Significance
Our studies demonstrate that Vip1 represents a rare class of bifunctional enzyme capable of synthesizing and destroying signaling molecules important for nutrient adaptation, cellular architecture, and organelle morphology. We find that Vip1 contains two tethered autonomous catalytic active sites, which modulate levels of 1-IP7 and 1,5-IP8 through 1-kinase and 1-pyrophosphatase domains. Each activity is critical for maintaining the highly dynamic anabolic and catabolic regulation of cellular pools of IP7 and IP8. That this occurs through a single gene product emphasizes that Vip1 is a key metabolic switch critical for cellular adaptation.
Keywords: inositol pyrophosphate, inositol phosphate, phosphatase, kinase, cell polarity
Abstract
Inositol diphosphates (PP-IPs), also known as inositol pyrophosphates, are high-energy cellular signaling codes involved in nutrient and regulatory responses. We report that the evolutionarily conserved gene product, Vip1, possesses autonomous kinase and pyrophosphatase domains capable of synthesis and destruction of D-1 PP-IPs. Our studies provide atomic-resolution structures of the PP-IP products and unequivocally define that the Vip1 gene product is a highly selective 1-kinase and 1-pyrophosphatase enzyme whose activities arise through distinct active sites. Kinetic analyses of kinase and pyrophosphatase parameters are consistent with Vip1 evolving to modulate levels of 1-IP7 and 1,5-IP8. Individual perturbations in kinase and pyrophosphatase activities in cells result in differential effects on vacuolar morphology and osmotic responses. Analogous to the dual-functional key energy metabolism regulator, phosphofructokinase 2, Vip1 is a kinase and pyrophosphatase switch whose 1-PP-IP products play an important role in a cellular adaptation.
Inositol pyrophosphates, also known as diphosphoryl inositol phosphates, are high-energy cellular messengers whose synthesis is required for a number of cellular processes ranging from phosphate sensing response to DNA metabolism (1–4). Along with inositol phosphates, they represent an ensemble of chemical codes required for a plethora of intracellular processes, including the following: calcium release; mRNA export; gene expression/transcription; telomere maintenance/DNA repair; embryogenesis; vesicular trafficking; stress responses: hypoxia, osmotic stress, and oxidative stress; translational control; RNA editing; cell morphology; and phosphate/cell cycle signaling.
Synthesis of inositol pyrophosphates occurs through two classes of evolutionarily conserved kinases. Class I gene products function to generate a D-5 position diphosphate and were identified as Kcs1 in yeast and IP6Ks in metazoans (5, 6). Class II gene products generate a D-1/3 diphosphate and were first cloned in yeast as Vip1 (also known as spAsp1 in Schizosaccharomyces pombe fission yeast) (7) and subsequently in metazoans as VIP1 and VIP2 (also known as PPIP5K1, 2, and in plants as VIH1, 2) (8, 9). The positional specificities of these two classes of enzyme generate inositol pyrophosphate molecules that include the conversion of IP6 to 1-IP7, 5-IP7, and 1,5-IP8 species. Destruction of inositol pyrophosphates is accomplished through the diphosphoryl inositol polyphosphate phosphatase DIPP class of pyrophosphatase (10, 11). Of note: The DIPP enzymes are more promiscuous and hydrolyze nucleotide dimers as well as polyphosphate molecules (12).
Analysis of the domain structure of the Vip1 class of enzymes reveals an evolutionarily conserved architecture with two distinct domains: an amino-terminal rimK/ATP GRASP fold and a histidine acid-phosphatase (HAP) or phytase-like domain (7–9). The dual-domain structure is conserved from yeast to mammals, with protein sequence alignments revealing conservation of key catalytic residues in the kinase domain; however, the phosphatase domain has several anomalies compared to the histidine motifs in the acid-phosphatase class of enzymes (8). In addition to the strict evolutionary conservation of this domain, phenotypic analysis of S. pombe mutants lacking the Asp1 suggested the existence of catalytic phosphatase activity (7, 13–15), and subsequently, bifunctionality was reported (16).
Our work, along with that of several other laboratories, provides evidence that the Vip1 class of enzymes is an evolutionarily conserved dual-functional protein with both kinase and pyrophosphatase activities that act to control levels of 1-IP7 and 1,5-IP8 in cells. Remarkably, distinct active sites in the kinase and pyrophosphatase domains are tethered by a linker region and encode the exquisite selectivity for addition and removal of the β-phosphate at the D-1 position. In addition to previously reported phenotypes, we find loss of either enzymatic function is important for cellular architecture and vacuolar morphology in fission yeast S. pombe. Vip1 represents an extremely rare class of bifunctional enzyme that is capable of synthesizing and destroying a signaling molecule. Ample evidence is accumulating that reinforces that the coordinated regulation of kinase and pyrophosphatase activities is critical for maintaining the highly dynamic production and breakdown of cellular pools of IP7 and IP8. That this occurs through a single gene product emphasizes that Vip1 is a key metabolic switch for inositol 1-pyrophosphate–mediated cellular signaling and adaptation.
Results
Note.
The vast majority of the work presented in this manuscript was first reported at international and Howard Hughes Medical Institute scientific meetings as well as universities beginning in the fall of 2009. While it was not published in a timely manner, which the senior author greatly regrets, at the suggestion of several colleagues in the field, the manuscript is presented here in an unorthodox retrospective manner. This is in no way meant to belittle or ignore the current state of the field, including published work of others; rather it is written in the historical context of our work in 2008 and 2009. The exceptions are as follows: 1) finalized enzymological parameters performed in February 2012, and 2) statistical analysis/quantification of the vacuole diameter, which was performed in 2019 by Kavi Mehta at Vanderbilt (Nashville, TN) on images generated in 2009.
The Vip1 Family of Kinases Phosphorylate the D-1 Phosphate of IP6.
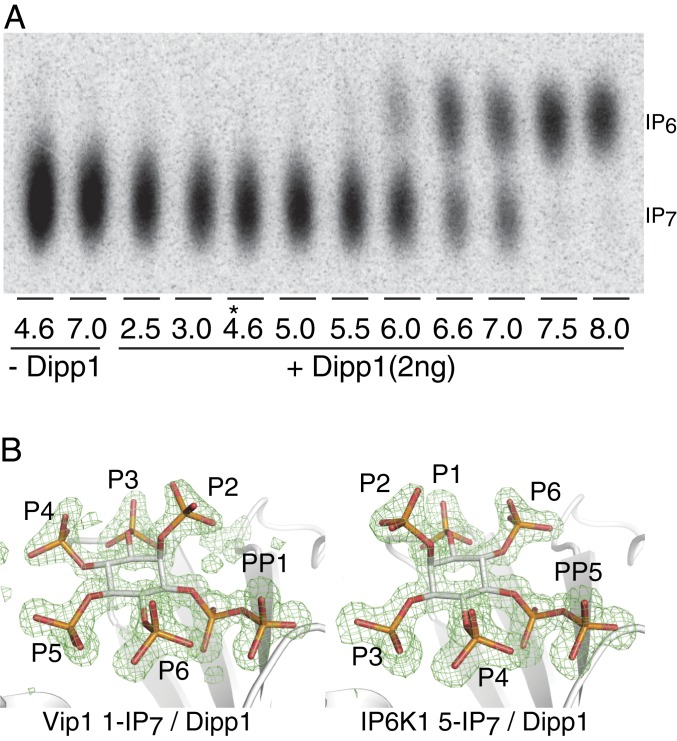
Previous studies using NMR methods aimed at identification of the product(s) of the Saccharomyces cerevisiae Vip1 (scVip1) kinase determined that the enzyme phosphorylated either the 1- and/or 3-phosphate positions (17). However, due to the 2/5 symmetry axis of the inositol ring, these studies could not differentiate between the 1- or 3- stereoisomer. We therefore initiated a structural approach in the context of a protein cocrystal to resolve chirality and isomer ambiguities. The cocrystal structures of inositol pyrophosphatase Dipp1 in complex with the IP7 species produced by scVip1 and hIP6K1 were determined at near atomic 1.2-Å resolution (Table 1 and Fig. 1). Since these IP7 species are substrates for Dipp1, low pH conditions for crystallization were used in which binding would still occur but that Dipp1 active was greatly diminished (Fig. 1A). Unambiguous electron density was found near the active site of DIPP in the scVip1 IP7 cocrystal, which clearly revealed a 1-IP7 molecule with no evidence of any other species or conformations (Fig. 1B). The high-resolution structure of the IP6K1 IP7 product/DIPP complex confirms the stereomer as unambiguously 5-IP7 (Fig. 1B). Both structures clearly define the axial D-2 phosphate and the relative position of the pyrophosphate. Our data demonstrate that yeast Vip1 harbors 1-kinase activity and confirms human IP6K1 is indeed a distinct 5-kinase enzyme.
Table 1.
Crystallographic data collection and refinement statistics for Dipp1/IP7 complexes
| Diffraction data | Dipp1/1-IP7 complex | Dipp1/5-IP7 complex |
| PDB ID code | 6PCK | 6PCL |
| Space group | P212121 | P212121 |
| a,b,c, Å | 46.41, 59.61, 62.56 | 45.46, 59.58, 62.36 |
| Wavelength, Å | 1.0000 | 1.0000 |
| Resolution limit, Å* | 1.20 | 1.30 |
| Unique reflections | 51,322 | 40,069 |
| Completeness, %, (last shell) | 93.9 (68.1) | 93.7 (65.2) |
| Average I/σI (last shell) | 70.7 (5.6) | 58.2 (3.5) |
| Redundancy (last shell) | 12.7 (8.4) | 12.5 (4.9) |
| Rsym,† % (last shell) | 6.1 (37.5) | 6.0 (36.5) |
| Crystallographic refinement | ||
| Resolution range, Å | 27.4–1.2 | 27.6–1.3 |
| Reflections | 47,532 | 38,468 |
| Rms deviation from ideality | ||
| Bond lengths, Å | 0.022 | 0.011 |
| Bond angles, ° | 1.898 | 1.374 |
| Rotamer outliers | 0.86% | 0.85% |
| Ramachandran | ||
| Outliers | 0.0% | 0.0% |
| Allowed | 100% | 100% |
| Favored | 100% | 100% |
| R value,‡ % | 15.7 | 14.3 |
| Rfree, % | 17.3 | 16.1 |
Resolution limit was defined as the highest-resolution shell where the average I/σI was >1.2 and Rsym <50%.
Rsym = ∑hkl∑i∣Ii(hkl) − <I(hkl)>∣/∑hkl∑II(hkl).
R = ∑∣Fo − Fc∣/∑Fo. Five percent of reflections were used to calculate Rfree.
Fig. 1.
Identification of the product of scVIP1 as 1-IP7 through structure analysis of a cocrystal with hDipp1. (A) pH dependence of the Dipp1 1-IP7 pyrophosphatase reaction. The 7.5 µM Vip1 IP7 was incubated with 2 ng of hDipp1 at varying pH values, for 20 min, at 37 °C. At pH 4.6 used for crystallization of the Dipp1/IP7 complexes (noted by an asterisk [*]), no Dipp1 phosphatase activity was observed. (B) Structural determination of Vip1 and IP6K IP7 products at near 1.2-Å resolution. Vip1 product may be unambiguously defined as D-1-IP7 as the protein complex has left-handed α-helices and the pyrophosphate density is clearly one-carbon clockwise of the D-2 axial phosphate. The product of the IP6K product is defined as D-5-IP7. Difference density contoured at 3σ (shown in green) represents Fo − Fc difference Fourier maps using phases calculated from the final model with the ligand omitted (composite omit).
Vip1 Is Evolutionarily Conserved Pyrophosphatase Selective for the D-1 Position.
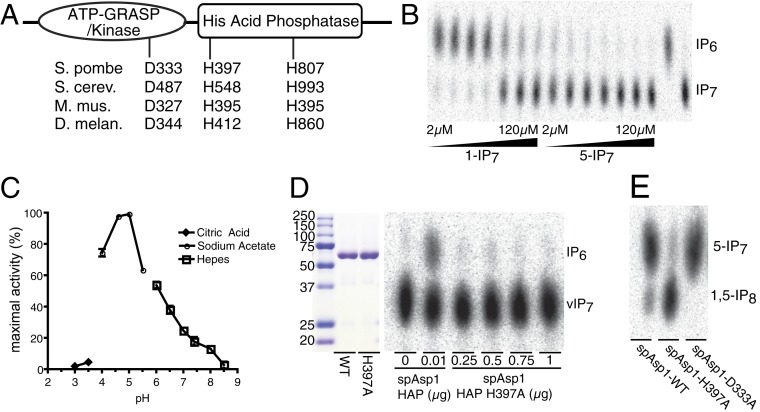
The two-domain topology of the Vip1 class of enzymes is evolutionarily conserved and has a consistent topology of an N-terminal kinase domain of ∼350 amino acids followed by a HAP-like domain of ∼450 amino acids (Fig. 2A). Among the HAP family members are the phytase cluster of phosphomonoesterase enzymes that hydrolyze IP6. Initially, we tested a variety of species of the Vip1 class of HAP domains for activity toward IP6 without success. We next tested S. pombe spAsp1 HAP domain (spAsp1-HAP, residues 377 to 920) for in vitro activity against a collection of additional inositol phosphates and pyrophosphates. Pyrophosphatase activity was detected that converted IP7 to IP6, and titration of IP7 species against spAsp1-HAP indicated that the enzyme showed strong selectivity for the 1-IP7 isomer produced by spAsp1 over the 5-IP7 isomer produced by IP6K (Fig. 2B). No phosphomonoesterase activity was observed toward other IPs, including IP6, indicating that the pyrophosphatase activity of spAsp1 is specific for cleaving the disphosphate bond; thus, we refer to it as a 1-pyrophosphatase (E.C. 3.6.1). Maximal pyrophosphatase activity was observed at pH 5.0 and nearly 30% activity was observed at physiological pH of 6.8 (Fig. 2C). To demonstrate that the 1-pyrophosphatase activity was not due to contamination, a catalytic point mutant (spAsp1-HAP-H397A) was cloned, expressed, and purified through the same procedure as the wild-type spAsp1 pyrophosphatase domain (Fig. 2D), and evaluated for the ability to hydrolyze inositol diphosphate (PP-IP) substrates. While the wild-type spAsp1-HAP hydrolyzed 1-IP7 to produce IP6, no pyrophosphatase activity was detected even up to a 500-fold excess of the mutant enzyme (Fig. 2D).
Fig. 2.
spAsp1 is a 1-pyrophosphatase. (A) Schematic of the Vip1 class of dual ATP-GRASP kinase and histidine acid phosphatase domains. Conserved catalytic residues, including aspartic acid and histidine residues in the kinase and His acid phosphatase domains, are noted. (B) spAsp1 acts as a selective 1-IP7 pyrophosphatase. Reactions were performed with 50 ng of spAsp1-HAP at range of substrate from 2 to 120 µM for Vip1 produced 1-IP7 and IP6K1 produced 5-IP7. (C) pH dependence of Asp1-HAP. Twenty micromolar 1-IP7 was incubated with 25 ng of Asp1-HAP at varying pH values. Each reaction buffer was 100 mM of the indicated buffer, pH = X, 50 mM NaCl. Reaction performed for 15 min at 37 °C. (D) Asp1 pyrophosphatase activity is dependent on a conserved histidine residue. (Left) Coomassie-stained sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) gel of 2 µg of each of spAsp1-HAP and spAsp1-HAP-H397A along with molecular weight standards (shown in kilodaltons). (Right) Pyrophosphatase assay using fixed substrate at 1.25 µM, varying the dose of spAsp1-HAP and spAsp1-HAP-H397A. Reaction was performed at pH 5.0 for 15 min at 37 °C. (E) spAsp1 in a 1,5-IP8 1-pyrophosphatase. Recombinant full-length spAsp1 wild-type or kinase-dead D333A proteins were incubated with radiolabeled 1,5-IP8 and robust conversion to 5-IP7 was observed compared to an H397A catalytic dead mutant.
To further characterize the spAsp1 1-pyrophosphatase activity, the kinetic parameters for the hydrolysis of 1-IP7, 5-IP7, and 1,5-IP8 were determined (Table 2). The spAsp1-HAP has a ∼12-fold higher turnover (Kcat) of 1-IP7 than for 5-IP7 with a ∼8-fold difference in catalytic efficiency (Kcat/Km). Similar differences in turnover and catalytic efficiency are seen when comparing the spAsp1-HAP pyrophosphatase activity toward 1,5-IP8 to that of 5-IP7. Additionally, we tested full-length recombinant spAsp1, spAsp1 H397A (HAP pyrophosphatase dead), and spAsp1-D333A (kinase dead mutation) protein for activity toward 1,5-IP8 (Fig. 2E). We observed robust 1-pyrophosphatase activity toward 1,5-IP8 substrate that was dependent on functional histidine catalytic residue (H397).
Table 2.
Kinetic parameters of Asp1/Vip1 pyrophosphatase activity
| Species | Construct | Substrate | Km, µM | Vmax, nmol/min/mg | kcat, s−1 | kcat/Km, 103 M−1⋅s−1 |
| S. pombe | Asp1-HAP | 1-IP7 | 17.4 | 227 | 0.24 | 13.5 |
| S. pombe | Asp1-HAP | 5-IP7 | 11.4 | 18.3 | 0.02 | 1.7 |
| S. pombe | Asp1-HAP | 1,5-IP8 | 14.0 | 123 | 0.13 | 9.1 |
| S. pombe | Asp1-FL-D333A | 1-IP7 | 15.0 | 93.2 | 0.16 | 10.9 |
| S. cerevisiae | Vip1-HAP | 1-IP7 | 5.0 | 106 | 0.12 | 23.4 |
| M. musculus | Vip2-FL | 1-IP7 | 5.9 | 51.2 | 0.11 | 18.6 |
| M. musculus | Vip2-FL | 5-IP7 | 10.8 | 7.7 | 0.017 | 1.6 |
| M. musculus | Vip2-FL | 1,5-IP8 | 6.8 | 44.2 | 0.095 | 14.0 |
Kinetic constants were determined by fitting enzyme activity determinations of at least three independent measurements by nonlinear regression to the following equation: Y = Vmax*X/(KM + X).
Comparison of the Catalytic Efficiency of the 1-Kinase and 1-Pyrophosphatase Reactions.
Given that the dual activity of Vip1 enzymes produce and destroy 1-PP-IPs at the expense of consuming ATP (a so-called futile cycle), we probed the catalytic efficiencies of the 1-kinase and 1-pyrophosphatase activities of full-length spAsp1 at near-physiological pH (Table 3). This shift in pH caused a threefold increase in the catalytic efficiency of the 1-pyrophosphatase activity observed at pH 5.0. Interestingly, the cause of this drop in catalytic efficiency was a 30-fold increase in the affinity of the enzyme for 1-IP7 offset by a 10-fold decrease in the reaction rate. At pH 7.0, the catalytic efficiency of the 1-pyrophosphatase reaction was 15 times greater than that of the kinase reaction.
Table 3.
Comparison of the catalytic efficiency of Asp1 kinase and pyrophosphatase activities
| Species | Construct | Reaction | Km, µM | Vmax, nmol/min/mg | kcat, s−1 | kcat/Km, 103 M−1⋅s−1 |
| S. pombe | Asp1-FL | Kinase | 4.6 | 5.8 | 0.010 | 2.2 |
| S. pombe | Asp1-FL | Pyrophosphatase | 0.47 | 9.3 | 0.016 | 33 |
Kinetic constants were determined by fitting enzyme activity determinations of at least three independent measurements by nonlinear regression to the following equation: Y = Vmax*X/(KM + X). Assays were performed at pH 7.0.
We next sought to determine whether the 1-pyrophosphatase is conserved in other species of Vip1 enzymes. Kinetic parameters for the hydrolysis of 1-IP7 by the pyrophosphatase domain of S. cerevisiae Vip1 (scVip1) and by full-length mouse Vip2 (mmVip2) were determined and demonstrate that both species have in vitro PP-IP pyrophosphatase activity (Table 2). A preference for removal of pyrophosphates at the 1-position of PP-IPs was observed with a ∼12-fold difference in catalytic efficiency for the mmVip2 hydrolysis of 1-IP7 compared to 5-IP7. The catalytic efficiencies seen for the hydrolysis of 1-IP7 and 1,5-IP8 are similar with Kcat/Km values of 18.6 and 14.0 (×103 M−1⋅s−1).
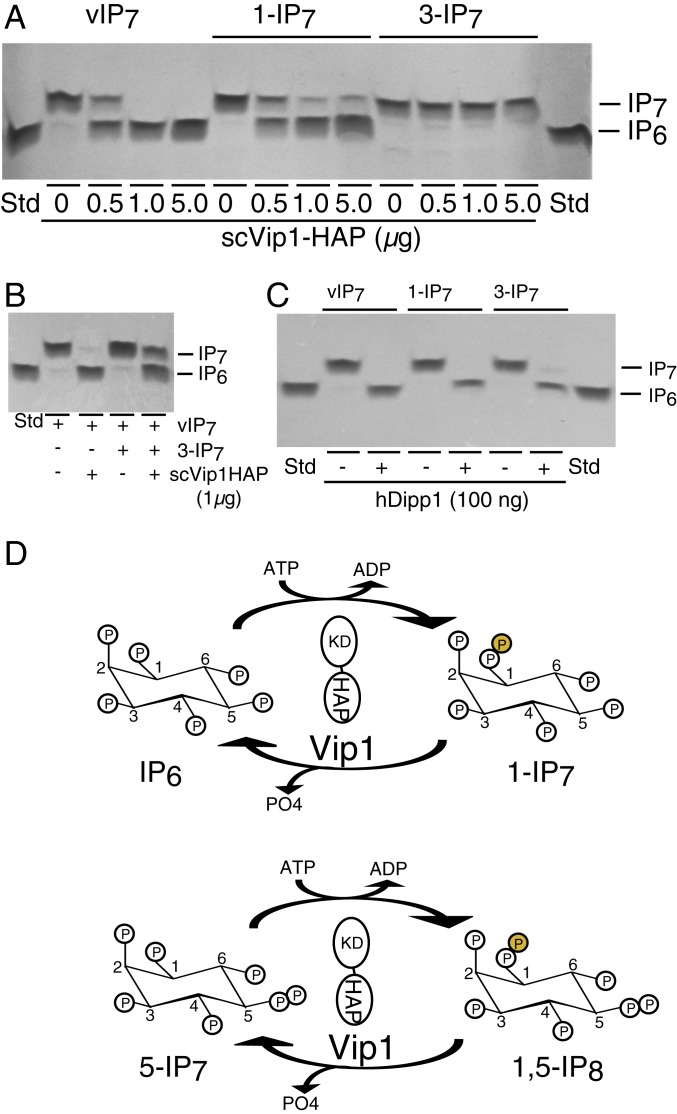
To examine the selectivity of the Vip1 pyrophosphatase activity, we tested synthetically prepared 1-IP7 and 3-IP7 using a polyacrylamide gel electrophoresis (PAGE)-based mass assay. The pyrophosphatase domain of scVIP1 readily utilized both enzymatically and synthetically prepared 1-IP7, but did not utilize synthetically prepared 3-IP7 (Fig. 3A). In contrast, hDIPP1 readily utilized both 1-IP7 and 3-IP7 (Fig. 3B), demonstrating a more promiscuous inositol pyrophosphatase activity. Additionally, these data serve as an important control validating the integrity of the synthetic 3-IP7 stereomer. Combination of 1-IP7 and 3-IP7 had no effect on scVIP1 pyrophosphatase activity (Fig. 3C). These data demonstrate that the Vip1 class of enzymes acts as exquisitely selective D-1-inositol pyrophosphatases.
Fig. 3.
scVip1 pyrophosphatase is selective for D-1 position of IP7. (A) scVip1-HAP does not utilize 3-IP7 as a substrate. Pyrophosphatase mass assay was performed with an escalating dose of scVip1-HAP, comparing substrate utilization of enzymatically prepared 1-IP7, purified synthetic 1-IP7, and purified synthetic 3-IP7. Reactions were performed with 1-nmol substrate at pH 6.8 for 40 min at 37 °C. Reaction products were resolved by SDS/PAGE and visualized by toluidine blue staining. (B) Synthetic 3-IP7 has no effect on the utilization of 1-IP7 by scVIP1-HAP. Reaction was performed with both substrates at 1 nmol at pH 6.8 for 40 min at 37 °C. Products were visualized by SDS/PAGE. (C) Synthetic 3-IP7 utilized as substrate by the nonselective inositol pyrophosphatase hDipp1. Pyrophosphatase assay was performed with 100 ng of hDipp1 using enzymatically prepared IP7, synthetic 1-IP7, and 3-IP7 as substrates. Reactions were performed with 1-nmol substrates at pH 7.5 for 20 min at 37 °C and were resolved by SDS/PAGE. (D) Vip1/Asp1 kinase/pyrophosphatase cycle. We depict Vip1 as a parallel homodimer based on the crystal structure of PFK2/FBPase2 (1K6M.pdb) via interactions through the N-terminal kinase domain (KD); whereas the C-terminal histidine acid pyrophosphatase (HAP) domains are independent of each other. The KD generates either 1-IP7 or 1,5-IP8 contributing a β-phosphate (shown as an orange sphere in the D-1 ring position). The HAP removes the β-phosphate generating IP6 and 5-IP7 products.
As mentioned earlier, due to a variety of limitations, our crystallographic analyses of the 1-IP7 or 1,5-IP8 products alone were not sufficient analysis to prove that Vip1 kinase activity exclusively produces D-1 stereomers. With the discovery that Vip1 functions as a selective 1-pyrophosphatase, we performed pyrophosphatase assays on the Vip1 IP7 product. We produced mass quantities of the Vip1 kinase product (Fig. 3A, designated vIP7) and postulated if vIP7 were a mixture of 1- and 3-IP7, then its hydrolysis by 1-pyrophosphatase activity would appear incomplete, leaving residual 3-IP7. Strikingly, we observed complete hydrolysis of vIP7, suggesting that if 3-IP7 is present it is at undetectable levels. Collectively, our structural and enzyme activity/selectivity data demonstrate that Vip1 encodes an evolutionarily conserved class of dual-functional enzyme switches that have 1-kinase activity toward IP6 and 5-IP7 substrates and 1-pyrophosphatase activities toward the kinase products 1-IP7 and 1,5-IP8 (schematic shown in Fig. 3D).
Vip1 Kinase and Pyrophosphatase Activities Regulate Cellular Levels of 1-IP7 and 1,5-IP8.
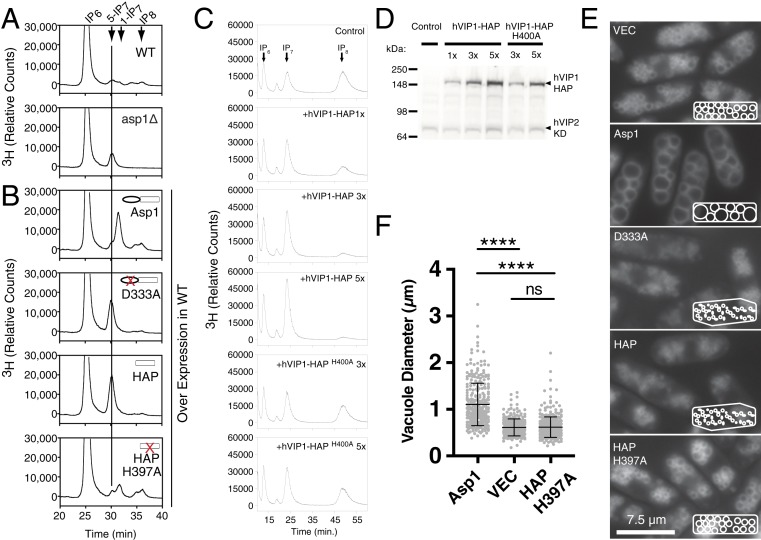
To investigate the relevance of the Vip1 class of kinase and pyrophosphatase activities in cells, we performed a series of high-resolution high-performance liquid chromatography (HPLC) on extracts produced from metabolic labeling of yeast strains expressing various spAsp1 mutants. After steady-state radiolabeling, cell extracts were prepared and separated by chromatography capable of separating individual IP species and, importantly, 5-IP7 and 1-IP7 stereoisomers (confirmed using enzymatically produced standards). Extracts from wild-type cells showed roughly equivalent levels of 5-IP7, 1-IP7, and 1,5-IP8 species; whereas the spAsp1-deficient cells showed a loss of 1-IP7 and 1,5-IP8 and increased 5-IP7 (Fig. 4 A, top and second panels). In addition, we examined the effects of overexpression of Asp1 and a series of mutants in wild-type yeast cells (Fig. 4B). Overexpression of full-length spAsp1 lead to an ∼10-fold increase in 1-IP7 and 2-fold increase in 1,5-IP8, while the levels of 5-IP7 remaining nearly unchanged (Fig. 4 B, top panel). Furthermore, we examined the effects of overexpression of either full-length kinase-dead spAsp1-D333A or a pyrophosphatase domain-only spAsp1-HAP (Fig. 4B, second and third panels), which led to the reduction of 1-IP7 to undetectable levels, and a ∼10-fold increase in 5-IP7. Overexpression of pyrophosphatase-dead point mutant spAsp1-HAP H397A, did not alter IP7 and IP8 levels (Fig. 4 B, bottom panel), confirming metabolic changes observed are attributable to 1-pyrophosphatase activity. Overall, our metabolic data are consistent with spAsp1 harboring endogenous 1-kinase and 1-pyrophosphatase activities that are essential for homeostasis of both 1-IP7 and 1,5-IP8 in cells.
Fig. 4.
In vivo analysis of inositide metabolism and vacuolar morphology in fission yeast and human cells. (A) Metabolic analysis of inositides from 3H-inositol–labeled WT and asp1-null (asp1Δ) fission yeast. (B) Inositide metabolism in radiolabeled fission yeast overexpressing Asp1 proteins. Increase in 1-IP7 and IP8 are observed when full-length Asp1 is overproduced (top trace). Loss of IP8 and increase 5-IP7 are observed in cells overexpressing Asp1 kinase-dead protein D333A mutant (second trace) or HAP domain along (third trace). No significant changes in IP7 or IP8 are observed in cells overproducing HAP pyrophosphatase dead H397A mutants (compare top trace in A to fourth trace in B). (C) Metabolic profiles of inositol pyrophosphates in human embryonic kidney HEK293 cells engineered to overproduce 1,5-IP8. Expression of human Vip1 pyrophosphatase domain (hVip1-HAP) but not catalytically dead mutant (hVip1-HAPH400A) results in decreased 1,5-IP8 levels. Transfections of 1×, 3×, and 5× reflect increased plasmid dosage during labeling. The isomer of IP7 observed is predominantly 5-IP7 which is the product of the 1,5-IP8 hydrolysis. For all metabolic labeling in A–C, experiments were performed using a minimum of three independent samples and representative HPLC traces are shown. y axis indicated relative counts (counts per minute) normalized to each sample’s total counts per minute (greater than 90% of which is derived from a peak that elutes at 5′ time corresponding to free 3H-inositol standard). (D) Western blot analysis of expression levels of human Vip1 pyrophosphatase domain (CFP-hVip1-HAP) and kinase domain (CFP-hVip2-KD). Transfections of 1×, 3×, and 5× reflect increased plasmid dosage during Western blotting. (E) Alterations in vacuolar morphology in osmotically shocked cells overexpressing Asp1 WT or kinase or pyrophosphatase dead proteins. Imaging in live cells induced to overexpress vector control (VEC), full-length (Asp1), full-length kinase-dead (D333A), Asp1 histidine acid pyrophosphate domain only (HAP), or HAP pyrophosphate dead (HAP-H397A). Cells were induced to overexpress proteins by removing thiamine for 30 h, osmotically shocked in H2O for 1 h, stained with vacuole specific dye FM4-64, and imaged. Cartoon Insets depict effect on cell shape, and vacuolar number/cell and diameter. (Scale bar, 7.5 μm.) (F) Vacuole diameter was quantified in fission yeast overexpressing Asp1 WT (n = 301), vector control (n = 267), or HAP pyrophosphatase dead HAP-H397A (n = 379) using Nikon Elements Analysis software. The quadruple asterisk indicates a value of P < 0.0001 as determined by ANOVA statistical analysis with Tukey’s multiple-comparison test.
We next examined the evolutionary conservation of the kinase and pyrophosphatase activities of Vip1 using metabolic studies in human embryonic kidney HEK 293T cell lines genetically engineered to up-regulate PP-IP synthesis (18). Of note, our engineered human line dramatically overproduces both IP7 and IP8 (Fig. 4 C, top panel); however, these cells preclude testing pyrophosphatase activity toward 1-IP7 as its levels are masked by massive amounts of 5-IP7. Expression of hVIP1-HAP reduced 1,5-IP8 levels alongside a corresponding mass increase in level of IP7, presumably 5-IP7 (Fig. 4C, second through fourth panels). The extent of the changes in IP8 are dependent on the dose of expression level of the pyrophosphatase domain as confirmed by antigen levels in 1×, 3×, and 5× overproducing samples (Fig. 4 C and D). Expression of increasing doses of hVIP1-HAPH400A catalytic site mutant had no effect on 1,5-IP8 levels, confirming that decreases in 1,5-IP8 are dependent on pyrophosphatase domain activity (Fig. 4C, lower two panels, and Fig. 4D).
Vip1 Kinase and Pyrophosphatase Activities Modulate Vacuolar Morphology.
Previous studies suggest links between spAsp1-deficient mutants and morphological abnormalities including vacuolar size, polarity, and actin cytoskeleton (7, 14, 19). Our results showing that Vip1 enzymes act as a kinase/pyrophosphatase switch provided an impetus to probe the function of each domain and their relative balance in contributing to these phenotypes. We transiently overexpressed a series of vector control, wild type, and spAsp1 mutants and used the acidic dye FM4-64 to examine vacuole size of cells after a 1-h hypotonic shock (Fig. 4E). Hypotonic treatment has previously been shown to activate vacuole fusion in S. pombe through a MAP kinase-dependent signaling pathway (20). Overexpression of full-length spAsp1 resulted in markedly increased average vacuole size and concomitant reduction in the average number per cell compared to vector control (Fig. 4E, top two panels). The diameter of the vacuoles was determined and quantified (Fig. 4F), demonstrating a nearly twofold change in average diameter. We observed significant variation in vacuole size and number within a cell as well as variability among cells within a given population, the latter of which correlated with the intensity of GFP fluorescence signal suggestive of a range of overexpression. These data are consistent with dose of cellular spAsp1 as a positive regulator of vacuole fusion and/or inhibitor of fission.
To identify which of spAsp1’s two domains, if any, and whether or not enzymatic activity is required for the effects observed, we overexpressed a full-length kinase and pyrophosphatase point mutant (spAsp1 H397A) and full-length kinase-dead pyrophosphatase active point mutant (spAsp1D333A). Prolonged overexpression of spAsp1H397A resulted in profound cytotoxicity. Therefore we used an inducible construct from a relatively weak promoter, which prevented cytotoxicity, and found that this construct produced the same vacuole phenotype as overexpression of full-length spAsp1. A truncation encoding only the kinase domain was lethal when overexpressed in cells, consistent with full-length kinase-only overexpression, whereas expression of full-length spAsp1 D333A H397A kinase-dead/pyrophosphatase-dead double point mutants produced no vacuolar phenotype. These data, coupled with our metabolic labeling studies, indicate elevation of 1-IP7 and/or 1,5-IP8 is sufficient to induce enlarged vacuoles and the effect is exacerbated by osmotic stress.
To determine whether spAsp1’s pyrophosphatase domain had any independent effect on vacuole morphology, we expressed a full-length kinase-dead point mutant spAsp1 D333A and remarkably observed a marked decrease in vacuole diameter alongside an increase in the number of vacuoles (Fig. 4E, third panel). Furthermore, these cells, when osmotically stressed in water, did not undergo any change in vacuole size or number. Cells expressing spAsp1-HAP or spAsp1-HAP also displayed defects in the actin cytoskeleton, similar to those previously reported in an asp1 knockout (7, 19).
To elucidate whether this apparent inhibition of vacuole fusion was attributable to Asp1’s pyrophosphatase domain alone, a truncation lacking the kinase domain was also expressed. Expression of this pyrophosphatase domain produced a similar phenotype, indicating that the mutant kinase domain is not responsible for these defects. Furthermore, expression of a pyrophosphatase domain catalytically dead point mutant led to normal vacuole morphology, confirming that enzymatic activity is required for the observed phenotype. This also demonstrates a biologically relevant role for Asp1 pyrophosphatase domain, its enzymatic activity, and defines key catalytic residues. The distinct vacuolar defects seen with overexpression of the pyrophosphatase domain alone were also rescued by simultaneous expression of the Asp1 full-length pyrophosphatase-dead mutant. This result, along with the opposite effects on vacuole fusion seen when independently expressing each activity, suggests that the two Asp1 domains have opposing signaling functions.
Discussion
In this long-overdue report, we present our historical data, which was not published in a timely manner, confirming that the Vip1 class of inositide metabolic enzymes harbors dual functionality. In addition to a well-documented kinase domain, Vip1 enzymes have an evolutionarily conserved biologically active pyrophosphatase domain. As Results is written as though it was a step back in time, it is important to note that several groups have published the existence of Vip1 pyrophosphatase activity and its conservation across the eukaryotic kingdom (13, 16, 21–23). Independently, we made these discoveries (as acknowledged in ref. 23), and importantly, our work provides unequivocal evidence that the Vip1 kinases are exclusive for placing a β-phosphate onto the D-1 phosphomonoester and do not possess observable D-3 kinase activity. This determination emerged from characterization of Vip1 IP7 product through X-ray crystallography in combination with biochemical assays using the Vip1 pyrophosphatase domain. X-ray crystallography of Vip1 products, alone, is not sufficient to make these conclusions (24).
The observed diphosphoinositol phosphohydrolase activity designated 1-pyrophosphatase represents a highly specialized activity for the HAP family of acid phosphatases on substrates other than phosphomonoesters. The HAP domain of Vip1, expressed either alone or in the context of the full-length protein alongside the kinase domain, selectively degrades 1-IP7 and 1,5-IP8, in both in vitro and cellular contexts. This dual-functional nature of Vip1 defines a regulatory homeostatic axis of the inositol phosphate and pyrophosphate metabolic pathway (25).
The tethering of kinase and pyrophosphatase domains on a single peptide that together regulate the synthesis and breakdown of a signaling axis is reminiscent of phosphofructokinase 2 (PFK2) and is a rare example in biology. To our knowledge, the only other known bifunctional futile cycle enzyme with two separate active sites is phosphofructokinase-2/fructose 2,6-bisphosphatase (PFK-2/FBPase-2). In a manner analogous to the Vip1 synthesis and degradation of 1-IP7 and 1,5-IP8, PFK-2/FBPase-2 catalyzes both the synthesis and degradation of fructose 2,6-P2. Fructose 2,6-P2 is a metabolite that allosterically affects the activity of phosphofructokinase 1 (PFK-1) and fructose 1,6-bisphosphatase (FBPase-1) and is a critical modulator of the switch between glycolysis and gluconeogenesis.
We, and others (23), note a number of key elements that enhance pyrophosphatase activity in the HAP domain. First, pyrophosphatase activity was susceptible to expression context: For example, pyrophosphatase activity of GST-mmVIP2 fusion protein was not evident until the GST domain was cleaved from mmVIP1. Additionally, we were unsuccessful in our attempts to express protein harboring pyrophosphatase activity in bacteria for either full length or the pyrophosphatase domains of hVIP1 or hVIP2, even though both proteins exhibited kinase activity, indicating at least partial correct folding of the protein. Rather, recombinant protein made from yeast or mammalian systems harbors pyrophosphatase activity. Of course, in vivo metabolic analysis presented here supports these conclusions. Finally, we note that 1-pyrophosphatase activity in vitro is inhibited by the chloromethylketone class of protease inhibitors, which are routinely used during enzyme purification from cell extracts.
The identification of 1-pyrophosphatase activity provides additional mechanistic insight into vacuolar and actin cytoskeletal defects previously seen in S. pombe strains either lacking or overexpressing an intact spAsp1 pyrophosphatase domain. Our further investigation of these S. pombe phenotypes here has also revealed a clear correlation between Asp1-regulated 1-IP7 levels and vacuole fusion activity. Modulating relative expression levels of Asp1 pyrophosphatase and kinase activities allows selective accumulation or degradation of 1-IP7, leading to activation or inhibition of vacuole fusion, respectively. It is also notable that osmotic stress, which has previously been linked to elevation of inositol pyrophosphate levels, appears to exacerbate the observed vacuolar defects (26, 27). Furthermore, as hypotonic stress has previously been shown to activate vacuole fusion in a MAPK-dependent fashion, it is possible that PP-IPs impinge on this signaling pathway (20). Similarly, our findings may be relevant to cAMP signaling regulation in S. pombe reported by Fleig and colleagues (14). Overall, a number of studies aimed at determining the regulation of the Vip1 class of dual-functional enzymes in a variety of cellular and organismal systems sheds light on how this fascinating enzyme works (2, 13, 14, 16, 21–25, 28–30). It remains an exciting mechanistic challenge to identify precisely how changes in 1-IP7 or 1,5-IP8 might interact with the vacuole fusion machinery, the actin cytoskeleton, or an uncharacterized pathway that influences cellular signaling, morphology, and architecture.
Our results that Vip1 is a dual-functional switch are especially relevant to the role of IP7 in the S. cerevisiae phosphate starvation response. Our high-resolution HPLC capable of separating 1-IP7 from 5-IP7 species obtained from cell extracts may be useful in resolving disagreement in the field regarding the role of which Vip1 products, 1-IP7 vs. 1,5-IP8, may be involved in regulation of phosphate (7, 12, 31–33). It has been reported that, in yeast, low phosphate conditions lead to an increase in IP7, which specifically binds and inactivates the Pho80/85/81 cyclin/CDK/CDK inhibitor complex, leading to the dephosphorylation and nuclear localization of the Pho4 transcription factor, and activation of PHO response genes (7, 32, 33). While this induction of IP7 was presumed to be the result of solely Vip1’s kinase activity, it is also possible that coordinated regulation and deactivation of Vip1’s 1-pyrophosphatase activity is required or modulatory for this signaling event.
Last, the dual-functional nature of the Vip1 enzyme prompts a fundamental question of how are the kinase and pyrophosphatase activities regulated, thereby avoiding a “futile cycle” of ATP consumption? Certainly, it is possible that posttranslational modifications such as phosphorylation of the individual domains may alter activity. Such is the case for PFK2 as it is regulated by cAMP and protein kinase A through phosphorylation of serine residues near the active sites (34). Studies of Asp1 indicate that a similar cAMP PKA switch regulates functionality in fission yeast (15). Additionally, we find that the kinase and pyrophosphatase activities are tuned to substrate concentrations within cells, suggesting that another form of regulation may be that high IP6/low IP7 ratios are conducive to favoring kinase activity; whereas as the IP7 levels build, then pyrophosphatase activity is favored, generating a feedback mechanism. Regardless, our studies provide additional steps forward in defining how the Vip1 class of kinase/pyrophosphatase enzymes are critically important for cellular signaling.
Materials and Methods
Plasmid Construction.
Generation of the pREP3X-GFP- asp1+, asp1H397A, and asp1D333A constructs were previously described (8). GFP-asp1+, GFP-asp1H397A, and GFP-asp1D333A were subcloned from pREP3X into pREP4X, pREP41X, and pUNI10 using XhoI and BamHI restriction sites. To clone the asp1+ pyrophosphatase domain (asp1-HAP) (residues 377 to 920) and the asp1+ pyrophosphatase domain with a catalytic point mutation (asp1-HAPH397A), primers 5′ CCTTCAGTCCAATAAGCTCGAGTTATGAATCCTCCGCCTAGAG-3′ and 5′-CTCTAGGCGGAGGATTCATAACTCGAGCTTaTTGGACTGAAGG-3′ were used to create an XhoI restriction site (underlined) in pREP3X-GFP- asp1+ and pREP3x-GFP-asp1H397A. An XhoI digest was used to remove the region encoding residues 1 to 376, and the plasmids were religated. Full-length asp1+ and asp1D333A were subcloned into pGEX-KG vector from their respective pUNI10-asp1 constructs through XhoI and SacI digestion, while asp1-HAP and asp1-HAPH397A were subcloned from pUNI10 constructs using EcoRI and SalI restriction sites.
The pyrophosphatase domain of S. cerevisiae [scVIP1(530–1106)] was cloned by PCR, generating an insert with flanking 5′ NcoI and 3′ XhoI sites. The previously reported full-length Vip1 construct (pGEX-KG-scVIP1) was used as a template with forward 5′-GGTACCATGGGTCGTGAAGAAAAGGAACAAAAGTGGGTATTC-3′ and reverse 5′-GGATCTCGAGCTACGTAATGTTTACTGGTGTAAATTTCGGAGGC-3′ primers (7). The PCR product was then cloned into a pGEX-KG GST-fusion vector after NcoI/XhoI digestion. cDNA for mouse VIP2 (mmVip2; accession number BC053396) was obtained from Invitrogen. The coding sequence of mmVip2 was cloned into a pGEX-KG GST fusion vector from the pSPORT-1 vector by successively subcloning fragments from XhoI–SacI and SacI–SacI restriction digests.
A pET-28–based expression plasmid (pNIC-Bsa4) encoding human diphosphoinositol polyphosphate phosphohydrolase 1 (Dipp1) (Nudt3; accession number AAH07727) residues 1–148 was kindly provided by the Structural Genomics Consortium, Karolinska Institutet (Stockholm, Sweden). Fission yeast strains used and generated: see Table 4.
Table 4.
Strains used in this study
| S. pombe strain | Genotype | Source |
| JYY841 | KGY246 h- leu1-32 ura4-D18 ade6-M210 | KGY246, from K. Gould (19) |
| JYY899 | JYY841 + pREP3X | Ref. 7 |
| JYY901 | JYY841 + pREP3X-GFP-Asp1 | Ref. 7 |
| JYY903 | JYY841 + pREP3X-GFP-asp1D333A | Ref. 7 |
| JYY905 | JYY841 + pREP3X-GFP-asp1H397A | Ref. 7 |
| JYY1083 | JYY841 + pREP3X-GFP-asp1-HAP | This work |
| JYY1097 | JYY841 + pREP3X-GFP-asp1-HAPH397A | This work |
| JYY1077 | JYY841 + pREP3X-GFP-asp1D333A + pREP41X | This work |
| JYY1078 | JYY841 + pREP3X-GFP-asp1D333A + pREP41X-GFP-asp1H397A | This work |
| JYY845 | KGY954 h- asp1::ura4 leu1-32 ura4-D18 ade6-M216 | KGY954, from K. Gould (19) |
| JYY847 | JYY845 + pREP3X | This work |
Recombinant Protein Expression and Purification.
Constructs of S. pombe asp1-HAP, asp1D333A, S. cerevisiae Vip1-HAP, and Mus musculus Vip2 were transformed and expressed as GST-fusion proteins in Escherichia coli. S. pombe and S. cerevisiae constructs were expressed in BL21(DE3) cells by growing cultures initially at 37 °C, and then reducing temperature to 18 °C at OD600 of 0.6 for a 16-h induction with a final concentration of 0.1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG). The murine Vip2-FL construct was expressed in ArcticExpress (DE3)RIL cells by growing cultures at 30 °C initially, and then reducing to 12 °C. The cells were induced at OD600 of 0.6 for 24 h with a final concentration of 0.25 mM IPTG.
Cells were collected by centrifugation and resuspended in lysis buffer containing 25 mM Tris, pH 8.0, 350 mM NaCl, 2 mM DTT, and 0.1 mM phenylmethylsulphonyl fluoride (PMSF). Cell resuspensions were lysed by serial passage through an EmulsiFlex-C5 high-pressure homogenizer at >15,000 psi. Lysis debris was cleared by centrifugation, and the resulting supernatant was applied to glutathione-Sepharose beads preequilibrated in lysis buffer. After a 20-column volume wash with lysis buffer, the proteins were eluted with 50 mM Tris, pH 8.0, 250 mM NaCl, 2 mM DTT, and 10 mM glutathione. The GST tag was removed from the fusions by digestion using a 1:2,000 (wt/wt) dilution of thrombin at 4 °C during dialysis into SP-Sepharose ion exchange buffer containing 25 mM Bis-Tris, pH 6.0, 150 mM NaCl, and 2 mM DTT. The digestions were loaded on a preequilibrated SP-Sepharose column and eluted using a 150 to 600 mM NaCl gradient in 25 mM Bis-Tris, pH 6.0, and 2 mM DTT. Fractions from the SP-Sepharose column were pooled, concentrated, and applied to an S-200 size exclusion column. The purified protein in 20 mM Hepes, pH 7.5, 150 mM NaCl, and 2 mM DTT was quantified by UV absorbance under denaturing condition and stored at −80 °C.
Hexa-histidine–tagged human Dipp1 residues 1 to 148 were produced by transformation and overexpression of the pNIC-Bsa-hDipp1 plasmid in BL21(DE3) cells similar to previously described (35). Cultures were grown at 37 °C to an OD600 of 0.5, after which the temperature was reduced to 18 °C and the cultures were induced with IPTG to a final concentration of 0.5 mM. After overnight induction, the cells were collected by centrifugation, and the resulting paste was resuspended in 20% (wt/vol) lysis buffer (50 mM Hepes, pH 7.5, 500 mM NaCl, 10% glycerol, 20 mM imidazole, 2 mM β-Me, and 0.1 mM PMSF) and lysed by multiple passes at ∼15,000 psi through an EmulsiFlex-C5 high-pressure homogenizer. The resulting lysate was cleared by centrifugation and applied to Ni-NTA agarose affinity column (Qiagen) in equilibrated in lysis buffer. The column was washed with 20 column volumes lysis buffer, and the protein was step eluted in 50 mM Hepes, pH 7.5, 500 mM NaCl, 10% glycerol, 400 mM imidazole, and 2 mM β-Me. The Ni-NTA protein fractions were pooled, concentrated with a Vivaspin 20 (10,000 MWCO), and injected onto a S-200 size-exclusion column (GE Biosciences) equilibrated with 20 mM Hepes, pH 7.5, 300 mM NaCl, 10% glycerol, and 0.5 mM tris(2-carboxyethyl) phosphine. The Dipp1-containing S200 fractions were pooled and concentrated to 12 mg/mL as determined by UV absorbance, ε280 = 19,643 1/M⋅cm, under denaturing conditions. Small aliquots were flash-frozen in liquid nitrogen and stored at –80 °C.
Preparation and Purification of Mass Quantities of Biological and Synthetic PP-IPs.
Biological isomers of IP7 and IP8 were enzymatically produced as previously described (7, 8). Lyophilized synthetic isomers of IP7 obtained from the laboratory of G. Prestwich (University of Utah, Salt Lake City, UT) were solubilized in 100 mM Tris, pH 8.8, and 6 mM EDTA prior to purification. Both the biological and synthetic IP7 and IP8 were purified identically using a GE Mono Q 5/50 GL column as described (8) except using the following gradient: 0 to 2 min, 100% buffer A (0.2 mM HCl, 2 mM EDTA, pH 8.0); 2 to 32 min, linear gradient 0 to 100% buffer B (0.5 M HCl, 2 mM EDTA, pH 8.0); 32 to 62 min, 100% buffer B; 62 to 72 min, 100% buffer A. One-milliliter fractions were collected and assayed for IP7 and IP8. IP-containing fractions were neutralized by addition of 100 mM Hepes and concentrated KOH to pH 7 to 7.5 and were quantified by phosphate ashing. Each IP fraction was dialyzed overnight against 4 L of water in 100- to 500-Da MWCO CE dialysis tubing (Spectra/Por). IP fractions were pooled and concentrated using a Savant DNA110 Speedvac under low heat to final concentrations of 30 to 50 mM.
Crystallization, Data Collection, and Refinement of Dipp1 PP-IP5 Complexes.
The Dipp1/IP7 complexes were cocrystallized by the hanging-drop vapor diffusion method. Prior to crystallization, a fivefold molar excess of MgCl2 and either Vip1 1-IP7 or IP6K1 5-IP7 was added to the Dipp1 protein solution at 12 mg/mL and incubated on ice for 15 to 30 min. The reservoir solutions for crystallization were based on published conditions and contained 12 to 18% (vol/vol) PEG8000, 150 to 300 mM LiSO4, and 0.1M sodium acetate, pH 4.6 (35). Hanging drops containing 2 µL each of ligand-bound protein and reservoir solution were incubated at 17 °C. Typically, crystals with dimensions of 600 × 30 × 200 µm appeared within 4 to 7 d. In preparation for data collection, crystals were equilibrated stepwise into cryoprotection solutions containing 12 to 18% (vol/vol) PEG8000, 0.1 M sodium acetate, pH 4.6, 300 mM LiCl, and increasing percentage (vol/vol) PEG400. Prior to flash cooling in liquid nitrogen, the final cryogenic solution (as determined by vitrification upon freezing) was supplemented with 2.5 mM of the crystallization ligand and 2.5 mM MgCl2 before soaking for 5 to 6 h (1-IP7) or overnight (5-IP7).
Data from the cryoprotected Dipp1/IP7 complexes were collected at the Advanced Photon Source, Argonne National Laboratory (Chicago, IL). Data for the Dipp1/1-IP7 complex were collected to 1.2 Å on a MAR300 charge-coupled detector (CCD) at 22-ID, and the data for the Dipp1/5-IP7 complex were collected to 1.3 Å on a MAR 225 CCD at 22-BM. Data for each complex were reduced and scaled using the program HKL2000 (36).
The Dipp1/1-IP7 structure was solved by molecular replacement with the program Phaser (37) using model phases derived from the protein component of a Dipp1/IP6 complex (PDB ID code: 2fvv) (35). The Dipp1/5-IP7 structure was solved by direct Fourier transform in Refmac (38) using phases calculated from the protein component of the Dipp1/1-PP-IP5 structure. Model building was completed by iterative rounds of building in Coot and automated refinement using Phenix1.4 including individual ADP, TLS, and occupancy refinements (39, 40). MolProbity was used for structure validation (41). Data collection and refinement statistics are presented in Table 1.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with the following ID codes: 6PCK (Dipp1/1-IP7 complex) and 6PCL (Dipp1/5-IP7 complex).
In Vitro IP7 and IP8 Pyrophosphatase Activity Assays.
Phosphatase activity assays of Vip1/Asp1 and Dipp1 were visualized by two methods: thin-layer chromatography (TLC) and PAGE analysis. Activity assays for TLC analysis were performed in 10-µL reactions containing the following: 100 mM sodium acetate, pH 5.0, 50 mM NaCl, with varying concentrations of enzyme and substrate, and 15,000 to 25,000 cpm of [d-2-32P]-1-IP7, [d-2-32P]-5-IP7, or [d-2-32P]-1,5-IP8. 32P-labeled substrates were prepared and purified similarly as above, except in each case [d-2-32P]-IP6 was prepared enzymatically using recombinant IPK1, IP5, and [α-32P]-ATP and used in place of IP6 (7). For kinetic analysis, substrate concentrations ∼10-fold above/below Km values were used. Enzyme dosage and time were adjusted to give linear conversion of substrates. Reactions were incubated at 37 °C for 15 to 20 min and stopped by addition of 1 µL of 2.5 M HCl. Quenched reactions were spotted onto polyethyleneimine cellulose TLC plates (JT Baker) and resolved in a tank equilibrated in 2.10 M HCl, 1.09 M KH2PO4, and 0.72 M K2HPO4. TLC plates were dried, exposed to a phosphor storage screen, and quantified using a 4500 SI PhosphorImager (Amersham Biosciences).
PAGE analysis using toluidine blue staining was used to visualize the pyrophosphatase activity of Dipp1 and Vip1/Asp1 against unlabeled biological and synthetic IP7 isomers (42). Ten-microliter reactions containing 1 nmol of PP-IP5 substrate, 100 mM buffer at various pH values, 50 mM NaCl, and either Vip1/Asp1 or Dipp protein at varying dosage. Enzyme assays were run 20 to 40 min at 37 °C and quenched by boiling for 5 min. After centrifugation, 6× loading dye (10 mM Tris, pH 6.8, 1 mM EDTA, 30% glycerol, and 0.25% bromophenol blue) was added to the supernatant and the samples were run at 300 V for ∼2.5 h on a 33.3% TBE-PAGE gel. The gels were stained and destained for visualization as described (42).
Fission Yeast Growth and Manipulation.
S. pombe strains were manipulated, propagated, and transformed using published standard procedures (43) and as described previously (7). For lithium acetate transformation of expression plasmids, 1 µg of DNA was used along with ∼1 × 108 log-phase cells as described (43), and the mixture was then plated onto MMA agar plate supplemented with 5 mg/L thiamine (Sigma Chemical Company) and 225 mg/L of the appropriate amino acids. Plates were incubated at 30 °C for 4 d. Individual colonies were picked and cultured in Edinburgh minimal medium (EMM2) liquid medium plus proper auxotrophy supplements at 30 °C. For the expression of asp1+ and its mutants, cells were cultured in liquid EMM2 medium with or without 5 mg/L thiamine for appropriate time periods.
Steady State Inositol Labeling and HPLC Analysis.
For in vivo [3H]-myo-inositol labeling, S. pombe strains were grown 48 h in 5 mL of EMM inositol minimal media (Sunrise Science Products) with appropriate auxotrophic nutrients, supplemented with 100 μCi/mL [3H]-myo-inositol (American Radiolabeled Chemicals). In strains overexpressing Asp1 and variants, thiamine was removed from the medium during labeling to relieve inhibition of the nmt1 thiamine regulated promotor. Soluble inositol phosphates were harvested by adapting previously reported methods (44). Briefly, washed cells were resuspended in 100 μL of 0.5 M HCl and 370 μL of chloroform/methanol (1:2). Cells were lysed by beating with 100 μL of glass beads twice for 30 s, with 125 μL of chloroform and 125 μL of 2 M KCl added between bead beatings. After a 10-min centrifugation at 16 × g at 4 °C, the supernatant was filtered and brought to 1 mL in 100 mM Tris, pH 8.8, 6 mM EDTA, pH 8.0. Samples were then loaded onto a Mono Q HR 5/5 FPLC column (GE Healthcare) and run under the following gradient: 0 to 2 min, 100% buffer A (0.2 mM HCl, 2 mM EDTA); 2 to 32 min, 0 to 100% buffer B (0.5 M HCl, 2 mM EDTA); 32 to 62 min, 100% buffer B. Radioactivity was measured using a BetaRAM in-line detector (IN/US Systems). PP-IP species were identified by coelution with enzymatically produced [32P]-IP7 and [32P]-IP8 standards. HEK 293T cells were radiolabeled and harvested as previously described (8). Inositol phosphates from these extracts were analyzed on a 4.6 × 125-mm Partisphere SAX HPLC column (Whatman) as previously reported (8). Experiments were performed using a minimum of three independent samples and representative HPLC traces are shown. y axis indicated relative counts (cpm) normalized to each sample’s total radioactivity.
Fission Yeast Vacuole Microscopy.
The membrane-selective dye FM 4-64 (Invitrogen) was used for vacuole staining of the S. pombe strains. For each strain, 400 µL of log-phase cells were pelleted by low-speed centrifugation and resuspended in yeast extracts with supplements (YES medium) plus FM 4-64 at a concentration of 16 µM. After a 30-min incubation at 30 °C, 1 mL of YES was added, and cells were cultured for an additional 30 min. For vacuole fusion analysis, FM 4-64–stained cells were washed to remove media and placed in water. Cells were visualized after 1-h incubation in water to measure vacuole diameter during hypotonic shock. Digital images were obtained using a Nikon Eclipse TE 2000-E microscope equipped with a 40× objective. The vacuole diameter measurements (in microns) were made with the Nikon Elements Analysis software package. Data were plotted using Prism software, and statistics were performed using ANOVA Tukey’s multiple-comparison test.
Data Availability Statement.
All materials, methods, and data are freely available upon request from the J.D.Y. laboratory. Structural data have been deposited in the PDB under ID codes 6PCL and 6PCK.
Acknowledgments
We thank members of the J.D.Y. laboratory for helpful discussions and comments. We thank Drs. Kathy Gould (Vanderbilt University) and Glenn Prestwich (University of Utah) for sharing strains, reagents, and for helpful discussions. This work was first presented at international meetings in 2009, 2011, and thereafter, and was supported by funds from the Howard Hughes Medical Institute and from National Institutes of Health Grants R01 HL055672 and GM124404 (all to J.D.Y.). J.D.Y. is an alumni investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Structural data have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6PCK and 6PCL).
References
- 1.Thota S. G., Bhandari R., The emerging roles of inositol pyrophosphates in eukaryotic cell physiology. J. Biosci. 40, 593–605 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Wilson M. S., Livermore T. M., Saiardi A., Inositol pyrophosphates: Between signalling and metabolism. Biochem. J. 452, 369–379 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty A., Kim S., Snyder S. H., Inositol pyrophosphates as mammalian cell signals. Sci. Signal. 4, re1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatch A. J., York J. D., SnapShot: Inositol phosphates. Cell 143, 1030–1030.e1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saiardi A., Caffrey J. J., Snyder S. H., Shears S. B., The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J. Biol. Chem. 275, 24686–24692 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Saiardi A., Erdjument-Bromage H., Snowman A. M., Tempst P., Snyder S. H., Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 9, 1323–1326 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Mulugu S., et al. , A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316, 106–109 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Fridy P. C., Otto J. C., Dollins D. E., York J. D., Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J. Biol. Chem. 282, 30754–30762 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Choi J. H., Williams J., Cho J., Falck J. R., Shears S. B., Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J. Biol. Chem. 282, 30763–30775 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safrany S. T., et al. , The diadenosine hexaphosphate hydrolases from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human diphosphoinositol polyphosphate phosphohydrolase. Overlapping substrate specificities in a MutT-type protein. J. Biol. Chem. 274, 21735–21740 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Safrany S. T., et al. , A novel context for the “MutT” module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 17, 6599–6607 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lonetti A., et al. , Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 286, 31966–31974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Topolski B., Jakopec V., Künzel N. A., Fleig U., Inositol pyrophosphate kinase Asp1 modulates chromosome segregation fidelity and spindle function in Schizosaccharomyces pombe. Mol. Cell. Biol. 36, 3128–3140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pöhlmann J., et al. , The Vip1 inositol polyphosphate kinase family regulates polarized growth and modulates the microtubule cytoskeleton in fungi. PLoS Genet. 10, e1004586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pöhlmann J., Fleig U., Asp1, a conserved 1/3 inositol polyphosphate kinase, regulates the dimorphic switch in Schizosaccharomyces pombe. Mol. Cell. Biol. 30, 4535–4547 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pascual-Ortiz M., et al. , Asp1 bifunctional activity modulates spindle function via controlling cellular inositol pyrophosphate levels in Schizosaccharomyces pombe. Mol. Cell. Biol. 38, e00047-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin H., et al. , Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J. Biol. Chem. 284, 1863–1872 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otto J. C., Kelly P., Chiou S. T., York J. D., Alterations in an inositol phosphate code through synergistic activation of a G protein and inositol phosphate kinases. Proc. Natl. Acad. Sci. U.S.A. 104, 15653–15658 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feoktistova A., McCollum D., Ohi R., Gould K. L., Identification and characterization of Schizosaccharomyces pombe asp1(+), a gene that interacts with mutations in the Arp2/3 complex and actin. Genetics 152, 895–908 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bone N., Millar J. B., Toda T., Armstrong J., Regulated vacuole fusion and fission in Schizosaccharomyces pombe: An osmotic response dependent on MAP kinases. Curr. Biol. 8, 135–144 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Yousaf R., et al. , Mutations in diphosphoinositol-pentakisphosphate kinase PPIP5K2 are associated with hearing loss in human and mouse. PLoS Genet. 14, e1007297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu C., et al. , KO of 5-InsP7 kinase activity transforms the HCT116 colon cancer cell line into a hypermetabolic, growth-inhibited phenotype. Proc. Natl. Acad. Sci. U.S.A. 114, 11968–11973 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H., et al. , Asp1 from Schizosaccharomyces pombe binds a [2Fe-2S]2+ cluster which inhibits inositol pyrophosphate 1-phosphatase activity. Biochemistry 54, 6462–6474 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H., Falck J. R., Hall T. M., Shears S. B., Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 8, 111–116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shears S. B., Baughman B. M., Gu C., Nair V. S., Wang H., The significance of the 1-kinase/1-phosphatase activities of the PPIP5K family. Adv. Biol. Regul. 63, 98–106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi K., Mollapour E., Shears S. B., Signal transduction during environmental stress: InsP8 operates within highly restricted contexts. Cell. Signal. 17, 1533–1541 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Pesesse X., Choi K., Zhang T., Shears S. B., Signaling by higher inositol polyphosphates. Synthesis of bisdiphosphoinositol tetrakisphosphate (“InsP8”) is selectively activated by hyperosmotic stress. J. Biol. Chem. 279, 43378–43381 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Wang H., et al. , Synthetic inositol phosphate analogs reveal that PPIP5K2 has a surface-mounted substrate capture site that is a target for drug discovery. Chem. Biol. 21, 689–699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pulloor N. K., et al. , Human genome-wide RNAi screen identifies an essential role for inositol pyrophosphates in Type-I interferon response. PLoS Pathog. 10, e1003981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weaver J. D., Wang H., Shears S. B., The kinetic properties of a human PPIP5K reveal that its kinase activities are protected against the consequences of a deteriorating cellular bioenergetic environment. Biosci. Rep. 33, e00022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wild R., et al. , Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Lee Y. S., Huang K., Quiocho F. A., O’Shea E. K., Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 4, 25–32 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee Y. S., Mulugu S., York J. D., O’Shea E. K., Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316, 109–112 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okar D. A., et al. , PFK-2/FBPase-2: Maker and breaker of the essential biofactor fructose-2,6-bisphosphate. Trends Biochem. Sci. 26, 30–35 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Thorsell A. G., et al. , Crystal structure of human diphosphoinositol phosphatase 1. Proteins 77, 242–246 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 37.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Winn M. D., Isupov M. N., Murshudov G. N., Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Adams P. D., et al. , PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Davis I. W., et al. , MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Losito O., Szijgyarto Z., Resnick A. C., Saiardi A., Inositol pyrophosphates and their unique metabolic complexity: Analysis by gel electrophoresis. PLoS One 4, e5580 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forsburg S. L., Rhind N., Basic methods for fission yeast. Yeast 23, 173–183 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stolz L. E., Kuo W. J., Longchamps J., Sekhon M. K., York J. D., INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J. Biol. Chem. 273, 11852–11861 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All materials, methods, and data are freely available upon request from the J.D.Y. laboratory. Structural data have been deposited in the PDB under ID codes 6PCL and 6PCK.