Significance
Bats represent 20% of all mammalian species and are an important reservoir of viruses that infect humans and other mammals. Retroviruses, such as HIV, are among the most important zoonotic viruses infecting humans, although little is known about their circulation in bat populations. We report the first exogenous retrovirus described in bats, the Hervey pteropid gammaretrovirus (HPG), a reproduction-competent retrovirus within northeast Australia. Koala populations are currently in severe decline and at risk from koala retrovirus (KoRV), which is closely related to HPG and whose origins remain unclear. The identification of bats as a source of diverse infectious retroviruses related to KoRV implicates bats as a reservoir of KoRV-related viruses that potentially can be transmitted to other mammalian species.
Keywords: bats, viruses, retroviruses, KoRV, GALV
Abstract
Bats are reservoirs of emerging viruses that are highly pathogenic to other mammals, including humans. Despite the diversity and abundance of bat viruses, to date they have not been shown to harbor exogenous retroviruses. Here we report the discovery and characterization of a group of koala retrovirus-related (KoRV-related) gammaretroviruses in Australian and Asian bats. These include the Hervey pteropid gammaretrovirus (HPG), identified in the scat of the Australian black flying fox (Pteropus alecto), which is the first reproduction-competent retrovirus found in bats. HPG is a close relative of KoRV and the gibbon ape leukemia virus (GALV), with virion morphology and Mn2+-dependent virion-associated reverse transcriptase activity typical of a gammaretrovirus. In vitro, HPG is capable of infecting bat and human cells, but not mouse cells, and displays a similar pattern of cell tropism as KoRV-A and GALV. Population studies reveal the presence of HPG and KoRV-related sequences in several locations across northeast Australia, as well as serologic evidence for HPG in multiple pteropid bat species, while phylogenetic analysis places these bat viruses as the basal group within the KoRV-related retroviruses. Taken together, these results reveal bats to be important reservoirs of exogenous KoRV-related gammaretroviruses.
Retroviruses are a widespread and diverse group of RNA viruses distinguished by their ability to integrate into the genome of their host cell (1). Several retroviruses cause immunodeficiency (e.g., HIV) (2) and malignancies such as leukemia (e.g., koala retrovirus [KoRV]) (3–5). When retroviruses integrate into germ line cells, they become “vertically” transmissible from parent to offspring and are referred to as endogenous retroviruses (ERVs) (1, 6). KoRV, for instance, is an infectious retrovirus currently undergoing endogenization in the koala gene pool (5). Through the course of evolutionary history, ERVs and related retroelements have become ubiquitous across metazoan genomes (6–8); for example, 8% of the human genome is derived from retroviruses (9). ERVs may or may not be capable of producing infectious viral particles.
Bats are reservoirs for many viruses from diverse viral families and are implicated in the transmission of numerous highly pathogenic viruses to humans and other mammals (10). Previous studies have revealed the presence of ERVs from the genera Betaretrovirus, Gammaretrovirus, and Deltaretrovirus within the genomes of bats (11–14). Analyses of the evolutionary relationships between these bat ERVs and those from other mammals imply that bats have played a key role in the transmission of retroviruses between different mammalian species (15, 16). Indeed, genomic analysis indicates that bats have served as hosts to retroviruses for most of their evolutionary history (11), and evidence of gene expansion and diversification in the antiretroviral APOBEC3 family of immune restriction factors suggests an ongoing relationship between bats and retroviruses (17). To date, however, no infectious, horizontally transmissible exogenous retroviruses (XRVs) have been identified and reported in bats.
KoRV and the gibbon ape leukemia virus (GALV) are closely related gammaretroviruses (77.5% nucleotide identity). However, the habitats of the hosts of these viruses (koalas in Australia and gibbons in Southeast Asia) do not overlap and are physically separated by the oceanic faunal boundary known as the Wallace line (18). It has been suggested that bats may have played a role in the transmission of gammaretroviruses between gibbons and koalas (19–21). In particular, the habitat of such bats as the black flying fox, Pteropus alecto, overlap and connect the habitats of both gibbons and koalas, with bats being capable of traversing the bodies of water that separate the islands of Australia and Southeast Asia (22). In addition, bat gammaretroviral ERVs are widely distributed across the broader gammaretroviral phylogeny (23), with one recently discovered bat gammaretroviral ERV reportedly falling between KoRV and GALV on phylogenetic trees (14).
To advance our understanding of the role of bats as hosts and potential transmitters of gammaretroviruses closely related to KoRV and GALV (herein referred to as KoRV-related viruses), between 2007 and 2014 we collected bat samples (feces, blood, urine, and oral swabs) from towns and the Daintree rainforest along the east coast of Australia to detect the presence of KoRV-related viruses. From this survey, we report the identification and characterization of a novel reproduction-competent bat retrovirus, the Hervey pteropid gammaretrovirus (HPG), from P. alecto. In addition, we have identified novel gammaretroviral sequences from two species of pteropid bats, Macroglossus minimus and Syconycteris australis, and two species of Yinpterochiropteran microbats from China, Hipposideros larvatus and Rhinolophus hipposideros. These gammaretroviral sequences are closely related to KoRV and GALV.
Results
Metagenomic Analyses Reveal the Presence of Novel KoRV-Related Gammaretroviruses in Australian and Asian Bats.
To identify KoRV-related viruses in bats, samples were collected from the east coast of Australia, including feces, oral swabs, blood, and urine. A total of 373 samples were collected from towns in New South Wales and Queensland, and 106 samples were collected from the Daintree rainforest in Queensland. The species of origin was determined by species-specific cytochrome B gene TaqMan RT-PCR. Metagenomic analysis of RNA extracted from the bat samples revealed the presence of KoRV-related viruses in samples collected from the pteropid bat species (subfamily Yinpterochiroptera) P. alecto (HPG), M. minimus (M. minimus gammaretrovirus [MmGRV]), and S. australis (S. australis gammaretrovirus [SaGRV]). To broaden our search, we probed the Sequence Read Archive (SRA) for the presence of KoRV-related viruses. This search revealed the presence of two additional viruses in metagenomic RNA extracted from samples obtained from the Asian microbat species (subfamily Yinpterochiroptera) H. larvatus (H. larvatus gammaretrovirus [HlGRV]) and R. hipposideros (R. hipposideros gammaretrovirus [RhGRV]). The identified KoRV-related viruses and their origins are summarized in SI Appendix, Table S1.
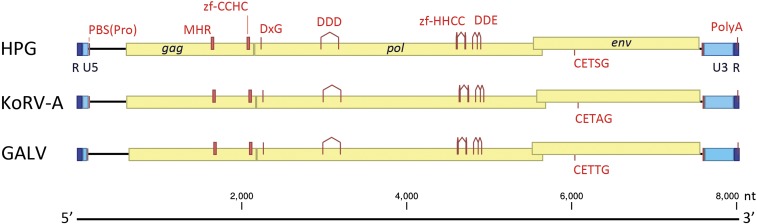
The complete genome sequence of HPG and partial genome sequences of MmGRV, SaGRV, HlGRV, and RhGRV were assembled and deposited in GenBank (SI Appendix, Table S2). The source of HPG was a fecal sample collected in 2011 from a single flying fox in Hervey Bay, Queensland. HPG viral particles in the sample were enriched using a sucrose gradient, total RNA was extracted, and genomic DNA was removed. The complete HPG genome sequence was generated from this total RNA sample via a modified single-cell whole-transcriptome amplification (WTA) procedure for detecting ultra-low-copy viral RNA and a de novo sequence assembly pipeline outlined in SI Appendix, Methods. The HPG genome is 8,030 nt in length, similar to KoRV-A and GALV (7,994 nt and 8,087 nt, respectively), and contains terminal repeats (R), 5′ and 3′ unique regions (U5/U3), and open reading frames (ORFs) encoding the canonical gammaretroviral genes gag, pol, and env, which do not contain any frameshift mutations or premature stop codons (Fig. 1). Other genomic elements essential for retroviral replication and reproduction, including the expected protease, polymerase, and integrase active site motifs; proline tRNA primer binding site; polypurine tract; and polyadenylation signal site, were also present (Fig. 1 and SI Appendix, Fig. S1A). The assembled partial genome sequences of MmGRV and SaGRV lacked coverage only at the terminal repeat and unique 5′ and 3′ regions, while ORFs encoding gag, pol, and env were intact and free from frameshift mutations or premature stop codons (SI Appendix, Fig. S2). For HlGRV and RhGRV, overall read coverage was low, and in both cases coverage dropped to zero at some locations within each of gag, pol, and env (SI Appendix, Fig. S2).
Fig. 1.
The genome of the HPG contains conserved functional motifs and is analogous to KoRV-A and GALV. R, terminal repeat sequence; U5/U3, unique 5′/3′ region; PBS(Pro), proline tRNA primer-binding site; gag, group-specific antigen; MHR, major homology region; zf, zinc finger; DxG, protease active site motif; DDD, reverse transcriptase active site motif; pol, polymerase; DDE, integrase active site motif; env, envelope; CET(S/A/T)G, pathogenicity motif; PolyA, polyadenylation signal.
HPG Sequences Were Not Detected in the Genomes of Pteropid Bats.
To exclude the possibility that HPG represents a fossilized ERV, we performed a BLAST analysis of the P. alecto and Pteropus vampyrus genomes. No sequences matching HPG were identified. The closest identified hit against the HPG sequence in this analysis was a 546-nt sequence within the genome of P. alecto, aligning to the pol gene of HPG, with an e-value of 5.0 × 10−46 and a nucleotide identity of 69%. We then performed a HPG-specific PCR analysis of the P. alecto genome, using genomic DNA extracted from two sources: P. alecto tissue from a male bat captured in Brisbane (Australia) and a P. alecto kidney cell line (24). This PCR analysis did not generate detectable amplicons, in contrast to amplification of a single-copy bat APOBEC3Z3 gene (17) (SI Appendix, Fig. S3). These data suggest that HPG has not integrated into the germ line of the P. alecto bats tested and is likely to be an XRV currently circulating among Australian bats.
Phylogenetic Analysis Reveals Close Relationships among Koala, Gibbon, and Bat Gammaretroviruses.
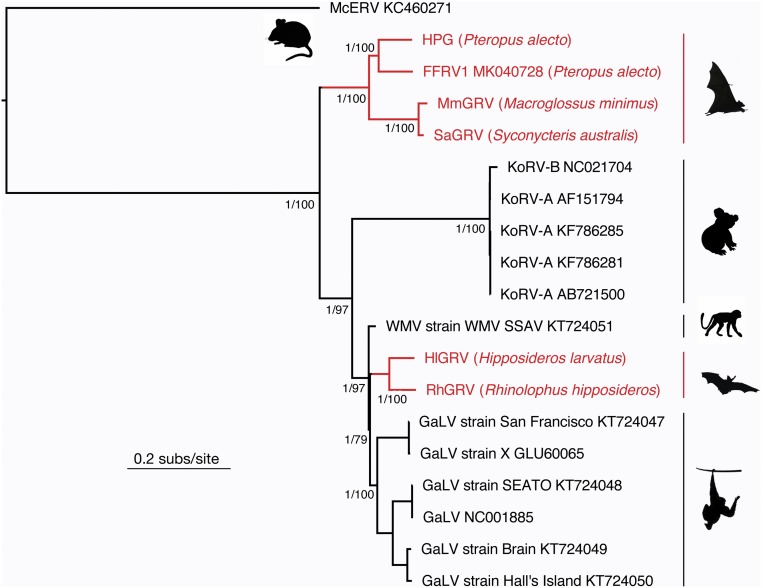
To determine the evolutionary relationships among the retroviruses that we identified here (SI Appendix, Table S1) with known gammaretroviruses (SI Appendix, Table S2), we performed a phylogenetic analysis of the full retroviral genome. Our maximum likelihood phylogenetic analysis revealed that HPG, MmGRV, and SaGRV form a distinct and well-supported clade that is basal to the KoRV and GALV groups (Fig. 2). In contrast, the Asian bat-derived HlGRV and RhGRV cluster as a sister group to the GALV clade. This finding is supported by phylogenetic analyses of the individual pol and env genes, which revealed the same branching pattern (SI Appendix, Fig. S4). While analysis of the gag gene resulted in a slightly different branching pattern, this is likely a result of low phylogenetic resolution, as indicated by low bootstrap support for key nodes on this tree (SI Appendix, Fig. S4). Thus, these data reveal that KoRV-related gammaretroviruses exist within multiple species of Australian and Asian bats, with those from Australia (HPG, MmGRV, SaGRV, and flying fox retrovirus 1 [FFRV1]) phylogenetically distinct from those from Asia. Although the presence of diverse and basal gammaretroviruses in bats suggests that they are a key reservoir species and may have transmitted viruses to other mammals, it is striking that those viruses sampled from bats (and other mammals) do not share close common ancestry with KoRV in koalas.
Fig. 2.
Evolutionary relationships among KoRV-related viruses. Maximum likelihood phylogeny of the complete (nucleotide) sequence genome of 19 gammaretroviruses. All branches are scaled according to the number of nucleotide substitutions per site, and branches representing bat retroviruses are shown in red. Support for key nodes on the phylogeny are shown in the form of SH-like branch support/bootstrap support. Silhouettes represent the host species: top left, mice; right (in descending order), pteropid bats, koalas, woolly monkeys, microbats, and gibbons. The tree was rooted using the McERV (Mus caroli endogenous retrovirus) KC460271 sequence.
HPG Is Reproduction-Competent in Human and Bat Cells In Vitro.
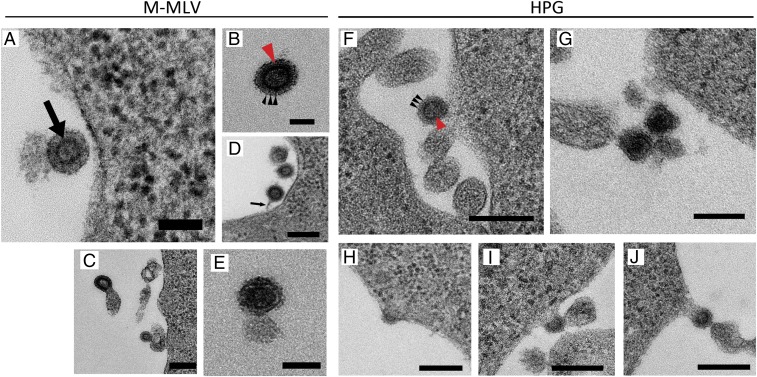
To assess the biological characteristics of KoRV-related bat viruses, we chemically synthesized the proviral genome of HPG (SI Appendix, Fig. S1B). Transfection of human 293T cells with a plasmid construct carrying the HPG provirus resulted in the generation and release of viral particles morphologically similar to ecotropic Moloney murine leukemia virus (M-MLV), as determined by electron microscopy (Fig. 3). In contrast, no virus was observed in untransfected 293T cells and mock control 293T cells that had been transfected with pcDNA3.1 (SI Appendix, Fig. S5). These data are supported by virion-associated reverse-transcriptase analysis of each sample analyzed in concert with electron microscopy analysis (SI Appendix, Fig. S5). Measurements of virion diameters indicate that HPG viral particles are smaller than M-MLV particles (mean ± SEM, 98.5 ± 2.5 nm vs. 130.8 ± 3.2 nm; P < 0.001, Mann–Whitney U test) (SI Appendix, Fig. S6).
Fig. 3.
Electron micrographs of M-MLV and HPG. (A) An extracellular, roughly spherical enveloped virus-like particle with a concentric icosahedral core (arrow). The cores have variable electron translucence ranging from lucent to dense, indicating variable stages of particle maturation. (B) An immature extracellular virus-like particle with tooth-like appearance of the viral envelope (red arrow) surrounding the double-layered shell of the core. Distinct banding can also be seen in the envelope of the particle (black arrowheads). (C) Virus-like particle exiting the cell demonstrating a type C budding profile, characteristic of viruses belonging to the genus Gammaretrovirus (59, 60). (D) Evidence of virus assembly and budding from the plasma membrane of the cell, including the presence of a tether-like structure connecting the cell membrane to the newly budded virus (black arrow). (E) A mature virus-like particle with an electron-dense core encapsulated in an envelope. (F) Immature virus-like particle exhibiting a tooth-like appearance of the viral envelope (black arrows) surrounding the double-layered shell of the core (red arrow). (G) A mature virus-like particle with an electron-dense core encapsulated in an envelope. (H–J) Evidence of virus-like particle assembly and budding from the plasma membrane of the cell. Budding begins with the formation of electron-dense material under the membrane (H), which progresses until the nascent virus-like particle pushes out from the membrane and is pinched off to form a free particle. (Scale bars: 50 nm in A, 100 nm in B, 250 nm in C, 200 nm in D, 100 nm in E, 200 nm in F, 200 nm in G, and 250 nm in H, I, and J.) Negative controls were untransfected cells and cells mock transfected with the empty vector pcDNA3.1. These controls were not observed to contain or produce viral particles (SI Appendix, Fig. S5).
To support our phylogenetic assessment indicating that HPG is a gammaretrovirus, we performed a virion-associated reverse-transcriptase assay using HPG, M-MLV, and HIV virions and including either manganese (Mn2+; utilized by gammaretroviruses) (25) or magnesium (Mg2+; utilized by lentiviruses) (26) as the cofactor for reverse-transcriptase DNA polymerase activity (SI Appendix, Fig. S7). The data show that HPG reverse-transcriptase has a preference for Mn2+ over Mg2+, typical of a gammaretrovirus (25).
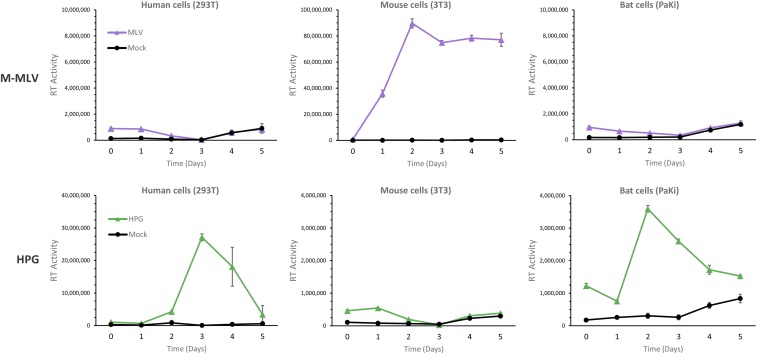
To determine whether the HPG virions generated by proviral-plasmid transfection of human 293T cells are reproduction-competent (i.e., capable of establishing a productive infection within the context of a cell culture system in vitro), we performed a replication kinetics assay (Fig. 4). We found that HPG was capable of entering and establishing a productive infection in human and bat cells, but not mouse cells (Fig. 4). In contrast, the ecotropic M-MLV infected the mouse cell line, but not human or bat cells (Fig. 4). HPG was confirmed to be capable of establishing successive rounds of replication through a secondary infection assay (SI Appendix, Fig. S8).
Fig. 4.
Replication kinetics of M-MLV and HPG in human, mouse, and bat cells. M-MLV and HPG virions were generated by transfection of human 293T cells with pNCS (61) and pCC1-HPG retroviral expression plasmids, respectively, and used to infect human 293T, mouse 3T3, and bat PaKi cell lines. Culture supernatants were collected daily for 5 d and assessed for the presence of virus by measuring virion-associated reverse transcriptase activity. Error bars represent the SEM (n = 6).
HPG Displays a Similar Pattern of Cell Tropism as GALV and KoRV-A.
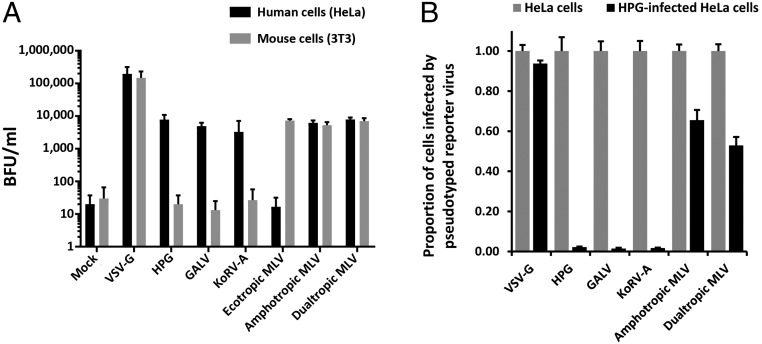
To investigate the cell tropism mediated by the HPG envelope (Env) protein, we performed a viral entry assay in which retroviral particles were pseudotyped with the Env protein of several gammaretroviruses that have distinct tropism for human and mouse cells (Fig. 5A). Our data show that HPG displays a similar pattern of cell tropism as GALV and KoRV-A, in that it is capable of entering human cells but not mouse cells. These data suggest that HPG likely uses the same cellular receptor, PiT-1 (SLC20A1), as GALV and KoRV-A (19, 27, 28).
Fig. 5.
Human and mouse cell tropism of pseudotyped gammaretroviral virus-like particles and HPG-induced resistance to cross-infection. (A) Gammaretrovirus Env or VSV-G pseudotyped retroviral particles, containing the lacZ reporter gene, were generated using the Retro-X packaging system. The infectivity of pseudotyped viral particles was determined in human HeLa cells and mouse 3T3 cells. Infected cells were quantified by counting blue cell-forming units (BFU) following incubation with X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Uninfected cells served as a control (Mock). (B) Human HeLa cells were persistently infected with HPG and then challenged with infection by a reporter virus pseudotyped with gammaretrovirus Env or VSV-G pseudotyped retroviral particles. Infected cells were quantified as in A, and infection in persistently HPG-infected cells is expressed as a proportion of the amount of uninfected HeLa cells infected by the same reporter virus.
Alignment of the binding motif within mammalian PiT-1 genes supports this result, as the binding sites within P. alecto and P. vampyrus PiT-1 share the permissive amino acid residues, which are distinct from the nonpermissive motif within mouse PiT-1 (29) (SI Appendix, Fig. S9). Some gammaretroviruses that use PiT-1 for cell entry also use the related protein, PiT-2; this has been attributed to subtle differences in the composition and length of amino acid sequences within the VRA and VRB regions of the viral Env protein (30, 31).
An alignment of the receptor-binding domain (RBD) (32) within the Env sequence of HPG against other KoRV-related viruses reveals numerous differences in the variable regions (VRA and VRB) within the RBD (SI Appendix, Fig. S10). Within this region, the pathologically important CETTG motif (33), which is conserved in all other bat KoRV-related viruses, contains a threonine-to-serine mutation in HPG, resulting in a CETSG motif. HPG is more similar to GALV than to KoRV across both the VRA and VRB, where the RBD amino acid identities for HPG compared with GALV and KoRV are 66% and 62%, respectively. However, all the KoRV-related bat gammaretroviruses analyzed contain a large insertion within the VRB of 10 and 16 amino acids, respectively, relative to GALV and KoRV. Amphotropic and dualtropic MLV also contain several insertions within the VRB, increasing the length of their VRB region by 17 and 23 amino acids, respectively, relative to GALV and KoRV. These insertions are not present within ecotropic MLV, which uses the mouse CAT1 (SLC7A1) cell receptor (34, 35).
To further investigate receptor use by HPG, we performed a superinfection interference assay (Fig. 5B). In this assay, human HeLa cells persistently infected with HPG (SI Appendix, Fig. S11) became strongly resistant to superinfection with a reporter virus pseudotyped with the envelope proteins of KoRV-A, GALV, or HPG (97.8% to 98.6% reduction in infectivity). Infections with viral particles pseudotyped with dual-tropic or amphotropic MLV Env were also moderately inhibited (34.5% and 47.1% reduction in infectivity, respectively). Dual-tropic MLV uses both PiT-1 and PiT-2 (SLC20A2) cell receptors (36), while amphotropic MLV uses PiT-2 exclusively (37). In contrast, superinfection by particles pseudotyped with the unrelated vesicular stomatitis virus (VSV) envelope G protein was not restricted. These data are consistent with HPG using the PiT-1 and possibly the PiT-2 receptors for cell entry. Taken together, these results indicate that HPG may share a similar host range as KoRV-A and GALV, with the caveat that the specific determinants of receptor use and cell tropism for PiT-1 and PiT-2 are complex (30, 31, 38) and further investigation is needed to more accurately delineate the host range and cell tropism of HPG.
Australian Bats Have Been Exposed to HPG and Closely Related Viruses.
To assess Australian bats for exposure to HPG or KoRV-related viruses, we tested bat sera for the presence of antibodies reactive against the HPG Env protein. We also tested for the presence of HPG-specific nucleic acid in bat fecal samples.
Bat sera (87 samples collected from nine bat species) were screened for the presence of antibodies reactive to the HPG Env trimer ectodomain (Glu38-Ser603) and a synthetic peptide of the HPG VRA region of Env in a solid-phase enzyme immunoassay. For sera reactive to HPG VRA, additional analyses were conducted against VRA peptides from KoRV-A, GALV, and ecotropic MLV (SI Appendix, Fig. S12).
A rabbit immune serum raised to the HPG Env trimer was used as a positive control and to determine cross-reactivity to KoRV, GALV, and MLV peptides. The immune sera reacted strongly to the HPG Env trimer and the HPG VRA peptide sequence but did not show reactivity to KoRV, GALV, or MLV peptides. In addition, immune serum raised to MLV reacted to the MLV peptide sequence but not to the HPG, KoRV, or GALV VRA peptide sequences (SI Appendix, Fig. S12).
Of the 87 bat samples, 27 (31%) showed reactivity to the HPG Env trimer, and of these, 19 (22%) were reactive to the HPG VRA peptide (P. alecto, n = 17; Pteropus conspicillatus, n = 1; Rhinolopus megaphyllus, n = 1). Of the 19 HPG VRA-positive sera, 8 showed additional reactivity to KoRV-A and 4 were also reactive to both KoRV-A and GALV peptides. One serum, #20 P. alecto, was more strongly reactive toward the GALV VRA peptide than toward the HPG or KoRV-A VRA peptide or the HPG Env protein. Two samples, #7 P. alecto and #8 P. alecto, were reactive against the KoRV-A and GALV VRA peptide, respectively, but not against the HPG VRA peptide. No bats demonstrated reactivity to the MLV VRA peptide (SI Appendix, Fig. S12).
These results reveal that 32% of bat samples were seropositive to HPG or other KoRV-related protein sequences. Within the species P. alecto, 83% were seropositive to HPG and/or other KoRV-related protein sequences and 27% were seropositive only to HPG protein sequences.
A nucleic acid analysis by RT-qPCR was performed on 373 bat fecal samples using both “broad” primers designed to amplify the HPG-related pteropid viruses (HPG, SaGRV, MmGRV, and FFRV1) (Fig. 2) and “specific” primers designed to amplify only HPG (SI Appendix, Table S3). Notably, the HPG-specific forward primer binds to a site that is not present (i.e., lost through a deletion event) within the closely related ERV FFRV1 (14) and contains multiple nucleotide differences at the 3′ end of the primer compared with MmGRV and SaGRV. We first performed the RT-qPCR assay in such a way that both DNA and RNA would be amplified. This was followed by a second RT-qPCR assay performed in the absence of reverse transcriptase so that only DNA could be amplified, allowing us to discriminate between amplification from retroviral DNA and RNA. Of note, this analysis does not discriminate between germ line and somatic viral genomic DNA. The results of the first RT-qPCR assay revealed that 57 of 373 samples (15.3%) contained HPG-related nucleic acid (either DNA or RNA), and that 25 of those 57 (6.7% of the total) contained HPG-specific nucleic acid (SI Appendix, Table S4). In the second RT-qPCR assay, all 25 HPG-specific samples were revealed to have been amplified from RNA (SI Appendix, Table S4), suggesting active infection with HPG. Interestingly, only 13 of the 25 samples were positive for HPG-related RNA.
These data indicate that the remaining 12 of the 25 samples were positive for HPG-specific RNA but not for “broad” HPG-related RNA and instead were positive for “broad” HPG-related DNA. While we cannot rule out the possibility that some or all of these samples may have contained a quantity of HPG-related RNA below the limit of detection of this assay, these data suggest that 12 bat samples were actively infected with HPG and were either latently infected with other HPG-related viruses or contained endogenous HPG-related sequences. Of the 57 samples positive for HPG-related nucleic acid, 32 were positive only for HPG-related DNA, suggesting evidence of endogenization or latent infection with HPG-related viruses. Taken together, these serologic and PCR results indicate that HPG and closely related viruses have infected multiple individuals across several species of Australian pteropid bats.
Discussion
To determine whether KoRV-related viruses are present in Australian bats, we collected samples from bats along the east coast of Australia. Metagenomic analyses of these samples revealed the presence of three KoRV-related viruses— HPG, MmGRV, and SaGRV—from the pteropid bat species P. alecto, M. minimus, and S. australis. A search of the public SRA (39) also revealed two additional KoRV-related viruses, HlGRV and RhGRV, from the Asian microbat species H. larvatus and R. hipposideros. Phylogenetic analysis of the complete viral genome (Fig. 2) revealed that the microbat viruses (HlGRV and RhGRV) clustered within a broad GALV/woolly monkey virus (WMV) clade, while the pteropid viruses (HPG, FFRV1, MmGRV, and SaGRV) form a more divergent clade that is basal to the KoRV and GALV/WMV clades. There is overlap between the habitats of all of the aforementioned bats except R. hipposideros (with habitats between Europe and West Asia) (22). Thus, in theory bat communities could provide a route of transmission for KoRV-related viruses between Asia and Australia, although the immediate ancestor of KoRV remains uncertain, and additional animal species need to be sampled. Indeed, there are likely to be other currently unidentified species infected with KoRV-related viruses linking the habitats of R. hipposideros and Australian bats. The long phylogenetic branch length linking the KoRV clade to its closest known relatives in the GALV/WMV clade indicates that the phylogenetic picture remains incomplete, with additional, as-yet unknown viruses and host species existing between the KoRV and GALV/WMV lineages of gammaretroviruses.
Other non-bat species, particularly rodents, have been suggested as intermediary hosts for the transmission of KoRV-related viruses between Asia and Australia (20, 21). Of particular note is Melomys burtoni, an Australian rodent. Short nucleotide sequences representing KoRV-related viruses, including the M. burtoni retrovirus (MbRV) and the Melomys woolly monkey virus (MelWMV), have been identified in M. burtoni (20, 40), both of which cluster closely with the WMV within the GALV clade and thus are no closer to KoRV than the bat viruses identified here (Fig. 2) (20, 40); sequences of these viruses were omitted from our phylogenetic analysis due to insufficient genome sequence coverage. However, because the habitat of M. burtoni does not extend past the Wallace line or overlap with the habitat of gibbons (19, 22), this species is unlikely to be responsible for the direct transmission of KoRV-related viruses between Australia and Asia.
KoRV and GALV use the PiT-1 receptor for cell entry (19, 27, 28). This receptor is almost ubiquitously expressed throughout the mammalian body at variable levels (41–44) and is highly expressed in many tissues, including the colon, breast, testes, bladder, placenta, and brain (41, 42). KoRV and GALV have been detected in numerous tissues and body fluids, including blood, sperm, breast milk, feces, and urine (5, 27, 45–50). Given the wide distribution of PiT-1 expression and the detection of KoRV and GALV in body fluids including blood, urine, and feces, it is possible that interspecies transmission might occur along routes including blood during fighting/predation and contamination of food sources by feces and urine.
We carefully searched for the presence of HPG in the genomes of P. alecto and P. vampyrus using molecular analyses and more broadly for KoRV-related viruses in the SRA, but were unable to detect these viral sequences in any currently available bat genome sequence. While these data suggest that bat KoRV-related viruses are not endogenous, we cannot exclude this possibility, as we have sampled only a small proportion of bats within each species. In this regard, KoRV endogenization in koalas is relatively recent and, accordingly, is not represented across the entire koala gene pool (45), existing in both endogenous and exogenous forms (5, 27, 51). Thus, given that HPG-specific sequences have been identified across several bat species, HPG is either an exogenous virus or is undergoing endogenization in real time. A possible example of the latter is FFRV1 (14), which was recently discovered in the brain tissue of a P. alecto bat but has not been identified within the genome of P. alecto or other bats. Serologic and nucleic acid analyses revealed that numerous individual bats across several species had been exposed to HPG and HPG-related viruses (SI Appendix, Fig. S12 and Table S4), and that 6.7% (25 of 373) of the bat scat samples analyzed contained HPG-specific RNA, suggesting that these bats were actively infected with HPG and, more generally, that HPG-related viruses are currently circulating among the communities of multiple species of Australian pteropid bats. The close evolutionary relationships among the bat KoRV-related viruses from several species of Australasian pteropid bats, Asian microbats, and the gibbon, koala, and rodent viruses suggest that bat populations in Australia and Asia play an important role in the transmission of KoRV-related viruses between bats and possibly other mammals.
The genome of HPG is typical of gammaretroviruses (Fig. 1), and while HPG virions are morphologically similar to M-MLV virions, possessing a spherical, electron-dense core (Fig. 3), measurements revealed a smaller HPG virion diameter compared with M-MLV virions (P < 0.001; SI Appendix, Fig. S6). This may be attributed to the smaller diameter of the viral core (P < 0.001). These data may indicate a difference in the quaternary structure of the HPG capsid compared with that of M-MLV, resulting in a more compact structure.
We generated HPG virions from a synthetic proviral expression construct to assess the reproduction capacity of HPG in cell culture. These HPG virions were capable of infecting human and bat cell lines (Fig. 4) but not a mouse cell line, as shown by the production of new virions and their release into the cell culture supernatant over the course of several days. It is important to note that while the complete genome of HPG was assembled from RNA extracted from a single bat, and virions generated from this sequence are reproduction-competent in vitro, the infectious molecular clone was engineered from the consensus sequence of the assembled reads. Accordingly, the HPG molecular clone represents the average of the HPG population contained in the extracted RNA, rather than the exact sequence of a single viral isolate. Similar consideration should be given to the other bat KoRV-related viruses reported here, which also represent the consensus of assembled sequence data.
An assessment of the cell tropism of HPG revealed that HPG Env-pseudotyped retroviral particles were able to enter human cells, but not mouse cells (Fig. 5A). GALV and KoRV-A are similarly restricted from entering mouse NIH 3T3 cells due to their use of the PiT-1 cellular receptor for viral entry (28). This inhibition is attributed to mouse PiT-1 containing differences in the binding sites of GALV and KoRV-A (29, 52) that are not present in P. alecto or P. vampyrus PiT-1 (SI Appendix, Fig. S7).
Infection of cells with a retrovirus can restrict the subsequent superinfection by viruses that use the same receptor by various mechanisms, including down-regulation of the receptor and blocking of the binding site on the cell receptor, preventing penetration or adsorption of the virus (53, 54). This method has been used to demonstrate the shared use of the PiT-1 receptor between KoRV-A and GALV (55). We performed a superinfection interference assay that demonstrated that infection with HPG restricts superinfection by a reporter virus pseudotyped with the envelope protein of KoRV-A, GALV, amphotropic MLV, and dual-tropic MLV. KoRV-A and GALV use the PiT-1 receptor (19, 27, 28), while amphotropic MLV uses PiT-2 (37) and dual-tropic MLV uses both PiT-1 and PiT-2 (36). These results are consistent with HPG using the PiT-1 receptor and possibly the PiT-2 receptor for cell entry.
Variations in receptor use can occur between closely related gammaretroviruses. For example, KoRV-B, although closely related to KoRV-A, uses the THTR1 receptor (56), possibly as the result of a recombination event within the RBD between an ancestral KoRV and an unknown retrovirus (27). This is particularly important to consider in light of the alignment of the RBD of HPG and other bat KoRV-related viruses (SI Appendix, Fig. S6), which reveals a large insertion within the hypervariable VRB region. Amphotropic and dual-tropic MLVs similarly contain a large insertion within the VRB relative to KoRV, GALV, and ecotropic MLV. The VRB region of amphotropic MLV is essential for interaction with the PiT-2 cell receptor (57), and the large insertion within the VRB of HPG may be involved in its possible use of the PiT-2 receptor demonstrated by the superinfection assay.
Interestingly, HPG contains a modification within the CETTG motif within the RBD (SI Appendix, Fig. S10) that is important for viral pathogenicity (33). Mutations within the CETTG attenuate cytopathology, as is the case for KoRV-A, which has a CETAG motif (58). HPG contains a CETSG motif (SI Appendix, Fig. S10), which is also found in 27% of KoRV-D proviruses and is hypothesized to attenuate syncytia formation-related pathogenicity (58). However, other bat KoRV-related viruses analyzed in this study have the pathogenic CETTG motif. The identification of bats as a source of infectious retroviruses related to KoRV and GALV implicates bats as a reservoir of KoRV-related viruses that can potentially be transmitted between Australia and Asia to other mammalian species.
Materials and Methods
Supplemental figures, tables, and details of the materials and methods used in this study, including all experimental procedures, are provided in SI Appendix.
Data Availability.
All data are included in the main text and the SI Appendix file. Sequences of HPG, MmGRV, RhGRV, SaGRV, and HlGRV have been deposited in the GenBank database; accession numbers are provided in SI Appendix, Table S2.
Supplementary Material
Acknowledgments
We thank the NIH AIDS Research and Reference Reagent program for providing HeLa cells, Damian Purcell for providing NIH 3T3 cells, and Richard Axel for providing HEK 293T cells. We also thank Amy Burroughs, Bronwyn Clayton, Huajun Zhang, Jennifer Barr, Johanna Dups, Kate Baker, Peng Zhou, Shawn Todd, Hugh Spencer, and Andreas Kurth for their roles in the collection of bat samples; Vicky Boyd for her assistance with the serologic analysis; and Reuben Klein for extracting viral nucleic acids from bat samples. We wish to acknowledge Microscopy Australia and the National Collaborative Research Infrastructure Strategy for funding the Commonwealth Scientific and Industrial Research Organisation (CSIRO) electron microscopy capability used in this study, and the Pathology and Pathogenesis team at the Australian Animal Health Laboratory (CSIRO) for their technical assistance. We also gratefully acknowledge the contribution to this work of the Victorian Operational Infrastructure Support Program received by the Burnet Institute. This work was supported by National Health and Medical Research Council (NHMRC) Grant GNT1121077 (to G.T., M.L.B., and G.A.M.) and NHMRC Senior Research Fellowship GNT1117748 (to G.T.). J.A.H. was funded by NHMRC Grant GNT1121077. H.E.D. is supported by NHMRC Grants GNT1041897 and GNT1146082. E.C.H. is supported by the Australian Research Council’s Australian Laureate Fellowship FL170100022. L-F.W. is funded by Singapore National Research Foundation Grants NRF2012NRF-CRP001-056 and NRF2016NRF-NSFC002-013.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Sequences of HPG, MmGRV, RhGRV, SaGRV, and HIGRV have been deposited in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/); accession numbers are provided in SI Appendix, Table S2.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915400117/-/DCSupplemental.
References
- 1.Vogt V. M., “Retroviral virions and genomes” in Retroviruses, Coffin J. M., Hughes S. H., Varmus H. E., Eds. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1997). [PubMed] [Google Scholar]
- 2.Gao F., et al. , Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436–441 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Tarlinton R., Meers J., Hanger J., Young P., Real-time reverse transcriptase PCR for the endogenous koala retrovirus reveals an association between plasma viral load and neoplastic disease in koalas. J. Gen. Virol. 86, 783–787 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Ito N., et al. , Clinical and pathological characteristics of acute myelogenous leukemia in a female koala with diabetes mellitus. J. Vet. Med. Sci. 81, 1229–1233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarlinton R. E., Meers J., Young P. R., Retroviral invasion of the koala genome. Nature 442, 79–81 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Stoye J. P., Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 10, 395–406 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Herniou E., et al. , Retroviral diversity and distribution in vertebrates. J. Virol. 72, 5955–5966 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke J. D., Symer D. E., “An everlasting war dance between retrotransposons and their metazoan hosts” in Retroviruses, Kurth R., Bannert N., Eds. (Caister Academic Press, Berlin, 2010), pp. 1–34. [Google Scholar]
- 9.Griffiths D. J., Endogenous retroviruses in the human genome sequence. Genome Biol. 2, reviews1017 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayman D. T., Bats as viral reservoirs. Annu. Rev. Virol. 3, 77–99 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Hayward J. A., et al. , Identification of diverse full-length endogenous betaretroviruses in megabats and microbats. Retrovirology 10, 35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui J., et al. , Identification of diverse groups of endogenous gammaretroviruses in mega- and microbats. J. Gen. Virol. 93, 2037–2045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farkašová H., et al. , Discovery of an endogenous Deltaretrovirus in the genome of long-fingered bats (Chiroptera: Miniopteridae). Proc. Natl. Acad. Sci. U.S.A. 114, 3145–3150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMichael L., et al. , A novel Australian flying-fox retrovirus shares an evolutionary ancestor with koala, gibbon, and Melomys gamma-retroviruses. Virus Genes 55, 421–424 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Cui J., Tachedjian G., Wang L.-F., Bats and rodents shape mammalian retroviral phylogeny. Sci. Rep. 5, 16561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayward A., Grabherr M., Jern P., Broad-scale phylogenomics provides insights into retrovirus-host evolution. Proc. Natl. Acad. Sci. U.S.A. 110, 20146–20151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayward J. A., et al. , Differential evolution of antiretroviral restriction factors in pteropid bats as revealed by APOBEC3 gene complexity. Mol. Biol. Evol. 35, 1626–1637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riddle B. R., Hafner D. J., Integrating pattern with process at biogeographic boundaries: The legacy of Wallace. Ecography 33, 321–325 (2010). [Google Scholar]
- 19.Denner J., Transspecies transmission of gammaretroviruses and the origin of the gibbon ape leukaemia virus (GALV) and the koala retrovirus (KoRV). Viruses 8, 336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simmons G., Clarke D., McKee J., Young P., Meers J., Discovery of a novel retrovirus sequence in an Australian native rodent (Melomys burtoni): A putative link between gibbon ape leukemia virus and koala retrovirus. PLoS One 9, e106954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denner J., Young P. R., Koala retroviruses: Characterization and impact on the life of koalas. Retrovirology 10, 108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International Union for the Conservation of Nature , The IUCN Red List of Threatened Species 2016, version 2016-1. https://www.iucn.org/zh-hans/node/24442. Accessed 17 March 2019.
- 23.Cui J., et al. , Discovery of retroviral homologs in bats: Implications for the origin of mammalian gammaretroviruses. J. Virol. 86, 4288–4293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crameri G., et al. , Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS One 4, e8266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaniv A., et al. , Biochemical characterization of the type C retrovirus associated with lymphoproliferative disease of turkeys. J. Virol. 30, 351–357 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Billamboz M., et al. , Magnesium chelating 2-hydroxyisoquinoline-1,3(2H,4H)-diones, as inhibitors of HIV-1 integrase and/or the HIV-1 reverse-transcriptase ribonuclease H domain: Discovery of a novel selective inhibitor of the ribonuclease H function. J. Med. Chem. 54, 1812–1824 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Xu W., et al. , An exogenous retrovirus isolated from koalas with malignant neoplasias in a US zoo. Proc. Natl. Acad. Sci. U.S.A. 110, 11547–11552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Hara B., et al. , Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1, 119–127 (1990). [PubMed] [Google Scholar]
- 29.Schneiderman R. D., Farrell K. B., Wilson C. A., Eiden M. V., The Japanese feral mouse Pit1 and Pit2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: Implications for virus binding motif. J. Virol. 70, 6982–6986 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boomer S., Eiden M., Burns C. C., Overbaugh J., Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J. Virol. 71, 8116–8123 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugai J., et al. , Identification of envelope determinants of feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 or Pit2. J. Virol. 75, 6841–6849 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fass D., et al. , Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 Angstrom resolution. Science 277, 1662–1666 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Oliveira N. M., Satija H., Kouwenhoven I. A., Eiden M. V., Changes in viral protein function that accompany retroviral endogenization. Proc. Natl. Acad. Sci. U.S.A. 104, 17506–17511 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albritton L. M., Tseng L., Scadden D., Cunningham J. M., A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57, 659–666 (1989). [DOI] [PubMed] [Google Scholar]
- 35.Yoshimoto T., Yoshimoto E., Meruelo D., Identification of amino acid residues critical for infection with ecotropic murine leukemia retrovirus. J. Virol. 67, 1310–1314 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller A. D., Chen F., Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J. Virol. 70, 5564–5571 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feldman S. A., Farrell K. B., Murthy R. K., Russ J. L., Eiden M. V., Identification of an extracellular domain within the human PiT2 receptor that is required for amphotropic murine leukemia virus binding. J. Virol. 78, 595–602 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han J.-Y., Zhao Y., Anderson W. F., Cannon P. M., Role of variable regions A and B in receptor binding domain of amphotropic murine leukemia virus envelope protein. J. Virol. 72, 9101–9108 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Z., et al. , Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 86, 10999–11012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alfano N., et al. , An endogenous gibbon ape leukemia virus (GALV) identified in a rodent (Melomys burtoni subsp.) from Wallacea (Indonesia). J. Virol. 90, 8169–8180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petryszak R., et al. , Expression Atlas update—an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 44, D746–D752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bastian F., et al. , “Bgee: Integrating and comparing heterogeneous transcriptome data among species” in International Workshop on Data Integration in the Life Sciences, A. Bairoch, S. Cohen-Boulakia, C. Froidevaux, Eds., (Springer, Berlin, Heidelberg, 2008), pp. 124–131. [Google Scholar]
- 43.Johann S. V., Gibbons J. J., O’Hara B., GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J. Virol. 66, 1635–1640 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kavanaugh M. P., et al. , Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc. Natl. Acad. Sci. U.S.A. 91, 7071–7075 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simmons G. S., et al. , Prevalence of koala retrovirus in geographically diverse populations in Australia. Aust. Vet. J. 90, 404–409 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Waugh C. A., et al. , Infection with koala retrovirus subgroup B (KoRV-B), but not KoRV-A, is associated with chlamydial disease in free-ranging koalas (Phascolarctos cinereus). Sci. Rep. 7, 134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris K. M., et al. , Characterisation of the immune compounds in koala milk using a combined transcriptomic and proteomic approach. Sci. Rep. 6, 35011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wedrowicz F., Saxton T., Mosse J., Wright W., Hogan F. E., A non-invasive tool for assessing pathogen prevalence in koala (Phascolarctos cinereus) populations: Detection of Chlamydia pecorum and koala retrovirus (KoRV) DNA in genetic material sourced from scats. Conserv. Genet. Resour. 8, 511–521 (2016). [Google Scholar]
- 49.Kawakami T. G., Sun L., McDowell T. S., Infectious primate type-C virus shed by healthy gibbons. Nature 268, 448–450 (1977). [DOI] [PubMed] [Google Scholar]
- 50.Kawakami T. G., Sun L., McDowell T. S., Natural transmission of gibbon leukemia virus. J. Natl. Cancer Inst. 61, 1113–1115 (1978). [PubMed] [Google Scholar]
- 51.Miyazawa T., Shojima T., Yoshikawa R., Ohata T., Isolation of koala retroviruses from koalas in Japan. J. Vet. Med. Sci. 73, 65–70 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Farrell K. B., Russ J. L., Murthy R. K., Eiden M. V., Reassessing the role of region A in Pit1-mediated viral entry. J. Virol. 76, 7683–7693 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steck F. T., Rubin H., The mechanism of interference between an avian leukosis virus and Rous sarcoma virus, II: Early steps of infection by RSV of cells under conditions of interference. Virology 29, 642–653 (1966). [DOI] [PubMed] [Google Scholar]
- 54.Nethe M., Berkhout B., van der Kuyl A. C., Retroviral superinfection resistance. Retrovirology 2, 52 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oliveira N. M., Farrell K. B., Eiden M. V., In vitro characterization of a koala retrovirus. J. Virol. 80, 3104–3107 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shojima T., et al. , Identification of a novel subgroup of koala retrovirus from koalas in Japanese zoos. J. Virol. 87, 9943–9948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tailor C. S., Kabat D., Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J. Virol. 71, 9383–9391 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quigley B. L., et al. , Changes in endogenous and exogenous koala retrovirus subtype expression over time reflect koala health outcomes. J. Virol. 93, e00849–e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gelderblom H. R., Fine Structure of HIV and SIV (Los Alamos Laboratory, Los Alamos, NM, 1997). [Google Scholar]
- 60.Lefkowitz E. J., et al. , Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 46, D708–D717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bacharach E., Gonsky J., Lim D., Goff S. P., Deletion of a short, untranslated region adjacent to the polypurine tract in Moloney murine leukemia virus leads to formation of aberrant 5′ plus-strand DNA ends in vivo. J. Virol. 74, 4755–4764 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the main text and the SI Appendix file. Sequences of HPG, MmGRV, RhGRV, SaGRV, and HlGRV have been deposited in the GenBank database; accession numbers are provided in SI Appendix, Table S2.