Abstract
Background
The adhesion molecule, FAT4, has a tumor suppressor function with a critical role in the epithelial-to-mesenchymal-transition (EMT) and anti-malignant growth in several cancers. No study has investigated yet its role in epithelial ovarian cancer (EOC) progression. In the present study, we examined the role of FAT4 in proliferation and metastasis, and its mechanisms of interaction in these processes.
Methods
We have performed cell viability, colony formation, and invasion assays in ovarian cancer cells treated with siRNA to knockdown FAT4 gene expression. The regulatory effects of FAT4 on proteins involved in apoptotic, Wnt, Hippo, and retinoblastoma signaling pathways were evaluated by Western blotting following FAT4 repression. Also, 426 ovarian tumor samples and 88 non-tumor samples from the Gene Expression Profiling Interactive Analysis (GEPIA) database were analyzed for the expression of FAT4. Pearson’s correlation was performed to determine the correlation between FAT4 and the E2F5, cyclin D1, cdk4, and caspase 9 expressions.
Results
Lower expression of FAT4 was observed in ovarian cancer cell lines and human samples as compared to non-malignant tissues. This down-regulation seems to enhance cell viability, invasion, and colony formation. Silencing FAT4 resulted in the upregulation of E2F5, vimentin, YAP, β-catenin, cyclin D1, cdk4, and Bcl2, and in the downregulation of GSK-3-β, and caspase 9 when compared to control. Furthermore, regulatory effects of FAT4 on the EMT and aggressive phenotype seem to occur through Hippo, Wnt, and cell cycle pathways.
Conclusion
FAT4 downregulation promotes increased growth and invasion through the activation of Hippo and Wnt-β-catenin pathways.
Keywords: FAT4, Silencing, Invasion, YAP, Ovarian cancer
Background
Epithelial Ovarian Cancer (EOC) is a malignant disease originating from the outer surface cells covering the ovary [1]. It represents 90% of all types of ovarian cancer [2] and is considered to be the 8th leading cause of cancer-related deaths among women worldwide [3]. Due to its asymptomatic nature and the absence of reliable biological markers for early detection [4], only 20% of ovarian tumors are diagnosed at early stages I/II, while the majority are diagnosed at advanced stages (III to IV) [5]. An increase in the incidence rate, and also the shift towards a younger affected population was recently noticed [3].
Despite the broad advances in gynecological research, the cascade of events leading to epithelial ovarian cancer remains ambiguous, and efforts are needed to elucidate the underlying mechanisms. To identify candidate genes with a potential role in the pathogenesis of ovarian cancer, we performed chromatin immunoprecipitation (ChIP) using E2F5, a transcription factor highly expressed in early stages of EOC [6]. ChIP data revealed that FAT4 was one of the immunoprecipitated downstream genes regulated by E2F5, and thus can be suspected to have a role in EOC pathogenesis.
FAT tumor suppressor homolog 4 (FAT4), is a member of the FAT family that consists of (FAT1–4) and encodes a single transmembrane protein containing 32–34 extracellular cadherin repeats, a transmembrane domain and a cytoplasmic domain [7]. FAT4 was identified as a tumor suppressor in mouse mammary epithelial cell line and triple-negative breast cancer [8–11].
There is increasing evidence of a possible relation between the FAT4 downregulation and the pathogenesis of several malignancies, including breast, colorectal, and gastric cancers [8, 12, 13]. Also, previous mutational screening studies revealed missense and nonsense mutations of FAT4 in hepatocellular (10%) [14], pancreatic (8%) [15], head-and-neck squamous cell cancers (6%) [16], endometrioid, and mucinous primary ovarian tumors (15%) [17].
In endometrial cancer, FAT4 downregulation was attributed to the silencing of USP51, a de-ubiquitinating enzyme, suggested as a direct interacting partner of FAT4, contributing to its tumor suppressor role [18]. While in colorectal cancer, FAT4 was found to inhibit tumorigenesis by regulating the PI3K activity in the PI3K/AKT/mTOR signaling pathway and to play a significant role in preventing the epithelial-to-mesenchymal transition (EMT) [13]. The EMT is a crucial step for several developmental processes and a genuine hallmark for aggressive phenotype and invasion [19, 20]. Moreover, in gastric cancer, FAT4 silencing stimulated cell proliferation, migration, and cell cycle progression through the nuclear translocation of YAP [21]. Hence, FAT4 was found to regulate the downstream effectors of the Hippo pathway, YAP/TAZ [18, 21, 22]. Alternatively, YAP activity is regulated by the core Hippo kinases. Phosphorylation of YAP results in its cytoplasmic retention and inactivation, while un-phosphorylated YAP is in its active mode, and are freely translocated into the nucleus to promote transcription of cell proliferation and anti-apoptotic genes [23]. In ovarian cancer, activated YAP was associated with poor survival by promoting cell proliferation, EMT, anchorage-independent growth, and resistance to cisplatin-induced apoptosis [24].
In the present study, we examined the role of FAT4 downregulation in the tumorigenesis of EOC cells and its consequent impact on the expression of key proteins involved in Hippo, Wnt-β-catenin, apoptotic, EMT, and cell cycle pathways. The obtained data shed some light on the role of the FAT4 adhesion molecules in ovarian cancer tumorigenesis through different pathways, namely, Hippo, and Wnt-β-catenin.
Methods
Cell culture
The human ovarian cancer cell lines: MCAS and OVSAHO (JCRB cell bank, Osaka, Japan, catalog no. JCRB0240 and no. JCRB1046 respectively) were kindly provided by Prof. Aikou Okamoto (Jikei University School of Medicine, Japan), in 2016. The cisplatin sensitive A2780 (The European Collection of Authenticated Cell, ECACC catalog no. 93112519) and cisplatin-resistant A2780-cis (ECACC catalog no. 93112517) cell lines were a generous gift from Dr. Benjamin Tsang (University of Ottawa, Canada), in 2018. The transformed normal epithelial ovarian cell line HOSE6–3 (RRID: CVCL_7673), established by Prof. GSW Tsao (School of Biomedical Sciences, The University of Hong Kong), was kindly provided by his laboratory in 2018. To avoid contaminations, our cell-culture laboratories, including hoods and incubators, are systemically fumigated every year, and any new cells are tested upon arrival, for the presence of mycoplasma using the “Mycoplasma Detection Kit” (Lonza, Catalog #: LT07–118). None of the cells used for this study were tested positive.
MCAS, A2780, A2780 cis and HOSE6–3 cells were propagated in DMEM (Gibco, NY, USA), while OVSAHO was cultured in RPMI-1640 media supplemented with 10% FBS (Gibco, NY, USA) and 1% penicillin-streptomycin antibiotic (Gibco, NY, USA) in a humidified incubator at 37 °C and 5% CO2.
RNA extraction and qRT-PCR
Total RNA was extracted from the ovarian cancer cell lines using PureLink RNA mini kit (Invitrogen, CA, USA) according to the manufacturer’s protocol, and subsequently reverse transcribed to complementary DNA using high capacity reverse transcription kit (Invitrogen, CA, USA). qRT-PCR was performed using TaqMan pre-optimized probes (FAT4: Catalogue No. Hs01570499_m1, E2F5: Catalogue No.Hs00231092_m1, GAPDH: Catalogue No. 402869; Thermo Scientific Fisher, USA) and run on ABI 7500 Fast real-time PCR machine (Applied Biosystems, Austin, TX). All the messenger RNA data were normalized to GAPDH expression, and relative expression was computed by the comparative Ct method.
Cell transfection with siRNA
1.5 × 105 to 3 × 105 cells (MCAS and OVSAHO) were seeded in six-well plate and incubated until they reached 70% confluency. The cells were then transfected with commercially available pre-designed anti-FAT4 expression siRNA (Catalogue No. sc-88,877, Santa Cruz Biotechnology Inc., USA), using lipofectamine RNAiMAX reagent (Invitrogen; Thermofisher Scientific, USA) and Opti-MEM medium (Invitrogen, USA) according to the manufacturers protocol. The siRNA used consists of a pool of 3 specific probes of 19–25 nucleotides long designed and pre-optimized to act on different positions along the mRNA with no off-target effects. FAT4 siRNA (35 pmol) and 7.5 μl of lipofectamine RNAiMAX reagent were used per ml of the media per well. The cells were then incubated for 36 h at 37 °C and 5% CO2 atmosphere before RNA and protein extraction. Cells treated with scrambled siRNA were used as a negative control.
In vitro cell proliferation assay
Cell viability was assessed using Alamar blue reagent (Invitrogen, CA, USA). MCAS and OVSAHO cells were seeded in 96 well culture plates at a density of 104 cells per well, and transfected 1 day later with FAT4 siRNA, and incubated for 24–96 h at 37 °C and 5% CO2. Cell proliferation was measured by adding 10 μl Alamar blue dye to the culture medium and incubating for 4 h at 37 °C. The absorbance was measured at 570 nm using Multiscan spectrum spectrophotometer (Thermo Fisher Scientific Inc., MA, USA).
Colony-forming assay
1 × 105 MCAS, and OVSAHO cells treated with FAT4-siRNA were seeded in a six-well plate and incubated at 37 °C in a 5% CO2 atmosphere for 14 days; medium was refreshed every 72 h. Cells were stained with crystal violet and observed under the microscope. For soft agar colony formation assay, 105 cells per well were treated with FAT4 siRNA, and resuspended in 0.3% soft agar in culture medium and layered onto 0.6% solidified agar in a six-well plate. After incubation for 2 weeks, colonies were stained with crystal violet and counted in three random fields at 40X magnification.
Cell invasion assay
The invasive potential of MCAS and OVSAHO cells, following the silencing of FAT4, was evaluated using QCM™ Collagen cell invasion assay kit 24-well 8 μm, colorimetric (Sigma Aldrich) according to the manufacturer’s protocol. Briefly, 1.25 × 105FAT4-siRNA transfected cells suspended in 250 μl of serum-free media was added to the upper chamber of the insert. 500 μl of the medium supplemented with 10% FBS was added to the lower chamber. After incubation for 48 h, non-invading cells on the upper surface were removed with a cotton swab and cells invading to the lower chamber were stained with crystal violet, and treated with commercially provided extraction solution (QCM™ Collagen cell invasion assay kit 24-well 8 μm, colorimetric, Sigma Aldrich). The absorbance was measured at 560 nm using Multiscan spectrophotometer (Thermofisher Scientific Inc., MA, USA).
Western blotting
FAT4 knocked-down MCAS cells (36 h post transfection) were subjected to proteins extraction using RIPA lysis buffer (Santa Cruz Biotechnology Inc., USA) supplemented with protease and phosphatase inhibitor (Thermofisher, USA). The lysate was centrifuged, and the supernatant was recovered and stored at -80 °C. After protein quantification, 40 μg were resolved on 8% SDS-PAGE and electro-transferred onto a nitrocellulose membrane which was incubated in a blocking reagent (5% BSA in tris buffered saline/Tween-20 buffer (TBST)) for 1 h at room temperature. Then incubated with the primary antibodies directed against the following proteins Rb (sc-50, Santa Cruz Biotechnology), actin (sc-1616, Santa Cruz Biotechnology), cdk 4 (sc-260, Santa cruz Biotechnology), E2F5 (sc-999, Santa Cruz Biotechnology, USA); caspase 9 (PA5–22252, Invitrogen, Thermofisher), β-catenin (PA5–77934, Invitrogen, Thermofisher), phospho-YAP (PA5–17481, Invitrogen, Thermofisher), phospho-β-catenin (pβ-catenin; PA5–67504, Invitrogen, Thermofisher), phospho-retinoblastoma (pRb; PA5–37715, Invitrogen, Thermofisher), E cadherin (PA5–32178, Invitrogen, Thermofisher), vimentin (PA5–27231, Invitrogen, Thermofisher, USA), FAT4 (ab130076, abcam), cyclin D1 (ab134175, Abcam), GSK-3-β (#9315, Cell Signaling Technology), and phospho-GSK-3-β (pGSK-3-β; #5558, Cell Signaling Technology). All the primary antibodies were used at a dilution of 1/200. Antibody against beta-actin was used as a loading control. After washing 3X5 minutes in TBST, membranes were incubated with horseradish peroxidase-conjugated secondary antibody (dilution 1:5000) for 2 h at room temperature and revealed using ECL western blotting detection reagent (Pierce Biotechnology, Illinois, USA).
Bioinformatics & Statistical analysis
We explored “Gene Expression Profiling Interactive Analysis” (GEPIA) (http://gepia.cancer-pku.cn/), available public database to examine the expression of genes of interest at the RNA levels. We analyzed 426 ovarian tumor samples and 88 non-tumor samples for the expression of FAT4. Pearson’s correlation was performed to determine the correlation between FAT4 and the E2F5, cyclin D1, cdk4, and caspase 9 expressions.
Student’s t-test was performed using SPSS software (version 23, SPSS, Inc., Chicago, Il, USA) as well as in GraphPad Prism (version 8.1.2 GraphPad Software Inc., San Diego, Ca). Statistical significance was set at p < 0.05 [p < 0.05(*), p < 0.01(**), p < 0.001(***) and p < 0.0001 (****)].
Results
FAT4expression is reduced in ovarian cancer cell lines
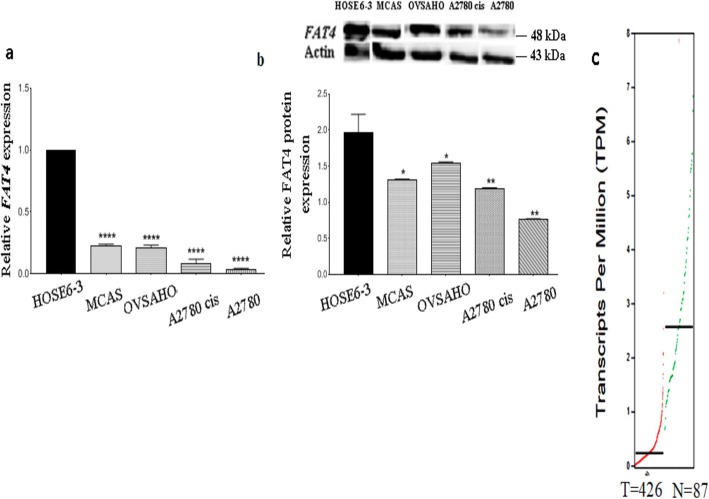
We evaluated the expression of FAT4 in ovarian cancer cell lines (MCAS, OVSAHO, A2780, A2780 cis), and normal (HOSE 6–3) cell line, using qRT-PCR and Western Blotting. As shown in Fig. 1a, FAT4 was significantly down-regulated at the mRNA level in A2780 and A2780 cis cell lines (p = 0.0001 and p = 0.0002 respectively). MCAS and OVSAHO displayed lower FAT4 expression (p = 0.0001) as compared to the normal HOSE 6–3 cell line. Similar results were obtained by western blot analysis of FAT4 protein expression in ovarian cancer cells (Fig. 1b and supplementary figure S1). FAT4 expression was the highest in HOSE 6–3 and moderately expressed in OVSAHO (p = 0.0459), MCAS (p = 0.0107), A2780 cis (p = 0.006) and A2780 (p = 0.0012) cell lines.
Fig. 1.
a. Relative expression of FAT4 at mRNA levels in ovarian cancer cell lines. MCAS and OVSAHO show high FAT4 expression as compared to A2780 and A2780 cis (p = 0.0001). b. Relative expression of FAT4 at protein levels in MCAS and OVSAHO cells. FAT4 was highly expressed in OVSAHO (p = 0.0459), and MCAS (p = 0.0107), cells while, it was least expressed in A2780 cis (p = 0.006) and A2780 (p = 0.0012) cells. Full-length blots are presented in Supplementary figure S1. c. Differential expression of FAT4 in tumor and healthy ovarian tissue sample from GEPIA database. FAT4 was significantly downregulated in 426 tumor samples as compared to 87 healthy samples. Red line represents tumor samples and green line represents healthy ovarian samples. Data represent mean and standard deviation from at least three independent experiments performed in triplicates
Analysis of FAT4 expression in 426 ovarian tumors and 87 non-malignant samples from TCGA and GEPIA public databases revealed that FAT4 expression was lower in ovarian tumors than in healthy tissues (Fig. 1c), which is consistent with our findings.
FAT4downregulation promotes proliferation and colony formation
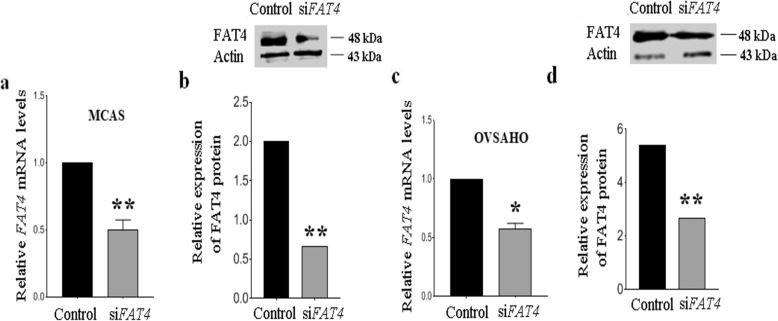
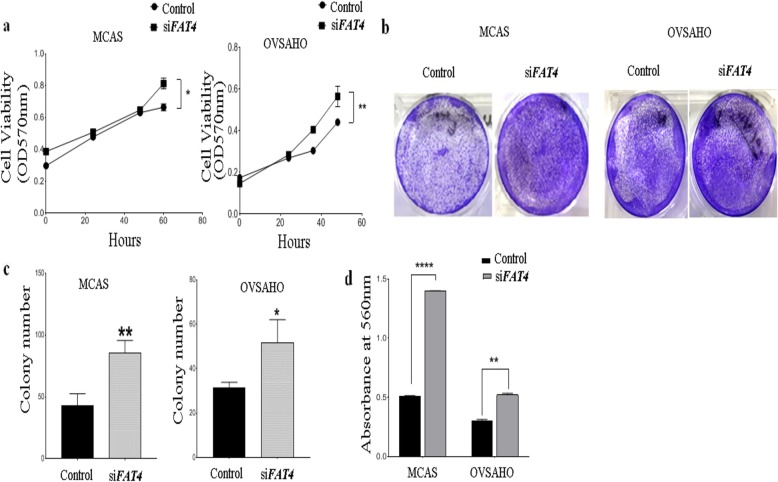
To evaluate the role of FAT4 on ovarian cancer cell proliferation, we knocked-down FAT4 in MCAS and OVSAHO ovarian cancer cell lines, using siRNA (Figs. 2a-d and S1). The effect of silencing FAT4 on cell growth was assessed by Alamar blue assay in MCAS and OVSAHO cells, which showed that transient inhibition of FAT4 significantly enhanced cell proliferation (p = 0.0286 and p = 0.0054 respectively) (Fig. 3a) and colony formation ability in MCAS and OVSAHO cells compared to control (Fig. 3b and c).
Fig. 2.
a. Relative expression of FAT4 in MCAS cells following its knockdown at a. mRNA level, and b. protein levels. FAT4 was knocked down significantly (p = 0.0016) in MCAS. Full-length blots are presented in Supplementary figure S1. c. Relative expression of FAT4 in OVSAHO cells following its knockdown at a. mRNA level, and d. protein levels. Full-length blots are presented in Supplementary figure S1. FAT4 was knocked down significantly in OVSAHO (p = 0.0055). Data represent mean and standard deviation from at least three independent experiments performed in triplicates
Fig. 3.
Functional role of FAT4 on cell proliferation, colony formation, and invasive ability. a. Alamar blue assay displaying enhanced cell proliferation in FAT4 knocked MCAS and OVSAHO cells (p = 0.0001 and p = 0.0054 respectively) as compared to control. b. Colony formation assay in control and FAT4 siRNA treated MCAS, and OVSAHO cells. The assay shows a higher number of colonies in FAT4 siRNA treated cells. The cells were stained with crystal violet. c. Soft agar colony formation assay in control and FAT4 siRNA treated MCAS and OVSAHO cells. Anchorage-independent growth was significantly higher in cells treated with siFAT4 MCAS cells (p = 0.0055) and OVSAHO cells (p = 0.0302). d. Silencing FAT4 by siRNA promoted cell invasion in vitro in MCAS and OVSAHO cells (p = 0.0001 in MCAS and p = 0.003 in OVSAHO). MCAS and OVSAHO cells treated with scrambled siRNA were used as control. Data represent the mean and the standard deviation from at least three independent experiments performed in triplicates
FAT4downregulation promotes invasion in ovarian cancer cells
We performed cell invasion assay to investigate whether FAT4 regulates invasiveness in ovarian cancer cell lines. Results showed that the invasive ability of MCAS and OVSAHO ovarian cancer cell lines increased significantly following the FAT4 knockdown (p = 0.0001 and p = 0.003 respectively), suggesting its tumor-suppressive ability to prevent invasion in vitro as shown in Fig. 3d.
FAT4role in the epithelial-mesenchymal transition (EMT)
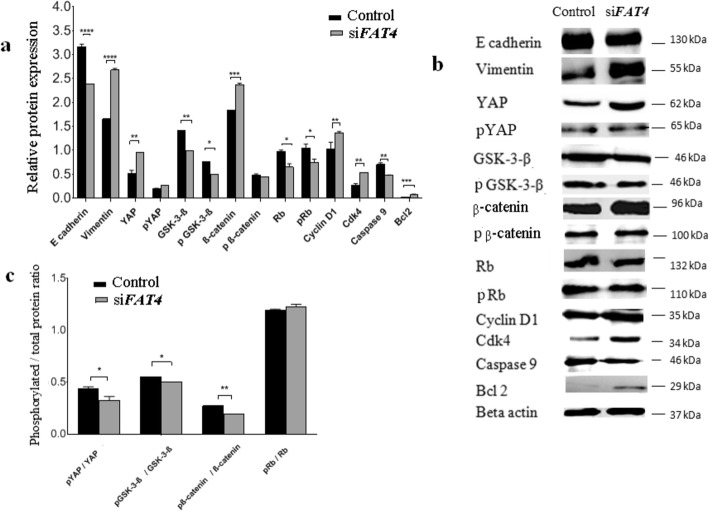
We investigated the role of FAT4 on EMT, a process which plays a crucial role in cell transformation, invasion, and metastasis by Western Blotting. Relative protein expression variation and the protein bands in control and FAT4-siRNA treated MCAS cells are shown in Figs. 4a-b and S1 respectively, while the ratio of phosphorylated to total proteins are displayed in Fig. 4c. In cells treated with FAT4-siRNA, we observed a reduced expression of the epithelial marker, E-cadherin. In contrast, the expression of the mesenchymal marker, vimentin was enhanced in FAT4-siRNA treated cells as compared to non-treated cells (p < 0.0001).
Fig. 4.
Regulatory effects of FAT4 on the expression of proteins involved in EMT, Hippo, Wnt-β-catenin, apoptotic, and retinoblastoma pathways by Western blotting. a. Relative expression variation of proteins. The expression of Vimentin (p = 0.0001), YAP (p = 0.0018), β-catenin (p = 0.001), Bcl2 (p = 0.0001), cyclin D1 (p = 0.0017) and cdk4 (p = 0.0025) was higher in FAT4 siRNA treated cells as compared to control. β-actin was used as an internal control. b. Western blot demonstrating bands for protein expression in control and FAT4 knocked cells. Full-length blots are presented in Supplementary figure S1. c. The ratio of phosphorylated to total YAP, GSK-3β, β-catenin, and retinoblastoma proteins following FAT4 repression. The pYAP/ YAP ratio was lower in FAT4 siRNA treated cells as compared to the control (p = 0.0286). Similarly, pGSK-3-β/GSK-3-β ratio, and pβ-catenin/β-catenin ratio was lower in FAT4 siRNA treated cells (p = 0.018, and p = 0.001 respectively) as compared to control. There was no significant difference in pRb/Rb ratio between FAT4 siRNA treated cells and control. MCAS cells treated with scrambled siRNA was used as control. Data represent mean and standard deviation from at least three independent experiments performed in triplicates
FAT4regulatory effects on hippo, apoptotic, cell cycle, and Wnt signaling pathways
We hypothesized that FAT4 exerts its effect on the tumor characteristics of ovarian cancer cells through Hippo, apoptotic, retinoblastoma, and Wnt pathways. Based on this, we aimed to detect the expression of crucial proteins involved in each of these pathways through western blotting (Fig. 4), performed on MCAS cells treated with FAT4-siRNA. The expression of YAP (p = 0.0018), β-catenin (p = 0.001), cyclin D1 (p = 0.0025), cdk4 (p = 0.0017) and Bcl2 (p = 0.0001), were significantly higher in FAT4-siRNA treated cells as compared to the control. In contrast, the expression of GSK-3-β (p = 0.0013), Rb (p = 0.0028), pRb (p = 0.0491), and caspase 9 (p = 0.001) was lower when compared to the control.
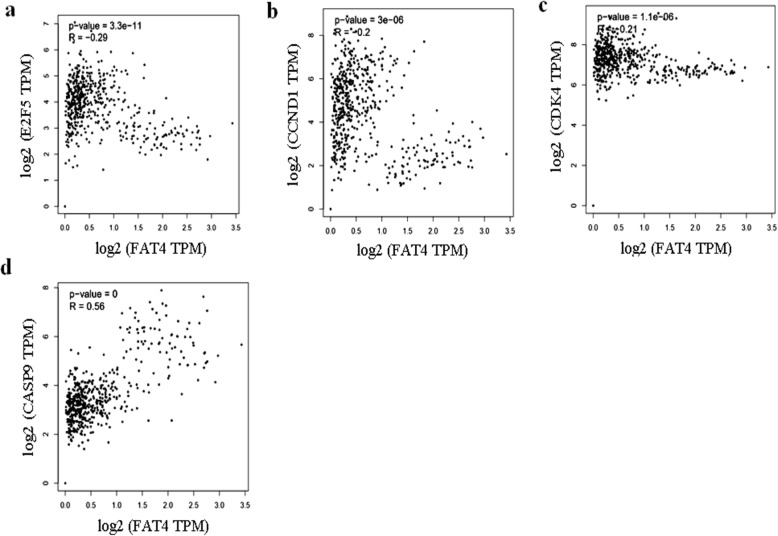
Consistent with the western blotting results, Pearson’s correlation showed a negative correlation between FAT4 and E2F5 (p = 3.3 e− 11, r = − 0.29); (Fig. 5a), Cyclin D1 (p = 3e− 6, r = − 0.2) (Fig. 5b); Cdk4 (p = 1.1 e− 6, r = − 0.21) (Fig. 5c); while a positive correlation was observed between caspase 9 and FAT4 (p < 0.05, r = 0.56) (Fig. 5d).
Fig. 5.
Pearson’s correlation between different gene expressions using samples from GEPIA database. a. FAT4 and E2F5 is negatively correlated (p = 3.3e− 11, r = − 0.29). b. Cyclin D1 expression was negatively correlated with FAT4 (p = 3e− 6, r = − 0.2). c. Pearson’s correlation demonstrating negative correlation between cdk4 and FAT4 (p = 1.1e− 6, r = − 0.21). d. FAT4 was positively correlated with caspase 9 (p < 0.05, r = 0.56)
FAT4 knockdown promotes E2F5 upregulation
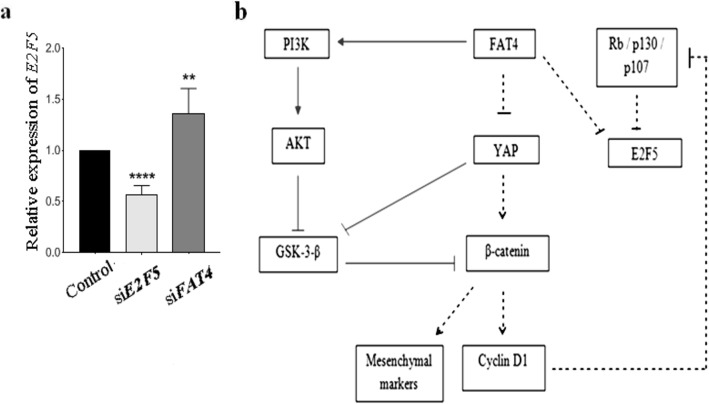
FAT4-knockdown cells express a significantly higher amount of E2F5 suggesting a link between the two genes (Fig. 6a), hence a pathway speculating the mechanism of EOC pathogenesis via FAT4, and E2F5 has been proposed (Fig. 6b).
Fig. 6.
a. FAT4 knockdown upregulated the expression of E2F5 suggesting a link between the two genes. b. A suggested mechanism of EMT and cell cycle regulation by FAT4 based on Western blot obtained data. Inhibition of FAT4 regulates the expression of EMT markers and promotes cell cycle progression via the suggested pathway. The solid lines indicate the results obtained from the previous study [13], while the dotted lines indicate results of our western blotting experiments
Discussion
In this study, we examined the role and associated signaling mechanism of FAT4 gene in EOC tumorigenesis. FAT4 expression was weaker in ovarian cancer cell lines, with MCAS and OVSAHO displaying higher expression as compared to A2780 and A2780 cis ovarian cancer cell lines (Fig. 1a and b). Although OVSAHO represents high grade serous ovarian cancer, the expression of FAT4 was similar to that of MCAS, a low-grade ovarian cancer cell line [25]. The non-correlation between the MCAS cells genotype/phenotype could be an explanation for this contradiction. Indeed, it has recently been shown that, although OVSAHO cells represent high grade serous ovarian cancer, they displayed less potential in invasion, migration, and soft agar colony formation assays [26]. Bioinformatics analysis of Human samples revealed similar results, showing downregulation of FAT4 expression in Human ovarian cancer samples as compared to normal tissues (Fig. 1c).
In a previous study, immunohistochemical analysis revealed lower FAT4 expression in gastric [27], colorectal [13], and endometrial cancer [18], when compared to adjacent non-cancerous tissues. Moreover, exome sequencing of FAT4 in gastric cancer revealed hyper-methylation of the FAT4 promoter, suggesting that epigenetics is a likely mechanism behind FAT4 downregulation [28]. Interestingly, FAT4 downregulation was inversely associated with the tumor grade, and higher repression corresponds to advanced grades [29]. Downregulation of FAT4 expression was significantly associated with lymph node invasion and poor survival in gastric [12] and endometrial cancer [18]. However, the role of FAT4 in the pathogenesis of EOC remains still unclear.
To gain further insight into the functional role of FAT4 on cell viability, colony formation ability and invasiveness, we transfected MCAS and OVSAHO with FAT4-siRNA (Fig. 2a, b, c and d) and assessed the proliferation, colony formation, and invasiveness of the cells. We observed that transient reduction of FAT4 significantly promoted cell growth (Fig. 3a), increased the colony-forming (Fig. 3b and c), and invasive ability (Fig. 3d). These results are in agreement with a previous study in gastric and breast cancer cell lines [8, 12], showing that silencing FAT4 expression increased invasion and migration of MDA-MB-436 and BT-549 breast cancer cell lines [8], and promoted cell viability, colony-forming ability of BGC-823 and HGC-27 gastric cancer cells [12]. To elucidate the cell proliferation mechanism(s) in FAT4 siRNA treated gastric cancer cells Ma et al. performed a flow cytometry to reveal an increase in the number of cells in S and G2/M phases, as well as an upregulation of proliferation-associated markers such as cyclin D1, c myc [21] in gastric cancer, and cdk 1 and cdk 2 [18] in endometrial cancer.
Tumor progression involves changes in cell-cell and cell-matrix interactions, allowing the tumor cells to leave the primary tumor and metastasize to distant sites. This complex process seems to be attributed to the loss of epithelial and gain of mesenchymal cadherins, which are considered as a hallmark of EMT [30]. Our results revealed that silencing FAT4 in EOC cells upregulated the mesenchymal marker (vimentin), and downregulated the epithelial marker (E-cadherin) as compared to control (Figs. 3a-c). These results are in agreement with the reported data obtained in breast and gastric cancer cell lines [8, 12]. In gastric cancer, the expression of MMP-14 and MMP-16 was found to be upregulated, thereby promoting EMT through Wnt/β-catenin pathway [12]. Similarly, in breast cancer, FAT4 silencing resulted in the upregulation of N-cadherin, MMP-7, and Cyr61 expression and the downregulation of E cadherin, indicating the role of FAT4 in the EMT process [8]. Furthermore, FAT4 silencing promoted metastasis in vivo, in gastric cancer xenograft mouse model [12].
The Hippo pathway, regulates cell growth, proliferation, and apoptosis [31]. The downstream effector of this pathway (YAP), is a crucial ovarian cancer oncogene promoting increased cell proliferation, invasion, and anchorage-independent growth [24]. Our Western blotting results revealed higher level of active, unphosphorylated YAP in FAT4 siRNA treated cells, which promotes proliferation by active transcription of cell growth and anti-apoptotic genes (Fig. 4). Further studies on the nuclear localization of YAP following FAT4 repression are required. The levels of pYAP did not change in our study similar to the results demonstrated by Ito et al., where consistent phosphorylated YAP levels in FAT4 siRNA treated cells and control MCF-10A breast cancer cells was observed [22], in contrast, pYAP expression was lower in FAT4 siRNA treated endometrial cancer [18] suggesting the differences in mechanisms ruling these different cancers. In gastric [21] and endometrial cancer [18] cells higher level of nuclear YAP was observed following FAT4 repression.
Wnt/β-catenin pathway plays a crucial role in regulating cell growth and tissue homeostasis, and its activation is a hallmark of several cancers [32]. In this study, the tumor suppressor role of FAT4 was shown to be mediated by the regulation of β-catenin activity (Fig. 4). Similarly, a previous finding revealed that FAT4 knocked-down gastric cancer cells [12] displayed higher levels of nuclear β-catenin accumulation as compared to the cytosolic fraction, thereby increasing cell growth over 6 days, while β-catenin repression reduced the growth [12].
GSK-3-β, a component of the Wnt/β-catenin pathway, is known to phosphorylate β-catenin in the cytoplasm, which becomes then targeted for degradation when the pathway is inactive [33]. In a previous study, inhibition of GSK-3-β activity was observed upon phosphorylation at serine-21 in GSK-3-α and serine-9 in GSK-3- β [34–36] while phosphorylation at tyrosine 216 is required for maximal activity [37]. In this study, knocking down FAT4 downregulated the level of GSK-3-β phosphorylated at serine-9 residue, suggesting further investigation on the levels of GSK-3-β phosphorylated at 216 tyrosine residue, considered as the most active form, are necessary to be able to draw relevant conclusions about its activity following FAT4 repression. In line with our findings, in colorectal cancer, the downregulation of GSK-3-β upon FAT4 repression, while upregulation of its phosphorylated form, was observed, which was attributed to the PI3K-AKT pathway in colorectal cancer [13]. Furthermore, YAP overexpression was shown to increase pGSK-3-β and β-catenin, thereby upregulating cyclin D1 expression in glioma [33]. Hence, Wang et al. suggested the ability of YAP to modulate and increase transcriptional activity of β-catenin, demonstrating a link between the Hippo and Wnt pathway [33].
We found that knockdown of FAT4 resulted in an upregulation of Bcl2 levels. In a previous study, Bcl2 expression was upregulated on YAP overexpression, which could be a possible mechanism of Bcl2 upregulation in our result [38].
Retinoblastoma pathway is a tumor suppressor pathway that plays a key role in regulating the cell cycle, differentiation, and apoptosis [39]. Rb phosphorylation (pRb) by cyclin-dependent kinases (cdks) during G1 phase cause Rb to dissociate from E2F transcription factors resulting in transcription of genes responsible for promoting entry into S phase [40]. In this study, FAT4 repression increased cyclin D1, and cdk4 expression, promoting progression into the G1/S phase of the cell cycle (Fig. 4a-c), consistent with results obtained in a previous study on gastric cancer cells [21].
In agreement with results obtained on cell lines, bioinformatics analysis also revealed an inverse correlation between FAT4 and E2F5 (Fig. 5a). The correlation between FAT4 and caspase-9, cyclin D1, cdk4 was consistent with that observed in cell lines (Fig. 5b, c, and d). However, there was no significant correlation observed between Rb, and FAT4, suggesting an alternative mechanism of regulating cell cycle by FAT4 through indirect regulation of p107 and p130, members of the retinoblastoma protein family.
Interestingly, FAT4-knockdown cells express a significantly higher amount of E2F5 (Fig. 6a), suggesting the tight relationship between these two genes. These data suggest a cross-talk between the two genes (FAT4, E2F5) and confirm the results obtained by ChIP analysis where FAT4 was found to be a real target of the transcription factor E2F5. E2F5 seems to be suppressed by gene FAT4, and its upregulation occurs in cells treated with FAT4-siRNA suggesting a link between the two genes.
Based on our western blot results, a plausible role of FAT4 on the mechanism of EMT, and the cell cycle regulation is proposed (Fig. 6b). Silencing FAT4 upregulated the expression of unphosphorylated active YAP, which in turn, increases the level of non-phosphorylated active β-catenin. YAP is also known to directly enhance the level of active β-catenin by inhibiting its degradation. Nuclear translocation of β-catenin leads to transcription of cyclin D1, c-myc, and mesenchymal markers such as twist, vimentin, hence promoting cell growth and EMT. Alternatively, FAT4 can inhibit GSK-3-β and activate β-catenin in part through PI3K-AKT pathway [13].
Furthermore, cyclin D1, a transcriptional target of β-catenin, is known to phosphorylate the retinoblastoma protein family, Rb, p130 and p107 resulting in its inability to associate with E2F5, and increase the level of E2F5 protein. p130 and p107, members of Rb family, are known to block the transcription of E2F regulated genes by interacting exclusively with E2F5 and E2F4 respectively, which suggests an alternative mechanism of E2F activity regulation by retinoblastoma [40, 41]. p107 and p130 are inactivated through phosphorylation by cyclin D1 [42] and cyclin D dependent kinases [43] such as cdk4 [44] at multiple sites, resulting in the transcription of the genes involved in the cell cycle to promote proliferation.
Conclusions
To the best of our knowledge, this is the first time that FAT4 tumor suppressor gene is reported in EOC pathogenesis. Our results demonstrated the involvement of FAT4 in EMT and cell cycle through the regulation of E2F5, YAP, and β-catenin activity. This study provided a better understanding of the anticancer molecular mechanisms of FAT4, and we suggest considering FAT4 gene as a potential target for anticancer therapy by targeting any component involved in its downregulation. These might include epigenetic therapies such as reverting DNA methylation, known to inactivate several tumor suppressor genes, including FAT4.
Supplementary information
Additional file 1: Supplementary Figure S1. The original Western blot of fig 1b/2b/2d/4b are displayed.
Acknowledgments
Not Applicable.
Consent to publish
No consent was required in this publication.
Abbreviations
- FAT4
FAT tumor suppressor homolog 4
- EMT
Epithelial-to-mesenchymal-transition
- EOC
Epithelial Ovarian cancer
- ChIP
Chromatin immunoprecipitation
- TCGA
The Cancer Genome Atlas
- qRT-PCR
Quantitative real time polymerase chain reaction.
Authors’ contributions
MM conceived the idea of initiating the work on FAT4 in ovarian cancer. YT designed the project and the experimental setup. SH performed the experiments, interpreted the data, and wrote the drafts of the manuscript under the supervision of YT. AO, IB, MK, and RL contributed to constructive critiques and discussion related to the clinical section of the project. They also contributed to the manuscript reviewing. All the authors approved the final version of this manuscript.
Funding
This study was financially supported by the grant from His Majesty Trust Fund (SR/MED/BIOC/14/01). The authors would like to thank His Majesty’s Trust Fund (HMTF) grants (SR/MED/BIOC/14/01) for supporting the initiation and the progression of this study. The role of the funding body was to provide the financial support for successful proposals from SQU.
Availability of data and materials
All the data generated and/or analyzed during this study are included in this published article (and its supplementary information files), and other datasets will be available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The cell lines used in this study do not require ethics approval for their use.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12885-020-06900-7.
References
- 1.Cho KR, Shih Ie M. Ovarian cancer. Annu Rev Pathol. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sankaranarayanan R, Ferlay J. Worldwide burden of gynaecological cancer: the size of the problem. Best Pract Res Clin Obstet Gynaecology. 2006;20(2):207–225. doi: 10.1016/j.bpobgyn.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Q, Zhang F, He Z, Zuo MZ. E2F2/5/8 serve as potential prognostic biomarkers and targets for human ovarian Cancer. Front Oncol. 2019;9:161. doi: 10.3389/fonc.2019.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J Clin. 2011;61(3):183–203. doi: 10.3322/caac.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kothandaraman Narasimhan BVB, Brendan Pang NK, Huak Chan Y, Keow Peh B, Khalil R, Manuel S-T, Mahesh C. E2F5 status significantly improves malignancy diagnosis of epithelial ovarian cancer. BMC Cancer. 2010;10:1. doi: 10.1186/1471-2407-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadeqzadeh E, de Bock CE, Thorne RF. Sleeping giants: emerging roles for the fat cadherins in health and disease. Med Res Rev. 2014;34(1):190–221. doi: 10.1002/med.21286. [DOI] [PubMed] [Google Scholar]
- 8.Hou L, Chen M, Zhao X, Li J, Deng S, Hu J, Yang H, Jiang J. FAT4 functions as a tumor suppressor in triple-negative breast cancer. Tumour Biol. 2016. [DOI] [PubMed]
- 9.Ishiuchi T, Misaki K, Yonemura S, Takeichi M, Tanoue T. Mammalian fat and Dachsous cadherins regulate apical membrane organization in the embryonic cerebral cortex. J Cell Biol. 2009;185(6):959–967. doi: 10.1083/jcb.200811030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katoh M. Function and cancer genomics of FAT family genes (review) Int J Oncol. 2012;41(6):1913–1918. doi: 10.3892/ijo.2012.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvey K, Tapon N. The Salvador-warts-hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7(3):182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 12.Cai J, Feng D, Hu L, Chen H, Yang G, Cai Q, Gao C, Wei D. FAT4 functions as a tumour suppressor in gastric cancer by modulating Wnt/beta-catenin signalling. Br J Cancer. 2015;113(12):1720–1729. doi: 10.1038/bjc.2015.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei R, Xiao Y, Song Y, Yuan H, Luo J, Xu W. FAT4 regulates the EMT and autophagy in colorectal cancer cells in part via the PI3K-AKT signaling axis. J Exp Clin Cancer Res. 2019;38(1):112. doi: 10.1186/s13046-019-1043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43(9):828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crobach S, Ruano D, van Eijk R, Fleuren GJ, Minderhout I, Snowdowne R, Tops C, van Wezel T, Morreau H. Target-enriched next-generation sequencing reveals differences between primary and secondary ovarian tumors in formalin-fixed, paraffin-embedded tissue. J Mol Diagnostics. 2015;17(2):193–200. doi: 10.1016/j.jmoldx.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Che Xiaoxia JF, Nan J, Ya Z, Yahui J, Weiwei F. FAT4-USP51 complex regulates the proliferation and invasion of endometrial cancer via hippo pathway. Am J Transl Res. 2019;11(5):2784–2800. [PMC free article] [PubMed] [Google Scholar]
- 19.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou D, Kannappan V, Chen X, Li J, Leng X, Zhang J, Xuan S. RBP2 induces stem-like cancer cells by promoting EMT and is a prognostic marker for renal cell carcinoma. Exp Mol Med. 2016;48:e238. doi: 10.1038/emm.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma L, Cui J, Xi H, Bian S, Wei B, Chen L. Fat4 suppression induces yap translocation accounting for the promoted proliferation and migration of gastric cancer cells. Cancer Biology Ther. 2016;17(1):36–47. doi: 10.1080/15384047.2015.1108488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito Takao TH, Kousuke F, Shota O, Akira K. Inhibitory Mechanism of FAT4 Gene Expression in Response to Actin Dynamics during Src-Induced Carcinogenesis. 2013;10(2):e0118336. [DOI] [PMC free article] [PubMed]
- 23.Yu FX, Guan KL. The hippo pathway: regulators and regulations. Genes Dev. 2013;27(4):355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia Y, Chang T, Wang Y, Liu Y, Li W, Li M, Fan HY. YAP promotes ovarian cancer cell tumorigenesis and is indicative of a poor prognosis for ovarian cancer patients. PLoS One. 2014;9(3):e91770. doi: 10.1371/journal.pone.0091770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haley James TS, Nicholas P, Xiong S, Susan MP, Adam RK, Mitra S, Kenneth PN, Anirban KM. Functional characterization of a panel of high-grade serous ovarian cancer cell lines as representative experimental models of the disease. Oncotarget. 2016;7:22. doi: 10.18632/oncotarget.9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Xiaoting LZ, Yingjie X, Luo J, Xu J, He X, Low TH. FAT4 expression is associated with a poor prognosis in gastric cancer patients. Oncotarget. 2018;9:4. doi: 10.18632/oncotarget.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida S, Yamashita S, Niwa T, Mori A, Ito S, Ichinose M, Ushijima T. Epigenetic inactivation of FAT4 contributes to gastric field cancerization. Gastric Cancer. 2017;20(1):136–145. doi: 10.1007/s10120-016-0593-5. [DOI] [PubMed] [Google Scholar]
- 29.Pilehchian Langroudi M, Nikbakhsh N, Samadani AA, Fattahi S, Taheri H, Shafaei S, Amirbozorgi G, Pilehchian Langroudi R, Akhavan-Niaki H. FAT4 hypermethylation and grade dependent downregulation in gastric adenocarcinoma. J Cell Commun Signaling. 2017;11(1):69–75. doi: 10.1007/s12079-016-0355-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4(2):118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 31.Bae SJ, Luo X. Activation mechanisms of the Hippo kinase signaling cascade. Biosci Rep. 2018;38:4. doi: 10.1042/BSR20171469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theka I, Sottile F, Cammisa M, Bonnin S, Sanchez-Delgado M, Di Vicino U, Neguembor MV, Arumugam K, Aulicino F, Monk D, et al. Wnt/beta-catenin signaling pathway safeguards epigenetic stability and homeostasis of mouse embryonic stem cells. Sci Rep. 2019;9(1):948. doi: 10.1038/s41598-018-37442-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Pan P, Wang Z, Zhang Y, Xie P, Geng D, Jiang Y, Yu R, Zhou X. beta-catenin-mediated YAP signaling promotes human glioma growth. J Exp Clin Cancer Res. 2017;36(1):136. doi: 10.1186/s13046-017-0606-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calum Sutherland IAL, Cohen P. Inactivation of glycogen synthase kinase-3fl by phosphorylation: new kinase connections in insulin and growth-factor signalling. Biochem J. 1993;296:15–19. doi: 10.1042/bj2960015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DRA DAEC, Vandenheede JR, McDowell HE, Hundal HS, Cohen P. The inhibition of glycogen synthase kinase-3 by insulin or insulin-like growth factor 1 in the rat skeletal muscle cell line L6 is blocked by wortmannin, but not by rapamycin: evidence that wortmannin blocks activation of the mitogen-activated protein kinase pathway in L6 cells between Ras and Raf. Biochemical J. 1994;303(1):21–26. doi: 10.1042/bj3030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vuk Stambolic JRW. Mitogen inactivation of glycogen synthase kinase-3p8 in intact cells via serine 9 phosphorylation. Biochem J. 1994;303:4. doi: 10.1042/bj3030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beurel E, Grieco SF, Jope RS. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol Ther. 2015;148:114–131. doi: 10.1016/j.pharmthera.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Y, Fu J, Zhou M, Xiao L, Feng X, Chen H, Huang W. Activated hippo/yes-associated protein pathway promotes cell proliferation and anti-apoptosis in endometrial stromal cells of endometriosis. J Clin Endocrinol Metab. 2016;101(4):1552–1561. doi: 10.1210/jc.2016-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.W. Du JS: The Rb pathway and Cancer therapeutics. Curr Drug Targets 2009, 10(7):581–589. [DOI] [PMC free article] [PubMed]
- 40.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12(15):2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 41.Marta M, Lipinski TJ. The retinoblastoma gene family in differentiation and development. Oncogene. 1999;18:7873–7882. doi: 10.1038/sj.onc.1203244. [DOI] [PubMed] [Google Scholar]
- 42.De Falco M, Fedele V, Cobellis L, Mastrogiacomo A, Giraldi D, Leone S, De Luca L, Laforgia V, De Luca A. Pattern of expression of cyclin D1/CDK4 complex in human placenta during gestation. Cell Tissue Res. 2004;317(2):187–194. doi: 10.1007/s00441-004-0880-z. [DOI] [PubMed] [Google Scholar]
- 43.Jacqueline L, Bruce RKH, Classon M, Koh J, Dyson N. Requirements for cell cycle arrest by p16INK4a. Mol Cell. 2000;6(3):737–742. doi: 10.1016/s1097-2765(00)00072-1. [DOI] [PubMed] [Google Scholar]
- 44.Donato Tedesco JL, Reed SI. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein–ubiquitin ligase SCFSkp2. Genes Dev. 2002;16(22):2946–2957. doi: 10.1101/gad.1011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure S1. The original Western blot of fig 1b/2b/2d/4b are displayed.
Data Availability Statement
All the data generated and/or analyzed during this study are included in this published article (and its supplementary information files), and other datasets will be available from the corresponding author on reasonable request.