In this study, Yue et al. describe a therapeutic strategy termed “targeted chemotherapy” that involves depleting PP2A or inhibiting it using a small molecule inhibitor, phendione, in drug-resistant melanoma. The authors show phendione induces DNA damage response without causing DNA breaks or inducing cellular dormancy, therefore blocking tumor growth of BRAF mutant and NRAS mutant melanomas.
Keywords: DNA damage response, drug resistance, cellular dormancy, MAP kinase signaling, protein phosphatase 2A, small molecule inhibitor
Abstract
The emergence of drug resistance is a major obstacle for the success of targeted therapy in melanoma. Additionally, conventional chemotherapy has not been effective as drug-resistant cells escape lethal DNA damage effects by inducing growth arrest commonly referred to as cellular dormancy. We present a therapeutic strategy termed “targeted chemotherapy” by depleting protein phosphatase 2A (PP2A) or its inhibition using a small molecule inhibitor (1,10-phenanthroline-5,6-dione [phendione]) in drug-resistant melanoma. Targeted chemotherapy induces the DNA damage response without causing DNA breaks or allowing cellular dormancy. Phendione treatment reduces tumor growth of BRAFV600E-driven melanoma patient-derived xenografts (PDX) and diminishes growth of NRASQ61R-driven melanoma, a cancer with no effective therapy. Remarkably, phendione treatment inhibits the acquisition of resistance to BRAF inhibition in BRAFV600E PDX highlighting its effectiveness in combating the advent of drug resistance.
The mitogen-activated protein kinase (MAPK) pathway is the most frequently mutated signaling cascade in cancer (Davies et al. 2002). The MAPK pathway is initiated by binding of growth factors such as epidermal growth factor (EGF) to their receptor tyrosine kinase (Peyssonnaux and Eychène 2001). The signal is propagated through the RAS GTPase and the RAF-MEK and ERK kinases, which leads to a potent induction of immediate early genes (IEGs) (Murphy et al. 2002). A large number of cancers including lung, colon, skin, pancreas, and multiple forms of leukemia harbor oncogenic mutations that promotes constitutively active MAPK signaling. These include mutations in FLT3, EGFR, KRAS, HRAS, NRAS, and BRAF (Davies et al. 2002; Malumbres and Barbacid 2003; Muzumdar et al. 2017; Patnaik 2017). The constitutive activation of the MAPK pathway promotes the proliferation and survival of cells and paves the path for cancer initiation and progression (McCubrey et al. 2007).
Tremendous strides have been made in the development of compounds that specifically target mutant kinases, and in particular, components of the MAPK pathway (Sawyers 2004; Gharwan and Groninger 2016). Targeting RAS has proven to be extremely challenging and consequently very few therapeutic options exist for RAS mutant cancer, which includes ∼30% of melanomas (Devitt et al. 2011; Jakob et al. 2012). Inhibitors of oncogenic BRAF, which occurs in ∼50% of melanoma patients (Cancer Genome Atlas 2015), have led to unprecedented clinical responses in patients with BRAF mutant metastatic melanoma. However, the clinical benefit of such agents is limited by genetic and/or nongenetic adaptive mechanisms that lead to intrinsic or acquired resistance (Manzano et al. 2016). Additionally, cancer cells can escape the effects of conventional chemotherapeutic agents through differentiation, de-differentiation into a slow cycling cell population, and/or entering a senescence state, which are collectively referred to as cellular dormancy (Yeh and Ramaswamy 2015). These clinical observations highlight the need for improved molecular strategies, combinations and/or sequential therapies that target cancer cells without allowing growth arrest. Here we show that protein phosphatase 2A (PP2A) inhibition provides a unique opportunity for cancer therapy by inducing the DNA damage response (DDR) without causing DNA breaks or promoting cellular dormancy.
Results
Targeting DNA damage response phosphatases in melanoma
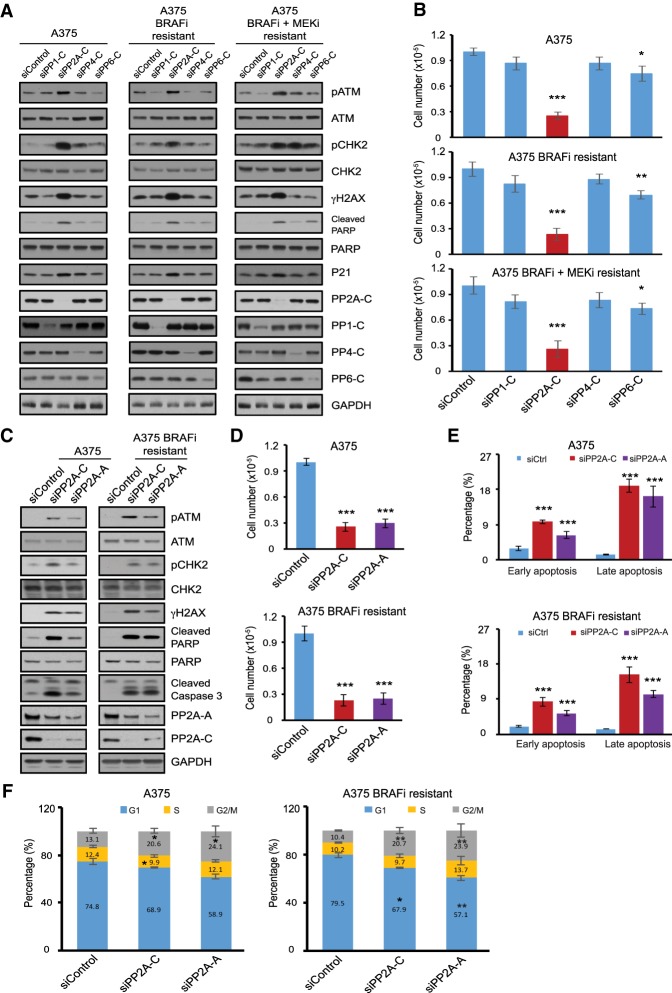
While conventional chemotherapy is effective for a variety of cancers, its utilization has been hampered by the development of a growth arrest commonly known as tumor dormancy (Gao et al. 2017). Paradoxically, melanomas that acquire resistance to BRAF inhibition display increased sensitivity to conventional chemotherapeutic DNA-damaging agents upon drug discontinuation (Kong et al. 2017). We sought to devise a strategy that exploits the DNA damage vulnerability of melanoma cells resistant to MAPK therapy while evading cell cycle arrest. We reasoned that targeting the protein phosphatases that regulate DNA damage signaling (Freeman and Monteiro 2010) may provide such an opportunity. We assessed the role of key phosphatases that regulate the DDR by the activation of ATM and its downstream targets, CHK2 and γH2AX, the main components of DNA damage signaling. We also monitored poly(ADP-ribose) polymerase (PARP) cleavage, a measure of cellular apoptosis (Ivana Scovassi and Diederich 2004). We depleted catalytic subunits of a number of protein phosphatases, including PP1-C (the catalytic subunit of protein phosphatase 1 [PP1]), PP2A-C (the catalytic subunit of PP2A), PP4-C (the catalytic subunit of protein phosphatase 4 [PP4]), and PP6-C (the catalytic subunit of protein phosphatase 6 [PP6]) and examined the induction of DNA damage signaling in three cell lines, including the parental A375 melanoma cell line harboring BRAFV600E mutation, the previously described A375-derived cell line (Yue et al. 2017) with resistance to BRAF inhibitor (A375 BRAFi-resistant) and another A375-derived cell line with resistance to both BRAF inhibitor and MEK inhibitor (trametinib [MEKi]; A375 BRAFi + MEKi-resistant) (Supplemental Fig. S1A–D). Intriguingly, depletion of PP2A catalytic subunits (PP2A-C, both α and β isoforms) led to a specific and potent activation of ATM and induction of the DDR pathway (pCHK2 and γH2AX) as well as PARP cleavage in all three cell lines (Fig. 1A). PP2A-C depletion also displayed the greatest decrease in cellular proliferation across the three cell lines tested, consistent with activation of DNA damage signaling (Fig. 1B). Importantly, these observations were recapitulated by knockdown of the PP2A regulatory subunit (PP2A-A [the scaffolding regulatory subunit of PP2A] both α and β isoforms) (Fig. 1C,D). Finally, reduction of PP2A catalytic and regulatory subunits increased the proportion of cells in apoptosis and in G2/M phase of the cell cycle (Fig. 1E,F; Supplemental Fig. S1E,F). These results attest to the critical role of PP2A in the activation of the DNA damage response, inhibition of cell proliferation, and cell cycle arrest in both parental and drug-resistant A375 melanoma cell lines.
Figure 1.
Targeting DDR phosphatases in melanoma activates DNA damage signaling and inhibits cell proliferation. (A) Immunoblots of cell lysates collected 3 d after siRNA transfection from parental A375, A375 BRAFi-resistant, and A375 BRAFi + MEKi-resistant cells. The cells were transfected with control siRNA or siRNA targeting the catalytic subunits of protein phosphatases PP1-C (siRNA pool against α, β, and γ isoforms), PP2A-C (siRNA pool against α and β isoforms), PP4-C or PP6-C. (B) Proliferation assay of parental A375, A375 BRAFi-resistant, and A375 BRAFi + MEKi-resistant cells. The number of viable cells was counted 3 d after siRNA transfection as shown in A. Data are shown as average ± SD of three independent experiments. (C) Immunoblots of cell lysates collected 3 d after siRNA transfection from parental A375 and A375 BRAFi-resistant cells. The cells were transfected with control siRNA or siRNA targeting the catalytic subunits of protein phosphatases PP2A-C (siRNA pool against α and β isoforms) or scaffolding regulatory subunit PP2A-A (siRNA pool against α and β isoforms). (D) Proliferation assay of parental A375 and A375 BRAFi-resistant cells. The number of viable cells was counted 3 d after siRNA transfection as shown in C. Data are shown as average ± SD of three independent experiments. (E) Quantification of cell apoptosis measured by Annexin V staining flow cytometry. Parental A375 and A375 BRAFi-resistant cells were transfected with siRNAs as described in C and apoptotic cells were measured 3 d after transfection. Results were presented as average ± SEM (n = 2). (F) Cell cycle analysis for A375 and A375 BRAFi-resistant cells performed 3 d after transfection with siRNAs as described in C. Graphs show the percentage of cells in each phase of the cell cycle. Data shown are average ± SD of three independent experiments. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, by unpaired two-tailed Student's t-test.
Phendione is a small molecule inhibitor of PP2A
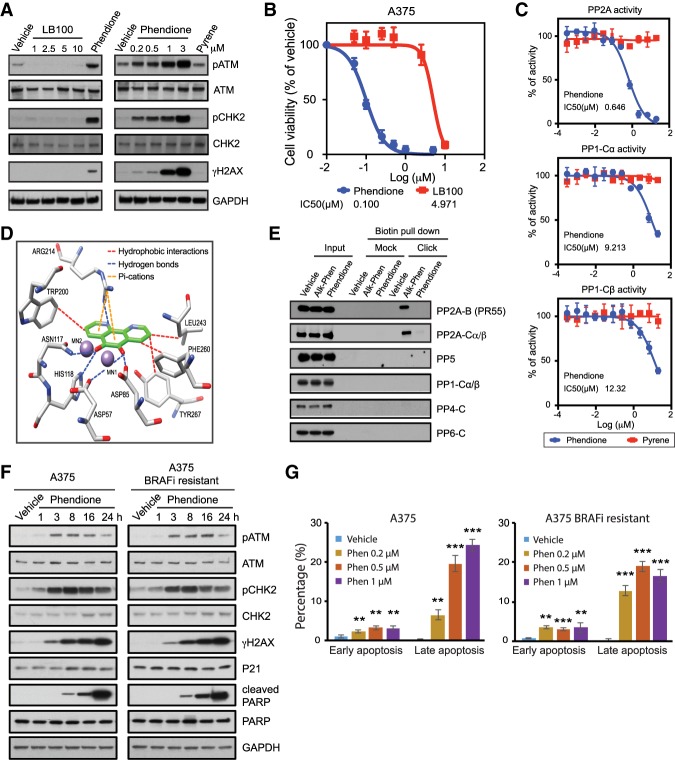
We next aimed to find a small molecule inhibitor of PP2A to induce the DNA damage response and inhibit melanoma cellular proliferation. LB100 was previously reported as a small molecule inhibitor of PP2A (Lu et al. 2009). Treatment of A375 cells with LB100 failed to induce the DDR at concentrations up to 10 µM, and the IC50 of LB100 for inhibition of A375 proliferation was 5 µM (Fig. 2A,B). In search of a small molecule inhibitor of PP2A that mimics PP2A depletion and subsequent DDR, we examined phendione (1,10-phenanthroline-5,6-dione), a phenanthroline derivative reported to inhibit phosphatase activity (Urbanek et al. 2001). Notably, a number of studies identified that phenanthroline and its derivatives promote cytotoxicity in cancer cells (Deegan et al. 2006; Pivetta et al. 2014). Indeed, phendione specifically inhibited recombinant PP2A (a core two-subunit enzyme of PP2A-Cα and PP2A-Aα), but not PP1-Cα or PP1-Cβ, in an in vitro phosphatase assay. Pyrene, a flat aromatic molecule, was used as a control (Fig. 2C). Molecular modeling supported the association of phendione with key residues of PP2A-Cα catalytic cleft with an estimated free energy of binding of −6.24 kcal/mol (Fig. 2D; Supplemental Fig. S2A). To confirm the direct association of phendione with PP2A, we developed alkyne-phendione to perform copper-catalyzed Click chemistry for biotin conjugation (Supplemental Fig. S2B). These experiments revealed that alkyne–phendione specifically interacts with PP2A following the Click reaction and biotin pull-down, confirming a direct association of phendione with PP2A (Fig. 2E). Importantly, in contrast to LB100, nanomolar concentrations of phendione were sufficient to induce the DDR and diminish the proliferation of A375 cells (Fig. 2A,B). Additionally, a time course and dose-escalation analysis of phendione revealed that BRAFi-resistant A375 cells displayed a similar induction of DDR, PARP cleavage, and apoptotic response to that of the parental line (Fig. 2F,G; Supplemental Figs. S1D, S2C–F). While we found differences in efficacy between the enzyme- and cell-based assays, such differences have been observed previously and are generally attributed to limitations of the in vitro assay's reliance on recombinant enzymes that may be partially folded or need additional regulatory subunits to display the full extent of their enzymatic activity (Krzyzosiak et al. 2018). Taken together, these results pinpoint phendione as an inhibitor of PP2A whose effects manifest in increased ATM phosphorylation and activation of the DNA damage response, leading to impaired growth of melanoma cells with acquired resistance to MAPK inhibitors.
Figure 2.
Phendione is a small molecule inhibitor targeting PP2A. (A) Immunoblots of A375 cell lysates collected after 3 h of drug treatment as indicated; i.e., increasing concentrations of LB100 compared with 0.2 µM phendione (left) and increasing concentrations of phendione compared with 3 µM pyrene (right). DMSO was used as negative control (vehicle). (B) Growth inhibition curves of A375 cells and IC50 values after treatment with phendione or LB100 for 96 h. Data are presented as mean ± SD of three independent experiments. (C) Phosphatase activity assay for PP2A core enzyme (complex of regulatory subunit Aα and catalytic subunit Cα; top), PP1 catalytic subunit Cα (middle) and PP1 catalytic subunit Cβ (bottom) incubated with phendione or pyrene at indicated concentrations. Data shown are average ± SD of two independent experiments. IC50s (micromolar) were calculated and is shown as an inset in each graph. (D) Binding model of phendione to PP2A. Docking pose of phendione (green) interacting with the catalytic residues of PP2A-Cα (PPP2CA) (white) within 5 Å around the ligand. Oxygens (red), nitrogens (blue), and manganese ions (purple) are shown. Dotted lines represent intermolecular polar contacts. Hydrogens are omitted for clarity. (E) Immunoblots for phendione derivative pull down. (F) Immunoblots of A375 and A375 BRAFi-resistant cell lysates after treatment with 0.2 µM phendione or vehicle for indicated time. (G) Quantification of cell apoptosis measured by Annexin V staining flow cytometry. Parental A375 and A375 BRAFi-resistant cells were treated with vehicle or increasing concentration of phendione as indicated in the panels for 24 h before apoptotic cells were measured by flow cytometry. Results were presented as average ± SD (n = 3). (**) P < 0.01; (***) P < 0.001, by unpaired two-tailed Student's t-test.
Phendione inhibits the proliferation of melanoma with intrinsic resistance to MAPK inhibitors
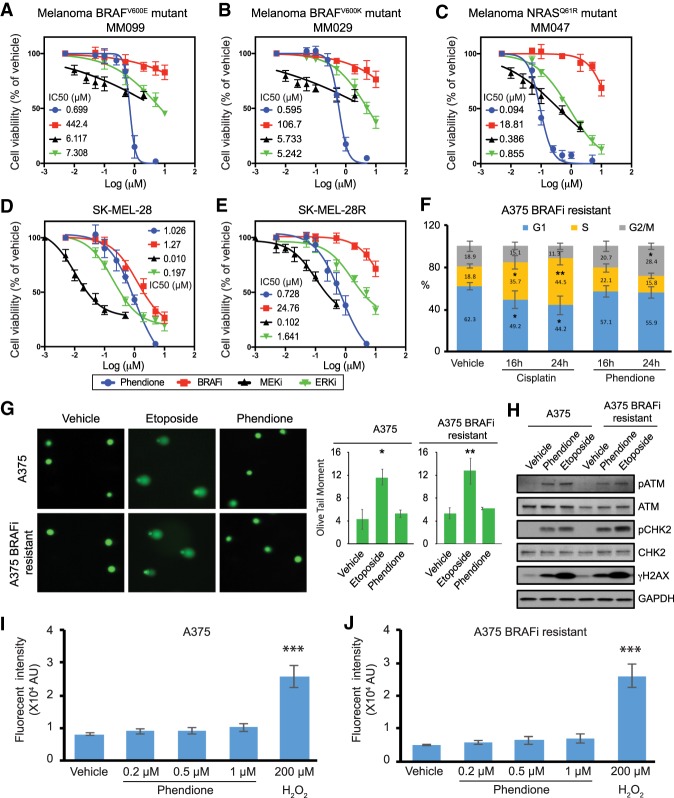
We next assessed phendione growth inhibitory activity in a variety of primary patient-derived cell lines exhibiting a mesenchymal-like/invasive phenotype, which confers intrinsic resistance to inhibitors of the MAPK pathway (Kemper et al. 2014; Verfaillie et al. 2015; Titz et al. 2016; Shaffer et al. 2017). MM099 BRAFV600E mutant melanoma cells were resistant to MAPK inhibition as expected, though they showed a strong decrease in cell viability following phendione treatment (Fig. 3A). Similarly, other invasive cell lines such as MM029 (BRAFV600K) and MM047 (NRASQ61R) were exquisitely sensitive to phendione while their responsiveness to MAPK inhibitors was muted (Fig. 3B,C). BRAFV600E SK-MEL-28R cells were resistant to BRAFi, MEKi, and ERKi yet showed a robust response to phendione that was comparable with the parental SK-MEL-28 cell line (Fig. 3D,E). Therefore, phendione can overcome both intrinsic and acquired drug resistance in melanoma resulting in potent inhibition of tumor cell proliferation.
Figure 3.
Phendione inhibits the proliferation of melanoma with intrinsic resistance to MAPK inhibitors. (A–E) Panels show growth inhibition curves and IC50 values. Primary patient-derived melanoma cell lines with BRAFV600E, BRAFV600K, and NRASQ61R mutations (A–C), and human melanoma cell line SK-MEL28 (D) and its BRAF inhibitor (dabrafenib)-resistant line SK-MEL28R (E) were treated with phendione or inhibitors of the MAPK pathway for 96 h. Data are presented as mean ± SD of three independent experiments. (F) Cell cycle analysis for A375 BRAFi-resistant cells. Cells were treated with 2.5 µM cisplatin, 0.2 µM phendione, or vehicle for the indicated time. Graph shows the percentage of cells in each phase of the cell cycle. Data shown are average ± SD of three independent experiments. (G) Comet assay to measure DNA damage. Representative images are shown at the left and quantification results are shown at the right. Data are presented as mean ± SD of three independent experiments. (H) Immunoblots of whole-cell lysate collected from comet assay. (I,J) Quantification of reactive oxygen species (ROS) production measured by flow cytometry in A375 (I) and A375 BRAFi-resistant (J) cells. Hydroxyl peroxide (H2O2; 200 µM) was used as positive control. Data are presented as means ± SEMs of three independent experiments. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. Statistical analysis was performed using unpaired two-tailed Student's t-test.
Phendione mechanism of action is distinct from that of conventional chemotherapeutic agents
To assess mechanistic differences between phendione and other DNA break-inducing chemotherapeutics, we measured cell cycle progression of BRAFi-resistant cells treated with phendione or cisplatin. In contrast to cisplatin, which triggered a block in S phase progression, phendione treatment resulted in a small, yet significant, increase in the fraction of cells in the G2/M phase of the cell cycle (Fig. 3F; Supplemental Fig. S3A). The parental A375 and BRAFi/MEKi-resistant cell lines exhibited a similar response to phendione and also induced a small increase in the G2/M population (Supplemental Fig. S3A,B; data not shown). Comet assays confirmed that unlike etoposide, another DNA damage inducing agent, phendione did not cause DNA breaks, rather it activated DNA damage signaling (Fig. 3G,H). Importantly, a flow cytometry assay revealed that phendione treatment did not promote the generation of reactive oxygen species (Fig. 3I,J; Supplemental Fig. S3C). Finally, the cytotoxicity of phendione was assessed in a panel of melanoma cell lines and normal cells, including primary human melanocytes (HEMn). While nontransformed lines were relatively resistant to phendione, most melanoma cells exhibited low micromolar IC50 (Supplemental Fig. S3D). Additionally, primary human melanocytes required a higher dose of phendione to induce DNA damage signaling and apoptosis (Supplemental Fig. S3E). These results indicate that phendione activates ATM and the consequent DNA damage response pathway by a mechanism of action that is distinct from cisplatin and etoposide, both of which produce physical DNA breaks.
Phendione does not chelate intracellular cations
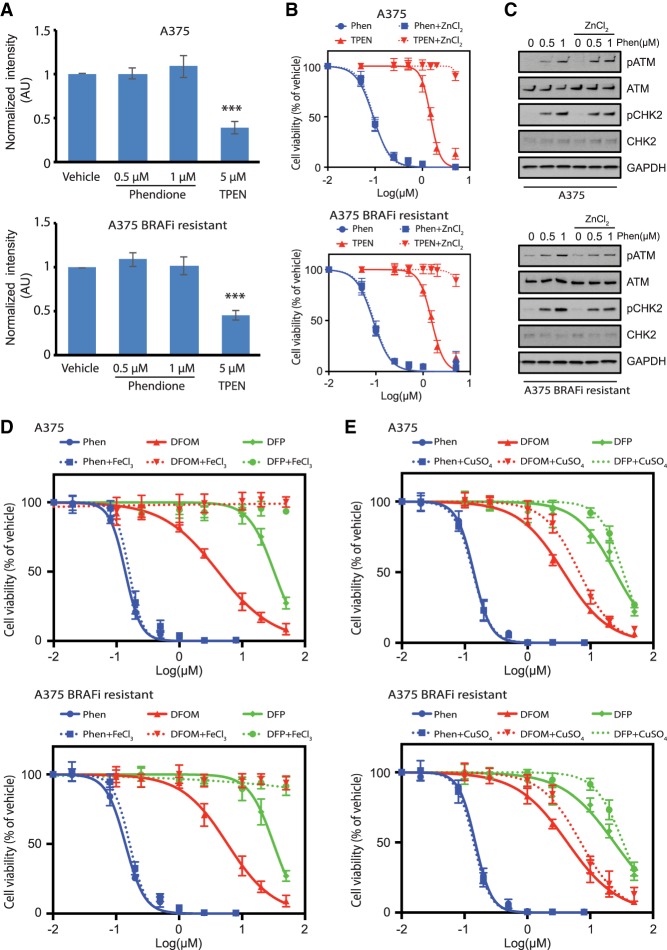
To assess whether the effects of phendione are mediated through zinc chelation, we measured intracellular zinc in parental and BRAFi-resistant cells using a flow cytomery assay. Unlike TPEN [N,N,N′,N′-tetrakis (2-pyridinylmethyl)-1,2-ethanediamine], a known membrane-permeable zinc chelator, phendione did not reduce intracellular zinc levels (Fig. 4A). Accordingly, in contrast to TPEN treatment, where the addition of ZnCl2 rescued cell viability, the DDR and cytotoxicity incurred by phendione treatment was not rescued with ZnCl2 (Fig. 4B,C). To evaluate whether impaired cellular viability was mediated through chelation of other metals, FeCl3 and CuSO4 were each administered alongside phendione. While the addition of these salts mitigated the cytotoxicity of DFOM (deferoxamine mesylate) and DFP (deferiprone) chelators, the loss of viability and induction of DNA damage signaling persisted in parental and BRAFi-resistant cells treated with phendione (Fig. 4D,E; Supplemental Fig. S4A,B). Moreover, cell viability was not restored by the addition of MgCl2, CaCl2, or NaCl to cells treated with phendione (Supplemental Fig. S4C,D). Taken together, these findings indicate that unlike conventional ion chelating agents, neither the activation of the DNA damage response nor the inhibition of cellular proliferation by phendione is mediated through the chelation of intracellular cations.
Figure 4.
Phendione does not chelate intracellular cations. (A) Quantification of fluorescent intensity in the cells labeled with zinc indicator by flow cytometry. A375 (top) and A375 BRAFi-resistant (bottom) cells were treated with vehicle, phendione or cell-permeable zinc chelator TPEN for 3 h. At the end of treatment, 20 µM zinquin ethyl ester was added into medium to stain Zn2+ in live cells. Unstained cells were used as negative control for flow cytometry. (***) P < 0.001 by unpaired two-tailed Student's t-test. (B) Growth inhibition curves of A375 (top) and A375 BRAFi-resistant (bottom) cells treated with phendione or zinc chelator TPEN prepared in regular medium or medium containing 15 µM ZnCl2. (C) Immunoblot of whole-cell lysate collected after the treatment of vehicle or phendione prepared in regular medium or medium containing 15 µM ZnCl2 as indicated. (D,E) Growth inhibition curves of A375 (top) and A375 BRAFi-resistant (bottom) cells after 96 h of treatment with phendione or ion chelators DFOM or DFP prepared in regular medium or medium containing 50 µM FeCl3 (D) or 10 µM CuSO4 (E).
PP2A inhibition activates MAPK response in BRAF resistant melanoma
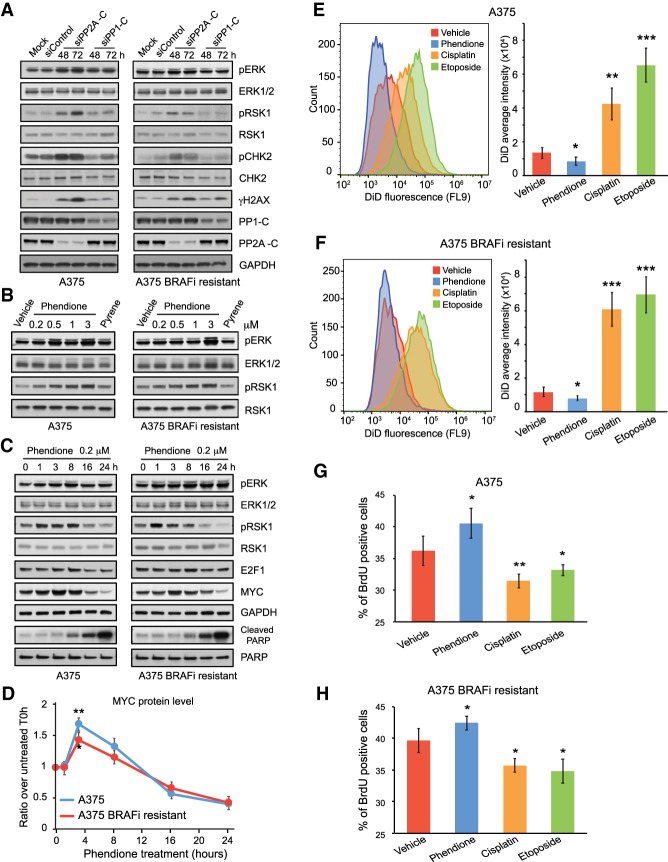
PP2A has been suggested to regulate MAPK signaling (Junttila et al. 2008). Therefore, we explored the possibility that phendione induces cell death by promoting cell cycle progression amidst elevated DNA damage signaling via hyperactivation of the MAPK cascade. Indeed, depletion of PP2A led to an enhancement of ERK1/2 activity in both the parental and BRAFi-resistant melanoma cells (Fig. 5A). Consistent with the effects observed after PP2A depletion, inhibition of PP2A using phendione similarly led to activation of ERK1/2 in a dose-dependent manner (Fig. 5B). Critically, a temporal analysis using 200 nM phendione treatment showed up-regulation of the cell cycle transcriptional activators MYC and E2F1 within the initial time course (1–8 h) followed by their degradation and induction of cell death at 24 h, as shown by PARP cleavage (Fig. 5C,D). Thus, in melanoma cells resistant to BRAF inhibition, phendione treatment or PP2A depletion activates the MAPK cascade and cell cycle entry concurrently with DNA damage signaling and apoptosis.
Figure 5.
PP2A inhibition transiently promotes cell proliferation leading to evasion from dormancy. (A–C) Immunoblots of cell lysate collected from A375 and A375 BRAFi-resistant cells after 2 and 3 d of siRNA transfection (A), 3 h of drug treatment (B), and time-course treatment with 0.2 µM phendione (C) as indicated in each panel, respectively. siRNAs used for transfection include control siRNA, siRNA targeting the catalytic subunits of protein phosphatases PP1-C (siRNA pool against α, β, and γ isoforms) and PP2A-C (siRNA pool against α and β isoforms). (D) Quantification of MYC protein level from immunoblots after time-course treatment with 0.2 µM phendione in A375 and A375 BRAFi-resistant cells. GAPDH protein level was used as internal control to perform normalization. Data shown are mean ± SEM of three independent experiments. (E,F) Histogram and quantification of fluorescent signal from Vybrant DiD cell-labeling dye detected by flow cytometry. (G,H) Quantification of incorporated bromodeoxyuridine (BrdU) measured by flow cytometry. Data shown are mean ± SD of two independent experiments. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. Unpaired two-tailed Student's t-test was used for statistical analysis.
To assess cellular dormancy, we asked whether 3 h of acute treatment with phendione or conventional chemotherapeutic agents affects the proliferative potential of cancer cells 14 d later. We measured proliferation of cells after treatment with phendione, cisplatin, etoposide, or vehicle using incorporation of the Vybrant DiD cytoplasmic membrane marker or BrdU (bromodeoxyuridine) labeling of DNA (Fig. 5E–H). Remarkably, while treatment with conventional chemotherapeutic agents induced a potent S-phase arrest, phendione treatment increased cell proliferation, suggesting a distinct molecular mechanism that prevents cell cycle arrest (Fig. 5E–H). Indeed, the BrdU incorporation rate demonstrates that acute treatment with phendione increased the proliferative capacity of melanoma cells (Fig. 5G,H). These results indicate that phendione, which activates the MAPK response and DNA damage signaling, maintains cell proliferation and enhances the window in which drug-resistant cells could succumb to DNA damage.
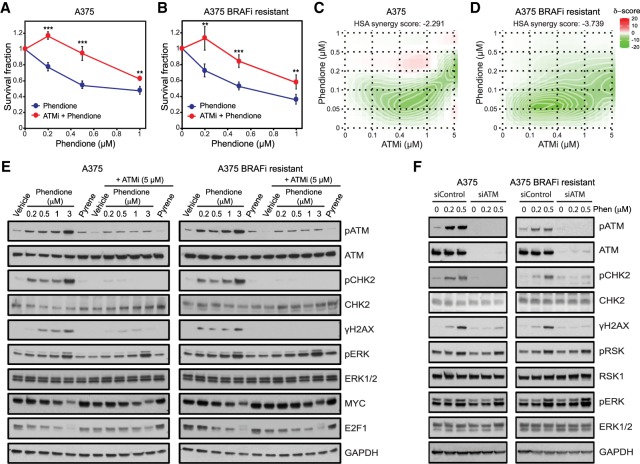
Phendione-induced growth inhibition is ATM-dependent
To establish whether ATM activation plays a role in phendione-mediated growth suppression, we measured the ability of A375 and A375 BRAFi-resistant cell lines to form colonies after treatment with a specific ATM inhibitor (ATMi, KU60019). While treatment with ATMi alone did not significantly affect cell growth, ATMi significantly attenuated the growth inhibition incurred by phendione treatment in both cell lines tested (Fig. 6A,B; Supplemental Fig. S5A,B). Additionally, combined dose response of phendione and ATMi revealed antagonism between the two drugs, pointing to activation of ATM by phendione as a key component of the cytotoxic response (Fig. 6C,D). Consistent with this contention, inhibition of ATM or its depletion abrogated the phendione-mediated ATM-induced phosphorylation of CHK2 and γH2AX in both cell lines (Fig. 6E,F). Importantly, inhibition of ATM or its depletion did not affect phendione-mediated induction of MAPK response (Fig. 6E,F). In summary, these experiments show that phendione, through its inhibition of PP2A-C, exerts a robust induction of ATM activity, triggering DNA damage signaling and ultimately the apoptotic cascade.
Figure 6.
Phendione-induced growth inhibition is ATM dependent. (A,B) Colony formation assay of A375 (A) and A375 BRAFi-resistant (B) cells treated with phendione or phendione in combination with ATMi (5 µM) for 3 h. Data are shown as mean ± SD of three independent experiments. (**) P < 0.01; (***) P < 0.001, by two-way ANOVA with Sidak's multiple comparisons test. (C,D) Synergy maps for A375 (C) and A375 BRAFi-resistant (D) cells showing the effect of combined treatment with phendione and ATMi at indicated concentrations for 96 h. The 2D synergy matrix was generated with SynergyFinder software using HAS model. (E) Immunoblots of A375 and A375 BRAFi-resistant cell lysates after treatment with phendione, pyrene (3 µM), or vehicle for 3 h in the presence or absence of ATM inhibitor (5 µM). (F) Immunoblots of A375 and A375 BRAFi-resistant cell lysates after 3 d of siRNA transfection as indicated. The cells were treated with vehicle or phendione for 3 h before collection.
Phendione inhibits growth of human tumors with NRAS or BRAF mutations
We next examined the therapeutic potential of the above findings by assessing phendione effectiveness in patient-derived melanoma xenograft (PDX) models. Phendione was administered to NMRI nude mice via intraperitoneal (i.p) injection at 5 mg/kg body weight every other day alone or in combination with BRAFi every day for 20 d. Histopathological analysis of trachea, larynx, lungs, heart, esophagus, stomach, liver, gall bladder, pancreas, bone marrow, brain, and small intestine revealed no adverse reactions, suggesting lack of toxicity. Moreover, no significant weight loss was observed during the treatment period, indicating that phendione was well tolerated (data not shown).
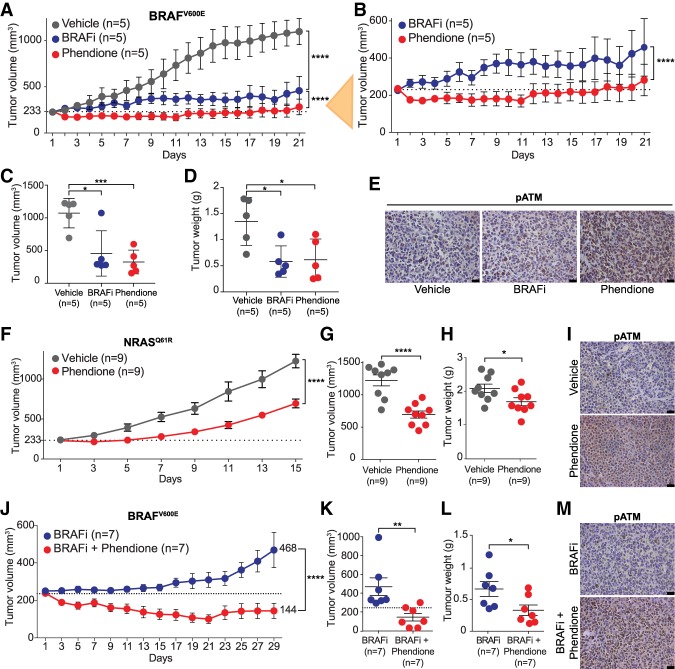
Next, we treated cohorts of melanoma PDXs harboring BRAFV600E mutation with 5 mg/kg phendione every other day or 30 mg/kg dabrafenib daily, a BRAFV600E inhibitor used in the clinic (Hauschild et al. 2012), and monitored tumor growth over time (Fig. 7A,B). Surprisingly, phendione treatment was significantly more effective in reducing tumor growth than the BRAF inhibitor (Fig. 7A–D). Importantly, phendione treatment resulted in the activation of ATM and induction of DNA damage pathway (Fig. 7E; Supplemental Fig. S6A). We next treated NRASQ61R driven melanoma PDXs with phendione, which resulted in a significant reduction in tumor volume and weight (Fig. 7F–H). Additionally, we observed a robust induction of ATM phosphorylation and the DDR (Fig. 7I; Supplemental Fig. S6B). These results attest to the effectiveness of phendione in reducing growth of melanomas with BRAF or NRAS mutations when used as a single agent in relevant preclinical models.
Figure 7.
Phendione inhibits growth of human tumors with BRAF or NRAS mutations and overcomes resistance to BRAF inhibition in vivo. (A,B) Tumor volume of BRAFV600E PDXs treated with BRAFi (dabrafenib) at 30 mg/kg daily by oral gavage, or phendione at 5 mg/kg every 2 d intraperitoneally (i.p.); vehicle was used as control in A. Zoomed-in curves of BRAFi and phendione treatments are shown in B. Data are mean ± SEM, n = 5. (****) P < 0.0001, by two-way ANOVA with Tukey's multiple comparisons test. Dotted line in A and B indicates the tumor volume on the day of treatment started. (C,D) Dot plots of BRAFV600E PDX tumor volume (C) and weight (D) at day 21. Data are mean ± SEM. (*) P < 0.05; (***) P < 0.001; (****) P < 0.0001, by unpaired two-tailed Student's t-test. (E) Representative images of immunohistochemistry staining with antibody against phospho-ATM (S1981) for BRAFV600E PDXs treated as indicated. (F) Tumor volume of NRASQ61R PDXs treated with phendione at 5 mg/kg every 2 d intraperitoneally (i.p.), or with vehicle. Data are mean ± SEM, n = 9. (****) P < 0.0001, by two-way ANOVA with Tukey's multiple comparisons test. (G,H) Dot plots of NRASQ61R tumor volume (G) and weight (H) at day 15. Data are mean ± SEM. (*) P < 0.05; (****) P < 0.0001, by unpaired two-tailed Student's t-test. (I) Representative images of immunohistochemistry staining with antibody against phospho-ATM (S1981) for NRASQ61R PDXs after indicated treatments. (J) Phendione prevents tumor relapse in BRAFV600E PDX model. Data are mean ± SEM, n = 7. (****) P < 0.0001, by two-way ANOVA with Tukey's multiple comparisons test. (K,L) Dot plots of tumor volume (K) and weight (L) of BRAFV600E PDXs. Data are mean ± SEM. (*) P < 0.05; (**) P < 0.01, by unpaired two-tailed Student's t-test. (M) Representative images of immunohistochemistry staining with antibody against phospho-ATM (S1981) of BRAFV600E PDXs treated as indicated. Scale bars, 25 µm.
Phendione overcomes resistance to BRAF inhibition in vivo
Finally, we evaluated whether combination treatment with a BRAF inhibitor and phendione could delay or prevent the development of resistance to BRAFi. We treated BRAFV600E mutant melanomas in PDX cohorts with either BRAFi, or a combination of BRAFi and phendione (Fig. 7J). Notably, the combined treatment of BRAFi and phendione had a significantly greater inhibitory effect on tumor growth than the BRAFi alone, highlighting the potential of phendione to increase the efficacy of BRAFi. Additionally, this analysis revealed that while stable tumors begin to grow following 25 d of treatment with BRAFi alone, dual treatment with phendione sustained inhibition of tumor growth during the 29 d of the treatment (Fig. 7J–L). Similar to treatment with phendione alone, we observed a strong induction of ATM phosphorylation in tumors treated with phendione and BRAF inhibitor (Fig. 7M; Supplemental Fig. S6C). Finally, we treated parental A375 cells or BRAFi-resistant cells with combination of phendione and either BRAF or ERK1/2 inhibitors (Supplemental Fig. S6D,E). Interestingly, inhibition of ERK1/2 significantly antagonized the antiproliferative response of phendione in both parental and BRAFi-resistant A375 cells, consistent with the role of phendione in the inhibition of PP2A and subsequent ERK1/2 activation (Supplemental Fig. S6E). However, we observed a synergistic response following a combination treatment with BRAFi and phendione (Supplemental Fig. S6D), in agreement with the cooperative effect of phendione and BRAFi treatment in mouse models of melanoma (Fig. 7J–L). These results support the transient activation of ERK1/2 by phendione in the presence of BRAFi (Supplemental Fig. S6C) and point to ERK1/2 as a key target of PP2A in the negative regulation of MAPK pathway. Taken together, these experiments underscore the utility of phendione in diminishing the resistance that develops during prolonged BRAFi treatment.
Discussion
The key findings of our study are as follows. First, we highlight the utility of a small molecule, phendione, as an inhibitor of PP2A, and support the importance of PP2A in the DNA damage response. Second, we demonstrate that PP2A depletion or inhibition by phendione induces ATM activation and the DNA damage response, leading to growth inhibition of melanoma that display intrinsic and acquired resistance to MAPK inhibitors. Third, we show that phendione, as a single agent, profoundly diminishes the growth of human melanoma tumors containing BRAF or NRAS mutations. Fourth, a combination treatment with phendione and BRAF inhibitor prevents the emergence of acquired resistance to BRAF inhibition in a PDX model of melanoma. Finally, through inhibition of PP2A, we usher in a cancer treatment strategy that subverts tumor dormancy through the activation of signaling pathways that promote cellular proliferation.
Our study highlights a unique strategy termed “targeted chemotherapy” in which the activation of DNA damage signaling through PP2A inhibition induces cell death in treatment-resistant cancer cells without physical DNA breaks. Remarkably, PP2A inhibition maintains proliferative capacity by hyperactivating the MAPK pathway and preventing escape from the apoptotic output of DNA damage signaling. Therefore, the key finding of our study is the discovery that while PP2A inhibition induces DNA damage signaling and promotes cancer cell death, unlike conventional chemotherapeutic agents, it does not result in cancer cell growth arrest. Indeed, in contrast to cisplatin or etoposide, in which their prolonged exposure promotes cellular dormancy, treatment of resistant melanoma cells with phendione activates proliferative pathways thereby preventing cell cycle exit and escape from cell death. Targeting PP2A with phendione uncovers the potential of a cancer therapy with a lower rate of tumor recurrence than current conventional chemotherapy due to the absence of cellular dormancy. Taken together, we propose phosphatase-directed therapy as an effective strategy to treat tumors that display resistance to targeted therapies and for those driven by constitutive RAS activation, for which there are no approved targeted therapies.
Materials and methods
Compounds
Vemurafenib (S1267), trametinib (S2673), PD0325901 (S1036), SCH772984 (S7101), etoposide (S1225), and cisplatin (S1166) were purchased from Selleck Chemicals. Phendione (1,10-phenanthroline-5,6-dione; #496383), N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN; #P4413), Deferoxamine mesylate (DFOM; #D9533), 3-hydroxy-1,2-dimethyl-4(1H)-pyridone (deferiprone [DFP] #379409), hydrogen peroxide solution (#95321), and pyrene (#82648) were purchased from Sigma. Ku-60019 (#17502) was purchased from Cayman Chemical. Dabrafenib was purchased from Biorbyt. LB-100 (#T2068) was purchased from TargetMol.
Cell line and antibody
Primary human melanocytes HEMn were ordered from Thermo Fisher Scientific (#C-102-5C) and cultured according to the manufacturer's protocol. Primary patent derived melanoma lines MM029, MM034, MM047, MM057, and MM099 were from Dr. Jean-Christophe Marine laboratory. All other cell lines were ordered from ATCC and cultured with the suggested protocol. The antibodies against pATM-S1981 (#13050), pCHK2-T68 (#2661 and #2197), cleaved PARP (Asp214; #5625), pERK-T202/Y204 (#4370), ERK1/2 (#4696), pRSK-S380 (#9335), RSK1 (8408), cleaved caspase-3 (#9664), MYC (#18583), E2F1 (#3742), P21 (#2947), PP2A-B (the regulatory subunit of PP2A [PR55]; #4953), PP2A-A (#2041), PP2A-C (#2159), and PP5 (#2289) were ordered from Cell Signaling Technology. The antibodies against PP4-C (PPP4C; #PA5-96059) and PP6-C (PPP6C; #PA5-28919) were ordered from Thermo Fisher Scientific. The antibodies against CHK2 (#05-649), pATM-S1981 (#05-740), and γH2AX (Ser139; #06-636-I) were ordered from Sigma. The antibody against PARP1 (#39561) was ordered from Active Motif. The antibodies against PP1-Cα isoform (PPP1CA; #A300-904A), PP1-Cβ isoform (PPP1CB; #A300-905A), and PP1-Cγ isoform (PPP1CC, #A300-906A) were ordered from Bethyl. The antibodies against ATM (sc-377293) and GAPDH (sc-25778) were ordered from Santa Cruz Biotechnology.
Immunofluorescence
Cells were seeded in 24-well plates on glass coverslips (Electron Microscopy Science) and treated as indicated. Before fixation with cold methanol for 20 min at −20°C, cells were washed twice with phosphate-buffered saline (PBS). Fixed cells were treated with cold acetone for 5 min at −20°C and then washed three times in PBS, and blocked with blocking solution (3% bovine serum albumin) for 30 min at room temperature. Fixed cells were incubated with primary antibodies as indicated in 1% bovine serum albumin overnight at 4°C, washed three times with PBS, and Alexa fluor 488- or Alexa fluor 568-labeled secondary antibody was added at a 1:1000 dilution for 1 h at room temperature). After three washes with PBS, the cells were mounted with ProLong Gold antifade mountant with DAPI (Thermo Fisher Scientific P36962). Samples were imaged with a Nikon A1plus Eclipse Ti2 microscope with a 60× plan apo 1.4 NA, at a 1-µm z depth interval, and a focused image was created using the z stack and Nikon Elements 5.02 software. Signals were quantified as number of foci per cell or integrated signal per cell. One-hundred-fifty cells from three independent experiments were analyzed.
Cell cycle analysis
To determine the effect of the compounds on cell cycle progression, cells were treated with vehicle, phendione at 0.2 µM, or cisplatin at 2.5 µM for the indicated time. Cells were collected and fixed with cold 70% ethanol. After two washes with PBS, cells were suspended in staining buffer (PBS with 100 μg/mL RNase A, 50 μg/mL propidium iodide, 0.1% Triton X-100). Cells were stained for 2 h at room temperature. Flow cytometric analyses were performed using BD LSR Fortessa cell analyzer (BD Biosciences). Data were analyzed using FlowJo software.
Protein and ligand preparation
We used the catalytic subunit of the protein phosphatase 2A holoenzyme as the target (PDB code 2iae chain C, region 4–308, protein length 309) (Cho and Xu 2007). We protonated the protein at pH 7, and added atom radii and Gasteiger partial charges per atom (net charge = −8, net charge including ions = −4) using PDB2PQR (v. 2.1.0) (Dolinsky et al. 2007) and the amber force field (Wang et al. 2000). The 3D structure of ligands were retrieved from ZINC15 (Sterling and Irwin 2015) (phendione code ZINC1580384, net charge = 0, HB acceptors = 4, and pyrene code ZINC1758808, net charge 0, HB acceptors = 0). We added Gasteiger charges to each ligand and refined the structures using the MMTK module (Hinsen 2000) in chimera (Pettersen et al. 2004) to obtain a planar conformation (number of steepest descent steps = 200, and conjugate gradient steps = 20). The catalytic site of PP2A was predicted to be druggable with Fpocket (Le Guilloux et al. 2009).
Molecular docking
We used AutoDock tools (Morris et al. 2009) to prepare the docking files (protein and ligand). Next, we generated precalculated grid maps with AutoGrid (Morris et al. 2009) using a grid box of 126 × 126 × 126. We performed the flexible docking of phendione with Autodock 4.2 (Morris et al. 2009) using the Lamarckian genetic algorithm with local search (number of independent docking runs set to 1000 for reaching convergence). The rest of Autodock parameters were kept as default. Docking solutions were clustered with a tolerance of 2 Å and a representative solution per cluster was selected. We prepared the molecular graphics of the complex with the lowest estimated free energy of binding using Chimera (Pettersen et al. 2004). Protein-ligand interactions were analyzed with the protein–ligand interaction profiler (PLIP) using default parameters (Salentin et al. 2015).
Phosphatase profiling
The effect of phendione on the activity of protein phosphatases was determined using a fluorescent-based in vitro enzymatic assay. The enzymatic reactions were conducted in duplicate at room temperature in a solution containing phosphatase enzyme and phendione at various concentrations in 25 mM HEPES (pH 7.5), 5 mM MgCl2, 0.01% Brij-35, 1 mM DTT, 1% DMSO, and 10 µM DiFMUP (6,8-difluoro-7-hydroxy-4-methylcoumarin). The indicated enzyme and substrate were prepared in fresh reaction buffer. Enzyme solution was delivered into the reaction well and then compounds in 100% DMSO were added to the enzyme solution using Echo 550 liquid handler (nanoliter range; Labcyte, Inc.). After 20 min of incubation at room temperature, the substrate solution was delivered into the reaction well to initiate the reaction. Phosphatase activity was monitored as a time-course measurement of the increase in fluorescence signal in the fluorescent substrate at an excitation of 355 nm and an emission of 460 nm using a microplate reader. The initial linear portion (0–40 min) of the slope (signal/min) was analyzed and the percentage of enzyme activity was calculated. Control in the absence of inhibitor was considered as 100% activity. Graphpad Prism was used to perform curve fitting and IC50 calculation. IC50 values were determined by the concentration causing a half-maximal percent activity. Pyrene was used as negative control.
Cell viability and proliferation assay
To measure the effect of the compounds on cell proliferation, cells were plated (3000–5000 per well) in 96-well black plates with clear bottom and incubated overnight at 37°C in 5% CO2 before the drug treatment. Cell viability was measured 96 h after drug treatment using PrestoBlue (Invitrogen A13261). GraphPad Prism software was used to generate dose response curves and calculate IC50 values. Each experiment was repeated at least three times. To determine the effect of RNAi on cell proliferation, cells were trypsinized and the cell number was counted using Moxi Z mini automated cell counter (Orflo Technologies).
Phendione derivative biotin labeling and pull-down
Phendione derivative (Alk-Phen) (structure shown in Supplemental Fig. S4) was custom-synthesized. Cells were treated with 5 µM Alk-Phen, 3 µM phendione, or DMSO for 3 h. Whole-cell lysate was obtained using lysis buffer containing 150 mM NaCl, 1.0% NP-40, and 50 mM Tris-HCl (pH 8.0) and dialyzed against buffer containing 10 mM NaCl, 1.0% NP-40, and 50 mM Tris-HCl (pH 8.0). The cell lysate was then clicked to biotin by adding 50 μM biotin picolyl azide (Click Chemistry Tools 1167), a preformed complex of CuSO4:THPTA (1.0 mM:5.0 mM), and 5 mM sodium ascorbate, sequentially. The click reaction was performed for 30 min at room temperature protected from light. The cell lysate was then dialyzed against lysis buffer to remove unclicked azidobiotin and pull-down was carried out with Dynabeads MyOne streptavidin C1 (Thermo Fisher Scientific 65002).
Comet assay
A375 and A375 BRAFi-resistant cells were treated with 0.5 μM phendione for 8 h before harvesting. Vehicle (DMSO) and etoposide at 20 μM were included as negative and positive controls, respectively. OxiSelect comet assay kit (Cell Biolabs, Inc., STA-352) was used to assess DNA damage in the cells. In brief, cell suspension at 1 × 105/mL was prepared in ice-cold PBS (without Mg2+ and Ca2+). Cell samples were prepared by mixing cell suspension with comet agarose (37°C) at 1:10 ration (v/v) and 75 µL was immediately transferred onto the top of comet agarose base layer. Cells were treated with prechilled lysis buffer for 45 min at 4°C and then treated by alkaline solution for 30 min. The comet agarose was subjected to alkaline electrophoresis for 15–30 min at 1 V/cm with 300 mA current and then washed three times with distilled water. The comet agarose was stained with diluted Vista Green DNA dye for 15 min at room temperature after being fixed with cold 70% ethanol for 5 min. Images were taken with an epifluorescent microscope using a FITC filter. Olive tail moment was calculated using automated ImageJ based plugin OpenComet (v1.3, available at http://www.cometbio.org). Data represent 150 cells analyzed from three independent experiments.
Drug combination effect
To test the effect of drug combination, cells were plated (3000–5000 per well) in 96-well black plates with clear bottom and incubated overnight at 37°C in 5% CO2 before the drug treatment. Cells were treated with various concentrations of phendione in combination with various concentrations of ATM, BRAF, or ERK inhibitor. Cell viability was measured 96 h after drug treatment using PrestoBlue (Invitrogen A13261). The SynergyFinder Web application (https://synergyfinder.fimm.fi) was used to calculate synergy scores and generate 2D synergy maps.
siRNA transfection
siRNA transfection was performed using RNAiMAX transfection reagent (#13778150) and Silencer Select predesigned siRNA from Thermo Fisher Scientific following manufacturer's instructions. Negative control no. 1 siRNA (AM4635), siRNAs against PP2A-Cα isoform (PPP2CA; ID s10959), PP2A-Cβ isoform (PPP2CB; ID s10962), PP2A-Aα isoform (PPP2R1A; ID s10964), PP2A-Aβ isoform (PPP2R1B; ID s10968), PP1-Cα isoform (PPP1CA; ID s10931), PP1-Cβ isoform (PPP1CB; ID s10934), PP1-Cγ isoform (PPP1CC; ID s720), PP4-C (PPP4C; ID s10999), PP6-C (PPP6C; ID s11016), and ATM (ID s530445) were used.
Colony formation assay
Log-phase cells were treated with vehicle, or with increasing concentrations of phendione in the absence or presence of 5 µM ATM inhibitor for 3 h. After treatment, a certain number of viable cells were plated into 100-mm culture dishes to yield 50–200 surviving colonies. Colonies were grown for 10–14 d, after which they were fixed with methanol and stained with 1% crystal violet. The number of colonies was normalized to the number of cells plated to calculate the surviving fraction. Each experiment was performed three times.
Detection of mitotically quiescent cells in vitro
Cells were labeled with Vybrant DiD cell-labeling solution (Thermo Fisher Scientific V-22887) for 20 min according to manufacturer's instructions and then treated with 0.2 µM phendione, 2.5 µM cisplatin, and 5 µM etoposide for 3 h. DMSO (vehicle) was used as control. Cells were maintained in growing medium after drug treatment and kept in incubator following routine cell culture protocols. On day 14 after drug treatment, the cells were pulse-chased with bromodeoxyuridine (BrdU) and immunofluorescent staining of incorporated BrdU was performed using FITC BrdU flow kit (BD BD Biosciences 559619) following manufacturer's protocol. Retention of Vybrant DiD cell-labeling dye and BrdU incorporation were measured using CytoFLEX LX flow cytometer (Beckman Coulter Life Sciences). Data were analyzed with FlowJo software.
Annexin V apoptosis flow cytometry
After trypsinized and washed with PBS, the cells were stained with Annexin V apoptosis detection kit according to the manufacturer's protocol (BD BD Biosciences 556547 and 559763), and then analyzed with CytoFLEX LX flow cytometer (Beckman Coulter Life Sciences). Data were processed with FlowJo software.
Intracellular zinc measurement
At the end of drug treatment, the cells were trypsinized and resuspended in Hank's balanced salt solution (HBSS). A final concentration of 20 µM zinquin ethyl ester (Cayman Chemical 15133) was added into cell suspension to stain Zn2+ in live cells. The cell suspension was incubated for 30 min at 37°C before flow cytometry analysis. Unstained cells were used as negative control.
ROS production measurement
Total reactive oxygen species (ROS) production was measured with ROS assay kit (Thermo Fisher Scientific 88-5930-74). In brief, the cell suspension was pretreated with ROS stain for 30 min, followed by 3 h of drug treatment. Fluorescent intensity was measured by flow cytometry using FITC channel. Hydroxyl peroxide (H2O2; 200 µM) was used as positive control.
In vivo tumor models (PDX)
Animals were housed under pathogen-free conditions. All procedures involving animals (NMRI nude, female, 4 wk old) were performed in accordance with the guidelines of the Catholic University of Leuven (KU Leuven) Animal Care and Use Ethical Committee (P147/2012). Mel018 and Mel006 PDX models were derived from two different metastatic melanoma lesions, carrying the NRASQ61R and BRAFV600E mutations, respectively. Written informed consent was obtained from both patients and all procedures involving human samples were approved by the UZ Leuven/KU Leuven Medical Ethical Committee (ML8713/S54185).
In order to test the efficacy of phendione on tumor growth inhibition, mice were implanted subcutaneously with patient-derived melanoma tumors harboring NRASQ61R (Mel018) or BRAFV600E (Mel006) mutation. Once tumors reached 200–300 mm3, mice were randomly divided into groups. For treatment with phendione only, Mel018 and Mel006 mice were treated with vehicle (DMSO) or phendione at 5 mg/kg every 2 d intraperitoneally (i.p.). For the combined treatment with the BRAF inhibitor, cohorts of Mel006 were treated with the BRAF inhibitor dabrafenib at 30 mg/kg, or vehicle administered daily by oral gavage, in addition to either vehicle or phendione at a concentration of 5 mg/kg, administered i.p. every 2 d. Tumor size was measured every 2 d and tumor volume was calculated using the formula l × w × h·π/6 (length × width × height × π/6). No specific randomization method was used. According to animal welfare guidelines, mice have to be sacrificed when tumors reach a volume of 2 × 103 mm3 or when their body weight decreases >20% from the initial weight. Mice used in this work did not reach or overcome these limits. The investigators were blinded for the evaluation of the results.
At end point of the experiment, tumor tissue was collected for paraffin embedding, and then 5-µm tissue sections were prepared. Immunohistochemistry staining was performed with antibodies against phospho-ATM (S1981), phospho-CHK2 (T68), and γH2AX using IHC kit (BioVision, Inc. K405) according to manufacturer's instructions.
Data availability
All data are available here or in the Supplemental Material. Additional data that support the findings of this study are available on request.
Supplementary Material
Acknowledgments
We thank the Shiekhattar laboratory for constructive discussions and suggestions for experimental design. We also thank Professor G. Ghanem (LOCE-Institut) and J. Bordet (Université Libre de Bruxelles) for providing us with the MM lines, and the patients and their families for consenting to establish the PDX models. Marguerite Stas and Veerle Boecxstaens provided samples for the PDX models. PDX experiments were conducted by the Leuven PDX platform (TRACE) with the assistance of Eleonora Leucci, Lara Rizzotto, Ellen Gommé, and Kimberly Crevits. David Nittner and Enrico Radaelli provided histopathology and immunohistochemistry support. R.V. received a research fellowship from the FWO (1S08316N). This work was supported by funding from the University of Miami Miller School of Medicine, Sylvester Comprehensive Cancer Center, and grants R01 GM078455, R01 GM105754, and DP1 CA228041 from the National Institutes of Health to R.S. Research reported here was also supported by the National Cancer Institute of the National Institutes of Health under award number P30CA240139. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions: J.Y. and R.S. conceived and designed the overall project. J.-C.M. designed the in vivo experiments. R.V. performed the in vivo experiments. J.Y. performed the phosphatase profiling, comet assay, cell viability assay, phospho-kinase array, and immunofluorescence. F.L. performed cell viability and FACS analyses. O.L. performed immunohistochemistry. H.G.D.S. performed protein modeling and molecular docking. G.G. acquired immunofluorescence images. M.G.V., F.B., and E.B. assisted with in vitro cell culture. E.B assisted with manuscript editing. R.S. wrote the manuscript with critical feedback from all coauthors. S.D.N. reviewed the data and helped write the manuscript. L.S. prepared the figures and edited the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.333864.119.
Freely available online through the Genes & Development Open Access option.
References
- Cancer Genome Atlas Network. 2015. Genomic classification of cutaneous melanoma. Cell 161: 1681–1696. 10.1016/j.cell.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho US, Xu W. 2007. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445: 53–57. 10.1038/nature05351 [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. 2002. Mutations of the BRAF gene in human cancer. Nature 417: 949–954. 10.1038/nature00766 [DOI] [PubMed] [Google Scholar]
- Deegan C, Coyle B, McCann M, Devereux M, Egan DA. 2006. In vitro anti-tumour effect of 1,10-phenanthroline-5,6-dione (phendione), [Cu(phendione)3](ClO4)2.4H2O and [Ag(phendione)2]ClO4 using human epithelial cell lines. Chem Biol Interact 164: 115–125. 10.1016/j.cbi.2006.08.025 [DOI] [PubMed] [Google Scholar]
- Devitt B, Liu W, Salemi R, Wolfe R, Kelly J, Tzen CY, Dobrovic A, McArthur G. 2011. Clinical outcome and pathological features associated with NRAS mutation in cutaneous melanoma. Pigment Cell Melanoma Res 24: 666–672. 10.1111/j.1755-148X.2011.00873.x [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. 2007. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res 35: W522–W525. 10.1093/nar/gkm276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman AK, Monteiro AN. 2010. Phosphatases in the cellular response to DNA damage. Cell Commun Signal 8: 27 10.1186/1478-811X-8-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XL, Zhang M, Tang YL, Liang XH. 2017. Cancer cell dormancy: mechanisms and implications of cancer recurrence and metastasis. Onco Targets Ther 10: 5219–5228. 10.2147/OTT.S140854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharwan H, Groninger H. 2016. Kinase inhibitors and monoclonal antibodies in oncology: clinical implications. Nat Rev Clin Oncol 13: 209–227. 10.1038/nrclinonc.2015.213 [DOI] [PubMed] [Google Scholar]
- Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, Rutkowski P, Blank CU, Miller WH, Kaempgen E, et al. 2012. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet 380: 358–365. 10.1016/S0140-6736(12)60868-X [DOI] [PubMed] [Google Scholar]
- Hinsen K. 2000. The molecular modeling toolkit: a new approach to molecular simulations. J Comput Chem 21: 79–85. [DOI] [Google Scholar]
- Ivana Scovassi A, Diederich M. 2004. Modulation of poly(ADP-ribosylation) in apoptotic cells. Biochem Pharmacol 68: 1041–1047. 10.1016/j.bcp.2004.04.023 [DOI] [PubMed] [Google Scholar]
- Jakob JA, Bassett RL, Ng CS, Curry JL, Joseph RW, Alvarado GC, Rohlfs ML, Richard J, Gershenwald JE, Kim KB, et al. 2012. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer 118: 4014–4023. 10.1002/cncr.26724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila MR, Li SP, Westermarck J. 2008. Phosphatase-mediated crosstalk between MAPK signaling pathways in the regulation of cell survival. FASEB J 22: 954–965. 10.1096/fj.06-7859rev [DOI] [PubMed] [Google Scholar]
- Kemper K, de Goeje PL, Peeper DS, van Amerongen R. 2014. Phenotype switching: tumor cell plasticity as a resistance mechanism and target for therapy. Cancer Res 74: 5937–5941. 10.1158/0008-5472.CAN-14-1174 [DOI] [PubMed] [Google Scholar]
- Kong X, Kuilman T, Shahrabi A, Boshuizen J, Kemper K, Song JY, Niessen HWM, Rozeman EA, Geukes Foppen MH, Blank CU, et al. 2017. Cancer drug addiction is relayed by an ERK2-dependent phenotype switch. Nature 550: 270–274. 10.1038/nature24037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzosiak A, Sigurdardottir A, Luh L, Carrara M, Das I, Schneider K, Bertolotti A. 2018. Target-based discovery of an inhibitor of the regulatory phosphatase PPP1R15B. Cell 174: 1216–1228.e19. 10.1016/j.cell.2018.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guilloux V, Schmidtke P, Tuffery P. 2009. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics 10: 168 10.1186/1471-2105-10-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Kovach JS, Johnson F, Chiang J, Hodes R, Lonser R, Zhuang Z. 2009. Inhibition of serine/threonine phosphatase PP2A enhances cancer chemotherapy by blocking DNA damage induced defense mechanisms. Proc Natl Acad Sci 106: 11697–11702. 10.1073/pnas.0905930106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. 2003. RAS oncogenes: the first 30 years. Nat Rev Cancer 3: 459–465. 10.1038/nrc1097 [DOI] [PubMed] [Google Scholar]
- Manzano JL, Layos L, Bugés C, de Los Llanos Gil M, Vila L, Martínez-Balibrea E, Martínez-Cardús A. 2016. Resistant mechanisms to BRAF inhibitors in melanoma. Ann Transl Med 4: 237 10.21037/atm.2016.06.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, et al. 2007. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta 1773: 1263–1284. 10.1016/j.bbamcr.2006.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. 2002. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol 4: 556–564. 10.1038/ncb822 [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Chen PY, Dorans KJ, Chung KM, Bhutkar A, Hong E, Noll EM, Sprick MR, Trumpp A, Jacks T. 2017. Survival of pancreatic cancer cells lacking KRAS function. Nat Commun 8: 1090 10.1038/s41467-017-00942-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik MM. 2017. The importance of FLT3 mutational analysis in acute myeloid leukemia. Leuk Lymphoma 59: 2273–2286. 10.1080/10428194.2017.1399312 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Peyssonnaux C, Eychène A. 2001. The Raf/MEK/ERK pathway: new concepts of activation. Biol Cell 93: 53–62. 10.1016/S0248-4900(01)01125-X [DOI] [PubMed] [Google Scholar]
- Pivetta T, Trudu F, Valletta E, Isaia F, Castellano C, Demartin F, Tuveri R, Vascellari S, Pani A. 2014. Novel copper(II) complexes as new promising antitumour agents. A crystal structure of [Cu(1,10-phenanthroline-5,6-dione)2(OH2)(OClO3)](ClO4). J Inorg Biochem 141: 103–113. 10.1016/j.jinorgbio.2014.08.011 [DOI] [PubMed] [Google Scholar]
- Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. 2015. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res 43: W443–W447. 10.1093/nar/gkv315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyers C. 2004. Targeted cancer therapy. Nature 432: 294–297. 10.1038/nature03095 [DOI] [PubMed] [Google Scholar]
- Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, Beqiri M, Sproesser K, Brafford PA, Xiao M, et al. 2017. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546: 431–435. 10.1038/nature22794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling T, Irwin JJ. 2015. ZINC 15-ligand discovery for everyone. J Chem Inf Model 55: 2324–2337. 10.1021/acs.jcim.5b00559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz B, Lomova A, Le A, Hugo W, Kong X, Ten Hoeve J, Friedman M, Shi H, Moriceau G, Song C, et al. 2016. JUN dependency in distinct early and late BRAF inhibition adaptation states of melanoma. Cell Discov 2: 16028 10.1038/celldisc.2016.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek RA, Suchard SJ, Steelman GB, Knappenberger KS, Sygowski LA, Veale CA, Chapdelaine MJ. 2001. Potent reversible inhibitors of the protein tyrosine phosphatase CD45. J Med Chem 44: 1777–1793. 10.1021/jm000447i [DOI] [PubMed] [Google Scholar]
- Verfaillie A, Imrichova H, Atak ZK, Dewaele M, Rambow F, Hulselmans G, Christiaens V, Svetlichnyy D, Luciani F, Van den Mooter L, et al. 2015. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat Commun 6: 6683 10.1038/ncomms7683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JM, Cieplak P, Kollman PA. 2000. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem 21: 1049–1074. [DOI] [Google Scholar]
- Yeh AC, Ramaswamy S. 2015. Mechanisms of cancer cell dormancy–another hallmark of cancer? Cancer Res 75: 5014–5022. 10.1158/0008-5472.CAN-15-1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Lai F, Beckedorff F, Zhang A, Pastori C, Shiekhattar R. 2017. Integrator orchestrates RAS/ERK1/2 signaling transcriptional programs. Genes Dev 31: 1809–1820. 10.1101/gad.301697.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available here or in the Supplemental Material. Additional data that support the findings of this study are available on request.