Abstract
The secondary structures of nucleic acids form a particularly important class of contact structures. Many important RNA molecules, however, contain pseudo-knots, a structural feature that is excluded explicitly from the conventional definition of secondary structures. We propose here a generalization of secondary structures incorporating ‘non-nested’ pseudo-knots, which we call bi-secondary structures, and discuss measures for the complexity of more general contact structures based on their graph-theoretical properties. Bi-secondary structures are planar trivalent graphs that are characterized by special embedding properties. We derive exact upper bounds on their number (as a function of the chain length n) implying that there are fewer different structures than sequences. Computational results show that the number of bi-secondary structures grows approximately like 2.35n. Numerical studies based on kinetic folding and a simple extension of the standard energy model show that the global features of the sequence-structure map of RNA do not change when pseudo-knots are introduced into the secondary structure picture. We find a large fraction of neutral mutations and, in particular, networks of sequences that fold into the same shape. These neutral networks percolate through the entire sequence space.
Keywords: Secondary Structure, Contact Structure, Neutral Network, Outerplanar Graph, Diagram Graph
Contributor Information
Christian Haslinger, Email: grisu@tbi.univie.ac.at.
Peter F. Stadler, Email: studla@tbi.univie.ac.at
References
- Abrahams J. P., van den Berg M., van Batenburg E., Pleij C. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucl. Acids Res. 1990;18:3035–3044. doi: 10.1093/nar/18.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart F., Kainen P. C. The book thickness of a graph. J. Comb. Theor. 1979;B27:320–331. doi: 10.1016/0095-8956(79)90021-2. [DOI] [Google Scholar]
- Bonhoeffer S., McCaskill J. S., Stadler P. F., Schuster P. RNA multi-structure landscapes. A study based on temperature dependent partition functions. Eur. Biophys. J. 1993;22:13–24. doi: 10.1007/BF00205808. [DOI] [PubMed] [Google Scholar]
- Brierley I., Rolley N. J., Jenner A. J., Inglis S. C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;229:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W. Structure and evolution of ribonuclease P RNA. Biochemie. 1991;73:689–697. doi: 10.1016/0300-9084(91)90049-7. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Wollenzien P. L., Hearst J. E. Structure and topology of 16S ribosomal RNA. an analysis of the pattern of psoralen crosslinking. Nucl. Acids Res. 1980;8:1855–1872. doi: 10.1093/nar/8.8.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro M., Parkin N., Varmus H. E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl. Acad. Sci. USA. 1992;89:713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H. S., Dill K. A. Interchain loops in polymers: effects of excluded volume. J. Chem. Phys. 1988;90:492–508. doi: 10.1063/1.456500. [DOI] [Google Scholar]
- Chartrand G., Harary F. Planar permutation graphs. Ann. Inst. Henri Poincaré. 1967;B3:433–438. [Google Scholar]
- Chen S.-J., Dill K. A. Statistical thermodynamics of double-stranded polymer molecules. J. Chem. Phys. 1995;103:5802–5808. doi: 10.1063/1.470461. [DOI] [Google Scholar]
- Chung, F. R. K., F. T. Leighton and A. L. Rosenberg (1987). Embedding graphs in books: A layout problem with applications to VLSI design. SIAM J. Alg. Disc. Math.8, 1987.
- Dinman J. D., Icho T., Wickner R. B. A-1 ribosomal frameshifting in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirac G. A. A property of 4-chromatic graphs and some remarks on critical graphs. J. London Math. Soc. 1952;27:85–92. [Google Scholar]
- Even S., Itai A. Queues, stacks, and graphs. In: Kohavi Z., Paz A., editors. Theory of Machines and Computation. New York: Academic Press; 1971. pp. 71–86. [Google Scholar]
- Felden B., Himeno H., Muto A., McCutcheon J. P., Atkins J., Gesteland R. F. Probing the structure of the Escherichia coli 10Sa RNA (tmRNA) RNA. 1997;3:89–103. [PMC free article] [PubMed] [Google Scholar]
- Fontana W., Griesmacher T., Schnabl W., Stadler P. F., Schuster P. Statistics of landscapes based on free energies, replication and degradation rate constants of RNA secondary structures. Mh. Chem. 1991;122:795–819. [Google Scholar]
- Fontana W., Konings D. A. M., Stadler P. F., Schuster P. Statistics of RNA secondary structures. Biopolymers. 1993;33:1389–1404. doi: 10.1002/bip.360330909. [DOI] [PubMed] [Google Scholar]
- Fontana W., Schuster P. Continuity in evolution: on the nature of transitions. Science. 1998;280:1451–1455. doi: 10.1126/science.280.5368.1451. [DOI] [PubMed] [Google Scholar]
- Fontana W., Stadler P. F., Bornberg-Bauer E. G., Griesmacher T., Hofacker I. L., Tacker M., Tarazona P., Weinberger E. D., Schuster P. RNA folding and combinatory landscapes. Phys. Rev. 1993;E47:2083–2099. doi: 10.1103/physreve.47.2083. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Altman S. Similar cage-shaped structures for the RNA component of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-W. [DOI] [PubMed] [Google Scholar]
- Fortsch I., Fritzsche H., Birch-Hirschfeld E., Evertsz E., Klement R., Jovin T. M., Zimmer C. Parallel-stranded duplex DNA containing dA·dU base pairs. Biopolymers. 1996;38:209–220. doi: 10.1002/(SICI)1097-0282(199602)38:2<209::AID-BIP7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci., USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Games R. Optimal book embeddings of FFT, benes, and barrel shifter networks. Algorithmica. 1986;1:233–250. doi: 10.1007/BF01840445. [DOI] [Google Scholar]
- Gluick T. C., Draper D. E. Thermodynamics of folding a pseudoknotted mRNA fragment. J. Mol. Biol. 1994;241:246–262. doi: 10.1006/jmbi.1994.1493. [DOI] [PubMed] [Google Scholar]
- Grüner W., Giegerich R., Strothmann D., Reidys C., Weber J., Hofacker I. L., Stadler P. F., Schuster P. Analysis of RNA sequence structure maps by exhaustive enumeration. I. Neutral networks. Monath. Chem. 1996;127:355–374. doi: 10.1007/BF00810881. [DOI] [Google Scholar]
- Grüner W., Giegerich R., Strothmann D., Reidys C., Weber J., Hofacker I. L., Stadler P. F., Schuster P. Analysis of RNA sequence structure maps by exhaustive enumeration. II. Structures of neutral networks and shape space covering. Monath. Chem. 1996;127:375–389. doi: 10.1007/BF00810882. [DOI] [Google Scholar]
- Gutell R. R., Larsen N., Woese C. R. Lessons from an evolving rRNA: 16S and 23S rRNA from a comparative perspective. Microbiol. Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas E. S., Morse D. P., Brown J. W., Schmidt J. F., Pace N. R. Long-range structure in ribonuclease P RNA. Science. 1991;254:853–856. doi: 10.1126/science.1719634. [DOI] [PubMed] [Google Scholar]
- Haslinger, Christian (1997). RNA secondary structures with pseudoknots. Master’s thesis, Inst. f. Theoretische Chemie, University of Vienna, 1997. http://www.tbi.univie.ac.at/papers/Masters_theses.html.
- Heath L. S., Leighton F. T., Rosenberg A. L. Comparing queues and stacks as mechanisms for laying out graphs. SIAM J. Discr. Math. 1992;5:398–412. doi: 10.1137/0405031. [DOI] [Google Scholar]
- Hofacker I. L., Fontana W., Stadler P. F., Bonhoeffer S., Tacke M., Schuster P. Fast folding and comparison of RNA econdary structures. Monath. Chem. 1994;125:167–188. doi: 10.1007/BF00818163. [DOI] [Google Scholar]
- Hofacker I. L., Schuster P., Stadler P. F. Combinatorics of RNA secondary structures. Discr. Appl. Math. 1999;89:177–207. [Google Scholar]
- Hogeweg P., Hesper B. Energy directed folding of RNA sequences. Nucl. Acid. Res. 1984;12:67–74. doi: 10.1093/nar/12.1part1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton D. A., Sheehan J. The Petersen Graph. Cambridge: Cambridge University Press; 1993. [Google Scholar]
- Hsieh W. N. Proportions of irreducible diagrams. Studies in Appl. Math. 1973;52:277–283. [Google Scholar]
- Huynen M. A. Exploring phenotype space through neutral evolution. J. Mol. Evol. 1996;43:165–169. doi: 10.1007/BF02338823. [DOI] [PubMed] [Google Scholar]
- Huynen M. A., Stadler P. F., Fontana W. Smoothness within ruggedness: the role of neutrality in adaptation. Proc. Natl. Acad. Sci. USA. 1996;93:397–401. doi: 10.1073/pnas.93.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson H., Stockmeyer W. H. Intramolecular reaction in polycondensations. J. Chem. Phys. 1950;18:1600–1606. doi: 10.1063/1.1747547. [DOI] [Google Scholar]
- Kleitman D. Proportions of irreducible diagrams. Studies in Appl. Math. 1970;49:297–299. [Google Scholar]
- Konings D. A. M., Gutell R. R. A comparison of thermodynamic foldings with comparatively derived structures of 16S and 16S-like rRNAs. RNA. 1995;1:559–574. [PMC free article] [PubMed] [Google Scholar]
- Kuratowski K. Sur le problème des courbes gauches en topologie. Fund. Math. 1930;15:271–283. [Google Scholar]
- Leydold J., Stadler P. F. Minimal cycle bases of outerplanar graphs. Elec. J. Comb. 1998;5:R16. [Google Scholar]
- Loria A., Pan T. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA. 1996;2:551–563. [PMC free article] [PubMed] [Google Scholar]
- Lovász, L. and A. Schrijver (1996). The Colin de Verdière number of linklessly embeddable graphs. preprint.
-
Malitz S. M. Genus g graphs have pagenumber
 J. Algorithms. 1994;17:85–109. doi: 10.1006/jagm.1994.1028. [DOI] [Google Scholar]
J. Algorithms. 1994;17:85–109. doi: 10.1006/jagm.1994.1028. [DOI] [Google Scholar] -
Malitz S. M. Graphs with e edges have pagenumber
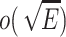 J. Algorithms. 1994;17:71–84. doi: 10.1006/jagm.1994.1027. [DOI] [Google Scholar]
J. Algorithms. 1994;17:71–84. doi: 10.1006/jagm.1994.1027. [DOI] [Google Scholar] - Mans R., Pleij C. W. A., Bosch L. Transfer RNA-like structures: structure, function and evolutionary significance. Eur. J. Biochem. 1991;201:303–324. doi: 10.1111/j.1432-1033.1991.tb16288.x. [DOI] [PubMed] [Google Scholar]
- Martinez H. M. An RNA folding rule. Nucl. Acid. Res. 1984;12:323–335. doi: 10.1093/nar/12.1part1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimmensional architecture of group I catalytic introns based on comparative sequence anaysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Obrenić Bojana. Embedding De Bruijn graphs and shuffle-exchange graphs in five pages. SIAM J. Discr. Math. 1993;6:642–654. doi: 10.1137/0406049. [DOI] [Google Scholar]
- Penner R. C., Waterman M. S. Spaces of RNA secondary structures. Adv. Math. 1993;101:31–49. doi: 10.1006/aima.1993.1039. [DOI] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucl. Acid. Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidys C. M. Random induced subgraphs of generalized n-cubes. Adv. Appl. Math. 1997;19:360–377. doi: 10.1006/aama.1997.0553. [DOI] [Google Scholar]
- Reidys C., Stadler P. F. Bio-molecular shapes and algebraic structures. Comp. & Chem. 1996;20:85–94. doi: 10.1016/S0097-8485(96)80010-6. [DOI] [PubMed] [Google Scholar]
- Reidys C., Stadler P. F., Schuster P. Generic properties of combinatory maps: Neural networks of RNA secondary structures. Bull. Math. Biol. 1996;59:339–397. doi: 10.1016/S0092-8240(96)00089-4. [DOI] [PubMed] [Google Scholar]
- Robertson N., Seymore P., Thomas R. Petersen family minors. J. Comb. Theory. 1995;B64:155–184. [Google Scholar]
- Robertson N., Seymore P., Thomas R. Sachs’ linkless embedding conjecture. J. Comb. Theory. 1995;B64:185–227. [Google Scholar]
- Saenger W. Principles of Nucleic Acid Structure. London: Springer Verlag; 1984. [Google Scholar]
- Schmitt W. R., Waterman M. S. Linear trees and RNA secondary structure. Discr. Appl. Math. 1994;12:412–427. [Google Scholar]
- Schuster P. How to search for RNA structures. Theoretical concepts in evolutionary biotechnology. J. Biotechnol. 1995;41:239–257. doi: 10.1016/0168-1656(94)00085-Q. [DOI] [PubMed] [Google Scholar]
- Schuster P., Fontana W., Stadler P. F., Hofacker I. L. From sequences to shapes and back: A case study in RNA secondary structures. Proc. R. Soc. 1994;B255:279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- Shapiro B. A., Zhang K. Comparing multiple RNA secondary structures using tree comparisons. CABIOS. 1990;6:309–318. doi: 10.1093/bioinformatics/6.4.309. [DOI] [PubMed] [Google Scholar]
- Singh R. K., Tropsha A., Vaisman I. I. Delaunay tessellation of proteins: Four body nearest neighbor propensities of amino acid residues. J. Comput. Biol. 1996;3:213–221. doi: 10.1089/cmb.1996.3.213. [DOI] [PubMed] [Google Scholar]
- Söler F., Jankowski K. Modeling RNA secondary structures I. Mathematical structural model of predicting RNA secondary structures. Math. Biosc. 1991;105:167–190. doi: 10.1016/0025-5564(91)90080-3. [DOI] [PubMed] [Google Scholar]
- Stein P. R. On a class of linked diagrams, I. Enumeration. J. Comb. Theory. 1978;A24:357–366. doi: 10.1016/0097-3165(78)90065-1. [DOI] [Google Scholar]
- Stein P. R., Everett C. J. On a class of linked diagrams. II. Asymptotics. Disc. Math. 1978;22:309–318. doi: 10.1016/0012-365X(78)90162-0. [DOI] [Google Scholar]
- Stein P. R., Waterman M. S. On some new sequences generalizing the Catalan and Motzkin numbers. Disc. Math. 1978;26:261–272. doi: 10.1016/0012-365X(79)90033-5. [DOI] [Google Scholar]
- Sysło M. M. Characterizations of outerplanar graphs. Discr. Math. 1979;26:47–53. doi: 10.1016/0012-365X(79)90060-8. [DOI] [Google Scholar]
- Tacker M., Fontana W., Stadler P. F., Schuster P. Statistics of RNA melting kinetics. Eur. Biophys. J. 1994;23:29–38. doi: 10.1007/BF00192203. [DOI] [PubMed] [Google Scholar]
- Tacker, M., P. F. Stadler, E. G. Bornberg-Bauer, I. L. Hofacker and P. Schuster. Algorithm independent properties of RNA structure prediction. Eur. Biophy. J.25, 115–130.
- Tang C. K., Draper D. E. An unusual mRNA pseudoknot structure is recognized by a protein translation repressor. Cell. 1989;57:531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Evidence for allosteric coupling between the ribosome and repressor binding sites of a translationally regulated mRNA. Biochemistry. 1990;29:4434–4439. doi: 10.1021/bi00470a025. [DOI] [PubMed] [Google Scholar]
- Ten Dam E., Brierly I., Inglis S., Pleij C. Identification and analysis od the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucl. Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Dam E. B., Pleij C. W. A., Bosch L. RNA pseudoknots and translational frameshifting on retroviral, coronaviral and luteoviral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchard J. Sur une problème de configurations et sur les fractions continues. Canad. J. Math. 1952;4:2–25. [Google Scholar]
- Tzeng T. H., Tu C. L., Bruenn J. A. Ribosomal frameshifting requires a pseudoknot in the saccharomyces cerevisiae double-stranded RNA virus. J. Virology. 1992;66:999–1006. doi: 10.1128/jvi.66.2.999-1006.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Verdière Y. C. Sur un novel invariant des graphes et un critère de planarité. J. Comb. Theory. 1990;B50:11–21. [Google Scholar]
- Vlassov V. V., Zuber G., Felden B., Behr J. P., Griege R. Cleavage of tRNA with imidazole and spermine imidazole constructs: a new approach for probing RNA structures. Nucl. Acid. Res. 1995;23:3161–3167. doi: 10.1093/nar/23.16.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner K., Bodendiek R. Graphentheorie II. Mannheim: B.I. Verlag; 1990. [Google Scholar]
- Walter A. E., Turner D. H., Kim J., Lyttle M. H., Müller P., Mathews D. H., Zuker M. Co-axial stacking of helixes enhances binding of oligoribonucleotides and improves predicions of RNA folding. Proc. Natl. Acad. Sci. USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman M. S. Secondary structure of single-stranded nucleic acids. Adv. Math. Suppl. Stud. 1978;1:167–212. [Google Scholar]
- Waterman M. S. Introduction to Computational Biology: Maps, Sequences, and Genomes. London: Chapman and Hall; 1995. [Google Scholar]
- Waterman M. S., Smith T. F. Combinatorics of RNA hairpins and cloverleaves. Studies Appl. Math. 1978;60:91–96. [Google Scholar]
- Waterman M. S., Smith T. F. RNA secondary structure: a complete mathematical analysis. Math. Biosc. 1978;42:257–266. doi: 10.1016/0025-5564(78)90099-8. [DOI] [Google Scholar]
- Weber, J. (1997). Dynamics on Neutral Evolution. PhD thesis, Friedrich Schiller University, Jena, January 1997. http://www.tbi.univie.ac.at/papers/PhD_theses.html.
- Westhof E., Jaeger L. RNA pseudoknots. Current Opinion Struct. Biol. 1992;2:327–333. doi: 10.1016/0959-440X(92)90221-R. [DOI] [Google Scholar]
- Wilf H. S. Generating functionology. San Diego, CA: Academic Press; 1994. [Google Scholar]
- Wills N., Gesteland R. F., Atkins J. F. Evidence that a downstream pseudoknot is required for translational readthrough of the moloney murine leukemia virus gag stop codon. Proc. Natl. Acad. Sci. USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt J. R., Puglisi J. D., Tinoco I. T. RNA pseudoknots stability and loop size requirements. J. Mol. Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- Yannakakis M. Embedding planar graphs in four pages. J. Comput. Syst. Sci. 1988;38:36–67. doi: 10.1016/0022-0000(89)90032-9. [DOI] [Google Scholar]
- Zipf G. K. Human Behaviour and the Principle of Least Effort. Reading, MA: Addison-Wesley; 1949. [Google Scholar]
- Zuker M., Sankoff D. RNA secondary structures and their prediction. Bull. Math. Biol. 1984;46:591–621. doi: 10.1016/S0092-8240(84)80062-2. [DOI] [Google Scholar]


