Abstract
In Alzheimer’s disease (AD), neurodegenerative processes are ongoing for years prior to the time that cortical atrophy can be reliably detected using conventional neuroimaging techniques. Recent advances in diffusion-weighted imaging have provided new techniques to study neural microstructure, which may provide additional information regarding neurodegeneration. In this study, we used neurite orientation dispersion and density imaging (NODDI), a multi-compartment diffusion model, in order to investigate cortical microstructure along the clinical continuum of mild cognitive impairment (MCI) and AD dementia. Using gray matter-based spatial statistics (GBSS), we demonstrated that neurite density index (NDI) was significantly lower throughout temporal and parietal cortical regions in MCI, while both NDI and orientation dispersion index (ODI) were lower throughout parietal, temporal, and frontal regions in AD dementia. In follow-up ROI analyses comparing microstructure and cortical thickness (derived from T1-weighted MRI) within the same brain regions, differences in NODDI metrics remained, even after controlling for cortical thickness. Moreover, for participants with MCI, gray matter NDI—but not cortical thickness—was lower in temporal, parietal, and posterior cingulate regions. Taken together, our results highlight the utility of NODDI metrics in detecting cortical microstructural degeneration that occurs prior to measurable macrostructural changes and overt clinical dementia.
Keywords: diffusion-weighted imaging, gray matter, neuroimaging, neurodegeneration, NODDI
Introduction
Over the last several decades, there has been substantial effort to characterize the neurodegenerative changes in Alzheimer’s disease (AD) in vivo using noninvasive neuroimaging techniques. Early studies using volumetric T1-weighted MRI focused on macrostructural atrophy in the hippocampus (Jack et al. 1992; Fox et al. 1996; Whitwell et al. 2007) and other medial temporal lobe structures (Mori et al. 1997; Krasuski et al. 1998; de Toledo-Morrell et al. 2000). With advances in automated segmentation techniques, recent studies have described widespread gross cortical atrophy in brain regions known to harbor AD pathology (Lerch et al. 2005; Apostolova et al. 2007; Dickerson et al. 2008; Bakkour et al. 2009; Frisoni et al. 2009; Becker et al. 2011; Chételat et al. 2012; LaPoint et al. 2017). While these studies have significantly contributed to our understanding of the disease-related regional patterns and progression of subcortical and cortical atrophy, it is well-known that AD pathology accumulates for years prior to the time when macrostructural changes can be reliably detected with conventional T1-weighted imaging (Villemagne et al. 2013). Given the irreversible nature of neuronal loss, sensitive detection of the earliest neurodegenerative changes occurring during this asymptomatic stage in AD is critical for improving diagnosis, staging, and monitoring response to therapeutic intervention.
In contrast to macrostructural T1-weighted MRI, diffusion-weighted imaging (DWI) provides unique quantitative information about neural microstructure through its sensitivity to the diffusion of water molecules within tissues (Bihan, 2003). Disruption or alteration of microscopic tissue barriers that normally restrict water molecules (e.g., breakdown of cell membranes, changes in cytoarchitecture, or loss of myelin) results in measurable changes to the diffusion signal. The most widely used technique to quantify the diffusion signal is the diffusion tensor imaging (DTI) model, which models the rate and direction of water diffusion in each voxel as a simple ellipsoid tensor (Alger, 2012). DTI metrics that quantify both directional diffusivity and overall degree of hindered diffusion are calculated from the tensor model and used to infer microstructural integrity and architecture within each voxel. While DTI analyses have been widely used to study white matter microstructure in AD, a much more limited number of studies have used these techniques to investigate subcortical and cortical gray matter microstructural changes (Weston et al. 2015). These studies have primarily assessed gray matter microstructure using the DTI metric of mean diffusivity (MD)—the average magnitude of diffusivity in all directions—and have reported that individuals with AD have increased MD in the hippocampus (Kantarci et al. 2005; Müller et al. 2005; Douaud et al. 2013) and cortical regions (Rose et al. 2008; Scola et al. 2010; Jacobs et al. 2013; Montal et al. 2018) where gross atrophy is also observed. However, there are several limitations to the DTI model, especially with respect to analyzing cortical gray matter. Notably, the close proximity of the cortex to cerebrospinal fluid (CSF) makes cortical DTI analyses particularly susceptible to bias from partial volume effects, whereby the tensor model in cortical voxels is contaminated with diffusion signal from adjacent CSF (Henf et al. 2018). Thus, concurrent cerebral atrophy may confound microstructural measurements and lead to overestimation of cortical MD changes in AD.
Advances in DWI acquisition, processing, and modeling have converged to facilitate more sophisticated techniques to study neural microstructure. Multi-shell DWI acquisitions allow for more complex and biologically relevant modeling of the diffusion signal using multi-compartment biophysical models. Neurite orientation dispersion and density imaging (NODDI) (Zhang et al. 2012) is a multi-compartment model that partitions the composite diffusion signal into three microstructural compartments: intracellular diffusion (restricted diffusion within axons or dendrites), extracellular diffusion (hindered diffusion outside of axons and dendrites), and isotropic diffusion (i.e., free water). The NODDI model provides three metrics of microstructure within each voxel: (1) the neurite density index (NDI), which reflects the proportion of water diffusion constrained within axons or dendrites; (2) the orientation dispersion index (ODI), which reflects the degree of neurite coherence; and (3) the volume fraction of isotropic diffusion (VISO), which reflects the proportion of free water (i.e., CSF) in a voxel. Importantly, by separating out the CSF component of the diffusion signal, the NODDI model (unlike DTI) is able to account for partial volume effects, which makes it a particularly well-suited tool for investigating cortical microstructure (Assaf et al. 2013; Fukutomi et al. 2018). Indeed, over the last several years, the NODDI model has been applied to study gray matter microstructure in a variety of conditions, including aging (Nazeri et al. 2015), schizophrenia and bipolar disorder (Nazeri et al. 2017), multiple sclerosis (Granberg et al. 2017), Parkinson’s disease (Kamagata et al. 2017), and young onset AD (Parker et al. 2018).
In this study, we used multi-shell DWI and the NODDI model to investigate cortical microstructural alterations in mild cognitive impairment (MCI) and AD dementia. The aim of this work was to characterize gray matter NODDI microstructure along the clinical continuum of dementia, with the goal of identifying whether regional patterns of microstructural changes provide unique information relative to conventional macrostructural T1-weighted MRI. Specifically, we applied a technique called gray matter-based spatial statistics (GBSS) (Nazeri et al. 2015, 2017), which takes advantage of the multi-compartment modeling of NODDI and allows for an unbiased gray matter-specific voxelwise statistical analysis of cortical microstructure. We then performed a follow-up region of interest (ROI) analysis in order to compare NODDI microstructural metrics and cortical thickness measures (from conventional T1-weighted MRI) within the same brain regions. Our findings suggest NODDI metrics are a sensitive marker of cortical microstructure and may provide early indication of gray matter neurodegenerative changes in MCI and AD dementia.
Materials and Methods
Participants
Participants included individuals with AD dementia, MCI, and cognitively unimpaired individuals from the Wisconsin Alzheimer’s Disease Research Center (ADRC) clinical core who had undergone both multi-shell DWI and T1-weighted structural MRI. For this study, participants with AD dementia and MCI were age- and sex-matched 1-to-1 with cognitively unimpaired participants using propensity score matching (MatchIt package in R v3.5.2), resulting in a final cohort of 112 participants: n = 56 cognitively unimpaired participants (control group), n = 30 MCI, and n = 26 AD dementia. All participants underwent a comprehensive neuropsychological battery to determine the cognitive status. Participants with MCI and AD dementia were diagnosed using available clinical and cognitive information in accordance with the updated 2011 National Institute on Aging-Alzheimer’s Association (NIA-AA) workgroup diagnostic criteria (Albert et al. 2011; McKhann et al. 2011). All participants with MCI in this study were of the amnestic subtype—22/30 participants had single-domain MCI (only memory impairment) and 8/30 individuals had multi-domain MCI (memory impairment plus impairment in one additional cognitive domain). All participants were discussed at a consensus review committee consisting of physicians, neuropsychologists, and nurse practitioners. General exclusion criteria for the ADRC studies include any significant neurologic disease (other than AD dementia), history of alcohol/substance dependence, major psychiatric disorders (including untreated major depression), or other significant medical illness. The University of Wisconsin Health Science Institutional Review Board approved all study procedures, and all experiments were performed in accordance with relevant guidelines and regulations. All participants provided written informed consent to be involved in this study.
Image Acquisition and Preprocessing
MRI data were acquired using a General Electric 3 T MR750 scanner (Waukesha, WI) with a 32-channel head coil. Diffusion-weighted images were acquired using a multi-shell spin-echo echo-planar imaging pulse sequence (6 × b = 0 s/mm2, 9 × b = 500 s/mm2, 18 × b = 800 s/mm2, and 36 × b = 2000 s/mm2; TR/TE = 8575/76.8 ms; 2 × 2 × 2 mm3 isotropic voxel resolution; 128 × 128 acquisition matrix). T1-weighted structural images were acquired using a 3D inversion recovery prepared fast spoiled gradient-echo FSPGR-BRAVO sequence (TI = 450 ms; TR/TE = 8.1/3.2 ms; flip angle = 12°; 1 × 1 × 1 mm3 isotropic voxel resolution; 256 × 256 acquisition matrix).
Diffusion-weighted images were denoised (Veraart et al. 2016) and corrected for Gibb’s ringing (Kellner et al. 2016) using MRtrix3 (Tournier et al. 2019), and then motion-corrected and eddy current distortion are corrected using the eddy tool (Andersson and Sotiropoulos 2016) in FSL (v5.0.11) (Jenkinson et al. 2012). Next, a brain mask was constructed using the Brain Extraction Tool (Smith 2002) in FSL, and diffusion tensor fitting was performed at each voxel within the brain mask using Diffusion Imaging in Python (Garyfallidis et al. 2014) to generate fractional anisotropy (FA) maps.
NDI, ODI, and VISO parameter maps were generated by fitting the NODDI model in Python using Accelerated Microstructure Imaging via Convex Optimization, which improves processing speed by approximating the NODDI model as a linear system (Daducci et al. 2015). Additionally, a gray matter-optimized intracellular intrinsic parallel diffusivity of 1.1 μm2/ms was used to improve the model fit in gray matter (Fukutomi et al. 2018).
Prior to further processing and statistical analysis, all images underwent visual inspection for quality control. Additionally, participant head motion during DWI acquisition was quantitatively evaluated using the root mean square movement summary from eddy, which showed no differences between groups with respect to average acquisition motion (ANOVA, F2,109 = 1.47, P = 0.24). No participants were excluded from analyses based on visual inspection or excessive motion.
Gray Matter-Based Spatial Statistics
GBSS is a statistical technique that adapts the tract-based spatial statistics (Smith et al. 2006) framework in order to allow voxelwise analysis of NODDI metrics in gray matter (Nazeri et al. 2015, 2017). Processing steps for GBSS are depicted in Figure 1. Briefly, gray matter fraction maps were generated by subtracting white matter fraction maps (estimated from FA maps using the Atropos segmentation tool (Avants et al. 2011a) in Advanced Normalization Tools [ANTs] v2.1.0) and CSF fraction maps (NODDI VISO parameter maps) from 1. Each tissue segmentation map was then multiplied by its respective tissue weighting (CSF = 0, white matter = 1, gray matter = 2) and summed to generate a “pseudo T1-weighted” image. Pseudo T1-weighted images from all participants were then used to create a population-specific template using the antsMultivariateTemplateConstruction2.sh script in ANTs (Avants et al. 2011b). Native diffusion space NODDI parameter maps (NDI and ODI) and gray matter fraction maps were then nonlinearly warped to the population template using the warp fields generated during template construction. Population-space gray matter fraction maps were averaged to create a mean gray matter image, which was then skeletonized using the tbss_skeleton tool in FSL. Finally, NDI and ODI were projected onto the gray matter skeleton from the local gray matter fraction maxima, and the gray matter skeleton was thresholded to only include voxels with gray matter fraction > 0.65 in > 70% of participants (Nazeri et al. 2017).
Figure 1.
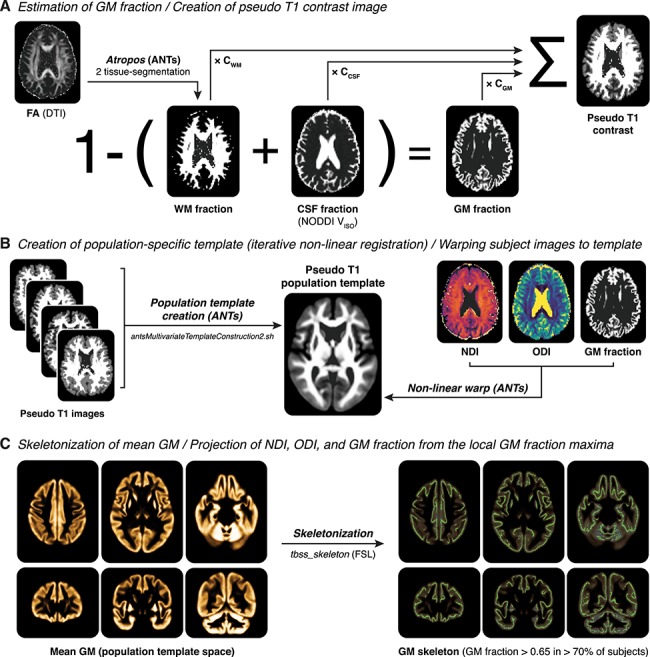
Processing steps of GBSS. (A) For each subject, a gray matter (GM) fraction map is generated by subtracting the white matter (WM) fraction (estimated from the subject’s FA map using Atropos) and the CSF fraction (estimated from the subject’s NODDI VISO parameter map) from 1. The GM, WM, and CSF fraction are then used to generate a pseudo T1-weighted contrast image. (B) Pseudo T1-weighted images from all subjects are used to create a population template via iterative nonlinear registration in ANTs (antsMultivariateTemplateConstruction2.sh). The warp fields generated from this step are then used to nonlinearly warp NDI, ODI, and GM fraction images for each subject into population template space. (C) GM fraction images in population template space are averaged to generate a mean GM image, which is then skeletonized using FSL’s tbss_skeleton tool. Finally, NDI and ODI are projected onto the GM skeleton from the local GM fraction maxima, and the GM skeleton is thresholded to only include voxels with GM fraction > 0.65 in > 70% of participants.
CAT12 Surface-Based Cortical Thickness Processing
T1-weighted structural images were processed using the Computational Anatomy Toolbox (CAT, v12.5) for SPM12 in MATLAB (http://www.neuro.uni-jena.de/cat/). Briefly, T1-weighted images underwent tissue segmentation followed by cortical thickness and central surface estimation using the projection-based thickness method (Dahnke et al. 2013). Surface data then underwent topological correction (Yotter et al. 2011b), spherical mapping (Yotter et al. 2011a), and spherical registration. Prior to performing whole-brain surface-based analyses, surface data were resampled into template space at 32-k mesh resolution (~2-mm average vertex spacing) (Glasser et al. 2013) and smoothed using a 25-mm FWHM Gaussian heat kernel (Chung et al. 2005).
Region of Interest Analysis
In order to interrogate NODDI microstructure and cortical thickness within the same brain regions, we performed a follow-up ROI analysis using eight bilateral cortical regions known to be affected in AD. ROIs were constructed from the Desikan atlas (Desikan et al. 2006; Klein and Tourville 2012) and included the following regions: “frontal” (composed of superior, rostral middle, and caudal middle frontal cortex), “temporal” (composed of middle and frontal temporal cortex), “parietal” (composed of inferior parietal and supramarginal cortex), “precuneus,” “anterior cingulate” (composed of rostral and caudal anterior cingulate cortex), “posterior cingulate” (composed of isthmus and posterior cingulate), “entorhinal,” and “parahippocampal.”
Mean cortical thickness for each ROI was extracted in the native T1-weighted space using CAT12’s ROI tools. In order to extract NODDI metrics in native diffusion space, a T1-weighted atlas-space image was nonlinearly warped to each subject’s pseudo T1 image using antsRegistration in ANTs. For each subject, the resulting warp field was then used to warp atlas-space ROIs to native diffusion space. In order to ensure ROIs only included gray matter voxels, we used each subject’s gray matter fraction map (thresholded at 0.7 and binarized) to mask each ROI. FSL’s fslstats was then used to extract mean values for NDI and ODI within each gray matter-masked ROI.
Statistical Analysis
GBSS voxelwise analyses on the final gray matter skeleton-masked population-space NDI and ODI images were performed with FSL’s randomise (Winkler et al. 2014) using nonparametric permutation inference (n = 10 000 permutations) followed by threshold-free cluster enhancement (Smith and Nichols 2009). Whole-brain surface-based cortical thickness analyses were performed in CAT12 using resampled and smoothed surface image data. For both GBSS and CAT12 surface-based analyses, separate contrasts tested groupwise comparisons (MCI group versus control group; AD dementia group versus control group; and MCI group versus AD dementia group), and all analyses included age and sex as covariates. Resulting statistical maps were family-wise error (FWE), corrected at PFWE < 0.05 and displayed as surfaces using Surf Ice (https://www.nitrc.org/projects/surfice/).
For ROI analyses, analysis of covariance (ANCOVA) models were used to test differences in both NODDI and cortical thickness measures between diagnostic groups (control, MCI, and AD dementia) while controlling for age and sex. To account for multiple comparisons across the eight ROIs, a Benjamini-Hochberg false discovery rate (FDR)-corrected significance threshold of PFDR < 0.05 was used. For visualization purposes of this analysis, NODDI and cortical thickness measures for MCI and AD dementia groups were expressed as percent change relative to the control group mean and plotted as mean percent change with bootstrapped 95% confidence intervals (10 000 iterations using the boot v1.3 package in R) for each group. In order to further investigate whether NODDI metrics provide unique microstructural information within an ROI, we reran the same ANCOVA models but controlled for macrostructural effects by including cortical thickness as a covariate.
Finally, we performed an exploratory logistic regression analysis to investigate and compare diagnostic accuracy of NODDI and cortical thickness measures for MCI and AD dementia. For this analysis, in order to generate single parameters for each metric, we first identified the most important ROIs by using recursive feature elimination (RFE) in the caret v6.0 package in R. Briefly, for each metric, RFE was run using values from all eight ROIs as predictors, and the most important ROIs were identified using classification accuracy with 10-fold cross validation. For cortical thickness, the most important ROIs were anterior cingulate, entorhinal, temporal, and posterior cingulate; for NDI, the most important ROIs were posterior cingulate, anterior cingulate, temporal, entorhinal, and precuneus; for ODI, the most important variables were parahippocampal, posterior cingulate, parietal, and entorhinal. We then averaged across these identified ROIs to generate single parameters for each metric, which were used to predict diagnosis (along with age and sex) in logistic regression models, running separate models for each diagnostic group. We ran one macrostructural model (using only cortical thickness), three microstructural models (using only NDI, only ODI, and then both NDI and ODI), and one multimodal model (using all three metrics: CT, NDI, and ODI). Model fit was evaluated using the Akaike information criteria (AIC). For each model, a smoothed receiver operating characteristic (ROC) curve and the area under the curve (AUC) with bootstrapped 95% confidence interval (10 000 iterations) were generated using the pROC package (v1.14) in R.
Results
Participant Characteristics
Participant characteristics are presented in Table 1. There were no differences between groups with respect to age, sex, APOE ε4 genotype prevalence, race, or years of education. As expected, Montreal Cognitive Assessment (MoCA) total scores were lower for the MCI and AD dementia groups.
Table 1.
Participant characteristics
| Control | MCI | AD | Statistic | |
|---|---|---|---|---|
| N | 56 | 30 | 26 | — |
| Age, years (mean ± SD) | 72.7 ± 9.9 | 73.0 ± 10.2 | 72.2 ± 10.2 | F = 0.05 P = 0.95 |
| Sex, % female (n) | 57.1% (32/56) | 46.7% (14/30) | 61.5% (16/26) | χ2 = 1.39 P = 0.50 |
| APOE ε4 genotype, % positive (n) | 46.2% (25/54) | 50% (15/30) | 58.3% (14/24) | χ2 = 0.96 P = 0.62 |
| Race, n (Caucasian/African American/Native American) | 42/11/3 | 25/5/0 | 23/2/1 | χ2 = 3.61 P = 0.46 |
| Education, years (mean ± SD) | 16.1 ± 2.6 | 15.3 ± 2.5 | 15.3 ± 3.3 | F = 1.33 P = 0.27 |
| MoCA, total score (mean ± SD) | 26.7 ± 2.3 | 21.0 ± 3.4 | 16.7 ± 3.4 | F = 116.8 P < 0.001 |
AD = Alzheimer’s disease dementia participants.
Whole-Brain GBSS Analysis
GBSS was used to investigate cortical patterns of altered NODDI microstructural metrics in participants with MCI and AD dementia. For both groups, we observed significantly lower cortical NDI across parietal, temporal, and frontal regions (Fig. 2A,B; Supplementary Fig. 1) relative to cognitively unimpaired participants. Specifically, the MCI group had lower NDI relative to the control group predominantly in the supramarginal and angular gyri, a majority of the temporal lobe including temporal pole, and the left frontal lobe including rostral middle frontal gyrus, pars triangularis, and lateral orbitofrontal cortex (Fig. 2A). The AD dementia group showed a similar pattern of lower NDI with additional involvement of the superior frontal gyrus as well as posterior cingulate and precuneus (Fig. 2B). There were no regions with significantly lower NDI in the AD dementia group relative to the MCI group. Finally, there were no cortical regions with significantly higher NDI in either the MCI or AD dementia groups, and there were no subcortical gray matter regions with significant NDI differences in any groupwise comparison.
Figure 2.
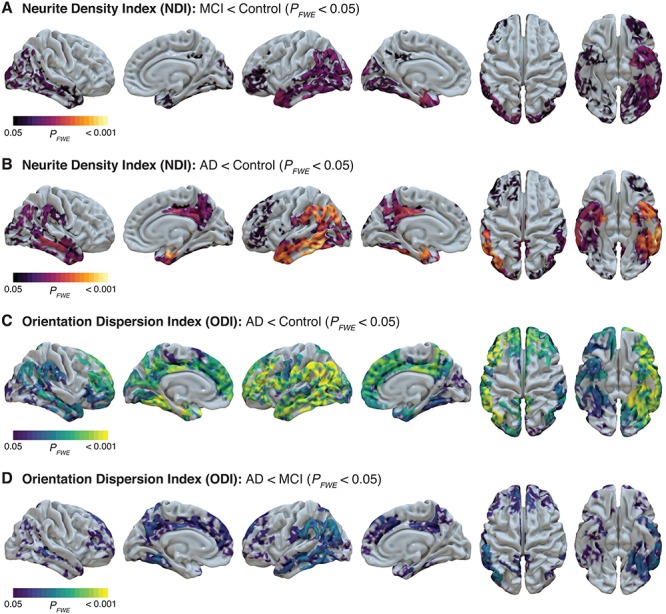
Decreased cortical gray matter NODDI metrics in MCI and AD dementia groups from whole-brain GBBS analysis. NDI in cortical gray matter is significantly decreased in both MCI (A) and AD dementia (B). ODI in gray matter is unchanged in MCI but significantly decreased in AD dementia relative to both the control group (C) and the MCI group (D). GBSS statistical maps were projected onto surfaces in Surf Ice and indicate areas with significant (FWE-corrected P < 0.05) differences. Representative axial slices of significant GBSS results can be found in Supplementary Figure 1.
With respect to ODI, while there were no differences in the MCI group compared to cognitively unimpaired participants, the AD dementia group demonstrated significantly lower cortical ODI in a majority of temporal, parietal, and frontal regions relative to the control group (Fig. 2C). This pattern of lower ODI was more widespread than the pattern observed for NDI and additionally included the lateral precentral and postcentral gyri, pars opercularis, medial superior frontal and orbitofrontal regions, and anterior cingulate. Additionally, the AD dementia groups showed lower cortical ODI relative to the MCI group in a similar distribution (Fig. 2D). There were no cortical regions with significantly higher ODI in the AD dementia group compared to cognitively unimpaired participants, and there were no subcortical gray matter regions with significant ODI differences in any groupwise comparison.
Surface-Based Cortical Thickness Analysis
CAT12 surface-based analysis demonstrated that, relative to the cognitively unimpaired group, the AD dementia group had significantly decreased cortical thickness in bilateral superior frontal regions, bilateral inferior parietal regions, bilateral inferior temporal gyri, entorhinal cortex, and temporal pole (Fig. 3A). Additionally, relative to the MCI group, the AD dementia group had significantly decreased cortical thickness in several small clusters in frontal and parietal regions (Fig. 3B). There were no regions with significantly lower cortical thickness in the MCI group relative to the control group.
Figure 4.
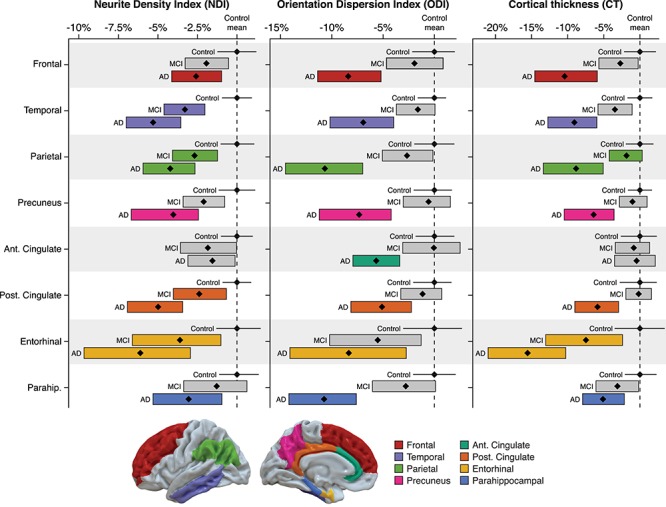
Region of interest (ROI) analysis of cortical gray matter NODDI metrics and cortical thickness within eight AD regions. NDI, ODI, and cortical thickness were extracted from eight ROIs in native imaging space using a common atlas, and ANCOVA models (controlling for age and sex) were run to determine differences in gray matter microstructure and macrostructure between diagnostic groups. Data for each ROI and each diagnostic group are presented as mean percentage change (black diamond) relative to control group mean, with bootstrapped 95% confidence intervals (bars for MCI and AD dementia groups; solid black line for control group). Bars are color-coded by ROI and indicate a significant (FDR-corrected P < 0.05) difference from control mean, while gray bars indicate no significant difference from control mean.
Figure 3.
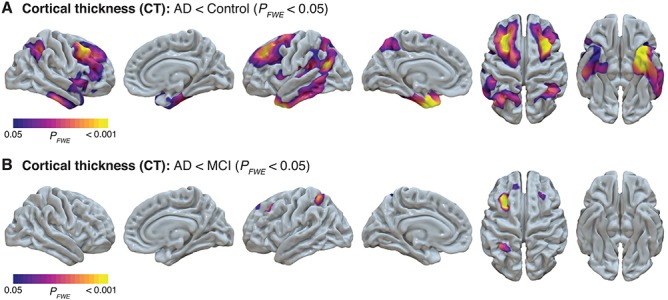
Decreased cortical thickness in AD dementia groups from whole-brain CAT surface-based analysis. CAT12 surface-based analysis demonstrated areas of decreased cortical thickness in the AD dementia group relative to the control group (A) and the AD dementia group relative to the MCI group (B). CAT12 statistical maps were projected onto surfaces in Surf Ice and indicate areas with significant (FWE-corrected P < 0.05) differences.
ROI Analysis
In order to investigate both microstructure and macrostructure within the same brain regions, we performed a follow-up ROI analysis that involved extracting NODDI metrics and cortical thickness in native space from eight bilateral gray matter ROIs using a common atlas. Overall, the AD dementia group tended to have greater differences in microstructure and macrostructure compared to control, relative to the differences observed between the MCI and control groups (Fig. 4). For the AD dementia group, NDI and cortical thickness were significantly lower in all ROIs except the anterior cingulate, while ODI was significantly lower in all eight ROIs (Supplementary Table 1 for summary of ANCOVA results). For the MCI group, there were no significant differences in ODI in any ROIs compared to the control group. Notably, in the MCI group, while cortical thickness was not significantly decreased in any ROIs, NDI was significantly lower in several ROIs including the temporal, parietal, and posterior cingulate regions.
Figure 5.
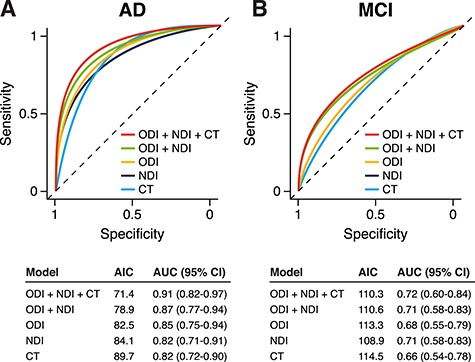
ROC curves from exploratory logistic regression models using NODDI microstructure and/or cortical thickness. For NDI, ODI, and cortical thickness (CT), per-subject parameters for each metric were used in logistic regression models (along with age and sex) to predict diagnosis of AD dementia (A) and MCI (B). Model summary tables below the ROC curves display AIC and AUC with bootstrapped 95% confidence intervals (CI).
To account for the effects of macrostructural atrophy on microstructural alterations in these ROIs, we performed the same statistical ROI analysis while controlling for cortical thickness. Results from this analysis are presented in Tables 2 and 3. After controlling for cortical thickness, NDI and ODI remained significantly lower in most of the same regions in MCI and AD dementia groups compared to cognitively unimpaired participants. Specifically, results for temporal, parietal, precuneus, and posterior cingulate regions were unchanged. Controlling for cortical thickness attenuated some effects—specifically, in the entorhinal region, both NDI and ODI were no longer significantly lower for the AD dementia group compared to the control group. Likewise, in the frontal and parahippocampal regions, NDI was no longer significantly lower in the AD dementia group after controlling for cortical thickness.
Table 2.
Summary of ANCOVA results for ROI analysis demonstrating differences in cortical NDI, with and without controlling for cortical thickness
| MCI vs. control | AD vs. control | |||||||
|---|---|---|---|---|---|---|---|---|
| NDI (uncorrected) | NDI (corrected for CT) | NDI (uncorrected) | NDI (corrected for CT) | |||||
| ROI | β | P FDR | β | P FDR | β | P FDR | β | P FDR |
| Frontal | −4.7E − 3 | 0.095 | −4.4E − 3 | 0.125 | −6.9E − 3 | 0.016 | −5.3E − 3 | 0.095 |
| Temporal | −7.7E − 3 | 4.1E − 3 | −6.7E − 3 | 0.019 | −1.3E − 2 | 2.4E − 7 | −1.1E − 3 | 2.5E − 4 |
| Parietal | −6.5E − 3 | 0.032 | −5.9E − 3 | 0.037 | −1.1E − 2 | 7.2E − 5 | −7.9E − 3 | 8.6E − 3 |
| Precuneus | −5.5E − 3 | 0.061 | −5.3E − 3 | 0.093 | −1.0E − 2 | 5.4E − 4 | −8.6E − 3 | 8.6E − 3 |
| Ant. cingulate | −4.1E − 3 | 0.095 | −4.2E − 3 | 0.115 | −4.0E − 3 | 0.106 | −4.1E − 3 | 0.095 |
| Post. cingulate | −6.0E − 3 | 0.032 | −5.9E − 3 | 0.037 | −1.3E − 2 | 4.8E − 6 | −1.2E − 2 | 4.3E − 5 |
| Entorhinal | −8.4E − 3 | 0.061 | −5.4E − 3 | 0.181 | −1.5E − 2 | 6.7E − 4 | −8.3E−3 | 0.077 |
| Parahippocampal | −3.4E − 3 | 0.258 | -3.0E − 3 | 0.330 | −7.5E − 3 | 0.022 | −6.6E − 3 | 0.070 |
ANCOVA models tested the effect of diagnosis on NDI, controlling for age, sex, and with and without cortical thickness; bold indicates PFDR < 0.05; CT = cortical thickness; AD = Alzheimer’s disease dementia group.
Table 3.
Summary of ANCOVA results for ROI analysis demonstrating differences in cortical ODI, with and without controlling for cortical thickness
| MCI vs. control | AD vs. control | |||||||
|---|---|---|---|---|---|---|---|---|
| ODI (uncorrected) | ODI (corrected for CT) | ODI (uncorrected) | ODI (corrected for CT) | |||||
| ROI | β | P FDR | β | P FDR | β | P FDR | β | P FDR |
| Frontal | −5.2E − 3 | 0.502 | −3.2E − 3 | 0.758 | −2.8E − 2 | 2.0E − 6 | −1.8E − 2 | 3.6E − 3 |
| Temporal | −5.3E − 3 | 0.502 | −2.6E − 3 | 0.758 | −2.4E − 2 | 4.4E − 6 | −1.6E − 2 | 3.6E − 3 |
| Parietal | −8.4E − 3 | 0.421 | −6.2E−3 | 0.698 | −3.8E−2 | 1.4E − 7 | −2.6E − 2 | 3.0E − 4 |
| Precuneus | −1.8E − 3 | 0.857 | −6.4E − 4 | 0.992 | −2.6E − 2 | 3.8E − 5 | −1.8E − 2 | 3.6E−3 |
| Ant. cingulate | −3.1E−5 | 0.995 | −5.5E − 5 | 0.992 | −1.9E − 2 | 1.1E − 3 | −1.9E − 2 | 2.3E−3 |
| Post. cingulate | −3.1E − 3 | 0.736 | −2.9E − 3 | 0.758 | −1.9E − 2 | 1.1E − 3 | −1.5E − 2 | 8.3E − 3 |
| Entorhinal | −1.8E − 2 | 0.421 | −1.4E−2 | 0.500 | −2.8E − 2 | 4.9E − 3 | 2.1E − 2 | 0.052 |
| Parahippocampal | −9.4E − 3 | 0.421 | −9.4E − 2 | 0.500 | −3.4E − 2 | 6.0E − 7 | −3.4E − 2 | 2.9E − 6 |
ANCOVA models tested the effect of diagnosis on ODI, controlling for age, sex, and with and without cortical thickness; bold indicates PFDR < 0.05; CT = cortical thickness; AD = Alzheimer’s disease dementia group.
Logistic Regression Analysis
An exploratory logistic regression analysis was performed using different combinations of NDI, ODI, and cortical thickness in order to test the diagnostic accuracy of these microstructural and macrostructural metrics. For distinguishing the AD dementia group from the control group, the macrostructural model using only cortical thickness performed the worst (AUC = 0.82), while the multimodal model, which combined macro- and microstructural metrics, performed the best (AUC = 0.91) (Fig. 5A). For distinguishing the MCI group from control group, cortical thickness once again performed the worst (AUC = 0.66), and the multimodal model performed the best (AUC = 0.72) (Fig. 5B), though all models performed more poorly than those for AD dementia versus control.
Discussion
In the present study, we used multi-shell DWI and the NODDI model to investigate cortical microstructure along the continuum of clinical cognitive impairment, including MCI and AD dementia. We report that participants with MCI had widespread decreases in cortical NDI (but no differences in ODI), while participants with AD dementia demonstrated widespread cortical areas of lower NDI and ODI. Moreover, our follow-up ROI analysis demonstrated that even after controlling for cortical thickness, NODDI metrics were still significantly lower in MCI and AD dementia groups. Additionally, for participants with MCI, gray matter NDI—but not cortical thickness—was decreased in temporal, parietal, and posterior cingulate regions.
While DWI has traditionally been applied to the study of white matter microstructure in AD, a small but growing number of studies have also used DWI techniques to interrogate gray matter microstructure (Weston et al. 2015). A majority of these studies have used the DTI model to investigate cortical MD and have reported increased MD in cortical regions that also show macrostructural atrophy, including medial and lateral temporal lobes, superior and medial frontal lobes, supramarginal gyrus, posterior cingulate, and precuneus (Rose et al. 2008; Montal et al. 2018). However, the relatively large voxel dimensions (~2 × 2 × 2 mm3) typically used in DWI makes gray matter DTI MD measurements particularly susceptible to partial volume effects from neighboring CSF or white matter tissue. In fact, a recent study showed that after correcting for partial volume effects in a voxelwise gray matter analysis, there were no longer any significant differences in MD between MCI and cognitively unimpaired participants, and the number of significant voxels between AD and cognitively unimpaired participants was drastically reduced (Henf et al. 2018).
In contrast to DTI, NODDI is a multi-compartment diffusion model that accounts for CSF partial volume effects, which makes it advantageous for analysis of cortical gray matter microstructure. In turn, GBSS can be used to capitalize on the NODDI model to broadly test for microstructural alterations across the brain using a voxelwise approach (Nazeri et al. 2015, 2017). In our study, GBSS analysis revealed extensive decreases in cortical NODDI metrics in the AD dementia group, with reductions in ODI being more widely distributed than reductions in NDI. These NODDI microstructural alterations occurred in regions, which have previously been shown to have cortical thinning (Dickerson et al. 2008; Bakkour et al. 2009) and increased MD (Rose et al. 2008; Montal et al. 2018) in AD. Additionally, for the MCI group, GBSS analysis highlighted significantly lower NDI throughout temporal, parietal, and portions of frontal cortex, while the CAT12 surface-based analysis did not show any regions with significantly decreased cortical thickness.
As a multi-compartment model, the metrics derived from the NODDI model may be able to capture microstructural properties and quantify neuronal cytoarchitecture better than the DTI model. Both ex vivo (Jespersen et al. 2010; Grussu et al. 2017) and in vivo (Fukutomi et al. 2018) evidence indicates that NDI reflects density of myelinated axons in gray matter, while ODI captures the heterogeneity of neurite fiber orientations, which may reflect complexity of dendritic arborization in gray matter. Thus, using these metrics may provide valuable information regarding specific gray matter microstructural changes in AD. In the present study, the MCI group showed significantly lower NDI in temporal, parietal, and frontal regions, suggesting that reduction in the density of myelinated axons in these regions is an early microstructural change that occurs prior to development of overt clinical dementia. Additionally, while the MCI group had no changes in ODI, the AD dementia group demonstrated prominent decreases in ODI across characteristic AD cortical regions, which provides neuroimaging evidence of an association between significant loss of dendritic architecture and the clinical dementia phenotype. While previous postmortem studies have demonstrated synaptic and dendritic loss in MCI and AD (Davies et al. 1987; Terry et al. 1991; Baloyannis et al. 2011; Scheff et al. 2011), additional studies combining ex vivo imaging and histopathological analysis at autopsy are needed in order to further characterize the relationship between NODDI metrics and neuropathological changes in AD.
Our follow-up ROI analysis used a common atlas in order to more thoroughly investigate changes in both NODDI microstructure and cortical thickness within regions affected by AD pathology. The results of this analysis highlight some intriguing findings. First, similar to the results observed at the whole-brain level in the GBSS and CAT12 surface-based analyses, the MCI group had significantly lower NDI in several ROIs where cortical thickness was unchanged, including temporal, parietal, and posterior cingulate (Fig. 4). Notably, these regions are some of the earliest areas of the brain where amyloid accumulates (Buckner et al. 2005; Sperling et al. 2009) and correspond to Stages I and II of a recently derived four-stage model of regional amyloid progression using in vivo amyloid PET imaging (Grothe et al. 2017). The regional overlap between these early stages of amyloid deposition (which are most prevalent in cognitively unimpaired and MCI individuals) and the current findings of reduced neurite density highlight the potential utility of NDI as an early biomarker of AD-related neurodegeneration. Our present results provide additional information regarding ongoing microarchitectural changes in these regions and, based on our stratification by clinical disease severity, suggest neurite density is reduced prior to cortical atrophy and development of dementia.
Second, by extracting both NODDI metrics and cortical thickness from the same ROI, we were able to test whether differences in NODDI metrics remained after controlling for cortical thickness within an ROI. Overall, a majority of regions with decreased NODDI metrics remained significant after accounting for cortical thickness, suggesting that changes in NDI and ODI represent unique microstructural alterations beyond simply gross atrophy. A notable exception was the entorhinal region—where controlling for cortical thickness accounted for the majority of microstructural alterations observed in the AD dementia group, possibly due to the relatively small size of this ROI and the fact that this region showed the greatest cortical thinning. The entorhinal cortex is well-known to be affected at the earliest stages of disease; thus, by the time AD dementia is diagnosed, this region is likely to have undergone substantial atrophy.
Finally, our exploratory logistic regression analysis used averaged microstructural and macrostructural parameters in order to test the diagnostic accuracy of NDI, ODI, and cortical thickness across these eight ROIs. This analysis showed that models combining macrostructural and microstructural information had the best accuracy for distinguishing the AD dementia and MCI groups from control, while models using only cortical thickness performed more poorly. These findings suggest that NODDI metrics may provide additional valuable diagnostic information regarding neurodegenerative changes in AD dementia.
While the NODDI model has been used to investigate gray matter microstructure in a variety of psychiatric and neurological conditions, only one previous study has investigated NODDI gray matter microstructure in the context of AD (Parker et al. 2018). That study, which took an ROI-based approach in individuals with young-onset AD (mean age 61.1 ± 4.9 years), demonstrated that NDI was significantly lower in all six ROIs studied (entorhinal cortex, inferior and middle temporal gyri, fusiform gyrus, precuneus, and precentral gyrus), while ODI was significantly lower in inferior and temporal gyri, fusiform gyrus, and precuneus. The results of our present study, which used an older population with sporadic AD dementia (mean age 72.2 ± 10.2 years), are broadly consistent with this previous report. Together, the previous and present studies suggest concordant microstructural alterations between young-onset and sporadic AD. Moreover, our present study extends findings of altered gray matter NODDI microstructure to the clinically intermediate stage of MCI, prior to the development of dementia.
A few limitations of the current study should be noted. First, participants in the AD dementia group were diagnosed based on clinical and cognitive criteria outlined in the updated NIA-AA guidelines (Albert et al. 2011; McKhann et al. 2011), but amyloid and tau biomarker data were not available to confirm the presence of Alzheimer’s pathology in all participants. Without these data, we cannot rule out the possibility that some participants in the AD dementia group do not have sufficient amyloid to meet the criteria of the new AT(N) Alzheimer’s research framework (Jack et al. 2018). Thus, interpretation of the present results is limited to clinically diagnosed cognitive impairment and AD dementia. Additional studies using CSF or PET imaging AD biomarkers are needed and may better inform the relationship between amyloid and tau deposition and cortical microstructure. Second, we are inherently limited by the voxel resolution of the DWI scans (2 × 2 × 2 mm3) relative to the thickness of the cortex (1.5–5 mm). Thus, while the NODDI model reduces CSF partial volume effects by specifically modeling free water diffusion, we acknowledge that NODDI metrics in cortical gray matter may be partially influenced by imperfect suppression of CSF signal or by the diffusion signal arising from underlying white matter, especially in thinner areas of the cortex. However, estimation and masking of the white matter fraction as well as the skeletonization step in GBSS are designed to restrict statistical analysis to areas of the cortex where risk of partial voluming is low. Finally, while we investigated cortical microstructure in diagnostic groups representing the continuum of progressive cognitive decline, this study was cross-sectional. Longitudinal studies will be needed to clarify the utility of NODDI metrics in predicting conversion from MCI to AD dementia or tracking the progression of disease. Furthermore, with larger longitudinal datasets, it will be possible to perform event-time modeling of cortical microstructural alterations (Marinescu et al. 2019), which may provide valuable insights into fine-grained patterns of regional progression of pathologic processes that are not apparent in cross-sectional analyses.
Conclusion
Overall, the present study demonstrates that microstructural neurodegenerative processes are widespread and robust in MCI and AD dementia and that these microstructural alterations are detectable in brain regions where cortical thinning is not detected. These findings suggest that NODDI metrics provide unique information regarding cortical neurodegeneration not previously detected using T1-weighted structural imaging or DTI-based approaches. Given that AD pathology accumulates for years prior to detectable macrostructural atrophy, future work investigating whether NODDI metrics are sensitive and early markers of altered cortical microstructure during the silent, prodromal stage will be vital toward furthering our understanding of the earliest neurodegenerative changes in AD pathogenesis.
Funding
National Institutes of Health (F30AG059346, R01AG037639, R01AG027161, P50AG033514); Geriatric Research, Education, and Clinical Center of William S. Middleton Memorial Veterans Hospital; National Institute of Child Health and Human Development (U54HD090256) to [N.A.]; the BRAIN Initiative (R01EB022883); University of Wisconsin Center for Predictive Computational Phenotyping (AI117924); University of Wisconsin Institute for Clinical and Translational Research KL2 Scholars Program (NCATS UL1TR002373) to [J.J.Y].
Notes
We would like to thank Paul Cary, Jennifer Oh, Chuck Illingworth, and the staff and researchers at the University of Wisconsin ADRC for their assistance in study organization, participant recruitment, and facilitating data availability. Additionally, we would like to thank Dr Arash Nazeri for making the GBBS processing scripts publicly available and for providing guidance on graphically summarizing the GBSS pipeline. We would also like to thank Dr Chris Rorden for his development of the Surf Ice visualization software and his assistance with making the software compatible with CAT12 surface-based analysis results. Finally, we would like to extend our thanks to the committed research participants at the UW ADRC who make this work possible. Conflict of Interest: None declared.
Supplementary Material
References
- Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, Gamst A, Holtzman DM, Jagust WJ, Peterson RC et al. 2011. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7:270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alger JR. 2012. The diffusion tensor imaging toolbox. J Neurosci. 32:7418–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J, Sotiropoulos SN. 2016. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage. 125:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Steiner CA, Akopyan GG, Dutton RA, Hayashi KM, Toga AW, Cummings JL, Thompson PM. 2007. Three-dimensional gray matter atrophy mapping in mild cognitive impairment and mild Alzheimer disease. Arch Neurol. 64:1489–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf Y, Alexander DC, Jones DK, Bizzi A, Behrens TE, Clark CA, Cohen Y, Dyrby TB, Huppi PS, Knoesche TR et al. 2013. The CONNECT project: combining macro- and micro-structure. NeuroImage. 80:273–282. [DOI] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. 2011a. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage. 54:2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Wu J, Cook PA, Gee JC. 2011b. An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics. 9:381–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkour A, Morris JC, Dickerson BC. 2009. The cortical signature of prodromal AD: regional thinning predicts mild AD dementia. Neurology. 72:1048–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloyannis SJ, Manolides SL, Manolides LS. 2011. Dendritic and spinal pathology in the acoustic cortex in Alzheimer’s disease: morphological estimation in Golgi technique and electron microscopy. Acta Otolaryngol. 131:610–612. [DOI] [PubMed] [Google Scholar]
- Becker AJ, Hedden T, Carmasin J, Maye J, Rentz DM, Putcha D, Fischl B, Greve DN, Marshall GA, Salloway S et al. 2011. Amyloid-β associated cortical thinning in clinically normal elderly. Ann Neurol. 69:1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihan D. 2003. Looking into the functional architecture of the brain with diffusion MRI. Nat Rev Neurosci. 4:469–480. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC et al. 2005. Molecular, structural, and functional characterization of Alzheimer’s disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci. 25:7709–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chételat G, Villemagne VL, Villain N, Jones G, Ellis KA, Ames D, Martins RN, Masters CL, Rowe CC. 2012. Accelerated cortical atrophy in cognitively normal elderly with high β-amyloid deposition. Neurology. 78:477–484. [DOI] [PubMed] [Google Scholar]
- Chung MK, Robbins SM, Dalton KM, Davidson RJ, Alexander AL, Evans AC. 2005. Cortical thickness analysis in autism with heat kernel smoothing. NeuroImage. 25:1256–1265. [DOI] [PubMed] [Google Scholar]
- Daducci A, Canales-Rodríguez EJ, Zhang H, Dyrby TB, Alexander DC, Thiran J-P. 2015. Accelerated microstructure imaging via convex optimization (AMICO) from diffusion MRI data. NeuroImage. 105:32–44. [DOI] [PubMed] [Google Scholar]
- Dahnke R, Yotter RA, Gaser C. 2013. Cortical thickness and central surface estimation. NeuroImage. 65:336–348. [DOI] [PubMed] [Google Scholar]
- Davies CA, Mann DMA, Sumpter PQ, Yates PO. 1987. A quantitative morphometric analysis of the neuronal and synaptic content of the frontal and temporal cortex in patients with Alzheimer’s disease. J Neurol Sci. 78:151–164. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT et al. 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 31:968–980. [DOI] [PubMed] [Google Scholar]
- de Toledo-Morrell L, Goncharova I, Dickerson F, Wilson RS, Bennett DA. 2000. From healthy aging to early Alzheimer’s disease: in vivo detection of entorhinal cortex atrophy. Ann N Y Acad Sci. 911:240–253. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Bakkour A, Salat DH, Feczko E, Pacheco J, Greve DN, Grodstein F, Wright CI, Blacker D, Rosas HD et al. 2008. The cortical signature of Alzheimer’s disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb Cortex. 19:497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douaud G, Menke RA, Gass A, Monsch AU, Rao A, Whitcher B, Zamboni G, Matthews PM, Sollberger M, Smith S. 2013. Brain microstructure reveals early abnormalities more than two years prior to clinical progression from mild cognitive impairment to Alzheimer’s disease. J Neurosci. 33:2147–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox NC, Warrington EK, Freeborough PA, Hartikainen P, Kennedy AM, Stevens JM, Rossor MN. 1996. Presymptomatic hippocampal atrophy in Alzheimer’s disease: a longitudinal MRI study. Brain. 119:2001–2007. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Prestia A, Rasser PE, Bonetti M, Thompson PM. 2009. In vivo mapping of incremental cortical atrophy from incipient to overt Alzheimer’s disease. J Neurol. 256:916–924. [DOI] [PubMed] [Google Scholar]
- Fukutomi H, Glasser MF, Zhang H, Autio JA, Coalson TS, Okada T, Togashi K, Van Essen DC, Hayashi T. 2018. Neurite imaging reveals microstructural variations in human cerebral cortical gray matter. NeuroImage. 182:488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garyfallidis E, Brett M, Amirbekian B, Rokem A, van der Walt S, Descoteaux M et al. 2014. Dipy, a library for the analysis of diffusion MRI data. Front Neuroinform. 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson AJ, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR et al. 2013. The minimal preprocessing pipelines for the human Connectome project. NeuroImage. 80:105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granberg T, Fan Q, Treaba CA, Ouellette R, Herranz E, Mangeat G, Louapre C, Cohen-Adad J, Klawiter EC, Sloane JA et al. 2017. In vivo characterization of cortical and white matter neuroaxonal pathology in early multiple sclerosis. Brain. 140:2912–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe MJ, Barthel H, Sepulcre J, Dyrba M, Sabri O, Teipel SJ. 2017. In vivo staging of regional amyloid deposition. Neurology. 89:2031–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grussu F, Schneider T, Tur C, Yates RL, Tachrount M, Ianuş A, Yiannakas MC, Newcombe J, Zhang H, Alexander DC et al. 2017. Neurite dispersion: a new marker of multiple sclerosis spinal cord pathology? Ann Clin Transl Neurol. 4:663–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henf J, Grothe MJ, Brueggen K, Teipel S, Dyrba M. 2018. Mean diffusivity in cortical gray matter in Alzheimer’s disease: the importance of partial volume correction. Neuroimage Clin. 17:579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, O’Brien PC, Tangalos EG. 1992. MR-based hippocampal volumetry in the diagnosis of Alzheimer’s disease. Neurology. 42:183–188. [DOI] [PubMed] [Google Scholar]
- Jack CR, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, Holtzman DM, Jagust W, Jessen F, Karlawish J et al. 2018. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 14:535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HIL, van Boxtel MPJ, Gronenschild EHBM, Uylings HBM, Jolles J, Verhey FRJ. 2013. Decreased gray matter diffusivity: a potential early Alzheimer’s disease biomarker? Alzheimers Dement. 9:93–97. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. 2012. FSL. NeuroImage. 62:782–790. [DOI] [PubMed] [Google Scholar]
- Jespersen SN, Bjarkam CR, Nyengaard JR, Chakravarty MM, Hansen B, Vosegaard T, Østergaard L, Yablonskiy D, Nielsen NC, Vestergaard-Poulsen P. 2010. Neurite density from magnetic resonance diffusion measurements at ultrahigh field: comparison with light microscopy and electron microscopy. NeuroImage. 49:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamagata K, Zalesky A, Hatano T, Ueda R, Biase MA, Okuzumi A, Shimoji K, Hori M, Caeyenberghs K, Pantelis C et al. 2017. Gray matter abnormalities in idiopathic Parkinson’s disease: evaluation by diffusional kurtosis imaging and neurite orientation dispersion and density imaging. Hum Brain Mapp. 38:3704–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarci K, Petersen RC, Boeve BF, Knopman DS, Weigand SD, O’Brien PC, Shiung MM, Smith GE, Ivnik RJ, Tangalos EG et al. 2005. DWI predicts future progression to Alzheimer disease in amnestic mild cognitive impairment. Neurology. 64:902–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner E, Dhital B, Kiselev VG, Reisert M. 2016. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magn Reson Med. 76:1574–1581. [DOI] [PubMed] [Google Scholar]
- Klein A, Tourville J. 2012. 101 labeled brain images and a consistent human cortical labeling protocol. Front Neurosci. 6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasuski JS, Alexander GE, Horwitz B, Daly EM, Murphy DGM, Rapoport SI, Schapiro MB. 1998. Volumes of medial temporal lobe structures in patients with Alzheimer’s disease and mild cognitive impairment (and in healthy controls). Biol Psychiatry. 43:60–68. [DOI] [PubMed] [Google Scholar]
- LaPoint MR, Chhatwal JP, Sepulcre J, Johnson KA, Sperling RA, Schultz AP. 2017. The association between tau PET and retrospective cortical thinning in clinically normal elderly. NeuroImage. 157:612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Pruessner JC, Zijdenbos A, Hampel H, Teipel SJ, Evans AC. 2005. Focal decline of cortical thickness in Alzheimer’s disease identified by computational neuroanatomy. Cereb Cortex. 15:995–1001. [DOI] [PubMed] [Google Scholar]
- Marinescu RV, Eshaghi A, Lorenzi M, Young AL, Oxtoby NP, Garbarino S, Crutch SJ, Alexander DC. 2019. DIVE: a spatiotemporal progression model of brain pathology in neurodegenerative disorders. NeuroImage. 192:166–177. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Kawas CH, Klunk WE, Koroshetz WJ, Manly JF, Mayeux R et al. 2011. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montal V, Vilaplana E, Alcolea D, Pegueroles J, Pasternak O, González-Ortiz S, Clarimón J, Carmona-Iragui M, Illán-Gala I, Morenas-Rodríguez E et al. 2018. Cortical microstructural changes along the Alzheimer’s disease continuum. Alzheimers Dement. 14:340–351. [DOI] [PubMed] [Google Scholar]
- Mori E, Yoneda Y, Yamashita H, Hirono N, Ikeda M, Yamadori A. 1997. Medial temporal structures relate to memory impairment in Alzheimer’s disease: an MRI volumetric study. J Neurol Neurosurg Psychiatry. 63:214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller MJ, Greverus D, Dellani PR, Weibrich C, Wille PR, Scheurich A, Stoeter P, Fellgiebel A. 2005. Functional implications of hippocampal volume and diffusivity in mild cognitive impairment. NeuroImage. 28:1033–1042. [DOI] [PubMed] [Google Scholar]
- Nazeri A, Chakravarty MM, Rotenberg DJ, Rajji TK, Rathi Y, Michailovich OV, Voineskos AN. 2015. Functional consequences of neurite orientation dispersion and density in humans across the adult lifespan. J Neurosci. 35:1753–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazeri A, Mulsant BH, Rajji TK, Levesque ML, Pipitone J, Stefanik L, Shahab S, Roostaei T, Wheeler AL, Chavez S et al. 2017. Gray matter neuritic microstructure deficits in schizophrenia and bipolar disorder. Biol Psychiatry. 82:726–736. [DOI] [PubMed] [Google Scholar]
- Parker TD, Slattery CF, Zhang J, Nicholas JM, Paterson RW, Foulkes AJ, Malone IB, Thomas DL, Modat M, Cash DM et al. 2018. Cortical microstructure in young onset Alzheimer’s disease using neurite orientation dispersion and density imaging. Hum Brain Mapp. 39:3005–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose SE, Janke AL, Chalk JB. 2008. Gray and white matter changes in Alzheimer’s disease: a diffusion tensor imaging study. J Magn Reson Imaging. 27:20–26. [DOI] [PubMed] [Google Scholar]
- Scheff SW, Price DA, Schmitt FA, Scheff MA, Mufson EJ. 2011. Synaptic loss in the inferior temporal gyrus in mild cognitive impairment and Alzheimer’s disease. J Alzheimers Dis. 24:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scola E, Bozzali M, Agosta F, Magnani G, Franceschi M, Sormani MP, Cercignani M, Pagani E, Falautano M, Filippi M et al. 2010. A diffusion tensor MRI study of patients with MCI and AD with a 2-year clinical follow-up. J Neurol Neurosurg Psychiatry. 81:798–805. [DOI] [PubMed] [Google Scholar]
- Smith SM. 2002. Fast robust automated brain extraction. Hum Brain Mapp. 17:143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM et al. 2006. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. 31:1487–1505. [DOI] [PubMed] [Google Scholar]
- Smith SM, Nichols TE. 2009. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. NeuroImage. 44:83–98. [DOI] [PubMed] [Google Scholar]
- Sperling RA, LaViolette PS, O’Keefe K, O’Brien J, Rentz DM, Pihlajamaki M, Marshall G, Hyman BT, Selkoe DJ, Hedden T et al. 2009. Amyloid deposition is associated with impaired default network function in older persons without dementia. Neuron. 63:178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R. 1991. Physical basis of cognitive alterations in Alzheimer’s disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 30:572–580. [DOI] [PubMed] [Google Scholar]
- Tournier J-D, Smith RE, Raffelt DA, Tabbara R, Dhollander T, Pietsch M, Christiaens D, Jeurissen B, Yeh C, Connelly A. 2019. MRtrix3: a fast, flexible and open software framework for medical image processing and visualisation. NeuroImage. 202: 116137. [DOI] [PubMed] [Google Scholar]
- Veraart J, Novikov DS, Christiaens D, Ades-aron B, Sijbers J, Fieremans E. 2016. Denoising of diffusion MRI using random matrix theory. NeuroImage. 142:394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemagne VL, Burnham S, Bourgeat P, Brown B, Ellis KA, Salvado O, Szoeke C, Macaulay SL, Martins R, Maruff P et al. 2013. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: a prospective cohort study. Lancet Neurol. 12:357–367. [DOI] [PubMed] [Google Scholar]
- Weston PSJ, Simpson IJA, Ryan NS, Ourselin S, Fox NC. 2015. Diffusion imaging changes in grey matter in Alzheimer’s disease: a potential marker of early neurodegeneration. Alzheimers Res Ther. 7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CR. 2007. 3D maps from multiple MRI illustrate changing atrophy patterns as subjects progress from mild cognitive impairment to Alzheimer’s disease. Brain. 130:1777–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE. 2014. Permutation inference for the general linear model. NeuroImage. 92:381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotter RA, Dahnke R, Thompson PM, Gaser C. 2011a. Topological correction of brain surface meshes using spherical harmonics. Hum Brain Mapp. 32:1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotter RA, Thompson PM, Gaser C. 2011b. Algorithms to improve the reparameterization of spherical mappings of brain surface meshes. J Neuroimaging. 21:e134–e147. [DOI] [PubMed] [Google Scholar]
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. 2012. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage. 61:1000–1016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


