Abstract
The Old World leaf-nosed bats (Hipposideridae) are aerial and gleaning insectivores that occur throughout the Paleotropics. Both their taxonomic and phylogenetic histories are confused. Until recently, the family included genera now allocated to the Rhinonycteridae and was recognized as a subfamily of Rhinolophidae. Evidence that Hipposideridae diverged from both Rhinolophidae and Rhinonycteridae in the Eocene confirmed their family rank, but their intrafamilial relationships remain poorly resolved. We examined genetic variation in the Afrotropical hipposiderids Doryrhina, Hipposideros, and Macronycteris using relatively dense taxon-sampling throughout East Africa and neighboring regions. Variation in both mitochondrial (cyt-b) and four nuclear intron sequences (ACOX2, COPS, ROGDI, STAT5) were analyzed using both maximum likelihood and Bayesian inference methods. We used intron sequences and the lineage delimitation method BPP—a multilocus, multi-species coalescent approach—on supported mitochondrial clades to identify those acting as independent evolutionary lineages. The program StarBEAST was used on the intron sequences to produce a species tree of the sampled Afrotropical hipposiderids. All genetic analyses strongly support generic monophyly, with Doryrhina and Macronycteris as Afrotropical sister genera distinct from a Paleotropical Hipposideros; mitochondrial analyses interpose the genera Aselliscus, Coelops, and Asellia between these clades. Mitochondrial analyses also suggest at least two separate colonizations of Africa by Asian groups of Hipposideros, but the actual number and direction of faunal interchanges will hinge on placement of the unsampled African-Arabian species H. megalotis. Mitochondrial sequences further identify a large number of geographically structured clades within species of all three genera. However, in sharp contrast to this pattern, the four nuclear introns fail to distinguish many of these groups and their geographic structuring disappears. Various distinctive mitochondrial clades are consolidated in the intron-based gene trees and delimitation analyses, calling into question their evolutionary independence or else indicating their very recent divergence. At the same time, there is now compelling genetic evidence in both mitochondrial and nuclear sequences for several additional unnamed species among the Afrotropical Hipposideros. Conflicting appraisals of differentiation among the Afrotropical hipposiderids based on mitochondrial and nuclear loci must be adjudicated by large-scale integrative analyses of echolocation calls, quantitative morphology, and geometric morphometrics. Integrative analyses will also help to resolve the challenging taxonomic issues posed by the diversification of the many lineages associated with H. caffer and H. ruber.
Keywords: cryptic species, mtDNA, nuclear introns, Paleotropical, phylogeny, species delimitation, systematics
Introduction
The Old World leaf-nosed bats, family Hipposideridae, currently include seven genera and 90 species of insectivorous bats distributed over much of the Paleotropics (Monadjem 2019; Simmons and Cirranello 2019). Both the taxonomic and phylogenetic histories of this family are confused. Throughout much of its history (e.g., Koopman 1989), Hipposideridae was considered either a subfamily of the Rhinolophidae (the horseshoe bats) or as its sister family within the Rhinolophoidea. Recently, however, the “trident bats” (Cloeotis, Paratriaenops, Rhinonicteris, and Triaenops) were shown to comprise a family-ranked group, the Rhinonycteridae, which is separate from and sister to the Hipposideridae (Foley et al. 2015; Armstrong et al. 2016). Even the genus Hipposideros Gray, 1831, as it was traditionally understood, appears paraphyletic with respect to the allied genera Asellia, Aselliscus, Coelops, and Anthops (Foley et al. 2015; Amador et al. 2018). Re-validation of Macronycteris Gray, 1866 and Doryrhina Peters, 1871 for groups of Afrotropical endemic species more closely related to each other than to African and Asian members of Hipposideros sensu stricto resolved a number of those issues (Foley et al. 2017).
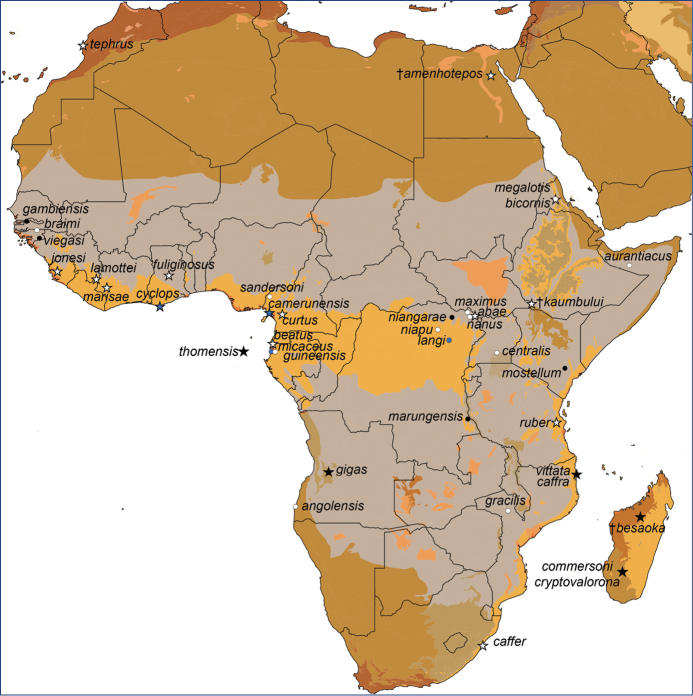
The species richness of Doryrhina, Macronycteris, and Hipposideros differs widely. Most authors recognize two species of Doryrhina (D. cyclops and D. camerunensis), five species of Macronycteris (M. commersoni, M. cryptovalorona, M. gigas, M. thomensis, and M. vittata), and 83 species of Hipposideros, 10 of which occur in Africa (Monadjem 2019; Simmons and Cirranello 2019). These are H. beatus, H. caffer, H. curtus, H. fuliginosus, H. lamottei, H. ruber, and H. tephrus in the bicolor group of Hipposideros; H. jonesi and H. marisae in the speoris group, and H. megalotis in the megalotis group (Hill 1963; Murray et al. 2012; Monadjem 2019). In addition, three extinct species of hipposiderid are known from the region: †Macronycteris besaoka (Madagascar), †Hipposideros amenhotepos (Egypt), and †H. kaumbului (Ethiopia). Type localities for valid species, subspecies, and synonyms for these three genera in Africa and Madagascar appear in Figure 1; after the removal of Doryrhina and Macronycteris taxa, group assignments for the species remaining in Hipposideros appear in Table 1.
Figure 1.
Type localities for Afrotropical hipposiderids: Doryrhina, blue symbols; Hipposideros, white symbols; Macronycteris, black symbols. Stars denote valid species, whereas circles indicate taxa considered as subspecies or synonyms. Localities are projected onto the biome map of Olson et al. (2001). Taxa depicted are: Hipposideros abae J. A. Allen,1917; †Hipposideros (Pseudorhinolophus) amenhotepos Gunnell, Winkler, Miller, Head, El-Barkooky, Gawad, Sanders & Gingerich, 2015; Phyllorhina angolensis Seabra, 1898; Hipposideros caffer var. aurantiaca De Beaux, 1924; Hipposideros beatus K. Andersen, 1906; †Hipposideros besaoka Samonds, 2007; Phyllorrhina bicornis Heuglin, 1861; Hipposideros braima Monard, 1939; Hipposideros caffer Sundevall, 1846; Phyllorhina caffra Peters, 1852; Hipposideros camerunensis Eisentraut, 1956; Hipposideros caffer centralis K. Andersen, 1906; Rhinolophus Commersonii É. Geoffroy, 1813; Hipposideros cryptovalorona Goodman, Schoeman, Rakotoarivelo & Willows-Munro, 2016; Hipposideros curtus G. M. Allen, 1921; Phyllorrhina cyclops Temminck, 1853; Phyllorrhina fuliginosa Temminck, 1853; Hipposideros gigas gambiensis K. Andersen, 1906; Rhinolophus gigas Wagner, 1845; Phyllorrhina gracilis Peters, 1852; Hipposideros caffer guineensis K. Andersen, 1906; Hipposideros jonesi Hayman, 1947; †Hipposideros kaumbului Wesselman, 1984; Hipposideros lamottei Brosset, 1985; Hipposideros langi J. A. Allen, 1917; Hipposideros marisae Aellen, 1954; Phyllorhina Commersoni, var. marungensis Noack, 1887; Hipposideros beatus maximus Verschuren, 1957; Phyllorrhina megalotis Heuglin, 1861; Rhinolophus micaceus de Winton, 1897; HipposiderosCommersoni mostellum Thomas, 1904; Hipposideros nanus J. A. Allen, 1917; Hipposideros gigas niangarae J. A. Allen, 1917; Hipposideros caffer niapu J. A. Allen, 1917; Phyllorrhina rubra Noack, 1893; Hipposideros sandersoni Sanderson, 1937; Hipposideros tephrus Cabrera, 1906; Phyllorhina Commersoni, var. thomensis Bocage, 1891; Hipposideros gigas viegasi Monard, 1939; Phyllorhina vittata Peters, 1852.
Table 1.
Species groups of Hipposideros (modified from Murray et al. 2012 to include newly recognized forms and to remove species now recognized in Doryrhina and Macronycteris).
| Armiger group | calcaratus subgroup | H. macrobullatus | H. lankadiva |
| H. alongensis | H. calcaratus c | H. maggietaylorae | H. lekaguli |
| H. armiger | H. cervinus c | H. nequam | H. pelingensis |
| H. griffini a | H. coxi c | H. obscurus | Larvatus group |
| H. pendelburyi a | H. galeritus c | H. orbiculus | H. grandis |
| H. turpis | ruber subgroup | H. papua | “H. khasiana” a,g |
| Bicolor group | H. abae d | H. pygmaeus | H. larvatus |
| ater subgroup | H. beatus e | Boeadii group | H. madurae |
| H. ater b | H. caffer e | H. boeadii | H. sorenseni |
| H. atrox a | H. fuliginosus e | Cyclops group f | H. sumbae |
| H. bicolor b | H. lamottei e | H. corynophyllus | Megalotis group |
| H. breviceps b | H. ruber e | H. edwardshilli | H. megalotis |
| H. cineraceus b | H. tephrus a,e | H. muscinus | Pratti group |
| H. coronatus b | subgroup uncertain | H. semoni | H. lylei |
| H. dyacorum b | H. cruminiferus | H. stenotis | H. pratti |
| H. einnaythu a,b | H. curtus | H. wollastoni | H. scutinares |
| H. halophyllus b | H. doriae | Diadema group | Speoris group |
| H. khaokhouayensis b | H. durgadasi | H. demissus | H. jonesi h |
| H. nicobarulae a,b | H. fulvus | H. diadema | H. marisae g |
| H. pomona b | H. gentilis a | H. dinops | H. speoris |
| H. ridleyi b | H. hypophyllus | H. inexpectatus | |
| H. rotalis b | H. kunzi a | H. inornatus |
(Endnotes) a Added to species list subsequent to Murray et al. (2012) b Recognized in the Ater species group by Monadjem (2019) c Recognized in the Calcaratus species group by Monadjem (2019) d Formerly listed in the Speoris group but transferred to the Ruber group by Monadjem (2019) e Recognized in the Ruber species group by Monadjem (2019) f H. cyclops and H. camerunensis are now recognized as members of Doryrhina; listed species were treated as Doryrhina inMonadjem (2019) on the basis of similar morphology but were recognized as the Muscinus group by Tate (1941); they might represent an unnamed genus or subgenus. g Invalid name accorded to what is likely a real biological entity (cf. Monadjem 2019) h Formerly in the Bicolor species group but transferred to the Speoris group by Monadjem (2019).
As suggested by their checkered taxonomic history, phylogenetic understanding of the Hipposideridae has slowly come into focus. Doryrhina and Macronycteris are two of a dozen generic-group names that were synonymized with Hipposideros for all of the 20th century (Miller 1907; Allen 1939; Koopman 1994). Instead of subgenera, taxonomists used the species groups delineated by Andersen (1918) and refined by Tate (1941) and Hill (1963) in their generic revisions based on morphology. Assessment of rhinolophoid relationships using an intron supermatrix (Eick et al. 2005) confirmed the early divergence of hipposiderids and rhinolophids (estimated at 41 Ma), thereby substantiating their rank as a separate families. Despite earlier suppositions that the area of origin for Hipposideridae was in Asia (Koopman 1970; Bogdanowicz and Owen 1998) or Australia (Hand and Kirsch 1998), Eick et al. (2005) clearly demonstrated the ancestry of the family (and superfamily) was in Africa. A recent supermatrix analysis with the most comprehensive taxonomic sampling (42 species; Amador et al. 2018) confirmed the early divergence of hipposiderids and rhinolophids at 41.3 Ma, but this analysis questioned the validity of both Doryrhina and Macronycteris. Amador et al. attributed the paraphyly of Hipposideros sensu lato documented by Foley et al. (2015) to their limited taxonomic sampling. Amador et al. (2018) also challenged the integrity of the commersoni, cyclops, speoris, and bicolor species groups, arguing that all African species save for H. jonesi belonged in a single, exclusively African species group.
Although new species of hipposiderids are regularly discovered and described in Asia (Robinson et al. 2003; Guillen-Servent and Francis 2006; Bates et al. 2007; Douangboubpha et al. 2011; Thong et al. 2012; Murray et al. 2018), the pace of discovery has been much slower in Africa. Only one extant species has been described since the recognition of Hipposideros lamottei (Brosset 1985 [“1984”]), and that one was from Madagascar (Goodman et al. 2016). Surveys of mitochondrial sequences from African hipposiderids have strongly suggested that supposedly widespread species such as Hipposideros caffer and H. ruber actually represent complexes of cryptic species (Vallo et al. 2008, 2011; Monadjem et al. 2013). Phylogenetic analyses (e.g., Vallo et al. 2008) show that these named species complexes are not monophyletic, resolving clades comprised of bats identified as both H. caffer and H. ruber. These studies have characterized the clades in both morphological and genetic terms, even establishing them in sympatry (see also Vallo et al. 2011). However, the uncertain relationship of the identified clades to the many names already proposed for Afrotropical hipposiderids, many based on incomplete or formalin-preserved specimens, has precluded formally naming them. Incomplete geographic sampling and the lack of evidence from nuclear genes for these populations has also clouded interpretations of this mitochondrial diversity.
Our field surveys in Eastern Africa and adjoining regions offer a new basis for considering the taxonomy and phylogenetics of Afrotropical hipposiderids. We sought to answer these questions: (1) Is there compelling evidence to support the recognition of Doryrhina and Macronycteris as distinct Afrotropical genera alongside the Paleotropical Hipposideros? (2) Which species belong to these groups? (3) Are the traditional species groups of African hipposiderids monophyletic? Using both mitochondrial and nuclear intron sequences, we also evaluate the question of cryptic species among African hipposiderids and the possibility of mitochondrial-nuclear discordance.
Material and methods
Selection of taxa and sampling
Our genetic dataset is based on 453 hipposiderid individuals, the vast majority being represented by museum vouchers. We generated original genetic data from 319 individuals collected at 102 georeferenced localities, and complemented them with 134 mitochondrial sequences from 90 localities downloaded from GenBank (we obtained new sequence data for five individuals with prior GenBank records; see Suppl. material 1: Figure S1 and Appendix I). All individuals were sequenced for Cytochrome-b (cyt-b) in order to maximize assessment of genetic diversity; however, redundant haplotypes were removed for subsequent phylogenetic analyses (see Appendix I for complete list of individuals sequenced). The bats newly sequenced for this study were obtained over several decades in the course of small mammal surveys across sub-Saharan Africa and Madagascar, with relatively dense sampling in East Africa. Initial assignment of East African individuals to species was determined using meristic, mensural, and qualitative characters published in the bat keys of Thorn et al. (2009) and Patterson and Webala (2012). Collection methods followed mammal guidelines for the use of wild mammals in research and education (Sikes and the Animal Care and Use Committee of the American Society of Mammalogists 2016) and the most recent collections were approved under Field Museum of Natural History’s IACUC #2012-003. Only GenBank records for cyt-b were available for records of the Arabian-North African hipposiderid Asellia, which was included for context in the phylogenetic analyses. Lacking information from nuclear introns, we draw no firm conclusions from their placement and do not discuss Asellia in this paper (see Benda et al. 2011; Bray and Benda 2016).
Appendix I contains the institutions and voucher numbers, GenBank accession numbers, and locality information for our samples. The fact that museum voucher specimens were used wherever possible for the genetic analyses permits the genetic analysis to serve as a foundation for integrative taxonomic analyses of dental, cranial, and skeletal variation, using the same specimens. To avoid adding to current taxonomic confusion, we take a conservative approach in assigning names to clades in our analyses. Where a clade’s taxonomic identity was ambiguous or unknown, we referred to it simply as a numbered clade. Integrative taxonomic diagnoses of the various clades supported by our analyses will be necessary to determine which, if any, existing names may apply to them. However, to relate our results to those of earlier studies of African Hipposideros (Vallo et al. 2008, 2011; Monadjem et al. 2013), we cross-referenced specimens used in two or more analyses to equate the various non-binomial names that have been applied to these cryptic lineages.
DNA extraction, amplification, and sequencing
Genomic DNA from preserved tissue samples was extracted using the Wizard SV 96 Genomic DNA Purification System (Promega Corporation, WI, USA). Fresh specimens were sequenced for mitochondrial cytochrome-b (cyt-b), using the primer pair LGL 765F and LGL 766R (Bickham et al. 1995; Bickham et al. 2004), and four unlinked autosomal nuclear introns: ACOX2 intron 3, COPS7A intron 4, ROGDI intron 7 (Salicini et al. 2011), and STAT5B (Matthee et al. 2001) for hipposiderid specimens and the sister group Triaenops afer (Rhinonycteridae; see Table 1 for primer information). PCR amplification, thermocycler conditions, and sequencing were identical to Patterson et al. (2018) and Demos et al. (2018). Sequences were assembled and edited using GENEIOUS PRO v.11.1.5 (Biomatters Ltd). Sequence alignments were made using MUSCLE (Edgar 2004) with default settings in GENEIOUS. Protein coding data from cyt-b were translated to amino acids to determine codon positions and confirm the absence of premature stop codons, deletions, and insertions. Several gaps were incorporated in the nuclear intron alignments, but their positions were unambiguous.
Sequence alignments used in this study have been deposited on the FIGSHARE data repository (https://doi.org/10.6084/m9.figshare.11936250). All newly generated sequences were deposited in GenBank with accession numbers MT149315–MT149893 (see also Appendix I).
Phylogenetic analyses
jMODELTEST2 (Darriba et al. 2012) on CIPRES Science Gateway v. 3.3 (Miller et al. 2010) was used to determine the sequence substitution models that best fit the data using the Bayesian Information Criterion (BIC) for cyt-b and the four nuclear introns. PARTITIONFINDER2 (Lanfear et al. 2016) on CIPRES was used to determine the sequence substitution models for the concatenated alignment of four nuclear introns using the Bayesian Information Criterion (BIC) with the ‘greedy’ search algorithm. Uncorrected sequence divergences (p-distances) between and within species/clades were calculated for cyt-b using MEGA X v. 10.0.5 (Kumar et al. 2018). Maximum-likelihood (ML) analyses were performed using the program IQ-TREE v. 1.6.10 (Chernomor et al. 2016; Nguyen et al. 2015) on the CIPRES portal for separate gene trees (cyt-b, ACOX2, COPS7A, ROGDI, and STAT5B) and a concatenated alignment, partitioned by gene, using the four nuclear introns. As in Hillis and Bull (1993), nodes supported by bootstrap values (BP) ≥ 70% were considered strongly supported. Gene tree analyses under a Bayesian Inference (BI) framework were inferred in MRBAYES v. 3.2.7 (Ronquist et al. 2012) on the CIPRES portal for the same set of genes as the ML analyses. Two independent runs were conducted in MrBayes, and nucleotide substitution models were unlinked across partitions for each nuclear locus in the concatenated alignment. Four Markov chains were run for 1 × 108 generations for individual gene trees, and 2 × 107 generations for the concatenated analysis, using default heating values and sampled every 1000th generation. A conservative 25% burn-in was applied and stationarity of the MRBAYES results was assessed in Tracer v. 1.7 (Rambaut et al. 2018). Majority-rule consensus trees were constructed for each Bayesian analysis. Following Erixon et al. (2003), nodes supported by posterior probabilities (PP) ≥0.95 were considered strongly supported.
Haplotype networks for cyt-b were inferred using the median-joining network algorithm in PopArt v. 1.7 (Leigh and Bryant 2015). Separate analyses were carried out for the following clades, each consisting of four subclades: (1) Doryhina (D. camerunensis, D. cf. camerunensis, D. cyclops1, and D. cyclops2); (2) Macronycteris (M. commersoni, M. cryptovalorona, M. gigas, and M. vittata); (3) Hipposideros caffer1–4; (4) Hipposideros caffer5–8; and (5) Hipposideros ruber1–4.
Hipposiderid taxa included in the species tree analyses were assigned to either species or numbered clades based on clade support in the ML and BI gene-tree analyses of the cyt-b dataset. This in turn identified populations to be used as ‘candidate species’ in a coalescent-based species-tree approach implemented in StarBEAST2 (Ogilvie et al. 2017), an extension of BEAST v. 2.5.1 (Drummond et al. 2012; Bouckaert et al. 2014). Species tree analysis was conducted using the four nuclear intron alignments. Substitution, clock, and tree models were unlinked across all loci. A lognormal relaxed-clock model was applied to each locus under a Yule tree prior and a linear with constant root population size model. Four independent replicates were run with random starting seeds, and chain lengths of 1 × 108 generations and parameters were sampled every 5,000 steps. For the StarBEAST2 analyses, evidence of convergence and stationarity of posterior distributions of model parameters was assessed based on ESS values >200 and examination of trace files in Tracer v. 1.7. The burn-in was set at 10% and separate runs were assembled using LOGCOMBINER v. 2.5.1 and TREEANNOTATOR v. 2.5.1 (Rambaut et al. 2018).
Coalescent lineage delimitation
Based on the well supported clades obtained in the cyt-b gene tree analyses and available intron samples, a lineage delimitation scenario with 18 candidate species was tested. We inferred the evolutionary isolation of their gene pools using the phased nuclear DNA dataset (ACOX2, COPS7A, ROGDI, and STAT5A; 104 individuals) for joint independent lineage delimitation and species-tree estimation evaluated under the multi-species coalescent model using the program BPP v. 3.3 (Yang and Rannala 2014; Rannala and Yang 2017). This analysis was carried out to guide future investigations of the species status of evolutionarily isolated lineages inferred here. Supported lineages will be examined using an integrative species taxonomic approach, including morphological, morphometric, and acoustic characters, as well as ectoparasite associations and distributional data. Species/clade memberships for BPP were identical to individuals assigned to lineages in the species tree analyses. The validity of our assignment of individuals to populations was tested using the guide-tree-free algorithm (A11) in BPP. Because the probability of delimitation by BPP is sensitive to selected parameters (Leaché and Fujita 2010; Yang 2015), we evaluated two independent runs for each of four different combinations of divergence depth and effective population sizes priors (τ and θ, respectively; Table 2). Two independent MCMC chains were run for 5 × 104 generations. The burn-in was 20% and samples drawn every 50th generation. In total, eight BPP runs were carried out using four phased nuclear intron alignments. Lineages were considered to be statistically well supported when the delimitation posterior probabilities generated were ≥0.95 under all four combinations of priors.
Table 2.
Primer information and chosen substitution models for regions amplified in this study. Substitution models before “/” are the best-supported models inferred by jMODELTEST2 and models after “/” indicate those inferred by PARTITIONFINDER2 for the concatenated intron alignment.
| Primer name | Sequence | Primer publication | Substitution model |
|---|---|---|---|
| ACOX2-3-F | 5’-CCTSGGCTCDGAGGAGCAGAT-3’ | Salicini et al. 2011 | K80+G / K81+G |
| ACOX2-3-R | 5’-GGGCTGTGHAYCACAAACTCCT-3’ | ||
| COPS7A-4-F | 5’-TACAGCATYGGRCGRGACATCCA-3’ | Salicini et al. 2011 | HKY / K80 |
| COPS7A-4-R | 5’-TCACYTGCTCCTCRATGCCKGACA-3’ | ||
| ROGDI-7-F | 5’-CTGATGGAYGCYGTGATGCTGCA-3’ | Salicini et al. 2011 | K80+G / K81+G |
| ROGDI-7-R | 5’-CACGGTGAGGCASAGCTTGTTGA-3’ | ||
| STAT5B-16-F | 5’--CTGCTCATCAACAAGCCCGA-3’ | Matthee et al. 2001 | GTR+G / K81+G |
| STAT5B-16-R | 5’-GGCTTCAGGTTCCACAGGTTGC-3’ | ||
| cyt-b-LGL-765-F | 5’-GGCTTCAGGTTCCACAGGTTGC-3’ | Trujillo et al. 2009 | GTR+I+G |
| cyt-b -LGL-766-R | 5’-GTTTAATTAGAATYTYAGCTTTGGG-3’ |
Table 3.
Prior Schemes (PS) used in BPP analyses. Prior distributions on τ represent two relative divergence depths (deep and shallow) and on θ represent two relative effective population sizes (large and small) scaling mutation rates.
| PS | Effective pop. size | Divergence depth | Gamma distribution for prior |
|---|---|---|---|
| 1 | Large | Deep | θ = Γ [1, 10] and τ = Γ [1, 10] |
| 2 | Large | Shallow | θ = Γ [1, 10] and τ = Γ [2, 2000] |
| 3 | Small | Shallow | θ = Γ [2, 2000] and τ = Γ [2, 2000] |
| 4 | Small | Deep | θ = Γ [2, 2000] and τ = Γ [1, 10] |
Results
In terms of cyt-b sequence divergence, clades within Doryrhina are separated by 3.0–5.7% genetic distances, whereas less than 3% separates the four recognized species of Macronycteris. Between Afrotropical Hipposideros, the greatest distances separate H. jonesi from other lineages (13.4–16.1%). The various numbered clades allied to Hipposideros caffer differ from one another in cyt-b sequences by 2.5–10.3% and clades allied to H. ruber differ by 3.0–8.2% (Table 4).
Table 4.
Uncorrected cyt-b p-distances between (off diagonal) and within (on diagonal) Afrotropical hipposiderid clades, showing the number of base differences per site averaged over all sequence pairs between groups. The analysis involved 386 nucleotide sequences and all ambiguous positions were removed; na (not available) reflects a sample size of one individual.
| [1] | [2] | [3] | [4] | [5] | [6] | [7] | [8] | [9] | [10] | [11] | [12] | [13] | [14] | [15] | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [1] | Doryrhina camerunensis | 0.003 | ||||||||||||||
| [2] | Doryrhina cf. camerunensis | 0.055 | na | |||||||||||||
| [3] | Doryrhina cyclops 1 | 0.057 | 0.048 | 0.008 | ||||||||||||
| [4] | Doryrhina cyclops 2 | 0.055 | 0.041 | 0.030 | 0.006 | |||||||||||
| [5] | Hipposideros abae | 0.176 | 0.173 | 0.168 | 0.166 | 0.033 | ||||||||||
| [6] | Hipposideros beatus 1 | 0.152 | 0.156 | 0.160 | 0.152 | 0.116 | 0.007 | |||||||||
| [7] | Hipposideros beatus 2 | 0.146 | 0.149 | 0.151 | 0.147 | 0.117 | 0.044 | 0.006 | ||||||||
| [8] | Hipposideros caffer 1 | 0.157 | 0.154 | 0.150 | 0.148 | 0.108 | 0.103 | 0.106 | 0.006 | |||||||
| [9] | Hipposideros caffer 2 | 0.153 | 0.150 | 0.150 | 0.147 | 0.106 | 0.096 | 0.108 | 0.045 | 0.01 | ||||||
| [10] | Hipposideros caffer 3 | 0.152 | 0.148 | 0.150 | 0.149 | 0.108 | 0.110 | 0.111 | 0.046 | 0.052 | 0.011 | |||||
| [11] | Hipposideros caffer 4 | 0.162 | 0.159 | 0.160 | 0.154 | 0.106 | 0.105 | 0.114 | 0.077 | 0.078 | 0.079 | 0.018 | ||||
| [12] | Hipposideros caffer 5 | 0.150 | 0.155 | 0.159 | 0.148 | 0.113 | 0.091 | 0.096 | 0.095 | 0.101 | 0.103 | 0.098 | 0.005 | |||
| [13] | Hipposideros caffer 6 | 0.155 | 0.156 | 0.164 | 0.153 | 0.112 | 0.093 | 0.102 | 0.090 | 0.097 | 0.099 | 0.094 | 0.028 | 0.011 | ||
| [14] | Hipposideros caffer 7 | 0.151 | 0.154 | 0.161 | 0.148 | 0.108 | 0.084 | 0.094 | 0.094 | 0.094 | 0.098 | 0.096 | 0.025 | 0.032 | 0.011 | |
| [15] | Hipposideros caffer 8 | 0.154 | 0.155 | 0.160 | 0.152 | 0.111 | 0.090 | 0.092 | 0.092 | 0.095 | 0.102 | 0.093 | 0.033 | 0.039 | 0.029 | 0.021 |
| [16] | Hipposideros fuliginosus | 0.155 | 0.149 | 0.154 | 0.142 | 0.101 | 0.095 | 0.096 | 0.078 | 0.084 | 0.087 | 0.094 | 0.088 | 0.086 | 0.080 | 0.085 |
| [17] | Hipposideros jonesi | 0.153 | 0.143 | 0.154 | 0.147 | 0.160 | 0.145 | 0.138 | 0.135 | 0.140 | 0.140 | 0.139 | 0.134 | 0.134 | 0.137 | 0.139 |
| [18] | Hipposideros lamottei | 0.171 | 0.173 | 0.174 | 0.163 | 0.106 | 0.097 | 0.118 | 0.094 | 0.086 | 0.095 | 0.097 | 0.057 | 0.059 | 0.052 | 0.060 |
| [19] | Hipposideros cf. lamottei | 0.158 | 0.158 | 0.156 | 0.149 | 0.107 | 0.103 | 0.107 | 0.091 | 0.099 | 0.095 | 0.097 | 0.053 | 0.058 | 0.052 | 0.054 |
| [20] | Hipposideros marisae | 0.177 | 0.180 | 0.178 | 0.172 | 0.159 | 0.153 | 0.153 | 0.144 | 0.148 | 0.140 | 0.152 | 0.148 | 0.144 | 0.154 | 0.159 |
| [21] | Hipposideros ruber 1 | 0.155 | 0.151 | 0.152 | 0.142 | 0.104 | 0.099 | 0.101 | 0.081 | 0.084 | 0.085 | 0.086 | 0.090 | 0.081 | 0.082 | 0.082 |
| [22] | Hipposideros ruber 2 | 0.155 | 0.155 | 0.156 | 0.142 | 0.103 | 0.097 | 0.102 | 0.081 | 0.082 | 0.089 | 0.089 | 0.086 | 0.078 | 0.083 | 0.081 |
| [23] | Hipposideros ruber 3 | 0.159 | 0.149 | 0.155 | 0.147 | 0.105 | 0.094 | 0.101 | 0.084 | 0.082 | 0.096 | 0.088 | 0.090 | 0.083 | 0.082 | 0.082 |
| [24] | Hipposideros ruber 4 | 0.153 | 0.147 | 0.151 | 0.141 | 0.100 | 0.094 | 0.097 | 0.073 | 0.073 | 0.085 | 0.082 | 0.083 | 0.077 | 0.078 | 0.078 |
| [25] | Hipposideros cf. ruber | 0.164 | 0.161 | 0.161 | 0.153 | 0.099 | 0.094 | 0.100 | 0.086 | 0.084 | 0.095 | 0.093 | 0.081 | 0.080 | 0.080 | 0.083 |
| [26] | Macronycteris commersoni | 0.157 | 0.156 | 0.155 | 0.145 | 0.164 | 0.159 | 0.154 | 0.164 | 0.169 | 0.168 | 0.171 | 0.166 | 0.170 | 0.167 | 0.162 |
| [27] | Macronycteris cryptovalorona | 0.147 | 0.143 | 0.147 | 0.142 | 0.157 | 0.152 | 0.146 | 0.156 | 0.162 | 0.161 | 0.165 | 0.159 | 0.164 | 0.158 | 0.157 |
| [28] | Macronycteris gigas | 0.151 | 0.149 | 0.154 | 0.144 | 0.164 | 0.163 | 0.158 | 0.162 | 0.168 | 0.165 | 0.170 | 0.165 | 0.171 | 0.164 | 0.165 |
| [29] | Macronycteris vittata | 0.147 | 0.148 | 0.149 | 0.140 | 0.162 | 0.158 | 0.147 | 0.160 | 0.161 | 0.166 | 0.165 | 0.164 | 0.169 | 0.164 | 0.163 |
Table 4.
Continued.
| [16] | [17] | [18] | [19] | [20] | [21] | [22] | [23] | [24] | [25] | [26] | [27] | [28] | [29] | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [16] | Hipposideros fuliginosus | 0.003 | |||||||||||||
| [17] | Hipposideros jonesi | 0.137 | 0.008 | ||||||||||||
| [18] | Hipposideros lamottei | 0.100 | 0.161 | 0.038 | |||||||||||
| [19] | Hipposideros cf. lamottei | 0.091 | 0.142 | 0.054 | 0.006 | ||||||||||
| [20] | Hipposideros marisae | 0.145 | 0.092 | 0.156 | 0.158 | na | |||||||||
| [21] | Hipposideros ruber 1 | 0.082 | 0.144 | 0.093 | 0.087 | 0.152 | 0.013 | ||||||||
| [22] | Hipposideros ruber 2 | 0.084 | 0.147 | 0.093 | 0.091 | 0.157 | 0.030 | 0.007 | |||||||
| [23] | Hipposideros ruber 3 | 0.082 | 0.142 | 0.085 | 0.088 | 0.158 | 0.052 | 0.057 | 0.022 | ||||||
| [24] | Hipposideros ruber 4 | 0.078 | 0.140 | 0.087 | 0.088 | 0.154 | 0.053 | 0.051 | 0.057 | na | |||||
| [25] | Hipposideros cf. ruber | 0.084 | 0.142 | 0.094 | 0.089 | 0.157 | 0.081 | 0.081 | 0.082 | 0.072 | 0.033 | ||||
| [26] | Macronycteris commersoni | 0.155 | 0.186 | 0.183 | 0.167 | 0.193 | 0.163 | 0.162 | 0.161 | 0.168 | 0.174 | 0.012 | |||
| [27] | Macronycteris cryptovalorona | 0.149 | 0.176 | 0.170 | 0.163 | 0.185 | 0.161 | 0.161 | 0.158 | 0.160 | 0.162 | 0.028 | 0.003 | ||
| [28] | Macronycteris gigas | 0.153 | 0.179 | 0.178 | 0.169 | 0.190 | 0.165 | 0.165 | 0.161 | 0.165 | 0.169 | 0.026 | 0.029 | 0.012 | |
| [29] | Macronycteris vittata | 0.150 | 0.181 | 0.180 | 0.168 | 0.192 | 0.158 | 0.159 | 0.158 | 0.159 | 0.164 | 0.026 | 0.029 | 0.027 | 0.006 |
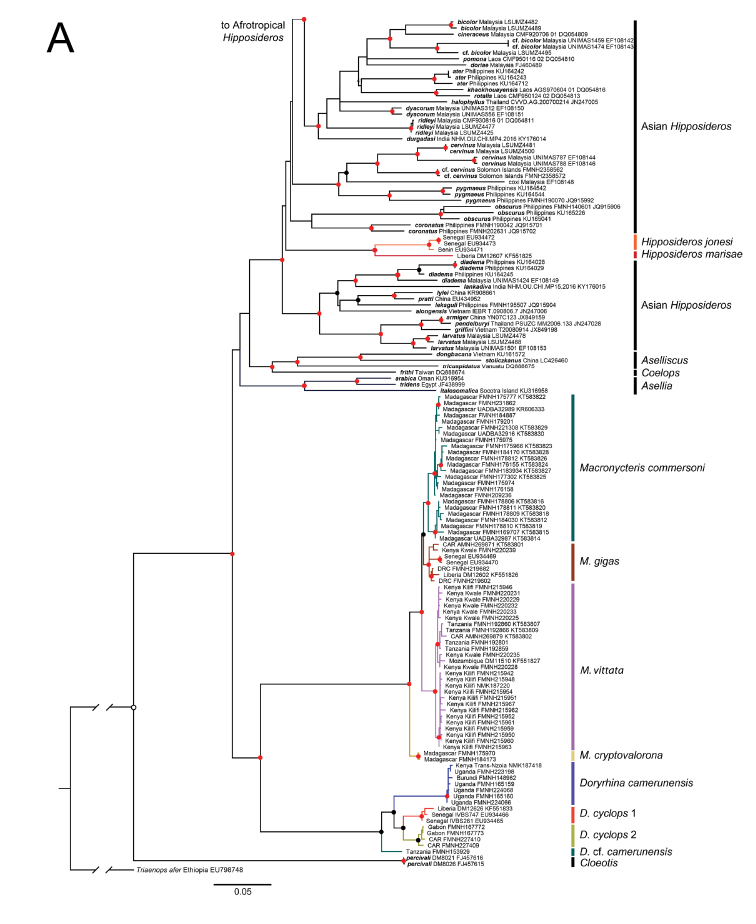
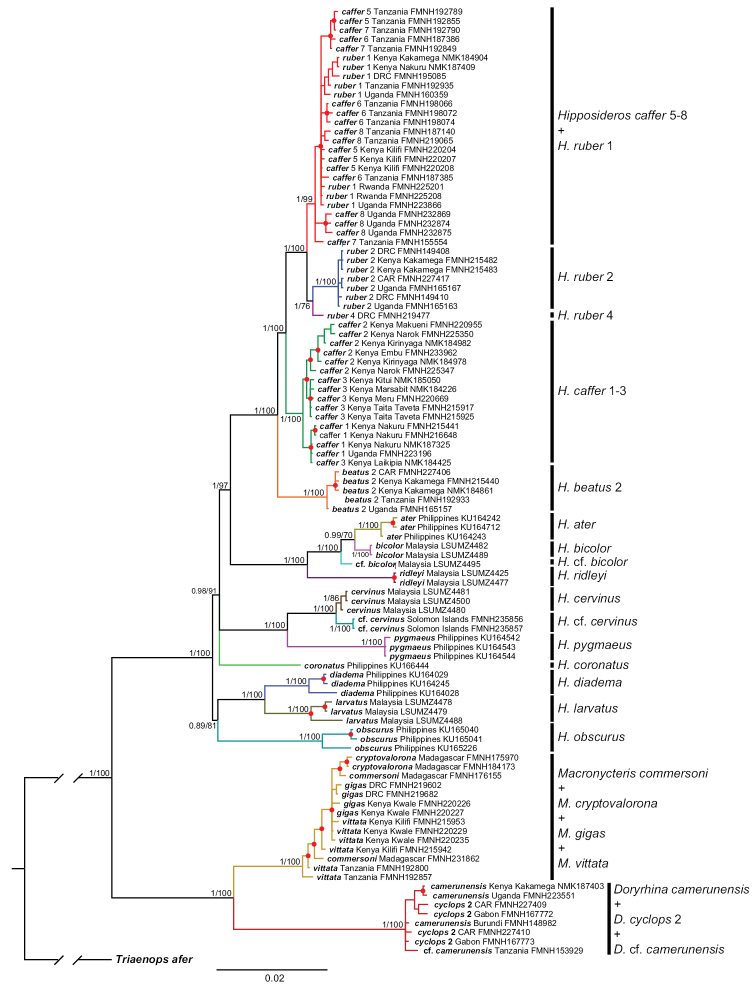
Maximum likelihood and Bayesian phylogenies from a 452-individual alignment of cyt-b are shown in Suppl. material 2: Figures S2, Suppl. material 3: Figures S3. Identical haplotypes were pruned from this tree to produce the 303 unique-haplotype alignment shown in Figure 2. The 303 haplotype alignment used in the ML and BI gene tree analyses ranged from 413 to 1140 base pairs (bp) in length (89.9% complete matrix). Only the Bayesian topology is shown, but both posterior probabilities and bootstrap values are depicted at common, well supported nodes. Multiple, geographically cohesive clades are evident for the three widely distributed Afrotropical Hippposideros, H. beatus, H. caffer, and H. ruber.
Figure 2.
Parts A and B. Phylogeny of Hipposideridae based on Bayesian analysis of 303 cyt-b sequences. Colored lines denote well supported clades and symbols denote nodal support: red circles, BS ≥ 70%, PP ≥ 0.95; black circles BS ≥ 70%, PP ≤ 0.95; open circles BS ≤ 70%, PP ≥ 0.95.
Figure 2.
Continued.
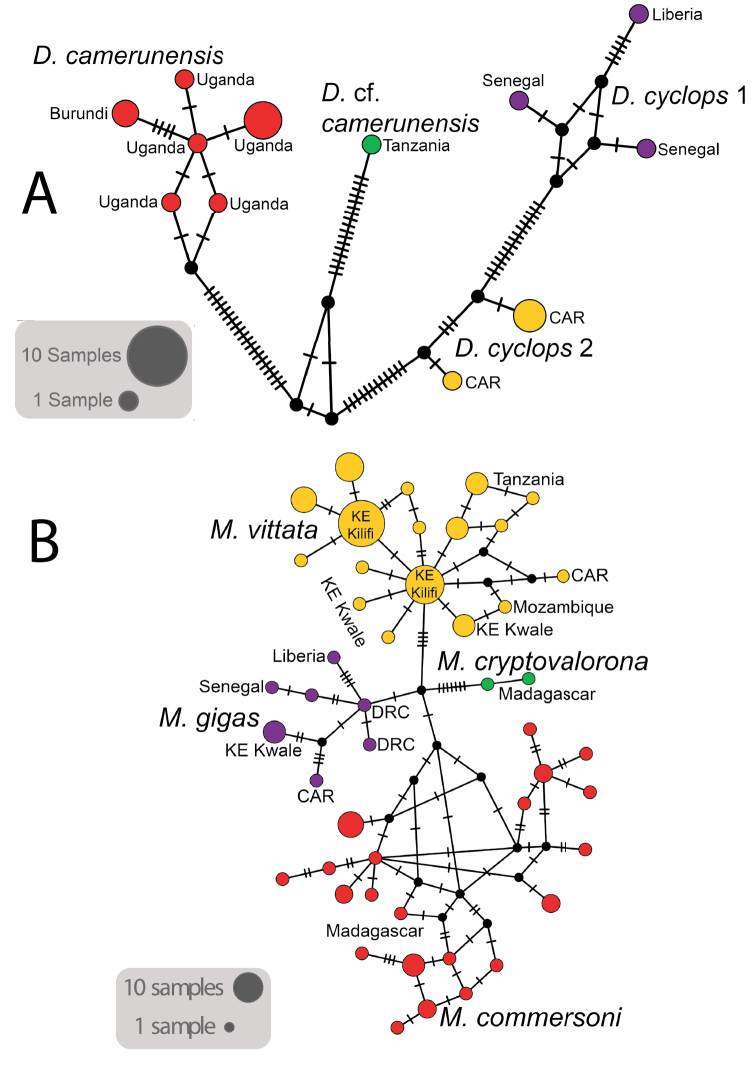
Substitution networks for cyt-b haplotypes for Doryrhina, Macronycteris, and Hipposideros are shown in Figures 3, 4, showing the genetic and geographic relationships of the clades identified in Figure 2.
Figure 3.
Substitution network plots for Afrotropical hipposiderids ADoryrhinaBMacronycteris.
Figure 4.
Substitution network plots for Afrotropical hipposiderids AHipposideros caffer clades 1–4 BHipposideros caffer clades 5–8 CH. ruber clades.
Maximum likelihood and Bayesian phylogenies from a 103-individual alignment of four concatenated introns for Doryrhina, Macronycteris, and Hipposideros are shown in Figure 5. Many of the numbered clades in Figures 2–4 are jumbled in Figure 5; they are not recovered as monophyletic units and the geographic structure evident in mtDNA analyses disappears.
Figure 5.
Phylogeny of Hipposideridae based on Bayesian analysis of 103 concatenated nuclear intron sequences. Numbers denote posterior probabilities (BI) and bootstrap percentages (ML); red circles at more terminal nodes indicate BS ≥ 70%, PP ≥ 0.95.
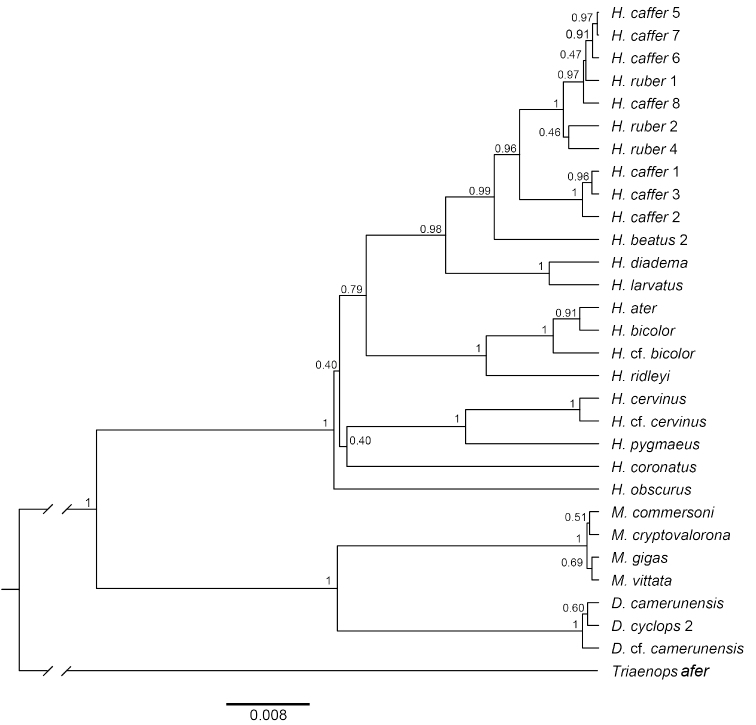
A species tree generated using StarBEAST from the four introns appears in Figure 6. It depicts well-supported relationships among the various clades allied with H. caffer, H. ruber, and H. beatus. Remarkably, and in contrast with the concatenated analyses, it shows support for the Asian dyad H. diadema and H. larvatus as sister to these ruber subgroup members, with the Asian ater subgroup outside this pairing. There is little support for the deeper phylogenetic nodes.
Figure 6.
Species tree Hipposideridae based on StarBEAST analysis of four introns. Posterior probabilities appear at all nodes.
Discussion
Overall genetic variability
The three Afrotropical hipposiderid genera differ substantially in terms of their internal genetic differentiation. Clades of Hipposideros are separated by cyt-b p-distances averaging 9.7% (2.5–16.1%), whereas Doryrhina clades average p-distances of 4.8% (3.0–5.7%) and Macronycteris clades 2.7% (2.6–2.9%). Distance values for these genera tend to fall at the lower end of values obtained with similar sampling intensity for species-ranked clades in other Afrotropical bat genera: 2.5% for Otomops (Patterson et al. 2018), 9.3% for Miniopterus (Demos et al. 2020), 10% for Scotophilus and Rhinolophus (Demos et al. 2018, 2019a), 13.5% for Myotis (Patterson et al. 2019), and 17% for Nycteris (Demos et al. 2019b). Fewer cyt-b substitutions on average for these hipposiderids does not limit support for individual clades, and because distances do not approach those characteristic of substitutional saturation, the cyt-b tree recovers much of the deeper phylogenetic structure evident with nuclear intron sequences (compare Figs 2, 5).
Phylogenetics
Both cyt-b and intron analyses securely recovered Doryrhina, Macronycteris, and Hipposideros as monophyletic. Doryrhina + Macronycteris are sister to the remaining hipposiderids. However, only the cyt-b analysis included the hipposiderid genera Aselliscus, Coelops, and Asellia alongside Hipposideros. That analysis recovered all four genera as monophyletic with strong support. Aselliscus and Coelops were recovered as sister to Hipposideros, with Asellia joining later, but these relationships lacked confident support.
Using a supermatrix approach on exemplars of 46 species of hipposiderids, Amador et al. (2018) found Hipposideros sensu stricto to be paraphyletic. They recovered a mostly Asian group of Hipposideros as sister to two subclades, Coelops + Aselliscus and Asellia + African hipposiderids excluding H. jonesi, which was recovered with the Asian taxa. Paraphyly in this molecular analysis echoed earlier indications of Hipposideros paraphyly from morphology (Bogdanowicz and Owen 1998; Hand and Kirsch 1998, 2003). In another supermatrix analysis of exemplars belonging to 49 hipposiderid species, Shi and Rabosky (2015) failed to recover Macronycteris as monophyletic; M. commersoni was sister to all remaining hipposiderids, but strangely it did not group with M. gigas. When the anomalous position of M. commersoni in their tree is ignored, their topology is highly similar to that of Figure 2, except that Asellia (Aselliscus, Coelops) become the sister of Hipposideros (Macronycteris, Doryrhina), rather than sister of just Hipposideros. Using both mitochondrial and nuclear loci, Lavery et al. (2014) found that 17 species of Asian, Oceanian and Australasian Hipposideros were monophyletic with respect to the genera Aselliscus, Coelops, and Anthops. Clearly, missing data and missing taxa compromise all of these phylogenetic appraisals, so that the question of hipposiderid and Hipposideros monophyly remains open. However, subject to its sampling limitations, there is clear support in our analyses of monophyly for Doryrhina, Macronycteris, and Hipposideros as we apply these names.
Despite employing different mitochondrial and nuclear loci and using different sets of taxa, the phylogeny recovered by Lavery et al. (2014) is largely congruent with that in Figure 5. Their earliest diverging species group of Hipposideros is the calcaratus group, not represented in our tree unless H. obscurus is a member (Table 1). Their next diverging unit is the diadema group, which is also positioned near the base of our tree. Their other two groups are paired: the galeritus group (which includes H. cervinus, indicating that this species is misclassified as calcaratus member) joined with the bicolor/ater group. In our intron analysis (Fig. 5), members of the larvatus and diadema groups join H. obscurus as sister to all remaining Hipposideros groups. The remainder form a trichotomy: H. coronatus, typically considered in the bicolor group; H. pygmaeus and H. cervinus, which are listed in different groups but were both considered members of the galeritus unit by Tate (1941); and the erstwhile bicolor group (sensu Hill 1963), which was subdivided into the ater subgroup (for Asian, Oceanian, and Australasian species) and the ruber subgroup (for Atrotropical ones) by Monadjem (2019).
The ater subgroup members included in our mitochondrial analysis (Fig. 2) form a well-supported clade consisting of H. bicolor, H. cineraceus, H. pomona, H. doriae, H. ater, H. khaokhouayensis, H. rotalis, H. halophyllusH. dyacorum, H. ridley, and H. durgadasi. This group is sister to all analyzed members of the ruber subgroup: the various clades allied with Hipposideros beatus, H. caffer, and H. ruber, as well as individuals of the Afrotropical species H. lamottei and H. fuliginosus. H. abae, which was previously considered in the speoris group (Simmons 2005; Murray et al. 2012), is clearly a member of the ruber group. Outside this pairing are the Asian species H. cervinus, H. coronatus, H. coxi, H. obscurus, and H. pygmaeus. Two Afrotropical species also lie outside the ruber + ater clade: H. jonesi and H. marisae, both thought to belong to the speoris group, appear as sisters in Figure 2A.
Parsimony, topological position, and the strong support of branching relationships in the mitochondrial and intron trees (Fig. 5; also Lavery et al. 2014) make it clear that the Afrotropical ruber group represents a comparatively recent colonization event from Asian ancestors–the ruber group is sister to the ater group and this pair has Asian sisters. However, although the basal dichotomy within Hipposideros includes an all Asian clade, lack of support for its sister(s) clouds the phylogenetic position of the H. jonesi-H. marisae clade–possibly sister to all sampled Hipposideros but more likely sandwiched between Asian clades. In any case, Figure 2 suggests that the H. jonesi-H. marisae clade resulted from an earlier African-Asian colonization event.
The lack of agreement in the phylogenetic position of H. diadema and H. larvatus between the concatenated intron tree (Fig. 5) and the species tree (Fig. 6) deserves comment, as both analyses were based on the same genetic dataset. The position of H. diadema-H. larvatus as sister to the ruber group (Fig. 6) runs counter to both our other genetic analyses (Figs 2, 5) and morphological assessments (Hill 1963; Murray et al. 2012; Table 1). This discrepancy is likely due to the generally weaker support for deep nodes within the tree; in the absence of saturation, this is often taken as evidence of rapid evolutionary radiations (e.g., Almeida et al. 2011). Lanier and Knowles (2014) used simulated data on deep phylogenies to show that species-tree methods do account for coalescent variance at deep nodes but that mutational variance among lineages poses the primary challenge for accurate reconstruction. In either case, vastly expanded genetic sampling via NGS techniques offers the most plausible avenues to clearer resolution.
However, the highly distinctive species H. megalotis belongs to its own species group (Table 1) and has not been included in any genetic analysis. Distributed in the Horn of Africa and the Arabian Peninsula, H. megalotis is the only hipposiderid with a fold of skin joining the base of the ear pinnae. Its uniquely specialized auditory system and derived dentition (e.g., loss of anterior premolars and enlargement of outer lower incisors), led Hill (1963) to regard it as a species that diverged early from the other groups of African Hipposideros. Including this species in future analyses would shed light on the group’s biogeography. Were there three colonizations of Africa by Asian groups of Hipposideros or could H. megalotis be sister to all Asian lineages of this genus? This information would greatly clarify ancestral geographic range inference.
Species limits
The lineage delimitation analyses indicate that a number of hipposiderid lineages are either unnamed or unidentified, and also that a number of recognized species may not be genetically and evolutionarily independent.
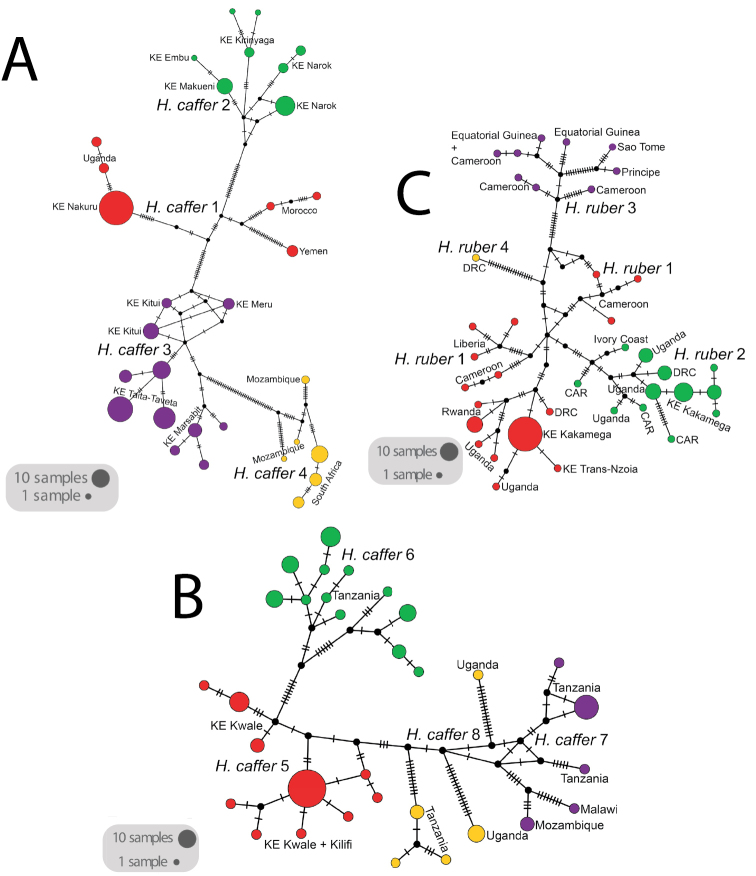
Previous studies had indicated that both Hipposideros caffer (Vallo et al. 2008) and H. ruber (Vallo et al. 2011) appear to be complexes of cryptic species. The two are traditionally distinguished on the basis of size and pelage color, H. ruber being the larger and more brightly colored form, but this distinction is clouded by geographic variation in size and the presence of both reddish and gray-brown phases in both species. Our mitochondrial analyses identified four H. ruber lineages and eight H. caffer lineages in two distinct groupings among the sampled populations (Fig. 2). Four of the caffer lineages and three of the ruber clades were identified as putative species by the BPP analyses (Table 5). The large number of clades in East Africa is remarkable: Kenya and Tanzania each support four of the eight clades allied with H. caffer, and all but one of the eight clades known from throughout the continent occur in one or the other East African country. This undoubtedly reflects the region’s great landscape diversity, where West and Central African rainforests reach their eastern limit, southern savannas reach their northern limits, the Sahel reaches its southern limits, and all are riven by the African Rift Valley. It also is a product of our sampling intensity (see Suppl. material 1: Fig. S1).
Table 5.
Lineage delimitation results from BPP based on the four intron dataset for mtDNA-supported clades of Afrotropical Hipposideridae. PS1-PS4 refer to four different prior schemes based on population size and age of divergence priors (see Table 3 for parameter details). Bold font indicates that the putative species was delimited under all parameter settings.
| Putative Species | PS1 | PS2 | PS3 | PS4 |
|---|---|---|---|---|
| Doryrhina camerunensis | 0.30 | 0.76 | 0.95 | 0.51 |
| D. cf. camerunensis | 0.32 | 0.73 | 0.97 | 0.79 |
| D. cyclops 2 | 0.23 | 0.68 | 0.95 | 0.51 |
| Hipposideros beatus 2 | 1 | 1 | 1 | 1 |
| H. caffer 1 | 0.99 | 0.99 | 0.99 | 0.99 |
| H. caffer 2 | 0.99 | 1 | 1 | 1 |
| H. caffer 3 | 0.99 | 0.99 | 0.99 | 0.99 |
| H. caffer 5 | 0.14 | 0.18 | 0.11 | 0.08 |
| H. caffer 6 | 0.56 | 0.61 | 0.85 | 0.82 |
| H. caffer 7 | 0.14 | 0.18 | 0.11 | 0.08 |
| H. caffer 8 | 0.99 | 0.99 | 0.99 | 0.99 |
| H. ruber 1 | 0.99 | 0.99 | 0.99 | 0.99 |
| H. ruber 2 | 1 | 0.99 | 1 | 0.99 |
| H. ruber 4 | 0.99 | 0.99 | 0.99 | 0.99 |
| Macronycteris commersoni | 0.24 | 0.72 | 0.94 | 0.52 |
| M. cryptovalorona | 0.35 | 0.81 | 0.97 | 0.76 |
| M. gigas | 0.09 | 0.43 | 0.91 | 0.38 |
| M. vittata | 0.11 | 0.44 | 0.90 | 0.34 |
Because some cyt-b sequences were used in multiple studies of this group, it is possible to relate our clade labels to those used by earlier studies (Table 6). Based on attributions made on morphological grounds by Vallo et al. (2008) and Monadjem et al. (2013), some well-supported but unnamed clades in our analysis can be identified. For instance, caffer1 has a distributional range and includes specimens previously identified as Hipposideros tephrus (Appendix I), while specimens of caffer4 come from near the type locality of H. caffer Sundevall, 1846, and may well represent that species. However, no samples confidently identified as H. ruber from the vicinity of its type locality have been sequenced, leaving the application of that name to clades in any of these trees purely conjectural. Applying formal names only after integrative taxonomic assessment is a responsible course as multispecies coalescent models like BPP can lead to over-splitting of species, especially when applied to geographically variable species complexes with parapatric distributions (Chambers and Hillis 2020).
Table 6.
Clade names and associated binomials (if used) for three analyses of cryptic lineages within the ruber species group of Hipposideros. No genetic analysis of this group has included type material; consequently, the application of binomials hinges on the robustness of ancillary morphological analyses, which were not conducted in our study. Boldfaced names denote clades supported by all four prior schemes in our BPP delimitation analyses.
| Vallo et al. (2008) | Monadjem et al. (2013) | This paper | ||
|---|---|---|---|---|
| A1 | H. caffer | caffer 4 | ||
| A1a | H. caffer | caffer 4 | ||
| A1b | H. caffer | caffer 4 | ||
| A2 | H. caffer | H. caffer tephrus | caffer 1 | |
| B | H. ruber | caffer 5 | ||
| B1 | H. ruber | cf. lamottei | ||
| B2 | H. ruber | caffer 7 | ||
| C1 | H. ruber | ruber 1, ruber 2 | ||
| C1a | H. cf. ruber | ruber 1 | ||
| C1b | H. cf. ruber | ruber 1 | ||
| C2 | H. ruber | C2 | H. cf. ruber | ruber 3 |
| ruber 4 | ||||
| D | H. ruber | H. cf. ruber | cf. ruber | |
| D1 | H. cf. ruber | cf. ruber | ||
| D2 | H. cf. ruber | cf. ruber | ||
| – | E1 | H. cf. ruber | cf. ruber | |
| – | E2 | H. cf. ruber | cf. ruber | |
| – | – | caffer 2 | ||
| – | – | caffer 3 | ||
| – | – | caffer 6 | ||
| – | – | caffer 8 | ||
| abae | H. abae | abae | H. abae | abae |
| beatus | H. beatus | beatus | H. beatus | beatus1, beatus 2 |
| fuliginosus | H. fuliginosus | fuliginosus | H. fuliginosus | fuliginosus |
| lamottei | H. lamottei | lamottei | H. lamottei | lamottei |
Doryrhina is a poorly known genus characterized morphologically by the peculiar club-shaped processes on the central and posterior nose leaves. This trait is shared by the two recognized African species, D. cyclops and D. camerunensis, which differ chiefly in size (the latter is larger, with forearm lengths >75 mm). Although D. cyclops is considered to be monotypic, mitochondrial sequences clearly separate West African populations in Liberia and Senegal (cyclops1) from Central African populations in Gabon and Central African Republic (cyclops2), and these are substantially separated from D. camerunensis and a specimen referred to that species from Tanzania (Figs 2, 3). However, both the intron analysis (Fig. 5) and the species tree (Fig. 6) show little or no geographic structure. The BPP analyses confirm that none of the mitochondrial clades is behaving as an independent evolutionary lineage (Table 5). Geographic structure in mtDNA but continent-wide admixture in the nuclear genome could result from either male-biased dispersal with female philopatry or highly structured seasonal migrations, which are known in other hipposiderids. In any case, the genetic patterns of Doryrhina are hard to reconcile with its space-use behavior; individuals appear to have very small home ranges, on the order of a few hectares (Monadjem 2019). An integrative taxonomic review of the genus Doryrhina is needed to determine the validity of D. cyclops and D. camerunensis. It would also shed light on whether six Australo-Papuan species tentatively allocated to that genus (cf. Monadjem 2019) belong there or elsewhere. Tate (1941) had earlier allocated those species to the Australasian muscinus group, convergent on but separate from his Afrotropical cyclops group, but Hill (1963) later united these groups.
Our analysis included four of the five recognized species of Macronycteris, lacking only M. thomensis, which is endemic to São Tomé Island in the Gulf of Guinea. Two species, M. gigas and M. vittata, occur on the African mainland and two others, M. commersoni and M. cryptovalorona, occur on Madagascar. Macronycteris cryptovalorona was named only in 2016, on the basis of its strong genetic divergence from M. commersoni; it appears in Figure 2 as sister to all three remaining species of Macronycteris. Despite a search for diagnostic characters, Goodman et al. (2016) could not distinguish it morphologically from M. commersoni. Both species are known to occur in the same caves in south central and southwestern Madagascar (Goodman et al. 2016; Rakotoarivelo et al. 2019). On the other hand, M. vittata and M. gigas are distinguished typically on the basis of size and pelage color (cf. Monadjem 2019). They are also known to occur together in the same cave (Shimoni Cave in Kwale, Kenya; Webala et al. 2019), where they utilize echolocation calls with different peak frequencies: vittata at 64–70 kHz and gigas at 53.4–54.8 kHz. Both in Africa and on Madagascar, these pairs of taxa appear to act as distinct species, but the monophyly evident in the cyt-b sequences (Figs 2, 4) disappears in the nuclear intron analyses. BPP analyses fail to resolve any of the Macronycteris species, and none appear as monophyletic in the concatenated intron analyses.
Our results clearly underscore the importance of using multilocus datasets to evaluate phylogenetic and phylogeographic relationships at the genus and species level in mammals. Use of a single genetic system may lead to widely divergent conclusions regarding species identity and distribution. Toews and Brelsford (2012) reviewed cases of mito-nuclear discordance in animals generally. Fully 18% of the cases they reviewed had discordant patterns of mitochondrial and nuclear DNA. In most cases, such patterns are attributable to adaptive introgression of mtDNA, demographic disparities, and sex-biased asymmetries; in some cases they found evidence for hybrid zone movement or human agency. Discordant patterns of variation between mitochondrial and nuclear DNA have been reported in at least six other families of bats (Nesi et al. 2011; Furman et al. 2014; Naidoo et al. 2016; Hassanin et al. 2018; Demos et al. 2019a; Gürün et al. 2019). Gürün et al. (2019) implicated the role of sex-biased dispersal in causing such discordance, male dispersal spreading nuclear variation farther and faster than the movement of mitochondria. This may be a more general pattern in bats (see also Demos et al. 2019b). To understand the processes responsible for these discordant patterns of genome evolution, extensive genomic sampling and far fuller knowledge of natural history will be required.
Acknowledgments
We thank Michael Bartonjo, Carl Dick, Ruth Makena, Beryl Makori, David Wechuli, Richard Yego, and Aziza Zuhura for help in obtaining specimens in the field. We acknowledge with special thanks the assistance of Simon Musila (National Museums of Kenya), Donna Dittman and Jacob Esselstyn (Louisiana State University) and Maria Eifler (University of Kansas) for loans of material. We also appreciate the efforts of curators and collection managers in all the institutions cited in Appendix I for maintaining the museum voucher specimens that enable integrative taxonomic studies to confidently name our numbered lineages. Fieldwork in eastern and southern Africa was funded by various agencies in cooperation with the Field Museum, especially the JRS Biodiversity Foundation. Field Museum’s Council on Africa, Marshall Field III Fund, and Barbara E. Brown Fund for Mammal Research were critical to fieldwork and analyses, as were supporters Bud and Onnolee Trapp and Walt and Ellen Newsom. The John D. and Catherine T. MacArthur Foundation, Fulbright Program of US Department of State, Wildlife Conservation Society, and the Centers for Disease Control and Prevention sponsored and/or assisted in providing samples from DRC, Malawi, Mozambique, and Uganda. WWF Gabon supported fieldwork in Gabon, as did the Partenariat Mozambique-Réunion dans la Recherche en Santé: pour une approche intégrée d’étude des maladies infectieuses à risque épidémique (MoZaR; Fond Européen de Développement Régional, Programme Opérationnel de Coopération Territoriale) in Mozambique. We thank Ara Monadjem and Daniela Rossoni for reviews of an earlier draft of this manuscript.
Appendix I
Genetic sampling of Hipposideridae. Wherever possible, the voucher numbers associated with the genetic samples are specified. Accession numbers identify sequences downloaded from GenBank or accessioned to Genbank for this study. The designation 'redundant' indicates a cyt-b sequence that was omitted from the 303 individual alignment because of its identity to another. Institutional acronyms are as follows: AMNH – American Museum of Natural History, New York; CM – Carnegie Museum, Pittsburgh; CVVD – ?; DM – Durban Natural Science Museum, Durban; EBD – Estación Biológica de Doñana, Sevilla; FMNH – Field Museum of Natural History, Chicago; IEBR-T – Thong collection at Institute of Ecology and Biological Resources, Hanoi; IVB – Institute of Vertebrate Biology, Brno; KU – Biodiversity Institute and Natural History Museum, University of Kansas, Lawrence; Czech Academy of Sciences, Prague; LSUMZ – Lousiana State University Museum of Natural Science-Mammal Tissues, Baton Rouge; NHMOU – Natural History Museum of Osmania University, Hyderabad; NMK – National Museums of Kenya, Nairobi; NMP – National Museum, Prague; PSUZC – Princess Maha Chakri Sirindhorn Natural History Museum, Songkhla; ROM – Royal Ontario Museum, Toronto; SMF – Senckenberg Museum, Frankfurt; TM – Transvaal Museum, Pretoria; TTU – Texas Tech University Museum, Lubbock; UADBA – Université d'Antananarivo, Département de Biologie Animale, Antananarivo; UNIMAS – University of Malaysia Sarawak Natural History Museum, Kuching.
| Voucher | cyt-b | ACOX2 | COPS | ROGDI | STAT5 | ScientificName | Country | Latitude | Longitude | |
|---|---|---|---|---|---|---|---|---|---|---|
| KU316954 | Asellia arabica | Oman | 17.100 | 54.080 | ||||||
| KU316958 | Asellia italosomalica | Yemen | 12.670 | 54.120 | ||||||
| JF438999 | Asellia tridens | Libya | 24.933 | 10.167 | ||||||
| IEBR-T | KU161572 | Aselliscus dongbacana | Vietnam | 22.360 | 105.395 | |||||
| LC426460 | Aselliscus stoliczkanus | China | ||||||||
| DQ888675 | Aselliscus tricuspidatus | Vanuatu | -15.307 | 166.926 | ||||||
| DM 8021 | FJ457616 | Cloeotis percivali | Swaziland | -25.817 | 31.283 | |||||
| DM 8026 | FJ457615 | Cloeotis percivali | Swaziland | -25.817 | 31.283 | |||||
| DQ888674 | Coelops frithi | Taiwan | 21.948 | 120.780 | ||||||
| FMNH 148981 | redundant | Doryrhina camerunensis | Burundi | -2.100 | 29.383 | |||||
| FMNH 148982 | MT149719 | MT149615 | MT149513 | MT149418 | MT149317 | Doryrhina camerunensis | Burundi | -2.850 | 29.400 | |
| NMK 187403 | redundant | MT149616 | MT149514 | MT149419 | MT149318 | Doryrhina camerunensis | Kenya | 0.344 | 34.857 | |
| NMK 187418 | MT149720 | Doryrhina camerunensis | Kenya | 0.344 | 34.857 | |||||
| FMNH 165159 | MT149721 | Doryrhina camerunensis | Uganda | 1.683 | 31.533 | |||||
| FMNH 165160 | MT149722 | Doryrhina camerunensis | Uganda | 1.683 | 31.533 | |||||
| FMNH 223198 | MT149723 | Doryrhina camerunensis | Uganda | 0.445 | 32.889 | |||||
| FMNH 223551 | redundant | MT149617 | MT149515 | MT149420 | MT149319 | Doryrhina camerunensis | Uganda | 0.445 | 32.889 | |
| FMNH 224066 | MT149724 | Doryrhina camerunensis | Uganda | 0.501 | 30.426 | |||||
| FMNH 224068 | MT149725 | Doryrhina camerunensis | Uganda | 0.501 | 30.426 | |||||
| FMNH 153929 | MT149726 | MT149618 | MT149516 | MT149421 | MT149320 | Doryrhina cf. camerunensis | Tanzania | -4.942 | 38.733 | |
| DM 12626 | KF551833 | Doryrhina cyclops1 | Liberia | 7.553 | -8.492 | |||||
| IVB S261 | EU934465 | Doryrhina cyclops1 | Senegal | 12.883 | -12.717 | |||||
| IVB S747 | EU934466 | Doryrhina cyclops1 | Senegal | 13.333 | -13.217 | |||||
| FMNH 227409 | MT149727 | MT149619 | MT149517 | MT149422 | MT149321 | Doryrhina cyclops2 | Central African Republic | 13.033 | 16.410 | |
| FMNH 227410 | MT149728 | MT149620 | MT149518 | MT149423 | MT149322 | Doryrhina cyclops2 | Central African Republic | 3.033 | 16.410 | |
| FMNH 167772 | MT149729 | MT149621 | MT149519 | MT149424 | MT149323 | Doryrhina cyclops2 | Gabon | -2.283 | 10.497 | |
| FMNH 167773 | MT149730 | MT149622 | MT149520 | MT149425 | MT149324 | Doryrhina cyclops2 | Gabon | -2.283 | 10.497 | |
| NMP 91850 | EU934446 | Hipposideros abae | Benin | 7.783 | 2.267 | |||||
| NMP 91851 | EU934447 | Hipposideros abae | Benin | 7.783 | 2.267 | |||||
| IVB S822 | EU934448 | Hipposideros abae | Senegal | 12.350 | -12.317 | |||||
| IEBR-T 90806.7 | JN247006 | Hipposideros alongensis | Vietnam | |||||||
| YN07C123 | JX849159 | Hipposideros armiger | China | 23.600 | 102.002 | |||||
| UNIMAS 729 | EF108140 redundant | Hipposideros ater | Malaysia | 1.407 | 110.169 | |||||
| UNIMAS 1577 | EF108139 redundant | Hipposideros ater | Malaysia | 3.801 | 113.785 | |||||
| KU 164242 | MT149731 | MT149623 | MT149521 | MT149426 | MT149325 | Hipposideros ater | Philippines | 13.796 | 120.159 | |
| KU 164243 | MT149732 | MT149624 | MT149522 | MT149427 | MT149326 | Hipposideros ater | Philippines | 13.796 | 120.159 | |
| KU 164712 | MT149733 | MT149625 | MT149523 | MT149428 | MT149327 | Hipposideros ater | Philippines | 19.085 | 121.241 | |
| ROM 100579 | FJ347975 | Hipposideros beatus1 | Ivory Coast | 6.930 | -7.217 | |||||
| DM 13241 | KF551829 | Hipposideros beatus1 | Liberia | 7.553 | -8.492 | |||||
| DM 13242 | KF551830 | Hipposideros beatus1 | Liberia | 7.553 | -8.492 | |||||
| FMNH 227406 | MT149734 | MT149613 | MT149524 | MT149429 | MT149328 | Hipposideros beatus2 | Central African Republic | 3.033 | 16.410 | |
| FMNH 149406 | FJ347976 | Hipposideros beatus2 | Democratic Republic of Congo | -1.417 | 28.583 | |||||
| FMNH 215440 | MT149735 | MT149626 | MT149525 | MT149430 | MT149329 | Hipposideros beatus2 | Kenya | 0.352 | 34.865 | |
| NMK 184861 | MT149736 | MT149627 | MT149526 | MT149431 | MT149330 | Hipposideros beatus2 | Kenya | 0.356 | 34.861 | |
| NMK 184864 | redundant | Hipposideros beatus2 | Kenya | 0.360 | 34.861 | |||||
| NMK 184870 | MT149737 | Hipposideros beatus2 | Kenya | 0.352 | 34.865 | |||||
| FMNH 192931 | MT149738 | Hipposideros beatus2 | Tanzania | -1.094 | 31.515 | |||||
| FMNH 192932 | MT149739 | Hipposideros beatus2 | Tanzania | -1.094 | 31.515 | |||||
| FMNH 192933 | redundant | MT149628 | MT149527 | MT149432 | MT149331 | Hipposideros beatus2 | Tanzania | -1.094 | 31.515 | |
| FMNH 164972 | MT149740 | Hipposideros beatus2 | Uganda | 1.733 | 31.467 | |||||
| FMNH 165157 | redundant | MT149629 | MT149528 | MT149433 | MT149332 | Hipposideros beatus2 | Uganda | 1.683 | 31.533 | |
| LSUMZ MT-4482 | MT149741 | MT149630 | MT149529 | MT149434 | MT149333 | Hipposideros bicolor | Malaysia | 1.970 | 103.500 | |
| LSUMZ MT-4489 | MT149742 | MT149631 | MT149530 | MT149334 | Hipposideros bicolor | Malaysia | 1.970 | 103.500 | ||
| FMNH 215441 | redundant | MT149632 | MT149531 | MT149435 | MT149335 | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |
| FMNH 215442 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 215443 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 215444 | MT149743 | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 215445 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 215446 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 215447 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 216628 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 216629 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 216630 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 216631 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 216632 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 216645 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 216646 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 216647 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 216648 | redundant | MT149633 | MT149532 | MT149436 | MT149336 | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |
| FMNH 225346 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| FMNH 225747 | redundant | Hipposideros caffer1 | Kenya | -0.564 | 36.254 | |||||
| FMNH 225748 | redundant | Hipposideros caffer1 | Kenya | -0.564 | 36.254 | |||||
| FMNH 225749 | redundant | Hipposideros caffer1 | Kenya | -0.564 | 36.254 | |||||
| FMNH 225750 | redundant | Hipposideros caffer1 | Kenya | -0.564 | 36.254 | |||||
| FMNH 225751 | redundant | Hipposideros caffer1 | Kenya | -0.564 | 36.254 | |||||
| NMK 184726 | MT149744 | Hipposideros caffer1 | Kenya | -0.564 | 36.254 | |||||
| NMK 184727 | redundant | Hipposideros caffer1 | Kenya | -0.564 | 36.254 | |||||
| NMK 184728 | redundant | Hipposideros caffer1 | Kenya | -0.564 | 36.254 | |||||
| NMK 184729 | redundant | Hipposideros caffer1 | Kenya | -0.564 | 36.254 | |||||
| NMK 184730 | MT149745 | Hipposideros caffer1 | Kenya | -0.564 | 36.254 | |||||
| NMK 184760 | redundant | Hipposideros caffer1 | Kenya | -0.430 | 36.174 | |||||
| NMK 184842 | redundant | Hipposideros caffer1 | Kenya | -0.539 | 36.294 | |||||
| NMK 184843 | redundant | Hipposideros caffer1 | Kenya | -0.539 | 36.294 | |||||
| NMK 187310 | MT149718 | Hipposideros caffer1 | Kenya | -0.539 | 36.294 | |||||
| NMK 187311 | redundant | Hipposideros caffer1 | Kenya | -0.539 | 36.294 | |||||
| NMK 187312 | redundant | Hipposideros caffer1 | Kenya | -0.539 | 36.294 | |||||
| NMK 187323 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| NMK 187324 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| NMK 187325 | redundant | MT149634 | MT149533 | MT149437 | MT149337 | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |
| NMK 187326 | redundant | Hipposideros caffer1 | Kenya | -0.346 | 36.119 | |||||
| EBD 23262 | FJ347977 | Hipposideros caffer1 | Morocco | 30.630 | 9.830 | |||||
| NMP | EU934449 | Hipposideros caffer1 | Morocco | 3.801 | 113.785 | |||||
| FMNH 223196 | MT149746 | MT149635 | MT149534 | MT149438 | MT149338 | Hipposideros caffer1 | Uganda | 0.445 | 32.889 | |
| FMNH 223197 | MT149747 | Hipposideros caffer1 | Uganda | 0.445 | 32.889 | |||||
| NMP | EU934463 | Hipposideros caffer1 | Yemen | 15.283 | 44.167 | |||||
| FMNH 220955 | redundant | MT149639 | MT149538 | MT149441 | MT149342 | Hipposideros caffer2 | Kenya | -2.203 | 37.714 | |
| FMNH 220956 | redundant | Hipposideros caffer2 | Kenya | -2.203 | 37.714 | |||||
| FMNH 220957 | MT149753 | Hipposideros caffer2 | Kenya | -2.203 | 37.714 | |||||
| FMNH 220958 | redundant | Hipposideros caffer2 | Kenya | -2.203 | 37.714 | |||||
| FMNH 225347 | redundant | MT149640 | MT149539 | MT149442 | MT149343 | Hipposideros caffer2 | Kenya | -1.547 | 35.306 | |
| FMNH 225348 | redundant | Hipposideros caffer2 | Kenya | -1.531 | 35.320 | |||||
| FMNH 225349 | MT149754 | Hipposideros caffer2 | Kenya | -1.531 | 35.320 | |||||
| FMNH 225350 | MT149755 | MT149641 | MT149540 | MT149443 | Hipposideros caffer2 | Kenya | -1.531 | 35.320 | ||
| FMNH 225351 | MT149756 | Hipposideros caffer2 | Kenya | -1.531 | 35.320 | |||||
| FMNH 225352 | redundant | Hipposideros caffer2 | Kenya | -1.531 | 35.320 | |||||
| NMK 184977 | MT149749 | Hipposideros caffer2 | Kenya | -0.117 | 34.541 | |||||
| NMK 184978 | MT149750 | MT149637 | MT149536 | MT149440 | MT149340 | Hipposideros caffer2 | Kenya | -0.117 | 34.541 | |
| NMK 184979 | MT149751 | Hipposideros caffer2 | Kenya | -0.117 | 34.541 | |||||
| NMK 184981 | redundant | Hipposideros caffer2 | Kenya | -0.117 | 34.541 | |||||
| NMK 184982 | MT149752 | MT149638 | MT149537 | MT149341 | Hipposideros caffer2 | Kenya | -0.117 | 34.541 | ||
| NMK 184999 | MT149748 | MT149636 | MT149535 | MT149439 | MT149339 | Hipposideros caffer2 | Kenya | -0.555 | 37.388 | |
| FMNH 215914 | MT149768 | Hipposideros caffer3 | Kenya | -3.706 | 38.776 | |||||
| FMNH 215915 | redundant | Hipposideros caffer3 | Kenya | -3.706 | 38.776 | |||||
| FMNH 215916 | MT149769 | Hipposideros caffer3 | Kenya | -3.706 | 38.776 | |||||
| FMNH 215917 | redundant | MT149646 | MT149545 | MT149448 | MT149348 | Hipposideros caffer3 | Kenya | -3.706 | 38.776 | |
| FMNH 215918 | redundant | Hipposideros caffer3 | Kenya | -3.706 | 38.776 | |||||
| FMNH 215921 | redundant | Hipposideros caffer3 | Kenya | -3.076 | 39.217 | |||||
| FMNH 215922 | MT149770 | Hipposideros caffer3 | Kenya | -3.076 | 39.217 | |||||
| FMNH 215923 | MT149771 | Hipposideros caffer3 | Kenya | -3.076 | 39.217 | |||||
| FMNH 215924 | redundant | Hipposideros caffer3 | Kenya | -3.076 | 39.217 | |||||
| FMNH 215925 | MT149772 | MT149647 | MT149546 | MT149449 | MT149349 | Hipposideros caffer3 | Kenya | -3.076 | 39.217 | |
| FMNH 220648 | MT149766 | Hipposideros caffer3 | Kenya | 0.170 | 38.194 | |||||
| FMNH 220669 | MT149767 | MT149645 | MT149544 | MT149447 | MT149347 | Hipposideros caffer3 | Kenya | 0.024 | 38.066 | |
| FMNH 234022 | MT149757 | Hipposideros caffer3 | Kenya | -1.019 | 38.326 | |||||
| FMNH 234023 | MT149758 | Hipposideros caffer3 | Kenya | -0.992 | 38.330 | |||||
| NMK 184226 | MT149762 | MT149644 | MT149543 | MT149446 | MT149346 | Hipposideros caffer3 | Kenya | 2.320 | 37.994 | |
| NMK 184238 | MT149763 | Hipposideros caffer3 | Kenya | 2.320 | 37.994 | |||||
| NMK 184284 | MT149764 | Hipposideros caffer3 | Kenya | 2.320 | 37.994 | |||||
| NMK 184287 | MT149765 | Hipposideros caffer3 | Kenya | 2.283 | 37.954 | |||||
| NMK 184425 | MT149761 | MT149643 | MT149542 | MT149445 | MT149345 | Hipposideros caffer3 | Kenya | 0.228 | 37.113 | |
| NMK 185050 | redundant | MT149642 | MT149541 | MT149444 | MT149344 | Hipposideros caffer3 | Kenya | -0.992 | 38.330 | |
| NMK 185051 | redundant | Hipposideros caffer3 | Kenya | -0.992 | 38.330 | |||||
| NMK 185052 | MT149759 | Hipposideros caffer3 | Kenya | -0.992 | 38.330 | |||||
| NMK 185053 | redundant | Hipposideros caffer3 | Kenya | -0.992 | 38.330 | |||||
| NMK 185054 | MT149760 | Hipposideros caffer3 | Kenya | -0.992 | 38.330 | |||||
| DM 8587 | KF551805 | Hipposideros caffer4 | Mozambique | -23.205 | 32.499 | |||||
| DM 8590 | KF551810 | Hipposideros caffer4 | Mozambique | -12.182 | 37.550 | |||||
| TM 48051 | EU934451 | Hipposideros caffer4 | Mozambique | -21.517 | 35.100 | |||||
| DM 11007 | KF551806 redundant | Hipposideros caffer4 | South Africa | -27.596 | 32.220 | |||||
| FJ347979 | Hipposideros caffer4 | South Africa | -27.660 | 32.251 | ||||||
| EU934452 | Hipposideros caffer4 | South Africa | -23.999 | 31.645 | ||||||
| DM 7920 | EU934458 | Hipposideros caffer4 | Swaziland | -26.870 | 31.463 | |||||
| FMNH 215941 | redundant | Hipposideros caffer5 | Kenya | -3.300 | 39.995 | |||||
| FMNH 220176 | MT149780 | Hipposideros caffer5 | Kenya | -4.590 | 39.331 | |||||
| FMNH 220177 | MT149781 | Hipposideros caffer5 | Kenya | -4.590 | 39.331 | |||||
| FMNH 220178 | redundant | Hipposideros caffer5 | Kenya | -4.590 | 39.331 | |||||
| FMNH 220179 | MT149782 | Hipposideros caffer5 | Kenya | -4.590 | 39.331 | |||||
| FMNH 220180 | MT149783 | Hipposideros caffer5 | Kenya | -4.590 | 39.331 | |||||
| FMNH 220182 | MT149784 | Hipposideros caffer5 | Kenya | -4.082 | 39.483 | |||||
| FMNH 220183 | MT149785 | Hipposideros caffer5 | Kenya | -4.082 | 39.483 | |||||
| FMNH 220184 | MT149786 | Hipposideros caffer5 | Kenya | -4.082 | 39.483 | |||||
| FMNH 220185 | redundant | Hipposideros caffer5 | Kenya | -4.082 | 39.483 | |||||
| FMNH 220186 | redundant | Hipposideros caffer5 | Kenya | -4.082 | 39.483 | |||||
| FMNH 220202 | MT149773 | Hipposideros caffer5 | Kenya | -3.300 | 39.995 | |||||
| FMNH 220203 | redundant | Hipposideros caffer5 | Kenya | -3.300 | 39.995 | |||||
| FMNH 220204 | redundant | MT149648 | MT149547 | MT149450 | MT149350 | Hipposideros caffer5 | Kenya | -3.300 | 39.995 | |
| FMNH 220205 | redundant | Hipposideros caffer5 | Kenya | -3.323 | 40.042 | |||||
| FMNH 220206 | MT149774 | Hipposideros caffer5 | Kenya | -3.323 | 40.042 | |||||
| FMNH 220207 | redundant | MT149649 | MT149548 | MT149451 | MT149351 | Hipposideros caffer5 | Kenya | -3.323 | 40.042 | |
| FMNH 220208 | redundant | MT149650 | MT149549 | MT149452 | MT149352 | Hipposideros caffer5 | Kenya | -3.323 | 40.042 | |
| FMNH 220209 | MT149775 | Hipposideros caffer5 | Kenya | -3.323 | 40.042 | |||||
| FMNH 233985 | redundant | Hipposideros caffer5 | Kenya | -3.335 | 40.031 | |||||
| NMK 187199 | MT149776 | Hipposideros caffer5 | Kenya | -3.323 | 40.042 | |||||
| NMK 187200 | redundant | Hipposideros caffer5 | Kenya | -3.323 | 40.042 | |||||
| NMK 187201 | MT149777 | Hipposideros caffer5 | Kenya | -3.323 | 40.042 | |||||
| NMK 187202 | MT149778 | Hipposideros caffer5 | Kenya | -3.323 | 40.042 | |||||
| NMK 187203 | MT149779 | Hipposideros caffer5 | Kenya | -3.323 | 40.042 | |||||
| CM 97957 | FJ347980 | Hipposideros caffer5 | Kenya | -4.250 | 39.383 | |||||
| FMNH 192789 | MT149787 | MT149651 | MT149550 | MT149453 | MT149353 | Hipposideros caffer5 | Tanzania | -4.902 | 39.688 | |
| FMNH 192855 | redundant | MT149652 | MT149551 | MT149454 | MT149354 | Hipposideros caffer5 | Tanzania | -4.902 | 39.688 | |
| FMNH 187385 | redundant | MT149653 | MT149552 | MT149455 | MT149355 | Hipposideros caffer6 | Tanzania | -8.003 | 39.762 | |
| FMNH 187386 | MT149788 | MT149654 | MT149553 | MT149456 | Hipposideros caffer6 | Tanzania | -8.003 | 39.762 | ||
| FMNH 187387 | MT149789 | Hipposideros caffer6 | Tanzania | -7.993 | 39.792 | |||||
| FMNH 187388 | redundant | Hipposideros caffer6 | Tanzania | -7.993 | 39.792 | |||||
| FMNH 187417 | MT149790 | Hipposideros caffer6 | Tanzania | -7.891 | 39.843 | |||||
| FMNH 187418 | MT149791 | Hipposideros caffer6 | Tanzania | -7.891 | 39.843 | |||||
| FMNH 187426 | MT149792 | Hipposideros caffer6 | Tanzania | -7.891 | 39.843 | |||||
| FMNH 187428 | redundant | Hipposideros caffer6 | Tanzania | -7.993 | 39.792 | |||||
| FMNH 198066 | MT149793 | MT149655 | MT149554 | MT149457 | MT149356 | Hipposideros caffer6 | Tanzania | -5.878 | 39.311 | |
| FMNH 198067 | MT149794 | Hipposideros caffer6 | Tanzania | -5.878 | 39.311 | |||||
| FMNH 198072 | redundant | MT149656 | MT149555 | MT149458 | MT149357 | Hipposideros caffer6 | Tanzania | -6.244 | 39.320 | |
| FMNH 198073 | redundant | Hipposideros caffer6 | Tanzania | -6.244 | 39.320 | |||||
| FMNH 198074 | MT149795 | MT149657 | MT149556 | MT149459 | MT149358 | Hipposideros caffer6 | Tanzania | -6.244 | 39.320 | |
| FMNH 198075 | MT149796 | Hipposideros caffer6 | Tanzania | -6.244 | 39.320 | |||||
| FMNH 198076 | redundant | Hipposideros caffer6 | Tanzania | -6.244 | 39.320 | |||||
| FMNH 198082 | redundant | Hipposideros caffer6 | Tanzania | -6.280 | 39.451 | |||||
| FMNH 198083 | MT149797 | Hipposideros caffer6 | Tanzania | -6.280 | 39.451 | |||||
| FMNH 198084 | MT149798 | Hipposideros caffer6 | Tanzania | -6.280 | 39.451 | |||||
| FMNH 198131 | redundant | Hipposideros caffer6 | Tanzania | -5.878 | 39.311 | |||||
| FMNH 198132 | MT149799 | Hipposideros caffer6 | Tanzania | -5.878 | 39.311 | |||||
| FMNH 198133 | MT149800 | Hipposideros caffer6 | Tanzania | -5.878 | 39.311 | |||||
| NMP | EU934460 | Hipposideros caffer6 | Tanzania | -5.998 | 39.187 | |||||
| NMP | EU934477 | Hipposideros caffer7 | Malawi | -16.033 | 35.500 | |||||
| DM 8528 | KF551817 | Hipposideros caffer7 | Mozambique | -13.401 | 34.870 | |||||
| DM 8550 | KF551816 redundant | Hipposideros caffer7 | Mozambique | -13.401 | 34.870 | |||||
| FMNH 155554 | MT149801 | MT149658 | MT149557 | MT149460 | MT149359 | Hipposideros caffer7 | Tanzania | -8.519 | 35.904 | |
| FMNH 192790 | redundant | MT149659 | MT149558 | MT149461 | MT149360 | Hipposideros caffer7 | Tanzania | -4.902 | 39.688 | |
| FMNH 192792 | MT149802 | Hipposideros caffer7 | Tanzania | -5.367 | 39.645 | |||||
| FMNH 192793 | redundant | Hipposideros caffer7 | Tanzania | -5.367 | 39.645 | |||||
| FMNH 192794 | redundant | Hipposideros caffer7 | Tanzania | -5.367 | 39.645 | |||||
| FMNH 192795 | redundant | Hipposideros caffer7 | Tanzania | -5.367 | 39.645 | |||||
| FMNH 192796 | MT149803 | Hipposideros caffer7 | Tanzania | -5.367 | 39.645 | |||||
| FMNH 192849 | MT149804 | MT149660 | MT149559 | MT149462 | MT149361 | Hipposideros caffer7 | Tanzania | -4.902 | 39.688 | |
| FMNH 187140 | MT149805 | MT149661 | MT149560 | MT149463 | MT149362 | Hipposideros caffer8 | Tanzania | -3.798 | 36.069 | |
| FMNH 219065 | MT149806 | MT149662 | MT149561 | MT149464 | MT149363 | Hipposideros caffer8 | Tanzania | -8.037 | 34.502 | |
| FMNH 219241 | MT149807 | Hipposideros caffer8 | Tanzania | -8.037 | 34.502 | |||||
| FMNH 219242 | MT149808 | Hipposideros caffer8 | Tanzania | -7.707 | 34.031 | |||||
| FMNH 232868 | redundant | Hipposideros caffer8 | Uganda | 2.240 | 31.688 | |||||
| FMNH 232869 | MT149809 | MT149663 | MT149562 | MT149465 | MT149364 | Hipposideros caffer8 | Uganda | 2.240 | 31.688 | |
| FMNH 232874 | redundant | MT149664 | MT149563 | MT149416 | MT149365 | Hipposideros caffer8 | Uganda | 2.240 | 31.688 | |
| FMNH 232875 | MT149810 | MT149665 | MT149564 | MT149366 | Hipposideros caffer8 | Uganda | 2.240 | 31.688 | ||
| LSUMZ MT-4480 | redundant | MT149666 | MT149466 | MT149367 | Hipposideros cervinus | Malaysia | 1.970 | 103.500 | ||
| LSUMZ MT-4481 | MT149811 | MT149667 | MT149565 | MT149467 | MT149368 | Hipposideros cervinus | Malaysia | 1.970 | 103.500 | |
| LSUMZ MT-4500 | MT149812 | MT149668 | MT149566 | MT149468 | MT149369 | Hipposideros cervinus | Malaysia | 1.970 | 103.500 | |
| UNIMAS 787 | EF108144 | Hipposideros cervinus | Malaysia | 3.316 | 113.125 | |||||
| UNIMAS 788 | EF108146 | Hipposideros cervinus | Malaysia | 3.316 | 113.125 | |||||
| LSUMZ MT-4495 | MT149813 | MT149669 | MT149469 | MT149370 | Hipposideros cf. bicolor | Malaysia | 1.970 | 103.500 | ||
| UNIMAS 1459 | EF108142 | Hipposideros cf. bicolor | Malaysia | 1.716 | 110.467 | |||||
| UNIMAS 1474 | EF108143 | Hipposideros cf. bicolor | Malaysia | 1.716 | 110.467 | |||||
| FMNH 235856 | MT149814 | MT149670 | MT149567 | MT149470 | MT149371 | Hipposideros cf. cervinus | Solomon Islands | -10.569 | 161.913 | |
| FMNH 235857 | MT149815 | MT149671 | MT149568 | MT149471 | MT149372 | Hipposideros cf. cervinus | Solomon Islands | -10.569 | 161.913 | |
| NMP 91848 | EU934474 | Hipposideros cf. lamottei | Benin | 7.783 | 2.267 | |||||
| NMP 91849 | EU934475 | Hipposideros cf. lamottei | Benin | 7.783 | 2.267 | |||||
| IVB S862 | EU934453 | Hipposideros cf. lamottei | Senegal | 12.350 | -12.317 | |||||
| IVB PV56 | HQ343266 | Hipposideros cf. ruber | Ghana | 7.668 | -1.962 | |||||
| DM 12598 | KF551812 | Hipposideros cf. ruber | Liberia | 7.553 | -8.492 | |||||
| DM 12620 | KF551811 | Hipposideros cf. ruber | Liberia | 7.553 | -8.492 | |||||
| IVB S119 | EU934478 | Hipposideros cf. ruber | Senegal | 13.050 | -13.083 | |||||
| IVB S132 | HQ343242 | Hipposideros cf. ruber | Senegal | 14.071 | -12.572 | |||||
| IVB S1374 | EU934479 | Hipposideros cf. ruber | Senegal | 13.250 | -13.217 | |||||
| IVB S8 | HQ343240 | Hipposideros cf. ruber | Senegal | 12.884 | -12.755 | |||||
| LSUMZ MT-4423 | DQ054809 | Hipposideros cineraceus | Malaysia | 3.717 | 102.167 | |||||
| FMNH 190042 | JQ915701 | Hipposideros coronatus | Philippines | 9.097 | 125.705 | |||||
| FMNH 202631 | JQ915702 | Hipposideros coronatus | Philippines | 9.764 | 124.266 | |||||
| KU 166444 | MT149672 | MT149472 | MT149373 | Hipposideros coronatus | Philippines | 11.813 | 125.278 | |||
| EF108148 | Hipposideros coxi | Malaysia | 1.378 | 110.120 | ||||||
| EF108147 redundant | Hipposideros coxi | Malaysia | 1.378 | 110.120 | ||||||
| UNIMAS 1424 | EF108149 | Hipposideros diadema | Malaysia | 5.531 | 118.072 | |||||
| KU 164028 | MT149816 | MT149673 | MT149569 | MT149473 | MT149374 | Hipposideros diadema | Philippines | 19.331 | 121.439 | |
| KU 164029 | MT149817 | MT149674 | MT149570 | MT149474 | MT149375 | Hipposideros diadema | Philippines | 19.331 | 121.439 | |
| KU 164245 | MT149818 | MT149675 | MT149571 | MT149475 | Hipposideros diadema | Philippines | 13.796 | 120.159 | ||
| FJ460489 | Hipposideros doriae | Malaysia | 1.117 | 110.217 | ||||||
| NHMOU.CHI MP4.2016 | KY176014 | Hipposideros durgadasi | India | 23.317 | 78.414 | |||||
| UNIMAS 312 | EF108150 | Hipposideros dyacorum | Malaysia | 4.401 | 117.889 | |||||
| UNIMAS 556 | EF108151 | Hipposideros dyacorum | Malaysia | 3.316 | 113.125 | |||||
| EU934468 | Hipposideros fuliginosus | Guinea Bissau | 11.117 | -14.933 | ||||||
| EU934467 | Hipposideros fuliginosus | Guinea Bissau | 11.333 | -13.900 | ||||||
| JX849198 | Hipposideros griffini | Vietnam | ||||||||
| CVVD AG | 200700214 | JN247005 | Hipposideros halophyllus | Thailand | ||||||
| NMP 91842 | EU934471 | Hipposideros jonesi | Benin | 7.783 | 2.267 | |||||
| IVB S804 | EU934472 | Hipposideros jonesi | Senegal | 12.350 | -12.317 | |||||
| EU934473 | Hipposideros jonesi | Senegal | 12.350 | -12.317 | ||||||
| EBD 23514 | DQ054816 | Hipposideros khaokhouayensis | Laos | 18.433 | 102.950 | |||||
| KF551824 | Hipposideros lamottei | Guinea | 7.570 | -8.471 | ||||||
| KF551823 | Hipposideros lamottei | Guinea | 7.570 | -8.471 | ||||||
| NHMOU.CHI MP15.2016 | KY176015 | Hipposideros lankadiva | India | 23.317 | 78.414 | |||||
| LSUMZ MT-4478 | MT149819 | MT149676 | MT149572 | MT149476 | MT149376 | Hipposideros larvatus | Malaysia | 1.970 | 103.500 | |
| LSUMZ MT-4479 | redundant | MT149677 | MT149573 | MT149377 | Hipposideros larvatus | Malaysia | 1.970 | 103.500 | ||
| LSUMZ MT-4488 | MT149820 | MT149678 | MT149574 | MT149477 | MT149378 | Hipposideros larvatus | Malaysia | 1.970 | 103.500 | |
| UNIMAS 1485 | EF108152 redundant | Hipposideros larvatus | Malaysia | 1.717 | 110.467 | |||||
| UNIMAS 1501 | EF108153 | Hipposideros larvatus | Malaysia | 1.717 | 110.467 | |||||
| FMNH 195507 | JQ915904 | Hipposideros lekaguli | Philippines | 16.314 | 121.394 | |||||
| KR908661 | Hipposideros lylei | China | 25.603 | 99.752 | ||||||
| DM 12607 | KF551825 | Hipposideros marisae | Liberia | 7.553 | -8.492 | |||||
| FMNH 140601 | JQ915906 | Hipposideros obscurus | Philippines | 13.767 | 124.350 | |||||
| KU 165040 | redundant | MT149679 | MT149575 | MT149379 | Hipposideros obscurus | Philippines | 11.434 | 122.079 | ||
| KU 165041 | MT149821 | MT149680 | MT149576 | MT149478 | MT149380 | Hipposideros obscurus | Philippines | 11.434 | 122.079 | |
| KU 165226 | MT149717 | MT149577 | MT149479 | MT149381 | Hipposideros obscurus | Philippines | 13.447 | 120.426 | ||
| PSUZC MM2006.129 | JN247029 | Hipposideros pendelburyi | Thailand | 7.565 | 99.624 | |||||
| DQ054810 | Hipposideros pomona | Laos | 18.250 | 104.517 | ||||||
| EU434952 | Hipposideros pratti | China | 27.729 | 115.734 | ||||||
| FMNH 190070 | JQ915992 | Hipposideros pygmaeus | Philippines | 9.097 | 125.705 | |||||
| KU 164542 | MT149822 | MT149681 | MT149578 | MT149480 | MT149382 | Hipposideros pygmaeus | Philippines | 14.823 | 121.968 | |
| KU 164543 | redundant | MT149682 | MT149579 | MT149481 | MT149383 | Hipposideros pygmaeus | Philippines | 14.823 | 121.968 | |
| KU 164544 | MT149716 | MT149683 | MT149580 | MT149482 | MT149384 | Hipposideros pygmaeus | Philippines | 14.823 | 121.968 | |
| LSUMZ MT-4425 | MT149715 | MT149684 | MT149581 | MT149483 | MT149385 | Hipposideros ridleyi | Malaysia | 3.557 | 102.761 | |
| LSUMZ MT-4477 | MT149823 | MT149582 | MT149484 | MT149386 | Hipposideros ridleyi | Malaysia | 3.557 | 102.761 | ||
| SMF 83828 | DQ054811 | Hipposideros ridleyi | Malaysia | 3.717 | 102.167 | |||||
| DQ054813 | Hipposideros rotalis | Laos | 18.250 | 104.517 | ||||||
| FJ347996 | Hipposideros ruber1 | Cameroon | 3.150 | 13.000 | ||||||
| FJ347995 | Hipposideros ruber1 | Cameroon | 4.451 | 11.571 | ||||||
| FJ347993 | Hipposideros ruber1 | Cameroon | 3.564 | 13.408 | ||||||
| FJ347992 | Hipposideros ruber1 | Cameroon | 3.564 | 13.408 | ||||||
| FJ347989 | Hipposideros ruber1 | Cameroon | 5.385 | 11.688 | ||||||
| FMNH 195085 | MT149824 | MT149685 | MT149583 | MT149485 | MT149387 | Hipposideros ruber1 | D. R. Congo | -4.991 | 29.080 | |
| FMNH 215448 | MT149826 | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| FMNH 215449 | MT149827 | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| FMNH 215450 | redundant | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| FMNH 215451 | redundant | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| FMNH 215452 | redundant | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| FMNH 215453 | redundant | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| FMNH 215476 | MT149828 | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| FMNH 215477 | redundant | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| FMNH 215478 | redundant | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| NMK 184904 | MT149825 | MT149686 | MT149584 | MT149486 | Hipposideros ruber1 | Kenya | 0.212 | 34.899 | ||
| NMK 184905 | redundant | Hipposideros ruber1 | Kenya | 0.212 | 34.899 | |||||
| NMK 187407 | redundant | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| NMK 187408 | MT149829 | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| NMK 187409 | MT149830 | MT149687 | MT149585 | MT149487 | MT149388 | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |
| NMK 187410 | redundant | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| NMK 187412 | MT149831 | Hipposideros ruber1 | Kenya | 1.036 | 34.753 | |||||
| DM 12603 | KF551819 | Hipposideros ruber1 | Liberia | 7.553 | -8.492 | |||||
| DM 13245 | KF551815 | Hipposideros ruber1 | Liberia | 7.553 | -8.492 | |||||
| DM 13246 | KF551820 | Hipposideros ruber1 | Liberia | 7.553 | -8.492 | |||||
| FMNH 225201 | MT149832 | MT149688 | MT149586 | MT149488 | MT149389 | Hipposideros ruber1 | Rwanda | -2.485 | 29.199 | |
| FMNH 225202 | MT149833 | Hipposideros ruber1 | Rwanda | -1.504 | 29.613 | |||||
| FMNH 225203 | redundant | Hipposideros ruber1 | Rwanda | -1.504 | 29.613 | |||||
| FMNH 225204 | redundant | Hipposideros ruber1 | Rwanda | -1.506 | 29.615 | |||||
| FMNH 225205 | redundant | Hipposideros ruber1 | Rwanda | -1.506 | 29.615 | |||||
| FMNH 225206 | MT149834 | Hipposideros ruber1 | Rwanda | -1.506 | 29.615 | |||||
| FMNH 225207 | redundant | Hipposideros ruber1 | Rwanda | -1.506 | 29.615 | |||||
| FMNH 225208 | MT149835 | MT149689 | MT149587 | MT149489 | MT149390 | Hipposideros ruber1 | Rwanda | -1.506 | 29.615 | |
| FMNH 225209 | redundant | Hipposideros ruber1 | Rwanda | -1.506 | 29.615 | |||||
| FMNH 192935 | MT149836 | MT149690 | MT149588 | MT149490 | MT149391 | Hipposideros ruber1 | Tanzania | -1.094 | 31.515 | |
| FMNH 137629 | FJ347987 | Hipposideros ruber1 | Uganda | 32.283 | -0.005 | |||||
| FMNH 160358 | MT149837 | Hipposideros ruber1 | Uganda | -0.989 | 29.614 | |||||
| FMNH 160359 | MT149838 | MT149691 | MT149589 | MT149491 | MT149392 | Hipposideros ruber1 | Uganda | -0.989 | 29.614 | |
| FMNH 160361 | MT149839 | Hipposideros ruber1 | Uganda | -1.041 | 29.580 | |||||
| FMNH 161040 | MT149840 | Hipposideros ruber1 | Uganda | -0.245 | 29.819 | |||||
| FMNH 223866 | redundant | MT149692 | MT149590 | MT149492 | MT149393 | Hipposideros ruber1 | Uganda | -0.342 | 31.966 | |
| FMNH 223867 | MT149841 | Hipposideros ruber1 | Uganda | -0.342 | 31.966 | |||||
| FMNH 227415 | MT149842 | Hipposideros ruber2 | Central African Republic | 3.033 | 16.410 | |||||
| FMNH 227416 | MT149843 | Hipposideros ruber2 | Central African Republic | 3.033 | 16.410 | |||||
| FMNH 227417 | MT149844 | MT149693 | MT149591 | MT149394 | Hipposideros ruber2 | Central African Republic | 3.033 | 16.410 | ||
| FMNH 149408 | FJ347986 | MT149694 | MT149592 | MT149493 | MT149395 | Hipposideros ruber2 | D. R. Congo | -1.417 | 28.583 | |
| FMNH 149409 | redundant | Hipposideros ruber2 | D. R. Congo | -1.417 | 28.583 | |||||
| FMNH 149410 | redundant | MT149695 | MT149593 | MT149494 | MT149396 | Hipposideros ruber2 | D. R. Congo | -1.417 | 28.583 | |
| FMNH 149412 | redundant | Hipposideros ruber2 | D. R. Congo | -1.417 | 28.583 | |||||
| ROM 100546 | FJ347978 | Hipposideros ruber2 | Ivory Coast | 6.930 | -7.217 | |||||
| FMNH 215479 | redundant | Hipposideros ruber2 | Kenya | 0.244 | 34.907 | |||||
| FMNH 215480 | MT149845 | Hipposideros ruber2 | Kenya | 0.244 | 34.907 | |||||
| FMNH 215481 | MT149846 | Hipposideros ruber2 | Kenya | 0.244 | 34.907 | |||||
| FMNH 215482 | redundant | MT149696 | MT149594 | MT149495 | MT149397 | Hipposideros ruber2 | Kenya | 0.244 | 34.907 | |
| FMNH 215483 | redundant | MT149697 | MT149595 | MT149496 | MT149398 | Hipposideros ruber2 | Kenya | 0.244 | 34.907 | |
| NMK 184878 | MT149847 | Hipposideros ruber2 | Kenya | 0.248 | 34.906 | |||||
| NMK 184880 | MT149848 | Hipposideros ruber2 | Kenya | 0.248 | 34.906 | |||||
| NMK 184882 | MT149849 | Hipposideros ruber2 | Kenya | 0.248 | 34.906 | |||||
| NMK 184883 | MT149850 | Hipposideros ruber2 | Kenya | 0.248 | 34.906 | |||||
| NMK 184884 | MT149851 | Hipposideros ruber2 | Kenya | 0.248 | 34.906 | |||||
| NMK 187383 | redundant | Hipposideros ruber2 | Kenya | 0.212 | 34.899 | |||||
| NMK 187384 | MT149852 | Hipposideros ruber2 | Kenya | 0.212 | 34.899 | |||||
| FMNH 165161 | redundant | Hipposideros ruber2 | Uganda | 1.733 | 31.467 | |||||
| FMNH 165162 | MT149853 | Hipposideros ruber2 | Uganda | 1.683 | 31.533 | |||||
| FMNH 165163 | redundant | MT149698 | MT149596 | MT149497 | MT149399 | Hipposideros ruber2 | Uganda | 1.683 | 31.533 | |
| FMNH 165164 | MT149854 | Hipposideros ruber2 | Uganda | 1.683 | 31.533 | |||||
| FMNH 165165 | MT149855 | Hipposideros ruber2 | Uganda | 1.683 | 31.533 | |||||
| FMNH 165166 | redundant | Hipposideros ruber2 | Uganda | 1.750 | 31.583 | |||||
| FMNH 165167 | redundant | MT149699 | MT149597 | MT149498 | MT149400 | Hipposideros ruber2 | Uganda | 1.733 | 31.467 | |
| FMNH 224069 | redundant | Hipposideros ruber2 | Uganda | 0.501 | 30.426 | |||||
| FMNH 224071 | redundant | Hipposideros ruber2 | Uganda | 0.501 | 30.426 | |||||
| FMNH 224074 | MT149856 | Hipposideros ruber2 | Uganda | 0.501 | 30.426 | |||||
| FMNH 224075 | MT149857 | Hipposideros ruber2 | Uganda | 0.501 | 30.426 | |||||
| FJ347994 | Hipposideros ruber3 | Cameroon | 3.564 | 13.408 | ||||||
| FJ347991 | Hipposideros ruber3 | Cameroon | 2.941 | 9.911 | ||||||
| FJ347990 | Hipposideros ruber3 | Cameroon | 2.941 | 9.911 | ||||||
| FJ347988 | Hipposideros ruber3 | Cameroon | 4.913 | 9.241 | ||||||
| EBD 18240 | FJ347984 | Hipposideros ruber3 | Equatorial Guinea | 1.889 | 9.793 | |||||
| EBD 18266 | FJ347985 | Hipposideros ruber3 | Equatorial Guinea | 1.889 | 9.793 | |||||
| EBD 18511 | FJ347983 | Hipposideros ruber3 | Equatorial Guinea | 3.747 | 8.750 | |||||
| EBD 18942 | FJ347981 | Hipposideros ruber3 | Principe | 1.615 | 7.404 | |||||
| EBD 18926 | FJ347982 | Hipposideros ruber3 | São Tomé | 0.219 | 6.727 | |||||
| FMNH 219477 | MT149858 | MT149700 | MT149598 | MT149499 | MT149401 | Hipposideros ruber4 | D. R. Congo | -5.290 | 14.871 | |
| FMNH 169707 | KT583815 | Macronycteris commersoni | Madagascar | -12.932 | 49.057 | |||||
| FMNH 175777 | KT583822 | Macronycteris commersoni | Madagascar | -16.380 | 45.345 | |||||
| FMNH 175966 | KT583823 | Macronycteris commersoni | Madagascar | -22.486 | 45.392 | |||||
| FMNH 175974 | MT149859 | Macronycteris commersoni | Madagascar | -22.317 | 45.293 | |||||
| FMNH 175975 | MT149860 | Macronycteris commersoni | Madagascar | -22.317 | 45.293 | |||||
| FMNH 176155 | KT583824 | MT149701 | MT149599 | MT149500 | MT149402 | Macronycteris commersoni | Madagascar | -22.778 | 43.523 | |
| FMNH 176158 | MT149861 | Macronycteris commersoni | Madagascar | -22.217 | 43.330 | |||||
| FMNH 176277 | redundant | Macronycteris commersoni | Madagascar | -12.942 | 49.055 | |||||
| FMNH 177302 | KT583825 | Macronycteris commersoni | Madagascar | -16.315 | 46.810 | |||||
| FMNH 178803 | redundant | Macronycteris commersoni | Madagascar | -12.712 | 49.474 | |||||
| FMNH 178806 | KT583816 | Macronycteris commersoni | Madagascar | -12.712 | 49.474 | |||||
| FMNH 178808 | KT583817 redundant | Macronycteris commersoni | Madagascar | -12.712 | 49.474 | |||||
| FMNH 178809 | KT583818 | Macronycteris commersoni | Madagascar | -12.712 | 49.474 | |||||
| FMNH 178810 | KT583819 | Macronycteris commersoni | Madagascar | -12.712 | 49.474 | |||||
| FMNH 178811 | KT583820 | Macronycteris commersoni | Madagascar | -12.712 | 49.474 | |||||
| FMNH 178812 | KT583826 | Macronycteris commersoni | Madagascar | -12.712 | 49.474 | |||||
| FMNH 178815 | KT583821 redundant | Macronycteris commersoni | Madagascar | -12.712 | 49.474 | |||||
| FMNH 179201 | MT149862 | Macronycteris commersoni | Madagascar | -17.280 | 49.420 | |||||
| FMNH 183934 | KT583827 | Macronycteris commersoni | Madagascar | -24.050 | 43.750 | |||||
| FMNH 183980 | KT583813 redundant | Macronycteris commersoni | Madagascar | -12.337 | 49.385 | |||||
| FMNH 184030 | KT583812 | Macronycteris commersoni | Madagascar | -14.966 | 47.308 | |||||
| FMNH 184170 | KT583828 | Macronycteris commersoni | Madagascar | -24.650 | 43.963 | |||||
| FMNH 184887 | MT149863 | Macronycteris commersoni | Madagascar | -15.904 | 46.598 | |||||
| FMNH 209236 | MT149864 | Macronycteris commersoni | Madagascar | -18.063 | 44.541 | |||||
| FMNH 217940 | KT583831 redundant | Macronycteris commersoni | Madagascar | -22.632 | 45.338 | |||||
| FMNH 221308 | KT583829 | Macronycteris commersoni | Madagascar | -12.932 | 49.057 | |||||
| FMNH 231862 | MT149865 | MT149702 | MT149600 | MT149501 | MT149315 | Macronycteris commersoni | Madagascar | -17.889 | 49.203 | |
| UADBA 32916 | KT583830 | Macronycteris commersoni | Madagascar | 15.538 | 46.886 | |||||
| UADBA 32987 | KT583814 | Macronycteris commersoni | Madagascar | -13.932 | 49.057 | |||||
| UADBA 32989 | KR606333 | Macronycteris commersoni | Madagascar | -12.917 | 49.143 | |||||
| FMNH 175970 | MT149866 | MT149703 | MT149601 | MT149502 | MT149403 | Macronycteris cryptovalorona | Madagascar | -22.317 | 45.293 | |
| FMNH 184173 | MT149867 | MT149704 | MT149602 | MT149503 | MT149404 | Macronycteris cryptovalorona | Madagascar | -24.650 | 43.963 | |
| AMNH 269871 | KT583801 | Macronycteris gigas | Central African Republic | |||||||
| FMNH 219602 | MT149868 | MT149705 | MT149603 | MT149504 | MT149405 | Macronycteris gigas | D. R. Congo | 0.241 | 20.883 | |
| FMNH 219682 | MT149869 | MT149706 | MT149604 | MT149415 | MT149406 | Macronycteris gigas | D. R. Congo | 0.241 | 20.883 | |
| FMNH 220226 | redundant | MT149707 | MT149605 | MT149505 | MT149407 | Macronycteris gigas | Kenya | -4.647 | 39.380 | |
| FMNH 220227 | redundant | MT149708 | MT149606 | MT149506 | MT149408 | Macronycteris gigas | Kenya | -4.647 | 39.380 | |
| FMNH 220239 | MT149870 | Macronycteris gigas | Kenya | -4.215 | 39.451 | |||||
| DM 12602 | KF551826 | Macronycteris gigas | Liberia | 7.553 | -8.492 | |||||
| IVB S1032 | EU934469 | Macronycteris gigas | Senegal | 12.350 | -13.217 | |||||
| IVB S1044 | EU934470 | Macronycteris gigas | Senegal | 12.350 | -13.217 | |||||
| AMNH 269879 | KT583802 | Macronycteris vittata | Central African Republic | 3.500 | 16.000 | |||||
| FMNH 215942 | MT149871 | MT149709 | MT149607 | MT149507 | MT149409 | Macronycteris vittata | Kenya | -3.300 | 39.995 | |
| FMNH 215943 | redundant | Macronycteris vittata | Kenya | -3.300 | 39.995 | |||||
| FMNH 215944 | redundant | Macronycteris vittata | Kenya | -3.300 | 39.995 | |||||
| FMNH 215945 | redundant | Macronycteris vittata | Kenya | -3.300 | 39.995 | |||||
| FMNH 215946 | MT149872 | Macronycteris vittata | Kenya | -3.287 | 39.982 | |||||
| FMNH 215947 | redundant | Macronycteris vittata | Kenya | -3.287 | 39.982 | |||||
| FMNH 215948 | MT149873 | Macronycteris vittata | Kenya | -3.287 | 39.982 | |||||
| FMNH 215949 | redundant | Macronycteris vittata | Kenya | -3.287 | 39.982 | |||||
| FMNH 215950 | MT149874 | Macronycteris vittata | Kenya | -3.287 | 39.982 | |||||
| FMNH 215951 | MT149875 | Macronycteris vittata | Kenya | -3.282 | 39.971 | |||||
| FMNH 215952 | MT149876 | Macronycteris vittata | Kenya | -3.282 | 39.971 | |||||
| FMNH 215953 | redundant | MT149710 | MT149608 | MT149410 | Macronycteris vittata | Kenya | -3.282 | 39.971 | ||
| FMNH 215954 | MT149877 | Macronycteris vittata | Kenya | -3.282 | 39.971 | |||||
| FMNH 215955 | redundant | Macronycteris vittata | Kenya | -3.282 | 39.971 | |||||
| FMNH 215959 | MT149878 | Macronycteris vittata | Kenya | -3.309 | 40.018 | |||||
| FMNH 215960 | MT149879 | Macronycteris vittata | Kenya | -3.309 | 40.018 | |||||
| FMNH 215961 | MT149880 | Macronycteris vittata | Kenya | -3.309 | 40.018 | |||||
| FMNH 215962 | MT149881 | Macronycteris vittata | Kenya | -3.309 | 40.018 | |||||
| FMNH 215963 | MT149882 | Macronycteris vittata | Kenya | -3.309 | 40.018 | |||||
| FMNH 215967 | MT149883 | Macronycteris vittata | Kenya | -3.303 | 39.999 | |||||
| FMNH 215968 | redundant | Macronycteris vittata | Kenya | -3.305 | 39.937 | |||||
| FMNH 215969 | redundant | Macronycteris vittata | Kenya | -3.305 | 39.937 | |||||
| FMNH 220224 | redundant | Macronycteris vittata | Kenya | -4.647 | 39.378 | |||||
| FMNH 220225 | MT149885 | Macronycteris vittata | Kenya | -4.647 | 39.380 | |||||
| FMNH 220228 | MT149886 | Macronycteris vittata | Kenya | -4.647 | 39.380 | |||||
| FMNH 220229 | MT149887 | MT149711 | MT149609 | MT149508 | MT149411 | Macronycteris vittata | Kenya | -4.647 | 39.380 | |
| FMNH 220231 | MT149888 | Macronycteris vittata | Kenya | -4.614 | 39.354 | |||||
| FMNH 220232 | MT149889 | Macronycteris vittata | Kenya | -4.614 | 39.354 | |||||
| FMNH 220233 | MT149890 | Macronycteris vittata | Kenya | -4.614 | 39.354 | |||||
| FMNH 220234 | redundant | Macronycteris vittata | Kenya | -4.614 | 39.354 | |||||
| FMNH 220235 | MT149891 | MT149712 | MT149610 | MT149509 | MT149412 | Macronycteris vittata | Kenya | -4.614 | 39.354 | |
| FMNH 220236 | redundant | Macronycteris vittata | Kenya | -4.614 | 39.354 | |||||
| FMNH 220237 | redundant | Macronycteris vittata | Kenya | -4.614 | 39.354 | |||||
| FMNH 220238 | redundant | Macronycteris vittata | Kenya | -4.614 | 39.354 | |||||
| FMNH 220240 | redundant | Macronycteris vittata | Kenya | -3.300 | 39.995 | |||||
| NMK 187219 | redundant | Macronycteris vittata | Kenya | -3.335 | 40.031 | |||||
| NMK 187220 | MT149884 | Macronycteris vittata | Kenya | -3.335 | 40.031 | |||||
| NMK 187221 | redundant | Macronycteris vittata | Kenya | -3.335 | 40.031 | |||||
| NMK 187222 | redundant | Macronycteris vittata | Kenya | -3.335 | 40.031 | |||||
| DM 8645 | KF551828 redundant | Macronycteris vittata | Mozambique | -18.565 | 32.220 | |||||
| DM 11510 | KF551827 | Macronycteris vittata | Mozambique | -18.978 | 34.176 | |||||
| FMNH 192800 | redundant | MT149713 | MT149611 | MT149510 | MT149413 | Macronycteris vittata | Tanzania | -4.902 | 39.688 | |
| FMNH 192801 | MT149892 | Macronycteris vittata | Tanzania | -4.902 | 39.688 | |||||
| FMNH 192857 | redundant | MT149714 | MT149612 | MT149511 | MT149414 | Macronycteris vittata | Tanzania | -4.902 | 39.688 | |
| FMNH 192858 | redundant | Macronycteris vittata | Tanzania | -4.902 | 39.688 | |||||
| FMNH 192859 | MT149893 | Macronycteris vittata | Tanzania | -4.902 | 39.688 | |||||
| FMNH 192860 | KT583807 | Macronycteris vittata | Tanzania | -4.902 | 39.688 | |||||
| FMNH 192865 | KT583808 redundant | Macronycteris vittata | Tanzania | -4.902 | 39.688 | |||||
| FMNH 192866 | KT583809 | Macronycteris vittata | Tanzania | -4.902 | 39.688 | |||||
| FMNH 220268 | MT149614 | MT149512 | MT149417 | MT149316 | Triaenops afer | Kenya | -4.590 | 39.331 |
Citation
Patterson BD, Webala PW, Lavery TH, Agwanda BR, Goodman SM, Kerbis Peterhans JC, Demos TC (2020) Evolutionary relationships and population genetics of the Afrotropical leaf-nosed bats (Chiroptera, Hipposideridae). ZooKeys 929: 117–161. https://doi.org/10.3897/zookeys.929.50240
Funding Statement
JRS Biodiversity Foundation; Field Museum’s Council on Africa, Marshall Field III Fund, and Barbara E. Brown Fund for Mammal Research; The John D. and Catherine T. MacArthur Foundation, Fulbright Program of US Department of State, Wildlife Conservation Society, and the Centers for Disease Control and Prevention; WWF Gabon; MoZaR-Fond Européen de Développement Régional, Programme Opérationnel de Coopération Territoriale.
Supplementary materials
Figure S1. Geographic distribution of voucher specimens used in this analysis
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bruce D. Patterson, Paul W. Webala, Tyrone H. Lavery, Bernard R. Agwanda, Steven M. Goodman, Julian C. Kerbis Peterhans, Terrence C. Demos
Data type
occurrence
Figure S2. Phylogeny of Hipposideridae based on maximum likelihood analysis of cyt-b based on 452 individuals
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bruce D. Patterson, Paul W. Webala, Tyrone H. Lavery, Bernard R. Agwanda, Steven M. Goodman, Julian C. Kerbis Peterhans, Terrence C. Demos
Data type
phylogenetic tree
Figure S3. Phylogeny of Hipposideridae based on Bayesian inference analysis of cyt-b based on 452 individuals
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bruce D. Patterson, Paul W. Webala, Tyrone H. Lavery, Bernard R. Agwanda, Steven M. Goodman, Julian C. Kerbis Peterhans, Terrence C. Demos
Data type
phylogenetic tree
References
- Aellen V. (1954) Description d’un nouvel Hipposideros (Chiroptera) de la Côte d’Ivoire. Revue Suisse de Zoologie 61: 473–483. 10.5962/bhl.part.75404 [DOI] [Google Scholar]
- Allen GM. (1921) A new horseshoe bat from West Africa. Revue de Zoologie Africaine 9: 193–196. 10.5962/bhl.part.24480 [DOI] [Google Scholar]
- Allen GM. (1939) A checklist of African mammals. Bulletin of the Museum of Comparative Zoology at Harvard College 83: 1–763. [Google Scholar]
- Allen JA, Lang H, Chapin JP. (1917) The American Museum Congo Expedition collection of bats. Bulletin of the American Museum of Natural History 37: 405–563. 10.5962/bhl.title.82385 [DOI] [Google Scholar]
- Almeida F, Giannini N, DeSalle R, Simmons N. (2011) Evolutionary relationships of the Old World fruit bats (Chiroptera, Pteropodidae): another star phylogeny? BMC Evolutionary Biology 11: 1–281. 10.1186/1471-2148-11-281 [DOI] [PMC free article] [PubMed]
- Amador LI, Moyers Arévalo RL, Almeida FC, Catalano SA, Giannini NP. (2018) Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. Journal of Mammalian Evolution 25: 37–70. 10.1007/s10914-016-9363-8 [DOI] [Google Scholar]
- Andersen K. (1906a) On Hipposideros caffer, Sund. and its closest allies; with some notes on H. fuliginosus, Temm. Annals and Magazine of Natural History (Series 7) 17: 269–282. 10.1080/00222930608562523 [DOI]
- Andersen K. (1906b) On the bats of the Hipposideros armiger and commersoni types. Annals and Magazine of Natural History (Series 7) 17: 35–48. 10.1080/00222930608562488 [DOI]
- Andersen K. (1918) Diagnoses of new bats of the families Rhinolophidæ and Megadermatidæ. Annals and Magazine of Natural History (Series 9) 2: 374–384. 10.1080/00222931808562380 [DOI]
- Armstrong KN, Goodman SM, Benda P, Hand SJ. (2016) A common name for the bat family Rhinonycteridae–the Trident Bats. Zootaxa 4179: 115–117. 10.11646/zootaxa.4179.1.7 [DOI] [PubMed] [Google Scholar]
- Bates PJJ, Rossiter SJ, Suyanto A, Kingston T. (2007) A new species of Hipposideros (Chiroptera: Hipposideridae) from Sulawesi. Acta Chiropterologica 9: 13–26. 10.3161/1733-5329(2007)9[13:ANSOHC]2.0.CO;2 [DOI]
- Benda P, Vallo P, Reiter A. (2011) Taxonomic revision of the genus Asellia (Chiroptera: Hipposideridae) with a description of a new species from southern Arabia. Acta Chiropterologica 13: 245–270. 10.3161/150811011X624749 [DOI] [Google Scholar]
- Bickham JW, Patton JC, Schlitter DA, Rautenbach IL, Honeycutt RL. (2004) Molecular phylogenetics, karyotypic diversity, and partition of the genus Myotis (Chiroptera: Vespertilionidae). Molecular Phylogenetics and Evolution 33: 333–338. 10.1016/j.ympev.2004.06.012 [DOI] [PubMed] [Google Scholar]
- Bickham JW, Wood CC, Patton JC. (1995) Biogeographic implications of cytochrome b sequences and allozymes in sockeye (Oncorhynchus nerka). Journal of Heredity 86: 140–144. 10.1093/oxfordjournals.jhered.a111544 [DOI] [PubMed] [Google Scholar]
- Bocage JVBd. (1891) Sur une variété de “Phyllorhina commersoni” de l’île St. Thomé. Jornal de Sciencias Mathematicas, Physicas e Naturaes, Academia Real das Sciencias de Lisboa (serie 2) 2: 1–88.
- Bogdanowicz W, Owen RD. (1998) In the Minotaur’s labrinth: phylogeny of the bat family Hipposideridae. In: Kunz TH, Racey PA. (Eds) Bat Biology and Conservation.Smithsonian Institution Press, Washington, 27–42.
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: e1003537. 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed]
- Bray TC, Benda P. (2016) Distribution of Asellia tridens (Chiroptera: Hipposideridae) lineages including representatives from Saudi Arabia. Zoology in the Middle East 62: 283–287. 10.1080/09397140.2016.1250708 [DOI] [Google Scholar]
- Brosset A (1985 [“1984”]) Chiroptères d’altitude du Mont Nimba (Guinée). Description d’une espèce nouvelle, Hipposideros lamottei Mammalia 48: 545–556. 10.1515/mamm.1984.48.4.545 [DOI]
- Cabrera A. (1906) Mamiferos de Mogador. Boletín de la Real Sociedad Española de Historia Natural 6: 357–368. [Google Scholar]
- Chambers EA, Hillis DM. (2020) The multispecies coalescent over-splits species in the case of geographically widespread taxa. Systematic Biology 69: 184–193. 10.1093/sysbio/syz042 [DOI] [PubMed] [Google Scholar]
- Chernomor O, von Haeseler A, Minh BQ. (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65: 997–1008. 10.1093/sysbio/syw037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772–772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beaux O. (1924) Mammiferi della Somalia Italiana. Atti della Società Italiana di Scienze Naturali Milano 62: 1–254. [Google Scholar]
- De Winton WE. (1897) Descriptions of two new mammals from West Africa. Annals and Magazine of Natural History (Series 6) 20: 1–524. 10.1080/00222939709487396 [DOI]
- Demos TC, Webala PW, Bartonjo M, Patterson BD. (2018) Hidden diversity of African yellow house bats (Vespertilionidae, Scotophilus): insights from multilocus phylogenetics and lineage delimitation. Frontiers in Ecology and Evolution 6: 1–86. 10.3389/fevo.2018.00086 [DOI] [Google Scholar]
- Demos TC, Webala PW, Goodman SM, Kerbis Peterhans JC, Bartonjo M, Patterson BD. (2019a) Molecular phylogenetics of the African horseshoe bats (Chiroptera: Rhinolophidae): expanded geographic and taxonomic sampling of the Afrotropics BMC Evolutionary Biology 19: 1–166. 10.1186/s12862-019-1485-1 [DOI] [PMC free article] [PubMed]
- Demos TC, Webala PW, Kerbis Peterhans JC, Goodman SM, Bartonjo M, Patterson BD. (2019b) Molecular phylogenetics of slit-faced bats (Chiroptera: Nycteridae) reveals deeply divergent African lineages. Journal of Zoological Systematics and Evolutionary Research 57: 1019–1038. 10.1111/jzs.12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demos TC, Webala PW, Lutz HL, Peterhans JCK, Goodman SM, Cortés-Delgado N, Bartonjo M, Patterson BD. (2020) Multilocus phylogeny of a cryptic radiation of Afrotropical long-fingered bats (Chiroptera, Miniopteridae). Zoologica Scripta 49: 1–3. 10.1111/zsc.12388 [DOI] [Google Scholar]
- Douangboubpha B, Bumrungsri S, Satasook C, Soisook P, Bu SSH, Aul B, Harrison DL, Pearch MJ, Thomas NM, Bates PJJ. (2011) A new species of small Hipposideros (Chiroptera: Hipposideridae) from Myanmar and a revaluation of the taxon H. nicobarulae Miller, 1902 from the Nicobar Islands. Acta Chiropterologica 13: 61–78. 10.3161/150811011X578624 [DOI] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick GN, Jacobs DS, Matthee CA. (2005) A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera). Molecular Biology and Evolution 22: 1869–1886. 10.1093/molbev/msi180 [DOI] [PubMed] [Google Scholar]
- Eisentraut M. (1956) Beitrage zur Chiropteran-Fauna von Kamerun (Westafrika). Zeitschrift für Morphologie und Ökologie der Tiere 84: 525–540. [Google Scholar]
- Erixon P, Svennblad B, Britton T, Oxelman B. (2003) Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Systematic Biology 52: 665–673. 10.1080/10635150390235485 [DOI] [PubMed] [Google Scholar]
- Foley NM, Goodman SM, Whelan CV, Puechmaille SJ, Teeling E. (2017) Towards navigating the Minotaur’s labyrinth: cryptic diversity and taxonomic revision within the speciose genus Hipposideros (Hipposideridae). Acta Chiropterologica 19: 1–18. 10.3161/15081109ACC2017.19.1.001 [DOI] [Google Scholar]
- Foley NM, Thong VD, Soisook P, Goodman SM, Armstrong KN, Jacobs DS, Puechmaille SJ, Teeling EC. (2015) How and why overcome the impediments to resolution: lessons from rhinolophid and hipposiderid bats. Molecular Biology and Evolution 32: 313–333. 10.1093/molbev/msu329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman A, Çoraman E, Çelik YE, Postawa T, Bachanek J, Ruedi M. (2014) Cytonuclear discordance and the species status of Myotis myotis and Myotis blythii (Chiroptera). Zoologica Scripta 43: 549–561. 10.1111/zsc.12076 [DOI] [Google Scholar]
- Geoffroy Saint-Hilaire É. (1813) Sur un genre de chauve-souris, sous le nom de Rhinolophes. Annales du Muséum National d’Histoire Naturelle, Paris 20: 254–266. [Google Scholar]
- Goodman SM, Schoeman MC, Rakotoarivelo A, Willows-Munro S. (2016) How many species of Hipposideros have occurred on Madagascar since the Late Pleistocene? Zoological Journal of the Linnean Society 177: 428–449. 10.1111/zoj.12368 [DOI]
- Guillen-Servent A, Francis CM. (2006) A new species of bat of the Hipposideros bicolor group (Chiroptera: Hipposideridae) from Central Laos, with evidence of convergent evolution with Sundaic taxa. Acta Chiropterologica 8: 39–61. 10.3161/1733-5329(2006)8[39:ANSOBO]2.0.CO;2 [DOI]
- Gunnell GF, Butler PM, Greenwood M, Simmons NB. (2015) Bats (Chiroptera) from Olduvai Gorge, early Pleistocene, bed I (Tanzania). American Museum Novitates 3846: 1–36. 10.1206/3846.1 [DOI] [Google Scholar]
- Gürün K, Furman A, Juste J, Ramos Pereira MJ, Palmeirim JM, Puechmaille SJ, Hulva P, Presetnik P, Hamidovic D, Ibáñez C. (2019) A continent-scale study of the social structure and phylogeography of the bent-wing bat, Miniopterus schreibersii (Mammalia: Chiroptera), using new microsatellite data. Journal of Mammalogy 100: 1865–1878. 10.1093/jmammal/gyz153 [DOI] [Google Scholar]
- Hand SJ, Kirsch JAW. (1998) A southern origin for the Hipposideridae (Microchiroptera)? Evidence from the Australian fossil record. In: Kunz TH, Racey PA. (Eds) Bat Biology and Conservation.Smithsonian Institution Press, Washington, 72–90.
- Hand SJ, Kirsch JAW. (2003) Archerops, a new annectent hipposiderid genus (Mammalia: Microchiroptera) from the Australian Miocene. Journal of Paleontology 77: 1139–1151. [DOI] [Google Scholar]
- Hassanin A, Colombo R, Gembu GC, Merle M, Tu VT, Görföl T, Akawa PM, Csorba G, Kearney T, Monadjem A, Ing RK. (2018) Multilocus phylogeny and species delimitation within the genus Glauconycteris (Chiroptera, Vespertilionidae), with the description of a new bat species from the Tshopo Province of the Democratic Republic of the Congo. Journal of Zoological Systematics and Evolutionary Research 56: 1–22. 10.1111/jzs.12176 [DOI] [Google Scholar]
- Hayman RW. (1947) A new Hipposideros from Sierra Leone. Annals and Magazine of Natural History (Series 14) 109: 71–73. 10.1080/00222934708654611 [DOI]
- Heuglin T. (1861) Beiträge zur fauna der Säugetheire N.O.-Afrika’s. I. Chiroptera. Nova acta Academiae Caesareae Leopoldino-Carolinae Germanicae Naturae Curiosorum 29: 1–18. [Google Scholar]
- Hill JE. (1963) A revision of the genus Hipposideros. Bulletin of the British Museum (Natural History): Zoology 11: 1–129. 10.5962/bhl.part.4716 [DOI] [Google Scholar]
- Hillis DM, Bull JJ. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Koopman KF. (1970) Zoogeography of bats. In: Slaughter BH, Walton DW. (Eds) About Bats: A Chiropteran Symposium.Southern Methodist University Press, Dallas, 29–50.
- Koopman KF. (1989) Systematic notes on Liberian bats. American Museum Novitates 2946: 1–11. http://hdl.handle.net/2246/5100 [Google Scholar]
- Koopman KF. (1994) Chiroptera: Systematics. Handbuch der Zoologie (Vol. 8). Mammalia, part 60. Walter de Gruyter, Berlin, 217 pp. [Google Scholar]
- Kruskop SV, Benda P, Vasenkov DA, Lavrenchenko LA. (2016) First records of bats from the Alatish National Park, north-western Ethiopia (Chiroptera). Lynx (Series Nova) 47: 51–69. 10.2478/lynx-2016-0004 [DOI] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. (2016) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- Lanier HC, Knowles LL. (2014) Applying species-tree analyses to deep phylogenetic histories: challenges and potential suggested from a survey of empirical phylogenetic studies. Molecular Phylogenetics and Evolution 83: 191–199. 10.1016/j.ympev.2014.10.022 [DOI] [PubMed] [Google Scholar]
- Lavery TH, Leung LKP, Seddon JM. (2014) Molecular phylogeny of hipposiderid bats (Chiroptera: Hipposideridae) from Solomon Islands and Cape York Peninsula, Australia. Zoologica Scripta 43: 429–442. 10.1111/zsc.12068 [DOI] [Google Scholar]
- Leaché AD, Fujita MK. (2010) Bayesian species delimitation in West African forest geckos (Hemidactylus fasciatus). Proceedings of the Royal Society B 277: 3071–3077. 10.1098/rspb.2010.0662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JW, Bryant D. (2015) POPART: Full-feature software for haplotype network construction. Methods in Ecology and Evolution 6: 1110–1116. 10.1111/2041-210X.12410 [DOI] [Google Scholar]
- Matthee CA, Burzlaff JD, Taylor JF, Davis SK. (2001) Mining the mammalian genome for artiodactyl systematics. Systematic Biology 50: 367–390. 10.1080/10635150119683 [DOI] [PubMed] [Google Scholar]
- Miller Jr GS. (1907) The Families and Genera of Bats. Government Printing Office, Washington, 282 pp [+214 pls.] 10.5962/bhl.title.55695 [DOI] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE). IEEE, New Orleans. 10.1109/GCE.2010.5676129 [DOI]
- Monadjem A. (2019) Family Hipposideridae (Old World leaf-nosed bats). In: Wilson DE, Mittermeier RA. (Eds) Handbook of the Mammals of the World (Vol.9). Bats. Lynx Ediciones, Barcelona, 210–227.
- Monadjem A, Richards L, Taylor PJ, Denys C, Dower A, Stoffberg S. (2013) Diversity of Hipposideridae in the Mount Nimba massif, West Africa, and the taxonomic status of Hipposideros lamottei. Acta Chiropterologica 15: 341–352. 10.3161/150811013X678964 [DOI] [Google Scholar]
- Monard A. (1939) Résultats de la Mission Scientifique du Dr. Monard en Guinée Portugaise 1937–1938. III. Chiroptères. Arquivos do Museu Bocage 10: 49–80. [Google Scholar]
- Murray SW, Campbell P, Kingston T, Zubaid A, Francis CM, Kunz TH. (2012) Molecular phylogeny of hipposiderid bats from Southeast Asia and evidence of cryptic diversity. Molecular Phylogenetics and Evolution 62: 597–611. 10.1016/j.ympev.2011.10.021 [DOI] [PubMed] [Google Scholar]
- Murray SW, Khan FA, Kingston T, Zubaid A, Campbell P. (2018) A new species in the Hipposideros bicolor group (Chiroptera: Hipposideridae) from Peninsular Malaysia. Acta Chiropterologica 20: 1–29. 10.3161/15081109ACC2018.20.1.001 [DOI] [Google Scholar]
- Naidoo T, Schoeman MC, Goodman SM, Taylor PJ, Lamb JM. (2016) Discordance between mitochondrial and nuclear genetic structure in the bat Chaerephon pumilus (Chiroptera: Molossidae) from southern Africa. Mammalian Biology-Zeitschrift für Säugetierkunde 81: 115–122. 10.1016/j.mambio.2015.11.002 [DOI] [Google Scholar]
- Nesi N, Nakoune E, Cruaud C, Hassanin A. (2011) DNA barcoding of African fruit bats (Mammalia, Pteropodidae). The mitochondrial genome does not provide a reliable discrimination between Epomophorus gambianus and Micropteropus pusillus. Comptes Rendus Biologies 334: 544–554. 10.1016/j.crvi.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack T. (1893) Neue Beiträge zur Kenntniss der Säugethier-Fauna von Ostafrika. Zoologische Jahrbücher Abteilung für Systematik Oekologie und Geographie der Tiere 7: 523–594. 10.5962/bhl.part.24067 [DOI] [Google Scholar]
- Ogilvie HA, Bouckaert RR, Drummond AJ. (2017) StarBEAST2 brings faster species tree inference and accurate estimates of substitution rates. Molecular Biology and Evolution 34: 2101–2114. 10.1093/molbev/msx126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson DM, Dinerstein E, Wikramanayake ED, Burgess ND, Powell GVN, Underwood EC, D’Amico JA, Itoua I, Strand HE, Morrison JC, Loucks CJ, Allnutt TF, Ricketts TH, Kura Y, Lamoreux JF, Wettengel WW, Hedao P, Kassem KR. (2001) Terrestrial ecoregions of the world: a new map of life on Earth BioScience 51: 933–938. 10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2 [DOI]
- Patterson BD, Webala PW. (2012) Keys to the bats (Mammalia: Chiroptera) of East Africa. Fieldiana: Life and Earth Sciences 6: 1–60. 10.3158/2158-5520-12.6.1 [DOI] [Google Scholar]
- Patterson BD, Webala PW, Bartonjo M, Nziza J, Dick CW, Demos TC. (2018) On the taxonomic status and distribution of African species of Otomops (Chiroptera: Molossidae). PeerJ 6: e4864. 10.7717/peerj.4864 [DOI] [PMC free article] [PubMed]
- Patterson BD, Webala PW, Kerbis Peterhans JC, Goodman SM, Bartonjo M, Demos TC. (2019) Genetic variation and relationships among Afrotropical species of Myotis (Chiroptera: Vespertilionidae). Journal of Mammalogy 100: 1130–1143. 10.1093/jmammal/gyz087 [DOI] [Google Scholar]
- Peters WCH. (1852) Naturwissenschaftliche Reise nach Mossambique: auf Befehl Seiner Majestät des Königs Friedrich Wilhelm IV, in den Jahren 1842 bis 1848 ausgeführt. Zoologie. G. Reimer, Berlin, 202 pp 10.5962/bhl.title.48863 [DOI] [Google Scholar]
- Rakotoarivelo AR, Goodman SM, Schoeman MC, Willows-Munro S. (2019) Phylogeography and population genetics of Macronycteris commersonii s.s. (Chiroptera: Hipposideridae), an endemic Malagasy bat. PeerJ 7: e5866. 10.7717/peerj.5866 [DOI] [PMC free article] [PubMed]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67: 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang Z. (2017) Efficient Bayesian species tree inference under the multispecies coalescent. Systematic Biology 66: 823–842. 10.1093/sysbio/syw119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MF, Jenkins PD, Francis CM, Fulford AJC. (2003) A new species of the Hipposideros pratti group (Chiroptera, Hipposideridae) from Lao PDR and Vietnam. Acta Chiropterologica 5: 31–48. 10.3161/001.005.0103 [DOI] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salicini I, Ibáñez C, Juste J. (2011) Multilocus phylogeny and species delimitation within the Natterer’s bat species complex in the Western Palearctic. Molecular Phylogenetics and Evolution 61: 888–898. 10.1016/j.ympev.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Samonds KE. (2007) Late Pleistocene bat fossils from Anjohibe Cave, northwestern Madagascar. Acta Chiropterologica 9: 39–65. 10.3161/1733-5329(2007)9[39:LPBFFA]2.0.CO;2 [DOI]
- Sanderson I. (1937) Animal Treasure. Viking Press, New York, 325 pp 10.2307/1436145 [DOI] [Google Scholar]
- Seabra AF. (1898) Sobre um caracter importante para a determinação dos generos e especies dos “Microchiroptera” e lista da especies d’este grupo existantes nas colleçoes de Museo Nacional. Jornal de Sciencias, Mathematicas, Physicas e Naturaes Lisboa (Serie 2) 5: 247–258.
- Shi JJ, Rabosky DL. (2015) Speciation dynamics during the global radiation of extant bats. Evolution 69: 1528–1545. 10.1111/evo.12681 [DOI] [PubMed] [Google Scholar]
- Sikes RS, Animal Care and Use Committee of the American Society of Mammalogists. (2016) 2016 Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. Journal of Mammalogy 97: 663–688. 10.1093/jmammal/gyw078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons NB. (2005) Chiroptera. In: Wilson DE, Reeder DAM. (Eds) Mammal Species of the World: A Taxonomic and Geographic Reference (3rd edn.). Johns Hopkins University Press, Baltimore, 312–529.
- Simmons NB, Cirranello AL. (2019) Bat Species of the World: A Taxonomic and Geographic Database. http://www.batnames.org [Accessed on: 2019-8-21]
- Sundevall CJ. (1846) Nya Mammalia från Sydafrika. Öfversigt. Kungliga Svenska Vetenskapsakademiens Handlinger 3(5): 118–121. [Google Scholar]
- Tate GHH. (1941) Results of the Archbold Expeditions. No. 35. A review of the genus Hipposideros with special reference to Indo-Australian species. Bulletin of the American Museum of Natural History 78: 353–393. http://hdl.handle.net/2246/1780 [Google Scholar]
- Temminck CJ. (1853) Esquisses zoologiques sur la côte de Guiné. I. Mammifères. C. C. Vander Hoek, Leiden, 256 pp 10.5962/bhl.title.14828 [DOI] [Google Scholar]
- Thomas O. (1904) Three new bats, African and Asiatic. Annals and Magazine of Natural History (Series 7) 13: 384–388. 10.1080/00222930408562462 [DOI]
- Thong VD, Puechmaille SJ, Denzinger A, Dietz C, Csorba G, Bates PJJ, Teeling EC, Schnitzler HU. (2012) A new species of Hipposideros (Chiroptera: Hipposideridae) from Vietnam. Journal of Mammalogy 93: 1–11. 10.1111/j.1365-2907.2011.00202.x [DOI] [Google Scholar]
- Thorn E, Kerbis Peterhans JC, Baranga J. (2009) Chiroptera. In: Thorn E, Kerbis Peterhans JC. (Eds) Small mammals of Uganda.Bonner Zoologische Monographien 55: 12–75.
- Toews DP, Brelsford A. (2012) The biogeography of mitochondrial and nuclear discordance in animals. Molecular Ecology 21: 3907–3930. 10.1111/j.1365-294X.2012.05664.x [DOI] [PubMed] [Google Scholar]
- Vallo P, Benda P, Martínková N, Kanuch P, Kalko EKV, Cervený J, Koubek P. (2011) Morphologically uniform bats Hipposideros aff. ruber (Hipposideridae) exhibit high mitochondrial genetic diversity in southeastern Senegal. Acta Chiropterologica 13: 79–88. 10.3161/150811011X578633 [DOI] [Google Scholar]
- Vallo P, Guillén-Servent A, Benda P, Pires DB, Koubek P. (2008) Variation of mitochondrial DNA in the Hipposideros caffer complex (Chiroptera: Hipposideridae) reveals high cryptic diversity Acta Chiropterologica 10: 193–206. 10.3161/150811008X414782 [DOI]
- Verschuren J. (1957) Introduction á ľecologie, biologie et systématique de cheiroptères. In: de Saeger ZH. (Ed.) Exploration du Parc National de la Garamba.Fascicule 7. Institut Parcs Nationaux du Congo Belge, Bruxelles, 25–65.
- Wagner A. (1845) Diagnosen einiger neuen Arten von Nagern und Handflügern. Archiv für Naturgeschichte 11: 145–149. [Google Scholar]
- Webala PW, Rydell J, Dick CW, Musila S, Patterson BD. (2019) Echolocation calls of high duty-cycle bats (Hipposideridae and Rhinonycteridae) from Kenya. Journal of Bat Research and Conservation 12: 10–20. 10.14709/BarbJ.12.1.2019.02 [DOI] [Google Scholar]
- Wesselman HB. (1984) The Omo micromammals: systematics and paleoecology of Early Man sites from Ethiopia. S. Karger, Basel, 222 pp. [Google Scholar]
- Yang Z. (2015) The BPP program for species tree estimation and species delimitation. Current Zoology 61: 854–865. 10.1093/czoolo/61.5.854 [DOI] [Google Scholar]
- Yang Z, Rannala B. (2014) Unguided species delimitation using DNA sequence data from multiple loci. Molecular Biology and Evolution 31: 3125–3135. 10.1093/molbev/msu279 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Geographic distribution of voucher specimens used in this analysis
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bruce D. Patterson, Paul W. Webala, Tyrone H. Lavery, Bernard R. Agwanda, Steven M. Goodman, Julian C. Kerbis Peterhans, Terrence C. Demos
Data type
occurrence
Figure S2. Phylogeny of Hipposideridae based on maximum likelihood analysis of cyt-b based on 452 individuals
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bruce D. Patterson, Paul W. Webala, Tyrone H. Lavery, Bernard R. Agwanda, Steven M. Goodman, Julian C. Kerbis Peterhans, Terrence C. Demos
Data type
phylogenetic tree
Figure S3. Phylogeny of Hipposideridae based on Bayesian inference analysis of cyt-b based on 452 individuals
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bruce D. Patterson, Paul W. Webala, Tyrone H. Lavery, Bernard R. Agwanda, Steven M. Goodman, Julian C. Kerbis Peterhans, Terrence C. Demos
Data type
phylogenetic tree