Significance
As the terminal electron acceptor of our mitochondrial respiratory chains, complex IV drives and regulates oxidative phosphorylation, the process by which most of our ATP is produced. Complex IV forms supercomplexes (SCs) of different stoichiometries with other respiratory proteins, interacting via its subunits with tissue-specific or oxygen level-dependent expression isoforms, suggesting a link between SC assembly and metabolic/disease state. We investigated the effect of complex IV subunit isoform exchange in yeast using cryo-EM and biochemical assays and found no significant differences in overall SC formation, architecture, or catalytic activities. However, our structural work unexpectedly revealed the presence of a Hig1 protein which we propose is a stoichiometric subunit of complex IV, at least when within a SC with complex III.
Keywords: bioenergetics, electron transport chain, cytochrome c oxidase, Hig1 proteins, respiratory supercomplexes
Abstract
The organization of the mitochondrial electron transport chain proteins into supercomplexes (SCs) is now undisputed; however, their assembly process, or the role of differential expression isoforms, remain to be determined. In Saccharomyces cerevisiae, cytochrome c oxidase (CIV) forms SCs of varying stoichiometry with cytochrome bc1 (CIII). Recent studies have revealed, in normoxic growth conditions, an interface made exclusively by Cox5A, the only yeast respiratory protein that exists as one of two isoforms depending on oxygen levels. Here we present the cryo-EM structures of the III2-IV1 and III2-IV2 SCs containing the hypoxic isoform Cox5B solved at 3.4 and 2.8 Å, respectively. We show that the change of isoform does not affect SC formation or activity, and that SC stoichiometry is dictated by the level of CIII/CIV biosynthesis. Comparison of the CIV5B- and CIV5A-containing SC structures highlighted few differences, found mainly in the region of Cox5. Additional density was revealed in all SCs, independent of the CIV isoform, in a pocket formed by Cox1, Cox3, Cox12, and Cox13, away from the CIII–CIV interface. In the CIV5B-containing hypoxic SCs, this could be confidently assigned to the hypoxia-induced gene 1 (Hig1) type 2 protein Rcf2. With conserved residues in mammalian Hig1 proteins and Cox3/Cox12/Cox13 orthologs, we propose that Hig1 type 2 proteins are stoichiometric subunits of CIV, at least when within a III-IV SC.
To live, we need a permanent supply of energy and most of this is provided to our cells in the form of adenosine triphosphate (ATP). ATP is continuously synthesized in our mitochondria by the action of five large protein complexes, named complexes I-V (CI-V), in a process called oxidative phosphorylation. CI-IV reduce the oxygen we breathe using electrons provided by the food we eat and store the energy of the reaction as a transmembrane proton motive force that drives ATP synthesis by CV (1). Despite earlier assumptions that CI-V were randomly distributed within the inner mitochondrial membrane (IMM), it is now widely accepted that they form higher-order structures in vivo (2, 3). The arrangement of CV in rows of V-shaped dimers has been linked to membrane bending and cristae formation (4–6), and a functional role for CV tetramers has recently been suggested (7). However, the roles of the supercomplexes (SCs) formed by CI-IV, as well as their assembly process, remain to be established (8–10).
Major complications in assigning functional roles to SCs are the heterogeneity of SC populations that coexist in the IMM and the inherent difficulty in separating SC assembly intermediates from their mature forms. This is further complicated by the presence of tissue-specific isoforms of the respiratory complexes, which may only be assembled into SCs in specific conditions. Several high-resolution structures of CI-containing mammalian SCs obtained by single-particle cryogenic electron microscopy (cryo-EM) are available. These include structures of “respirasomes” formed by CI, III, and IV in varying stoichiometries (11–14) and I–III2 SCs (11, 14). Overall, they reveal a conserved arrangement of CI and CIII across species, including strong protein–protein interactions. Slightly different orientations of CIII relative to CI within the respirasome have been reported that have been linked to instability of the SC during the purification procedure, although the possibility of a subpopulation of respirasomes in which CI is in the deactivated state could not be excluded (9, 14). However, when present, CIV was found in very different positions, making it more difficult to determine any conserved pattern of interaction. Similarly in the bovine, porcine, and tight ovine respirasome structures, CIV interacts with CIII via subunit COX7A (11, 12, 14, 15), albeit in very different positions, whereas in the loose ovine and human structures, the same CIV subunit interacts with CI instead (11, 13).
The existence of homologous proteins for COX7A may provide a rationale for the different CIV orientation/COX7A interactions observed. COX7A1 and COX7A2 are differentially expressed isoforms, and a third is the SC assembly factor 1 (SCAF1) incriminated in III-IV SC assembly/stability (16, 17). Their coexistence in varying ratios in different tissue types might have contributed to the difficulty in identifying specific isoforms in most of the reported structures, with only COX7A2 assigned with confidence in respirasomes purified from human epithelial kidney cells (18), and the possibility that different COX7A isoforms assemble into different SCs cannot be excluded. To determine whether the observed heterogeneity of SC structures are artifacts of SC purification or cryo-EM sample preparation, or whether physiologically relevant isoforms of the mammalian respirasome mediate changes in SC stoichiometry/architecture, new methods are needed that selectively isolate specific SC isoforms and SC assembly intermediates.
Saccharomyces cerevisiae is an ideal system for investigating mitochondrial respiratory protein organization. It lacks CI, but its CII-V are remarkably similar to its mammalian counterparts in terms of subunit composition, existence of isoforms and SC association, and the genetic amenability of its nuclear and mitochondrial DNA allows alterations to any part of the respiratory proteins to comprehensively test hypotheses. The structures of the mitochondrial III2-IV2 and III2-IV1 SCs formed in S. cerevisiae have recently been published (19, 20); however, the reason for the existence of two different SCs remains to be determined. The mild detergents used to purify the yeast SCs allowed the first full structure determination of both CIII and CIV at atomic resolution, including Qcr10 of CIII and Cox26 of CIV, and their presence was also confirmed by mass spectrometry (19). The EM maps revealed similar interactions between CIII and CIV, irrespective of the III-IV stoichiometry, with an interface formed exclusively by subunit Cox5A of CIV. Curiously, Cox5 is the only subunit in yeast that exists as one of two isoforms depending on oxygen levels (21), Cox5A or Cox5B, expressed in normoxic or hypoxic conditions, respectively. Despite different CIV subunits interacting with CIII within the mammalian respirasomes (COX7A) and the yeast III-IV SCs (Cox5), in all structures the CIII–CIV interface is formed by a CIV subunit with isoforms that have also been implicated in modulation of CIV activity (22–24).
Along with the respiratory complexes that form the mitochondrial SCs, several proteins have been implicated to have roles in SC assembly or stability; however, no SC structure has been solved with an assembly factor/scaffold protein present, so these roles remain undefined. In yeast, the proteins most closely associated with SC assembly are the respiratory SC factors Rcf1-3 (25–28). Mass spectrometry revealed the presence of both Rcf1 and Rcf2 in the purified III2-IV2 SC sample used for structure determination, although no density for additional proteins could be seen in the EM maps published (19).
In this study, we used three yeast variants that differentially assemble CIV with its Cox5A or Cox5B isoform and investigated the effect of CIV isoform exchange on SC formation, structure, and activity. We found that the change of Cox5 isoform did not affect III-IV SC formation or their overall CIII-CIV arrangement, but that the III-IV SC stoichiometry was governed by the varying CIV expression levels seen in the different strains used. Atomic models of the different assemblies revealed few differences between them, although additional density was revealed in all III-IV SCs, independent of the isoform. In the two SCs containing CIV5B, this could be confidently assigned to the C-terminal fragment of Rcf2, calling into question the proposed role of Rcf2 as a late CIV assembly factor (26, 29, 30) and suggesting that it is a stoichiometric subunit of CIV within the III-IV SCs.
Results
Yeast Strains.
When cultured in normoxic conditions, S. cerevisiae wild-type (WT) mainly assembles CIV with the Cox5A isoform. However, a small proportion of Cox5B is also expressed, leading to a heterogeneous population of CIV and, as a result, a heterogeneous population of III-IV SCs. To avoid this and ensure SC homogeneity, we used a series of S. cerevisiae mutant strains that selectively express Cox5A or Cox5B (23). In Δcox5b, the gene encoding the Cox5B protein was knocked out from the nuclear genome so that it only assembled CIV with Cox5A (CIV5A). Two mutant strains were used to produce CIV with Cox5B (CIV5B). The first strain was a combination knockout of Δcox5a Δrox1 (henceforth referred to as Δrox1) in which the genes encoding Cox5A and Rox1, a repressor of COX5B expression, were knocked out, with the effect of elevating the expression level of Cox5B when cultured in normoxic conditions. This resulted in assembly of slightly higher amounts of CIV5B compared with a single Δcox5a knockout mutant, albeit still at much lower levels than CIV5A in the WT (or Δcox5b) strain. In the second mutant strain, cox5ab, both the COX5A and COX5B genes were knocked out, and the sequence encoding Cox5B was placed downstream of the COX5A promoter, leading to the assembly of CIV5B at CIV5A WT levels, with no additional change. All mutant strains carried an additional 6-histidine tag on the C-terminal domain of Cox13 for facile CIV/SC purification as described previously (31).
The III-IV SC Stoichiometry Is Governed by the Relative Amounts of CIII and CIV within the Mitochondrial Membrane.
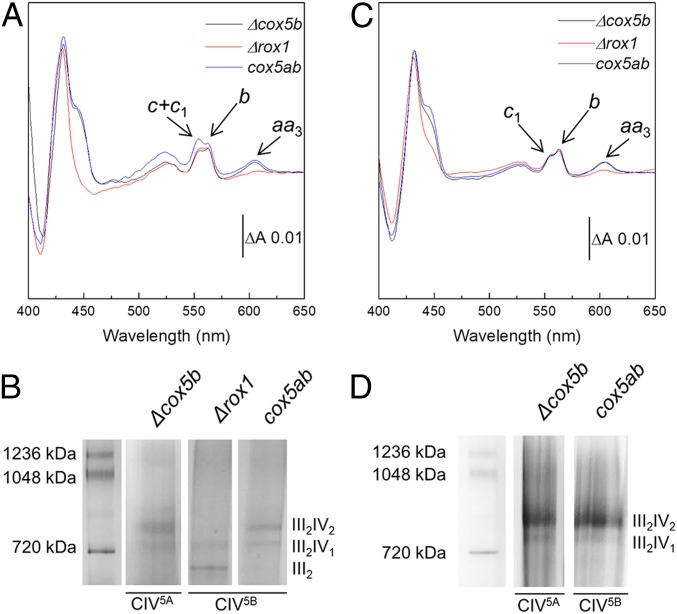
The three yeast variants were cultured under the same conditions and harvested in the late exponential phase of growth, and mitochondrial membranes were prepared as described in Materials and Methods. CIII and CIV were then quantitated in each preparation from the specific absorption of their B- and A-type hemes, respectively, in dithionite-reduced minus oxidized difference spectra, recorded in the visible range (Fig. 1A). Very similar difference spectra were recorded for all three mutant strains, with only the ratio of CIII over that of CIV differing between strains. The Δcox5b and cox5ab strains displayed a CIII:CIV ratio in mitochondrial membranes of close to 3, whereas in Δrox1, this ratio was estimated to be as high as 12. This is a consequence of the low level of Cox5B in Δrox1 and the resulting low CIV5B abundance in the membrane, with the amount of CIII being constant across the three strains.
Fig. 1.
The stoichiometry of the III-IV SC is governed by the level of CIV biogenesis. (A) Reduced minus oxidized visible absorption spectra recorded on mitochondrial membrane preparations from all three S. cerevisiae strains. The absorption bands specific to cyt aa3 of CIV (445 and 604 nm), cyt b of CIII (432 and 562 nm), and cyt c1 of CIII and cyt c (554 nm) are indicated with arrows. Traces are normalized on their ΔA562–578 nm. (B) BN-PAGE gel of solubilized mitochondrial membranes from all three S. cerevisiae strains. (C) Reduced minus oxidized visible absorption spectra recorded on nickel-affinity chromatography eluates from all three S. cerevisiae strains. Absorption bands are as in A. (D) BN-PAGE gel of the nickel-affinity chromatography eluates obtained from those S. cerevisiae strains containing both III2-IV2 and III2-IV1 SCs.
To assess whether this change in CIII:CIV ratio had an impact on SC formation, the mitochondrial membranes from each strain were solubilized with the mild detergent glyco-diosgenin (GDN), and the migration profiles of the resulting respiratory assemblies were observed using blue native (BN)-PAGE (Fig. 1B). III2-IV2 and III2-IV1 SCs were detected in both Δcox5b and cox5ab mitochondrial membranes, with approximately equal contributions, but only III2-IV1 SCs appeared to have formed in Δrox1. This observation and the fact that Δrox1 contained only one-fourth of the mature CIV contained in the two other strains suggest that the protein stoichiometry of the III-IV SCs is governed by the relative amounts of CIII and CIV within the mitochondrial membrane, independent of whether CIV has assembled with the Cox5A (Δcox5b) or Cox5B (cox5ab and Δrox1) isoform. It is noteworthy that a significant amount of free III2 was detected in Δrox1 mitochondrial membranes, whereas none could be seen in the two other strains (Fig. 1B).
Selection of SCs Containing Fully Assembled CIV.
The III-IV SCs from all three mutant strains were then purified by metal affinity chromatography, exploiting a 6-histidine tag on Cox13 of CIV, as previously described for Δcox5b (19). In the case of Δcox5b and cox5ab, the BN-PAGE migration profiles of the resulting chromatography eluates highlighted a change in the relative contributions of the III2-IV2 and III2-IV1 forms of the SC from an estimated 50:50 to 80:20 after metal affinity chromatography (Fig. 1D). This shift in contribution most likely arises from the biased selection of the SCs with two copies of CIV, due to the presence of two copies of the histidine-tagged subunit Cox13.
It has recently been proposed that CIII could serve as a platform for the completion of CIV biogenesis (32). As Cox13 is one of the last subunits (along with Cox12) to assemble into CIV (33), it is interesting to speculate that the metal affinity chromatography step could have promoted the selection of SCs containing mature forms of CIV (i.e., those CIVs that have at least assembled Cox13) and thus maximized the homogeneity of the SC populations.
Purified III-IV SCs Are Active but Do Not Differ in Their Catalytic Activity.
The III-IV5B SCs from Δrox1 and cox5ab were further purified by gel filtration chromatography (SI Appendix, Fig. S1). The final samples were active and reduced the molecular oxygen in the presence of exogenous cytochrome c at a rate of 10.8 ± 1.4 e.s−1 for Δrox1 (n = 3) and 9.7 ± 0.2 e.s−1 for cox5ab (n = 3), using decylubiquinol as a substrate. Compared with the rate of 10.3 ± 0.9 e.s−1 previously reported for the III2-IV5A2 Δcox5b SC (19), this suggests similar activity of the three SCs tested, independent of the presence of CIV5A or CIV5B or of the CIII:CIV stoichiometry. All rates were expressed as turnover numbers (TNs) relative to the concentration of the rate-limiting CIII.
The CIII–CIV Interface Is Unaffected by the Change of the Cox5 Isoform in CIV.
The purified III2-IV5B1 Δrox1 and III2-IV5B2 cox5ab SCs were prepared for structural analysis by cryo-EM. Cryo-EM maps (SI Appendix, Figs. S2 and S3 and Table S1) were obtained at 3.4-Å resolution for Δrox1 and at 2.8-Å resolution for cox5ab and revealed a CIII dimer flanked by either one or two copies of CIV in the III2-IV5B1 or the III2-IV5B2 SC. Apart from the change in CIII:CIV stoichiometry, the overall interaction of CIII and CIV5B appeared to be very similar to that in the III-IV5A SCs reported previously (19, 20).
As previously described for the III2-IV5A2 SC (19), the resolution of CIV5B was lower than that of the CIII dimer, particularly for those subunits at the periphery of the SC. Thus, for each dataset, we followed the same procedure of using subtracted experimental particle images and performing focused refinement for each CIV5B and the CIII of the III2-IV5B1 Δrox1 SC (Materials and Methods). This approach effectively increased the resolution of CIV5B and CIII to 3.4 Å and 3.3 Å, respectively, within the III2-IV5B1 SC (SI Appendix, Figs. S2 and S4). For the cox5ab III2-IV5B2 SCs, the resolution of each CIV5B was increased to 2.8 Å (CIV5B-a) and 3.0 Å (CIV5B-b); no additional refinement was performed on CIII (SI Appendix, Figs. S3 and S4).
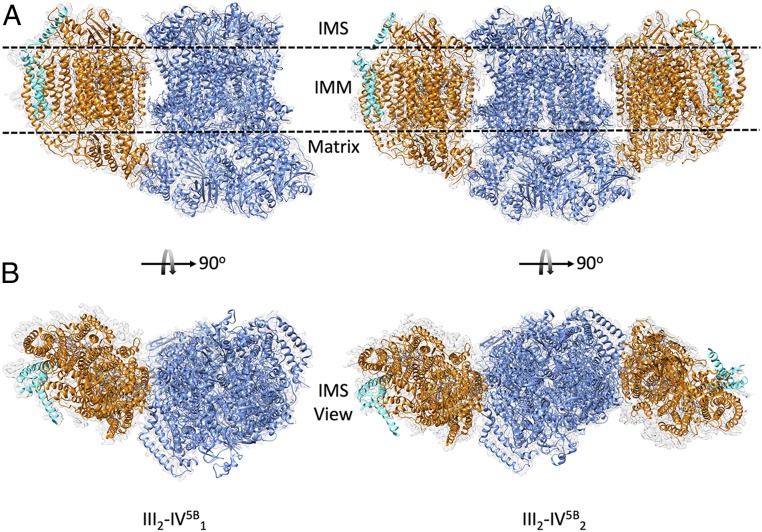
From the merged maps (Fig. 2) obtained for the III2-IV5B1 (Δrox1) and the III2-IV5B2 (cox5ab) SCs, all expected subunits could be confidently modeled, namely 20 protein subunits for the CIII dimer, including Qcr10, and 12 subunits in each CIV5B, including Cox5B and Cox26 (19). An additional protein, Rcf2, was unambiguously identified in the CIV5B of the III2-IV5B1 SC structure (see below). All expected metal cofactors and prosthetic groups were well defined except for the calcium cation, which was clearly present in the CIV5B of the III2-IV5B2 SC but absent in that of the III2-IV5B1 SC (19, 34). A total of 30 lipids were modeled in the III2-IV5B1 (41 in the III2-IV5B2) SC structure, including seven cardiolipins (eight in the III2-IV5B2 SC). This compares to the 44 lipids in the previously reported III2-IV5A2 SC structure, including eight cardiolipins. Among the lost lipids in the III2-IV5B1 SC are a cardiolipin and a phosphocholine lost from the accessible side of CIII where no CIV is bound.
Fig. 2.
Cryo-EM maps and fitted models of the III2-IV5B1 and III2-IV5B2 SCs. Membrane view (A) and IMS view (B) of the Δrox1 III2-IV5B1 and cox5ab III2-IV5B2 merged maps with overall dimensions of 224 × 165 × 114 Å and 289 × 165 × 114 Å, respectively. The transmembrane region is suggested by two dashed lines. CIII is represented in blue and CIV in orange in both views. Rcf2 is highlighted in cyan.
Alignment of the final models revealed little to no difference in the CIII dimers between the III2-IV5B1 and III2-IV5B2 SCs, or with the III2-IV5A2 SC previously published, with rmsd values <0.5 Å (with an average of 0.4 Å for the core domains and values of 0.8 to 1 Å for the less-ordered Rip1 head domain and Qcr10), highlighting the conserved assembly of CIII within all III-IV SCs irrespective of protein stoichiometry. In addition, no apparent differences were observed between the two CIII monomers in the III2-IV5B1 SC structure. Specifically, the subunits of the CIII monomer that interact with CIV5B (Cor1, Cyt c1, Rip1, and Qcr8) were virtually identical to the same subunits in the CIII monomer that is free of these interactions. This suggests that there are no conformational changes in CIII to accommodate interactions with CIV. Similarly, alignment of the three CIV5B monomers described here revealed only a few differences, most of which were observed in CIV5B-b of the III2-IV5B2 SC and most certainly arise from the lower quality of the map in these regions. Finally, irrespective of the SC stoichiometry, CIV5B displayed the bow-shaped conformation of Cox13 (19) that would preclude dimerization of the enzyme, as seen in the bovine X-ray structure (35).
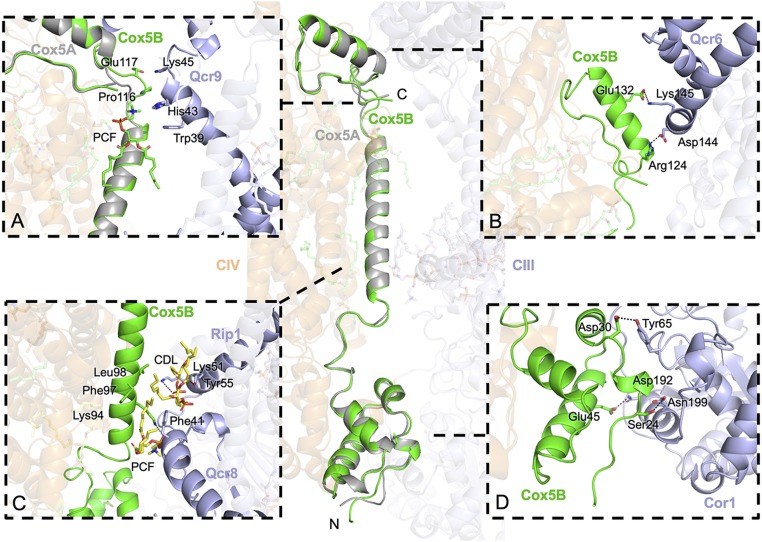
The interface between CIII and CIV also appeared to be well conserved in the two CIV5B-containing SC structures. As in the CIV5A-containing SC (19), Cox5B is the only CIV subunit contributing to the III-IV interface with apparent interactions on both sides of the IMM with multiple subunits of CIII and with clear stabilizing contributions from lipids, including a cardiolipin, in the membrane domain. In the matrix, Cox5B adopts the same conformation as Cox5A (Fig. 3), with its N-terminal domain also exhibiting the conformational shift seen compared with the X-ray structure of the bovine ortholog (35, 36), which most certainly arises due to the conserved interactions between Cox5 and Cor1 observed with both Cox5 isoforms (Fig. 3D).
Fig. 3.
Protein–protein interactions between Cox5B and CIII. The atomic models of the CIV5B-containing SC purified from Δrox1 and the CIV5A-containing SC purified from Δcox5b (PDB ID code 6HU9) were aligned on their Cox5 subunit. Around the main panel are zoom-in views of the interface between CIV (orange) and CIII (blue) in the presence of Cox5A (gray) or Cox5B (green). Hydrogen bond and salt bridge interactions are shown as dashed lines. (A) Conformational changes at the C-terminal loop of Cox5B promote interactions with a phosphocholine (PCF) molecule and Qcr9 from CIII. (B) Electrostatic interactions within the IMS between the C-terminal α-helix of Cox5B and Qcr6 of CIII. (C) IMM interactions between Cox5B and Rip1 via the conserved bridging cardiolipin. (D) Interactions in the matrix between the N-terminal region of Cox5B and Cor1.
Interactions within the IMM are also maintained, with a conserved lysine (Lys94 in Cox5B, Lys97 in Cox5A) interacting with Qcr8 via conserved bridging lipids (Fig. 3C). In the inner membrane space (IMS), interactions with cytochrome c1 and Qcr6 are preserved despite differences in the two isoform sequences at their interface with Qcr6 (Fig. 3B). While the C-terminal Lys153 in Cox5A forms an H-bond with Glu83 of Cyt c1, the corresponding Lys151 in Cox5B is >6 Å apart. Instead, the backbone carbonyl oxygen of Pro116 of Cox5B interacts with the side chain of Lys45 of Qcr9 (Fig. 3A). These findings highlight a conserved interaction of CIII and CIV in all S. cerevisiae III-IV SCs.
Rcf2 Is Revealed in the III2-IV5B1 SC Structure.
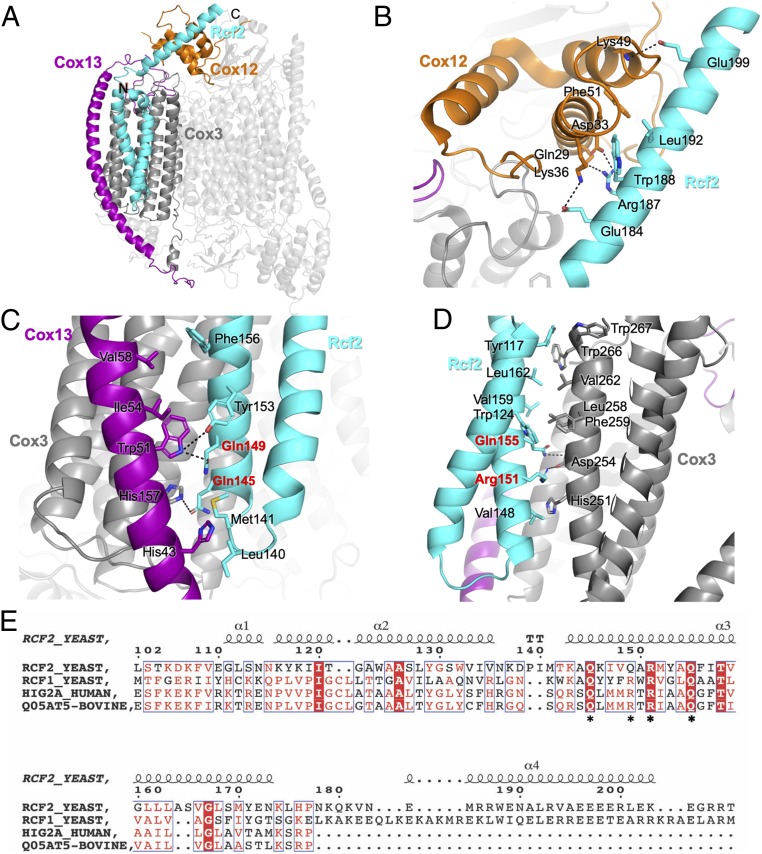
Strong additional density was revealed in the CIV5B of the III2-IV5B1 SC away from the interface with CIII, in a pocket formed by Cox1, Cox3, Cox13, and Cox12. It could be modeled as a single polypeptide chain, with both N and C termini in the IMS folded into two transmembrane helices and an additional helix that extends in the IMS, a pattern reminiscent of previous reports on respiratory SC-associated factors Rcf1 and Rcf2 (25–28). This density was at a relatively low resolution, in the range of 7 to 10 Å, precluding assignment. We thus collected a second dataset of the same sample, and by increasing the number of aligned particles, we were able to increase the local resolution in this region to 4 to 5 Å (SI Appendix, Fig. S4A). This allowed us to confidently assign the new density to the C-terminal fragment of Rcf2 (residues 107 to 205; SI Appendix, Fig. S5), which comprises a short N-terminal α-helix (residues 107 to 114), followed by a transmembrane helix-hairpin-helix motif in the IMM (residues 118 to 175) and a final C-terminal α-helix in the IMS (residues 182 to 203). This assignment is further supported by a previous report showing stable association of a C-terminal fragment of Rcf2 with CIV after proteolytic processing of the full-length protein (28). Interactions of Rcf2 with multiple subunits of CIV were revealed (Fig. 4A, cyan). Within the IMM, the HIG1 domain of Rcf2 forms extensive hydrophobic/Van der Waals interactions with Cox3 and the transmembrane domain of Cox13 toward the matrix (Fig. 4 C and D). The C-terminal α-helix of Rcf2 that extends into the IMS makes core hydrophobic and flanked electrostatic interactions with Cox12 (Fig. 4B), supporting a role of Rcf2 in the recruitment/stabilization of Cox12 at a late stage of CIV assembly (28).
Fig. 4.
Rcf2 is revealed in the Δrox1 III2-IV5B1 SC structure. (A) Structure of CIV in the Δrox1 III2-IV5B1 SC with Rcf2 (cyan), Cox3 (dark gray), Cox12 (orange), and Cox13 (purple) highlighted. (B) Interactions between the C-terminal helix of Rcf2 and Cox12 in the IMS. Residues that form interactions are labeled. (C) Interactions within the IMM between Rcf2 and Cox13 and Cox3. (D) Interactions within the IMM between Rcf2 and Cox3. Residues of the conserved Q(R/Q)RQ motif of Hig1 type 2 proteins are highlighted in red. (E) Sequence alignment of yeast Rcf2 (Uniprot: P53721), Rcf1 (Q03713), human Hig2a (Q9BW72), and bovine Higd2a (Q05AT5). The numbers above the sequences correspond to the Rcf2 sequence. The alignment shows only the fraction of the determined Rcf2 structure, highlighting the HIG1 domain (residues 110 to 177) and the conserved Q(R/Q)RQ motif (stars underneath the sequences). The secondary structure elements for Rcf2 are displayed above the sequence. The image was generated in ESPript (53).
Similar, although weaker, density was found on both CIVs in the III2-IV5B2 SC map (SI Appendix, Fig. S6 B and C). The density in both CIVs was sufficient to trace the main chain of a protein with very similar topology as Rcf2. The local resolution allowed side chain assignment to Rcf2 in CIV5B-a, but more data are needed to definitively determine the additional density in CIV5B-b. Nonetheless, these data suggest that Rcf2 is not specific to the III2-IV1 SC. In our previous study of the Cox5A-containing SC (purified from the Δcox5b strain), mass spectrometry analysis detected the presence of Rcf1 and Rcf2 in the final protein sample used for structure determination, even though neither protein was detected in the final map (19). Reanalysis of this dataset with focused refinement of both III2-IV5A2 and III2-IV5A1 SCs (80% and 20% of particles, respectively) using a mask that includes Rcf2 led to the observation of additional density (SI Appendix, Fig. S6 D–F) where the transmembrane helices of Rcf2 were found in the Cox5B-containing SCs.
Overall, our model of Rcf2 fits well in these extra densities, but again, more data are needed to completely model and identify the new protein within the Cox5A-containing SCs. The resolution of this region of the map is stronger and better defined in the III2-IV1 SCs compared with the III2-IV2 SCs (SI Appendix, Fig. S6), most likely resulting from an increased heterogeneity of CIV populations within the two halves of the III2-IV2 SCs. While this could be explained by the presence of both assembly intermediates and mature forms of CIV coexisting within these SCs, or the presence of different hypoxia-induced gene 1 (Hig1) type 2 proteins in each CIV monomer, the possibility that Rcf2 starts to dissociate after full maturation of the III2-IV2 SCs cannot be excluded.
No Effect of Cox5 Isoform on CIV Activity.
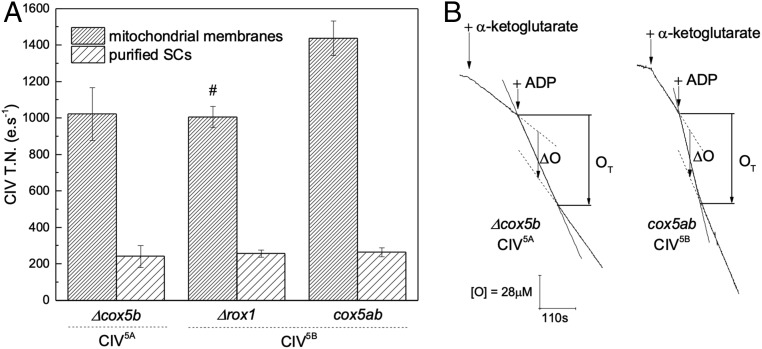
A twofold to threefold increase in activity of CIV5B compared with CIV5A has been reported in yeast (37) as well as in a mammalian system for the equivalent COX4-2/1 isoform pair (38), in both cases suggesting faster activity of the hypoxic isoform. An extensive steady-state analysis of the oxygen reduction rates of yeast CIV isoforms within mitochondrial membrane fragments, including comparison of TNs using horse heart cytochrome c or yeast iso-1 or iso-2 cytochrome c, has been published previously (23). Based on that work, we measured the oxygen reduction rates of CIV in conditions of maximum turnover, with horse heart cytochrome c, N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD), and ascorbate for the three strains used in this study. In mitochondrial membrane fragments, TNs were high at 1,022 ± 145 e.s−1 for CIV5A and 1,006 ± 145 and 1,438 ± 94 e.s−1 for CIV5B in Δrox1 and cox5ab, respectively (Fig. 5A). This is consistent with the previous characterization of the Δcox5b and cox5ab strains (both with TNs at ∼1,400 e.s−1) but contrasts with the significant increase in activity reported for Δrox1 (23). However, the accuracy of TN measurements depends on the accurate quantitation of CIV in the membrane preparations, which is difficult at the low levels of CIV biogenesis seen in Δrox1 (Fig. 1A) and carries a greater error than the difference of activity reported for the isoenzymes. Accurate quantitation of CIV5B within Δrox1 can be achieved only after metal affinity chromatography (Fig. 1C). TNs measured at this stage of purification were very similar, at ∼250 e.s−1 in each strain (Fig. 5A), again suggesting no difference in activity between CIV5A and CIV5B. Of note, 70 to 80% of the CIV activity was lost in this purification step, most likely due to delipidation of the enzyme on the column, although we cannot exclude the possibility that other types of interaction essential for full activity (involving other proteins, for instance) could have been lost as well.
Fig. 5.
Isoform exchange has no effect on CIV oxygen reduction rates or SC pumping activity. (A) CIV oxygen reduction activity measured as TNs in conditions of maximum turnover rate with horse heart cytochrome c, TMPD, and ascorbate in at least two independent preparations of mitochondrial membranes and purified SCs after metal affinity chromatography from Δcox5b (n = 6 and 3), Δrox1 (n = 3 and 4), and cox5ab (n = 4 and 3). #Indicates where quantitation of CIV within the sample bears an additionally large error due to its low abundance in the IMM. (B) Oxygen consumption rates of intact yeast mitochondria respiring on α-ketoglutarate. ADP/O values were calculated from the average of the ADP/OT and ADP/ΔO values determined after ADP addition on preparations of intact mitochondria from Δcox5b (n = 6) and cox5ab (n = 5).
No Effect of CIV Isoform on Overall Pumping Activity.
To assess whether isoform exchange had an effect on CIII and CIV (or the III-IV SC) proton translocation activity, we measured ADP/O ratios from preparations of intact mitochondria from the Δcox5b and cox5ab strains, which differ only in their unique expression of CIV5A and CIV5B, respectively, using α-ketoglutarate as a substrate. No difference was observed between the two variants with ADP/O ratios measured at 1.93 ± 0.24 for Δcox5b and 2.06 ± 0.11 for cox5ab, giving H+/e stoichiometries of 3.4 ± 0.5 and 3.6 ± 0.3, respectively. These values are close to the theoretical value of 3 expected in yeast S. cerevisiae when six protons are translocated for every two electrons from reduced ubiquinone passing through CIII and CIV to reduce 1/2 O2 to H2O. This definitely shows that replacement of Cox5A with Cox5B, in the absence of any additional change, has no effect on III-IV SC pumping stoichiometry and overall energy conservation efficiency.
Discussion
Over the past 20 y, research efforts have intensified to understand the role of mitochondrial respiratory SCs and, by association, the function of several proteins that have been implicated in their formation. To guide these investigations, several SC structures have been solved at near-atomic resolution that revealed the molecular details of the protein–protein interactions within the SCs, although to date none has revealed any SC assembly factors.
In all mitochondrial CIV-containing SC structures available, CIV interacts with CIII via subunits with expression isoforms. In S. cerevisiae, in normoxic conditions of growth, the CIII-CIV interface was shown to be made exclusively via Cox5A, the only yeast CIV subunit that exists as two isoforms (Cox5A and Cox5B) that are differentially expressed depending on oxygen levels. In this study, we investigated the effect of CIV isoform exchange on SC formation, structure, and activity using mutant strains designed to selectively assemble CIV5B in normoxic conditions of growth, albeit in very different amounts. We systematically compared our results with those for a strain that assembles only CIV5A and whose III2-IV5A2 SC structure was recently solved (19).
First, we showed that III-IV SCs form within the mitochondrial membrane of all mutant strains irrespective of the Cox5 isoform, even in Δrox1, where CIV is assembled only at very low levels. However, in Δrox1, no III2-IV2 SCs could be detected, only III2-IV1 SCs. This is consistent with previous reports (2, 30), and it now seems established that in yeast, III-IV SC stoichiometry is governed by the relative amounts of CIII and CIV within the IMM.
Next, we showed through cryo-EM that a change in Cox5 isoform had little effect on the overall structure of the III-IV SCs formed, even though Cox5 was the sole CIV subunit to form the III-IV interface. We measured similar O2 reduction rates and proton translocation activities for all III-IV SCs, independent of CIV isoform or SC stoichiometry. This calls into question the purpose of isoform evolution in yeast, as in eukaryotes. It might be found in, for instance, regulation of CIV assembly or activity in hypoxia, perhaps involving posttranslational modifications or allosteric effectors such as ATP (39). Further studies on yeast strains grown in low oxygen concentrations might be needed to reveal the effect of CIV isoforms in yeast.
Unexpectedly, the structures described above revealed a Hig1 type 2 protein associated with a respiratory SC. We could confidently detect the C-terminal fragment of Rcf2 within both CIV5B-containing SCs and have found evidence if not of its presence, then of a protein with very similar transmembrane topology in the CIV5A-containing SCs.
The Hig1 protein family was originally described as being strongly inducible by hypoxia in an HIF-1–dependent manner (40). Today, members of this family are defined by their HIG1 domain, a specific sequence pattern within two transmembrane helices and a loop. The family is further subdivided into two classes. The Hig1 type 1 proteins (HIG1A in mammals) are specific to higher eukaryotes, and the Hig1 type 2 proteins (Rcf1 and Rcf2 in yeast and HIG2A in mammals), characterized by their QX3(R/Q)XRX3Q motif, are found in all eukaryotes and α-proteobacteria. Hig1 type 2 proteins have been identified as essential components of the mitochondrial III-IV SCs in both yeast and mammals (25–27), with some speculation that they could form the interface between CIII and CIV. However, as neither Rcf1 nor Rcf2 were resolved in the first III-IV SC structures obtained in yeast (19, 20), it became clear that they are not essential for SC stability.
Evidence is mounting for a role of Hig1 proteins in supporting CIV assembly and activity (25, 26, 28–30, 41–43). It has been proposed that Hig1 type 1 proteins (specific to higher eukaryotes) are involved in the initial step of CIV assembly (42), and that Hig1 type 2 proteins (yeast Rcf1, Rcf2 and mammalian HIG2A) are involved in the later stages, for the assembly of subunits Cox13 and Cox12 in yeast. Specifically, Rcf1 has been implicated in the recruitment of Cox13 and possibly Rcf2 (26), but evidence suggests that it leaves CIV after stable assembly of Cox13 (29), consistent with a role of Rcf1 as an assembly factor. The position of the C-terminal fragment of Rcf2, as revealed in this work, is consistent with a role in the assembly of Cox12; however, its stable occurrence within the Cox5B-containing SC structures presented here strongly suggests that it remains associated to CIV within the III-IV SCs and serves as a stoichiometric subunit of CIV. More data are needed to definitively establish whether Rcf2 or another Rcf protein is present in both CIV5Bs of the III2-IV5B2 or the Cox5A-containing SCs.
Several studies of Rcf1 and/or Rcf2 gene knockout mutant strains have highlighted the importance of these proteins for optimal yeast CIV assembly and activity. The mutant Δrcf1, but not Δrcf2, resulted in a 50% decrease of CIV levels and reduced CIV TNs were observed in both mutant strains (30, 41). In addition, Rcf2 has extensive interactions with Cox13, Cox3, and Cox12, all of which contribute to the binding site of cytochrome c. Disruption or destabilization of this region of CIV in Δrcf1 and/or Δrcf2 strains would explain their reportedly increased reactive oxygen species production (26, 27).
Importantly, the residues of CIV described herein to interact with the HIG1 domain of Rcf2 are conserved in mammalian orthologs, suggesting that similar interactions could occur in mammalian systems. Mammalian Hig1 proteins are shorter, and sequence alignments suggest that they lack the C-terminal IMS helix of Rcf2 that interacts with Cox12 (Fig. 4E). Stabilization of this region could instead be provided by NDUFA4, as seen in the structure of CIV within the human respirasome (18). It is also noteworthy that as for NDUFA4 of CIV within the mammalian respirasome, a Hig1 type 2 protein at the position of Rcf2 would hinder CIV dimerization, as seen in the bovine X-ray structure (35). Whether Hig1 type 2 proteins are specific components of those CIVs that form only III-IV SCs remains to be investigated. To date, no evidence of Hig1 proteins or of NDUFA4 has been reported in structural studies of isolated mammalian CIV (35, 36), and knockdown of HIG2A in mammalian cells has been shown to cause depletion of all higher-order SCs that contain CIV (27).
Although our understanding of the organization of respiratory proteins within the dense environment of the IMM is rapidly advancing, continued efforts involving structural biology, genetic, proteomic, and functional studies will be needed to elucidate the role of SC formation.
Materials and Methods
Yeast Strain and Cell Growth.
Three strains of S. cerevisiae were used in this study, all constructed from W303-1B (Mat α ade2 leu2 trp1 ura3) (23, 31) and containing a 6-histidine tag at the C terminus of Cox13: a Δcox5a Δrox1 strain that expresses only Cox5B (rox1::KanMx4 cox5a::URA3), a cox5ab strain that expresses Cox5B under control of the Cox5A promoter (cox5a::COX5B), and a Δcox5b strain that expresses only Cox5A (cox5B::KanMx4). Yeast cells were grown aerobically at 28 °C in YPGal medium and harvested in late log phase by centrifugation as described by Meunier et al. (31). Cells were washed once in a resuspension/centrifugation cycle in 50 mM KPi (pH 7.0) and stored at −80 °C until use.
Preparation of Mitochondrial Membranes and SC Purification.
The thawed cells were disrupted by mechanical means using glass beads (425 to 600 μm diameter) and a bead-beater in a medium composed of 50 mM KPi, 650 mM d-mannitol, 5 mM EDTA, and 0.1 mM PMSF, pH 7.4 (31). Cell debris was removed by centrifugation at 5,600 × g for 20 min at 4 °C, and the mitochondrial membranes were subsequently pelleted by centrifugation at 120,000 × g for 50 min at 4 °C. They were washed once by a resuspension/centrifugation cycle in 50 mM KPi, 100 mM KCl, 10 mM MgCl2, 150 μM CaCl2, and 0.1 mM PMSF, pH 7.4, and twice in 50 mM KPi, 2 mM EDTA, and 0.1 mM PMSF, pH 7.4, as described previously (19), then stored at −80 °C in a minimal volume of 50 mM Hepes, pH 8.0. Respiratory SCs were purified after membrane solubilization with 1% GDN (Anatrace) by affinity chromatography (HisTrap HP column, 5 mL; GE Healthcare) and gel filtration (Superose 6 Increase column; GE Healthcare), using an Äkta Pure 25 chromatography system (GE Healthcare) operated at 4 °C, as described previously (19). In brief, for affinity chromatography, the column was equilibrated with 50 mM Hepes, 500 mM NaCl, 5 mM imidazole, and 0.05% GDN, pH 8.0, followed by overnight loading of the solubilized proteins. The column was then washed with the same buffer, and the SCs were subsequently eluted with 50 mM Hepes, 150 mM NaCl, 0.05% GDN, and 100 mM imidazole, pH 7.2. The same buffer but without imidazole was used for gel filtration. When required, samples were concentrated using 100-kDa MWCO centrifugal concentrators (GE Healthcare).
Analytical Methods.
The amounts of CIII and CIV within protein samples were determined from their sodium dithionite reduced minus oxidized difference absorption spectra recorded in 50 mM Hepes and 0.1% undecylmaltoside, pH 8.0, in the visible range, using extinction coefficients (ΔƐ) of 28 mM−1 cm−1 (ΔA562–578 nm) and 26 mM−1 cm−1 (ΔA604–621 nm) for CIII and CIV, respectively (19). Precast NativePAGE 3 to 12% Bis-Tris gels (Invitrogen) were used in accordance with the manufacturer’s instructions for BN-PAGE analysis, as described previously (19).
Activity Measurements.
Steady-state oxygen consumption rates were measured at 25 °C using a Clark-type oxygen electrode (Oxygraph; Hansatech) in a medium of 10 mM KPi and 50 mM KCl, pH 6.6, supplemented with 0.05% GDN and 50 μM equine heart cytochrome c as described previously (19). SC activity was measured in the presence of 500 units/mL SOD and 250 units/mL catalase; a baseline was recorded before initiation of the reaction by the addition of 40 μM decylubiquinol. CIV activity was measured in the presence of 40 μM TMPD; a baseline was recorded before initiation of the reaction by the addition of 2 mM Na+-ascorbate. TNs were calculated from linear fitting of the oxygen consumption rate with Origin (OriginLab) using the formula TN (e.s−1) = O2 consumption gradient (M.s−1) × 4/[CIII or CIV] (M) for SCs or CIV, respectively. The data presented are the average of several measurements performed on different SC preparations. Error bars were calculated from the SD from the mean.
Cryo-EM.
Grids for cryo-EM (UltrAuFoil R1.2/1.3; Quantifoil) were prepared using a Vitrobot Mark IV (Thermo Fisher Scientific) after 1:1 or 1:3 dilution of purified Δrox1 III2-IV5B1 SC (2.9 μM CIII; 0.6 μM CIV) or cox5ab III2-IV5B2 SC (6.9 μM CIII; 2.9 μM CIV) in 50 mM Hepes, 150 mM NaCl, and 0.05% GDN, pH 7.2 (12- to 14-s blotting time, 4 °C, and 100% humidity), as described previously (19). Data were collected with a Titan Krios microscope (Thermo Fisher Scientific) operated at 300 kV and equipped with a Quantum energy filter (Gatan), using EPU software (Thermo Fisher Scientific).
Two datasets were recorded for the Δrox1 III2-IV5B1 SC, one at the Electron Bio-Imaging Centre (eBIC; Diamond Light Source, Oxfordshire, UK) and one at Birkbeck College, in otherwise similar conditions so that the particles from the two datasets could be merged (see below). A post-GIF K2 Summit direct electron detector (Gatan), operating in counting mode at a nominal magnification of 130,000× (pixel size of 1.048 Å), was used for image collection, and an energy slit with a width of 20 eV was used during data collection. A total of 4,140 micrographs were collected at eBIC (dose rate of 5.14 e/Å2/s on the specimen and a total dose of 56.4 e/Å2 fractionated over 40 frames; nominal defocus range set from −1.6 μm to −3.6 μm) and 2,981 micrographs were collected at Birkbeck College (dose rate of 5.14 e/Å2/s on the specimen and a total dose of 51.4 e/Å2 fractionated over 40 frames; nominal defocus range set from −1.5 μm to −2.7 μm).
The cox5ab III2-IV5B2 SC data were collected at eBIC in “superresolution” mode using a post-GIF K3 Summit direct electron detector (Gatan) operating in counting mode at a nominal magnification of 81,000× (pixel size of 1.085 Å). An energy slit with a width of 20 eV was used during data collection. A total of 3,634 micrographs were collected with a dose rate of 22.4 e/pixel/s on the specimen and a total dose of 56.4 e/Å2 fractionated over 40 frames and a nominal defocus range set from −1.5 μm to −2.8 μm.
Image Processing.
The two Δrox1 III2-IV5B1 SC datasets were processed separately until particles were picked. In both cases, MOTIONCOR2 (44) was used for frame alignment and exposure weighting, while CTFFIND4.1 (45) was used to estimate the contrast transfer function (CTF) parameters. Micrographs with excessive specimen drift, overfocus, or ice defects were removed manually. Reference-free particle picking was performed with Gautomatch v0.53 (written by Kai Zhang; https://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/), using a 384-Å2 box size. This resulted in the selection of 44,268 particles from 3,648 micrographs in the first dataset and 44,968 particles from 2,541 micrographs in the second dataset. Particles were combined, resulting in a final particle number of 89,236, and 3D classification was performed in Relion (46) using a map generated from homogeneous refinement of the first dataset in cryoSPARC2 (47). This revealed a 3.41-Å resolution map, based on the Fourier shell correlation (FSC) gold standard. To improve resolution of the SC, we used a particle subtraction approach, as described previously (19). In brief, a mask was generated around CIII (or CIV) and used to subtract density from the particles, resulting in a new set of particles that was used in focused refinement to improve local resolution on both complexes (SI Appendix, Figs. S2 and S4 A and C). For real-space refinement, the two maps (CIV5B at 3.41 Å and CIII at 3.29 Å) were merged using UCSF Chimera (48).
Frame alignment and CTF estimation of the cox5ab III2-IV5B2 SC dataset were performed using MOTIONCOR2 in Relion and CTFFIND4.1 within the cryoSPARC v2 interface, respectively. The superresolution micrographs were binned 2 × 2 (1.085 Å/pixel) during frame alignment. An initial set of 892 particles was picked manually and then classified into 50 representative 2D classes, 12 of which were then used as templates for automatic particle picking in cryoSPARC v2. Autopicked particles (138,154) were then classified using multiple iterations of reference-free 2D classification, and an initial model was built using ab initio reconstruction from 65,999 particles. No III2-IV1 SCs were revealed within the final particles, so homogeneous refinement was used to increase the resolution of the map to 2.8 Å based on the FSC gold standard. As in the Δrox1 III2-IV5B1 SC, the cox5ab III2-IV5B2 map displayed high resolution at the core of CIII (up to 2.5 Å; SI Appendix, Fig. S4B), but lower resolution toward the periphery of the CIV monomers. To increase the resolution of the CIV monomers, particle subtraction was used to remove density for the CIII core dimer and one monomer of CIV, and then local refinement was used with a mask generated around the CIV monomer of interest. This resulted in three maps, one for the core of the SC (covering CIII and part of the two CIV monomers as well) and one each for the two monomers of CIV (CIV5B-a and CIV5B-b), with resolutions of 2.80, 2.83, and 3.01 Å, respectively (SI Appendix, Figs. S3 and S4 B and D). For real-space refinement, the maps were then merged using UCSF Chimera.
For the CIV5A within the Δcox5b III2-IV5A1 and III2-IV5A2 SCs, data collection, frame alignment, and CTF estimation of micrographs have been reported previously (19). For the present work, the micrographs were further processed in cryoSPARC v2. A total of 98,968 particles were picked, of which 57,132 were selected from the 2D classification for ab initio reconstruction, including particles of both III2-IV2 and III2-IV1 SCs. The generated ab initio models together with their particles (44,195 for the III2-IV2 SC and 12,937 for the III2-IV1 SC) were further used for nonuniform refinement. To generate focused refined maps on the CIVs, we followed the same procedure as above for signal subtraction, followed by refinement with a CIV mask that included Rcf2. The resulting maps obtained for CIV5A within the III2-IV1 SC (4.1 Å) and for CIV5A-a and CIV5A-b within the III2-IV2 SC (3.09 and 3.24 Å, respectively) are shown in SI Appendix, Fig. S6 E and F.
Model Building.
The two individual maps after focused refinement corresponding to CIII and CIV in the Δrox1 III2IV5B1 SC were aligned on the initial map and merged in Chimera using the “vop maximum” command. The dimeric CIII and one monomer of CIV from the previously determined yeast III2IV5A2 SC (PDB ID code 6HU9) (19) were fitted into this merged map using Chimera, and this model was further improved through several iterations of model building in Coot (49) and real-space refinement in Phenix (50). The map was of sufficient quality to fit all expected prosthetic groups, including heme groups and metal clusters, and enabled the assignment of amino acids in all chains, including Rcf2. Some regions displayed characteristic disorder, such as the C terminus of Rip1 (although its Fe-S cluster was clearly visible), and the N terminus of Qcr6 (1 to 72) was not resolved, a feature common to previously determined crystal structures of CIII. Additional densities resembling long carbon chains were modeled as palmitoyl-phosphatidylethanolamine (PEF), diacyl-glycero-phosphocholine (PCF), and cardiolipin based on similarities to previously published structures in which these ligands were observed. For the refine space refinement in Phenix, we used secondary structure restraints as well as Ramachandran and rotamer restraints. Geometry definitions for the ligands were defined from values in the CCP4 ligand library (51). Additional bond and distance restraints were implemented on specific molecules based on previously published high-resolution structures. No symmetry restraints were applied during real-space refinement. The model was then checked in Coot for any refinement errors and manually corrected. The final Δrox1 III2IV5B1 SC model contains 6,002 protein residues, eight heme groups (four B-type hemes in the two cytochrome b subunits of CIII, two C-type hemes in the two cytochrome c1 subunits of CIII, and one heme a and one heme a3 in Cox1 of CIV), two Fe-S clusters (one in each Rip1 subunit of CIII), and four noniron metal centers (CuB and a magnesium in Cox1, the dinuclear CuA in Cox2, and a zinc in Cox4). In addition, we modeled seven cardiolipin, 18 PEF, and six PCF molecules.
The same procedure was used to build the CIII dimer and the two CIV monomers of the cox5ab III2-IV2 SC in Coot individually, using the three refined maps described above. The models of the proteins in the cox5ab III2-IV2 SC share many features with the models that form the Δrox1 III2IV5B1 SC, namely the heme groups, metal centers, and number of chains present in each protein (including Rcf2 in each CIV monomer). The same regions of Rip1 and Qcr6 in CIII displayed increased disorder and reduced resolution. Additional densities for lipids were also observed, many in conserved positions. All three models were refined individually using the real-space refine tool in Phenix as described above. The models were then checked manually for errors before a final real-space refinement on the combined cox5ab III2-IV2 SC using the merged map. The final cox5ab III2-IV2 SC model contains 7,902 protein residues, 10 heme groups (the same distribution of hemes as above but with one additional heme a and one heme a3 in the second CIV monomer), two Fe-S clusters (one in each Rip1 subunit of CIII), and 10 noniron metal centers (CuB, a magnesium and a calcium in Cox1, the dinuclear CuA in Cox2, and a zinc in Cox4, duplicated in the second CIV monomer). There was also an increase in the number of lipids present; eight cardiolipin, 26 PEF, and seven PCF molecules were modeled into the cox5ab III2-IV2 SC.
Refinement and model statistics are summarized in SI Appendix, Table S1. Maps and molecule representations in all figures were prepared using PyMOL and UCSF Chimera.
Intact Mitochondria Preparation and ADP/O Ratio Measurements.
Intact mitochondria were prepared from 12 to 15 g of yeast cells (wet weight) grown aerobically at 28 °C in YPGal medium (yeast extract 1%, peptone 2%, galactose 2%) and harvested in log phase. Digestion of the yeast cell wall was performed enzymatically following the protocol of Guérin et al. (52). ADP/O measurements were performed with the oxygen-electrode setup described above in a medium of 0.65 M mannitol, 5 mM MgCl2, 3 mM KPi, 10 mM Tris-maleate, 17 mM KCl and 0.1% BSA (w:v) at pH 7.0 and 25 °C. Oxygen consumption rates were determined after successive additions of intact mitochondria (10 to 20 μL), 10 mM α-ketoglutarate (Na+ salt), and 100 μM ADP (Na+ salt). The latter induced state 3 of respiration, which spontaneously reverted to state 4 after all the ADP was transformed into ATP. Respiratory rate ratios, calculated from the ratio of the state 3 over state 4 oxygen consumption rates measured, ranged from 1.4 to 2.0, depending on preparations. The ADP/O values presented here are the average of the ADP/OT and ADP/ΔO values determined after ADP addition. Error bars are calculated from the SD from the mean.
Data Availability.
All relevant data are included in the paper and/or are available from the corresponding author on reasonable request. Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession nos. EMD-10317 (CIII2) and EMD-10318 (CIV5B) for Δrox1; EMD-10340 (III2IV5B2 SC), EMD-10335 (CIV5B-a), and EMD-10334 (CIV5B-b) for cox5ab; and EMD-10375 (CIV5A-a) and EMD-10376 (CIV5A-b) for Δcox5b. The coordinates of the atomic models of the CIV5B-containing SCs built from a combination of EM maps have been deposited in the Protein Data Bank under ID codes PDB ID 6T15 (Δrox1 III2IV5B1) and 6T0B (cox5ab III2IV5B2).
Supplementary Material
Acknowledgments
We thank the Diamond Light Source and Dr. A. Howe for access to and support from the cryo-EM facilities at the UK National electron bio-imaging center (eBIC; Proposal EM14704-36), funded by the Wellcome Trust, the Medical Research Council UK, and the Biotechnology and Biological Sciences Research Council. We also thank Dr. N. Lukoyanova from the EM laboratory at Birkbeck for cryo-EM data collection, as well as Dr. D. Houldershaw and the computer support group at Birkbeck. This work was supported by the Medical Research Council UK (Career Development Award MR/M00936X/1, to A.M.) and Wellcome Trust grants to the Birkbeck EM facility (202679/Z/16/Z and 206166/Z/17/Z).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession nos. EMD-10317 (CIII2) and EMD-10318 (CIV5B) for Δrox1; EMD-10340 (III2IV5B2 SC), EMD-10335 (CIV5B-a), and EMD-10334 (CIV5B-b) for cox5ab; and EMD-10375 (CIV5A-a) and EMD-10376 (CIV5A-b) for Δcox5b. The coordinates of the atomic models of the CIV5B-containing SCs built from a combination of EM maps have been deposited in the Protein Data Bank under PDB ID codes 6T15 (Δrox1 III2IV5B1) and 6T0B (cox5ab III2IV5B2).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920612117/-/DCSupplemental.
References
- 1.Rich P. R., Maréchal A., “Electron transfer chains: Structures, mechanisms and energy coupling” in Comprehensive Biophysics, Ferguson S. J., Ed. (Elsevier Inc., 2012), pp. 72–93. [Google Scholar]
- 2.Schägger H., Pfeiffer K., Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies K. M., Blum T. B., Kühlbrandt W., Conserved in situ arrangement of complex I and III2 in mitochondrial respiratory chain supercomplexes of mammals, yeast, and plants. Proc. Natl. Acad. Sci. U.S.A. 115, 3024–3029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paumard P., et al. , The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21, 221–230 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies K. M., Anselmi C., Wittig I., Faraldo-Gómez J. D., Kühlbrandt W., Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl. Acad. Sci. U.S.A. 109, 13602–13607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum T. B., Hahn A., Meier T., Davies K. M., Kühlbrandt W., Dimers of mitochondrial ATP synthase induce membrane curvature and self-assemble into rows. Proc. Natl. Acad. Sci. U.S.A. 116, 4250–4255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu J., et al. , Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science 364, 1068–1075 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Milenkovic D., Blaza J. N., Larsson N. G., Hirst J., The enigma of the respiratory chain supercomplex. Cell Metab. 25, 765–776 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Letts J. A., Sazanov L. A., Clarifying the supercomplex: The higher-order organization of the mitochondrial electron transport chain. Nat. Struct. Mol. Biol. 24, 800–808 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Fedor J. G., Hirst J., Mitochondrial supercomplexes do not enhance catalysis by quinone channeling. Cell Metab. 28, 525–531.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letts J. A., Fiedorczuk K., Sazanov L. A., The architecture of respiratory supercomplexes. Nature 537, 644–648 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Gu J., et al. , The architecture of the mammalian respirasome. Nature 537, 639–643 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Guo R., Zong S., Wu M., Gu J., Yang M., Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell 170, 1247–1257.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Sousa J. S., Mills D. J., Vonck J., Kühlbrandt W., Functional asymmetry and electron flow in the bovine respirasome. eLife 5, e21290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu M., Gu J., Guo R., Huang Y., Yang M., Structure of mammalian respiratory supercomplex I1III2IV1. Cell 167, 1598–1609.e10 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Lapuente-Brun E., et al. , Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Cogliati S., et al. , Mechanism of super-assembly of respiratory complexes III and IV. Nature 539, 579–582 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Zong S., et al. , Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 28, 1026–1034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartley A. M., et al. , Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome Bc1. Nat. Struct. Mol. Biol. 26, 78–83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rathore S., et al. , Cryo-EM structure of the yeast respiratory supercomplex. Nat. Struct. Mol. Biol. 26, 50–57 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Burke P. V., Raitt D. C., Allen L. A., Kellogg E. A., Poyton R. O., Effects of oxygen concentration on the expression of cytochrome c and cytochrome c oxidase genes in yeast. J. Biol. Chem. 272, 14705–14712 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Allen L. A., Zhao X.-J., Caughey W., Poyton R. O., Isoforms of yeast cytochrome c oxidase subunit V affect the binuclear reaction center and alter the kinetics of interaction with the isoforms of yeast cytochrome c. J. Biol. Chem. 270, 110–118 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Dodia R., Meunier B., Kay C. W. M., Rich P. R., Comparisons of subunit 5A and 5B isoenzymes of yeast cytochrome c oxidase. Biochem. J. 464, 335–342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukuda R., et al. , HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Strogolova V., Furness A., Robb-McGrath M., Garlich J., Stuart R. A., Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol. Cell. Biol. 32, 1363–1373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vukotic M., et al. , Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 15, 336–347 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Chen Y. C., et al. , Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 15, 348–360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Römpler K., et al. , Overlapping role of respiratory supercomplex factor Rcf2 and its N-terminal homolog Rcf3 in Saccharomyces cerevisiae. J. Biol. Chem. 291, 23769–23778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garlich J., Strecker V., Wittig I., Stuart R. A., Mutational analysis of the QRRQ motif in the yeast Hig1 type 2 protein Rcf1 reveals a regulatory role for the cytochrome c oxidase complex. J. Biol. Chem. 292, 5216–5226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strogolova V., Hoang N. H., Hosler J., Stuart R. A., The yeast mitochondrial proteins Rcf1 and Rcf2 support the enzymology of the cytochrome c oxidase complex and generation of the proton motive force. J. Biol. Chem. 294, 4867–4877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meunier B., Maréchal A., Rich P. R., Construction of histidine-tagged yeast mitochondrial cytochrome c oxidase for facile purification of mutant forms. Biochem. J. 444, 199–204 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Protasoni M., et al. , Respiratory supercomplexes act as a platform for complex III-mediated maturation of human mitochondrial complexes I and IV. EMBO J. 39, e102817 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timón-Gómez A., et al. , Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 76, 163–178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maréchal A., Iwaki M., Rich P. R., Structural changes in cytochrome c oxidase induced by binding of sodium and calcium ions: An ATR-FTIR study. J. Am. Chem. Soc. 135, 5802–5807 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Tsukihara T., et al. , The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 272, 1136–1144 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Shinzawa-Itoh K., et al. , Monomeric structure of an active form of bovine cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 116, 19945–19951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterland R. A., Basu A., Chance B., Poyton R. O., The isoforms of yeast cytochrome c oxidase subunit V alter the in vivo kinetic properties of the holoenzyme. J. Biol. Chem. 266, 4180–4186 (1991). [PubMed] [Google Scholar]
- 38.Hüttemann M., et al. , Cytochrome c oxidase subunit 4 isoform 2-knockout mice show reduced enzyme activity, airway hyporeactivity, and lung pathology. FASEB J. 26, 3916–3930 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnold S., The power of life: Cytochrome c oxidase takes center stage in metabolic control, cell signalling and survival. Mitochondrion 12, 46–56 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Denko N., et al. , Epigenetic regulation of gene expression in cervical cancer cells by the tumor microenvironment. Clin. Cancer Res. 6, 480–487 (2000). [PubMed] [Google Scholar]
- 41.Rydström Lundin C., von Ballmoos C., Ott M., Ädelroth P., Brzezinski P., Regulatory role of the respiratory supercomplex factors in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 113, E4476–E4485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidoni S., et al. , MR-1S interacts with PET100 and PET117 in module-based assembly of human cytochrom c oxidase. Cell Rep. 18, 1727–1738 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Schäfer J., Dawitz H., Ott M., Ädelroth P., Brzezinski P., Structural and functional heterogeneity of cytochrome c oxidase in S. cerevisiae. Biochim. Biophys. Acta Bioenerg. 1859, 699–704 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guérin B., Labbe P., Somlo M., Preparation of yeast mitochondria (Saccharomyces cerevisiae) with good P/O and respiratory control ratios. Methods Enzymol. 55, 149–159 (1979). [DOI] [PubMed] [Google Scholar]
- 53.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320-4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the paper and/or are available from the corresponding author on reasonable request. Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession nos. EMD-10317 (CIII2) and EMD-10318 (CIV5B) for Δrox1; EMD-10340 (III2IV5B2 SC), EMD-10335 (CIV5B-a), and EMD-10334 (CIV5B-b) for cox5ab; and EMD-10375 (CIV5A-a) and EMD-10376 (CIV5A-b) for Δcox5b. The coordinates of the atomic models of the CIV5B-containing SCs built from a combination of EM maps have been deposited in the Protein Data Bank under ID codes PDB ID 6T15 (Δrox1 III2IV5B1) and 6T0B (cox5ab III2IV5B2).