Abstract
Background
Host genetic polymorphisms may be important in determining susceptibility to Mycobacterium tuberculosis (Mtb) infection, but their role is not fully understood. Detection of microbial DNA and activation of type I interferon (IFN) pathways regulate macrophage responses to Mtb infection.
Methods
We examined whether seven candidate gene SNPs were associated with tuberculin skin test (TST) positivity in close contacts of microbiologically confirmed pulmonary TB patients in Brazil. Independent associations with TST positivity were tested using multivariable logistic regression (using genotypes and clinical variables) and genetic models.
Results
Among 482 contacts of 145 TB index cases, 296 contacts were TST positive. Multivariable regression analysis adjusted for population admixture, age, family relatedness, sex and clinical variables related to increased TB risk demonstrated that SNPs in PYHIN1-IFI16-AIM2 rs1101998 (adjusted OR [aOR]: 3.72; 95% CI = 1.15–12.0; p = 0.028) and in PYHIN1-IFI16-AIM2 rs1633256 (aOR = 24.84; 95%CI = 2.26–272.95; p = 0.009) were associated with TST positivity in a recessive model. Furthermore, an IRF7 polymorphism (rs11246213) was associated with reduced odds of TST positivity in a dominant model (aOR: 0.50, 95%CI: 0.26–0.93; p = 0.029).
Conclusions
Polymorphisms in PYHIN1-IFI16-AIM2 rs1633256, rs1101998 and in IRF7 rs11246213 were associated with altered susceptibility to Mtb infection in this Brazilian cohort.
Keywords: Single nucleotide polymorphism, Tuberculin skin test, Mycobacterium tuberculosis
Introduction
Tuberculosis (TB) is the leading cause of death from a single infectious agent (WHO, 2018). Approximately one-quarter of the global population is infected with Mycobacterium tuberculosis (Mtb) (Houben and Dodd, 2016). Latent tuberculosis infection (LTBI) is defined by immunological sensitization to Mtb antigens in the absence of clinical symptoms of disease and the diagnosis is based on the tuberculin skin test (TST) and/or Interferon-γ(IFN-γ) release assay (IGRA) (Robertson et al., 2012). Nevertheless, these tests do not discriminate between active disease and LTBI and, more importantly, have a low predictive value for progression to active TB (Rangaka et al., 2012). Many risk factors for developing active TB have been described, including HIV co-infection, diabetes, young age and recently acquired Mtb infection (Reid et al., 2019), but intriguingly some TB patients do not exhibit any known risk factors (Yan et al., 2015). TB occurs as the result of an intricate and dynamic relationship involving host genetics (van Tong et al., 2017) as well as immunological (Mahan et al., 2012; Tameris et al., 2013), and epidemiological (Shin et al., 2016) factors, in addition to characteristics of the Mtb strain itself (Koch and Mizrahi, 2018), that contribute to disease susceptibility (Pai et al., 2016).
Genetic factors are important for TB susceptibility, but the major genes involved remain unknown (van Tong et al., 2017). Candidate gene/pathway studies interrogate selected pathways that are important in the human host response to mycobacterial infection (Kinnear et al., 2017). Type I IFN pathways mediate an important role in TB pathogenesis. Whole blood RNA signatures dominated by Type I IFN-signaling identify individuals who will develop active disease (Berry et al., 2010). In Mtb-infected mice, increased expression of type I IFNs is deleterious for survival in association with reduced Th1 immunity (Manca et al., 2005). The Type I IFN pathway is activated by DNA (e.g. IFI16-PYHIN1-AIM2, cGAS, STING) and RNA sensors (e.g. IFIT1 and 5), and contains several important signaling molecules and transcription factors (e.g. IRF family). For example, the cytosolic DNA sensor cGAS regulates IFN production during Mtb infection of macrophages (Watson et al., 2015). Although these murine and cellular studies suggest an important role for Type I IFNs in TB pathogenesis, the human genetics of this pathway in the context of Mtb infection are poorly understood (Donovan et al., 2017).
In a longitudinal investigation examining TB contacts from Brazil, we recently found that polymorphisms in toll-like receptor 4 (TLR4) and tumor necrosis factor (TNFA) are associated with increased risk of TST conversion and development of active TB (Cubillos-Angulo et al., 2019). Here we investigated in this same cohort whether genetic variation of Type I IFN pathway genes were associated with susceptibility to Mtb infection by examining single nucleotide polymorphisms (SNPs) involved in DNA and RNA sensing: (rs1101998, rs1633256, rs866484 in IFI16-PYHIN1-AIM2 region, rs59633641 and rs10887959 in IFIT5), rs304478 and rs304498 in IFIT1 and the IFN signaling pathway (rs11246213 [IRF7]). The objective of this study was to identify potential genetic biomarkers of susceptibility to Mtb infection. We studied close contacts of microbiologically confirmed pulmonary TB patients to estimate factors associated with a positive versus negative TST.
Methods
Study design
The present study was based on analyses performed retrospectively on a cohort of contacts of pulmonary TB patients, recruited between November 1998 through March 2004. The parent study was reported previously (Cubillos-Angulo et al., 2019). The cases and controls were enrolled in the state of Rio de Janeiro, Brazil where the population is mostly white and brown (‘parda, mixed ethnic ancestries) (IBGE, 2012). Racial/ethnic background was self-reported and used the definitions/approaches employed by the Brazilian government for race documentation. TB index cases were diagnosed by acid-fast bacilli (AFB) smear and/or culture, according to Brazilian Ministry of Health Guidelines (Ministério da Saúde, Brasil, 2019). TB index case variables included cough, AFB sputum grade, and chest radiographs. TB contacts were defined as living in the same household or reporting contact with the TB index case for >20 h weekly for 2 months (Cubillos-Angulo et al., 2019). In the analyses presented here, we used data from a subgroup of 482 individuals, which were selected by such criteria and included contacts with TST-positive or TST-negative results. Patients who developed active TB were excluded from the analysis. Additional details on inclusion and exclusion criteria as well as patient characteristics have been described previously (Cubillos-Angulo et al., 2019).
A standardized questionnaire was administered to obtain demographic and clinical data, including a history of risk factors for TB (e.g., HIV, diabetes, hematologic malignancies, and use of immunosuppressant drugs) and duration of contact with the index case. Consanguinity was considered if a contact was a grandparent, parent or sibling of the index case, where as spouses or other relationships were not. At study baseline, a medical visit and chest radiograph were performed. BCG scar was assessed and TST reading was performed 48–72 h after administration at baseline, using 2 tuberculin units of the purified protein derivative RT 23 (Statens Serum Institute, Copenhagen, Denmark).
TST interpretation and TB diagnosis
A positive TST was defined as an induration larger than ≥5 mm induration, according to the Brazilian Ministry of Health (Ministério da Saúde, Brasil, 2019). Contacts with any TST ≥5 mm were not re-tested with TST. The Brazilian National TB Guidelines indicated that treatment of TST-positive individuals was systematically offered but implementation was not mandatory during the study period (Ministério da Saúde, Brasil, 2019). For the index case, active TB was diagnosed when ≥1 specimen yielded a positive microbiologic (AFB smear or culture) result by AFB smear and/or culture in Lowenstein Jensen (LJ) medium (Cubillos-Angulo et al., 2019).
Genotyping
Genomic DNA was extracted from peripheral blood collected from TB contacts at study enrollment. DNA extraction and genotyping were performed using the Flexi Gene kit (Qiagen, Germany). Genotypes of 8 gene polymorphisms were chosen for convenience since an RFLP assay was available: rs1101998 (IFI16-PYHIN1-AIM2), rs1633256 (IFI16-PYHIN1-AIM2), rs866484 (IFI16-PYHIN1-AIM2), rs304478 (IFIT1), rs304498 (IFIT1), rs11246213 (IRF7), rs59633641 (IFIT5) and rs10887959 (IFIT5) were detected using polymerase chain reaction restriction fragment length polymorphism (RFLP) method (Cubillos-Angulo et al., 2019). The primer sequences are in Supplementary Table S1. The PCR products were digested by the enzymes EcoRII for rs1101998 (IFI16), AgsI for rs1633256 (IFI16), AgsI for rs866484 (IFI16), AarI for rs304478 (IFIT1), TfiI for rs304498 (IFIT1), BsaAI for rs11246213 (IRF7), ApoI for rs59633641 (IFIT5) and AgsI for rs10887959 (IFIT5). Hardy-Weinberg equilibrium was tested for each SNP. We did not find significant deviation from Hardy Weinberg equilibrium except in rs304498 (IFIT1), and thus this SNP was excluded from further analysis. Linkage disequilibrium coefficients were calculated using Package “LDheatmap” (Shin et al., 2006) in the stats package in R 3.5.2 and using an R2 and D’ cutoff of 0.8. Haplotypes analysis were constructed in the stats package R 3.5.2 using the haplo.stats (version 1.6.0) R package (Sinnwell and Schaid, 2018).
Data analysis
Categorical data were presented as proportions and continuous data as medians and interquartile ranges (IQR). For clinical characteristics, a Fisher’s exact test was used to perform 2 × 2 comparisons. Continuous variables were compared using the Mann–Whitney U test. For genetic analysis, a Cochrane-Armitage test for trend was used initially to examine the association of genotypes with TST positivity. SNPs were then evaluated with a Fisher’s exact test using dominant (00 vs 01/11) and recessive (00/01 vs 11) models. We also estimated significant associations between indicated SNPs and TST positivity using multivariable logistic regression adjusted for race/ethnicity, family relatedness, gender and age in both dominant and recessive models. Finally, we also performed additional investigations with dominant and recessive models in a multivariable analysis with adjustment for age, gender, race/ethnicity, family relatedness, household contact status and characteristics of TB index case, such as cavities on chest X-ray, ≥ 2+ AFB sputum smear grade and positive sputum culture for Mtb. We also used the GTEx portal (https://gtexportal.org/home/) to evaluate the expression quantitative trait loci (eQTL) of the SNPs (Consortium, 2013). Furthermore, the likelihood of being a regulatory SNP was examined using the RegulomeDB dataset (http://www.regulomedb.org/snp/chr10/91150921 ) (Boyle et al., 2012).
Results
Characteristics of the study participants
We used a re trospective cohort study of contacts (N = 482) of pulmonary TB index cases (N = 145) to examine whether genetic variants of candidate genes were associated with TST positivity. Household contacts were more frequently observed in the group of individuals presenting with a positive TST result than in those with a negative TST (Table 1). Cavitary lesions as well as cough in the index TB cases were more frequent in participants who were TST positive compared to those who had negative results (p = 0.04 and p = 0.008, respectively). Other characteristics were similar between TST positive and TST negative individuals.
Table 1.
Clinical characteristics of the study participants and association with tuberculin skin test (TST) positivity.
| Characteristic | n | TST negative n = 219 | TST positive n = 263 | P-value |
|---|---|---|---|---|
| Age -median (IQR) | 482 | 34 (23–50) | 37 (24–49) | 0.40 |
| Male sex | 482 | 74 (34) | 87 (33) | 0.92 |
| First-degree relative of the index case | 482 | 138 (63) | 161 (61) | 0.71 |
| HIV infection | 21 | 2 (67) | 1 (6) | 0.04 |
| Race (% white) | 462 | 113 (53) | 144 (58) | 0.40 |
| Illicit drug usea | 412 | 3 (2) | 3 (1) | 1.00 |
| Prior tuberculosis | 411 | 0 (0) | 3 (1.4) | 0.25 |
| Household contact | 480 | 190 (88) | 244 (93) | 0.06 |
| Duration of contact (>20 h) | 482 | 202 (92) | 248 (94) | 0.46 |
| Comorbid conditionsb | 459 | 53 (26) | 65 (26) | 1.00 |
| Immunosuppressant drugsc | 414 | 0 (0) | 2 (1) | 0.50 |
| Cough (>4 weeks) | 481 | 5 (2) | 5 (2) | 0.76 |
| Characteristics of TB index case | ||||
| Cavities on chest X-ray | 473 | 24 (11) | 47 (18) | 0.04 |
| Cough (>4 weeks) | 481 | 86 (39) | 136 (52) | 0.008 |
| ≥2 AFB sputum smear | 443 | 77 (39) | 103 (42) | 0.44 |
| Mtb positive culture | 339 | 147 (94) | 178 (97) | 0.18 |
“n” is the number of TB contacts for whom such data were available, out of a total of 482 included in the study. Data represents no. (%) or median and interquartile range (IQR) and were compared using the Fisher’s exact test (categorical variables) or the Mann–Whitney U test (for age). TST: tuberculin skin test; AFB: acid-fast bacilli on sputum smear, CI: confidence interval; OR: odds ratio.
Illicit drugs: cannabis, cocaine, or crack.
Co-morbid conditions: diabetes mellitus, heart failure, chronic obstructive pulmonary disease, neoplasia, systemic lupus erythematous and hepatitis.
Immunosuppressant drugs: corticosteroids, tumor necrosis factor blockers, calcineurin inhibitors, or interleukin inhibitors.
The study population was mostly female (n = 321, 67%), with a high frequency of first degree relatives with the index case (n = 229, 62%) (Table 1). In addition, the vast majority of participants were household contacts (n = 434, 90%). There were low frequencies of HIV infection, illicit drug use, prior TB and use of immune suppressive drugs. Approximately 97% (n = 141) of the index cases had TB confirmed by culture. TB index cases frequently reported cough for more than 4 weeks (80%) and had high bacterial loads in sputum (60% had AFB grade ≥ +2;). In addition, 100 index TB patients had cavitary lesions on chest radiograph.
Association between polymorphisms and TST positivity
Two of seven polymorphisms were associated with TST positivity (rs1633256 and rs59633641 with an unadjusted genotypic trend test, Table 2). PYHIN1-IFI16-AIM2 SNPs rs1101998 allele C (p = 0.01) and rs1633256 allele A (p = <0.01) were more common in TST positive participants and fit a recessive model (Table 2). IFIT5 rs59633641 allele G (p = 0.04) was more common in TST positive individuals (trend test p = 0.04, Table 2). IFIT1 rs304478 and IFIT5 rs10887959 were also significantly associated with outcomes in recessive and dominant models, respectively.
Table 2.
Association between candidate gene polymorphisms and TST.
| SNP | Genotype Frequency in TST negative |
Genotype Frequency in TST positive |
P-value |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 00 | 01 | 11 | Total | 00 | 01 | 11 | Total | HWE | Genotypic Trendb | Dominant 00 vs 01/11 | Recessive 00/01 vs 11 | |
| rs1101998 - PYHIN1- IFI16-AIM2 (T/C) | 0.43 | 0.49 | 0.09 | 105 | 0.43 | 0.35 | 0.22 | 102 | 0.21 | 0.20 | 1.00 | 0.01 |
| rs1633256 - PYHIN1- IFI16-AIM2 (G/A) | 0.56 | 0.41 | 0.03 | 80 | 0.53 | 0.26 | 0.22 | 78 | 0.02 | 0.04 | 0.75 | <0.01 |
| rs866484 - PYHIN1- IFI16-AIM2 (C/G) | 0.55 | 0.36 | 0.09 | 120 | 0.49 | 0.39 | 0.13 | 127 | 0.1 | 0.27 | 0.37 | 0.42 |
| rs304478 - IFIT1 (T/G) | 0.37 | 0.47 | 0.17 | 131 | 0.44 | 0.48 | 0.08 | 135 | 0.66 | 0.06 | 0.26 | 0.04 |
| rs59633641 - IFIT5a (C/G) | 0.97 | 0.03 | 0 | 121 | 0.9 | 0.1 | 0 | 117 | 0.59 | 0.04 | – | – |
| rs10887959 - IFIT5 (C/T) | 0.64 | 0.3 | 0.06 | 115 | 0.46 | 0.46 | 0.08 | 119 | 1.00 | 0.06 | 0.009 | 0.80 |
| rs11246213 - IRF7 (A/G) | 0.48 | 0.4 | 0.12 | 126 | 0.37 | 0.46 | 0.17 | 133 | 0.25 | 0.07 | 0.08 | 0.29 |
Data represent genotype frequency of SNP TST: tuberculin skin test; SNP: single-nucleotide polymorphism; 00, homozygous common allele; 01, heterozygous allele; 11, homozygous rare allele. HWE: Hardy Weinberg equilibrium. Data were analyzed using the Fisher’s exact test (2 × 2 comparisons) or the chi-square trend test (3 × 2 comparisons).
No uncommon homozygous mutation; in this particular case, the test employed for the genotypic analysis was based on 2 × 2 comparison. P-value represents comparison of genotype frequencies without adjustment for any covariates.
Cochrane-Armitage trend test.
In a multivariable model that included adjustment for race/ethnicity, family relatedness, gender, and age (Figure 1), we observed in the recessive model that PYHIN1-IFI16-AIM2 rs1101998 (adjusted OR [aOR] = 2.90; 95%CI = 1.24–6.78; p = 0.014) and rs1633256 (aOR = 10.1; 95%CI = 2.20–46.28; p = 0.003) were associated with an increasedrisk of TSTpositivity. Moreover, in the dominant model, IFIT5 rs10887959 (aOR = 0.49; 95%CI = 0.28–0.84; p = 0.01) and IRF7 rs11246213 (aOR = 0.60; 95% CI = 0.36–1.00; p = 0.049) were also linked to a lower likelihood of positive TST.
Figure 1. Multivariable model of association between genetic variants and TST positivity.
Analysis in all study participants Data represent no. SNP: single-nucleotide polymorphism; OR: odds ratio, 95% CI: confidence interval; P-value represents comparison of genotype frequencies in a dominant and recessive model with adjustment for race/ethnicity, family relatedness, gender, and age. OR (Odds ratio) represents association of minor allele with risk of TST positivity.
We next used a multivariable regression analysis to adjust for household contact and characteristics of TB index case (cavities on chest X-ray, ≥2 AFB sputum smear and positive Mtb culture) as well as race/ethnicity, family relatedness, gender, and age (Figure 2). We confirmed in the recessive model that PYHIN1-IFI16-AIM2 rs1101998 (aOR = 3.72; 95%CI = 1.15–12.0; p = 0.028) and rs1633256 (aOR = 24.84; 95%CI = 2.26–272.95; p = <0.009) were independently associated with increased odds of a positive TST. In addition, in the dominant model, IRF7 rs11246213 was also independently associated with a lower likelihood of a positive TST (aOR: 0.50, 95%CI: 0.26–0.93; p = 0.029).
Figure 2. Multivariable model of association between genetic variants and TST positivity including clinical variables.
Analysis in all study participants Data represent no. SNP: single-nucleotide polymorphism; OR: odds ratio, 95% CI: confidence interval; P-value represents comparison of genotype frequencies in a dominant and recessive model with adjustment for age, gender, race/ethnicity, family relatedness, household contact and characteristics of TB index case: Cavities on chest X-ray, ≥ 2 AFB sputum smear and positive Mtb culture.
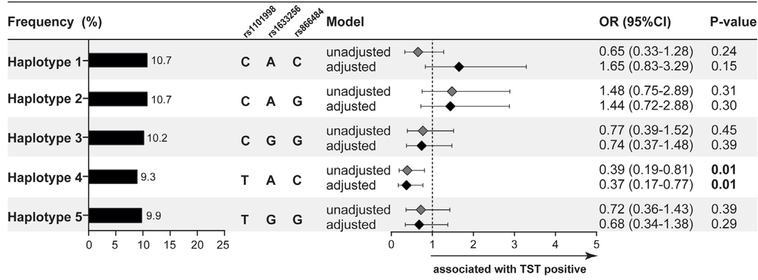
We next examined effects of linkage disequilibrium and SNP-SNP interactions in the PYHIN1-IFI16-AIM2 region on chromosome 1. PYHIN1-IFI16-AIM2 SNPs rs8666484, rs1101998 and rs1633256 were all in moderate to high linkage disequilibrium (Supplemental Figure S1). In a haplotype analysis of chromosome 1 adjusted for age, gender, race/ethnicity, family relatedness and household contact, the haplotypes containing allele C from rs1101998 and allele A from rs1633256 did not have a higher risk of TST positivity compared to single SNP analyses (Figure 3 compared to Figure 2).
Figure 3. Haplotype analysis chromosome 1.
Haplotype analysis chromosome 1 of SNPs rs1101998, rs1633256, rs866484- PYHIN1- IFI16-AIM2. P-value represents comparison of haplotype frequencies with TST conversion in an unadjusted and adjusted model for age, gender, race/ethnicity, family relatedness and household contact.
Using an in silico approach with data from the GTEx portal tool (see Methods for details and also in (Consortium, 013), we found that six polymorphisms (rs1101998, rs1633256, rs866484, rs304478, rs10887959 and rs11246213) were eQTLs in different tissues (Supplementary Table S2). Interestingly, three different SNPs were reported to be expressed in the spleen and/or lung, which are organs commonly affected by TB (Figure 4). The findings indicated that the PYHIN1-IFI16-AIM2 rs1101998 genotype CC was linked to decreased expression of AIM2 in spleen (Figure 4). The PYHIN1-IFI16-AIM2 rs1633256 genotype AA was also associated with dampened expression of AIM2 in spleen tissue (Figure 4). The IFIT5 rs10887959 genotype CC was associated with lower expression of IFIT5 in spleen and lung tissues (Figure 4). Finally, using a different online tool, the RegulomeDB dataset, we observed that PYHIN1-IFI16-AIM2 rs1101998 exhibited high likelihood of being a regulatory SNP for a DNAase I hypersensitivity peak or transcription factor binding. Moreover, IFIT5 rs10887959 displayed a high likelihood of being a regulatory SNP for transcription factor binding and a DNAase I hypersensitivity peak. Together, these data suggest that rs1101998, rs1633256, and rs10887959 are eQTLs.
Figure 4. In silico expression of SNPs rs1101998, rs1633256 and rs10887959 adapted from GTEx eQTL database.
Normalized expression values were obtained from the GTEx eQTL database and violin plots (with median and interquartile range values) were used to represent the trends in data variation between the different SNPs. The full list of the SNPs and tissues evaluated is described in the Supplementary Table S2. The Figures describe the SNPs that had publicly available data on expression in spleen and/or lungs, due to its importance in TB pathogenesis. Thus, data on the SNPs rs1101998, rs1633256 and rs10887959 are shown. Allele frequency was determined as the following: 00, homozygous common allele; 01, heterozygous allele; 11, homozygous rare allele. A summary of the results of the analysis from the present study testing the association between each indicated allele and a positive TST result is shown at the bottom of the graphs. A star denotes statistically significant associations with TST positivity in the following conditions: (i) Univariate analysis: a comparison of genotype frequencies without adjustment for any covariates; (ii) Adjusted model 1: analysis in a dominant and recessive model with adjustment for race/ethnicity, family relatedness, sex, and age; and (iii) Adjusted model 2: analysis in a dominant and recessive model with adjustment for age, sex, race/ethnicity, family relatedness, household contact and characteristics of TB index case (cavity on chest X-ray, 2 AFB sputum smear and positive Mtb culture). SNP: single-nucleotide polymorphism.
Discussion
In the present study, we tested associations between SNPs from related genes in different pathways of DNA and RNA sensing and the type I IFN pathway in a large number of TB contacts. The notable finding was that PYHIN1- IFI16-AIM2 rs1633256 and rs1101998 were associated with an increased risk of TST positivity where as IRF7 rs11246213 was associated with a lower probability of TST positivity. To our knowledge, SNPs in these genes have not previously been reported to be associated with the pathogenesis of Mtb infection in contacts.
Our results suggest that the PYHIN1- IFI16-AIM2 rs1633256 and rs1101998 polymorphisms are associated with increased susceptibility to Mtb infection (i.e., a positive TST). The two polymorphisms are in a 3-gene locus on chromosome 1q23.1; thus, it is not possible to know which specific gene is most likely to exert a functional effect related to these genetic variants. The gene encoding Interferon-γ-inducible protein 16 (IFI16) (Trapani et al., 1994) is a multifunctional and ubiquitous host protein (Trapani et al.,1992), and a member of the PyHIN (pyrin and HIN200 domain-containing) protein family that consists of four family members: PYHIN1 (alias IFIX), IFI16 (alias PYHIN2), MNDA (alias PYHIN3) and AIM2 (alias PYHIN4) (Thompson et al., 2011). During Mtb infection of macrophages, IFI16 is reported to be localized into the cytosolic compartment (Thompson et al., 2011) and Mtb DNA activates the cytosolic surveillance pathway. Mice genetically lacking IFI204 (a homolog gene of human IFI16) show reduced IFIT1 and IFN-β induction against Mtb infection (Manzanillo et al., 2012).Furthermore, mycobacterial infection of AIM2−/− (absent in melanoma 2) mice induces elevated IFN-γ and reduced IFN-γ responses, leading to higher infection burdens and more severe pathology (Yan et al., 2018). In addition, in vitro studies demonstrated that AIM2-deficient macrophages display impaired activation of the inflammasome and defective production of IL-1b and IL-18 upon Mtb infection, making such cells highly susceptible to bacterial proliferation and cell death (Saiga et al., 2012). To the best of our knowledge, there are no previously reported studies on the relationship of PYHIN1 and TB. PYHIN1 detects Herpes Simplex (HSV-1) DNA and contributes to the induction of interferon response in human fibroblasts (Diner et al., 2015). In the present study, the SNPs associated with TST positivity (rs1633256 and rs1101998) are part of a large locus; thus it is possible that at least these two SNPs could be associated with any one of the 3 genes described above (PYHIN1- IFI16-AIM2) and influence the detection of Mtb DNA during infection. Future studies are warranted to directly elucidate the molecular mechanisms underlying these associations.
The human IRF7 gene is located on chromosome 11p15.5 and is a member of the interferon regulatory factor family of transcription factors, comprised of nine members (IRF1 to 9) (Ning et al., 2011). This family is recognized by the regulation of many facets of innate and adaptive immune responses (Tamura et al., 2008). IRF7 is the central transcription factor that induces IFNA/B gene transcription in response to cytosolic vira l DNA and RNA in host cells (McNab et al., 2015). In addition, IRF7 is produced by murine bone marrow–derived macrophage infected with Mtb (Cheng and Schorey, 2018; Leisching et al., 2017). In a recent meta-analysis, Mtb infection of THP-1 macrophages induced differential expression of IRF7 (Zhang et al., 2019). Excessive type I IFN expression has been linked to increased TB-associated immunopathology and susceptibility to severe TB (Mayer-Barber et al., 2011; Mayer-Barber et al., 2014). IRF7 SNPs have been reported to significant reduce IFNα production by plasmacytoid dendritic cells following stimulation with HIV-1 (Chang et al., 2011). The effect of IRF7 SNPs on reduced IFNα production, if present also in exposure to Mtb, could be a factor explaining the decreased susceptibility to Mtb infection reported here.
We also found that IFIT5 rs59633641 was less frequently observed in individuals with a positive TST where as IFIT5 rs10887959 was more commonly detected in individuals with positive TST. IFIT5 (IFN-induced protein with tetratricopeptide repeats-5) is a member of an interferon-induced protein with tetratricopeptide repeats (IFIT) family with five members (IFIT1, IFIT2, IFIT3, IFIT1B and IFIT5) localized in chromosome 10q23 (Diamond, 2014). The multivariable model with adjustment for race/ethnicity, family relatedness, gender and age demonstrated associations between the IFIT5 rs10887959 and increased chance of negative TST. It has been recently demonstrated that IFIT5 physically interacts with MAP3K7/TAK1 and IκB kinase (IKK) to activate the transcription factor NF-κB, which is a key regulator of the expression of genes involved in immune responses, inflammation, cell survival and cancers (Zheng et al., 2015). IFIT5 is one of the main genes upregulated in active TB patients (Ahmed et al., 2016). The IFN-induced proteins regulate immune response against viruses. For example, it has been recently shown that IFIT3 has a protective role in response to dengue virus infection of human lung epithelial cells (Hsu et al., 2013).
Our study has several strengths such as systematic TST testing (currently recommended as the diagnostic test for LTBI in most resource-restrained countries) and inclusion criteria that ensure microbiological confirmation of TB index cases. However, it is also important to highlight potential limitations of our investigation, such as the cross-sectional nature of the analyses, which are not able to establish causal relationships. We have not performed functional validation of the findings; however, we showed an analysis of gene expression data in silico. In addition, we considered a common Mtb strain to be responsible for infections within a household, but it is possible that that may not have always been true. In addition, LTBI was only measured by TST with no IGRA assessments. These two tests are not perfectly concordant, so the TST negative group could probably include some individuals with positive IGRA results. Of note, IGRA was not available in Brazil at the time of the patient enrollment. The Food and Drug Administration (FDA) approved IGRA in 2001, and this test was introduced in Brazil in 2014, 10 years after the data collection of the present study was finalized. Regardless, our results clearly indicate associations between polymorphisms in innate immune genes linked to interferon responses and odds of Mtb infection assessed by TST positivity. Further translational studies are required to delineate the molecular events behind these associations.
Supplementary Material
Acknowledgments
The authors acknowledge study participants and also the staff of the Clementino Fraga Filho University Hospital of the Federal University of Rio de Janeiro. This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)/Instituto Nacional de Ciência e Tecnologia (INCT, grant number: 421703/2017–2) and Fundação de Amparo à Pesquisa do Rio de Janeiro (FAPERJ, grant number: E-26/110.974/2011). BBA, JRLS, and AK are senior investigators from CNPq and AK and JRLS receive senior fellowships from FAPERJ. The work from BBA and KFF was supported by intramural research program from FIOCRUZ and from the National Institutes of Health (U01AI115940). JMC-A was supported by the Organization of American States – Partnerships Program for Education and Training (OAS-PAEC) and his study was financed in part by the Coordenação de Aperfeiçoa-mento de Pessoal de Nível Superior - Brazil (CAPES) - Finance Code 001. MBA receives a fellowship from the Fundação de Amparo à Pesquisa da Bahia (FAPESB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethics statement
The study was approved by the Clementino Fraga Filho University Hospital (HUCFF), Federal University of Rio de Janeiro Ethics Review Board. Written informed consent was obtained from all participants or their legally responsible guardians, and all clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki. The anonymity of study subjects was preserved with a code created with a link to personal identifiers.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2019.12.013.
References
- Ahmed A, Rakshit S, Vyakarnam A. HIV-TB co-infection: mechanisms that drive reactivation of Mycobacterium tuberculosis in HIV infection. Oral Dis 2016;22 Suppl. 1:53–60. [DOI] [PubMed] [Google Scholar]
- Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010;466(7309):973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22(9):1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Lindsay RJ, Kulkarni S, Lifson JD, Carrington M, Altfeld M. Polymorphisms in interferon regulatory factor 7 reduce interferon-alpha responses of plasmacytoid dendritic cells to HIV-1. AIDS 2011;25(5):715–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Schorey JS. Mycobacterium tuberculosis-induced IFN-beta production requires cytosolic DNA and RNA sensing pathways. J Exp Med 2018;215 (11):2919–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45 (6):580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Angulo JM, Arriaga MB, Silva EC, Muller BLA, Ramalho DMP, Fukutani KF, et al. Polymorphisms in TLR4 and TNFA and risk of Mycobacterium tuberculosis infection and development of active disease in contacts of tuberculosis cases in Brazil: a prospective cohort study. Clin Infect Dis 2019;69(6):1027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS. IFIT1: a dual sensor and effector molecule that detects non-2’-O methylated viral RNA and inhibits its translation. Cytokine Growth Factor Rev 2014;25(5):543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner BA, Li T, Greco TM, Crow MS, Fuesler JA, Wang J, et al. The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Mol Syst Biol 2015;11(1):787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan ML, Schultz TE, Duke TJ, Blumenthal A. Type I interferons in the pathogenesis of tuberculosis: molecular drivers and immunological consequences. Front Immunol 2017;8:1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben RM, Dodd PJ. The global burden of latent tuberculosis infection: a reestimation using mathematical modelling. PLoS Med 2016;13(10)e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YL, Shi SF, Wu WL, Ho LJ, Lai JH. Protective roles of interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) in dengue virus infection of human lung epithelial cells. PLoS One 2013;8(11)e79518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IBGE. Instituto Brasileiro de Geografia e Estatística. Censo Brasileiro de 2010 Rio de Janeiro: IBGE; 2012. [Google Scholar]
- Kinnear C, Hoal EG, Schurz H, van Helden PD, Moller M. The role of human host genetics in tuberculosis resistance. Expert Rev Respir Med 2017;11(9):721–37. [DOI] [PubMed] [Google Scholar]
- Koch A, Mizrahi V. Mycobacteriu tuberculosis. Trends Microbiol 2018;26(6):555–6. [DOI] [PubMed] [Google Scholar]
- Leisching G, Pietersen RD, van Heerden C, van Helden P, Wiid I, Baker B. RNAseq reveals hypervirulence-specific host responses to M. tuberculosis infection. Virulence 2017;8(6):848–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan CS, Zalwango S, Thiel BA, Malone LL, Chervenak KA, Baseke J, et al. Innate and adaptive immune responses during acute M. tuberculosis infection in adult household contacts in Kampala, Uganda. Am J Trop Med Hyg 2012;86(4):690–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca C, Tsenova L, Freeman S, Barczak AK, Tovey M, Murray PJ, et al. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. J Interferon Cytokine Res 2005;25 (11):694–701. [DOI] [PubMed] [Google Scholar]
- Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 2012;11(5):469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Barber KD, Andrade BB, Barber DL, Hieny S, Feng CG, Caspar P, et al. Innate and adaptive interferons suppress IL-1alpha and IL-1beta production by distinct pulmonary myeloid subsets during Mycobacterium tuberculosis infection. Immunity 2011;35(6):1023–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Barber KD, Andrade BB, Oland SD, Amaral EP, Barber DL, Gonzales J, et al. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature 2014;511(7507):99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol 2015;15(2):87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministério da Saúde Brasil. Manual de Recomendações para o Controle da Tuberculose no Brasil. 2019. Available from: http://portalarquivos2.saude.gov.br/images/pdf/2019/marco/28/manual-recomendacoes.pdf. [Accessed 31 July 2019].
- Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun 2011;12(6):399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai M, Behr MA, Dowdy D, Dheda K, Divangahi M, Boehme CC, et al. Tuberculosis. Nat Rev Dis Primers 2016;2:16076. [DOI] [PubMed] [Google Scholar]
- Rangaka MX, Wilkinson KA, Glynn JR, Ling D, Menzies D, Mwansa-Kambafwile J, et al. Predictive value of interferon-gamma release assays for incident active tuberculosis: a systematic review and meta-analysis. Lancet Infect Dis 2012;12 (1):45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid MJA, Arinaminpathy N, Bloom A, Bloom BR, Boehme C, Chaisson R, et al. Building a tuberculosis-free world: the Lancet Commission on tuberculosis. Lancet 2019;393(10178):1331–84. [DOI] [PubMed] [Google Scholar]
- Robertson BD, Altmann D, Barry C, Bishai B, Cole S, Dick T, et al. Detection and treatment of subclinical tuberculosis. Tuberculosis (Edinb) 2012;92(6):447–52. [DOI] [PubMed] [Google Scholar]
- Saiga H, Kitada S, Shimada Y, Kamiyama N, Okuyama M, Makino M, et al. Critical role of AIM2 in Mycobacterium tuberculosis infection. Int Immunol 2012;24(10):637–44. [DOI] [PubMed] [Google Scholar]
- Sinnwell Jason P, Schaid Daniel J. Statistical methods for haplotypes when linkage phase is ambiguous. 2018. ed2018. [Google Scholar]
- Shin Ji-Hyung SB, McNeney Brad, Graham Jinko. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Softw 2006;16:. [Google Scholar]
- Shin SS, Modongo C, Zetola NM. The impact of mixed infections on the interpretation of molecular epidemiology studies of tuberculosis. Int J Tuberc Lung Dis 2016;20(3):423–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameris MD, Hatherill M, Landry BS, Scriba TJ, Snowden MA, Lockhart S, et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet 2013;381(9871):1021–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 2008;26:535–84. [DOI] [PubMed] [Google Scholar]
- Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011;3 (6):920–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapani JA, Browne KA, Dawson MJ, Ramsay RG, Eddy RL, Show TB, et al. A novel gene constitutively expressed in human lymphoid cells is inducible with interferon-gamma in myeloid cells. Immunogenetics 1992;36(6):369–76. [DOI] [PubMed] [Google Scholar]
- Trapani JA, Dawson M, Apostolidis VA, Browne KA. Genomic organization of IFI16, an interferon-inducible gene whose expression is associated with human myeloid cell differentiation: correlation of predicted protein domains with exon organization. Immunogenetics 1994;40(6):415–24. [DOI] [PubMed] [Google Scholar]
- van Tong H, Velavan TP, Thye T, Meyer CG. Human genetic factors in tuberculosis: an update. Trop Med Int Health 2017;22(9):1063–71. [DOI] [PubMed] [Google Scholar]
- Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, et al. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe 2015;17(6):811–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Global tuberculosis report 2018. World Health Organization; 2018. [Google Scholar]
- Yan S, Chen L, Wu W, Fu Z, Zhang H, Li Z, et al. Early versus delayed antiretroviral therapy for HIV and tuberculosis co-infected patients: a systematic review and meta-analysis of randomized controlled trials. PLoS One 2015;10(5)e0127645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Shen H, Lian Q, Jin W, Zhang R, Lin X, et al. Deficiency of the AIM2-ASC signal uncovers the STING-driven overreactive response of type I IFN and reciprocal depression of protective IFN-gamma immunity in mycobacterial infection. J Immunol 2018;200(3):1016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Lin Y, Yu HY, Tian RN, Li F. Characteristic genes in THP1 derived macrophages infected with Mycobacterium tuberculosis H37Rv strain identified by integrating bioinformatics methods. Int J Mol Med 2019;44(4):1243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Zheng Z, Zhang Z, Meng J, Liu Y, Ke X, et al. IFIT5 positively regulates NF-kappaB signaling through synergizing the recruitment of IkappaB kinase (IKK) to TGF-beta-activated kinase 1 (TAK1). Cell Signal 2015;27 (12):2343–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.