of +1 bases between Gh to nondamaged (dG), abasic site (AP), or CPD lesion (PDBs: 6BLO and 4A93) (30, 35). (Left) Canonical view of the active site, where Gh, abasic site, or CPD lesion is located at a common half-way translocation intermediate position (Right above the BH), while dG base is correctly loaded at the +1 canonical template position (fully across the BH). (Right) The 90° rotated view (along x axis) from Left, the hydrogen bonds between R337 and 5′ phosphate group of lesions are highlighted in dashed lines. The Gh base in state 1* is colored in white, while other +1 bases are colored in yellow.
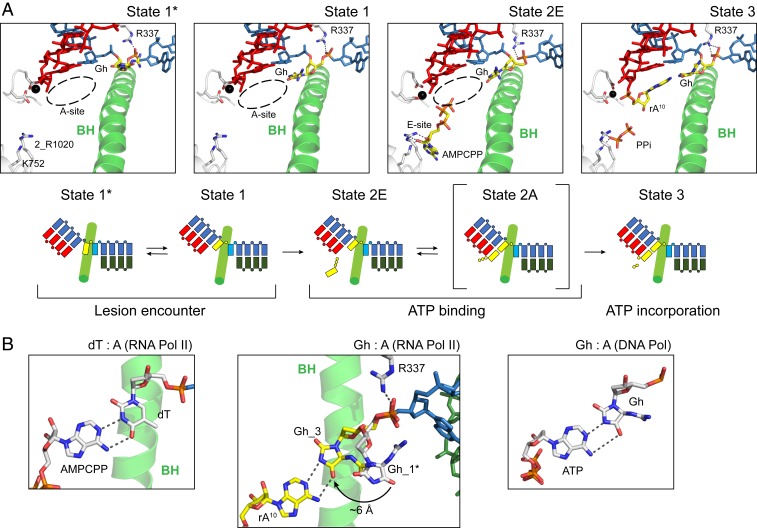
To investigate how the ATP substrate is bound and subsequently incorporated opposite the Gh lesion, we obtained Gh-AMPCPP and Gh-ATP complex structure by soaking with AMPCPP (nonhydrolyzable ATP) or ATP, respectively (Fig. 7). The Gh-AMPCPP represents the state of ATP binding (before chemistry, state 2E), whereas the Gh-ATP complex structure represents the state after ATP incorporation (postchemistry, state 3). In state 2, we found that AMPCPP binds to the entry site (E-site) instead of the addition site (A-site) (Fig. 7A and SI Appendix, Fig. S5 B, Bottom). In sharp contrast, in the ATP soaking structure (state 3) that allows chemistry, we found ATP occupies the addition site and is incorporated to the 3′ end of RNA, indicating chemistry drives the equilibrium from E-site to A-site. Pol II is in a pretranslocation state (Fig. 7 A, Right). We also observed clear electron density for pyrophosphate (PPi) in this structure (Fig. 8, Top). Intriguingly, in both nucleotide soaking structures (states 2 and 3), we found that the Gh base is loaded at the canonical template position equivalent to state 1 (Fig. 8 and SI Appendix, Fig. S5C). Therefore, binding of ATP stabilizes the loaded conformation of the Gh base (Fig. 7B). In state 3, we observed base pairing between Gh and AMP via hydrogen bonds that resembles base pairing between dT and ATP in Pol II or Gh and dATP in a DNA polymerase (36, 37), with the exception that the Gh lesion is facing much more toward the minor groove (SI Appendix, Fig. S5D). Interestingly, the hydrogen bonding interaction between R337 and the phosphate backbone is still maintained in these structures, which is absent in the structure containing the undamaged dG template (Fig. 6B and SI Appendix, Fig. S5D).
Fig. 7.
Structural basis of ATP binding and incorporation opposite the Gh lesion. (A) The overview structures of the Pol II EC active site at four different snapshots of ATP binding and incorporation (states 1*, 1, 2E, and 3). The Gh template and incoming ATP are colored in yellow. Mg2+ ion is represented as a black sphere. Residues that interact with nucleotide substrate or metal are indicated. Dashed circle indicates the A-site. Schematic representations of each state are shown at Bottom. Rpb2 R1020 is labeled as 2_R1020. All other labeled Pol II residues are from Rpb1 subunit. (B) Comparison of base pairing between dT: A or Gh: A in RNA pol II, and Gh: A in DNA polymerase (PDBs: 4A3L and 3NAE). The Gh base from state 3 and rA10 is colored in yellow, while other bases are colored in white. Dashed line indicates potential hydrogen bonding distance. Black arrow indicates movement of the hydantoin ring of the Gh lesion upon ATP binding.
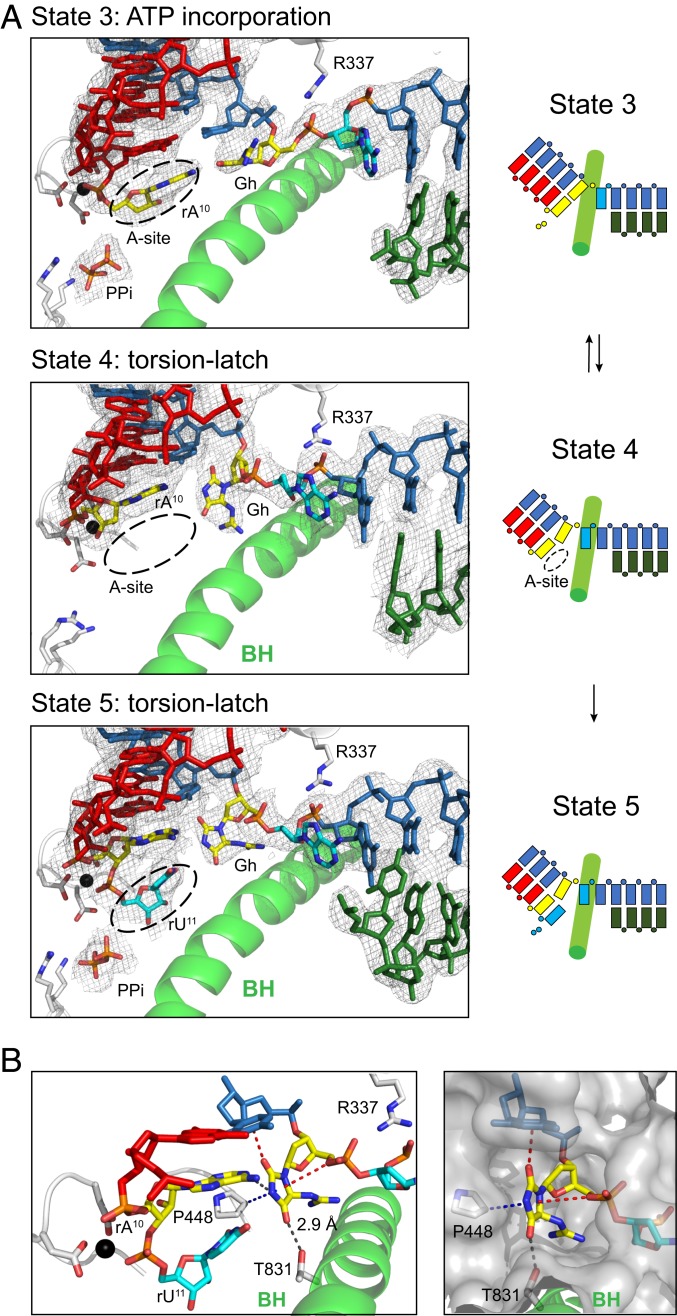
Fig. 8.
Unusual perpendicular Gh lesion reconfiguration impairs proper loading of the downstream undamaged 5′ template nucleobase during the extension step. (A) Structural analysis of consecutive translocation and extension of EC containing Gh lesion. (Left) Show canonical views of Pol II active sites in four states (states 3 to 6), with 2Fo-Fc electron density map of each scaffold contoured at 1.1 σ. In state 3, Gh lesion is located at canonical +1 template position and newly incorporated rA10 is located at the pretranslocated state. The pyrophosphate group is shown as PPi. In state 4, rA10 is translocated to −1 position, whereas the hydantoin ring of Gh rotates about 90°, thereby occupying both canonical −1 and +1 template positions. Crystal structures after UTP soaking were solved in two successive incorporations of UTP (SI Appendix, Fig. S5E) (states 5 and 6). In state 5, the rU11 is located at the pretranslocate state, whereas Gh occupies between the canonical −1 and +1 template positions. (Right) Schematic representation of extension which starts from state 3. (B) Detailed analysis of binding environment of rotated Gh base in state 5. Possible hydrogen bonding is indicated with black dashes. Lone pair–π and CH–π interactions are indicated with red and blue dashes, respectively. To indicate the binding pocket of rotated Gh, surface is shown in gray.
To understand how Pol II translocates after ATP incorporation, we solved a new structure containing the Gh template and a synthetic 10A RNA primer (state 4). This structure captures Pol II in a new translocation intermediate state: the RNA chain is translocated (along with its base pair partners in upstream DNA template strand) 1 nt upstream and leaves the active site empty, but the downstream DNA template strands fail to translocate to cross over the bridge helix (Fig. 8, second panel; Movie S3). Intriguingly, we found that the nucleobase of Gh is rotated about 90° which is perpendicular to RNA:DNA base pair planes. As a result, the Gh occupies both canonical −1 and +1 sites (Fig. 8, second panel) and the downstream 5′ A template base of Gh failed to be loaded into the canonical +1 template position. Furthermore, Arg337 interacts with the 5′ phosphate group of the next base. Therefore, our structure was in a “half posttranslocation” state, in which RNA and most of the upstream tsDNA was translocated, but the +1 base was not loaded. These results suggest that the Gh:A base pair at the −1 position is unstable and tends to fall apart. Our structure provides a structural explanation of slow bypass of the Gh lesion site observed in the transcription assay (Fig. 1B).
To investigate whether addition of the correct UTP substrate can induce loading of the +1 base to the active site, we soaked Gh-10A crystals with 10 mM UTP (Fig. 8 and SI Appendix, Fig. S5E). Consistent with transcription assay results, we observed slow incorporation of UTP to the RNA strand (state 5). The newly added UMP is accommodated at the addition site, resulting in Pol II EC in a pretranslocation state. However, we did not observe full translocation of the 5′ A base to the +1 site that forms a Watson–Crick pair with UMP. Rather, the Gh base is still occupying both −1 and +1 sites because of the unstable nature of the Gh:A pair at the −1 position. One of the crystals soaked with UTP for prolonged incubation revealed a backtracked state with two consecutive UTP additions (SI Appendix, Fig. S5E) (state 6). The Gh lesions in states 5 and 6 adopt very similar unusual perpendicular conformations observed in state 4.
A Torsion Latch Transcription Stalling Mechanism through Lesion Reconfiguration within the Active Site via Noncanonical Interactions.
The unusual perpendicular conformation of Gh observed in states 4 through 6 was totally unexpected, as it needs to break a π–π interaction between the hydantoin ring and upstream base pairs as well as hydrogen bonds within Gh:A base pairs. The loss of these interactions must be energetically compensated by new chemical interactions.
To take a closer look at how this unusual perpendicular Gh base is stabilized in the active site, we examined the surrounding environment of the hydantoin ring of Gh. We identified several important interactions that stabilize this unusual perpendicular configuration of the Gh lesion from above, below, left, and right (Fig. 8B). These interactions include noncanonical lone pair–π interaction, CH–π interaction, as well as canonical hydrogen bondings.
From the above, we found that the C2 carbonyl group of the Gh hydantoin ring faces toward the center of the nucleobase of the upstream template base and forms a lone pair–π interaction with the upstream base (Fig. 8B and SI Appendix, Fig. S6). The lone pair–π interaction is defined as the interaction between a neutral electron-rich molecule and an electron-poor π ring (38, 39). This type of noncovalent bond was previously reported in a few nucleic acid structures, such as left-handed Z-DNA duplex, uridine-turn motifs of RNAs, as well as a RNA pseudoknot (38, 40–44). However, to the best of our knowledge, there is no previous report for the involvement of lone pair–π interactions in a protein–nucleic acid complex like transcription machinery (over 600 kDa) that is essential for biological function (such as lesion processing and transcriptional stalling). From the bottom of the Gh lesion, we found the hydrogen bonding between the C4 carbonyl group of the Gh hydantoin ring and conserved Thr831 (Rpb1) from the bridge helix motif further fix the hydantoin ring of Gh lesion. Thr831 is a strictly conserved residue in the bridge helix and has an important role for Pol II translocation (45–47).
From the left of the Gh lesion, we identified that the conserved Pro448 (Rpb1) forms CH–π interaction with the perpendicular hydantoin ring of Gh. The Pro448 is located in a motif consisting of a connection loop between the two β-sheets (β14 and β15) in the Pol II active-site domain (48, 49). Previously we termed the P448-containing loop (Rpb1 440 to 460) as the “Pro-gate” loop, because it faces toward the minor groove of a RNA:DNA hybrid and acts as a “steric gate” to prevent or slow down the translocation of DNA with minor groove alkylation (50). The work presented here reveals a distinct function of this conserved Pro residue. From the right of the Gh lesion, the oxygen of the 5′ phosphodiester backbone of Gh faces toward the hydantoin ring of Gh and may potentially form additional lone pair–π interactions from the opposite side of the hydantoin ring (Fig. 8B). The flexible positively charged guanidinium group of Gh may also contribute to additional interactions with the nearby negatively charged template phosphodiester backbone or Pol II active site residues. Taken together, these coordinated interactions between Gh and the Pol II active site help to reconfigure and stabilize the Gh lesion in this unusual perpendicular configuration and lead to transcriptional stalling.
It is important to note that the Gh lesion per se does not lead to this reconfiguration, rather the specific interactions with the Pol II active site promote this unusual perpendicular conformation of Gh. We didn’t observe this unusual perpendicular conformation in other scenarios such as Gh in duplex DNA or in the DNA polymerase active site from previous reports (36, 37). The environment of the Gh lesion within the Pol II active site is strikingly different from that in the DNA duplex or DNA polymerases’ active site; it is surrounded by an upstream RNA:DNA hybrid (A-form), bridge helix, and other active site residues (48, 51). The reconfiguration of the Gh lesion promoted by the Pol II active site functions as a “molecular torsion latch” to “lock in” RNA polymerase from moving forward.
Discussion
Impact of Guanine-Derived Oxidative Lesions on Pol II Transcription Elongation Dynamics and Fidelity.
Successive oxidation of guanine leads to several chemically related oxidative lesions, including 8OG, Gh, R-Sp, and S-Sp lesions. Here we systematically investigated the effect of these guanine-derived oxidative lesions (8OG, Gh, R-Sp, and S-Sp) on RNA Pol II transcription using structural analysis and biochemical assays. First, we showed that these four lesions lead to a wide spectrum of distinct transcription elongation dynamics. While Pol II is able to effectively bypass 8OG lesions, it becomes virtually completely arrested at R-Sp and S-Sp lesions. Gh leads to strong stalling and allows a small portion of slow bypass. Second, we showed that while all lesions are highly transcriptionally mutagenic, the molecular mechanisms of these four lesions in modulating transcriptional fidelity are lesion specific. Nucleotide preference is also significantly changed among the four lesions. 8OG allows fast incorporation of C and A, which is consistent with previous reports (21, 31). Strikingly, the presence of 8OG greatly increases the efficiency of error-prone ATP incorporation by 2,100-fold. The discrimination between error-free and error-prone extension from the lesion is greatly diminished for 8OG. We found both 8OG:A and 8OG:C can be extended similarly. Intriguingly, we also showed that Pol II fails to detect the mismatched 8OG:A base pair via its cleavage proofreading mechanism. This explains the molecular mechanism of why the 8OG lesion is highly transcriptionally mutagenic in vivo (52, 53). In contrast to 8OG, Gh and Sp prefer incorporation of purines (A and G) rather than C, which are similar to prior results from DNA polymerase and reverse transcriptase studies (19, 20).
For the Gh lesion, we observed slow extension products from a Gh:A pair, but not from a Gh:C pair in the absence of TFIIS. We observed strong TFIIS-stimulated cleavage of the Gh:A pair in the absence of NTPs, indicating that the Gh:A pair is prone to backtracking. The net effect of the Gh lesion on transcription depends on the relative rates of nucleotide incorporation (Pol II polymerase activity for transcript extension), as well as intrinsic and TFIIS-stimulated cleavage activity (proofreading and shortened transcripts). Given the slow ATP incorporation opposite the Gh lesion and strong TFIIS cleavage activity, we expected to observe strong transcriptional stalling in the presence of TFIIS. Indeed, we observed the ratio of stalling bands to run-off product is much higher in the presence of TFIIS (Figs. 1C and 2 and SI Appendix, Fig. S1). The strong Gh-induced stalling is also consistent with a previous report using HeLa nuclear extract (23). For both R-Sp and S-Sp lesions, ATP and GTP incorporation are preferred and CTP incorporation is greatly inhibited in the first step. Pol II experiences almost complete blockage after one more nucleotide incorporation from the Sp:A base pair. Strong TFIIS-stimulated cleavage of the Sp:A pair suggests that the Sp:A pair is also highly prone to backtracking.
We further compared the catalytic efficiency of ATP incorporation opposite G-derived oxidative lesions (SI Appendix, Table S1). ATP incorporation efficiency opposite 8OG is 2,100-fold higher than that of undamaged G, presumably due to a proposed Hoogsteen base pair between 8OG and A for ATP incorporation (21, 22, 31, 52, 54). For the Gh lesion, the efficiency of ATP incorporation is about 140-fold more efficient than that of undamaged G. This relatively high efficiency may be attributed to potential hydrogen bonding between Gh and ATP (Fig. 7).
Structural Basis of Gh Lesion-Induced Pol II Stalling.
The impact of DNA lesions on transcriptional outcomes can vary dramatically from full bypass, to pausing, to full arrest. Some of the bulky, helix-distorting lesions induce prolonged pausing/arrest of EC and trigger transcription-coupled nucleotide excision repair (TC-NER), whereas other DNA lesions can be bypassed by translesion RNA synthesis (50, 55, 56). Sometimes Pol II can slowly bypass the lesion site in a nontemplated, error-prone mode called the “A-rule” (30, 32, 35). This slow error-prone bypass prefers to incorporate ATP or GTP in a nontemplate-dependent manner. We and others previously revealed the structural basis of the A-rule guiding the transcriptional processing of several DNA lesions (30, 32, 35). Interestingly, we revealed a striking structural similarity between completely different sizes of oxidized lesions, abasic site, and cyclobutane pyrimidine dimer (CPD) (30, 35). Both abasic site and CPD lesions were accommodated at a half-way translocation intermediate state above the bridge helix, and the 5′ phosphate group interacted with R337 of Rpb1 (Fig. 6 B and C). Intriguingly, our encounter structure containing the Gh lesion (state 1*) also shows similar features. The 5′ phosphate group of the Gh lesion interacted with R337, and this interaction remained in the ATP-incorporated pretranslocation state.
Our structures (states 2 and 3) provided insights into ATP binding and incorporation opposite the Gh lesion. First, we revealed the ATP substrate (AMPCPP) prefers to bind to the entry site (state 2E) rather than the addition site (before chemistry). In contrast, overnight soaking with ATP allowed the incorporation of AMP opposite the Gh lesion, suggesting that ATP is able to convert from the E-site to the A-site to allow a nucleotide addition reaction (state 3). We found that Gh can form hydrogen bonds with incorporated AMP in RNA but not free substrate AMPCPP, suggesting while the hydrogen bonding between Gh and AMP may facilitate ATP incorporation, it is not sufficient to keep ATP in the addition site before chemistry (Fig. 7B). We also successfully solved another crystal structure containing the Gh lesion with 10mer RNA in the posttranslocation state (state 4; Fig. 8, second panel). Taken together, we were able to summarize the snapshots of ATP incorporation opposite the Gh lesion (Fig. 7A). The incoming nucleoside triphosphate first binds the entry site (E-site) when it enters into the active site of Pol II (state 2E). Subsequently, it rotates toward the addition site and base pairs with template strand DNA (state 2A), leading to nucleotide incorporation (state 3) (51, 57, 58). Our soaking structure of AMPCPP suggests that although ATP is the most favorable substrate for the Gh lesion, it occupies the E-site predominantly (state 2E). This result is consistent with the apparent weak binding affinity of ATP measured by kinetic analysis. New structures also suggest that ATP incorporation opposite the Gh lesion is partially template dependent, which explains why the efficiency of ATP incorporation opposite the Gh lesion is about 30-fold higher than that of ATP incorporation opposite an abasic site (template independent).
We unraveled a distinct mechanism of nonbulky lesion-induced Pol II stalling. The most striking finding in this work is that we revealed a unique conformational change of the Gh lesion promoted by the Pol II active site. We found that the hydantoin ring of the Gh lesion rotates about 90° which is perpendicular to the base pair planes. As a result, Gh occupies two neighboring template positions (both −1 and +1 template positions) in the active site and causes the failure of loading of downstream template DNA into +1 template positions (Fig. 8). We suggest that this unusual 90° rotated base is the major reason for prolonged stalling and slow extension. Our crystallographic studies offer a structural explanation of a long-standing puzzle in the field, why nonbulky lesions like Gh cause strong transcriptional blockage similar to 1,2-intrastrand crosslinks such as CPD and cisplatin-DNA adduct. It is striking to find that the five-membered hydantoin ring is rotated 90° so that it is perpendicular to the base pair planes and therefore occupies two neighboring template positions, functioning like a 1,2-intrastrand crosslink, such as CPD (SI Appendix, Fig. S7).
Structural Insights into Sp and Other Hydantoin Lesions Induced Stalling.
The Sp lesions share the same five-membered hydantoin ring (A-ring) with Gh lesions; Sp lesions are much more rigid than the Gh lesion because the Sp lesions have two rigid five-membered rings essentially perpendicular to one another, whereas the Gh lesion has a flexible guanidinium group. To gain insights as to whether Sp lesions can be accommodated in the same way as the Gh lesion, we generated models of Sp lesions in both state 1 and state 1* for the encounter step. As shown in SI Appendix, Fig. S8A, both R-Sp and S-Sp lesions are able to be accommodated at the half-way point of translocation right above the bridge helix without any apparent steric clash (state 1*). The B-ring of R-Sp is facing toward the bridge helix, whereas the B-ring of S-Sp is facing away from the bridge helix. Given that Sp lesions are more rigid than the Gh lesion, we expected the crossover of the bridge helix would be much more difficult for Sp lesions.
To understand why nucleotide insertion opposite Sp lesions is much slower than that for the Gh lesion, we prepared models of Sp lesions in state 1 in which the template is fully loaded at the +1 position for the incoming NTP. In contrast to state 1*, we observe severe steric clashes in the template +1 position (state 1). We found that the B-ring of R-Sp clashes with the bridge helix backbone, whereas the B-ring of S-Sp clashes with the upstream base pair. We also modeled Sp lesions in state 3 to evaluate whether Sp lesions are able to form hydrogen bonds with an incoming ATP (SI Appendix, Fig. S8B). Similar to what we observed in state 1, there are severe steric clashes for both Sp lesions. These results suggest that both R-Sp and S-Sp lesions are much more difficult to cross over the bridge helix and become fully loaded at the template +1 position to support template-dependent nucleotide incorporation compared to the Gh lesion. Rather, Sp lesions likely stay in the half-way translocation state 1*, and ATP is incorporated in a nontemplate-dependent manner (A-rule). These models offer a structural explanation for why ATP incorporation efficiency for Sp lesions is similar to that of the abasic lesion, which is much slower than that of the Gh lesion.
As shown in Fig. 1, Sp lesions are much stronger blocks for Pol II transcription giving very few extension products beyond the insertion step. To gain structural insights as to why this is the case, we modeled the Pol II complex with Sp lesions in postinsertion states. As shown in SI Appendix, Fig. S9, both Sp lesions can form similar lone pair–π interactions observed for the Gh lesion using the common carbonyl group in the hydantoin ring. As a result, the hydantoin ring (A-ring) of Sp lesions is perpendicular to the base pair planes and consequently occupies both canonical −1 and +1 template positions. Our model prepared by superimposing the Sp lesions with N4 and the carbonyl group of the n-1 position of cytosine in the nondamaged structure (dG, Fig. 6B) further supports this prediction. The properly loaded state of Gh and Sp lesions clash with the template strand except for R-Gh (SI Appendix, Fig. S9B). The B-ring faces the major groove toward the bridge helix and serves as a stronger blockage than the flexible guanidinium group of the Gh lesion, preventing the 5′ downstream template nucleobase to cross over the bridge helix and load. In addition, we also noticed that the imine B-ring of R-Sp can form potential hydrogen bond with the 5′ phosphodiester backbone, whereas S-Sp cannot.
Hydantoin DNA lesions can be derived not only from purines, but also from pyrimidines. It was reported that hydroxyl radical oxidation of pyrimidines (thymine, cytosine, and the epigenetic marker 5-methylcytosine [5mC]) also can produce hydantoin lesions, namely, Hyd-T, Hyd-C, and Hyd-5mC, respectively (59). It is noteworthy that the major oxidation products of 5mC from hydroxy radical are 5-formylcytosine and Hyd-5mC.
To gain structural insights into hydantoin ring-induced Pol II stalling, we aligned the hydantoin ring of Gh in our Gh-stalled Pol II structures to the Sp A-ring (PDB: 4PPX), three hydantoin ring-containing lesions, Hyd-T, Hyd-C, and Hyd-5mC (59, 60). To this end, we generated stepwise models for Pol II complexes containing various hydantoin lesions to understand the encounter (lesion loading), insertion, extension, and bypass steps, respectively (SI Appendix, Figs. S8–S10). The mechanism of Pol II stalling reported herein differs from those previously reported stalling mechanisms by other DNA lesions and is likely to represent a distinct stalling mechanism shared by several ring-modified DNA lesions. Further studies to investigate whether these pyrimidine derived lesions also form the torsion latch would be interesting.
Transcription-Coupled Repair of G-Derived Oxidative Lesions.
Previous reports suggest that the Gh and Sp lesions can be targeted by both base excision repair and global-genome nucleotide excision repair in human cells as well as in cell extracts (13, 29, 33, 61–67). However, it is unknown whether these lesions can trigger the specific transcription-coupled repair pathways if located within transcribing strands of gene bodies. Current TC-NER models suggest that the prolonged arrested state of EC is the prerequisite of TC-NER (5, 7). Small lesions or naturally occurring pausing sequences can be bypassed in the presence of CSB by pushing EC forward. However, bulky, helix-distorting lesions such as CPD, cannot be overcome by CSB. This will lead to prolonged arrest of EC, which becomes a strong signal to initiate TC-NER (5, 7, 26).
Our results further support the possibility that Gh and Sp lesions can be processed by TC-NER. First, by combining transcriptional assays and structural analysis of Gh and Sp lesions, we showed that Gh, R-Sp, and S-Sp lesions severely block transcription, while a small portion of Gh can be slowly bypassed with misincorporation. Second, using an RNA transcript cleavage assay, we showed that RNA transcripts from Gh- or Sp-containing scaffolds were efficiently cleaved, regardless of the 3′ base pairing opposite the lesion (Fig. 5). Furthermore, a transcription assay using a full-bubble scaffold showed that the addition of Rad26 or TFIIS fails to rescue the stalled EC for Gh, R-Sp, and S-Sp lesions (Fig. 2). Our analysis provides a further structural basis for prolonged stalling induced by Gh lesions (Fig. 8). Taken together, we suggest that Gh and Sp lesions can trigger Pol II stalling and subsequent transcription-coupled repair pathways. It would be of great interest to test this with in vivo studies in the future.
Materials and Methods
Detailed information about damaged oligonucleotides synthesis, protein purification, transcription assay, crystallization, and structural determination and modeling is shown in SI Appendix.
Data Availability.
All atomic coordinates and structure factors are deposited at Protein Data Bank (PDB) (PDB accession codes: 6UPX, 6UPY, 6UPZ, 6UQ0, 6UQ1, 6UQ2, and 6UQ3). All other detailed information about methods and materials are deposited as SI Appendix. All of the model coordinates (Figs. S8–S10) are deposited as Datasets S1–15. All of the movies are deposited as Movies S1–S3.
Supplementary Material
Acknowledgments
We appreciate the beamline staff for their kind support during our remote data collection. This work was supported by NIH Grants R01 GM102362 (to D.W.) and R01 CA090689 (to C.J.B). This research used resources of the Advanced Light Source (beamlines 5.0.3 and 8.2.2), which is a Department of Energy Office of Science User Facility under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All atomic coordinates and structure factors are deposited at Protein Data Bank (PDB) (PDB accession codes: 6UPX, 6UPY, 6UPZ, 6UQ0, 6UQ1, 6UQ2, and 6UQ3).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919904117/-/DCSupplemental.
References
- 1.Bjelland S., Seeberg E., Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat. Res. 531, 37–80 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Lagerwerf S., Vrouwe M. G., Overmeer R. M., Fousteri M. I., Mullenders L. H., DNA damage response and transcription. DNA Repair (Amst.) 10, 743–750 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Ciccia A., Elledge S. J., The DNA damage response: Making it safe to play with knives. Mol. Cell 40, 179–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoeijmakers J. H., DNA damage, aging, and cancer. N. Engl. J. Med. 361, 1475–1485 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Hanawalt P. C., Spivak G., Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Svejstrup J. Q., Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem. Sci. 32, 165–171 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Spivak G., Transcription-coupled repair: An update. Arch. Toxicol. 90, 2583–2594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein C., Bernstein H., Payne C. M., Garewal H., DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: Fail-safe protection against carcinogenesis. Mutat. Res. 511, 145–178 (2002). [DOI] [PubMed] [Google Scholar]
- 9.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swenberg J. A., et al. , Endogenous versus exogenous DNA adducts: Their role in carcinogenesis, epidemiology, and risk assessment. Toxicol. Sci. 120 (suppl. 1), S130–S145 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steenken S., Jovanovic S. V., Bietti M., Bernhard K., The trap depth (in DNA) of 8-Oxo-7,8-dihydro-2‘deoxyguanosine as derived from electron-transfer equilibria in aqueous solution. J. Am. Chem. Soc. 122, 2373–2374 (2000). [Google Scholar]
- 12.Cadet J., Davies K. J. A., Medeiros M. H., Di Mascio P., Wagner J. R., Formation and repair of oxidatively generated damage in cellular DNA. Free Radic. Biol. Med. 107, 13–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKibbin P. L., et al. , Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair. J. Am. Chem. Soc. 135, 13851–13861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming A. M., Burrows C. J., Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic. Biol. Med. 107, 35–52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleming A. M., et al. , Reconciliation of chemical, enzymatic, spectroscopic and computational data to assign the absolute configuration of the DNA base lesion spiroiminodihydantoin. J. Am. Chem. Soc. 135, 18191–18204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye Y., et al. , Formation of 13C-, 15N-, and 18O-labeled guanidinohydantoin from guanosine oxidation with singlet oxygen. Implications for structure and mechanism. J. Am. Chem. Soc. 125, 13926–13927 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Mangerich A., et al. , Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. U.S.A. 109, E1820–E1829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson P. T., et al. , The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry 42, 9257–9262 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Alenko A., Fleming A. M., Burrows C. J., Reverse transcription past products of guanine oxidation in RNA leads to insertion of A and C opposite 8-Oxo-7,8-dihydroguanine and A and G opposite 5-guanidinohydantoin and spiroiminodihydantoin diastereomers. Biochemistry 56, 5053–5064 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aller P., Ye Y., Wallace S. S., Burrows C. J., Doublié S., Crystal structure of a replicative DNA polymerase bound to the oxidized guanine lesion guanidinohydantoin. Biochemistry 49, 2502–2509 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damsma G. E., Cramer P., Molecular basis of transcriptional mutagenesis at 8-oxoguanine. J. Biol. Chem. 284, 31658–31663 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tornaletti S., Maeda L. S., Kolodner R. D., Hanawalt P. C., Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst.) 3, 483–494 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Kolbanovskiy M., et al. , The nonbulky DNA lesions spiroiminodihydantoin and 5-guanidinohydantoin significantly block human RNA polymerase II elongation in vitro. Biochemistry 56, 3008–3018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kettenberger H., Armache K. J., Cramer P., Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114, 347–357 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Xu L., Plouffe S. W., Chong J., Wengel J., Wang D., A chemical perspective on transcriptional fidelity: Dominant contributions of sugar integrity revealed by unlocked nucleic acids. Angew. Chem. Int. Ed. Engl. 52, 12341–12345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J., et al. , Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature 551, 653–657 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selby C. P., Sancar A., Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 94, 11205–11209 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellinger M. W., et al. , 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 19, 831–833 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shafirovich V., Kropachev K., Kolbanovskiy M., Geacintov N. E., Excision of oxidatively generated guanine lesions by competing base and nucleotide excision repair mechanisms in human cells. Chem. Res. Toxicol. 32, 753–761 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W., Walmacq C., Chong J., Kashlev M., Wang D., Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 115, E2538–E2545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konovalov K. A., et al. , 8-Oxo-guanine DNA damage induces transcription errors by escaping two distinct fidelity control checkpoints of RNA polymerase II. J. Biol. Chem. 294, 4924–4933 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walmacq C., et al. , Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions. Proc. Natl. Acad. Sci. U.S.A. 112, E410–E419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnamurthy N., Zhao X., Burrows C. J., David S. S., Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry 47, 7137–7146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X., Fleming A. M., Muller J. G., Burrows C. J., Endonuclease and exonuclease activities on oligodeoxynucleotides containing spiroiminodihydantoin depend on the sequence context and the lesion stereochemistry. New J. Chem. 37, 3440–3449 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walmacq C., et al. , Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol. Cell 46, 18–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheung A. C., Sainsbury S., Cramer P., Structural basis of initial RNA polymerase II transcription. EMBO J. 30, 4755–4763 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beckman J., Wang M., Blaha G., Wang J., Konigsberg W. H., Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP and dGMP to be inserted opposite Guanidinohydantoin. Biochemistry 49, 8554–8563 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egli M., Sarkhel S., Lone pair-aromatic interactions: To stabilize or not to stabilize. Acc. Chem. Res. 40, 197–205 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Mooibroek T. J., Gamez P., Reedijk J., Lone pair–π interactions: A new supramolecular bond? J CrystEngComm 10, 1501–1515 (2008). [Google Scholar]
- 40.Egli M., Gessner R. V., Stereoelectronic effects of deoxyribose O4′ on DNA conformation. Proc. Natl. Acad. Sci. U.S.A. 92, 180–184 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi H., Moore P. B., The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: A classic structure revisited. RNA 6, 1091–1105 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bancroft D., Williams L. D., Rich A., Egli M., The low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove. Biochemistry 33, 1073–1086 (1994). [DOI] [PubMed] [Google Scholar]
- 43.Sarkhel S., Rich A., Egli M., Water-nucleobase “stacking”: H-pi and lone pair-pi interactions in the atomic resolution crystal structure of an RNA pseudoknot. J. Am. Chem. Soc. 125, 8998–8999 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Singh S. K., Das A., The n → π* interaction: A rapidly emerging non-covalent interaction. Phys. Chem. Chem. Phys. 17, 9596–9612 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Silva D. A., et al. , Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 111, 7665–7670 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Da L. T., et al. , Bridge helix bending promotes RNA polymerase II backtracking through a critical and conserved threonine residue. Nat. Commun. 7, 11244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ka Man Tse C., et al. , Intrinsic cleavage of RNA polymerase II adopts a nucleobase-independent mechanism assisted by transcript phosphate. Nat. Energy 2, 228–235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gnatt A. L., Cramer P., Fu J., Bushnell D. A., Kornberg R. D., Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science 292, 1876–1882 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Cramer P., Bushnell D. A., Kornberg R. D., Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292, 1863–1876 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Xu L., et al. , Mechanism of DNA alkylation-induced transcriptional stalling, lesion bypass, and mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 114, E7082–E7091 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D., Bushnell D. A., Westover K. D., Kaplan C. D., Kornberg R. D., Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell 127, 941–954 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saxowsky T. T., Meadows K. L., Klungland A., Doetsch P. W., 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 105, 18877–18882 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brégeon D., Peignon P. A., Sarasin A., Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 5, e1000577 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beard W. A., Batra V. K., Wilson S. H., DNA polymerase structure-based insight on the mutagenic properties of 8-oxoguanine. Mutat. Res. 703, 18–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang D., Zhu G., Huang X., Lippard S. J., X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct. Proc. Natl. Acad. Sci. U.S.A. 107, 9584–9589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kellinger M. W., Park G. Y., Chong J., Lippard S. J., Wang D., Effect of a monofunctional phenanthriplatin-DNA adduct on RNA polymerase II transcriptional fidelity and translesion synthesis. J. Am. Chem. Soc. 135, 13054–13061 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westover K. D., Bushnell D. A., Kornberg R. D., Structural basis of transcription: Nucleotide selection by rotation in the RNA polymerase II active center. Cell 119, 481–489 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Kaplan C. D., Larsson K. M., Kornberg R. D., The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol. Cell 30, 547–556 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Madugundu G. S., Cadet J., Wagner J. R., Hydroxyl-radical-induced oxidation of 5-methylcytosine in isolated and cellular DNA. Nucleic Acids Res. 42, 7450–7460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eckenroth B. E., Fleming A. M., Sweasy J. B., Burrows C. J., Doublié S., Crystal structure of DNA polymerase β with DNA containing the base lesion spiroiminodihydantoin in a templating position. Biochemistry 53, 2075–2077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shafirovich V., et al. , Base and nucleotide excision repair of oxidatively generated guanine lesions in DNA. J. Biol. Chem. 291, 5309–5319 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim N., Jinks-Robertson S., Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 30, 3206–3215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Torres-Ramos C. A., Johnson R. E., Prakash L., Prakash S., Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell. Biol. 20, 3522–3528 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leipold M. D., Muller J. G., Burrows C. J., David S. S., Removal of hydantoin products of 8-oxoguanine oxidation by the Escherichia coli DNA repair enzyme, FPG. Biochemistry 39, 14984–14992 (2000). [DOI] [PubMed] [Google Scholar]
- 65.Hazra T. K., et al. , Repair of hydantoins, one electron oxidation product of 8-oxoguanine, by DNA glycosylases of Escherichia coli. Nucleic Acids Res. 29, 1967–1974 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hailer M. K., Slade P. G., Martin B. D., Rosenquist T. A., Sugden K. D., Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst.) 4, 41–50 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Zhao X., Krishnamurthy N., Burrows C. J., David S. S., Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry 49, 1658–1666 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All atomic coordinates and structure factors are deposited at Protein Data Bank (PDB) (PDB accession codes: 6UPX, 6UPY, 6UPZ, 6UQ0, 6UQ1, 6UQ2, and 6UQ3). All other detailed information about methods and materials are deposited as SI Appendix. All of the model coordinates (Figs. S8–S10) are deposited as Datasets S1–15. All of the movies are deposited as Movies S1–S3.