Abstract
Recent advancements in computational power, machine learning, and artificial intelligence technology have enabled automated evaluation of medical images to generate quantitative diagnostic and prognostic biomarkers. Such objective biomarkers are readily available and have the potential to improve personalized treatment, precision medicine, and patient selection for clinical trials. In this article, we explore the merits of the most recent addition to the “-omics” concept for the broader field of head and neck cancer – “Radiomics”. This review discusses radiomics studies focused on (molecular) characterization, classification, prognostication and treatment guidance for head and neck squamous cell carcinomas (HNSCC). We review the underlying hypothesis, general concept and typical workflow of radiomic analysis, and elaborate on current and future challenges to be addressed before routine clinical application.
Keywords: Radiomics, Machine learning, Neck, Oropharynx, Squamous cell carcinoma
Background
Structural and functional imaging provide data that are integral for diagnosis, treatment response evaluation, and surveillance in patients with head and neck squamous cell carcinoma (HNSCC). The large amount of volumetric bioimaging information amassed in institutional archives constitutes an extensive database amenable to high-throughput, quantitative image analysis. Radiomics refers to automated extraction of high-dimensional sets of quantitative descriptors (“radiomic features”) from medical images (e.g. CT, MRI, PET etc.) for development of novel diagnostic and prognostic biomarkers. Machine learning (ML) algorithms and artificial intelligence (AI) are best suited for analysis of radiomics high-dimensional data. Radiomics provides fast, low-cost and non-invasive, yet comprehensive tissue and organ characterization, as features are extracted directly from (pre-processed) standard-of-care medical images. The generated features offer information complementary to traditional clinical predictors in numerous applications, which may help advance cancer care towards personalized precision medicine. Numerous recent radiomics studies have focused on classification, characterization, prognostication, and treatment guidance of HNSCC.
Paired with key clinical predictors, radiomic analysis can capture a large variety of HNSCC properties [1], enabling the predictive models to more accurately reflect the spatial, metabolic, and morphological heterogeneity of primary tumor lesions and metastatic lymph nodes. This review aims to provide an overview of recently published HNSCC radiomics studies focusing on (molecular) characterization, classification, prognostication and treatment guidance. The general principle of radio(geno)mic analysis and the typical radiomics workflow are introduced. We also discuss the applications of advanced machine learning for radiomics-based modelling. Finally, we summarize future challenges, barriers and limitations of individual radiomic applications, as well as the field of head and neck radiomics in general.
Radiomics
Over the last decade, advancements in high-throughput computing and machine learning algorithms have led to emergence of the “-omics” concept – referring to the collective characterization and quantification of pools of biologic information, such as genomics, proteomics or metabolomics. Radiomics refers to automated extraction of mathematically defined, numerical descriptors (“radiomics features”) from 2-dimensional – or more commonly – 3-dimensional medical images and subsequent application of data mining and analysis techniques. Over the past few years, there has been an increasing interest in application of radiomics in patients with HNSCC for prediction of molecular biomarkers, prognostication, and treatment response.
Radiomics features commonly describe shape, intensity (histogram) and texture characteristics. These features can be extracted from different imaging modalities, such as CT, MRI, or metabolic imaging like 18- fludeoxyglucose positron emission tomography (FDG-PET). The notion that certain characteristics of medical images – which are not reliably assessed by human visual inspection – can provide medically meaningful information for diagnostic and prognostic purposes as well as treatment guidance is the underlying hypothesis in the emerging field of radiomics [2]. Prior studies showed that radiomics features represent biological characteristics of the tissue such as cellularity, heterogeneity, and necrosis [3]; and frequently exhibit correlation with diagnostic and outcome variables [2]. Furthermore, certain features can be reflective of molecular and genetic characteristics of malignant tissue. The subfield of Radiogenomics focuses on the identification and scientific exploitation of relationships between quantitative bioimaging features and genomic characteristics of the tumor [4]. It is worth noting that radiomics analysis captures information from the whole volume of interest (VOI), and therefore may act as a quantitative descriptor of tumor spatial heterogeneity, whereas the diagnostic validity of localized tools like tissue sampling may be degraded in heterogeneous tumors [3, 5].
Radiomics workflow
Despite not being part of the radiomics workflow in a narrower sense, image acquisition is often considered the first step in radiomics analysis. Radiomics feature robustness and reproducibility against variation in scan acquisition protocols have been extensively investigated across imaging modalities and in various settings [6], including test-retest assessments [7–9], studies designed to evaluate the impact of scanner types/manufacturers using phantoms [10, 11], reconstruction algorithms /slice thickness [12, 13], and motion artifacts [14]. Traverso et al. [6] conducted a systematic review of 41 studies investigating the reproducibility and stability of radiomics features in phantoms and different cancers – including lung, HNSCC, and esophageal cancer – and found that only three studies investigated radiomics reproducibility in HNSCC. Bagher-Ebadian et al. [15] investigated the impact of smoothing and noise on CT and cone beam CT textural features and reported general feature robustness against low-power Gaussian noise and low pass filtering, whereas a high-pass filter significantly impacted textural features. Bogowicz et al. [16] focused on feature stability regarding CT perfusion calculation factors. Finally, Lu et al. [17] studied the effect of seven different segmentation methods and 5 forms of fixed-bin SUV-discretization on PET radiomic features, reporting 50 and 23% of 88 tested features were robust to FDG-PET segmentation and discretization, respectively (with robustness ascertained by an intraclass correlation coefficient ≥ 0.8). While there is as yet no consensus regarding stable radiomic feature sets, it is crucial to assess stability of radiomic features in each study – especially for generalization of findings and future comparison.
The next step in the radiomics workflow involves the delineation (“segmentation”) of the target area/volume in medical images, resulting in image sub-sections referred to as regions of interest (ROI) and volumes of interest (VOI) in 2- and 3-dimensional images, respectively. Manual and (semi-) automated segmentation have both been applied in recent radiomics studies, each with its inherent advantages and drawbacks. Manual segmentation is affected by observer variability; several studies investigated the inter- and intra-rater reproducibility of CT and PET radiomic features extracted from repeated manual segmentations of lung cancer lesions [8, 18, 19]. Lu et al. [17] assessed feature stability across manual and various automated segmentation techniques applied to oropharyngeal cancer lesions on PET scans and showed that 50% of features extracted from 18-FDG-PET achieved an intraclass correlation coefficient ≥ 0.8, which was considered sufficiently reproducible. Across all studies, individual radiomic features were found to exhibit varying degrees of robustness against observer variability, suggesting stability measures may be appropriated for feature dimensionality reduction. A multitude of (semi) automated segmentation methods have been proposed or adapted for Radiomics purposes [20]. An in-depth discussion of the various algorithmic approaches is beyond the scope of this review; however, reproducibility and observer variability are certainly a minor concern with (semi-) automated approaches. On the other hand, fully automated segmentation can only be “as good as” the expert-generated ground truth data used for development and may be impaired by artifacts, presence of multiple pathologic findings and other abnormalities not considered in the development process. The resulting imprecisions in segmentations will undoubtedly affect the quality and usefulness of extracted radiomic features, warranting thorough human validation.
Image pre-processing is usually applied as the next step following segmentation: Resampling voxels to uniform sizes is often necessary due to the heterogeneity of the available imaging data, originating from different scanners and reconstruction protocols. Additionally, resampling to isotropic voxels (i.e. voxel with identical edge lengths) should be considered as it guarantees rotational invariance of texture features [21]. While CT imaging uses a “real-valued” grey scale (the Hounsfield unit scale is an absolute representation of physical density), other imaging modalities require gray scale homogenization to facilitate inter-patient comparability of radiomic features; for example, PET scanners measure radioactivity concentrations [MBq/mL] which directly depend on the amount of injected radiotracer and patient weight [22]. To compensate for variability, the standardized uptake value (SUV) is calculated for each voxel as a relative measure of radiotracer uptake in clinical practice as well as radiomics studies [17, 19, 23–25]. MRI grey scales are expressed in arbitrary units unique to the hardware and reconstruction method used. Presence of heterogeneous image acquisition variables in an MRI dataset always necessitates image normalization before radiomic feature extraction [26–28]. Notably, in addition to the original image, radiomic features are often extracted from transformed or filtered images. A multitude of studies applied wavelet-decompositions to extract texture features from different frequency bands of the original image [7, 18, 24, 25, 29, 30]. Smoothing filters (e.g. Gaussian filters) or combined filters (e.g. Gaussian smoothing followed by Laplacian for edge enhancement) have also been implemented by some studies [20, 30–32].
Radiomic feature extraction represents the last step of common radiomics pipelines. Zwanenburg et al. published “The image biomarker standardisation initiative” (IBSI), which is the most recent attempt to standardize image pre-processing and radiomics feature sets across the field [21]. In brief, IBSI defines 11 feature families, assessing geometric aspects of the ROI/VOI shape, quantifying the grey scale intensity (distribution), and lesion texture. Feature extraction is usually performed by dedicated software in a fully automated fashion. Recent studies extracted their feature sets from original images and several derivatives thereof – generated by filtering, resampling and transformation. This approach commonly yields feature vectors in the magnitude of hundreds to several thousand data points per segmented ROI/VOI.
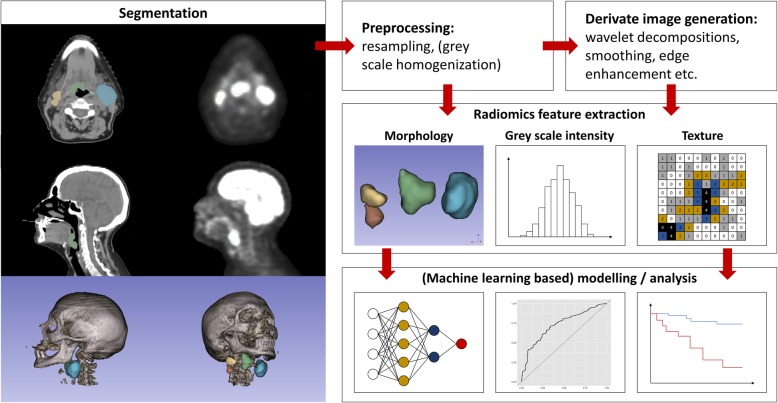
Both open source- and in house-developed feature libraries and radiomics extraction software have been utilized in recent radiomics studies. Two commonly used open-source solutions for radiomics feature extraction are the “Imaging Biomarker Explorer (IBEX)” [33], and “PyRadiomics” [34]. They represent adaptable, configurable platforms for image preprocessing and feature extraction and were applied in recent HNSCC radiomics studies (for example IBEX in refs [31, 35], .PyRadiomics in refs [36, 37].). The considerable methodological variability in HNSCC-related radiomics studies heralds the need for devising evidence-based consensus radiomics pipelines to improve reproducibility and generalizability. Fig. 1 summarizes the essential steps in common radiomics pipelines.
Fig. 1.
Typical radiomics workflow pipeline
Machine learning analysis of radiomics features
Radiomics pipelines extract high-dimensional, quantitative feature sets from medical images [2]. This bioimage-based information is most helpful when combined with clinical variables, serum markers, and other conventional prognostic biomarkers, creating the need for efficient analysis and development of predictive models based on high-dimensional data. Machine learning (ML) methods have proven to be statistically powerful tools for taking on such challenges [2, 38].
ML refers to a series of statistical algorithms driving their functionality from labelled or unlabeled training data, rather than applying predefined sets of rules and functions [38]. This property is ideal in the setting of radiomics research, where extensive numbers of bioimaging features are extracted, to predict molecular biomarkers, histopathological characteristics, clinical outcome, or treatment response [2].
To limit overfitting and augment generalizability, ML studies are ideally based on training, validation, and independent/external testing in separate datasets [38]. The training and validation datasets are used to iteratively fit the ML model (training data), assess its performance (validation data) and optimize model parameters (“tuning of hyperparameters”) [38]. Alternatively, cross validation may be applied to fit/assess/tune the model based on random subdivision and iterative rounds of training and validation [20]. The independent/external test cohort will be kept fully isolated from the model development process and is used to test the final ML model and confirm its performance and generalizability [20, 38].
Typically, in radiomics studies, a data dimensionality reduction strategy is combined with a ML classification or regression algorithm [20]. Dimensionality reduction usually aims to exclude redundant and unstable features and rank-orders the remaining features according to their predictive association with the target outcome. Then, ML algorithms combine the most predictive features into a meaningful, predictive model [20]. The model is next applied to the validation set, where its performance is assessed [38]. The process is iteratively repeated and hyperparameters are adjusted throughout [38].
In a study exploring the importance of feature selection in radiomics analysis, Parmar et al. [29] compared different combinations of 13 feature selection methods and 11 ML classifiers to predict overall survival based on a set of 440 radiomic features extracted from 231 HNSCC primary tumor lesions in contrast-enhanced CT images. Using multifactor analysis of variance (ANOVA) on the receiver operating characteristics (ROC) area under the curve (AUC), they assessed the effect of three ML framework variables (feature selection methods, classification methods, number of selected features). They found that while ML classification methods accounted for 29.02% of the total variance in classification accuracy, the feature selection methods explained 14.02%, and the interaction of classifier and feature selection explained 16.59%. These findings highlight the importance of selecting the appropriate combination of feature selection and ML models, for example by testing various combinations of algorithms with high performance in prior studies.
In the field of head and neck cancer radiomics, classification and (survival) regression models are frequently applied for prediction of molecular markers, identification of genomic signatures, diagnostic differentiation of suspected tissue, survival prognostication, and prediction of treatment response. Increasing numbers of publicly available mega-data and open-source machine-learning algorithms have paved the road for development of novel multivariate diagnostic and prognostic biomarkers integrating quantitative radiomics features and clinical variables for risk stratification, outcome prediction, and precision treatment planning in HNSCC.
Radiomics signatures of HNSCC molecular markers
Multiple recent radiomics studies reported the associations of bioimaging features with various molecular HNSCC traits, such as human papillomavirus (HPV) status, somatic mutations, methylation and gene expression subtypes and PD-L1 expression levels. Among all investigated HNSCC molecular traits, HPV has been evaluated the most:
Human papillomavirus status
The incidence of HPV-associated oropharyngeal SCC (OPSCC) has been rising in recent decades [39, 40]. The prevalence of HPV-associated forms among OPSCC in North America has increased from 50.7% before 2000, to 69.7% in the period from 2005 through 2010 [41]. HPV-positivity is a strong, independent prognostic factor for favorable outcome and overall survival (OS) in patients with OPSCC [42, 43]. HPV association in HNSCC is associated with distinct tumor morphology (smaller primary tumors, marked cervical adenopathy at presentation), younger patients’ age at presentation, and favorable response to radiation therapy [43]. Consequently, the latest release of the American Joint Committee on Cancer (AJCC), and Union for International Cancer Control (UICC) staging manuals have classified HPV-mediated OPSCC as a distinct tumor entity with different staging rules from the OPSCC-negative form [44, 45]. In addition, recent studies suggest that HPV association may analogously impact OS in non-oropharyngeal forms of the HNSSC [46, 47].
Since 2015, multiple studies have demonstrated the association of radiomic features with HPV status in HNSCC: While Buch et al. [48] and Fujita et al. [49] examined the association of individual texture features with HPV status, other groups have designed machine learning classification models for HPV prediction in HNSCC. Table 1 summarizes prior work in this field.
Table 1.
Prediction of HPV status based on radiomics features of HNSCC tumors
| Authors, year | Sample size, cancer type | Ground truth | Imaging modality | ML classifier | Metric: maximum performance a |
|---|---|---|---|---|---|
| Bogowicz et al. 2017 [50] |
Train: 93, HNSCC Test: 56, HNSCC |
p16 | Contrast CT | Logistic regression | Test-AUC: 0.78 |
| Buch et al. 2015 [48] | Total: 40, OPSCC | Not reported | Contrast CT | n/a b | n/a b |
| Fujita et al. 2016 [49] | Total: 46: non-OPSCC | Not reported | Contrast CT | n/a b | n/a b |
| Huang et al. 2019 [51] |
Train: 113, HNSCC Test: 53, HNSCC |
Train: HPV RNA c Test: p16 |
Contrast CT | LASSO-regularized logistic regression |
Nested CV-AUC: 0.73 Test-AUC: 0.76 |
| Leijenaar et al. 2018 [52] |
Train: 628, OPSCC Test:150, OPSCC |
p16 | Contrast CT | LASSO-regularized logistic regression | Test-AUC: 0.70–0.80 d |
| Mungai et al. 2019 [53] | Total: 50, OPSCC | Not reported | Contrast CT | Logistic regression | n/a e |
| Parmar et al. 2015 [54] |
Train: 136, OPSCC and LSCC Test:95, OPSCC |
Not reported | Contrast CT | Logistic regression | Test-AUC: 0.60 |
| Ranjbar et al. 2018 [55] | Total: 107, OPSCC | HPV DNA-ISH | Contrast CT | Diagonal quadratic discriminant analysis | LOOCV-AUC: 0.80 |
|
Yu et al. 2017 [56] |
Train: 150, OPSCC Test:165, OPSCC |
p16 | Contrast CT | Logistic regression |
CV-AUC: 0.75 test-AUC 1 f: 0.87 test-AUC 2 f: 0.92 |
|
Zhu et al. 2018 [57] |
Total: 126, HNSCC | Not reported | Contrast CT | Random forest | CV-AUC: 0.71 |
a The reported performance pertains to pure imaging feature-based HPV classification (i.e. models with clinical features were not considered)
b A t-test was used to evaluate differences in texture parameters between HPV-positive and HPV-negative cases
c The VirusSeq-software was used to detect strain-specific HPV RNA sequences in whole-transcriptome sequencing data [51, 58]
d This study evaluated the impact of CT artifacts on the HPV classification performance. A test set AUC performance of 0.8 was achieved after exclusion of all artifact-affected cases from both the training- and test set. The test AUC ranged between 0.70 and 0.80 for all evaluated dataset combinations, including those with artifacts, and was not significantly different for all tested models
e The logistic regression model was trained and tested on the same dataset without feature selection or cross validation, which is prone to overfitting, and overestimation of classification accuracy
f Study reports results of winning submission of radiomics competition, wherein 165 test cases were split into two test sets
AUC Area under the receiver operating characteristics curve, CV Cross validation (of total set or training dataset), DNA-ISH DNA in situ hybridization, HNSCC Head and neck SCC, LOOCV Leave one out cross validation of total set, LSCC Laryngeal SCC, OPSCC Oropharyngeal SCC, Test Independent test dataset, Total Only one dataset used, Train Training dataset
Of note, some studies did not report the details of the HPV test used for ground truth labeling [48, 49, 53, 54, 57], and some used p16 immunohistochemical surrogate testing to consequently predict p16 status [59]. Many studies evaluated their models’ generalizability in independent confirmation cohorts and confirmed similar performance as compared to the training datasets [50–52, 54, 56]. While the majority of studies to date have applied CT-based radiomics for HPV classification, Vallieres et al. [60] reported their preliminary results based on radiomics features from FDG-PET scans in 67 patients with HNSCC. In addition, quantitative diffusion MRI studies have shown the difference in apparent diffusion coefficient values between HPV-positive and HPV-negative OPSCC [61–63]; however, there has yet been no report of MR-based radiomics signatures for prediction of HPV status.
A potential application of radiomics-based biomarkers for HPV status would be to aid pathologists if standard p16 immunohistochemical staining is equivocal or to supplement the immunohistochemical tests in subjects requiring second-line testing. For routine clinical HPV-testing, the 2018 Guideline from the College of American Pathologists recommends p16 immunohistochemistry as a surrogate marker for HPV-association on samples from the primary tumor or cervical level II or III nodal metastases. However, they recommend using HPV-specific testing – such as in situ hybridization for HPV DNA – in certain p16-positive cervical nodes or multisite primary tumors [59]. In such cases, radiomics-based biomarkers may be an inexpensive substitute confirmatory test for HPV status.
In addition, radiomics signatures for HPV classification may serves as a prognostic biomarker in patients with OPSCC. Leijenaar et al. [52] used contrast-enhanced CT radiomic features from OPSCC primary tumors (628 subjects for training and 150 for validation) to devise a radiomic biomarker for HPV status. Using Kaplan-Maier survival analysis, they showed that both p16 (as a surrogate for HPV), and the radiomics-based classifier could differentiate low- versus high-risk patients in survival curve analysis. Future studies will likely explore the role of other imaging modalities such as MRI or FDG-PET as well as state-of-the-art ML classifiers to enhance classification performance. There is also a potential role for application of radiomics to detect HPV-association of metastatic nodes in carcinoma of unknow primary that may direct search for the tumor origin to the oropharynx.
Radiomics biomarkers of HNSCC molecular subtypes beyond HPV status
Several recent studies have proposed novel radiomics biomarkers for prediction of HNSCC molecular features and subtypes, aside from HPV status.
Zwirner et al. [64] hypothesized that frequently mutated HNSCC driver genes may correlate with radiomics features known to quantify intra-tumor heterogeneity. The analysis was thus focused on three radiomics features initially described by Aerts et al. [18]. A total of 20 patients with locally advanced SCC of the oral cavity, oropharynx or hypopharynx were recruited for a prospective study by [64]; next-generation tumor sequencing and radiomics analysis of corresponding non-contrast radiotherapy planning CTs was performed. The presence of mutations in known driver genes (TP53, FAT1 and KMT2D) were correlated with each of the three selected radiomics features; and showed significant association of all three tested radiomics features with FAT1 [64]. The authors suggested that these findings are likely related to lower heterogeneity in FAT1-mutated HNSCC tumors.
Huang et al. [51] studied a series of molecular HNSCC “phenotypes”: five DNA methylation subtypes, four previously identified HNSCC gene expression subtypes (transcriptomics-based [65]) and five common somatic gene mutations. DNA methylation aberrations were explored using the MethylMix algorithm [66], followed by consensus clustering for subtyping. Contrary to Zwirner et al. [64], Huang et al. used a large radiomics feature set comprised of 540 individual features extracted from pre-treatment contrast-enhanced CT scans of 113 patients [51]. Feature selection and LASSO-penalized logistic regression were applied in nested cross validation. Multi-class classification was facilitated using a “one-vs-all” approach (i.e. binary classifiers were trained to predict any given class against all others). The machine learning classifiers yielded moderate to good predictive performance in identification of the HNSCC molecular phenotypes, even exceeding models based on clinical variables only.
In a cohort of 126 HNSCC patients, Zhu et al. [57] examined the correlation of radiomic features extracted from contrast-enhanced CT-images with whole-genome multiomics data (microRNA expression, somatic mutations, transcriptional activity of pathways, copy number variations and promoter region DNA methylation changes of pathways). They identified over 5000 significant associations, suggesting widespread association of genomic markers and radiomic features from various feature families. Additionally, Zhu et al. trained random forest classifiers in 5-fold cross validation to predict HPV status (Table 1) and disruptive TP53 mutation status, with the most predictive model yielding an AUC of 0.641 (averaged across 30 cross validation repetitions).
In 2016, nivolumab and pembrolizumab were FDA-approved for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck with disease progression on or after a platinum-based therapy [67]. Expression of programmed cell death protein 1 ligand (PD-L1) is the single factor that is most strongly correlated with response to PD-1 blockers like nivolumab or pembrolizumab [68]. Since overall response rates to these agents are low, ranging from 13 to 18% [69, 70], quantification of PD-L1 expression by immunohistochemical staining has been applied to identify patients who are more likely to respond [71, 72]. Extracting textural features from the PET-portions of staging FDG-PET/CT scans, Chen et al. [73] reported significant association of several radiomics features with PD-L1 expression in 53 patients with oropharyngeal and hypopharyngeal SCC. Multivariate logistic regression analysis revealed one FDG-PET radiomics feature as an independent predictor for PD-L1 expression (PD-L1 staining cutoff of 5%) [73].
Thus far, exploratory studies show associations of CT- and FDG-PET-derived radiomic imaging features with genomic, transcriptomic and proteomic characteristics of HNSCC, suggesting that future “multiomic” investigations of HNSCC should incorporate radiomics-based biomarkers. Additional imaging modalities as well as molecular targets are the focus of future investigations.
Prediction of recurrence, treatment response, and survival in HNSCC
Despite major efforts in treatment and drug development, prognosis of HNSCC is generally poor, with five-year survival rates in Europe ranging from 25% in hypopharyngeal cancer to 59% for cancers of the larynx [74–76]. Additionally, the majority of patients with HNSCC presents with advanced-stage disease [76, 77].
More accurate risk stratification, treatment response prediction and prognostication may help clinicians to selectively plan treatment options, guide treatment intensity and ultimately tailor personalized cancer care for their patients. This notion triggered interest among scientists, making outcome prediction by means of bioimaging-features the most popular field within head and neck radiomics.
Table 2 summarizes recent studies focusing on prediction of survival, locoregional recurrence, distant metastasis, progression or treatment failure as well as several composite outcome endpoints. One study used radiomics for prediction of early response to induction chemotherapy [27]; another predicted response to chemoradiotherapy [28] – both in nasopharyngeal carcinoma. Oropharyngeal SCC, laryngeal SCC, hypopharyngeal SCC, nasopharyngeal cancer and combined HNSCC cohorts were investigated from 2013 through 2019, with a marked range in terms of cohort size: While exploratory studies used as few as 30 cases [92], others gathered expansive datasets. For example: 240 and 204 contrast CTs were used for model training and testing, respectively, by Zhai et al. study [93], reporting significantly better prognostic performance of a combined model (radiomics + clinical predictors) as compared to a clinical-variables-only model for disease-free survival in HNSCC. Using 542 oropharyngeal SCC cases from Canada, Leijenaar et al. [87] externally validated a radiomics signature previously devised by Aerts et al. [18] on 422 non-small cell lung cancer contrast-enhanced CTs, which showed significant prognostic differentiation in Kaplan-Meier overall survival analysis in all sub-cohorts. A similarly large dataset of pre-treatment contrast-enhanced CT scans (465 oropharyngeal SCC cases) was analyzed by the Head and Neck Quantitative Imaging Working Group of M.D. Anderson Cancer Center [31]; whose proposed 2-feature-signature could robustly discriminate between the high- versus low-recurrence probability groups. Individual radiomics features, radiomic signatures/scores (e.g. (linear) combinations of several features [18, 27, 31, 94]) as well as ML-generated models [29, 30, 95] showed significant predictive value in a multitude of HNSCC settings, including various HNSCC sub-entities, and outcomes (Table 2).
Table 2.
Prediction of locoregional recurrence, treatment response, and survival
| Author, year | Dataset: sample size, HNSCC type | Imaging modality | Therapy | Outcome, model, analysis type | (Endpoint:) variables: metric: maximum performance a |
|---|---|---|---|---|---|
|
Aerts et al. 2014 [18] |
Test: 231, HNSCC (HNSCC cohort used for validation only, training on 422 lung cancer primaries) |
Pre-treatment contrast CT | RT + chemotherapy or RT + chemotherapy + surgery or RT alone |
OS, multivariable Cox regression, regression |
Radiom: test-CI 1:0.69 test-CI 2: 0.69 Radiom+Clin: test-CI 1: 0.70 test-CI 2: 0.69 Clin: test-CI 1: 0.69 test-CI 2: 0.66 |
|
Bogowicz et al. 2017 [78] |
Train: 128, HNSCC Test: 50, HNSCC |
3-month post treatment FDG-PET | RT + cisplatin / cetuximab or RT + cisplatin + cetuximab |
LC, multivariable Cox regression, regression |
Radiom: CV-CI: 0.74–0.76 test-CI: 0.71–0.73 (study evaluated reproducibility of identical features using two different software - performance range is reported) |
|
Bogowicz et al. 2017 [25] |
Train: 121, HNSCC Test: 51, HNSCC |
Pre-treatment FDG-PET, contrast CT | RT + cisplatin / cetuximab or RT + cisplatin + cetuximab |
LC, 3 different regression methods, regression |
Radiom: CV-CI: 0.77 test-CI: 0.73 |
|
Bogowicz et al. 2017 [50] |
Train: 93, HNSCC Test: 56, HNSCC |
Pre-treatment contrast CT | RT + cisplatin / cetuximab |
LC, multivariable Cox regression, regression |
Radiom: train-CI: 0.75 test-CI: 0.78 Clin: train-CI: 0.79 test-CI: 0.73 Radiom + Clin: train-CI: 0.80 test-CI: 0.76 |
|
Bogowicz et al. 2019 [79] |
Train: 77, HNSCC Test: 51, HNSCC |
Pre-treatment contrast CT | RT + cisplatin / cetuximab or RT + cisplatin + cetuximab |
LC and LRC, multivariable Cox regression, regression |
LC: Radiom: CV-CI: 0.81 test-CI: 0.70 LRC: Radiom: CV-CI: 0.75 test-CI: 0.67 (addition of lymph node to primary tumor radiomics features was investigated –the best performance reported) |
|
Bogowicz et al. 2019 [80] |
Total: 45, HNSCC | Pre-treatment perfusion CT | IMRT + cisplatin / cetuximab or IMRT + cisplatin + cetuximab |
LC, multivariable Cox regression, regression |
Radiom: CV-CI: 0.79 Clin: CV-CI: 0.66 |
|
Cheng et al. 2013 [81] |
Total: 70, OPSCC | Pre-treatment FDG-PET | RT + platinum-based chemotherapy / cetuximab or RT alone |
DSS, OS and PFS, multivariate Cox regression, regression |
DSS: Radiom: HR: 0.28 (p = 0.001) OS: Radiom: HR: 0.46 (p = 0.017) PFS: Radiom: HR: 0.32 (p = 0.001) (Only 1 radiomics feature was tested in multivariate Cox regression, along with clinicopathological and FDG-PET variables) |
|
Cheng et al. 2015 [82] |
Total: 88, OPSCC | Pre-treatment FDG-PET | RT + chemotherapy / biotherapy or RT alone |
PFS and DSS, multivariate Cox regression, regression |
PFS: Radiom: HR: 4.38 (p = 0.002) DSS: Radiom: HR: 4.24 (p = 0.005) (single radiomics features were tested in multivariate Cox regression, along with clinicopathological and FDG-PET variables) |
|
Cozzi et al. 2019 [83] |
Train: 70, HNSCC Test: 40, HNSCC |
Pre-treatment contrast CT | RT + chemotherapy or RT + chemotherapy + induction-chemotherapy |
OS, PFS and LC, multivariable Cox regression, regression |
OS: Radiom: train-CI: 0.88 test-CI: 0.90 PFS: Radiom: train-CI: 0.72 test-CI: 0.80 LC: Radiom: train-CI: 0.72 test-CI: 0.80 |
| Feliciani et al. 2018 [84] | Total: 90, HNSCC | Pre-treatment FDG-PET | IMRT + platinum-based chemotherapy w/ or w/o adjuvant / neoadjuvant chemotherapy |
PFS and OS, multivariate Cox regression, regression |
PFS: Radiom + Clin: CV-CI: 0.76 Clin: CV-CI: 0.65 OS: Radiom + Clin: CV-CI: 0.76 Clin: CV-CI: 0.73 |
| Folkert et al. 2017 [85] |
Train: 174, OPSCC Test: 65, OPSCC |
Pre-treatment FDG-PET | RT + platinum-based chemotherapy / cetuximab / multidrug regimens |
ACM, LF and DM, multivariable logistic regression, classification |
ACM: Radiom + Clin: CV-AUC: 0.65 test-AUC: 0.60 LF: Radiom + Clin: CV-AUC: 0.73 test-AUC: 0.68 DM: Radiom + Clin: CV-AUC: 0.66 test-AUC: 0.65 |
|
Ger et al. 2019 [35] |
Train 1: 377, HNSCC (CT) Train 2: 345, HNSCC (PET) Test 1: 349, HNSCC (CT) Test 2: 341, HNSCC (PET) |
Pre-treatment contrast CT, pre-treatment FDG-PET (separately analyzed) | Not reported (definitive RT as part of treatment was inclusion criterion) |
OS, multivariable Cox regression, regression |
Radiom: test-AUC 1: 0.72 (CT) test-AUC 2: 0.59 (PET) Clin: test-AUC 1: 0.73 (CT) (AUC was calculated at 3 years post treatment, with patients with risk prediction > median assigned to the high-risk group) |
| Kuno et al. 2017 [86] | Total: 62, HNSCC | Pre-treatment contrast CT | IMRT + chemotherapy w/ or w/o induction chemotherapy or IMRT alone |
LF, multivariate Cox regression, regression |
Radiom: HR: 3.75–8.61 (8 features were significant after adjusting for clinical variables; the HR range is reported above) |
| Leger et al. 2017 [30] |
Train: 213, HNSCC Test: 80, HNSCC |
Pre-treatment non-contrast CT | RT + chemotherapy |
LRC and OS, 11 different ML algorithms, regression |
LRC: Radiom: test-CI: 0.71 OS: Radiom: test-CI: 0.64 |
| Leijenaar et al. 2015 [87] |
Test: 542, OPSCC (validation of radiomics signature by Aerts et al. [18]) |
Pre-treatment contrast CT | IMRT + chemotherapy or IMRT alone |
OS, multivariable Cox regression, regression |
Radiom: test-CI: 0.63 |
|
Liu et al. 2016 [28] |
Train: 42, NPC Test: 11, NPC |
Pre-treatment T2, contrast-enhanced T1 MRI, diffusion weighted MRI | RT + cisplatin |
Therapy response (complete/partial response vs. stable / progressive disease), k-nearest neighbors, neural network, classification |
Radiom: CV-acc: 0.95 CV-sens: 0.97 CV-spec: 091 test-acc: 0.91 test-sens: 0.88 test-spec: 1 |
|
Lv et al. 2019 [88] |
Total: 296, HNSCC (various partitions in train/test were evaluated) |
Pre-treatment non-contrast CT, FDG-PET | RT + chemotherapy or RT alone |
RFS, MFS and OS, multivariate Cox regression, regression |
RFS: Radiom: mean test-CI: 0.61 Radiom + Clin: mean test-CI: 0.60 Clin: mean test-CI: 0.58 MFS: Radiom: mean test-CI: 0.70 Radiom + Clin: mean test-CI: 0.71 Clin: mean test-CI: 0.61 OS: Radiom: mean test-CI: 0.62 Radiom + Clin: mean test-CI: 0.65 Clin: mean test-CI: 0.62 (the mean was calculated across all test partitions) |
|
Lv et al. 2019 [24] |
Train: 85, NPC Test: 43, NPC |
Pre-treatment CT, FDG-PET | IMRT + cisplatin or IMRT alone |
PFS, multivariate Cox regression, regression |
Radiom: train-CI: 0.76 test-CI: 0.62 Radiom + Clin: train-CI: 0.75 test-CI: 0.75 Clin: train-CI: 0.71 test-CI: 0.75 |
| M.D. Anderson C.C.H.a.N.Q.I.W.G. 2018 [31] |
Train: 255, OPSCC Tune: 165, OPSCC Test: 45, OPSCC |
Pre-treatment contrast CT | One or combinations of: IMRT / chemotherapy / induction chemotherapy / neck dissection |
LC, multivariate Cox regression, regression |
Overall performance evaluation of Cox models not reported |
|
Mo et al. 2019 [89] |
Train: 80, HYSCC Test: 33, HYSCC |
Pre-treatment non-contrast CT and contrast-CT | Laryngeal-preservation treatments (RT, chemotherapy, induction-chemotherapy, neck dissection) |
PFS, multivariable Cox regression, regression |
Radiom: train-CI: 0.79 test-CI: 0.76 Radiom + Clin: train-CI: 0.80 test-CI: 0.76 Clin: train-CI: 0.63 test-CI: 0.54 |
|
Ou et al. 2017 [90] |
Total: 120, HNSCC | Pre-treatment CT | CRT / IMRT + cisplatin / cetuximab |
OS and PFS, multivariable Cox regression, regression |
OS: Radiom: HR: 0.3 (p = 0.02) PFS: Radiom: HR: 0.3 (p = 0.01) |
| Ouyang et al. 2017 [91] |
Train: 70, NPC Test: 30, NPC |
Pre-treatment T2, contrast-enhanced T1 MRI | Not reported |
PFS, multivariable Cox regression, regression |
Radiom: train-HR: 5.14 (p < 0.001) test-HR: 7.28 (p = 0.015) |
| Parmar et al. 2015 [29] |
Train: 101, HNSCC Test: 95, HNSCC |
Pre-treatment contrast CT | RT + chemotherapy or RT + chemotherapy + surgery or RT alone |
OS, 12 different ML classifiers, classification |
Radiom: test-AUC: 0.79 |
| Parmar et al. 2015 [54] |
Train: 136, HNSCC Test: 95, HNSCC |
Pre-treatment contrast CT | RT + chemotherapy or RT + chemotherapy + surgery or RT alone |
OS, multivariable Cox regression, regression |
Radiom: test-CI: 0.63 |
| Ulrich et al. 2019 [92] | Total: 30, OPSCC and LSCC | Pre-treatment 18F-fluorothymidine PET | RT + platinum-based chemotherapy |
PFS, univariate Cox regression, regression |
Radiom: HR: 4.10 (p = 0.001) |
| Vallieres et al. 2017 [23] |
Train: 194, HNSCC Test: 106, HNSCC |
Pre-treatment FDG-PET, non-contrast CT | RT + platinum-based chemotherapy / cetuximab or RT alone |
LR, DM and OS, logistic regression, random forests, classification (regression analysis was performed for a subset of models; see publication) |
LR: Radiom: test-AUC: 0.64 Radiom + Clin: test-AUC: 0.69 DM: Radiom: test-AUC: 0.86 Radiom + Clin: test-AUC: 0.86 OS: Radiom: test-AUC: 0.62 Radiom + Clin: test-AUC: 0.74 |
| Wang et al. 2018 [27] | Total: 120, NPC | Pre-treatment T2, contrast-enhanced T1 MRI | Induction-chemotherapy (cisplatin + 5-fluorouracil + docetaxel) |
Early response to induction chemotherapy, “Rad-score”, classification |
Radiom: train-AUC: 0.82 internally bootstrap-validated train-AUC: 0.82 |
| Zhai et al. 2019 [93] |
Train: 240, HNSCC Test: 204, HNSCC |
Pre-treatment contrast CT | RT + chemotherapy / cetuximab or RT alone |
LC, RC, MFS and DFS, multivariate Cox regression, regression |
LC: Radiom: train-CI: 0.62 test-CI: 0.62 Radiom + Clin: train-CI: 0.66 test-CI: 0.64 Clin: train-CI: 0.64 test-CI: 0.62 RC: Radiom: train-CI: 0.78 test-CI: 0.80 Radiom + Clin: train-CI: 0.78 test-CI: 0.80 Clin: train-CI: 0.74 test-CI: 0.76 MFS: Radiom: train-CI: 0.73 test-CI: 0.68 Radiom + Clin: train-CI: 0.72 test-CI: 0.71 Clin: train-CI: 0.71 test-CI: 0.70 DFS: Radiom: train-CI: 0.66 test-CI: 0.65 Radiom + Clin: train-CI: 0.69 test-CI: 0.70 Clin: train-CI: 0.66 test-CI: 0.66 |
| Zhang et al. 2017 [94] |
Train: 80, NPC Test: 33, NPC |
Pre-treatment T2, contrast-enhanced T1 MRI | Not reported |
PFS, “Rad-score”, classification |
Radiom: train-AUC: 0.89 test-AUC: 0.82 |
| Zhang et al. 2017 [95] |
Train: 70, NPC Test: 40, NPC |
Pre-treatment T2, contrast-enhanced T1 MRI | Not reported |
LF and DF, 9 different ML classifiers, classification |
LF and DF: Radiom: test-AUC: 0.85 |
| Zhang et al. 2017 [96] |
Train: 88, NPC Test: 30, NPC |
Pre-treatment T2, contrast-enhanced T1 MRI | Not reported |
PFS, univariate / multivariable Cox regression, regression |
Radiom: train-CI: 0.76 test-CI: 0.74 Clin: train-CI: 0.65 test-CI: 0.63 Radiom + Clin: train-CI: 0.78 test-CI: 0.72 |
a The reported performance pertains to the maximum observed performance among all models of each respective category (i.e. we are reporting the highest achieved performance, in case different radiomics features / models / signatures or clinical predictors / models were tested). For radiomics-based models, the performance of the purest imaging feature-based model is reported. (i.e. the model with fewest or no other predictors)
acc Accuracy, ACM All-cause mortality, AUC Area under the receiver operating characteristics curve, CI Concordance index, Clin Non-radiomic predictor(s) or model(s) (“clinical”), CRT Conformal radiotherapy, DF/DM Distant failure/metastasis, DFS Disease-free survival, DSS Disease-specific survival, HNSCC Head and neck SCC, HR Hazard ratio, HYSCC Hypopharyngeal SCC, IMRT Intensity-modulated radiotherapy, LC/LF Local tumor control/failure, LR Locoregional recurrence, LRC Locoregional control, LSCC Laryngeal SCC, MFS Metastasis-free survival, NPC Nasopharyngeal carcinoma, OPSCC Oropharyngeal SCC, OS Overall survival, PFS Progression-free survival, Radiom Radiomics model, radiomic feature(s) or feature combinations (“signature”, “Rad-score”), RC Regional control, RFS Recurrence-free survival, RT Radiotherapy, sens Sensitivity, spec Specificity, test Independent test dataset, total Only one dataset used, train Training dataset, tune Validation set used for hyperparameter tuning
The complimentary value of radiomics analysis in addition to conventional “clinical” predictors has been emphasized by several groups [18, 93, 96]. However, using multi-institutional and multi-national dataset of 726 pre-treatment contrast CT scans and 686 FDG-PET scans, Ger et al. [35] were unable to improve HNSCC overall survival prediction using multivariate Cox proportional hazard models incorporating only two and one radiomic features in separate CT-based and FDG-PET-based analysis, respectively. These findings suggest more complex analysis strategies may help improve predictive performance. Leger et al. [30] applied 11 ML algorithms combined with 12 feature selection methods in a proof-of-technology study and identified several promising combinations which may be applied in future time-to-event modelling. Combining large HNSCC cohorts with advanced ML analysis may eventually enable radiomics to more consistently improve prognostic models.
Contrast-enhanced and non-contrast CT, (contrast-enhanced) T1 and T2 MRI sequences and FDG-PET imaging were all applied for radiomics based outcome prediction (Table 2) as well as some less common imaging techniques including diffusion-weighted MRI [28], 18F-fluorothymidine-PET [92], and perfusion CT [80]. Studies listed in Table 2 applied different analytical strategies, such as using single feature, feature combinations (“signatures”, “scores”) or more complex combined models; such analytical heterogeneity limits direct comparison of studies [97], and cannot be fully reflected in Table 2. The majority of studies, however, applied multivariate Cox proportional hazard models, the results of which are summarized in the table. The performance of radiomics, clinical or combined models with regards to the respective outcome(s) prediction is expressed in the Cox-model hazard ratio, and the concordance index typically reflects the overall accuracy of models in survival prediction.
Detection of extra-nodal extension of metastasis
Extra-nodal extension (ENE) of metastasis in cervical lymph nodes is a poor prognostic factor and is associated with higher risk of developing recurrent disease [98–101]. Thus, the presence of ENE warrants addition of chemotherapy to adjuvant irradiation [98–101], requiring tri-modality treatment with increased toxicity and patient morbidity [102, 103]. Reliable detection of ENE prior to the therapy, could help guide treatment choices, reduce morbidity, and avoid surgery in patients likely requiring adjuvant chemoradiation. In clinical practice, ENE is ascertained by pathology review after neck dissection, whereas radiographical identification remains challenging [104, 105]. Kann et al. developed [106] and validated [107] quantitative imaging tools for pre-operative detection of ENE: the group segmented 653 nodes in total (380 negative, 153 without ENE and 120 nodes with ENE) on contrast-enhanced CT scans and extracted 99 radiomic features [106]. Random forest ML classifiers were trained and yielded an AUC (95% confidence interval) of 0.88 (0.81–0.95) for the detection of ENE and 0.91 (0.86–0.97) for nodal metastasis detection in an independent test set of 131 lymph nodes; whereas – being the methodological focus of the study – a deep neural network yielded an AUC performance of 0.91 (0.85–0.97) and 0.91 (0.86–0.96) for ENE and metastasis detection, respectively [106]. The deep neural network model generalized well to an external test set, outperforming radiologists in ENE classification [107].
Of note [106, 107], there was no significant difference in performance of deep neural networks (exploratory radiomics) over (preset conventional) radiomic analysis in detection of ENE. They highlight the potential quantitative imaging may possess for augmenting radiologist performance and guiding HNSCC treatment.
Predicting post chemoradiotherapy complications
Radiotherapy combined with chemotherapy (chemoradiotherapy, CRT) is the mainstay treatment regimen for many patients with HNSCC [101]. However, patients not uncommonly suffer from treatment-related side effects such as xerostomia, trismus, hearing loss, mucositis and dermatitis. Identification of those patients who are at risk of developing specific side effects may guide oncologists to plan personalized treatment strategies and adopt preventive remedies to improve therapy tolerance. Several groups have devised radiomics biomarkers to predict the occurrence or severity of treatment-related toxicities based on bioimaging features of at-risk organs.
Xerostomia
Radiation-induced xerostomia is a common side effect of radiation therapy for HNSCC and remains a challenge in long-term patient management [108, 109]. The dose-dependent increased risk of xerostomia after irradiation of the salivary glands is well established [109]. Four separate groups designed radiomics-based models to predict post-radiation xerostomia in patients with HNSCC with or without concurrent chemotherapy (Table 3). Imaging features were extracted from salivary glands – either the parotid gland(s) or parotid glands and submandibular glands. A heterogeneous set of xerostomia endpoints was investigated: Sheikh et al. [110] predicted a binary xerostomia-endpoint 3 month post radiotherapy; Liu et al. [111] applied regression analysis for acute xerostomia prediction; and van Dijk et al. [112–114] used three different imaging modalities (CT, MRI, FDG-PET) for long-term binary xerostomia outcome classification. Furthermore, the xerostomia assessment methods varied: Liu et al. used objective saliva amount measurements over 5 min [111], whereas other groups used patients-filled questionnaires [112–114]. While these results appear promising, their clinical application is limited by the lack of external validation, heterogeneity in image processing, statistical analysis, and treatment outcome measures.
Table 3.
Prediction of post-radiation xerostomia based on salivary gland radiomics features
| Authors, year | Dataset: sample size, cancer type | Time of xerostomia assessment, endpoint/scale | Imaging modality | VOI | Classifier / regression model(s) | Metric: maximum performance a |
|---|---|---|---|---|---|---|
| Sheikh et al. 2019 [110] |
Train:216, HNSCC Test:50, HNSCC |
3-month post-RT, CTCAE v4.0 b grade ≥ 2 vs. grade 0/1 | Pre-treatment CT, T1-weighted MRI | Parotid and submandibular glands (bilateral) | Multivariable logistic regression |
CV-AUC: 0.75 test-AUC: 0.70 |
|
Liu et al. 2019 [111] |
Train:35, NPC Test:4, NPC |
day of 10th and 30th RT, saliva amount (ml) over 5 min (a regression analysis) |
CT at start and day of 10th RT fraction | Parotid glands (bilateral) | 8 different regression models |
CV-MSE: 0.9042 (10th fraction), 0.0569 (30th fraction) test-MSE: 0.0233 (30th fraction) |
| van Dijk et al. 2018 [112] |
Train:68, HNSCC Test:25, HNSCC |
12 moth post-RT, patient-rated moderate-to-severe xerostomia present vs. not present | Pre-treatment T1-weighted MRI | Parotid glands, (bilateral) | Multivariable logistic regression | n/a c |
| van Dijk et al. 2017 [113] | Total: 249, HNSCC | 12 moth post-RT, EORTC QLQ-H, N35 questionnaire d moderate-to-severe xerostomia vs. not present | Pre-treatment contrast CT | Parotid and submandibular glands (bilateral) | Multivariable logistic regression | n/a c |
| van Dijk et al. 2018 [114] | Total: 161, HNSCC |
12-month post-RT, EORTC QLQ-H questionnaire d moderate-to-severe xerostomia present vs. not present |
Pre-treatment FDG PET | Contralateral parotid gland | Multivariable logistic regression | n/a c |
a The reported performance pertains to the maximum observed performance among the purest imaging feature-based models reported (i.e. the best model with fewest or no other predictors is reported)
b Common Terminology Criteria for Adverse Events Version 4.0 [115]
c “Pure” radiomics models were not built. Instead, the contribution of individual radiomics features to baseline models was investigated in terms of performance (gains)
d European Organization for Research and Treatment of Cancer questionnaire module for quality of life assessments in head and neck cancer patients [116]
AUC Area under the receiver operating characteristics curve, CV Cross validation (of total set or training data set), HNSCC Head and neck SCC, MSE Mean squared error, NPC Nasopharyngeal carcinoma, RT Radiotherapy, Test Independent test data set, Total Only one data set used, Train Training data set
Trismus
Trismus in HNSCC patients may result from involvement of masticatory muscles in radiotherapy treatment fields, surgery or cancerous invasion into mastication structures or the neural innervation of masticatory muscles [117, 118]. Defining trismus ≥ Grade 1 by CTCAE v4.0 (Common Terminology Criteria for Adverse Events Version 4.0 [115]) criteria 1 year following completion of intensity-modulated radiotherapy (IMRT), Thor et al. [119] compared 24 imaging features from four masticatory muscles on contrast-enhanced post-treatment T1-weighted MRI scans in 10 patients with radiation-induced trismus, versus 10 control subjects. The best discriminative ability among radiomics predictors was observed for the Haralick Correlation GLCM-matrix feature of the medial pterygoid muscle VOI (logistic regression p = 0.12, AUC = 0.78). Their result was not significant, but may be indicative of a potential of radiomics biomarkers for prediction of post-radiation trismus. Studies in larger cohorts may be the focus of future research, to devise radiomics signature predictive of post-radiotherapy trismus.
Hearing loss
Abdollahi et al. [120] explored the potential application of cochlear radiomics for prediction of chemoradiotherapy-induced hearing loss. Using radiomics features extracted from the cochlea on pre-treatment CT scans, they evaluated 47 cancer patients (brain, nasopharynx, parotid, other) treated with 3-dimensional conformal radiation therapy, 23 of whom also received cisplatin-chemotherapy. They showed that combination of radiomic features with clinical and dosimetric variables may predict radiotherapy-induced sensory neural hearing loss.
Future directions, challenges and barriers
The next leap forward in radiomic analysis undoubtedly lies in developing decision support and prognostic tools for day-to-day clinical usage. However, several key barriers and challenges in the field of quantitative imaging should be addressed first:
While exploratory radiomics studies have achieved promising results throughout, independent large-scale validation is lagging [121, 122]. A recent publication by Kim et al. [122] reported on design characteristics of 516 studies applying AI algorithms for diagnostic analysis of medical images. Only 31 studies (6%) have validated their proposed models in external test cohorts – i.e. cohorts from institutions other than the one providing the training data, as well as cohorts obtained from the same institution but a different time period as the training data. On the other hand, usage of homogenous, single-institution or even single-scanner training data may limit the generalizability of radiomics-based models [121]. These limitations highlight the importance of multi-institutional, multi-national medical imaging archives for development of radiomics tools for future clinical usage. Data sharing may help mitigate the shortage of diverse imaging data [2]; hence, platforms like “The Cancer Imaging Archive” (TCIA) were created. TCIA publicly hosts de-identified imaging collections with corresponding clinical data and provides digital infrastructure for data sharing [123]. As of December 2019, nine head and neck cancer collections are available comprising CT, MRI and FDG-PET imaging data [123].
Further challenges lie in the implementation of the radiomics pipeline (including image acquisition) as outlined in this article. Forghani et al. [1] described sources of variation impairing generalizability and reproducibility of radiomics studies, including:
Scan acquisition parameters
Variability in post-contrast images – such as the degree of enhancement achieved, depending on timing of a contrast agent administration, patients’ circulatory dynamics, anatomical location of the VOI
Wear and tear of scanners
Differences in manufacturer, model, type of scanner
Reconstruction parameters
VOI/ROI segmentation
Radiomics feature set / feature extraction
Preprocessing steps like resampling and filtering (Fig. 1) may help mitigate some variation. However, standardization of reconstruction and acquisition parameters across providers as well as scanner components among manufacturers should be pursued as the field moves towards clinical application of AI-driven image analysis.
Inter- and intra-observer VOI/ROI delineation variability could be addressed by using semi-automated or automated segmentation tools. In addition, there have been efforts to standardize radiomics features – the most recognized being the “The image biomarker standardisation initiative” (IBSI) [21]. Moreover, open-source feature libraries and radiomics extraction software packages like “PyRadiomics” [34], or the “Imaging Biomarker Explorer” [33] allow for reproducible feature extraction as well as easy reporting of radiomics feature definitions and are increasingly adopted by recent publications.
Conclusions
Precision prognostication and treatment personalization is considered the next major evolution in cancer care, and the “-omics”-concept has been postulated as key enabler thereof. Numerous studies have established radiomics as powerful addition to the “-omics”-toolbox, and ongoing research provides incremental upgrades. Radiomics has indeed revolutionized the landscape of quantitative imaging research: In the future, fast, low-cost and comprehensive tumor and tissue characterization facilitated by radiomic analysis may constitute a compelling augmentation – or even alternative – for traditional clinical testing and prognostication, if adequate performance and stability is attained. Numerous studies in the past 6 years have reported potential applications of radiomics analysis for molecular classification, prognostic characterization, and treatment response prediction in patients with HNSCC. While recent exploratory studies yield promising results in the field of HNSCC radiomics, independent large-scale validation is lagging behind as access to multi-institutional, multi-national imaging data is restricted. Standardization of radiomics pipelines, image acquisition protocols, and outcome targets can pave the road towards engineering of radiomics tools for day to day clinical usage, and ultimately superior outcomes and reduced treatment-related toxicities in the field of head and neck cancer.
Acknowledgements
Not applicable.
Abbreviations
- AI
Artificial intelligence
- AJCC
American Joint Committee on Cancer
- AUC
Area under the curve
- FDG-PET
18- fludeoxyglucose positron emission tomography
- HNSCC
Head and neck squamous cell carcinoma
- HPV
Human papillomavirus
- ML
Machine learning
- OPSCC
Oropharyngeal SCC
- OS
Overall survival
- ROC
Receiver operating characteristics
- SCC
Squamous cell carcinoma
- TCIA
The Cancer Imaging Archive
- UICC
Union for International Cancer Control
Authors’ contributions
SPH and SP searched literature. SPH, BB, WY, and SP wrote the manuscript. All authors read and approved the final manuscript.
Funding
There was no funding for this review article.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Barbara Burtness and Seyedmehdi Payabvash contributed equally to this work.
References
- 1.Forghani R, Savadjiev P, Chatterjee A, Muthukrishnan N, Reinhold C, Forghani B. Radiomics and artificial intelligence for biomarker and prediction model development in oncology. Comput Struct Biotechnol J. 2019;17:995–1008. doi: 10.1016/j.csbj.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, They Are Data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016;61(13):R150–R166. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinker K, Shitano F, Sala E, Do RK, Young RJ, Wibmer AG, et al. Background, current role, and potential applications of radiogenomics. J Magn Reson Imaging. 2018;47(3):604–620. doi: 10.1002/jmri.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of Radiomic features: a systematic review. Int J Radiat Oncol Biol Phys. 2018;102(4):1143–1158. doi: 10.1016/j.ijrobp.2018.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balagurunathan Y, Kumar V, Gu Y, Kim J, Wang H, Liu Y, et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging. 2014;27(6):805–823. doi: 10.1007/s10278-014-9716-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leijenaar RT, Carvalho S, Velazquez ER, van Elmpt WJ, Parmar C, Hoekstra OS, et al. Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol. 2013;52(7):1391–1397. doi: 10.3109/0284186X.2013.812798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiset S, Welch ML, Weiss J, Pintilie M, Conway JL, Milosevic M, et al. Repeatability and reproducibility of MRI-based radiomic features in cervical cancer. Radiother Oncol. 2019;135:107–114. doi: 10.1016/j.radonc.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Mackin D, Fave X, Zhang L, Fried D, Yang J, Taylor B, et al. Measuring computed tomography scanner variability of Radiomics features. Investig Radiol. 2015;50(11):757–765. doi: 10.1097/RLI.0000000000000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bologna M, Corino V, Mainardi L. Technical note: virtual phantom analyses for preprocessing evaluation and detection of a robust feature set for MRI-radiomics of the brain. Med Phys. 2019;46(11):5116–5123. doi: 10.1002/mp.13834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao B, Tan Y, Tsai WY, Qi J, Xie C, Lu L, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016;6:23428. doi: 10.1038/srep23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan J, Chu-Shern JL, Loi HY, Khor LK, Sinha AK, Quek ST, et al. Impact of Image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med. 2015;56(11):1667–1673. doi: 10.2967/jnumed.115.156927. [DOI] [PubMed] [Google Scholar]
- 14.Oliver JA, Budzevich M, Zhang GG, Dilling TJ, Latifi K, Moros EG. Variability of Image features computed from conventional and respiratory-gated PET/CT images of lung Cancer. Transl Oncol. 2015;8(6):524–534. doi: 10.1016/j.tranon.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bagher-Ebadian H, Siddiqui F, Liu C, Movsas B, Chetty IJ. On the impact of smoothing and noise on robustness of CT and CBCT radiomics features for patients with head and neck cancers. Med Phys. 2017;44(5):1755–1770. doi: 10.1002/mp.12188. [DOI] [PubMed] [Google Scholar]
- 16.Bogowicz M, Riesterer O, Bundschuh RA, Veit-Haibach P, Hullner M, Studer G, et al. Stability of radiomic features in CT perfusion maps. Phys Med Biol. 2016;61(24):8736–8749. doi: 10.1088/1361-6560/61/24/8736. [DOI] [PubMed] [Google Scholar]
- 17.Lu L, Lv W, Jiang J, Ma J, Feng Q, Rahmim A, et al. Robustness of Radiomic features in [(11)C]choline and [(18)F]FDG PET/CT imaging of nasopharyngeal carcinoma: impact of segmentation and discretization. Mol Imaging Biol. 2016;18(6):935–945. doi: 10.1007/s11307-016-0973-6. [DOI] [PubMed] [Google Scholar]
- 18.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Velden FH, Kramer GM, Frings V, Nissen IA, Mulder ER, de Langen AJ, et al. Repeatability of Radiomic features in non-small-cell lung Cancer [(18)F]FDG-PET/CT studies: impact of reconstruction and delineation. Mol Imaging Biol. 2016;18(5):788–795. doi: 10.1007/s11307-016-0940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larue RT, Defraene G, De Ruysscher D, Lambin P, van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol. 2017;90(1070):20160665. doi: 10.1259/bjr.20160665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. arXiv e-prints. 2016;2016 Available from: https://ui.adsabs.harvard.edu/abs/2016arXiv161207003Z.
- 22.Kinahan PE, Fletcher JW. Positron emission tomography-computed tomography standardized uptake values in clinical practice and assessing response to therapy. Semin Ultrasound CT MR. 2010;31(6):496–505. doi: 10.1053/j.sult.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallieres M, Kay-Rivest E, Perrin LJ, Liem X, Furstoss C, Aerts H, et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep. 2017;7(1):10117. doi: 10.1038/s41598-017-10371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv W, Yuan Q, Wang Q, Ma J, Feng Q, Chen W, et al. Radiomics analysis of PET and CT components of PET/CT imaging integrated with clinical parameters: application to prognosis for nasopharyngeal carcinoma. Mol Imaging Biol. 2019;21(5):954–964. doi: 10.1007/s11307-018-01304-3. [DOI] [PubMed] [Google Scholar]
- 25.Bogowicz M, Riesterer O, Stark LS, Studer G, Unkelbach J, Guckenberger M, et al. Comparison of PET and CT radiomics for prediction of local tumor control in head and neck squamous cell carcinoma. Acta Oncol. 2017;56(11):1531–1536. doi: 10.1080/0284186X.2017.1346382. [DOI] [PubMed] [Google Scholar]
- 26.Shinohara RT, Sweeney EM, Goldsmith J, Shiee N, Mateen FJ, Calabresi PA, et al. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin. 2014;6:9–19. doi: 10.1016/j.nicl.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, He L, Yuan C, Huang Y, Liu Z, Liang C. Pretreatment MR imaging radiomics signatures for response prediction to induction chemotherapy in patients with nasopharyngeal carcinoma. Eur J Radiol. 2018;98:100–106. doi: 10.1016/j.ejrad.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Mao Y, Li Z, Zhang D, Zhang Z, Hao S, et al. Use of texture analysis based on contrast-enhanced MRI to predict treatment response to chemoradiotherapy in nasopharyngeal carcinoma. J Magn Reson Imaging. 2016;44(2):445–455. doi: 10.1002/jmri.25156. [DOI] [PubMed] [Google Scholar]
- 29.Parmar C, Grossmann P, Rietveld D, Rietbergen MM, Lambin P, Aerts HJ. Radiomic machine-learning classifiers for prognostic biomarkers of Head and neck Cancer. Front Oncol. 2015;5:272. doi: 10.3389/fonc.2015.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leger S, Zwanenburg A, Pilz K, Lohaus F, Linge A, Zophel K, et al. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep. 2017;7(1):13206. doi: 10.1038/s41598-017-13448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Group MDACCHaNQIW Investigation of radiomic signatures for local recurrence using primary tumor texture analysis in oropharyngeal head and neck cancer patients. Sci Rep. 2018;8(1):1524. doi: 10.1038/s41598-017-14687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zdilar L, Vock DM, Marai GE, Fuller CD, Mohamed ASR, Elhalawani H, et al. Evaluating the effect of right-censored end point transformation for Radiomic feature selection of data from patients with Oropharyngeal Cancer. JCO Clin Cancer Inform. 2018;2:1–19. doi: 10.1200/CCI.18.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys. 2015;42(3):1341–1353. doi: 10.1118/1.4908210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational Radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e1e7. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ger RB, Zhou S, Elgohari B, Elhalawani H, Mackin DM, Meier JG, et al. Radiomics features of the primary tumor fail to improve prediction of overall survival in large cohorts of CT- and PET-imaged head and neck cancer patients. PLoS One. 2019;14(9):e0222509. doi: 10.1371/journal.pone.0222509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang ZG, Tan HQ, Zhang F, Rui Tan LK, Lin L, Lenkowicz J, et al. Comparison of radiomics tools for image analyses and clinical prediction in nasopharyngeal carcinoma. Br J Radiol. 2019;92(1102):20190271. doi: 10.1259/bjr.20190271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Dong D, Li H, Tian J, Ouyang F, Mo X, et al. Development and validation of a magnetic resonance imaging-based model for the prediction of distant metastasis before initial treatment of nasopharyngeal carcinoma: a retrospective cohort study. EBioMedicine. 2019;40:327–335. doi: 10.1016/j.ebiom.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choy G, Khalilzadeh O, Michalski M, Do S, Samir AE, Pianykh OS, et al. Current applications and future impact of machine learning in radiology. Radiology. 2018;288(2):318–328. doi: 10.1148/radiol.2018171820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mourad M, Jetmore T, Jategaonkar AA, Moubayed S, Moshier E, Urken ML. Epidemiological trends of Head and neck Cancer in the United States: a SEER population study. J Oral Maxillofac Surg. 2017;75(12):2562–2572. doi: 10.1016/j.joms.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of human papillomavirus-positive Head and neck squamous cell carcinoma. J Clin Oncol. 2015;33(29):3235–3242. doi: 10.1200/JCO.2015.61.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehanna H, Beech T, Nicholson T, El-Hariry I, McConkey C, Paleri V, et al. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer--systematic review and meta-analysis of trends by time and region. Head Neck. 2013;35(5):747–755. doi: 10.1002/hed.22015. [DOI] [PubMed] [Google Scholar]
- 42.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benson E, Li R, Eisele D, Fakhry C. The clinical impact of HPV tumor status upon head and neck squamous cell carcinomas. Oral Oncol. 2014;50(6):565–574. doi: 10.1016/j.oraloncology.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lydiatt WM, Patel SG, O'Sullivan B, Brandwein MS, Ridge JA, Migliacci JC, et al. Head and neck cancers-major changes in the American joint committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(2):122–137. doi: 10.3322/caac.21389. [DOI] [PubMed] [Google Scholar]
- 45.Glastonbury CM, Mukherji SK, O'Sullivan B, Lydiatt WM. Setting the stage for 2018: how the changes in the American joint committee on Cancer/Union for International Cancer Control Cancer staging manual eighth edition impact radiologists. AJNR Am J Neuroradiol. 2017;38(12):2231–2237. doi: 10.3174/ajnr.A5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wookey VB, Appiah AK, Kallam A, Ernani V, Smith LM, Ganti AK. HPV status and survival in non-Oropharyngeal squamous cell carcinoma of the Head and neck. Anticancer Res. 2019;39(4):1907–1914. doi: 10.21873/anticanres.13299. [DOI] [PubMed] [Google Scholar]
- 47.Burr AR, Harari PM, Ko HC, Chen S, Yu M, Baschnagel AM, et al. HPV impacts survival of stage IVC non-oropharyngeal HNSCC cancer patients. Otorhinolaryngol Head Neck Surg.2018;3(1):1–7. 10.15761/OHNS.1000160. [DOI] [PMC free article] [PubMed]
- 48.Buch K, Fujita A, Li B, Kawashima Y, Qureshi MM, Sakai O. Using texture analysis to determine human papillomavirus status of Oropharyngeal squamous cell carcinomas on CT. AJNR Am J Neuroradiol. 2015;36(7):1343–1348. doi: 10.3174/ajnr.A4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujita A, Buch K, Li B, Kawashima Y, Qureshi MM, Sakai O. Difference between HPV-positive and HPV-negative non-Oropharyngeal Head and neck Cancer: texture analysis features on CT. J Comput Assist Tomogr. 2016;40(1):43–47. doi: 10.1097/RCT.0000000000000320. [DOI] [PubMed] [Google Scholar]
- 50.Bogowicz M, Riesterer O, Ikenberg K, Stieb S, Moch H, Studer G, et al. Computed tomography Radiomics predicts HPV status and local tumor control after definitive Radiochemotherapy in Head and neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2017;99(4):921–928. doi: 10.1016/j.ijrobp.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Huang C, Cintra M, Brennan K, Zhou M, Colevas AD, Fischbein N, et al. Development and validation of radiomic signatures of head and neck squamous cell carcinoma molecular features and subtypes. EBioMedicine. 2019;45:70–80. doi: 10.1016/j.ebiom.2019.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leijenaar RT, Bogowicz M, Jochems A, Hoebers FJ, Wesseling FW, Huang SH, et al. Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: a multicenter study. Br J Radiol. 2018;91(1086):20170498. doi: 10.1259/bjr.20170498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mungai F, Verrone GB, Pietragalla M, Berti V, Addeo G, Desideri I, et al. CT assessment of tumor heterogeneity and the potential for the prediction of human papillomavirus status in oropharyngeal squamous cell carcinoma. Radiol Med. 2019;124(9):804–811. doi: 10.1007/s11547-019-01028-6. [DOI] [PubMed] [Google Scholar]
- 54.Parmar C, Leijenaar RT, Grossmann P, Rios Velazquez E, Bussink J, Rietveld D, et al. Radiomic feature clusters and prognostic signatures specific for lung and Head & Neck cancer. Sci Rep. 2015;5:11044. doi: 10.1038/srep11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ranjbar S, Ning S, Zwart CM, Wood CP, Weindling SM, Wu T, et al. Computed tomography-based texture analysis to determine human papillomavirus status of Oropharyngeal squamous cell carcinoma. J Comput Assist Tomogr. 2018;42(2):299–305. doi: 10.1097/RCT.0000000000000682. [DOI] [PubMed] [Google Scholar]
- 56.Yu K, Zhang Y, Yu Y, Huang C, Liu R, Li T, et al. Radiomic analysis in prediction of human papilloma virus status. Clin Transl Radiat Oncol. 2017;7:49–54. doi: 10.1016/j.ctro.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu Y, Mohamed ASR, Lai SY, Yang S, Kanwar A, Wei L, et al. Imaging-genomic study of Head and neck squamous cell carcinoma: associations between Radiomic phenotypes and genomic mechanisms via integration of the Cancer genome atlas and the Cancer imaging archive. JCO Clin Cancer Inform. 2019;3:1–9. doi: 10.1200/CCI.18.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Yao H, Thompson EJ, Tannir NM, Weinstein JN, Su X. VirusSeq: software to identify viruses and their integration sites using next-generation sequencing of human cancer tissue. Bioinformatics. 2013;29(2):266–267. doi: 10.1093/bioinformatics/bts665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis JS, Jr, Beadle B, Bishop JA, Chernock RD, Colasacco C, Lacchetti C, et al. Human papillomavirus testing in Head and neck carcinomas: guideline from the College of American Pathologists. Arch Pathol Lab Med. 2018;142(5):559–597. doi: 10.5858/arpa.2017-0286-CP. [DOI] [PubMed] [Google Scholar]
- 60.Vallieres M, Kumar A, Sultanem K, El Naqa I. FDG-PET Image-Derived Features Can Determine HPV Status in Head-and-Neck Cancer. Int J Radiation Oncol Biol Phys. 2013;87(2):S467. doi: 10.1016/j.ijrobp.2013.06.1236. [DOI] [Google Scholar]
- 61.Payabvash S. Quantitative diffusion magnetic resonance imaging in head and neck tumors. Quant Imaging Med Surg. 2018;8(10):1052–1065. doi: 10.21037/qims.2018.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Payabvash S, Brackett A, Forghani R, Malhotra A. Differentiation of lymphomatous, metastatic, and non-malignant lymphadenopathy in the neck with quantitative diffusion-weighted imaging: systematic review and meta-analysis. Neuroradiology. 2019;61(8):897–910. doi: 10.1007/s00234-019-02236-7. [DOI] [PubMed] [Google Scholar]
- 63.Payabvash S, Chan A, Jabehdar Maralani P, Malhotra A. Quantitative diffusion magnetic resonance imaging for prediction of human papillomavirus status in head and neck squamous-cell carcinoma: a systematic review and meta-analysis. Neuroradiol J. 2019;32(4):232–240. doi: 10.1177/1971400919849808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zwirner K, Hilke FJ, Demidov G, Socarras Fernandez J, Ossowski S, Gani C, et al. Radiogenomics in head and neck cancer: correlation of radiomic heterogeneity and somatic mutations in TP53, FAT1 and KMT2D. Strahlenther Onkol. 2019;195(9):771–779. doi: 10.1007/s00066-019-01478-x. [DOI] [PubMed] [Google Scholar]
- 65.Walter V, Yin X, Wilkerson MD, Cabanski CR, Zhao N, Du Y, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One. 2013;8(2):e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gevaert O, Tibshirani R, Plevritis SK. Pancancer analysis of DNA methylation-driven genes using MethylMix. Genome Biol. 2015;16:17. doi: 10.1186/s13059-014-0579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moskovitz J, Moy J, Ferris RL. Immunotherapy for Head and neck squamous cell carcinoma. Curr Oncol Rep. 2018;20(2):22. doi: 10.1007/s11912-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20(19):5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferris RL, Blumenschein G, Jr, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the Head and neck. N Engl J Med. 2016;375(19):1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larkins E, Blumenthal GM, Yuan W, He K, Sridhara R, Subramaniam S, et al. FDA approval summary: Pembrolizumab for the treatment of recurrent or metastatic Head and neck squamous cell carcinoma with disease progression on or after platinum-containing chemotherapy. Oncologist. 2017;22(7):873–878. doi: 10.1634/theoncologist.2016-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oliva M, Spreafico A, Taberna M, Alemany L, Coburn B, Mesia R, et al. Immune biomarkers of response to immune-checkpoint inhibitors in head and neck squamous cell carcinoma. Ann Oncol. 2019;30(1):57–67. doi: 10.1093/annonc/mdy507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in Cancer immunotherapy. Mol Cancer Ther. 2015;14(4):847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 73.Chen RY, Lin YC, Shen WC, Hsieh TC, Yen KY, Chen SW, et al. Associations of tumor PD-1 ligands, Immunohistochemical studies, and textural features in (18)F-FDG PET in squamous cell carcinoma of the Head and neck. Sci Rep. 2018;8(1):105. doi: 10.1038/s41598-017-18489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 75.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 76.Gatta G, Botta L, Sanchez MJ, Anderson LA, Pierannunzio D, Licitra L, et al. Prognoses and improvement for head and neck cancers diagnosed in Europe in early 2000s: the EUROCARE-5 population-based study. Eur J Cancer. 2015;51(15):2130–2143. doi: 10.1016/j.ejca.2015.07.043. [DOI] [PubMed] [Google Scholar]
- 77.Brouha XD, Tromp DM, De Leeuw JR, Hordijk GJ, Winnubst JA. Increasing incidence of advanced stage head and neck tumours. Clin Otolaryngol Allied Sci. 2003;28(3):231–234. doi: 10.1046/j.1365-2273.2003.00696.x. [DOI] [PubMed] [Google Scholar]
- 78.Bogowicz M, Leijenaar RTH, Tanadini-Lang S, Riesterer O, Pruschy M, Studer G, et al. Post-radiochemotherapy PET radiomics in head and neck cancer - the influence of radiomics implementation on the reproducibility of local control tumor models. Radiother Oncol. 2017;125(3):385–391. doi: 10.1016/j.radonc.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 79.Bogowicz M, Tanadini-Lang S, Guckenberger M, Riesterer O. Combined CT radiomics of primary tumor and metastatic lymph nodes improves prediction of loco-regional control in head and neck cancer. Sci Rep. 2019;9(1):15198. doi: 10.1038/s41598-019-51599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bogowicz M, Tanadini-Lang S, Veit-Haibach P, Pruschy M, Bender S, Sharma A, et al. Perfusion CT radiomics as potential prognostic biomarker in head and neck squamous cell carcinoma. Acta Oncol. 2019;58(10):1514–1518. doi: 10.1080/0284186X.2019.1629013. [DOI] [PubMed] [Google Scholar]
- 81.Cheng NM, Fang YH, Chang JT, Huang CG, Tsan DL, Ng SH, et al. Textural features of pretreatment 18F-FDG PET/CT images: prognostic significance in patients with advanced T-stage oropharyngeal squamous cell carcinoma. J Nucl Med. 2013;54(10):1703–1709. doi: 10.2967/jnumed.112.119289. [DOI] [PubMed] [Google Scholar]
- 82.Cheng NM, Fang YH, Lee LY, Chang JT, Tsan DL, Ng SH, et al. Zone-size nonuniformity of 18F-FDG PET regional textural features predicts survival in patients with oropharyngeal cancer. Eur J Nucl Med Mol Imaging. 2015;42(3):419–428. doi: 10.1007/s00259-014-2933-1. [DOI] [PubMed] [Google Scholar]
- 83.Cozzi L, Franzese C, Fogliata A, Franceschini D, Navarria P, Tomatis S, et al. Predicting survival and local control after radiochemotherapy in locally advanced head and neck cancer by means of computed tomography based radiomics. Strahlenther Onkol. 2019;195(9):805–818. doi: 10.1007/s00066-019-01483-0. [DOI] [PubMed] [Google Scholar]
- 84.Feliciani G, Fioroni F, Grassi E, Bertolini M, Rosca A, Timon G, et al. Radiomic profiling of Head and neck Cancer: (18)F-FDG PET texture analysis as predictor of patient survival. Contrast Media Mol Imaging. 2018;2018:3574310. doi: 10.1155/2018/3574310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Folkert MR, Setton J, Apte AP, Grkovski M, Young RJ, Schoder H, et al. Predictive modeling of outcomes following definitive chemoradiotherapy for oropharyngeal cancer based on FDG-PET image characteristics. Phys Med Biol. 2017;62(13):5327–5343. doi: 10.1088/1361-6560/aa73cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuno H, Qureshi MM, Chapman MN, Li B, Andreu-Arasa VC, Onoue K, et al. CT texture analysis potentially predicts local failure in Head and neck squamous cell carcinoma treated with Chemoradiotherapy. AJNR Am J Neuroradiol. 2017;38(12):2334–2340. doi: 10.3174/ajnr.A5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leijenaar RT, Carvalho S, Hoebers FJ, Aerts HJ, van Elmpt WJ, Huang SH, et al. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol. 2015;54(9):1423–1429. doi: 10.3109/0284186X.2015.1061214. [DOI] [PubMed] [Google Scholar]
- 88.Lv W, Ashrafinia S, Ma J, Lu L, Rahmim A. Multi-level multi-modality fusion radiomics: application to PET and CT imaging for prognostication of head and neck cancer. IEEE J Biomed Health Inform. 2019. [Epub ahead of print]. [DOI] [PubMed]
- 89.Mo X, Wu X, Dong D, Guo B, Liang C, Luo X, et al. Prognostic value of the radiomics-based model in progression-free survival of hypopharyngeal cancer treated with chemoradiation. Eur Radiol. 2020;30:833–843. 10.1007/s00330-019-06452-w. [DOI] [PubMed]
- 90.Ou D, Blanchard P, Rosellini S, Levy A, Nguyen F, Leijenaar RTH, et al. Predictive and prognostic value of CT based radiomics signature in locally advanced head and neck cancers patients treated with concurrent chemoradiotherapy or bioradiotherapy and its added value to human papillomavirus status. Oral Oncol. 2017;71:150–155. doi: 10.1016/j.oraloncology.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 91.Ouyang FS, Guo BL, Zhang B, Dong YH, Zhang L, Mo XK, et al. Exploration and validation of radiomics signature as an independent prognostic biomarker in stage III-IVb nasopharyngeal carcinoma. Oncotarget. 2017;8(43):74869–74879. doi: 10.18632/oncotarget.20423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ulrich EJ, Menda Y, Boles Ponto LL, Anderson CM, Smith BJ, Sunderland JJ, et al. FLT PET Radiomics for response prediction to Chemoradiation therapy in Head and neck squamous cell Cancer. Tomography. 2019;5(1):161–169. doi: 10.18383/j.tom.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhai TT, Langendijk JA, van Dijk LV, Halmos GB, Witjes MJH, Oosting SF, et al. The prognostic value of CT-based image-biomarkers for head and neck cancer patients treated with definitive (chemo-)radiation. Oral Oncol. 2019;95:178–186. doi: 10.1016/j.oraloncology.2019.06.020. [DOI] [PubMed] [Google Scholar]
- 94.Zhang B, Ouyang F, Gu D, Dong Y, Zhang L, Mo X, et al. Advanced nasopharyngeal carcinoma: pre-treatment prediction of progression based on multi-parametric MRI radiomics. Oncotarget. 2017;8(42):72457–72465. doi: 10.18632/oncotarget.19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang B, He X, Ouyang F, Gu D, Dong Y, Zhang L, et al. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. 2017;403:21–27. doi: 10.1016/j.canlet.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 96.Zhang B, Tian J, Dong D, Gu D, Dong Y, Zhang L, et al. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res. 2017;23(15):4259–4269. doi: 10.1158/1078-0432.CCR-16-2910. [DOI] [PubMed] [Google Scholar]
- 97.Guha A, Connor S, Anjari M, Naik H, Siddiqui M, Cook G, et al. Radiomic analysis for response assessment in advanced head and neck cancers, a distant dream or an inevitable reality? A systematic review of the current level of evidence. Br J Radiol. 2019;93:20190496. doi: 10.1259/bjr.20190496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bernier J, Cooper JS, Pajak TF, van Glabbeke M, Bourhis J, Forastiere A, et al. Defining risk levels in locally advanced head and neck cancers: a comparative analysis of concurrent postoperative radiation plus chemotherapy trials of the EORTC (#22931) and RTOG (# 9501) Head Neck. 2005;27(10):843–850. doi: 10.1002/hed.20279. [DOI] [PubMed] [Google Scholar]
- 99.Cooper JS, Zhang Q, Pajak TF, Forastiere AA, Jacobs J, Saxman SB, et al. Long-term follow-up of the RTOG 9501/intergroup phase III trial: postoperative concurrent radiation therapy and chemotherapy in high-risk squamous cell carcinoma of the head and neck. Int J Radiat Oncol Biol Phys. 2012;84(5):1198–1205. doi: 10.1016/j.ijrobp.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bernier J, Domenge C, Ozsahin M, Matuszewska K, Lefebvre JL, Greiner RH, et al. Postoperative irradiation with or without concomitant chemotherapy for locally advanced head and neck cancer. N Engl J Med. 2004;350(19):1945–1952. doi: 10.1056/NEJMoa032641. [DOI] [PubMed] [Google Scholar]
- 101.Oosting SF, Haddad RI. Best practice in systemic therapy for Head and neck squamous cell carcinoma. Front Oncol. 2019;9:815. doi: 10.3389/fonc.2019.00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sethia R, Yumusakhuylu AC, Ozbay I, Diavolitsis V, Brown NV, Zhao S, et al. Quality of life outcomes of transoral robotic surgery with or without adjuvant therapy for oropharyngeal cancer. Laryngoscope. 2018;128(2):403–411. doi: 10.1002/lary.26796. [DOI] [PubMed] [Google Scholar]
- 103.Mermod M, Tolstonog G, Simon C, Monnier Y. Extracapsular spread in head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2016;62:60–71. doi: 10.1016/j.oraloncology.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 104.Patel MR, Hudgins PA, Beitler JJ, Magliocca KR, Griffith CC, Liu Y, et al. Radiographic imaging does not reliably predict macroscopic Extranodal extension in human papilloma virus-associated Oropharyngeal Cancer. ORL J Otorhinolaryngol Relat Spec. 2018;80(2):85–95. doi: 10.1159/000487239. [DOI] [PubMed] [Google Scholar]
- 105.Maxwell JH, Rath TJ, Byrd JK, Albergotti WG, Wang H, Duvvuri U, et al. Accuracy of computed tomography to predict extracapsular spread in p16-positive squamous cell carcinoma. Laryngoscope. 2015;125(7):1613–1618. doi: 10.1002/lary.25140. [DOI] [PubMed] [Google Scholar]
- 106.Kann BH, Aneja S, Loganadane GV, Kelly JR, Smith SM, Decker RH, et al. Pretreatment identification of Head and neck Cancer nodal metastasis and Extranodal extension using deep learning neural networks. Sci Rep. 2018;8(1):14036. doi: 10.1038/s41598-018-32441-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kann BH, Hicks DF, Payabvash S, Mahajan A, Du J, Gupta V, et al. Multi-Institutional Validation of Deep Learning for Pretreatment Identification of Extranodal Extension in Head and Neck Squamous Cell Carcinoma. J Clin Oncol. 2019:JCO1902031. [Epub ahead of print]. [DOI] [PubMed]
- 108.Dirix P, Nuyts S. Evidence-based organ-sparing radiotherapy in head and neck cancer. Lancet Oncol. 2010;11(1):85–91. doi: 10.1016/S1470-2045(09)70231-1. [DOI] [PubMed] [Google Scholar]
- 109.Hawkins PG, Lee JY, Mao Y, Li P, Green M, Worden FP, et al. Sparing all salivary glands with IMRT for head and neck cancer: longitudinal study of patient-reported xerostomia and head-and-neck quality of life. Radiother Oncol. 2018;126(1):68–74. doi: 10.1016/j.radonc.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 110.Sheikh K, Lee SH, Cheng Z, Lakshminarayanan P, Peng L, Han P, et al. Predicting acute radiation induced xerostomia in head and neck Cancer using MR and CT Radiomics of parotid and submandibular glands. Radiat Oncol. 2019;14(1):131. doi: 10.1186/s13014-019-1339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Y, Shi H, Huang S, Chen X, Zhou H, Chang H, et al. Early prediction of acute xerostomia during radiation therapy for nasopharyngeal cancer based on delta radiomics from CT images. Quant Imaging Med Surg. 2019;9(7):1288–1302. doi: 10.21037/qims.2019.07.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.van Dijk LV, Thor M, Steenbakkers R, Apte A, Zhai TT, Borra R, et al. Parotid gland fat related magnetic resonance image biomarkers improve prediction of late radiation-induced xerostomia. Radiother Oncol. 2018;128(3):459–466. doi: 10.1016/j.radonc.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van Dijk LV, Brouwer CL, van der Schaaf A, Burgerhof JGM, Beukinga RJ, Langendijk JA, et al. CT image biomarkers to improve patient-specific prediction of radiation-induced xerostomia and sticky saliva. Radiother Oncol. 2017;122(2):185–191. doi: 10.1016/j.radonc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 114.van Dijk LV, Noordzij W, Brouwer CL, Boellaard R, Burgerhof JGM, Langendijk JA, et al. (18)F-FDG PET image biomarkers improve prediction of late radiation-induced xerostomia. Radiother Oncol. 2018;126(1):89–95. doi: 10.1016/j.radonc.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 115.U.S. Department of Health and Human Services NIH NCI . Common Terminology Criteria for Adverse Events (CTCAE) Version 4.0. 2010. [Google Scholar]
- 116.Bjordal K, Ahlner-Elmqvist M, Tollesson E, Jensen AB, Razavi D, Maher EJ, et al. Development of a European Organization for Research and Treatment of Cancer (EORTC) questionnaire module to be used in quality of life assessments in head and neck cancer patients. EORTC quality of life study group. Acta Oncol. 1994;33(8):879–885. doi: 10.3109/02841869409098450. [DOI] [PubMed] [Google Scholar]
- 117.Epstein JB, Thariat J, Bensadoun RJ, Barasch A, Murphy BA, Kolnick L, et al. Oral complications of cancer and cancer therapy: from cancer treatment to survivorship. CA Cancer J Clin. 2012;62(6):400–422. doi: 10.3322/caac.21157. [DOI] [PubMed] [Google Scholar]
- 118.Rapidis AD, Dijkstra PU, Roodenburg JL, Rodrigo JP, Rinaldo A, Strojan P, et al. Trismus in patients with head and neck cancer: etiopathogenesis, diagnosis and management. Clin Otolaryngol. 2015;40(6):516–526. doi: 10.1111/coa.12488. [DOI] [PubMed] [Google Scholar]
- 119.Thor M, Tyagi N, Hatzoglou V, Apte A, Saleh Z, Riaz N, et al. A magnetic resonance imaging-based approach to quantify radiation-induced normal tissue injuries applied to trismus in head and neck cancer. Phys Imaging Radiat Oncol. 2017;1:34–40. doi: 10.1016/j.phro.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Abdollahi H, Mostafaei S, Cheraghi S, Shiri I, Rabi Mahdavi S, Kazemnejad A. Cochlea CT radiomics predicts chemoradiotherapy induced sensorineural hearing loss in head and neck cancer patients: a machine learning and multi-variable modelling study. Phys Med. 2018;45:192–197. doi: 10.1016/j.ejmp.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 121.Langlotz CP, Allen B, Erickson BJ, Kalpathy-Cramer J, Bigelow K, Cook TS, et al. A roadmap for foundational research on artificial intelligence in medical imaging: from the 2018 NIH/RSNA/ACR/the academy workshop. Radiology. 2019;291(3):781–791. doi: 10.1148/radiol.2019190613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim DW, Jang HY, Kim KW, Shin Y, Park SH. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol. 2019;20(3):405–410. doi: 10.3348/kjr.2019.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.