Summary
The outbreaks of 2019 novel coronavirus disease (COVID-19) caused by SARS-CoV-2 infection have posed a severe threat to global public health. It is unclear how the human immune system responds to this infection. Here, we used metatranscriptomic sequencing to profile immune signatures in the bronchoalveolar lavage fluid of eight COVID-19 cases. The expression of proinflammatory genes, especially chemokines, was markedly elevated in COVID-19 cases compared to community-acquired pneumonia patients and healthy controls, suggesting that SARS-CoV-2 infection causes hypercytokinemia. Compared to SARS-CoV, which is thought to induce inadequate interferon (IFN) responses, SARS-CoV-2 robustly triggered expression of numerous IFN-stimulated genes (ISGs). These ISGs exhibit immunopathogenic potential, with overrepresentation of genes involved in inflammation. The transcriptome data was also used to estimate immune cell populations, revealing increases in activated dendritic cells and neutrophils. Collectively, these host responses to SARS-CoV-2 infection could further our understanding of disease pathogenesis and point toward antiviral strategies.
Keywords: COVID-19, SARS-CoV-2, Coronavirus, Innate immune response, Interferon response, Chemokines, Interferon-stimulated genes, Hypercytokinemia, Bronchoalveolar lavage fluid, Metatranscriptomic sequencing
Graphical Abstract
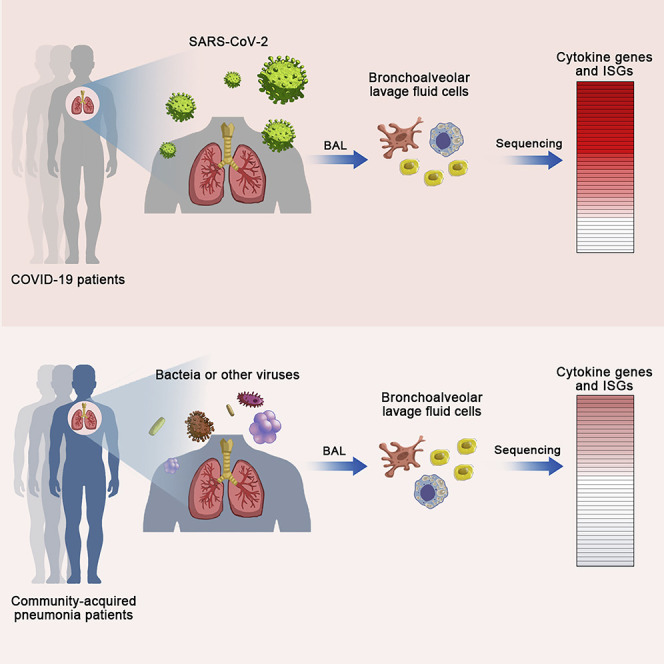
Highlights
-
•
BALF cell transcriptome indicates robust innate immune responses in COVID-19 patients
-
•
COVID-19 patients exhibit chemokine-dominant hypercytokinemia
-
•
ISGs are highly expressed in COVID-19 patients and exhibit pathogenic potential
Zhou et al. use metatranscriptomic sequencing to profile immune signatures in the bronchoalveolar lavage fluid of COVID-19 patients. COVID-19 patients exhibit robust innate immune responses with notable hypercytokinemia and increased expression of IFN-stimulated genes. This analysis provides timely information for understanding SARS-CoV-2-host interactions and COVID-19 pathogenesis.
Introduction
The COVID-19 outbreak caused by SARS-CoV-2 infection (Lu et al., 2020, Ren et al., 2020, Zhou et al., 2020, Zhu et al., 2020) has been declared as a global pandemic. Typical clinical symptoms of COVID-19 are fever, cough, myalgia, and shortness of breath (Huang et al., 2020, Wang et al., 2020). Severe cases often develope acute lung injury, or the fatal form, acute respiratory distress syndrome (ARDS) (Chen et al., 2020, Huang et al., 2020, Wang et al., 2020). To date, no specific antiviral treatment is available for COVID-19.
The infection of certain respiratory viruses has often resulted in a robust inflammatory response (de Jong et al., 2006, Huang et al., 2005). This immune dysregulation, termed hypercytokinemia or “cytokine storm,” is often associated with detrimental outcomes such as ARDS (de Jong et al., 2006). To date, it remains elusive how pathogen infections disrupt the host immune homeostasis and stimulate a hyperactive proinflammatory response. A proposed model has suggested that viral evasion of the innate immune response plays a critical role (D’Elia et al., 2013): the virus can effectively blind the immune system, causing an inadequate or delayed response. This immune escape gives rise to the unrestrained virus replication, which eventually results in a hyperactivated proinflammatory response.
Type I IFN (IFN-I) response plays a critical role in combating virus infection, mainly by inducing the expression of interferon-stimulated genes (ISGs) that exert antiviral functions (Samuel, 2001). Meanwhile, viruses have developed strategies to counteract IFNs. For example, SARS-CoV can efficiently suppress IFN induction and antagonize IFN’s effect by harnessing its structural and non-structural proteins (de Wit et al., 2016). SARS-CoV infection failed to induce competent IFN response in cultured cells (Cheung et al., 2005, Spiegel and Weber, 2006, Ziegler et al., 2005) and SARS patients (Reghunathan et al., 2005). The inadequate IFN response may account for the progressive increase of viral load in SARS patients, the concomitant hypercytokinemia, and the ultimate fatal outcomes.
Clinical study on COVID-19 has suggested the hypercytokinemia in severe cases (Huang et al., 2020). However, a more comprehensive investigation of the host immune response is needed. Here, we collected the bronchoalveolar lavage fluid (BALF) of eight COVID-19 patients, 146 community-acquired pneumonia patients, and 20 healthy controls; this is followed by metatranscriptome sequencing and functional analysis. The molecular signatures of the host response, especially the innate immune response to SARS-CoV-2 infection, are explicitly depicted, showing distinct patterns from those seen in SARS and pneumonia caused by other pathogens.
Results
Sample Collection, Metatranscriptome Sequencing, Differential Expression Gene Analysis
BALFs were obtained from laboratory-confirmed COVID-19 patients (SARS2) (n = 8), community-acquired pneumonia patients (CAP) (n = 146), and healthy controls without known respiratory diseases (Healthy) (n = 20). The demographic and clinical characteristics of the cohorts are detailed in Table S1. By metatranscriptome sequencing, more than 20 million reads were generated for each BALF sample from SARS2, CAP, and Healthy. Among them, 56% of the reads could be mapped to the human genome and were used for further analysis. As metatranscriptome data enabled us to capture the transcriptionally active microbes, we further classified CAP into two categories: Virus-like CAP (determined by at least 100 viral reads and 10-fold higher reads than those in the negative controls) and Non-viral CAP. The read counts for SARS-CoV-2 in each COVID-19 case were listed in Table S2.
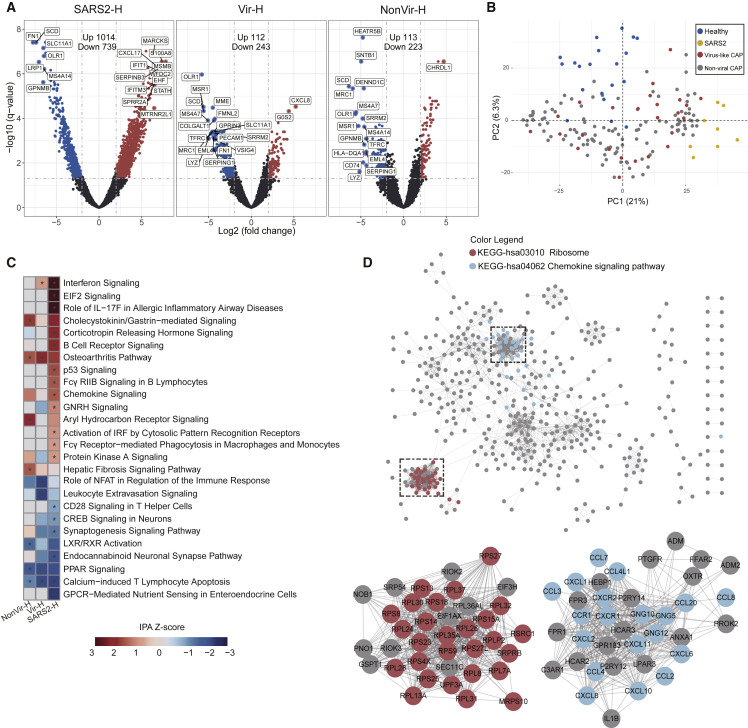
Differentially expressed genes (DEGs) were then identified by comparing pneumonia transcriptomes with the healthy control transcriptomes (SARS2 versus Healthy [SARS2-H], Virus-like CAP versus Healthy [Vir-H], and Non-viral CAP versus Healthy [NonVir-H]). An adjusted p value (q-value < 0.05) and fold change (FC) ratio (|log2FC| ≥ 2) were used to determine the DEGs (Tables S3A–S3C). The volcano plot showed that the number of DEGs in SARS2-H was markedly higher than that in Vir-H and NonVir-H, suggesting that SARS-CoV-2 infection strongly perturbed transcriptome homeostasis of cells in the lung (Figure 1 A). The principal component analysis revealed that SARS2 formed distinct clusters, while Virus-like CAP and Non-viral CAP clusters were more widely dispersed and overlapped (Figure 1B), showing the disparate characteristics of DEGs in SARS2 from others.
Figure 1.
Analysis of DEGs in BALF of COVID-19 and CAP Patients Compared to Healthy Controls
(A) Volcano plot of DEGs comparing SARS2 versus Healthy (SARS2-H), Virus-like CAP versus Healthy (Vir-H), and Non-viral CAP versus Healthy (NonVir-H). The names of DEGs with the top 20 absolute FC are shown.
(B) PCA loading plot based on all DEGs. Autoscaling of data was performed.
(C) Functional enrichment analysis of DEGs with IPA. Asterisks (∗) indicate q-values < 0.05 and absolute Z score ≥ 1.
(D) PPI network of upregulated DEGs in SARS2 comparing to Healthy. Each node represents a protein, and interactions with confidence score > 0.9 are presented.
See also Figure S2.
We next collated genes whose expression is mostly regulated (|log2FC| ≥ 5) in SARS2-H (Figure S1A). Expression levels of proinflammatory cytokine and chemokine genes (IL-1B, CXCL17, CXCL8, and CCL2), typical antiviral ISGs (IFIT and IFITM family genes), and calgranulin genes that exert pleiotropic functions in inflammatory disorders (S100A8, S100A9, and S100A12) were upregulated in SARS2 as compared to Healthy (Figure S1A). Meanwhile, we observed marked upregulation of IL1RN and SOCS3 (Figure S1A), both of which encode cytokine signaling antagonists, suggesting that the negative feedback loops were elicited. Genes involved in morphogenesis and migration of immune cells (NCKAP1L, DOCK2, SPN, and DOCK10) were underexpressed (Figure S1A). A host of DEGs was also observed when comparing CAPs to Healthy, albeit with much lower fold changes (Figure S1A).
Global Functional Analyses
We then performed global functional analyses. For pathway analysis, we applied the Ingenuity Pathway Analysis (IPA) and KEGG analyses on DEGs. In IPA, “Interferon Signaling” ranked first in the upregulated pathways in SARS2 (Z score = 3.3, Figure 1C). This pathway is also upregulated, though to a lesser extent, in Virus-like CAP (Z score = 1), whereas not in the Non-viral CAP (Figure 1C), indicating robust IFN response in SARS-CoV-2 infection. The pathways involved in the inflammatory response in SARS2 was revealed by both analyses, with upregulated “Role of IL-17F in Allergic Inflammatory Airway Diseases” (Z score = 3.0) and “Chemokine Signaling” (Z score = 1.6) in IPA (Figure 1C), along with “Chemokine signaling pathway,” “IL-17 signaling pathway,” “TNF signaling pathway,” and “NF-κB signaling pathway” in KEGG (q < 0.05, Figure S1B). Moreover, both IPA and KEGG analyses showed that DEGs from SARS2-H were strongly and specifically enriched in mRNA translation-related categories, with upregulated “EIF2 Signaling” (Z score = 3.3, Figure 1C) in IPA and “Ribosome” (q < 0.05, Figure S1B) in KEGG. Intriguingly, several neuron function-related pathways were specifically downregulated in SARS2 by IPA analysis, including “CREB Signaling in Neurons” (Z score = −1.1), “Synaptogenesis Signaling Pathway” (Z score = −1.4), and “Endocannabinoid Neuronal Synapse Pathway” (Z score = −2.1, Figure 1C). This unexpected observation awaits further investigation.
Next, we build protein-protein interaction (PPI) networks with the upregulated DEGs in SARS2 by STRING (Figures 1D and S2). A dominant network comprising several subnetworks was generated using the confidence score of 0.9. Genes in the top two enriched KEGG categories (Figure S1B)— namely, “Ribosome” and “Chemokine signaling pathway”—formed two most densely connected subnetworks, which are mainly composed of ribosomal proteins and chemokines, respectively (Figure 1D). Overall, our global functional analysis revealed a highly responsive state against noxious stimuli in COVID-19 cases, characterized by the potent defense responses and hyperactive ribosome biogenesis.
Cytokine Profiles
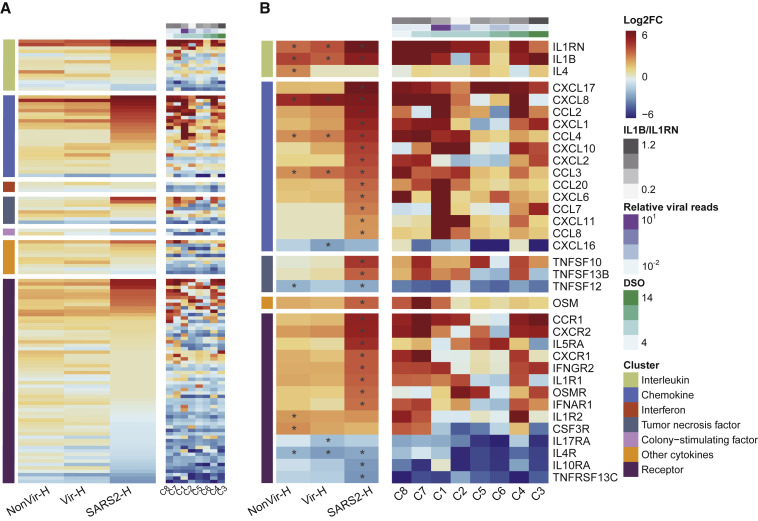
To understand the cytokine profile in BALF from the COVID-19 patients, we assigned DEGs into seven categories of cytokine-related genes (Figure 2 A). In categories of “Chemokine” and “Receptor,” multiple genes were more remarkably upregulated in SARS2 than those in CAPs. Several genes were regulated in the “Interleukin” and “Tumor necrosis factor” categories, and few were modulated in other categories (Figure 2A). To examine potential cytokine expression dynamics, we aligned individual cases according to the increasing days between sampling and symptom onset. Expression levels of cytokine-related genes seem to decrease over time, with one patient (C4) who eventually deceased being the outlier (Figure 2A). These observations suggested that the exuberant inflammation in COVID-19 could be progressively resolved, and unquenched inflammation may result in detrimental outcomes.
Figure 2.
Cytokine-Related Gene Expressions in COVID-19 and CAP Patients
(A) Heatmap of 218 genes encoding cytokines and receptors.
(B) Heatmap of DEGs encoding cytokines and receptors. SARS2 samples (n = 8) were ordered by days after symptom onset (DSO) in the right panel of (A) and (B). Asterisks (∗) indicate significant DEGs (absolute log2FC ≥ 2, q-value < 0.05). Relative viral reads (calculated by the ratio of SARS-CoV-2 reads to human reads) and the ratios of IL1B to IL1RN are shown in (A) and (B).
Chemokines are predominant among upregulated cytokine-related genes in SARS2 (Figure 2A). CXCL17 ranked first in the upregulated chemokines. CXCL17 upregulation is specific to SARS2 and was observed in all eight cases (Figure 2B), suggesting a role of CXCL17 in COVID-19 pathogenesis. CXCL8, the archetypal neutrophil chemoattractant, together with CXCL1 and CXCL2, was upregulated (Figure 2B). These chemokines are thought to be critical for recruiting neutrophils into the inflamed lung (Donnelly et al., 1993, Frevert et al., 1995, Miller et al., 1992). CXCR2, the receptor for CXCL8, CXCL1, and CXCL2, was also markedly upregulated (Figure 2B). Moreover, we observed the upregulation of CCL2 and CCL7, both of which are vital for monocyte recruitment (Shi and Pamer, 2011) (Figure 2B). By presenting individual cases in the heatmap, one case (C1) with ultra-high viral reads (Table S2) exhibited the most pronounced chemokine upregulation (Figure 2B), suggesting that higher virus replication resulted in a more robust proinflammatory response. Corroborating this, in three cases (C1, C2, and C5) sampled at the same time after symptom onset (day 8), the increases of viral load corresponded with the upregulations of critical chemokines, including CXCL8, CCL2, CCL7, etc. (Figure 2B). Both CAPs showed less DEGs and lower expression of cytokine genes as compared to SARS2 (Figure 2B).
IL1RN and ILB are interleukin genes significantly upregulated in SARS2 (Figure 2B), and these upregulations are mirrored by elevated protein levels of IL-1Ra and IL-1β in plasma of COVID-19 patients (Huang et al., 2020). Because IL-1β was recognized as the key cytokine driving proinflammatory response in BALF from patients with ARDS (Pugin et al., 1996), it is possible that the ratio of IL-1β to its inhibitor, IL-1Ra, may correlate to inflammation intensity in COVID-19 patients; however, we did not observe clear correlation in the current study (Figure 2B).
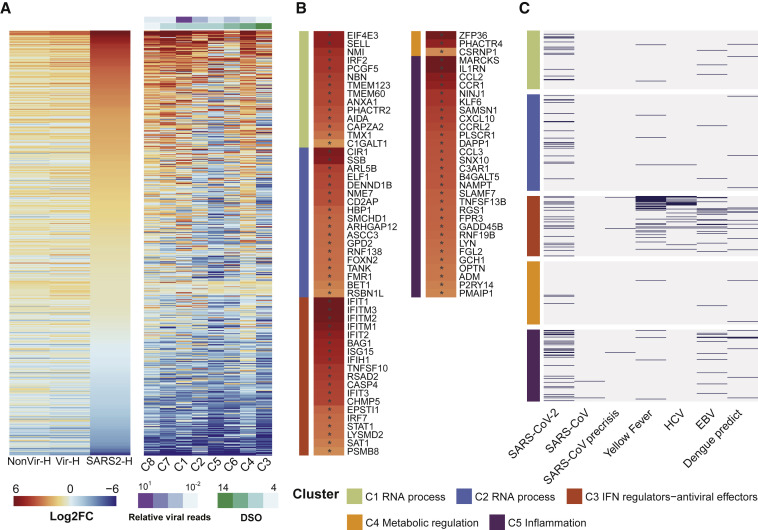
Signatures of IFN Response
We then examined the IFN response in COVID-19 patients. By intersecting our data with a list composed of 628 ISGs (Mostafavi et al., 2016), we found that SARS2 showed more markedly elevated expression of ISGs than CAPs, and the expression seems decreased over time, again with the fatal case (C4) being the outlier (Figure 3 A). Eighty-three ISGs were significantly elevated in SARS2, suggesting the robust IFN response (Table S4). ISGs known to exert direct antiviral activity were upregulated, including IFIT and IFITM genes that exert broad-spectrum antiviral functions (IFIT1, -2, -3, and IFITM1, -2, -3) and others (ISG15 and RSAD2) (Diamond and Farzan, 2013, Helbig and Beard, 2014, Perng and Lenschow, 2018) (Figure 3B). Among those, IFITMs have been shown to inhibit cellular entry of SARS-CoV and MERS-CoV (Huang et al., 2011, Wrensch et al., 2014). Moreover, we observed the increased expression of IFIH1, TANK, IRF7, and STAT1, which may further potentiate IFN signaling (Figure 3B).
Figure 3.
Expression of ISGs in COVID-19 and CAP Patients
(A) Heatmap of 628 ISGs. SARS2 samples (n = 8) were ordered by days after symptom onset (DSO) in the right panel.
(B) Heatmap of 83 upregulated ISGs in SARS2 comparing to Healthy. Asterisks (∗) indicate significant DEGs (absolute log2FC ≥ 2, q-value < 0.05).
(C) Upregulated ISGs in SARS-CoV-2 infection identified in this study, as well as in SARS-CoV and other viral infections (see Table S4).
In (B) and (C), ISGs were assigned into five biclusters.
In addition to antiviral activity, ISGs may exert diverse functions. We assigned these ISGs to five previously established functional clusters (Mostafavi et al., 2016), including RNA processing (C1 and C2), IFN regulation andantiviral function (C3), metabolic regulation (C4), and inflammation regulation (C5) (Table S4). Surprisingly, ISGs are substantially enriched in the cluster of C5 (29 out of 83) (Figure 3B), which mainly comprises inflammation mediators or regulators. CCL2 and CXCL10, two IFN-inducible chemokine genes found in C5 (Figure 3B), were also elevated in peripheral blood of patients with SARS (Cameron et al., 2007). These results pointed to the immunopathological role of IFN response in COVID-19 patients. Further, using a curated ISG list from patient blood transcriptomes (Mostafavi et al., 2016) and data from peripheral blood of SARS patients (Cameron et al., 2007, Reghunathan et al., 2005), we compared the ISG distributions in the five clusters between SARS-CoV-2 and other viruses. ISGs identified in infections of yellow fever virus (live attenuated vaccine), HCV, EBV, and dengue virus were primarily assigned to C3 with less involvement of C5, while SARS-CoV-2 exhibited overly high C5 distribution (Figure 3C, Table S4). Compared to other viruses, SARS-CoV infections induced significantly fewer ISGs (Figure 3C, Table S4). Together, our results unveiled the unique signatures of IFN response in SARS-CoV-2 infection.
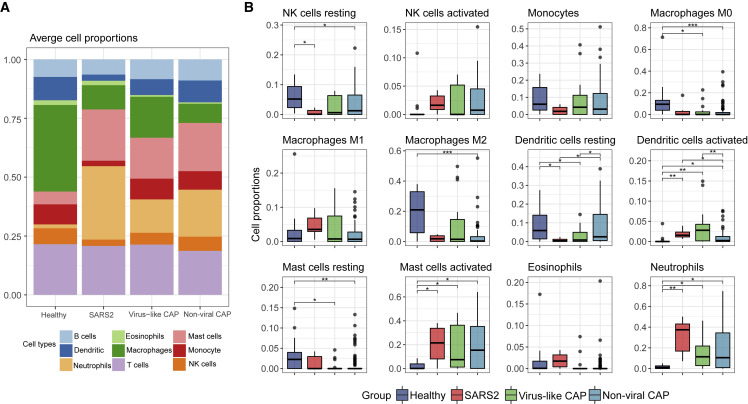
Cell Composition Analyses
The compositions of BALF cells could reflect immune cell profiles in the affected lung (Costabel and Guzman, 2001). Using the transcriptome data, we estimated immune cell types and their proportions by CIBERSORT (Newman et al., 2015). Similar to findings in BALF from healthy adults (Meyer et al., 2012), the BALF cell components in Healthy were mainly macrophages and lymphocytes (Figure 4 A). This result supported the accuracy of the BALF cell sequencing and the effectiveness of our analytical approach. Among innate immune cells, activated dendritic cells, activated mast cells, and neutrophils were more abundant in viral pneumonia groups than those in Healthy (Figure 4B). SARS2 showed a more pronounced increase in neutrophils than other pneumonia (Figure 4B). Compared to innate immune cells, the composition of T cells and B cells was less varied among pneumonia groups and Healthy (Figure S3A). A recent study suggested that the neutrophil-to-lymphocyte (NLR) ratio was associated with the disease severity of COVID-19 (Liu et al., 2020). Our analysis also showed a significantly higher NLR in COVID-19 cases (Figure S3A). The highest NLR was observed in the case with ultra-high viral reads and robust cytokine andISG expression (Figures S3B and S3C).
Figure 4.
Composition of Immune Cells in BALF Predicted from Transcriptome Data
(A) The proportion of nine major immune cell types.
(B) The proportion of 12 innate-immunity-related cell subtypes. Asterisks represent significant differences between groups (∗q-value < 0.05, ∗∗q-value < 0.01, ∗∗∗q-value < 0.001, Mann-Whitney test).
See also Figure S3.
Discussion
Many of the transcriptomic profiling studies have been conducted by using intermittent samples such as peripheral blood (Chaussabel, 2015). However, some infections, such as avian influenza or SARS, occur in the lower respiratory tract (Gu and Korteweg, 2007, van Riel et al., 2006), where the immune response was triggered and regulated. Thus, blood transcriptome may not be optimal for profiling the immunopathological features in those scenarios. In the current study, we sampled COVID-19 patients with bronchoalveolar lavage (BAL), a method for retrieving cells and solutes from different areas of the lung (Meyer et al., 2012). By sequencing and analyzing the BAL fluid cells from eight COVID-19 patients and control cohorts, we obtained gene expression profiles of both host and virus, which directly reflected the in situ host response against SARS-CoV-2 infection. We thus provided a valuable opportunity to garner insights into COVID-19 pathogenesis.
In COVID-19 patients, we observed the upregulation of a plethora of proinflammatory cytokines, suggesting the pathogenic role of hypercytokinemia. The most salient feature of the cytokine profile is the chemokine response, as seen from the elevated expression of multiple chemokines and their receptors. Among those, various neutrophil chemoattractants were upregulated. Consonant with this, the higher neutrophil amount was observed in SARS2 by cell composition analysis. Also, chemoattractants for monocytes and other immune cells were significantly upregulated. In patients with ARDS, the alveolar spaces are occupied by the infiltrating neutrophils and monocytes (Matute-Bello et al., 2008), suggesting the pathogenic role of those cells. Thus, our data unraveled a possible chemokine-dominant hyperactive cytokine response in COVID-19 cases.
In contrast to SARS-CoV, SARS-CoV-2 triggered a robust IFN response, hallmarked by the expression of numerous ISGs, including IFITMs shown to counteract SARS-CoV and MERS-CoV (Huang et al., 2011, Wrensch et al., 2014). This protective potential of ISGs may account for the lower proportion of severe cases and the case-fatality rate in COVID-19 as compared to SARS (Wu and McGoogan, 2020). Nevertheless, high SARS-CoV-2 loads were detected very early after symptom onset (Zou et al., 2020), suggesting that the virus may have developed countermeasures against the IFN system such as delaying the IFN response by inhibiting innate immune signaling. Moreover, although robust ISG induction was observed, we failed to detect significant upregulation of IFNs. This discrepancy needs further investigation.
IFN-I could also be pathogenic. In the mouse model, depletion of IFN-α/β receptor protects mice from lethal infection of SARS-CoV (Channappanavar et al., 2016). Moreover, delayed IFN-β treatment failed to inhibit virus replication and aggravated pulmonary inflammation in the mice infected with MERS-CoV (Channappanavar et al., 2019). These data suggested that the timing of IFN therapy against SARS and MERS is critical. In the ISGs identified in COVID-19 cases, we found a subset of genes with proinflammatory activities, which is relatively underrepresented in other viral infections. These “proinflammatory ISGs” may assist the antiviral protection by amplifying inflammatory signals to the environment. However, they could also be deleterious in infections, especially when hypercytokinemia has already been triggered. As IFN treatment has been adopted as an antiviral therapy against SARS-CoV-2 infection (ChiCTR: ChiCTR2000029600), the timing and dose should be carefully considered.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Human bronchoalveolar lavage fluid samples | Peking University People’s Hospital, Shenzhen Third People’s Hospital, Fujian Provincial Hospital, the Second Affiliated Hospital of Harbin Medical University, the First Affiliated Hospital of Xi’an Jiaotong University, and hospitals in Wuhan | NA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trizol LS | Thermo Fisher Scientific | Cat#10296028 |
| Critical Commercial Assays | ||
| Direct-zol RNA Miniprep Kit | Zymo Research | Cat#R2051 |
| Trio RNA-Seq Library Preparation Kit | NuGEN | Cat#0357-32 |
| Deposited Data | ||
| Reads mapped to human genome GRCh38 extracted from the raw sequencing data | This paper | Genome Warehouse in National Genomics Data Center https://bigd.big.ac.cn/gsa; Project number: PRJCA002273 |
| Software and Algorithms | ||
| HISAT2 v2.1.0 | Kim et al., 2019 | http://ccb.jhu.edu/software/hisat2/ |
| FeatureCounts 2.0.0 | Liao et al., 2014 | http://subread.sourceforge.net/ |
| Fastp 0.20.0 | Chen et al., 2018 | https://github.com/OpenGene/fastp |
| Quantile normalization (Bioconductor limma package v3.42.2) | Ritchie et al., 2015 | https://www.bioconductor.org/packages/release/bioc/html/limma.html |
| Ingenuity Pathway Analysis, December 2019 release (v499329) | Krämer et al., 2014 | https://digitalinsights.qiagen.com/ |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Bioconductor clusterProfiler package v3.14.3) | Yu et al., 2012 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| STRING v11 | Szklarczyk et al., 2019 | https://string-db.org |
| Cytoscape v3.7.1 | Shannon et al., 2003 | https://cytoscape.org |
| CIBERSORT algorithm v1.06 | Newman et al., 2015 | https://cibersort.stanford.edu/ |
| Samtools 1.9 | Li et al., 2009 | http://samtools.sourceforge.net/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jianwei Wang (wangjw28@163.com).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The accession number for the reads mapped to human genome GRCh38 extracted from the raw sequencing data reported in this paper is [Genome Warehouse in National Genomics Data Center (National Genomics Data Center Members and Partners, 2020)]: [PRJCA002273] that is publicly accessible at https://bigd.big.ac.cn/gsa.
Experimental Model and Subject Details
BALF samples from eight laboratory-confirmed COVID-19 patients (SARS2) were collected from hospitals in Wuhan in January 2020. BALFs from 146 community-acquired pneumonia patients (CAP) and 20 healthy controls from volunteers without any known pulmonary diseases (Healthy) were collected from Peking University People’s Hospital, Shenzhen Third People’s Hospital, Fujian Provincial Hospital, the Second Affiliated Hospital of Harbin Medical University, and the First Affiliated Hospital of Xi’an Jiaotong University between 2014 and 2018. CAP was diagnosed following guidelines of the Infectious Diseases Society of America and the American Thoracic Society (Mandell et al., 2007). The known respiratory pathogens in CAP patients were screened by using FTD Respiratory pathogens kit (Fast Track Diagnostics, Luxembourg). The study was approved by the Institutional Review Board of hospitals where sampling was carried out. The data collection for the COVID-19 patients were deemed by the National Health Commission of the People’s Republic of China as the contents of the public health outbreak investigation. The written informed consent was obtained from all subjects before inclusion. Of the COVID-19 cases, five were male and three were female. Of the CAP cases, 75 were male and 54 were female. Eight COVID-19 cases had an average age of 49 years (range 40-61), and 112 CAP cases had an average of 56.4 years (range 22-91). The healthy subjects were pre-screened, and 20 had no known respiratory diseases were included as healthy control. Currently or potentially immunocompromised subjects with active cancer, primary immunodeficiency disorders, HIV infection, or tuberculosis infection were excluded from the study. Subjects with a recent medical record of taking immunosuppressive medications were excluded from the study. The detailed information of the subjects could be found in the demographic and clinical Table (Table S1).
Method Details
Sample Preparation and Sequencing
BALF samples were obtained during bronchoscopies done as part of clinical management. In the biosafety level (BSL)-III laboratory, a 200 ul aliquot of each BALF sample from COVID-19 patients were lysed in Trizol LS (Thermo Fisher Scientific, Carlsbad, CA, USA), followed by RNA extraction using a Direct-zol RNA Miniprep kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions. RNA extractions from other samples were conducted following the same protocol in the BSL-II laboratory. 10 ul of purified RNA was used for cDNA generation and library preparation using an Ovation Trio RNA-Seq Library Preparation Kit (NuGEN, CA, USA) according to the manufacturer’s instructions. Three samples, including two saline solutions passing through the bronchoscope and one nuclease-free water, were included as the negative controls. Metatranscriptome sequencing was performed on an Illumina HiSeq 2500/4000 platform (Illumina, United Kingdom).
Data Processing and Analysis
Raw sequencing reads were quality controlled as previously described (Shen et al., 2020). Briefly, the FASTQ files were subjected to adaptor trimming, low quality reads removal, and short reads removal using Fastp 0.20.0 (Chen et al., 2018). All clean data were mapped to the human genome GRCh38 using HISAT2 v2.1.0 (Kim et al., 2019) with default parameters. Bam files were sorted by Samtools 1.9 (Li et al., 2009).
Gene counts were summarized using the featureCounts program (Liao et al., 2014) as part of the Subread package release 2.0.0 (http://subread.sourceforge.net/). To identify differentially expressed genes (DEGs) between two groups, genes that were present in less than 50% of samples in both groups and genes with average counts per million (CPM) lower than five in both groups were removed. Remained gene counts were then normalized using the quantile method and the voom method. DEGs were determined with the threshold adjusted p value < 0.05 and absolute logged fold-change (Log2FC) ≥ 2 using the Bioconductor limma package v3.42.2 (Ritchie et al., 2015). In presenting the individual case of SARS2, the fold change of each gene was calculated as the ratio of normalized gene expression of each SARS2 individual to the mean expression of Healthy.
For the functional enrichment of DEGs, Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Inc.) (Krämer et al., 2014) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Kanehisa and Goto, 2000) were applied via IPA December 2019 release (v499329) and the Bioconductor clusterProfiler package v3.14.3 (Yu et al., 2012), respectively. Protein-protein interaction (PPI) networks of upregulated DEGs were built using STRING v11 (Szklarczyk et al., 2019) with a confidence score threshold of 0.9 and plotted with Cytoscape v3.7.1 (Shannon et al., 2003).
To infer the composition of immune cells in BALF, raw gene counts were normalized as transcripts per million (TPM) and processed using the CIBERSORT algorithm v1.06 (Newman et al., 2015) with the original CIBERSORT gene signature file LM22 and 100 permutations. Twenty-two immune cell subtypes were obtained and further summarized into nine major immune cell types. Samples with p-value < 0.05, which reflects the statistical significance of the deconvolution results across all cell subsets, were included.
Quantification and Statistical Analysis
Fisher’s exact test was used for categorical variables, and the Mann-Whitney test was used for continuous variables that do not follow a normal distribution. P values from multiple testing were adjusted (q-value) using the Benjamini-Hochberg false discovery rate (FDR) with a significance level of 0.05 (Benjamini and Hochberg, 1995).
Acknowledgments
We thank Dr. Xue Yongbiao (China National Center for Bioinformation), Dr. Wang Jianbin (Tsinghua University, Beijing, China), and Dr. Yanyi Huang (Peking University) for technical assistance and helpful discussions. This work was supported by grants from the National Major Science & Technology Project for Control and Prevention of Major Infectious Diseases in China (2017ZX10204401, 2018ZX10301401, 2017ZX10103004, 2018ZX10305409), the Innovation Fund for Medical Sciences (2016-I2M-1-014), the National Natural Science Foundation of China (31670169, 31470267, 31701155), the National Key Research and Development Program of China (2017YFC0908403, 2020YFA0707600), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2019PT310029), the Strategic Priority CAS Project (XDB38000000), Chinese Academy of Sciences, and the Beijing Advanced Innovation Center for Genomics (ICG).
Author Contributions
Conceptualization, J.W., M.L., L.R., and Q.J.; Lab work, L.R., Y.X., L.G., and J.D.; Data, Q.C., J.Z., M.L., Jian Yang, and B.L.; High-throughput sequencing, X.Z.; Transcriptomic analysis, Z.Z., L.Z., J.Z., Z.J., Jing Yang, C.W., S.J., W.W., and K.H.; Clinical sample management, L.R., Y.X., D.Y., G.Z., H.L., F.C., Y.X., M.C., and Z.G.; Writing - Original Draft, Z.Z., L.Z., J.Z., M.L., and J.W.; Writing - Review & Editing, J.W., M.L., Z.Z., L.Z., and L.R.
Declaration of Interests
The authors declare no competing interests.
Published: May 4, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2020.04.017.
Contributor Information
Mingkun Li, Email: limk@big.ac.cn.
Jianwei Wang, Email: wangjw28@163.com.
Supplemental Information
Demographic and clinical information of all subjects were shown. The criterion for virus infection (Virus) is at least supported by 100 viral reads and should be 10 times higher than the abundance in the negative control by sequencing.
DEGs identified by comparing SARS2 to Healthy (Table S3A), Virus-like CAP to Healthy (Table S3B), and Non-viral CAP to Healthy (Table S3C) were presented. An adjusted p value (q-value < 0.05) and fold change (FC) ratio (|log2FC| ≥ 2) were used to determine the DEGs.
ISGs identified in viral infections were assigned to five previously established functional clusters, including RNA processing (C1 and C2), IFN regulation/antiviral function (C3), metabolic regulation (C4), and inflammation regulation (C5).
References
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M., Canadian SARS Research Network Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Jr., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;130:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussabel D. Assessment of immune status using blood transcriptomics and potential implications for global health. Semin. Immunol. 2015;27:58–66. doi: 10.1016/j.smim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C.Y., Poon L.L., Ng I.H., Luk W., Sia S.F., Wu M.H., Chan K.H., Yuen K.Y., Gordon S., Guan Y., Peiris J.S. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costabel U., Guzman J. Bronchoalveolar lavage in interstitial lung disease. Curr. Opin. Pulm. Med. 2001;7:255–261. doi: 10.1097/00063198-200109000-00002. [DOI] [PubMed] [Google Scholar]
- D’Elia R.V., Harrison K., Oyston P.C., Lukaszewski R.A., Clark G.C. Targeting the “cytokine storm” for therapeutic benefit. Clin. Vaccine Immunol. 2013;20:319–327. doi: 10.1128/CVI.00636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M.D., Simmons C.P., Thanh T.T., Hien V.M., Smith G.J., Chau T.N., Hoang D.M., Chau N.V., Khanh T.H., Dong V.C. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly S.C., Strieter R.M., Kunkel S.L., Walz A., Robertson C.R., Carter D.C., Grant I.S., Pollok A.J., Haslett C. Interleukin-8 and development of adult respiratory distress syndrome in at-risk patient groups. Lancet. 1993;341:643–647. doi: 10.1016/0140-6736(93)90416-e. [DOI] [PubMed] [Google Scholar]
- Frevert C.W., Huang S., Danaee H., Paulauskis J.D., Kobzik L. Functional characterization of the rat chemokine KC and its importance in neutrophil recruitment in a rat model of pulmonary inflammation. J. Immunol. 1995;154:335–344. [PubMed] [Google Scholar]
- Gu J., Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 2007;170:1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig K.J., Beard M.R. The role of viperin in the innate antiviral response. J. Mol. Biol. 2014;426:1210–1219. doi: 10.1016/j.jmb.2013.10.019. [DOI] [PubMed] [Google Scholar]
- Huang K.J., Su I.J., Theron M., Wu Y.C., Lai S.K., Liu C.C., Lei H.Y. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., Zhang M., Tan J., Xu Y., Song R. Neutrophil-to-Lymphocyte Ratio Predicts Severe Illness Patients with 2019 Novel Coronavirus in the Early Stage. medRxiv. 2020 doi: 10.1186/s12967-020-02374-0. 2020.2002.2010.20021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell L.A., Wunderink R.G., Anzueto A., Bartlett J.G., Campbell G.D., Dean N.C., Dowell S.F., File T.M., Jr., Musher D.M., Niederman M.S., Infectious Diseases Society of America. American Thoracic Society Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 2007;44(Suppl 2):S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute-Bello G., Frevert C.W., Martin T.R. Animal models of acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008;295:L379–L399. doi: 10.1152/ajplung.00010.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.C., Raghu G., Baughman R.P., Brown K.K., Costabel U., du Bois R.M., Drent M., Haslam P.L., Kim D.S., Nagai S., American Thoracic Society Committee on BAL in Interstitial Lung Disease An official American Thoracic Society clinical practice guideline: the clinical utility of bronchoalveolar lavage cellular analysis in interstitial lung disease. Am. J. Respir. Crit. Care Med. 2012;185:1004–1014. doi: 10.1164/rccm.201202-0320ST. [DOI] [PubMed] [Google Scholar]
- Miller E.J., Cohen A.B., Nagao S., Griffith D., Maunder R.J., Martin T.R., Weiner-Kronish J.P., Sticherling M., Christophers E., Matthay M.A. Elevated levels of NAP-1/interleukin-8 are present in the airspaces of patients with the adult respiratory distress syndrome and are associated with increased mortality. Am. Rev. Respir. Dis. 1992;146:427–432. doi: 10.1164/ajrccm/146.2.427. [DOI] [PubMed] [Google Scholar]
- Mostafavi S., Yoshida H., Moodley D., LeBoité H., Rothamel K., Raj T., Ye C.J., Chevrier N., Zhang S.Y., Feng T., Immunological Genome Project Consortium Parsing the Interferon Transcriptional Network and Its Disease Associations. Cell. 2016;164:564–578. doi: 10.1016/j.cell.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Genomics Data Center Members and Partners Database Resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 2020;48(D1):D24–D33. doi: 10.1093/nar/gkz913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng Y.C., Lenschow D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018;16:423–439. doi: 10.1038/s41579-018-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugin J., Ricou B., Steinberg K.P., Suter P.M., Martin T.R. Proinflammatory activity in bronchoalveolar lavage fluids from patients with ARDS, a prominent role for interleukin-1. Am. J. Respir. Crit. Care Med. 1996;153:1850–1856. doi: 10.1164/ajrccm.153.6.8665045. [DOI] [PubMed] [Google Scholar]
- Reghunathan R., Jayapal M., Hsu L.Y., Chng H.H., Tai D., Leung B.P., Melendez A.J. Expression profile of immune response genes in patients with Severe Acute Respiratory Syndrome. BMC Immunol. 2005;6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L.L., Wang Y.M., Wu Z.Q., Xiang Z.C., Guo L., Xu T., Jiang Y.Z., Xiong Y., Li Y.J., Li X.W. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J (Engl) 2020;133:1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., Zhou Z., Yang J., Zhong J., Yang D. Genomic diversity of SARS-CoV-2 in Coronavirus Disease 2019 patients. Clin. Infect. Dis. 2020:ciaa203. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Pamer E.G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M., Weber F. Inhibition of cytokine gene expression and induction of chemokine genes in non-lymphatic cells infected with SARS coronavirus. Virol. J. 2006;3:17. doi: 10.1186/1743-422X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D., Munster V.J., de Wit E., Rimmelzwaan G.F., Fouchier R.A., Osterhaus A.D., Kuiken T. H5N1 Virus Attachment to Lower Respiratory Tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrensch F., Winkler M., Pöhlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014;6:3683–3698. doi: 10.3390/v6093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler T., Matikainen S., Rönkkö E., Osterlund P., Sillanpää M., Sirén J., Fagerlund R., Immonen M., Melén K., Julkunen I. Severe acute respiratory syndrome coronavirus fails to activate cytokine-mediated innate immune responses in cultured human monocyte-derived dendritic cells. J. Virol. 2005;79:13800–13805. doi: 10.1128/JVI.79.21.13800-13805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demographic and clinical information of all subjects were shown. The criterion for virus infection (Virus) is at least supported by 100 viral reads and should be 10 times higher than the abundance in the negative control by sequencing.
DEGs identified by comparing SARS2 to Healthy (Table S3A), Virus-like CAP to Healthy (Table S3B), and Non-viral CAP to Healthy (Table S3C) were presented. An adjusted p value (q-value < 0.05) and fold change (FC) ratio (|log2FC| ≥ 2) were used to determine the DEGs.
ISGs identified in viral infections were assigned to five previously established functional clusters, including RNA processing (C1 and C2), IFN regulation/antiviral function (C3), metabolic regulation (C4), and inflammation regulation (C5).
Data Availability Statement
The accession number for the reads mapped to human genome GRCh38 extracted from the raw sequencing data reported in this paper is [Genome Warehouse in National Genomics Data Center (National Genomics Data Center Members and Partners, 2020)]: [PRJCA002273] that is publicly accessible at https://bigd.big.ac.cn/gsa.