Significance
In a technical tour de force, we have created a framework demonstrating the underlying fundamental principles of bidirectional coupling of neuronal and neurotransmitter dynamical systems. Specifically, in the present study, we combined multimodal neuroimaging data to causally explain the functional effects of specific serotoninergic receptor (5-HT2AR) stimulation with psilocybin in healthy humans. Longer term, this could provide a better understanding of why psilocybin is showing considerable promise as a therapeutic intervention for neuropsychiatric disorders including depression, anxiety, and addiction.
Keywords: serotonin, PET, psilocybin, neurotransmitter, whole-brain modeling
Abstract
Remarkable progress has come from whole-brain models linking anatomy and function. Paradoxically, it is not clear how a neuronal dynamical system running in the fixed human anatomical connectome can give rise to the rich changes in the functional repertoire associated with human brain function, which is impossible to explain through long-term plasticity. Neuromodulation evolved to allow for such flexibility by dynamically updating the effectivity of the fixed anatomical connectivity. Here, we introduce a theoretical framework modeling the dynamical mutual coupling between the neuronal and neurotransmitter systems. We demonstrate that this framework is crucial to advance our understanding of whole-brain dynamics by bidirectional coupling of the two systems through combining multimodal neuroimaging data (diffusion magnetic resonance imaging [dMRI], functional magnetic resonance imaging [fMRI], and positron electron tomography [PET]) to explain the functional effects of specific serotoninergic receptor (5-HT2AR) stimulation with psilocybin in healthy humans. This advance provides an understanding of why psilocybin is showing considerable promise as a therapeutic intervention for neuropsychiatric disorders including depression, anxiety, and addiction. Overall, these insights demonstrate that the whole-brain mutual coupling between the neuronal and the neurotransmission systems is essential for understanding the remarkable flexibility of human brain function despite having to rely on fixed anatomical connectivity.
Human connectomics has been very successful in revealing how function arises from structure (1, 2), and by showing how anatomy can give rise to a complex dynamical neuronal system as measured with multimodal neuroimaging (3–5). Despite the attractiveness of the idea that function is shaped by anatomy, it is also clear that the single fixed structure of the anatomical connectome should not be able to give rise to the full palette and complexity of brain function. However, evolution has found a solution to this apparent paradox by dynamically modulating the connectome over time through neuromodulatory systems, enabling the richness of behaviors needed for survival. Indeed, the necessary dynamics of human brain function can be obtained by modulating the effective connectivity of the coupling over time, as proposed by Friston and many others (6, 7). Still, a principled and mechanistic description of the dynamic connectome must bring together the anatomical, neuronal, and neurotransmitter systems at the whole-brain level (8).
Here, we show how the mutual coupling between the neuronal and neurotransmitter systems is fundamental to understanding the dynamic connectome. This can be achieved through whole-brain modeling of multimodal neuroimaging data using a mutually coupled neuronal–neurotransmitter whole-brain model where the structural anatomical connectivity can be measured using diffusion magnetic resonance imaging (dMRI), the functional connectivity with functional magnetic resonance imaging (fMRI), and neurotransmission (receptor density) with positron electron tomography (PET). In this model, the synaptic/neuronal activity and the neurotransmitter diffusive system are portrayed in a realistic biophysical way by a set of separate dynamical equations, which are mutually coupled through the receptor maps and synaptic dynamics (neurotransmitters to neuronal), and the excitation of the projections from the brain regions producing the neurotransmitters (neuronal to neurotransmitters). The explicit linkage between this dual-coupled dynamical system yields a deeper understanding of the crucial mutual coupling between neuronal and neurotransmitters systems at the whole-brain level. We perceive this as a significant improvement compared to our previous unique whole-brain study, which had only one neuronal dynamical system influenced by static neurotransmitter concentrations modulating the neuronal gain (9). Specifically, we demonstrate the explanatory and predictive power of this mutually coupled whole-brain model by investigating the effects of psychedelics on brain activity.
Psilocybin, the prodrug of psilocin (4-OH-dimethyltryptamine), is a good model system to demonstrate the power of a mutually coupled whole-brain model, since it has been shown to act mainly through the serotonin 2A receptor (5-HT2AR) (10), rather than more complex interactions between many receptors. The serotonin system works through the projections of the raphe nucleus. In addition, conveniently, the 5-HT receptor density maps have recently been mapped with PET (11). Here, we were interested in revealing the effects of mutual coupling of both neuronal and neurotransmitter systems on brain repertoire and specifically the effects of psilocybin on resting-state activity on healthy human participants.
Specifically, the bidirectional coupling of the neuronal and neurotransmitter systems was modeled in the following way: For the placebo condition, we used a standard whole-brain model to simulate the neuronal system, i.e., modeling spontaneous brain activity at the whole-brain level (measured with blood oxygen level-dependent [BOLD] fMRI), where each node represents a brain area and the links between them are represented by white matter connections (measured with dMRI). For the psilocybin condition, we mutually coupled the whole-brain neuronal and neurotransmitter systems by including an explicit description of the neurotransmitter dynamical system and the mutual coupling with the neuronal system. This was done by modeling the dynamics of the neurotransmitter system through simulating the release-and-reuptake dynamics, where the serotonin receptor density of each brain area is measured with PET. The neurotransmitter dynamics are then in turn coupled with the neuronal activity through the firing rate activity of the raphe nucleus, source of the serotonin neurotransmitter.
The relevance of the whole-brain modeling developed here is strongly supported by recent studies that have started to demonstrate the functional neuroanatomy underlying the experience of unconstrained cognition and enhanced mind-wandering reported following psilocybin (12–15). Due to its therapeutic action for the treatment of neuropsychiatric disorders such as depression, anxiety, and addiction (16–18), psilocybin has begun to elicit significant interest from the neuropsychiatric community as a potential treatment (19). Long term, the in silico framework presented here for investigating the dynamical connectome has the potential to bring insights and help design interventions for brain disease including neuropsychiatric disorders, which are otherwise difficult to study with traditional animal models.
Results
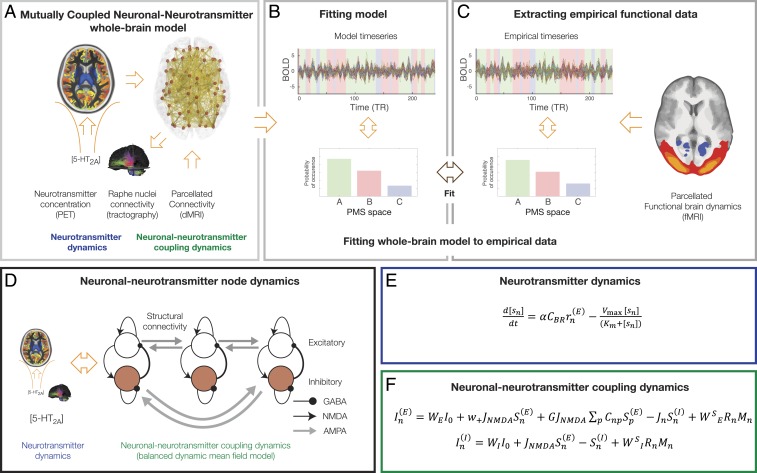
The main aim of the mutually coupled neuronal–neurotransmitter whole-brain model (shown in Fig. 1) is to investigate the tight interactions between these two different but mutually coupled dynamical whole-brain systems (Fig. 1A), which are fitted (Fig. 1B) to the empirical neuroimaging BOLD data (Fig. 1C). In other words, neuronal-only models that fit neuroimaging data using local node dynamics constrained by anatomical structural connectivity (20–23) are now described by a balanced dynamic mean field model that expresses consistently the time evolution of the ensemble activity of the different neural populations building up the spiking network (24, 25). The spontaneous activity of each single brain region is described by a network consisting of two pools of excitatory and inhibitory neurons (Fig. 1D and Eqs. 2–7 in Materials and Methods). The neurotransmitter system, on the other hand, uses a separate set of differential equations describing the dynamics of the neurotransmitter concentration level, given by the well-known Michaelis–Menten release-and-reuptake dynamics (Eq. 13 in Materials and Methods and shown in Fig. 1E) (26–28). We couple the neurotransmitter and neuronal dynamical systems by means of the anatomical connectivity between the raphe nucleus and the rest of the brain, estimated using Human Connectome Project (HCP) dMRI (Materials and Methods). The explicit coupling between the neurotransmitter and the neuronal system is shown in Fig. 1F (and in Eqs. 14 and 15 in Materials and Methods). As can be seen, the neurotransmitter currents are applied to each region’s excitatory and inhibitory pools of neurons using the effectivity/conductivity parameters (WE and WI, respectively). In each region, the neurotransmitter currents are also scaled by each region’s receptor density (measured in vivo using PET). The reverse coupling from the neuronal to the neurotransmitter system is given by inserting in the Michaelis–Menten release-and-reuptake equation the neuronal population firing rate of the source brain region of the neurotransmitter spread from the raphe nucleus.
Fig. 1.
Overview of the coupled neuronal–neurotransmitter whole-brain model. (A) We studied the mutual coupling of two different mutually coupled dynamical whole-brain systems (neuronal and neurotransmitter). (B) This system is fitted to the empirical neuroimaging data, which is described by probabilistic metastable substate (PMS) space (C), which is extracted from the empirical BOLD data. (D) We achieve this by adding a coupled neurotransmitter system (blue) to modulate and interact with the neuronal system (green), which was modeled using a balanced dynamic mean field model that expresses consistently the time evolution of the ensemble activity of the different neural populations building up the spiking network (24, 25). (E) The neurotransmitter system (blue) is modeled by a set of differential equations describing the dynamics of the neurotransmitter concentration level, which is given by the well-known Michaelis–Menten release-and-reuptake dynamics (Materials and Methods) (26–28). (F) The neuronal coupling dynamics (green) is modeled by another set of differential equations describing the spontaneous activity of each single brain region consisting of two pools of excitatory and inhibitory neurons (Materials and Methods). We couple the neurotransmitter and neuronal dynamical systems through the anatomical connectivity between the raphe nucleus and the rest of the brain, estimated using dMRI from the HCP (Materials and Methods). The explicit coupling between the neurotransmitter and the neuronal system is given in Eqs. 16–17 (shown here and described in Materials and Methods). As can be clearly seen, the neurotransmitter currents are applied to each region’s excitatory and inhibitory pools of neurons using the effectivity/conductivity parameters (WE and WI, respectively). In each region, the neurotransmitter currents are also scaled by the each region’s receptor density (measured in vivo using PET). The reverse coupling from the neuronal to the neurotransmitter system is given by inserting in the Michaelis–Menten release-and-reuptake equation the neuronal population firing rate of the source brain region of the neurotransmitter spread from the raphe nucleus.
As a demonstration of the power of this general framework, we show how it can used to explain the specific case of modulation of the serotonin system by psilocybin by analyzing the neuroimaging data from a group of healthy participants being administered psilocybin i.v. (see details in Materials and Methods). This allowed us to study the relevant role of the coupling between the neuronal and neurotransmitter systems to understand the specificity of the 5-HT2A receptor densities [mapped with PET in vivo (11)]. Specifically, the main source of serotonin neurotransmission is the raphe nucleus, which is essential for the correct coupling between both dynamical systems. We therefore used the structural connectivity from this region estimated using dMRI tractography (Materials and Methods).
We fitted the mutually coupled whole-brain model using our framework of describing a brain state as an ensemble or probabilistic “cloud” in a given state space (29). This cloud is of course not clustered into discrete states (30), but it has been shown that clustering can nevertheless be useful in providing so-called “metastable substates” that can significantly distinguish between brain states (31, 32). A brain state is determined by a collection of metastable substates, i.e., of time-varying pseudo-states resulting from the clustering process (33–35).
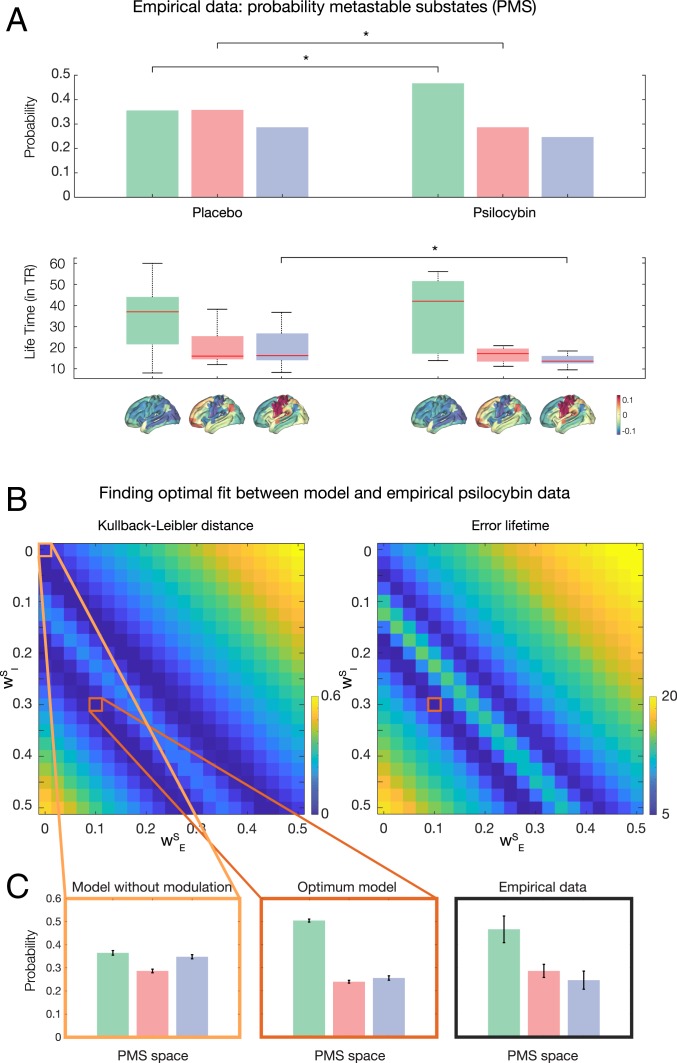
We extracted this probabilistic metastable substate (PMS) space for the empirical psilocybin data (placebo and active condition) using the leading eigenvector dynamics analysis (LEiDA) method (described in detail in Materials and Methods and schematized in Fig. 2) (31). Briefly, we define a time-resolved dynamic FC (dFC) matrix by using BOLD phase coherence connectivity. In order to reduce the dimensionality of the problem, we compute the corresponding time-resolved leading eigenvector, which captures the dominant connectivity pattern of dFC(t) at time t. Then we detect a discrete number of metastable substates by clustering the dFC(t) across time points and subjects. The obtained k cluster centroids define the PMS space, for which we compute the probabilities, lifetimes, and transition probability between them for both the placebo and active conditions of psilocybin. The placebo and the active conditions of the psilocybin can be significantly distinguished by three substates. Fig. 3A shows three different substates of the PMS methodology with associated probabilities and transition probabilities between them for the placebo and active conditions of the psilocybin. As can be seen in the subplots, two substates (1 and 3) are significantly different between the two conditions (P < 10−4) in terms of probability and substate 3 is significantly different for lifetimes (P < 10−2). This demonstrates that the clustering approach is indeed useful for distinguishing brain states.
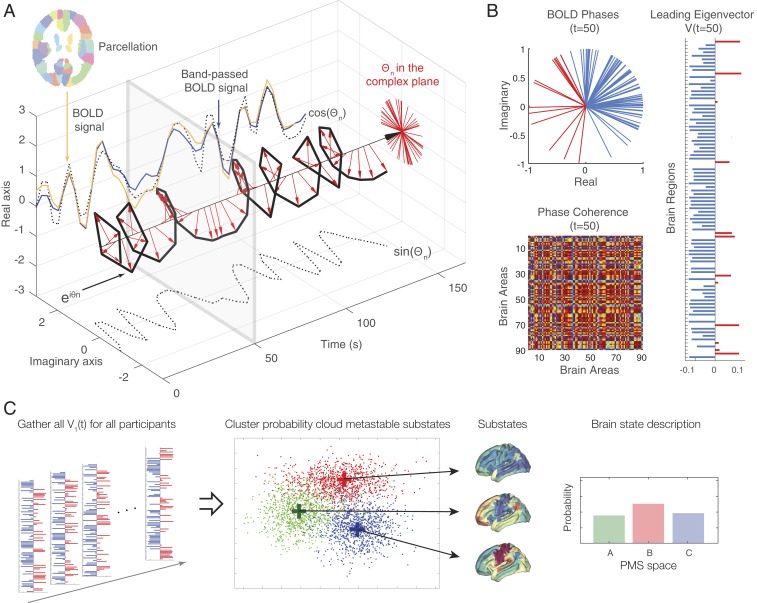
Fig. 2.
Computing the probabilistic metastable substate (PMS) space for whole-brain activity. For illustrative purposes in the following, we sketch the full process of computing the PMS space. For all parcellated brain regions of each participant (A), we extract the original BOLD signal (orange line), bandpass filter between 0.02 and 0.1 Hz (blue line), and then use the Hilbert transform to compute the time-varying amplitude A and its phase θ (with real and imaginary components) of the BOLD signal (black dotted lines). The red arrows represent the BOLD phase at each TR, and, as can be seen, much of the original BOLD signal is captured by the BOLD phase, cos(θ). (B) We compute the BOLD phase coherence matrix between brain regions (Lower) and extract its leading eigenvector V1(t) for every timepoint t (Top, colored red and blue according to their sign). (C) Finally, to determine the PMS space, we gather all leading eigenvectors V1(t) for all participants for every time point as a low-resolution representation of the BOLD phase coherence patterns (leftmost panel). We apply a clustering algorithm (k-means), which divides the data into a predefined number of clusters k (here k = 3). Each cluster is represented by a central vector (green, red, and blue), which represents a recurrent pattern of phase coherence, or substate, which occurs with a given probability (rightmost panel). Any given brain state can thus be represented by this PMS space.
Fig. 3.
Finding the optimum fit of mutually coupled whole-brain model as a function of excitatory and inhibitory coupling parameters. (A) We computed the three probability metastable substate (PMS) spaces that significantly distinguish the placebo and psilocybin conditions (each shown rendered on the human brain). As shown, substates 1 and 2 are significantly different in terms of probability of occurrence and substate 3 in terms of lifetimes. (B) We first fitted the whole-brain model to the PMS space of the placebo condition using only the neuronal system (and thus without coupling the neurotransmitter system). We then studied the coupled neuronal–neurotransmission whole-brain model (using the same centroids found in the neuroimaging empirical data) and a 2D matrix of coupling parameters WE and WI for generating the modeled PMS spaces. We show the resulting matrices for the symmetrized Kullback–Leibler distance (KLD) and average error distance between the lifetimes of the substates. (C) First, we show the PMS when the neurotransmitter system is disconnected ( and , leftmost orange box). However, the most significantly optimal fit to the empirical PMS (rightmost black panel) is found at and (Middle, red box, P < 10−4).
We first fitted the whole-brain model to the PMS space of the placebo condition of psilocybin using only the neuronal system (and thus without coupling the neurotransmitter system). We did this by fitting the minimum of the symmetrized Kullback–Leibler distance (KLD) between the empirical placebo condition PMS and modeled PMS (Materials and Methods). This yielded a global neuronal coupling parameter G = 1.6. Equally, we measured the Markov entropy distance (ME) (Materials and Methods). The optimal values were found to be KLD = 0.002 and ME = 0.05.
Second, we proceeded to study the role of the neurotransmitter dynamical system by coupling both systems in the model. This allowed us to predict the changes in brain dynamics under the effects of psilocybin. Specifically, the mutually coupled whole-brain model used the same centroids found in the neuroimaging empirical data and a two-dimensional (2D) matrix of coupling parameters and for generating the modeled PMS (Fig. 3B). We searched this matrix for the optimal fitting (global minimum) to the active condition of the psilocybin data by using the optimal symmetrized KLD between the empirical active psilocybin condition PMS and the modeled PMS. Nevertheless, it is clear from the deep diagonal ridge that there is an invariant ratio between and that yields equivalent significantly different results with respect to and , which is the relevant comparison. Fig. 3B shows the matrices of the KLD and the error lifetimes of the substates in the empirical active psilocybin condition PMS and the modeled PMS as a function of coupling parameters and (Materials and Methods). The optimal description of the active psilocybin condition is found at the minimum of the KLD at and , while the full disconnection of the neurotransmitter system is found at and . Fig. 3C shows the PMS spaces for the uncoupled system ( and , leftmost panel), the optimal model fit ( and , middle panel), and the empirical data (rightmost panel).
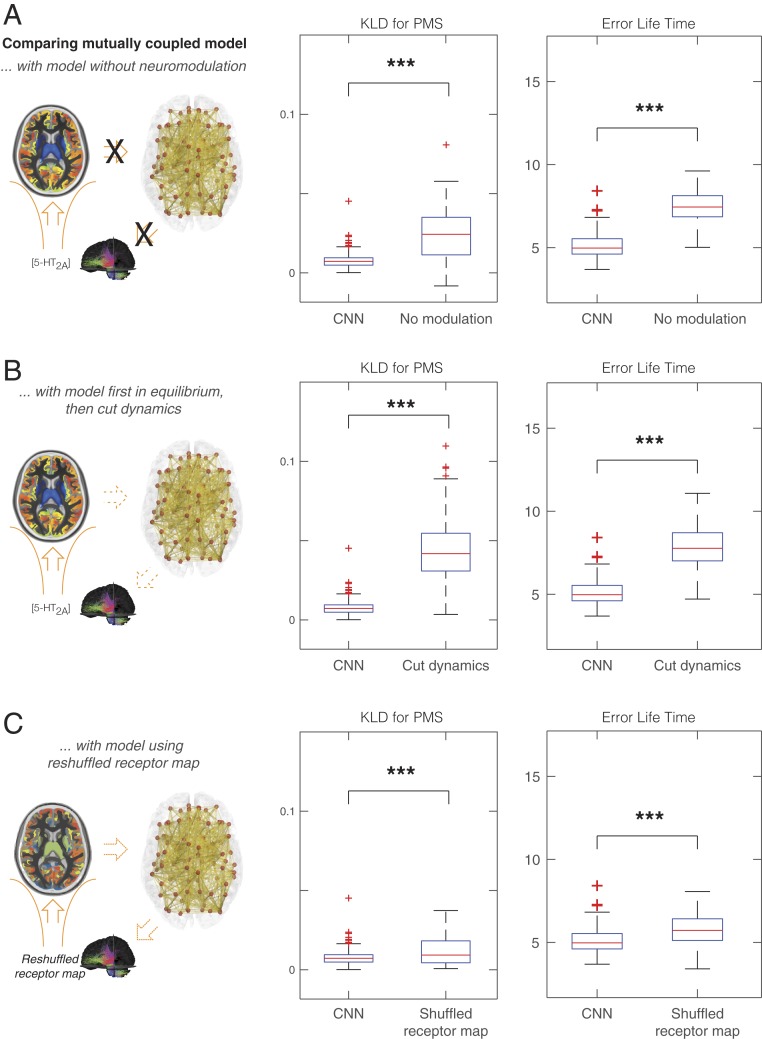
At this optimal fit, the mutually coupled whole-brain model allows us to obtain further insights into the underlying dynamics of neurotransmission involved in psilocybin (in this case for serotonin). Fig. 4A shows a significant difference between the optimal fit and the uncoupled system in KLD (P < 10−6). This clearly demonstrates the significance of coupling the neuronal and neurotransmitter systems. Fig. 4B further dissects this finding by showing a significant difference between the optimal fit and the optimal fit but where we have frozen the feedback dynamics from the neurotransmitter system to the neuronal system (P < 10−6). This was done by allowing the coupling until the steady state is achieved, and then we take the average of the neurotransmitter variables and freeze these and cancel the feedback dynamics. With this uncoupling, we consider a simpler coupled system, where the neuronal dynamics are preserved but unable to influence the neurotransmitter dynamics that are frozen.
Fig. 4.
Optimal dynamical coupled neuronal–neurotransmission (CNN) whole-brain model is significantly better then alternative models. Further insights into the underlying dynamics of neuromodulation involved in psilocybin were obtained by comparing the Kullback–Leibler distance (KLD) for the PMS and the error lifetimes between the empirical data and the whole-brain model undergoing various manipulations. (A) For the optimal fit of the mutually coupled whole-brain model, we found a very significant difference between the optimal fit and the uncoupled system (i.e., without neuromodulation) (P < 10−6). (B) We also found a significant difference when removing the feedback dynamics (P < 10−6). We tested this by allowing optimal model coupling until steady state, at which point we kept just the average of the neurotransmitter variables while cancelling all feedback dynamics. (C) We found a significant difference between using the empirical 5-HT2A receptor densities across the regions at the optimal fit compared with randomly shuffling the receptor densities (P < 10−4). This demonstrates the causal importance of the 5-HT2A receptor densities.
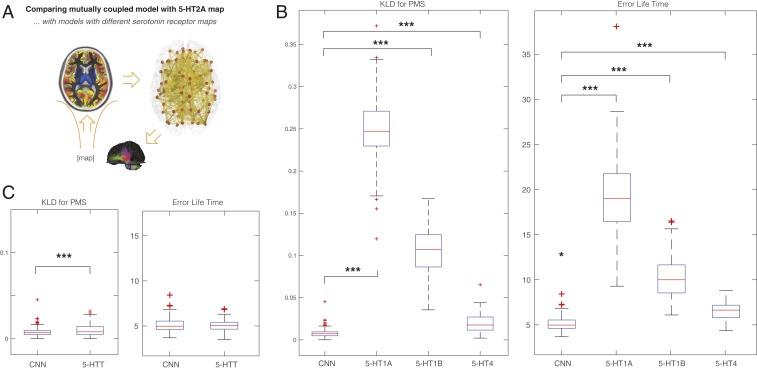
Fig. 4C shows the significant difference between using the empirical 5-HT2A receptor densities across the regions at the optimal fit compared with the results of randomly shuffling the receptor densities (P < 10−4). Similarly, as shown in Fig. 5A, we also tested the specificity of the receptor binding maps by comparing (at the optimal fit) the 5-HT2A receptor with the other serotonin receptors, namely 5-HT1A, 5-HT1B, and 5-HT4, which showed significant differences between 5-HT2A and 5-HT1A (P < 10−6), 5-HT1B (P < 10−6), and 5-HT4 (P < 10−6). This strongly confirms the main role this receptor plays in the effects of psilocybin (10). Finally, in Fig. 5B, we show the comparison to the 5-HTT, which is also significant (P < 0.02). 5-HTT is not a receptor but has been shown to play an important role for the treatment of depression.
Fig. 5.
Optimality of mutually coupled neuronal–neurotransmission (CNN) whole-brain model causally depends on the 5-HT2A receptor map. (A) We studied the effect of replacing the 5-HT2A receptor map with other serotonin receptor binding maps and 5-HTT. (B) The results show significant differences at the optimal fit of the impact of 5-HT2A receptor compared with the other serotonin receptors: 5-HT1A (P < 10−6), 5-HT1B (P < 10−6), and 5-HT4 (P < 10−6). (C) We also found a significant difference when comparing with 5-HTT (P < 0.02 for KLD, but not significant for error lifetimes).
Discussion
Here, we have shown how a dynamic mutually coupled whole-brain model can address the major challenge in neuroscience, which is to explain the paradoxical flexibility of human brain function despite the reliance on a fixed anatomical connectome. One of the most important elements in this flexibility is the way that the fixed connectome can be modulated by neuromodulators to selectively change the balance of the excitation and inhibition of individual brain regions. Here, we modeled the dynamical mutual coupling between neuronal and neuromodulator systems at the whole-brain level. In particular, we implemented a mutually coupled dynamical system, which couples at the whole-brain level the neuronal system, which was modeled using a balanced dynamic mean field model (24, 25) and the neuromodulator system describing the dynamics of the neurotransmitter concentration levels (measured in vivo using PET) and modeled by the well-known Michaelis–Menten release-and-reuptake dynamics (26–28). Here, as proof of principle, we consider the effects of psilocybin on the serotonin system and therefore use anatomical connectivity between the raphe nucleus and the rest of the brain to couple the two systems.
Overall, the results show that the interaction between these dynamical systems is fundamental for explaining the empirical data. In other words, the dynamic mutual interaction between neuronal and neuromodulator systems at the whole-brain level is important to fully explain the functional modulation of brain activity by psilocybin, a powerful psychedelic drug, acting on the serotonin system. This is especially important given the demonstrated ability of psilocybin to rebalance the human brain in treatment-resistant depression. The results provide evidence for how the integration of dMRI (anatomy), fMRI (functional neuronal activity), and PET (neurotransmitter system) at the whole-brain level is necessary for predicting properly brain dynamics as a result of the mutual coupling between a dual dynamical system. This expands on the rich existing experimental and theoretical research on the local effects of neurotransmitters (e.g., refs. 36–40).
Specifically, the results provide insights into the underlying dynamics of neurotransmission involved in psilocybin. In terms of dynamics, we first uncoupled the neuromodulators from the neuronal systems and found that this produced a highly significant breakdown in fitting the empirical data (Fig. 4A). We then ran further simulations to investigate the role of specific parts of the dynamic coupling. The full mutually coupled whole-brain model ran until a steady state was achieved, upon which we then froze the feedback dynamics from neuromodulators to neuronal system by removing coupling through the raphe nucleus. This again produced a highly significant breakdown in fitting the empirical data (Fig. 4B).
In further sets of experiments designed to investigate the importance of the receptor distribution, we changed the distribution of regional receptor densities by 1) randomly shuffling the 5-HT2A (Fig. 4C) and 2) replacing them with those of other serotonin receptors, known to be much less sensitive to psilocybin, namely 5-HT1A, 5-HT1B, and 5-HT4. Again, this produced a highly significant breakdown in the ability to explain the empirical data (Fig. 5B). This result confirms the crucial, causative role of the precise anatomical location and density of 5-HT2A.
Interestingly, as mentioned above, psilocybin has been demonstrated to play a role in rebalancing the human brain in treatment-resistant depression (19). The therapeutic action of psilocybin in depression is thought to depend on activation of serotonin 2A receptors, thereby initiating a multilevel plasticity (41). This is different from selective serotonin reuptake inhibitors, which are the most frequently prescribed antidepressant drug class and whose therapeutic action is thought to begin with reuptake blockade at the 5-HTT; psilocybin has no appreciable affinity or action at the 5-HTT. We were interested in further investigating this by replacing the receptor densities with the 5-HTT densities. We found that the fit with 5-HTT was significantly worse than 5-HT2A (Fig. 5C) and supports the potential for psilocybin in rebalancing brain function in treatment-resistant depression (19).
More broadly, the mutually coupled whole-brain model and results shed important light on our understanding of human brain function. Over the last decade, the discovery of whole-brain modeling has led to important new insights for explaining the principles of human brain function based on human neuroimaging data. The initial breakthrough was the recognition that the anatomical connectivity of the human brain shapes function at the whole-brain level (1, 2). However, it also became clear relatively quickly that this was a by-product of the close link between anatomy and function, given that it is possible to describe static spontaneous activity (i.e., grand average functional spatial connectivity over longer periods) even in the absence of neuronal dynamics, i.e., combining the underlying anatomy with noise (42). Still, it was shown by further investigations that capturing the dynamics of the spatiotemporal whole-brain activity requires more sophisticated dynamical modeling (3, 35, 43). All of these whole-brain studies were initially solely based on neuronal modeling, but recently it was shown that even better results can be obtained when the dynamics of the neuronal system (through the neuronal gain of excitatory neurons) is influenced by static neuromodulation (9).
Here, we have demonstrated the importance of having a biophysically realistic dynamic neuromodulator system, and more importantly the need for full mutual coupling between the full dynamics of both neuronal and neuromodulation systems. In other words, this framework for mutually coupled whole-brain modeling involves the underlying anatomical connectivity and two mutually interacting dynamical neuronal and neuromodulator systems.
Overall, the framework put forward here is likely to be essential for accurately modeling and explaining mechanisms of human brain function in health and disease. The framework combines multimodal neuroimaging from dMRI (anatomy), fMRI (functional neuronal activity), and PET (neurotransmitter system) at the whole-brain level. This offers unique opportunities for investigating how to rebalance human brain activity in disease through pharmacological manipulation that can be discovered and tested in silico in the mutually coupled whole-brain model proposed here. This whole-brain modeling can thus be used to make predictions that can be tested causally with electromagnetic stimulation (deep brain stimulation [DBS] and transcranial magnetic stimulation) or pharmacological interventions. In the future, this framework could further extended by measuring even faster whole-brain dynamics (measured with magnetoencephalography) (44) and incorporating gene expression at the whole-brain level, offering even more exciting avenues for discovering new and effective ways of rebalancing the human brain in disease.
Materials and Methods
We provide here the details on the pipeline used to integrate structural and functional connectivity (dMRI and fMRI) with neurotransmission (PET) in a model of the placebo and psilocybin response in healthy participants (summarized in Fig. 1). Please note that all structural, functional and neuromodulation data are integrated into the Automated Anatomical Labeling (AAL) parcellation:
-
1)
Structural connectivity: probabilistic tractography derived from the dMRI;
-
2)
Functional dynamics: functional dynamics described as probability metastable substates were estimated from the fMRI data obtained in the placebo and psilocybin condition;
-
3)
Neurotransmitter receptor density: estimation of the density of the 5HT-2A receptors that has been obtained using PET;
-
4)
Whole-brain neuronal model: description of the (uncoupled) neuronal dynamic mean field model used to fit the placebo condition;
-
5)
Mutually coupled neuronal and neurotransmission whole-brain model: dynamic coupling of the neuronal and neurotransmission models to dynamically fit psilocybin condition;
-
6)
Empirical fitting of mutually coupled whole-brain model to neuroimaging data: measuring the fit of model to the empirical neuroimaging data.
Parcellation.
Based on our previous whole-brain studies, we used the AAL atlas but considered only the 90 cortical and subcortical noncerebellar brain regions (45). All structural, functional, and neuromodulation data were integrated using this atlas.
Structural connectivity.
Ethics.
The study was approved by the Research Ethics Committee of the Central Denmark Region (De Videnskabsetiske Komitéer for Region Midtjylland). Written informed consent was obtained from all participants prior to participation.
Participants and acquisition.
The dMRI data used in the study was collected from 16 healthy right-handed participants at Aarhus University, Denmark (11 men; mean age, 24.8 ± 2.5) (46, 47). We recorded the imaging data in a single session on a 3-T Siemens Skyra scanner at Center of Functionally Integrative Neuroscience, Aarhus University, Denmark. The structural MRI T1 scan used the following parameters: reconstructed matrix size, 256 × 256; voxel size of 1 mm3; echo time (TE) of 3.8 ms; and repetition time (TR) of 2,300 ms. We collected dMRI using the following parameters of TE = 84 ms, TR = 9,000 ms, flip angle = 90°, reconstructed matrix size of 106 × 106, voxel size of 1.98 × 1.98 mm with slice thickness of 2 mm and a bandwidth of 1,745 Hz/Px. The data were recorded with 62 optimal nonlinear diffusion gradient directions at b = 1,500 s/mm2. Approximately one nondiffusion weighted image (b = 0) per 10 diffusion-weighted images was acquired. We collected two dMRI datasets: one with anterior-to-posterior phase encoding direction and the other acquired in the opposite direction.
Tractography.
We used the structural connectivity between 90 AAL regions in the dMRI dataset described above. First, we used the linear registration tool from the FSL toolbox (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki; Functional Magnetic Resonance Imaging of the Brain [FMRIB], Oxford, UK) (48) to coregister the echo-planar imaging (EPI) image to the T1-weighted structural image. In turn, the T1-weighted image was coregistered to the T1 template of ICBM152 in Montreal Neurological Institute (MNI) space (49). We concatenated and inversed the resulting transformations, which were further applied to warp the AAL template (45) from MNI space to the EPI native space; ensured discrete labeling values were preserved by interpolation using nearest-neighbor method. The brain parcellations were thus conducted in each individual’s native space. The structural connectivity maps were generated for each participant using the dMRI data acquired. We combined the two datasets acquired with different phase encoding to optimize signal in difficult regions. Constructing these structural connectivity maps or structural brain networks involved a three-step process: 1) Regions of the whole-brain network were defined using the AAL template as used in the functional MRI data. 2) Probabilistic tractography was used to estimate the connections between nodes in the whole-brain network (i.e., edges). 3) Data were averaged across participants.
The FSL diffusion toolbox (Fdt) in FSL was used to carry out the various processing stages of the dMRI dataset. The default parameters of this imaging preprocessing pipeline were used for all participants. We then estimated the local probability distribution of fiber direction at each voxel (50). The probtrackx tool in Fdt was used to provide automatic estimation of crossing fibers within each voxel, which significantly improves the tracking sensitivity of nondominant fiber populations in the human brain (51).
We defined the connectivity probability from a given seed voxel i to another voxel j by the proportion of fibers passing through voxel i that reach voxel j when using a sampling of 5,000 streamlines per voxel (51). We then extended from the voxel level to the regional level, i.e., to a parcel in the AAL consisting of n voxels, i.e., 5,000 × n fibers were sampled. This allowed us to compute the connectivity probability from region n to region p as the number of sampled fibers in region n that connect the two regions divided by 5,000 × v, where v is the number of voxels in a given AAL region i.
We computed the connectivity probability from a given brain region to each of the other 89 regions within the AAL. It is important to note that the dependence of tractography on the seeding location, i.e., the probability from voxel i to voxel j is not necessarily equivalent to that from j to i. Still, these two probabilities are highly correlated across the brain for all participants (the least Pearson r = 0.70, P < 10−50). We defined the unidirectional connectivity probability between regions n and p by averaging these two connectivity probabilities since directionality of connections cannot be determined based on dMRI. This unidirectional connectivity can be thought of as a measure of the structural connectivity between the two areas, with . The computation of the regional connectivity probability was implemented using in-house Perl scripts. For both phase encoding directions, we constructed 90 × 90 symmetric weighted networks, based on the AAL90 parcellation, and normalized by the number of voxels in each AAL region. This represents the structural connectivity network organization of the brain.
In order to consider the serotonin source region, we included in our model the connectivity between the raphe nucleus region and the rest of the brain. In this way, the neuronal activity across the whole brain excites through the existing fibers the raphe neurons, and this information is used in the neurotransmitter system (see below). In brief, we used the estimate of the raphe nucleus region from the Harvard Ascending Arousal Network Atlas (Athinoula A. Martinos Center for Biomedical Imaging; https://www.nmr.mgh.harvard.edu/resources/aan-atlas) (52). We then used LeadDBS Mapper (https://www.lead-dbs.org/) within Matlab to estimate the whole-brain tractography from this mask of the raphe nucleus to the rest of the brain (53, 54) using the Structural group connectome based on 32 Adult Diffusion HCP subjects GQI (55, 56). This allowed us to estimate the projections from the raphe nucleus to each AAL region, represented as a vector of 90 values.
It is important to note that the AAL parcellation has been shown to be less than optimal, in terms of being the least homogeneous parcellation scheme compared to a number of other parcellation schemes (57). Still, it is not clear whether the methodology used for this comparison is particularly meaningful. A recent paper by Eickhoff et al. (58) reviewed the literature on the topographic organization of the brain and concluded that there is no current consensus about what is the right spatial parcellation scheme. It is also important to note that, in contrast to these two papers, which are mostly concerned with the spatial organization of the brain, here we focus on the spatiotemporal global dynamics. As such, the AAL parcellation would seem a good choice given that AAL yields excellent significant results in the whole-brain literature in general (20, 35, 59), and, crucially, the relative low number of parcels in the AAL makes it highly suitable for our very extensive computational demands.
Functional dynamics.
The functional data are described in detail in a previously published study (13), but here we briefly summarize the participants, study setting, and acquisition protocol.
Ethics.
The study was approved by a National Health Service research ethics committee. Written informed consent was obtained from all participants prior to participation.
Participants in psilocybin study.
fMRI data from nine healthy subjects were included in this study (seven men; age, 32 ± 8.9 SD y of age). Study inclusion criteria were: at least 21 y of age, no personal or immediate family history of a major psychiatric disorder, substance dependence, cardiovascular disease, and no history of a significant adverse response to a hallucinogenic drug. All of the subjects had used psilocybin at least once before, but not within 6 wk of the study.
All subjects underwent two 12-min eyes-closed resting-state fMRI scans over separate sessions, at least 7 d apart. In each session, subjects were injected i.v. with either psilocybin (2 mg dissolved in 10 mL of saline, 60-s i.v. injection) or a placebo (10 mL of saline, 60-s i.v. injection) in a counterbalanced design. The injections were given manually by a medical doctor within the scanning suite. The infusions began exactly 6 min after the start of the 12-min scans and lasted 60 s. The subjective effects of psilocybin were felt almost immediately after injection and sustained for the remainder of the scanning session.
Anatomical scans.
Imaging was performed on a 3-T GE HDx system. These were 3D fast spoiled gradient echo scans in an axial orientation, with field of view = 256 × 256 × 192 and matrix = 256 × 256 × 192 to yield 1-mm isotropic voxel resolution. TR/TE, 7.9/3.0 ms; inversion time, 450 ms; flip angle, 20°.
BOLD fMRI data acquisition.
Two BOLD-weighted fMRI data were acquired using a gradient echo planer imaging sequence, TR/TE = 2,000/35 ms, field of view = 220 mm, 64 × 64 acquisition matrix, parallel acceleration factor = 2, 90° flip angle. Thirty-five oblique axial slices were acquired in an interleaved fashion, each 3.4 mm thick with zero slice gap (3.4-mm isotropic voxels). The precise length of each of the two BOLD scans was 7:20 min.
Preprocessing.
The fMRI data were preprocessed using MELODIC 3.14 (60), part of FSL (FMRIB’s Software Library; https://fsl.fmrib.ox.ac.uk/fsl/fslwiki) with standard parameters and without discarding any ICA components. For each participant and for each brain state (i.e., placebo and psilocybin), we used FSL tools to extract and average the BOLD time courses from all voxels within each region defined in the AAL atlas (considering only the 90 cortical and subcortical noncerebellar brain regions) (45).
Extracting PMSs.
Extracting the PMSs for the neuroimaging data relies on conducting the LEiDA analysis (31), summarized in Fig. 2. Similar to the procedure described in Deco et al. (29), we compute a phase coherence matrix to capture the amount of interregional BOLD signal synchrony at each time point, for all subject and conditions (placebo and psilocybin conditions). The phase coherence between each pair of brain regions is given by the following:
| [1] |
where the BOLD phases, , at a region is estimated using the Hilbert transform for each BOLD regional time course. The Hilbert transform expresses a given signal in polar coordinates as . Using the cosine function, two regions and with temporarily aligned BOLD signals (i.e., with similar angles) at a given TR will have a phase coherence value close to 1 [since ]. On the other hand, time points where the BOLD signals are orthogonal (for instance, one increasing at 45° and the other decreasing at 45°) will have close to 0 [since ]. The resulting for each participant in each condition is a 3D matrix with size N×N×T, where N = 90 is the number of brain regions and T is the total number of time points (here different for each subject and each condition). It is important to note that the phase coherence matrix is undirected and, as such, for each time , the N×N matrix is symmetric across the diagonal. In order to characterize the evolution of the dFC matrix over time, we reduced the dimensionality of the problem by focusing on the evolution of the leading eigenvectors of the matrices. The leading eigenvector, , is a N×1 vector that captures the dominant connectivity pattern of the at each time , given in matrix format by its outer product . This approach substantially reduces the dimensionality of the data when compared to more traditional approaches considering all of the values in the connectivity matrix (43, 61, 62). Further details about LEiDA can be found in the work of Cabral et al. (31).
After computing the leading eigenvector of the phase coherence matrix for each TR, we identified PMSs, i.e., recurrent FC patterns in the data, first described by Deco et al. (29). We detected a discrete number of FC patterns by clustering the leading eigenvectors from the collapsed pre/post-psilocybin fMRI data (two conditions) including all participants. We ran the k-means clustering algorithm with values of k from 2 to 20. Clustering solutions output a k number of cluster centroids in the shape of N×1 VC vectors, which represent recurrent PMSs. The outer product of each cluster centroid vector is a N×N matrix representing the dominant connectivity pattern and the elements of weight the contribution of each brain area to the community structure established by the signs of the corresponding vector elements. We used HCP Workbench to render the cluster centroid vectors onto a cortical surface facilitate to visualization and interpretation of FC states. Here, we found that the optimal number of clusters was k = 3 according many criteria including Silhouette analysis and the minimal P value for significant differences between probabilities between conditions.
Upon identifying PMSs, we computed the probability of occurrence and lifetime of each metastable substate in each condition. The probability of occurrence (or fractional occupancy) is given by the ratio of the number of epochs assigned to a given cluster centroid divided by the total number of epochs (TRs) in each experimental condition. The lifetime is defined as the amount of time spent in each metastable substate (cluster centers). The probabilities and lifetimes were computed for each participant, in each experimental condition, and across the whole range of clustering solutions explored.
Furthermore, to capture the trajectories of FC dynamics in a directional manner, we computed the switching matrix. This matrix contains the probabilities of transitioning from a given FC state (rows) to any of the other FC states (columns). Differences in probabilities of transition and probabilities of occurrence were statistically assessed between conditions using a permutation-based paired t test. This nonparametric test uses permutations of group labels to estimate the null distribution (computed independently for each experimental condition). To compare populations, we applied a t test to each of 1,000 permutations and used a significance threshold of α = 0.05.
Neurotransmitter receptor density.
A previous study has carefully described the methods used to obtain the 5-HT2A receptor density distribution (11), but here we briefly summarize the main methods.
Ethics.
The study was approved by the Ethics Committee of Copenhagen and Frederiksberg, Denmark. Written informed consent was obtained from all participants prior to participation.
Participants.
The participants were healthy male and female controls from the freely available Cimbi database (63). The present data analysis was restricted to include individuals aged between 18 and 45 y. Acquisition of PET data and structural MRI scans were taken from 210 individual participants, yielding a total of 232 PET scans; of which 189 participants had only one scan, 20 participants had two scans, and a single had three scans.
PET and structural MRI.
Full details on PET and structural MRI acquisition parameters can be found in the original study (63) and in abbreviated form in ref. 9.
Extracting receptor density maps.
Similar to Deco et al. (9), we extracted the average receptor density for each individual AAL region 5-HT1A, 5-HT1B, 5-HT2A, and 5-HT4 as well as 5-HTT using standard FSL tools on the freely available receptor density maps in MNI space.
Whole-brain model neuronal system.
Whole-brain neuronal model (anatomy-plus-neuronal activity).
First, we modeled the system without any coupling between systems, i.e., a pure neurodynamical system (9). For this, we simulated the spontaneous brain activity at the level of the whole brain in a network model where each node represents a brain region and the links between nodes represent white matter connections. As proposed by Deco et al. (25), a dynamic mean field (DMF) model is used to simulate the activity in each brain region. In brief, based on the original reduction of Wong and Wang (64), this DMF model uses a reduced set of dynamical equations describing the activity of coupled excitatory (E) and inhibitory (I) pools of neurons to describe the activity of large ensembles of interconnected excitatory and inhibitory spiking neurons. The inhibitory currents, , are mediated by GABAA receptors, while excitatory synaptic currents, , are mediated by NMDA receptors. In terms of connectivity, each brain region, n, consists of reciprocally connected excitatory and inhibitory pools of neurons, whereas coupling between two areas n and p occurs only at the excitatory-to-excitatory level where it is scaled by the underlying anatomical connectivity (Materials and Methods, Structural Connectivity).
The following system of coupled differential equations expresses the DMF model at the whole-brain level:
| [2] |
| [3] |
| [4] |
| [5] |
| [6] |
| [7] |
As can be seen, for each inhibitory (I) or excitatory (E) pool of neurons in each brain area n, the vector represents the total input current (in nanoamperes), the firing rate is denoted by the vector (in hertz), while the synaptic gating is denoted by the vector . The total input currents received by the E and I pools is converted by the neuronal response functions, , into firing rates, , using the input–output function of Abbott and Chance (65), where the gain factors nC−1 and nC−1 determine the slope of . Above the threshold currents of nA, and nA, the firing rates increase linearly with the input currents. The shape of the curvature of around is given by the constants and . GABA receptors with s controls the average synaptic gating in inhibitory pools. NMDA receptors with a decay time constant s and (the factor 1,000 is for expressing everything in milliseconds) control the synaptic gating variable of excitatory pools, . All excitatory synaptic couplings is weighted by nA, while the weight of recurrent excitation is . The overall effective external input is nA with and . Note that in Eqs. 6 and 7, the uncorrelated standard Gaussian noise, , has an amplitude of nA.
Emulating the resting-state condition, we used parameters in the DMF model based on Wong and Wang (64) such that each isolated node exhibited the typical noisy spontaneous activity with low firing rate observed in electrophysiology experiments (66–69). Furthermore, similar to Deco et al. (25), for each node n, we adjusted the inhibition weight, , such that the firing rate of the excitatory pools remained clamped at 3 Hz—even when receiving excitatory input from connected areas. The algorithm for achieving feedback inhibition control is described by Deco et al. (25), where it was shown that using this leads to a better prediction of the resting-state FC and to a more realistic evoked activity.
The whole-brain network model used parcellated structural and functional MRI data from 90 cortical and subcortical brain regions (9), where each brain region n receives excitatory input from all structurally connected regions p into its excitatory pool, weighted by the underlying anatomical connectivity matrix, , obtained from dMRI (9) (Materials and Methods, Structural Connectivity). Importantly, we used a global coupling factor G to equally scale all interarea E-to-E connections. Finding the optimal working point of the system simply requires finding the optimal global scaling factor, such that the simulated activity by the model is maximally fitting the empirical resting-state activity of participants in the placebo conditions.
The simulations were run with a time step of 1 ms for a range of G between 0 and 2.5 (with increments of 0.025). In order to emulate the empirical resting-state scans from nine participants, for each value of G, we ran 200 simulations of 435 s each.
Please note that to transform the simulated mean field activity from our DMF model into a BOLD signal, the generalized hemodynamic model of Stephan et al. (70) was used. This involves computing the BOLD signal in each brain area, n, arising from the firing rate of the excitatory pools , such that an increase in the firing rate causes an increase in a vasodilatory signal, , subject to autoregulatory feedback. In proportion to this vasodilatory signal, the blood inflow induces changes in deoxyhemoglobin content and blood volume .
These biophysical variables are described by these equations:
| [8] |
| [9] |
| [10] |
| [11] |
where is a time constant, is the resting oxygen extraction fraction, and the resistance of the veins is represented by α.
For each area, n, the BOLD signal, , is a static nonlinear function of deoxyhemoglobin content, , and volume, , that comprises a volume-weighted sum of extravascular and intravascular signals:
| [12] |
We used all biophysical parameters stated by Stephan et al. (70) and concentrated on the most functionally relevant frequency range for resting-state activity, i.e., both simulated and empirical and BOLD signals were bandpass filtered between 0.1 and 0.01 Hz (71–74).
Mutually coupled whole-brain neuronal and neurotransmitter system.
Finally, in order to fully couple the whole-brain neuronal and neurotransmitter systems, we have to include an explicit description of the neurotransmitter dynamical system and the mutual coupling. We do this by modeling the dynamics of the neurotransmitter system through simulating the release-and-reuptake dynamics. These dynamics are then in turn coupled with the neuronal activity through the firing rate activity of the raphe brain region, source of the serotonin neurotransmitter.
Specifically, the concentration of serotonin is modeled through the Michaelis–Menten equations (26–28):
| [13] |
where is a vector, denoting the dynamics of the neurotransmitter concentration level in a brain region n, and where the corresponding density of a serotonin receptor is measured with PET. This is defining the interaction between the neuronal and neurotransmitter system through the first term on the right-hand side in Eq. 13 (i.e., brain raphe coupling). The is outer product of the fiber density connectivity vector between the whole brain and the raphe nucleus, obtained through dMRI probabilistic tractography, and is a factor normalizing the activity such that the current generated by the neurotransmitter (Eqs. 14–16) is working in the nonlinear central sigmoidal part. For the second term on the right-hand side of Eq. 13, nM⋅s−1 is the maximum reuptake rate and is the substrate concentration where the uptake proceeds at one-half of the maximum rate (26–28).
The reverse coupling, i.e., the effect of the neurotransmitter system on the neuronal system, is described in Eqs. 14–16. Specifically, the neuronal activity is modeled as a dynamical system in Eqs. 14 and 15 (as a coupled variation of Eqs. 2 and 3) by generating an extra current on the excitatory pyramidal and GABAergic inhibitory neurons in the following way:
| [14] |
| [15] |
where and are the excitatory and inhibitory feedback coupling parameters from the neurotransmitter system to the neuronal activity. The density of a serotonin receptor (as measured with PET) weighs the serotonin modulated current vector (current for each brain region), which is given by the sigmoid-like function commonly used in pharmacology and current equation (26):
| [16] |
where , , and (26). The last terms on the right-hand side of Eqs. 14 and 15 describe the coupling of the neuronal to the neurotransmitter system. In this way, both neuronal and neurotransmitter dynamical system are explicitly expressed and mutually coupled.
Empirical fitting of mutually coupled whole-brain model to neuroimaging data.
We used the following measurements to measure the empirical fit of the mutually coupled whole-brain system.
Comparing empirical and simulated PMS space measurements.
We computed the symmetrized KLD between the simulated and empirical corresponding probabilities of the metastable substates, i.e., the probabilities of the extracted empirical centers after clusterization:
| [17] |
where and are the empirical and simulated probabilities on the same empirical extracted metastable substates i.
Comparing empirical and simulated transition probabilities between metastable substates.
We computed the entropy rate of a Markov chain, with states and transition matrix . The rate entropy is given by the following:
| [18] |
where
| [19] |
The stationary probability of state is given by . For long realizations of the Markov chain, the probabilities of each state converge to the stationary distribution , which is the solution of the following equation:
| [20] |
The stationary distribution is the eigenvector of the transpose of the transition matrix with associated eigenvalue equal to 1. A Markov model with a lot of transitions will have a large rate entropy, while low entropy will be found in a Markov model with minimal transitions. For each transition matrix, we obtained the stationary distribution and, then, computed the entropy rate. Comparing the two transition probability matrices is defined by the absolute value of the difference between both respective Markov entropies.
Data Availability.
The code to run the analysis and multimodal neuroimaging data from the experiment are available on GitHub (https://github.com/decolab/pnas-neuromod).
Acknowledgments
G.D. is supported by the Spanish Research Project PSI2016-75688-P (Agencia Estatal de Investigación/Fondo Europeo de Desarrollo Regional, European Union); by the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreements 720270 (Human Brain Project [HBP] SGA1) and 785907 (HBP SGA2); and by the Catalan Agencia de Gestión de Ayudas Universitarias Programme 2017 SGR 1545. M.L.K. is supported by the European Research Council Consolidator Grant CAREGIVING (615539); Center for Music in the Brain, funded by the Danish National Research Foundation (DNRF117); and Centre for Eudaimonia and Human Flourishing, funded by the Pettit and Carlsberg Foundations. J. Cabral is supported by the Portuguese Foundation for Science and Technology (CEECIND/03325/2017), Portugal.
Footnotes
The authors declare no competing interest.
Database deposition: The code to run the analysis and the multimodal neuroimaging data (dMRI, fMRI, and PET) from the experiment are available on GitHub (https://github.com/decolab/pnas-neuromod).
See online for related content such as Commentaries.
References
- 1.Sporns O., Tononi G., Kötter R., The human connectome: A structural description of the human brain. PLoS Comput. Biol. 1, e42 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atasoy S., Donnelly I., Pearson J., Human brain networks function in connectome-specific harmonic waves. Nat. Commun. 7, 10340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deco G., Cruzat J., Kringelbach M. L., Brain songs framework used for discovering the relevant timescale of the human brain. Nat. Commun. 10, 583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sporns O., Network attributes for segregation and integration in the human brain. Curr. Opin. Neurobiol. 23, 162–171 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Deco G., Tononi G., Boly M., Kringelbach M. L., Rethinking segregation and integration: Contributions of whole-brain modelling. Nat. Rev. Neurosci. 16, 430–439 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Friston K. J., Functional and effective connectivity: A review. Brain Connect. 1, 13–36 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Pallarés V., et al. , Extracting orthogonal subject- and condition-specific signatures from fMRI data using whole-brain effective connectivity. Neuroimage 178, 238–254 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopell N. J., Gritton H. J., Whittington M. A., Kramer M. A., Beyond the connectome: The dynome. Neuron 83, 1319–1328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deco G., et al. , Whole-brain multimodal neuroimaging model using serotonin receptor maps explains non-linear functional effects of LSD. Curr. Biol. 28, 3065–3074.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Nichols D. E., Psychedelics. Pharmacol. Rev. 68, 264–355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beliveau V., et al. , A high-resolution in vivo atlas of the human brain’s serotonin system. J. Neurosci. 37, 120–128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lord L. D., et al. , Dynamical exploration of the repertoire of brain networks at rest is modulated by psilocybin. Neuroimage 199, 127–142 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Carhart-Harris R. L., et al. , Neural correlates of the psychedelic state as determined by fMRI studies with psilocybin. Proc. Natl. Acad. Sci. U.S.A. 109, 2138–2143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tagliazucchi E., Carhart-Harris R., Leech R., Nutt D., Chialvo D. R., Enhanced repertoire of brain dynamical states during the psychedelic experience. Hum. Brain Mapp. 35, 5442–5456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schartner M. M., Carhart-Harris R. L., Barrett A. B., Seth A. K., Muthukumaraswamy S. D., Increased spontaneous MEG signal diversity for psychoactive doses of ketamine, LSD and psilocybin. Sci. Rep. 7, 46421 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grob C. S., et al. , Pilot study of psilocybin treatment for anxiety in patients with advanced-stage cancer. Arch. Gen. Psychiatry 68, 71–78 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Johnson M. W., Garcia-Romeu A., Cosimano M. P., Griffiths R. R., Pilot study of the 5-HT2AR agonist psilocybin in the treatment of tobacco addiction. J. Psychopharmacol. 28, 983–992 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carhart-Harris R. L., et al. , Psilocybin with psychological support for treatment-resistant depression: An open-label feasibility study. Lancet Psychiatry 3, 619–627 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Carhart-Harris R. L., et al. , Psilocybin for treatment-resistant depression: fMRI-measured brain mechanisms. Sci. Rep. 7, 13187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabral J., Kringelbach M. L., Deco G., Functional connectivity dynamically evolves on multiple time-scales over a static structural connectome: Models and mechanisms. Neuroimage 160, 84–96 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Deco G., Kringelbach M. L., Great expectations: Using whole-brain computational connectomics for understanding neuropsychiatric disorders. Neuron 84, 892–905 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Deco G., Jirsa V. K., McIntosh A. R., Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat. Rev. Neurosci. 12, 43–56 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Honey C. J., Kötter R., Breakspear M., Sporns O., Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc. Natl. Acad. Sci. U.S.A. 104, 10240–10245 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deco G., et al. , Resting-state functional connectivity emerges from structurally and dynamically shaped slow linear fluctuations. J. Neurosci. 33, 11239–11252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deco G., et al. , How local excitation-inhibition ratio impacts the whole brain dynamics. J. Neurosci. 34, 7886–7898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi A., et al. , An integrated modelling framework for neural circuits with multiple neuromodulators. J. R. Soc. Interface 14, 20160902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.John C. E., Budygin E. A., Mateo Y., Jones S. R., Neurochemical characterization of the release and uptake of dopamine in ventral tegmental area and serotonin in substantia nigra of the mouse. J. Neurochem. 96, 267–282 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Michaelis L., Menten M. L., Die Kinetik der Invertinwirkung. Biochem. Z. 49, 333–369 (1913). [Google Scholar]
- 29.Deco G., et al. , Awakening: Predicting external stimulation to force transitions between different brain states. Proc. Natl. Acad. Sci. U.S.A. 116, 18088–18097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laumann T. O., et al. , On the stability of BOLD fMRI correlations. Cereb. Cortex 27, 4719–4732 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabral J., et al. , Cognitive performance in healthy older adults relates to spontaneous switching between states of functional connectivity during rest. Sci. Rep. 7, 5135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker A. P., et al. , Fast transient networks in spontaneous human brain activity. eLife 3, e01867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tognoli E., Kelso J. A., The metastable brain. Neuron 81, 35–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deco G., Kringelbach M. L., Metastability and coherence: Extending the communication through coherence hypothesis using a whole-brain computational perspective. Trends Neurosci. 39, 125–135 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Deco G., Kringelbach M. L., Jirsa V. K., Ritter P., The dynamics of resting fluctuations in the brain: Metastability and its dynamical cortical core. Sci. Rep. 7, 3095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deco G., Thiele A., Attention: Oscillations and neuropharmacology. Eur. J. Neurosci. 30, 347–354 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris K. D., Thiele A., Cortical state and attention. Nat. Rev. Neurosci. 12, 509–523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiele A., Bellgrove M. A., Neuromodulation of attention. Neuron 97, 769–785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sajedin A., Menhaj M. B., Vahabie A. H., Panzeri S., Esteky H., Cholinergic modulation promotes attentional modulation in primary visual cortex—a modeling study. Sci. Rep. 9, 20186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Kempen J., Panzeri S., Thiele A., Cholinergic control of information coding. Trends Neurosci. 40, 522–524 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Carhart-Harris R. L., How do psychedelics work? Curr. Opin. Psychiatry 32, 16–21 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Messé A., Rudrauf D., Benali H., Marrelec G., Relating structure and function in the human brain: Relative contributions of anatomy, stationary dynamics, and non-stationarities. PLoS Comput. Biol. 10, e1003530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen E. C., Battaglia D., Spiegler A., Deco G., Jirsa V. K., Functional connectivity dynamics: Modeling the switching behavior of the resting state. Neuroimage 105, 525–535 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Pfeffer T., et al. , Catecholamines alter the intrinsic variability of cortical population activity and perception. PLoS Biol. 16, e2003453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tzourio-Mazoyer N., et al. , Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15, 273–289 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Deco G., et al. , Single or multiple frequency generators in on-going brain activity: A mechanistic whole-brain model of empirical MEG data. Neuroimage 152, 538–550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deco G., Van Hartevelt T. J., Fernandes H. M., Stevner A., Kringelbach M. L., The most relevant human brain regions for functional connectivity: Evidence for a dynamical workspace of binding nodes from whole-brain computational modelling. Neuroimage 146, 197–210 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Jenkinson M., Bannister P., Brady M., Smith S., Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Collins D. L., Neelin P., Peters T. M., Evans A. C., Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J. Comput. Assist. Tomogr. 18, 192–205 (1994). [PubMed] [Google Scholar]
- 50.Behrens T. E., et al. , Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn. Reson. Med. 50, 1077–1088 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Behrens T. E., Berg H. J., Jbabdi S., Rushworth M. F., Woolrich M. W., Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage 34, 144–155 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edlow B. L., et al. , Neuroanatomic connectivity of the human ascending arousal system critical to consciousness and its disorders. J. Neuropathol. Exp. Neurol. 71, 531–546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horn A., Kühn A. A., Lead-DBS: A toolbox for deep brain stimulation electrode localizations and visualizations. Neuroimage 107, 127–135 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Horn A., et al. , Lead-DBS v2: Towards a comprehensive pipeline for deep brain stimulation imaging. Neuroimage 184, 293–316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Setsompop K., et al. , Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. Neuroimage 80, 220–233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horn A., et al. , Probabilistic conversion of neurosurgical DBS electrode coordinates into MNI space. Neuroimage 150, 395–404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordon E. M., et al. , Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex 26, 288–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eickhoff S. B., Constable R. T., Yeo B. T. T., Topographic organization of the cerebral cortex and brain cartography. Neuroimage 170, 332–347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cabral J., et al. , Exploring mechanisms of spontaneous functional connectivity in MEG: How delayed network interactions lead to structured amplitude envelopes of band-pass filtered oscillations. Neuroimage 90, 423–435 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Beckmann C. F., Smith S. M., Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. Imaging 23, 137–152 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Hutchison R. M., et al. , Dynamic functional connectivity: Promise, issues, and interpretations. Neuroimage 80, 360–378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Preti M. G., Bolton T. A., Ville D. V., The dynamic functional connectome: State-of-the-art and perspectives. Neuroimage 160, 41–54 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Knudsen G. M., et al. , The center for integrated molecular brain imaging (Cimbi) database. Neuroimage 124, 1213–1219 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Wong K. F., Wang X. J., A recurrent network mechanism of time integration in perceptual decisions. J. Neurosci. 26, 1314–1328 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abbott L. F., Chance F. S., Drivers and modulators from push-pull and balanced synaptic input. Prog. Brain Res. 149, 147–155 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Burns B. D., Webb A., The spontaneous activity of neurones in the cat’s cerebral cortex. Proc. R. Soc. Lond. B Biol. Sci. 194, 211–223 (1976). [DOI] [PubMed] [Google Scholar]
- 67.Koch K. W., Fuster J. M., Unit activity in monkey parietal cortex related to haptic perception and temporary memory. Exp. Brain Res. 76, 292–306 (1989). [DOI] [PubMed] [Google Scholar]
- 68.Softky W. R., Koch C., The highly irregular firing of cortical cells is inconsistent with temporal integration of random EPSPs. J. Neurosci. 13, 334–350 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shadlen M. N., Newsome W. T., The variable discharge of cortical neurons: Implications for connectivity, computation, and information coding. J. Neurosci. 18, 3870–3896 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stephan K. E., Weiskopf N., Drysdale P. M., Robinson P. A., Friston K. J., Comparing hemodynamic models with DCM. Neuroimage 38, 387–401 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Glerean E., Salmi J., Lahnakoski J. M., Jääskeläinen I. P., Sams M., Functional magnetic resonance imaging phase synchronization as a measure of dynamic functional connectivity. Brain Connect. 2, 91–101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Biswal B., Yetkin F., Haughton V., Hyde J., Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 34, 537–541 (1995). [DOI] [PubMed] [Google Scholar]
- 73.Buckner R. L., et al. , Cortical hubs revealed by intrinsic functional connectivity: Mapping, assessment of stability, and relation to Alzheimer’s disease. J. Neurosci. 29, 1860–1873 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Achard S., Salvador R., Whitcher B., Suckling J., Bullmore E., A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J. Neurosci. 26, 63–72 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The code to run the analysis and multimodal neuroimaging data from the experiment are available on GitHub (https://github.com/decolab/pnas-neuromod).