Here, Beebe et al. investigated the molecular mechanisms underlying the link between metabolism and development. They show that the Drosophila estrogen-related receptor (dERR) directs a transcriptional switch in mid-pupae that promotes glucose oxidation and lipogenesis in young adults.
Keywords: glycolysis, gene regulation, lipogenesis, nuclear receptors, metabolism, development
Abstract
Metabolism and development must be closely coupled to meet the changing physiological needs of each stage in the life cycle. The molecular mechanisms that link these pathways, however, remain poorly understood. Here we show that the Drosophila estrogen-related receptor (dERR) directs a transcriptional switch in mid-pupae that promotes glucose oxidation and lipogenesis in young adults. dERR mutant adults are viable but display reduced locomotor activity, susceptibility to starvation, elevated glucose, and an almost complete lack of stored triglycerides. Molecular profiling by RNA-seq, ChIP-seq, and metabolomics revealed that glycolytic and pentose phosphate pathway genes are induced by dERR, and their reduced expression in mutants is accompanied by elevated glycolytic intermediates, reduced TCA cycle intermediates, and reduced levels of long chain fatty acids. Unexpectedly, we found that the central pathways of energy metabolism, including glycolysis, the tricarboxylic acid cycle, and electron transport chain, are coordinately induced at the transcriptional level in mid-pupae and maintained into adulthood, and this response is partially dependent on dERR, leading to the metabolic defects observed in mutants. Our data support the model that dERR contributes to a transcriptional switch during pupal development that establishes the metabolic state of the adult fly.
Studies over the past century have demonstrated that metabolism and development are closely intertwined. For example, it has long been known that pyruvate and not glucose is the primary energy source for the early mouse zygote (Clough 1985). This state changes at the postimplantation stage when metabolism is characterized by high rates of aerobic glycolysis and pentose phosphate pathway activity. These signatures have been classically associated with tumor metabolism and are ideally placed to support the growth demands of early mammalian development (Vander Heiden and DeBerardinis 2017). Over the past decade, these correlative observations have been confirmed and extended at the molecular level providing initial insights into the mechanisms that couple development and metabolism (e.g., Miyazawa et al. 2017; Nagaraj et al. 2017). Most progress has been made in characterizing elegant cell type-specific switches in metabolic state, demonstrating that proliferating cells are often highly glycolytic, with low levels of mitochondrial activity, while differentiated cells display increased oxidative metabolism (Krejci and Tennessen 2017; Gándara and Wappner 2018; Miyazawa and Aulehla 2018). In contrast, relatively few studies have addressed how changes in systemic physiology are coupled with different stages of development to meet the changing needs of each stage in the life cycle.
Here we focus on the transcriptional regulation of metabolism by ERR in Drosophila. The ERRs comprise a subfamily of orphan nuclear receptors that act as transcriptional activators (Huss et al. 2015; Misra et al. 2017; Xia et al. 2019). There are three mammalian ERR paralogs: ERRα, ERRβ, and ERRγ. ERRβ functions as a pluripotency factor in stem cells and is required for the growth of extraembryonic tissues as well as neuronal regulation of feeding behavior and systemic energy balance (Huss et al. 2015). In contrast, extensive studies have defined metabolic functions for ERRα and ERRγ in multiple tissues, including the liver, muscle, and adipose tissue (Huss et al. 2015; Misra et al. 2017; Xia et al. 2019). These have revealed central roles for these receptors in oxidative energy metabolism and mitochondrial function, in part through their close association with the PGC-1α coactivator. Genetic studies of ERR family members, however, have been complicated by their functional redundancy. ERRα and ERRγ are expressed widely in overlapping patterns in the mouse (Bookout et al. 2006; Huss et al. 2015) and ERRγ expression is elevated in the absence of ERRα, indicating compensatory control (Liu et al. 2005; Dufour et al. 2007). A recent study in brown adipose tissue demonstrated the majority of the adaptive response to adrenergic stimulation is dependent on the combined functions of these two ERR paralogs (Brown et al. 2018). Similarly, ERRβ and ERRγ are expressed in overlapping patterns in the mouse hindbrain and ERRγ levels are elevated in ERRβ mutant neurons with downstream effects on target gene expression (Byerly et al. 2013). A current goal for the field is to dissect these redundant ERR activities through the use of double mutants.
Our studies are focused on functional analysis of the single ERR ortholog in Drosophila, dERR, with the goal of better understanding its ancestral functions in the absence of genetic redundancy. Our initial work showed that dERR null mutants go through early development normally but die as mid-second instar larvae with hyperglycemia and hypolipidemia (Tennessen et al. 2011). GC/MS metabolomics revealed that wild-type larvae normally display aerobic glycolytic metabolism similar to cancer cells, with abundant lactate and 2-hydroxyglutarate, and this progrowth metabolic profile is dependent on dERR function (Tennessen et al. 2011; Li et al. 2017). Interestingly, dERR establishes this metabolic state midway through embryogenesis when it coordinately induces genes that encode enzymes in glycolysis, the pentose phosphate pathway (PPP), and lactate dehydrogenase (Ldh) (Tennessen et al. 2011, 2014b). These observations support the model that dERR links metabolism with developmental progression by establishing a metabolic state in late embryos that is optimized for the rapid and massive growth that occurs during larval development.
Here we extend our functional analysis of dERR to the adult fly when there is no developmental growth. Using a conditional allele, we show that, similar to dERR mutant larvae, mutant adults display reduced expression of genes in the glycolytic and pentose phosphate pathways, many of which appear to be directly induced by the receptor. In contrast to larvae, however, this results in reduced levels of citrate and downstream lipogenesis, leading to an almost complete absence of stored triglycerides in the adult fat body. Transcriptional profiling of staged control and dERR mutant pupae and adults revealed that this metabolic state is normally induced during mid-pupal development and the glycolytic transcriptional switch is disrupted in dERR mutants. We conclude that dERR acts during pupal stages to contribute to the physiological state of the adult fly by up-regulating metabolic pathways that support glucose oxidation and lipogenesis. This study provides insights into the molecular mechanisms that allow the animal to transition from an immobile pupa into a highly motile and reproductively active adult fly.
Results
dERR is required for normal adult locomotor activity, longevity, and starvation resistance
We engineered a conditional dERR mutant allele using CRISPR/Cas9 in order to bypass its early essential role in larval development and study its functions in adults (Fig. 1A). Two FRTs were inserted into the endogenous dERR locus, flanking the majority of the protein-coding region, along with a Pax3-DsRed reporter at the 3′ end to identify recombinants (Supplemental Fig. S1). This conditional allele was carried in combination with the dERR2 deletion allele and animals with and without a hsFLP transgene were subjected to sequential heat treatments as late third instar larvae and prepupae. This results in a hsFLP-dependent loss of dERR genomic sequences between the FRTs as well as greatly reduced levels of dERR mRNA in adults (Supplemental Fig. S2A–C). We conclude that FRT-mediated excision of dERR coding sequences results in a strong loss-of-function for dERR. From here on, we refer to these animals as dERR mutants, with controls representing animals that carry the same dERR allelic combination without the hsFLP transgene.
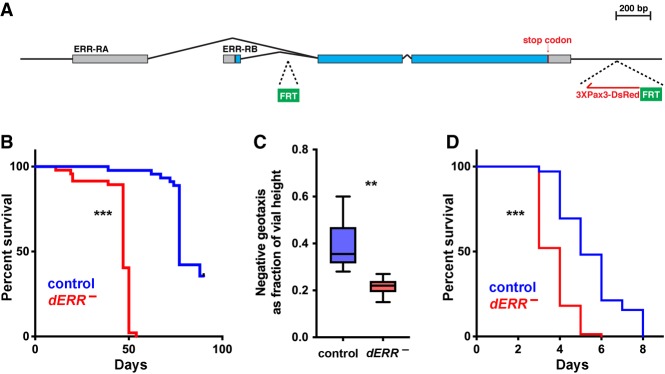
Figure 1.
dERR is required for maximal life span, starvation resistance, and adult locomotor activity. (A) The genomic locus for the dERR conditional allele is depicted with the two FRT insertion sites (green boxes) and the 3XPax3-dsRed marker gene (red). The two isoforms for dERR are shown (ERR-RA and ERR-RB), including protein-coding sequences (blue boxes) and UTRs (gray boxes). (B,D) Life span was scored for controls (blue) and dERR mutants (red) on a normal diet (B) or following water-only starvation initiated on day 6 of adulthood (D). Median life spans: controls: normal diet, 77 d (n = 76); water, 5 d (n = 141); dERR mutants: normal diet, 47 d (n = 140); water, 4 d (n = 144). (C) Locomotor activity was measured as a fraction of total vial height achieved for controls (blue) and dERR mutants (red) using the RING assay. Animals were fed a normal diet and assayed on day 6 of adulthood. n = 8 independent samples with 10 animals per sample. (***) P ≤ 0.001; (**) P ≤ 0.01.
dERR mutant adults eclose at a normal rate indicating that dERR exerts no essential functions during metamorphosis (Supplemental Fig. S2D). Mutant adults, however, display a noticeable decrease in motility compared with controls (Fig. 1C). In addition, dERR mutants have a significantly shortened life span, with a median life span of 47 d compared with 77 d in controls (Fig. 1B). Interestingly, dERR mutants are also sensitive to humid starvation, with mutant animals dying at almost twice the rate as controls (Fig. 1D).
dERR supports adult metabolic homeostasis
The decreased locomotor activity and sensitivity to starvation in dERR mutants raises the possibility that dERR functions to maintain energy homeostasis in young adults. To test this hypothesis, we assayed the major forms of available and stored energy in controls and mutants. Similar to our earlier study of dERR mutant larvae, we found that glucose levels are increased while glycogen levels are not significantly affected, although they appear to be more variable in dERR mutants relative to controls (Fig. 2A,B; Tennessen et al. 2011). Also similar to dERR mutant larvae, triglyceride levels are reduced in mutant adults; however, this effect is much more dramatic, in some cases approaching the lower detection limit of the assay (Fig. 2C). Interestingly, protein levels are also reduced in dERR mutants, suggesting that this may reflect a compensatory response to the reduced levels of stored energy (Fig. 2D). Consistent with dERR mutants undergoing sustained energy depletion, ATP levels are lower in these animals relative to controls (Supplemental Fig. S2E). Staining with the lipophilic dye Oil Red O confirmed our triglyceride assays, revealing greatly reduced lipid stores in dissected abdomens as well as in isolated fat tissue (Fig. 2E–H). Taken together, we conclude that dERR is required for adult metabolic homeostasis, with a major role for this receptor in maintaining appropriate levels of stored triglycerides.
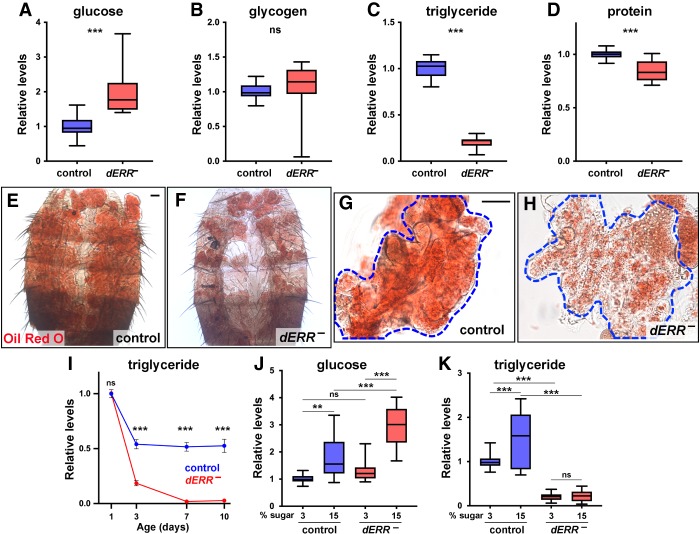
Figure 2.
dERR is required for adult metabolic homeostasis and establishing adult triglyceride stores. (A–D) Whole animal homogenates from controls (blue) and dERR mutants (red) at day 6 of adulthood were analyzed for glucose (A), glycogen (B), triglyceride (C), and protein (D) levels. Data is normalized to the controls. Animals were fed a normal diet; n = 17–23 independent samples with five animals per sample. (E–H) Bright-field images of adult fat bodies from six day-old controls (E,G) and dERR mutants (F,H) stained with Oil Red O to detect neutral lipids. Representative images show the dorsal fat body attached to the abdominal cuticle (E,F), or dissected dorsal fat bodies (G,H). n = 12 animals analyzed. Scale bar, 50 µm. (I) Triglyceride levels were measured from whole animal homogenates of controls (blue) and dERR mutants (red) collected at 1, 3, 7, and 10 d of adulthood. Animals were fed a normal diet and all data is normalized to 1-d control levels. n = 6 independent samples with five animals per sample. Points indicate mean ± SEM. (J,K) Whole animal homogenates from 6-d-old controls (blue) and dERR mutants (red) were analyzed for glucose and triglyceride levels following exposure to a 3% or 15% sugar diet from days 2 to 6 of adulthood. Data were normalized to controls on a 3% sugar diet. n = 18 independent samples with five animals per sample. (***) P ≤ 0.001; (**) P ≤ 0.01; (ns) P > 0.05.
To gain a better understanding of when dERR mutants become hypolipidemic, we assayed triglyceride levels in staged animals during the first 10 days of adulthood (Fig. 2I). As expected, controls display an approximately ∼twofold drop in triglyceride levels by day 3 and then remain flat for the rest of the time course (Fig. 2I, blue line). This is consistent with earlier studies showing that flies normally eclose with significant amounts of persisting larval lipids that are depleted during the first three days of adulthood followed by de novo lipogenesis that maintains adult lipid stores (Aguila et al. 2007; Storelli et al. 2019). In contrast, dERR mutants display an approximately fivefold drop in triglycerides during the first 3 d of adulthood followed by almost undetectable lipid levels at days 7 and 10 (Fig. 2I, red line). Interestingly, this rapid rate of lipid depletion is similar to the kinetics of triglyceride loss seen in control adults starved at eclosion, suggesting that this arises from a block in adult lipogenesis (Storelli et al. 2019). In addition, triglyceride levels are normal in newly eclosed dERR mutants indicating that the observed defects in lipid metabolism are adult-specific. Similar adult-specific roles for dERR are evident in protein and glucose homeostasis, with a gradual onset of the mutant phenotypes during the first week of adulthood (Supplemental Fig. S2F,G).
The rapid decline in triglyceride levels seen in dERR mutants raises the possibility that dERR is required for de novo lipogenesis from dietary sugar (Musselman et al. 2011). To test this hypothesis, we assayed glucose and triglyceride levels in controls and dERR mutants fed a low (3%) and high (15%) sugar diet. As expected, control animals displayed diet-dependent increases in both glucose and triglycerides (Fig. 2J,K; Musselman et al. 2011). In contrast, dERR mutants showed a greater susceptibility to diet-induced hyperglycemia and no change in triglyceride levels in response to high dietary sugar (Fig. 2J,K). Taken together, these findings indicate that dERR mutants are resistant to diet-induced obesity and unable to establish triglyceride stores from excess dietary carbohydrate.
dERR regulates glycolytic gene expression in adults
To gain a molecular understanding of dERR function in adults, we performed RNA-seq analysis on controls and dERR mutants at day 6 of adulthood. RNA-seq analysis revealed a significant change in 1004 transcripts, with 748 transcripts expressed at reduced levels and 256 transcripts at elevated levels (Supplemental Table S1). We focused our attention on the down-regulated gene set since these are likely to represent direct targets of dERR due to its role as a transcriptional activator (Huss et al. 2015; Misra et al. 2017; Xia et al. 2019). GO analysis of these genes revealed significant enrichment of metabolic processes, with carbohydrate metabolism as a top category (Fig. 3A; Supplemental Tables S1, S2). Consistent with this, almost every gene encoding an enzyme in glycolysis is expressed at reduced levels in dERR mutants along with many genes in the PPP (Fig. 3B). RT-qPCR was used to validate the reduced expression of key targets in the glycolytic and pentose phosphate pathways, as well as the gene encoding ATP citrate lyase, which converts cytoplasmic citrate into acetyl CoA—the first step in lipogenesis (Fig. 3C). We also confirmed that Gpdh is expressed at reduced levels in dERR mutants (Fig. 3C). The enzyme encoded by this gene converts the glycolytic intermediate DHAP into glycerol 3-phosphate, which is an important precursor for triglyceride synthesis. Genes involved in both glycogen synthesis and glycogenolysis are also reduced in dERR mutants (Fig. 3C; Supplemental Table S1). These include glycogen synthase and UDP-glucose pyrophosphorylase (UGP) in glycogen synthesis and glycogen phosphorylase (GlyP) and phosphoglucomutase (Pgm) in glycogenolysis, all of which play central roles in these pathways. These divergent effects on glycogen metabolism may contribute to the increased variability in glycogen levels seen in dERR mutants (Fig. 2B).
Figure 3.
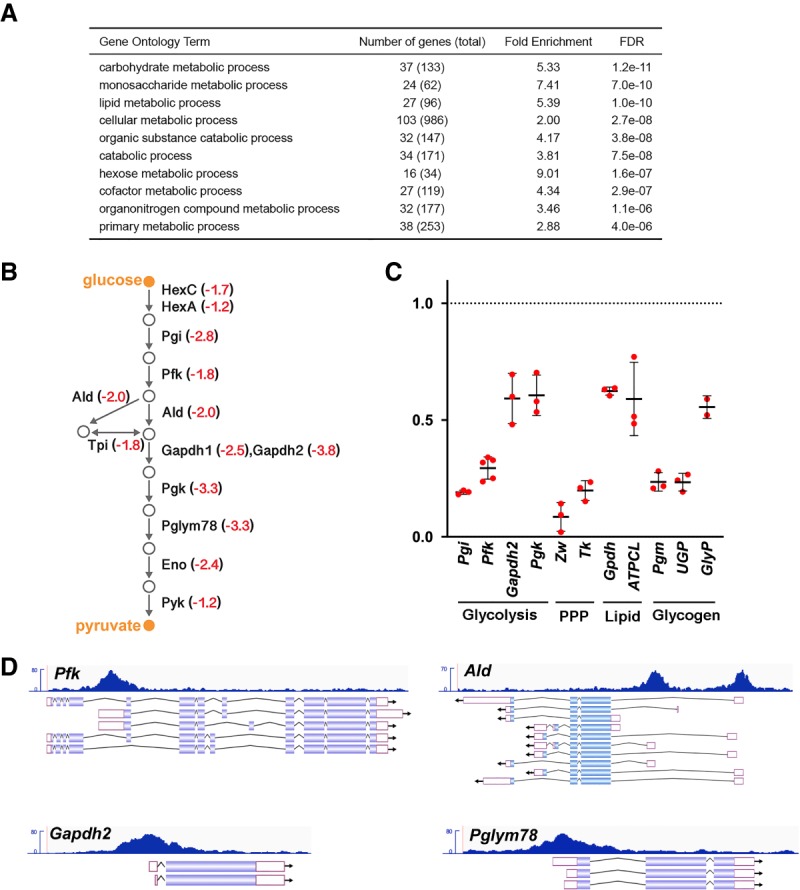
dERR directly maintains glycolytic gene expression in adults. (A) Gene ontology (GO) enrichment analysis of genes expressed at a significantly reduced level in dERR mutants relative to controls as determined by RNA-seq. GO term analysis was performed using the Biological Processes designation in PANTHER GO-slim, and categories are listed in order by FDR. (B) A schematic is shown depicting the glycolytic pathway in Drosophila with the average fold-change in transcript levels in dERR mutants as determined by RNA-seq (red). (C) RT-qPCR measurement of mRNA levels corresponding to key genes in glycolysis, the pentose phosphate pathway (PPP), lipid metabolism, and glycogen metabolism in dERR mutants relative to controls. mRNA values were normalized to rp32 expression and fold-change values were calculated using the comparative CT method. n = 2–5 independent samples with 10–15 animals per sample. Lines indicate mean ± SD. (D) ChIP-seq analysis performed on adult animals for dERR-3xFlag genomic binding shows direct association with genes involved in glycolysis. Data tracks display fragment pileup from MACS2 (Y-axis) at four significantly bound dERR target loci. The read accumulation histogram (dark blue) is aligned to the isoforms of each gene, with protein coding sequences (filled boxes), introns (lines), and UTRs (open boxes) shown.
To identify potential direct transcriptional targets of dERR, we performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq). Analysis of reads revealed 61 genes as high-confidence targets by selecting for dERR binding sites within 1 kb of the promoter and a Q-value < 0.05 (Supplemental Table S3). Interestingly, 11 of the 12 genes that encode glycolytic enzymes had significant ChIP-seq peaks within 1 kb of the promoter defining them as possible direct targets of the receptor. Similar results were seen in a ChIP-seq analysis of dERR binding sites in S2 tissue culture cells, confirming that these represent targets for dERR regulation (Kovalenko et al. 2019). Of these, Pfk, Pgk, Tpi, Pglym78, Gapdh2, and Ald are within the top 15 genes in the data set, representing a higher frequency of dERR occupancy relative to other target loci (Fig. 3D; Supplemental Table S3). In addition, zwischenferment, which encodes glucose 6-phosphate dehydrogenase, the rate-limiting step in the PPP, as well as transketolase, are possible direct targets of dERR (Supplemental Tables S3, S4). In addition to its role in supporting nucleotide biosynthesis, this pathway is a critical source of NADPH for lipogenesis. Taken together, these studies define a central role for dERR in supporting glycolytic and PPP gene expression in young adults.
GC/MS metabolomic analysis of dERR mutants confirmed and extended our understanding of its roles in carbohydrate metabolism and lipogenesis. Consistent with a reduction in glycolysis, dERR mutants display increased levels of glycolytic intermediates, including glucose-6-phosphate, fructose-6-phosphate, 2-phosphoglycerate, and 3-phosphoglycerate, along with accumulations of glucose, fructose, and mannitol (Fig. 4). In addition, pyruvate levels are reduced in dERR mutants, along with the six tricarboxylic acid (TCA) cycle intermediates that we detected: citrate, isocitrate, α-ketoglutarate, succinate, fumarate, and malate (Fig. 4). The amino acids alanine and proline are also significantly reduced, likely reflecting their depletion by anaplerosis in an effort to replenish the TCA cycle. These results are consistent with the reduced levels of total protein in dERR mutants (Fig. 2D). In parallel, GC/MS analysis of fatty acid methyl esters (FAMEs) revealed reduced levels of the major long chain fatty acids in dERR mutants: lauric acid, myrisitic acid, palmitic acid, and oleic acid (Fig. 4). Taken together with the reduced levels of citrate, which is required for the first step in lipogenesis, and reduced expression of ATP citrate lyase, our metabolomic analysis supports the model that the block in glycolysis in dERR mutants leads to reduced de novo lipogenesis, resulting in the dramatic effects we see on total stored lipids in adults.
Figure 4.
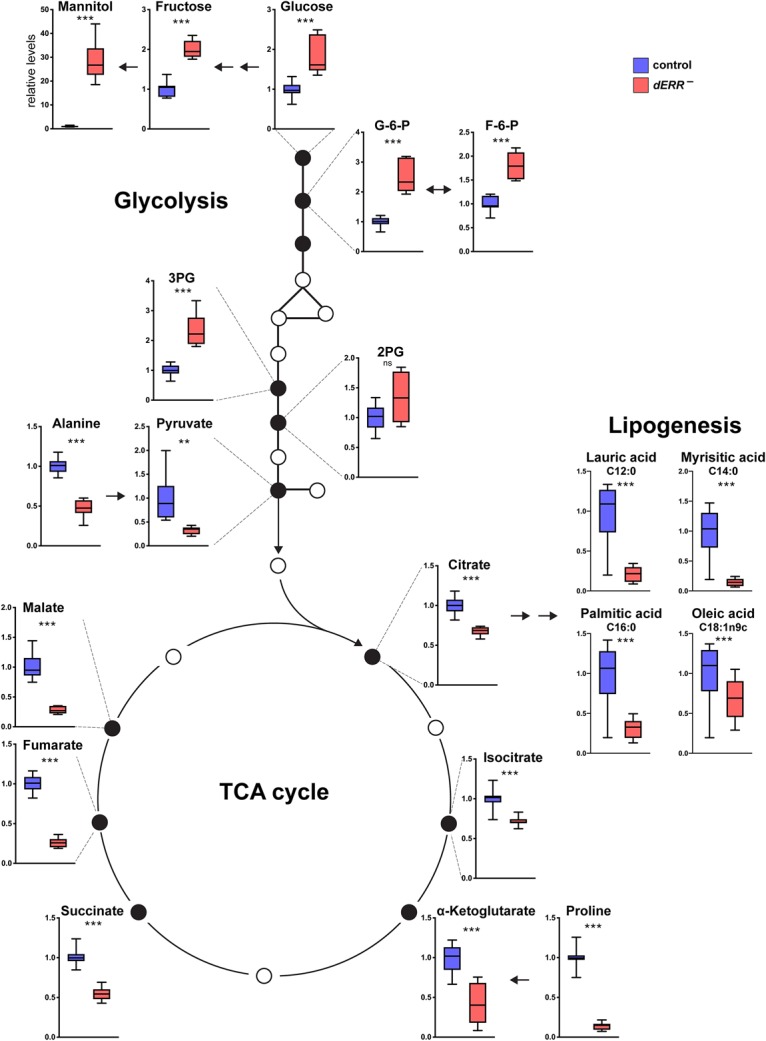
dERR mutants display reduced glycolytic oxidation. GC/MS metabolomic profiling of 6-d-old controls (blue) and dERR mutants (red) raised to adulthood on a normal diet (9% sugar) and transferred to a 15% sugar diet at eclosion. n = 8–12 independent samples with 20 animals per sample. Metabolite data are from three independent GC/MS experiments, with the exception of the fatty acid measurements, which were performed in one GC/MS experiment. Values were normalized to control levels for each metabolite. (***) P ≤ 0.001; (**) P ≤ 0.01; (ns) P > 0.05.
dERR is required in the adult fat body to maintain lipid stores
The fat body and intestine play central roles in Drosophila lipogenesis (Palm et al. 2012; Musselman et al. 2013). To determine whether dERR acts in these tissues to maintain normal levels of stored lipid, we used the GAL4/UAS system to direct RNAi against dERR in the adult fat body or intestine and examined the effects on whole-body glucose and triglyceride levels (Fig. 5A,B). This study showed that fat body-specific loss of dERR function recapitulates the major phenotypes seen in our mutant studies, including hyperglycemia, hypolipidemia, and reduced levels of neutral lipids as demonstrated by Oil Red O staining (Fig. 5A–D). Similar effects were observed using adult fat body-specific RNAi against Pfk, which encodes the rate-limiting enzyme in glycolysis and appears to be a direct target of dERR regulation (Fig. 5A,B; Supplemental Table S4). In contrast, adult-specific disruption of dERR or Pfk function in the intestine had no significant effects on glucose or triglyceride levels (Fig. 5A,B). We conclude that dERR and glycolysis are required in the adult fat body to maintain normal systemic levels of glucose and triglycerides.
Figure 5.
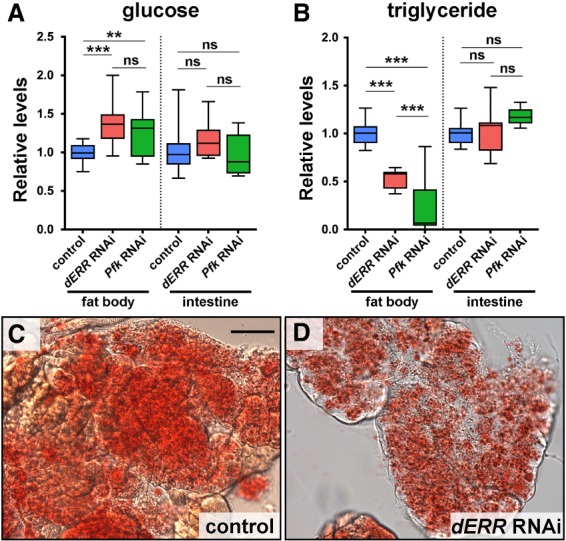
dERR functions in the adult fat body to maintain lipid stores. (A,B) Whole-animal homogenates were prepared from controls (blue) or flies with adult-specific RNAi targeting dERR (red) or Pfk (green) in either the fat body (cg-GAL4, Tub-GAL80ts) or intestine (Myo1a-GAL4, Tub-GAL80ts). These samples were analyzed for glucose (A) and triglyceride (B) levels. Animals were maintained for the first 11 d of development at 18°C and subsequently transferred for the remainder of the experiment to 29°C. Data were normalized to the controls. Animals were fed a normal diet; n = 9–29 independent samples with five animals per sample. (***) P ≤ 0.001; (**) P ≤ 0.01; (ns) P > 0.05. (C,D) Bright-field images of adult fat bodies stained with Oil Red O to detect neutral lipids. Samples were collected from 6-d-old controls (C) or animals with adult-specific RNAi targeting dERR in the fat body (cg-GAL4, Tub-GAL80ts) (D). Scale bar, 50 µm.
dERR directs a transcriptional switch in glycolytic gene expression at the onset of adulthood
Our previous study of dERR function demonstrated that it triggers a global switch in gene expression midway through embryogenesis, establishing an aerobic glycolytic metabolic state that supports larval growth (Tennessen et al. 2011, 2014b). This glycolytic transcriptional program is subsequently repressed at the end of larval development, a few hours before metamorphosis (White et al. 1999). This raises the interesting possibility that dERR might act again, at the onset of adulthood, to direct the coordinated induction of glycolytic gene expression. Examination of modENCODE time course expression data in wild-type animals supports this hypothesis, revealing a coordinate increase in glycolytic transcripts in late pupae and maintained expression into adulthood (Supplemental Fig. S3; Graveley et al. 2011). To determine whether this switch is dERR dependent, we performed an RNA-seq time course analysis of controls and dERR mutants staged at 1-d intervals during metamorphosis, day 1 adults, and day 6 adults (Supplemental Table S5). Similar to the modENCODE data, control animals display coordinated glycolytic gene induction in 2- to 3-d-old pupae with an expression peak in day 1 adults, followed by decreased but sustained expression in day 6 adults (Fig. 6A, left panel). In contrast, dERR mutants display delayed and reduced glycolytic gene expression in late pupae and early adults, with lower expression at day 6 (Fig. 6A, right panel). Examining the temporal profiles of individual glycolytic genes confirms this observation and shows that Pyk is less affected, while Hex-C (encoding a homolog of glucokinase) is unaffected by a loss of dERR (Fig. 6B). We conclude that dERR is required in mid-pupae to direct a switch in metabolism that supports adult glycolysis.
Figure 6.
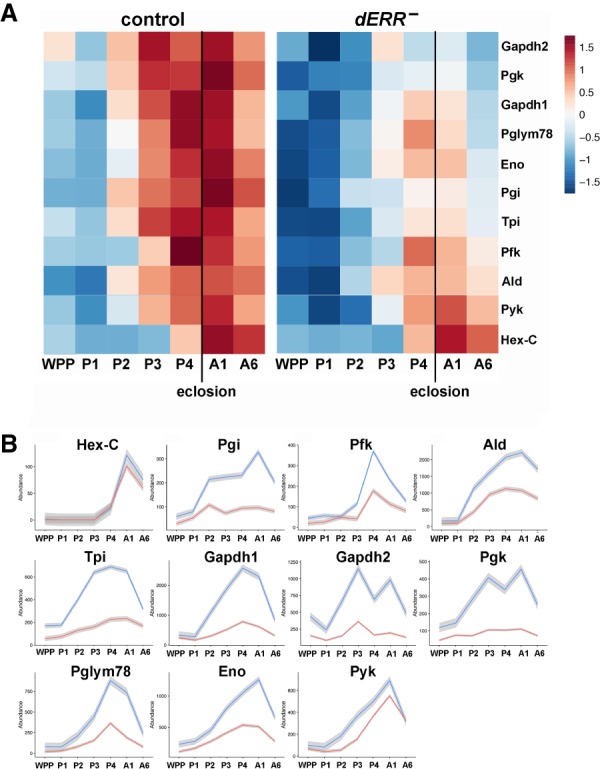
dERR is required for the induction of glycolytic gene expression in late pupae. (A) Heat maps of relative gene expression are depicted for transcripts encoding central enzymes in glycolysis from staged controls (left) and dERR mutants (right) using RNA-seq data. Genes are ordered vertically by median expression value over time. Eclosion is marked with a vertical line. (B) Expression plots through time are depicted for transcripts that encode glycolytic enzymes in controls (blue) and dERR mutants (red). Mean values at each time point are plotted to make the line, with the gray border representing the standard error bounds using a LOESS fitting. Expression levels are shown as transcripts per million (TPM) normalized expression values to account for gene size and total RNA in the sample. Animals were staged as: white prepupae (WPP), day 1 pupae (P1), day 2 pupae (P2), day 3 pupae (P3), day 4 pupae (P4), day 1 adult (A1), day 6 adult (A6). RNA-seq data was derived from triplicate samples with 15 animals per sample.
The onset of adulthood is accompanied by large-scale switches in metabolic gene expression
The broad induction of glycolytic gene expression in mid-pupae raises the possibility that other metabolic genes might be coordinately regulated at this stage in development. This would be similar to our earlier study of gene expression in staged embryos, which showed that the dERR-dependent glycolytic switch is part of a much wider transcriptional response during mid-embryogenesis (Tennessen et al. 2014b). As a first step toward determining whether a similar global transcriptional transition is occurring at the onset of adulthood, we used the Mfuzz package in R to perform a clustering analysis of metabolic genes (Supplemental Table S2) to identify those that display similar temporal expression profiles in staged pupae and early adults (Supplemental Table S5). We initially evaluated cluster assignments using multiple cluster values ranging from two to 16 and found that at values larger than four most of the clusters resemble each other in their overall expression profiles as well as in their gene identities. Accordingly, we focused our attention on four clusters that represent distinct patterns of expression at the onset of adulthood (Fig. 7A). Interestingly, the majority of metabolic genes (66%, 942 genes) had transcript expression profiles that peaked after mid-pupariation, similar to the temporal profiles of glycolytic gene induction (Fig. 7A,B, up 1 and up 2). As expected, these sets included genes involved in glycolysis, although genes involved in the PPP, pyruvate metabolism, and fructose and mannose metabolism are also in these up-regulated gene sets (Fig. 7B). Remarkably, most genes involved in the TCA cycle and electron transport are also up-regulated at the onset of adulthood, indicating that transcriptional induction of oxidative metabolism is coordinated with increased glycolysis at this stage in development (Fig. 7B; Supplemental Figs. S4, S5). The remaining 34% percent of metabolic genes (495 genes) displayed profiles of expression that peaked in either early pupae or mid-pupal stages (Fig. 7A, down 1 and down 2). These pupal down-regulated metabolic genes act in several metabolic pathways including 20-hydroxyecdysone metabolism, tRNA synthesis, and pyrimidine metabolism.
Figure 7.
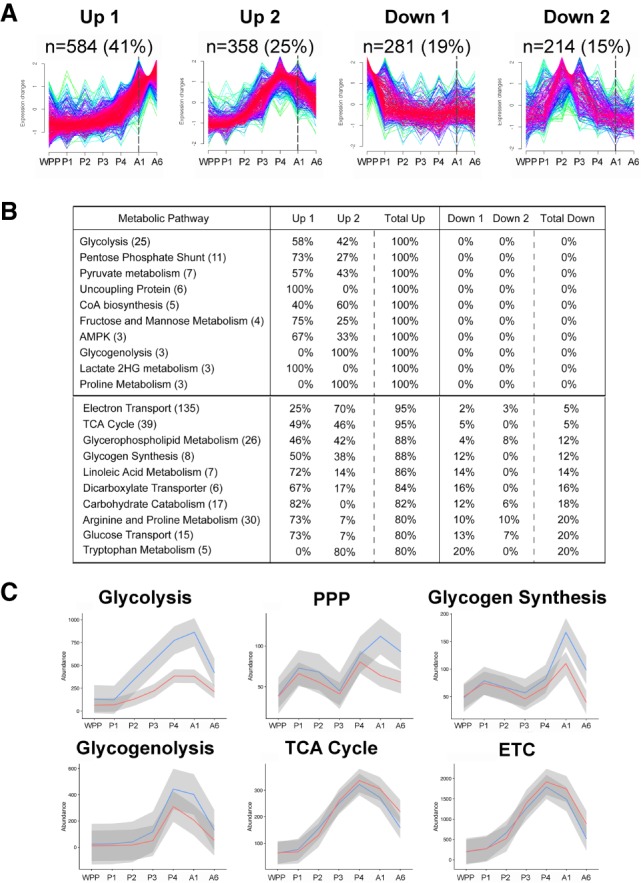
Coordinate transcriptional induction of metabolic pathways in late pupae. (A) Soft-clustering analysis was performed on transcripts from RNA-seq data for genes representing the major metabolic pathways in Drosophila using the Mfuzz package in R (Supplemental Table S2). FPKM normalized reads were used to account for gene size, with number of clusters set to four (up 1, up 2, down 1, and down 2). Temporal profiles with low membership value (yellow, green) and high membership value (red, purple) are depicted. The number of genes assigned to a given cluster is shown (n=) along with its percentage relative to the total set of metabolic genes analyzed. The first day of adulthood is marked with a dashed line. (B) Table of metabolic pathways with highest representation in the up clusters. Metabolic pathways are ordered vertically first by the percent total up, and then by the total number of genes in that pathway, shown in parentheses. The distribution of genes for each pathway in the four clusters is shown as a percentage of the total. Pathways in which every gene is in the up clusters (100%) are separated by a line from the top pathways where most genes are up (80%–95%). (C) Gene expression plots through time are shown for RNA-seq data from controls (blue) and dERR mutants (red) for six metabolic pathways: glycolysis, the pentose phosphate pathway (PPP), glycogenolysis, the electron transport chain (ETC), TCA cycle, and glycogen synthesis (n = 3 with 15 animals per sample). Mean expression values of the replicates of all genes at each time point are plotted to make the line, and the gray border represents the standard error bounds using a LOESS fitting. Expression levels are shown as transcripts per million (TPM) normalized expression values to account for gene size and total RNA in the sample. Animals were staged as white prepupae (WPP), day 1 pupae (P1), day 2 pupae (P2), day 3 pupae (P3), day 4 pupae (P4), day 1 adult (A1), and day 6 adult (A6).
The observation that a majority of metabolic genes are up-regulated in late pupae raises the question of how broadly dERR might impact metabolism at this stage. To determine this, we examined the temporal profiles of gene expression for several metabolic categories in staged controls and dERR mutants (Fig. 7C). As expected, the dERR-dependent glycolytic switch can be readily identified using this approach (Fig. 7C). Indeed, 8 of the 12 genes that act in glycolysis are expressed at reduced levels at all time points examined in our RNA-seq analysis (Supplemental Table S5). In addition, we found that genes involved in the pentose phosphate pathway are largely up-regulated in young adults in a dERR-dependent manner (Fig. 7C). Glycogenolysis and glycogen synthesis are also affected at this stage. The average effect of the dERR mutation on these gene sets, however, is relatively small because only a few genes are expressed at significantly reduced levels in the mutant background, including glycogen synthase, UGP, GlyP, and Pgm (Supplemental Table S5). UGP, GlyP, and Pgm are also bound by dERR, suggesting that they are direct regulatory targets of the receptor (Supplemental Table S3). Interestingly, although genes that act in the TCA cycle and electron transport are globally up-regulated at the onset of adulthood, this response occurs independently of dERR regulation. These widespread switches in oxidative metabolic gene expression are also readily apparent in the time course data from modENCODE (Supplemental Fig. S4). Our data thus indicate that other temporal regulators must act together with dERR at the onset of adulthood to direct the adult metabolic transition toward glucose oxidation and lipogenesis in Drosophila.
Discussion
Metabolism changes dramatically as animals progress through development. How these systemic metabolic states are coordinated with the life cycle, however, remains unclear. Although some transitions can occur slowly and thus could involve complex compensatory and feedback regulation, other metabolic transitions occur within a period of a few hours or days and require a more rapid and direct form of control. Here we show that the onset of adulthood in Drosophila is accompanied by a widespread switch in gene expression that occurs midway through metamorphosis. This transition involves the central pathways of energy metabolism, glycolysis, the TCA cycle, and electron transport chain, and thus is ideally designed to meet the increased energy demands of adult motility. We show that the dERR nuclear receptor is required for part of this transcriptional response, inducing the expression of key genes in glycolysis, the PPP, and lipogenesis. Below we describe this regulatory response in more detail and propose a model for how dERR contributes to the pupal-to-adult metabolic transition.
dERR is required for glycolysis and lipogenesis in adults
We used CRISPR/Cas9 to generate a conditional dERR mutant allele in order to facilitate functional studies during later stages of development. Although widely used in mice, conditional alleles are rarely implemented in Drosophila largely due to the ease of RNAi studies and clonal analysis. Conditional alleles, however, provide an ideal way to characterize organism-wide responses at any stage of development as well as a genetic alternative to cell type- and tissue-specific RNAi, which can be complicated by partial knockdown and off-target effects.
dERR mutant adults eclose at a normal rate and with normal metabolites (Fig. 2I; Supplemental Fig. S2D,F,G). This implies that metamorphosis can proceed with basal levels of glycolysis. This is perhaps not surprising given that insects do not feed during pupal stages but instead depend on stored energy and nutrients recycled from the autophagic death of larval tissues. In addition, our time course analysis of metabolic gene expression reveals an unexpected role for dERR in enhancing glycolysis at puparium formation (Fig. 6A). dERR mutants show a modest but significant reduction in glycolytic transcripts during early stages of metamorphosis, which in control animals are already at relatively low levels. Interestingly, this correlates with the transcriptional induction of dERR mRNA in apparent response to the late larval pulse of 20-hydroxyecdysone (Sullivan and Thummel 2003). Further studies are required to determine the possible significance of this regulatory connection with ecdysone signaling and roles for glycolysis during early metamorphosis.
The accumulation of lipid in the newly formed adult fat body appears to be entirely dependent on de novo lipogenesis, as reflected by the absence of triglycerides and neutral lipids in adults starved at eclosion (Storelli et al. 2019). We propose that this efficient conversion of dietary nutrients into stored fat is regulated by dERR at the onset of adulthood. Several lines of evidence support this model. First, dERR mutants rapidly become hypolipidemic, leading to an almost complete absence of lipids by 1 wk of age (Fig. 2I), resembling the hypolipidemia seen in controls starved at eclosion (Storelli et al. 2019). In addition, unlike controls, a high sugar diet has no effect on the low triglyceride levels in dERR mutant adults (Fig. 2K). This is consistent with the reduced expression of genes that encode glycolytic enzymes (Fig. 3B,C), many of which appear to be directly regulated by dERR (Supplemental Tables S3, S4). This reduced rate of glycolysis leads to an accumulation of glucose along with reduced downstream TCA cycle intermediates, including citrate, which is the key precursor for lipogenesis (Fig. 4). In addition, ATP citrate lyase (ATPCL) is expressed at reduced levels in dERR mutants (Fig. 3C). This gene encodes the enzyme that converts cytoplasmic citrate into the critical precursor for lipid synthesis, acetyl CoA (Fig. 8). Lipogenesis is also dependent on NADPH, which is largely derived from the PPP, another pathway that appears to be under direct control by dERR (Fig. 3C; Supplemental Tables S3, S4). In addition, Gpdh, which encodes a key enzyme that supports triglyceride biosynthesis, is expressed at reduced levels in dERR mutants (Fig. 3C; Supplemental Table S1). Taken together, these effects of dERR on glycolysis, the PPP, citrate levels, Gpdh expression, and ATPCL expression, can account for the reduced levels of fatty acids seen in dERR mutants along with the reduced levels of stored fat (Figs. 4, 8).
Figure 8.
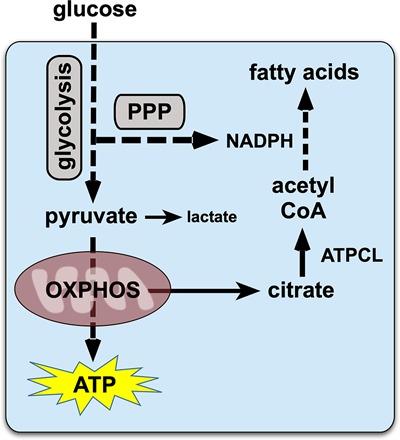
A model for dERR function in adult metabolism. dERR directly induces genes in glycolysis and the pentose phosphate pathway (PPP), highlighted with gray boxes. The end product of glycolysis, pyruvate, enters the TCA cycle to undergo oxidative phosphorylation for ATP production. Relatively little pyruvate is converted to lactate due to low levels of Ldh expression in adults. Cytoplasmic citrate, derived from the TCA cycle, is converted into the critical precursor for lipogenesis, acetyl CoA, by ATP citrate lyase (ATPCL), which is up-regulated by dERR in adults. NADPH from the PPP provides the reducing power for efficient lipogenesis, supporting the synthesis of fatty acids, the precursor for stored triglycerides. Mitochondrial activity is independently up-regulated in adults by the dHNF4 nuclear receptor.
In addition to hypolipidemia, dERR mutant adults rapidly develop hyperglycemia and hypoproteinemia (Supplemental Fig. S2F,G). As mentioned above, the hyperglycemia is likely due, at least in part, to reduced glycolysis, which leads to an accumulation of glucose along with fructose and mannitol (Fig. 4). Similarly, the reduced protein levels seen in dERR mutants are consistent with the fundamental block in energy metabolism in these animals. It is likely that dERR mutants are breaking down protein in order to compensate for the reduced entry of pyruvate into the TCA cycle. This possibility is supported by the major reductions in alanine and proline in dERR mutants seen by metabolomic analysis (Fig. 4). In addition, a shift from glucose oxidation to protein catabolism is a hallmark of the starved state and thus could contribute to the shortened life span and reduced motility seen in dERR mutant adults (Fig. 1B,C). It is also interesting to note that dERR mutant larvae do not display a reproducible effect on total protein levels. Consistent with this, proline is only reduced about twofold in mutant larvae and alanine levels are only slightly affected (Tennessen et al. 2011). Similarly, citrate, isocitrate, and succinate levels are all unaffected in dERR mutant larvae and significantly reduced in dERR mutant adults. These stage-specific differences appear to reflect a more significant reduction in TCA cycle activity in mutant adults and thus could explain why triglycerides are more severely affected at this stage.
Previous studies have demonstrated that the adult fat body is a critical tissue for de novo lipogenesis (DiAngelo and Birnbaum 2009; Musselman et al. 2013). Our results support this conclusion and show that dERR and glycolysis are both required in this tissue for proper lipid accumulation. Disruption of either dERR or Pfk in the adult fat body leads to reduced levels of triglycerides and reduced neutral lipids in adult fat tissue (Fig. 5). These phenotypes are similar to the hypolipidemia seen in dERR mutants, supporting the proposal that efficient glycolysis in the adult fat body is required to maintain appropriate levels of stored lipid. However, it is important to note that dERR also has effects on multiple metabolic pathways that act in tissues outside the fat body (Supplemental Table S1). Further studies are required to dissect these activities and determine how they might contribute to adult physiology and health.
Interestingly, roles for ERR in maintaining stored lipid reserves appear to be conserved through evolution. ERRα mutant mice appear normal and are fully viable but display reduced body weight, reduced peripheral adipose mass, and are resistant to diet-induced obesity (Luo et al. 2003). More recent studies of ERRα mutant mice have confirmed and extended these original observations and also shown that pharmacological inhibition of ERRα leads to reduced body weight as well as reduced hepatic lipid accumulation and reduced gonadal and inguinal fat mass (Kim et al. 2013; B'Chir et al. 2018). To date, however, no studies have demonstrated a direct role for ERRα in supporting normal glycolytic oxidation or lipogenesis and thus it remains unclear whether the molecular mechanisms that we describe here for dERR are conserved through evolution. However, it is important to note that glycolytic genes are expressed at reduced levels in ERRγ mutant hearts and ERRα is bound to almost every gene in the glycolytic pathway in the mouse liver (Alaynick et al. 2007; Charest-Marcotte et al. 2010). In addition, dERR has been shown to support glycolytic Warburg metabolism in cancer cells (Deblois and Giguère 2013). Further studies are required to address possible roles for mammalian ERR family members in supporting glucose oxidation for energy production and lipogenesis.
dERR and dHNF4 contribute to efficient glucose oxidation in adults
Although our studies demonstrate that dERR is required to establish adult lipid reserves, its central role in glycolysis is also consistent with the high energetic demands of adult motility. Indeed, at peak capacity, Drosophila flight sustains remarkable mechanical power driven by highly efficient energy production (Wigglesworth 1949; Lehmann and Dickinson 1997). Adult flies must therefore maintain high levels of pyruvate to fuel ATP synthesis through mitochondrial oxidative phosphorylation. Unlike larvae, which efficiently convert pyruvate into lactate to support aerobic glycolysis, Ldh expression is relatively low during pupal and adult development (Supplemental Fig. S3). In addition, previous studies from our lab have shown that the dHNF4 nuclear receptor triggers a transcriptional switch in newly emerged adults that supports mitochondrial function and maximal ATP production (Barry and Thummel 2016). We propose that it is the coordinated activities of dERR and dHNF4 that establish the efficient energetic state that characterizes adult life. dERR is required for a transcriptional switch during mid-pupal stages that up-regulates glycolysis along with other pathways. This is followed by the induction of dHNF4 at the end of metamorphosis, activating downstream target genes that promote mitochondrial oxidative metabolism (Barry and Thummel 2016). Taken together, these two nuclear receptors can direct the efficient conversion of glucose into ATP, promoting oxidative energy production. In addition, this nuclear receptor mediated metabolic switch provides a molecular context to understand the so-called “U-shaped curve” of oxygen consumption that occurs during insect metamorphosis (Bodine and Orr 1925; Merkey et al. 2011). Studies in multiple insect species have shown that their overall metabolic rate drops at the onset of metamorphosis and increases prior to adult eclosion, as represented by a U-shaped curve of oxygen consumption. This profile correlates with the relatively low levels of metabolic gene expression that we observed during metamorphosis. In addition, our results support the proposal that dERR and dHNF4, acting together at the onset of adulthood, end the U-shaped curve of low metabolic rate during metamorphosis and establish the efficient physiological state that sustains adult energetic needs (Fig. 8).
Interestingly, previous studies have shown that nuclear receptors can cooperate to link developmental progression with metabolism. Both ERRγ and PPARα contribute to a postnatal change in fuel utilization in the mouse heart, from primarily glucose during fetal stages to fatty acids following birth (Lehman and Kelly 2002; Alaynick et al. 2007). This switch in energy substrate preference allows the newborn pup to effectively utilize dietary lipids from maternal milk. Taken together with our studies of dERR and dHNF4 in Drosophila, these studies in mammals highlight the importance of nuclear receptors in coupling developmental progression with metabolism and raise the possibility that other metabolic switches may be discovered through future studies of these transcriptional regulators.
Major metabolic pathways are coordinated with developmental maturation
Transcriptional profiling of staged pupae and young adults revealed that the central glycolytic genes are coordinately induced during mid-pupal stages in a dERR-dependent manner (Fig. 7; Supplemental Table S5). Unexpectedly, we also found that most genes in the TCA cycle and ETC are globally up-regulated in synchrony with this switch (Supplemental Fig. S4). The transcription factors that mediate this regulation, however, remain unknown. Only a few genes in the TCA cycle and ETC display a small change in their expression in dERR and dHNF4 mutants (Supplemental Table S1; Barry and Thummel 2016). Similarly, although dHNF4 plays an important role in supporting adult mitochondrial activity, the global transcriptional induction we observed in mid-pupae precedes the dHNF4-regulated switch at the onset of adulthood (Barry and Thummel 2016; Storelli et al. 2019). These observations highlight the importance of transcriptional switches as a molecular mechanism that links developmental progression with changes in metabolic state, and raise the question of what other regulators feed into these pathways. A number of transcription factors have been identified that play central roles in metabolism; however, most of these, such as Myc, DHR38, Mondo, Sugarbabe, and Cabut, regulate carbohydrate metabolism, with no clear effect on the TCA cycle or ETC (Mattila and Hietakangas 2017). Further studies are required to identify the key regulator(s) that establish adult mitochondrial oxidative metabolism. It will also be interesting to see whether studies in vertebrates will continue to generalize the observations we report here and provide further insights into the molecular mechanisms that couple switches in metabolic state with the changing demands of developmental progression through the life cycle.
Materials and methods
Drosophila strains and media
Genetic studies of dERR in adults used a transheterozygous combination of a dERR null allele (dERR2) (Tennessen et al. 2011) and a conditional allele for dERR (described below). For most experiments, control and experimental animals differed by the presence or absence of a hsFLP transgene on the X chromosome (y w hsFLP12, BSC 1929) and were subjected to heat treatments as described below. GAL4 lines used: cg-GAL4 (BSC 7011), Myo1a-GAL4 (a gift from B. Edgar); both of which were combined with Tub-GAL80ts (BSC 7018) to provide temporal control. RNAi lines: UAS-dERRRNAi (TRiP 50686), UAS-PfkRNAi (VDRC 105666), and UAS-RNAi control (VDRC 60100). Flies were maintained on a diet containing 8% yeast, 1% agar, 0.05% MgSO4, 0.05% CaCl2, and either 3%, 9%, or 15% dietary sugar (2:1 ratio of glucose to sucrose, percentages represent weight/final food volume). Tegosept (10 mL/L) and propionic acid (6 mL/L) were added prior to pouring. Unless noted otherwise, 6-d-old males were used for all studies, maintained at 25°C on a 9% sugar diet, and transferred to fresh food every 2–3 d. For RNAi experiments, embryos maintained at 18°C were transferred to 29°C on day 11 after egg lay (AEL). At eclosion, adults were maintained on fresh food every 2 d until collection.
Generation of the dERR conditional allele
CRISPR-Cas9 technology was used to introduce two FRT sequences into the dERR locus (Gratz et al. 2014). We targeted a genomic region extending from nucleotides 8,182,007 to 8,186,262 on the left arm of the third chromosome. The sequence of this region is depicted in Supplemental Figure S1, with the FRT sequences highlighted in yellow and the 3xPax3-DsRed reporter insertion in red font. The 3xPax3-DsRed reporter is positioned such that it is transcribed off the opposite strand from dERR so that it will not interfere with dERR transcript elongation. A plasmid carrying this sequence was injected into y1 M(vas-Cas9.RFP)ZH-2A w1118 embryos (Gratz et al. 2014) along with plasmids expressing the following guide RNAs: 5′ guide GAAAATGATTGATTAGATAGAGG and 3′ guide GCACTTTTAAAATACGATAGTGG. Integrants were selected based on DsRed expression in the adult eye. Stocks were established from individual males and sequenced to confirm the accurate integration of both FRT sequences. The efficient FLP-mediated excision of the intervening sequences was assayed by genomic PCR as well as RT-qPCR analysis of dERR mRNA levels (Supplemental Fig. S2A–C). Unless otherwise stated, 24- to 40-h egg lays were allowed to develop until 5–6 d AEL and then subjected to two sequential 1-h heat pulses at 37°C separated by 24 h at 25°C. Adults that eclosed by day 11 AEL were used for experiments. For the time course RNA-seq data in Figures 6 and 7, the same method was followed except that 24-h egg lays were used and animals were subjected to heat treatment at days 4–5 AEL.
Locomotor assay
Locomotor activity was measured using the RING assay (Gargano et al. 2005). Ten flies (n = 8 vials) were transferred to a vial and allowed to recover for 24 h. The next day, animals were assayed for height achieved four seconds after being tapped to the bottom of an empty vial. The procedure was repeated five times at 30-sec intervals and the average height of all flies over all trials was scored per vial.
Life span studies and humid starvation
Flies were maintained on a 9% sugar diet and adults were allowed to mature and mate for 3 d. For life span analysis, 10 males/vial were selected and transferred to fresh food every 2–5 d. Lethality was assayed at the time of transfer. For humid starvation, 10 males/vial were selected on day 6 of adulthood and transferred to vials in which a dense weave cellulose acetate stopper (Flug; Genesee Scientific 49-101) was pushed to the bottom of the vial and water was added to saturation. Animals were maintained at 25°C and transferred to fresh vials every 2–3 d. Lethality was scored daily.
Metabolite assays
Metabolite assays were performed as described with minor modifications (Tennessen et al. 2014a). Briefly, samples of five animals were homogenized and glucose and glycogen levels were determined using a hexokinase assay kit (Sigma GAHK20) and amyloglucosidase (Sigma A1602-25MG). Whole-body triglyceride levels were determined using triglyceride reagent (Sigma T2449-10ML) and free glycerol reagent (Sigma F6428-40ML). Protein levels were determined using a Bradford assay (Bio-Rad 5000006). Metabolites levels were calculated using a standard curve. Glucose, glycogen and triglyceride levels were not normalized to protein due to the depletion of protein in dERR mutants. ATP assays were performed on day 6 adult males as described (Tennessen et al. 2014a). ATP levels were analyzed using a luminescence assay kit (Molecular Probes ATP kit A22066).
Oil Red O stains
Oil Red O stains were performed as described (Tennessen et al. 2014a; Storelli et al. 2019) with the modification of incubation for 45 min at 60°C in the 0.5% Oil Red O/propylene glycol solution. Samples were observed by bright-field microscopy, and pictures were taken with a Zeiss Axioskop Plus microscope equipped with an Infinity 3 Lumenera camera and the Infinity Analyze software. Independent stains were performed twice on sets of six cuticles (n = 12), with representative images shown in each figure.
RNA-seq
For RNA-seq data from 6-d-old dERR mutants (Fig. 3; Supplemental Table S1), hsFLP; ERRcond/ERR2 animals were either not heat treated (controls) or heat-treated (dERR mutants) on days 5 and 6 AEL, and RNA was isolated from samples of 20 6-d-old flies using TriPure reagent (Roche). Samples were pooled in triplicate (60 flies/sample) and further purified using RNeasy columns (Qiagen) with DNase treatment. Three independent control and mutant samples were submitted for sequencing. RNA quality was analyzed using an Agilent Bioanalyzer RNA 6000. Poly(A)-selected RNAs from each biological replicate were assembled into barcoded libraries and pooled into a single-flow cell lane for Illumina HighSeq2000 50-cycle single-read sequencing, which produced 15 million to 17 million reads per sample. Standard replicate RNA-seq analysis was performed using USeq and DESeq analysis packages with alignment to the Drosophila melanogaster dm3 genome assembly. Transcripts meeting a cutoff of padj ≤ 0.001 were considered as differentially expressed genes, identifying 1004 genes (Supplemental Table S1). RNA quality control, library preparation, sequencing, and data analysis were performed at the University of Utah High-Throughput Genomics and Bioinformatics Core Facilities. RNA-seq data from this study can be accessed at NCBI GEO (accession no. GSE144762).
For the time-course RNA-seq analysis (Figs. 6, 7; Supplemental Table S5), control ERRcond/ERR2 animals and hsFLP; ERRcond/ERR2 experimental animals were heat treated in parallel as third instar larvae on days 4 and 5 AEL. Animals were selected as white prepupae, allowed to develop, and frozen in liquid nitrogen at the following time points: white prepupae (WPP), day 1 pupae (P1), day 2 pupae (P2), day 3 pupae (P3), day 4 pupae (P4), day 1 adult males (A1), and day 6 adult males (A6). RNA was isolated from triplicate samples with 10 animals per sample using the Direct-zol RNA Miniprep kit (Zymo Research), including DNase treatment. RNA quality was analyzed using an Agilent Bioanalyzer RNA 6000. Poly(A)-selected RNAs from each biological replicate were assembled into barcoded libraries and pooled into a single-flow cell lane for Illumina NovaSeq 2 × 50-bp sequencing 100 million read pairs, which produced 5.2 million to 8.6 million reads per sample. Optical duplicates were removed with clumpify, adapter trimming was performed with cutadapt (version 1.16), and alignment was done using STAR (version 2.7.0f). Transcript expression was quantified with RSEM v1.3.1 (Li and Dewey 2011) following alignment to the dm6 genome (Ensembl release 96) with STAR v2.7.0f (Dobin et al. 2013). Differential gene expression modeling with DESeq2 v1.22.2 (Love et al. 2014) tested for the effects of genotype at each time point using a genotype x time interaction design. Specifically, at each time, mutant versus control were compared and differences (log2 fold-changes different from zero) were reported and controlled with a Benjamini-Hochberg FDR of 0.001. The log2 fold changes were reduced to zero with ashr methodology (Stephens 2017). Transcripts meeting a cutoff of padj ≤ 0.001 were considered as differentially expressed genes. Temporal line graphs (Figs. 6B, 7C) were generated using the R package ggplot2 (Wickham 2016). Mfuzz (Kumar and Futschik 2007) was used to perform soft clustering analysis of the temporal patterns of RNA-seq data (Figs. 6A, 7). GO term analysis was conducted using PANTHER (http://pantherdb.org) by performing the “Statistical overrepresentation test” with “PANTHER GO-slim Biological Process” on the list of significantly reduced transcripts in dERR mutants. RNA-seq data from this study can be accessed at NCBI GEO (accession no. GSE144764).
RT-qPCR
RNA was extracted from samples of 10–15 males using the Direct-zol RNA Miniprep kit (Zymo Research). Reverse transcription was performed on 1 µg of RNA using the high-capacity cDNA reverse transcription kit (Thermo Fisher 4368814) with random primers. cDNA was used as a template for RT-qPCR using an Applied Biosystems QuantStudio 3 system. PowerUp SYBR Green master mix (Thermo Fisher Scientific A25742) was used for RT-qPCR reactions. mRNA fold changes were calculated using the comparative CT method (Schmittgen and Livak 2008). Transcript levels were normalized to rpl32. All RT-qPCR experiments included three technical replicates, three to five independent samples, and 10–15 animals per sample. Primer sequences are listed in Supplemental Table S6 and have an efficiency between 90% and 110%.
ChIP-seq
ChIP-seq was performed using dERR1/dERR2 mutants that carry a BAC genomic rescue construct spanning the dERR locus with an in-frame fusion of the coding sequence for a 3XFlag tag immediately upstream of the stop codon. The BAC construct was engineered using BAC CH322-54A09, which carries a 20,644-bp genomic fragment including dERR (Venken et al. 2009). Bacterial recombineering was used to insert the 3XFlag coding sequence at the 3′ end of the dERR coding region, along with two FRTs that can be used to remove dERR coding sequences and the 3′ Flag tag in response to hsFLP expression. The BAC construct was inserted into the VK22 attP landing site on the second chromosome and maintained as a homozygous stock. ChIP-seq was performed on four samples each of 6-d-old w1118 controls and w1118; dERR-3XFlag-BACVK22; dERR1/dERR2 adults essentially as described (Barry and Thummel 2016) using Flag antibodies (Sigma F1804, M2 monoclonal). Barcoded libraries were generated by the University of Utah High-Throughput Genomics Core Facility and sequenced in a single-flow lane for Illumina HighSeq2000 50-cycle single-read sequencing. Data analysis was performed by the Bioinformatics Core at the University of Utah School of Medicine using model-based analysis for ChIP-seq 2.0 (MACS2) (Zhang et al. 2008) with a threshold Q-value of 0.05. Peaks were annotated with the R ChIPseeker package, and nearest neighboring genes were compiled using USeq FindNeighboringGenes with UCSC dm3 EnsGenes gene tables. ChIP-seq data from this study can be accessed at NCBI GEO (accession no. GSE144763).
GC/MS metabolomic analysis
Animals were reared on a 9% sugar diet until eclosion, after which they were maintained on a 15% sugar diet for 6 d prior to collection. Twenty adult males per sample were snap-frozen in liquid nitrogen and prepared for analysis by gas chromatography-mass spectrometry (GC/MS) as described (Tennessen et al. 2014a). GC/MS data are presented from three independent experiments, each consisting of three to four biological replicates per condition. FAMES GC/MS samples were run in one experiment with 12 replicates per condition to detect fatty acids. Sample preparation and GC/MS analysis were performed by the Metabolomics Core Research Facility at the University of Utah School of Medicine.
Statistics
Graphical representation and statistical analysis of most data (Figs. 1–5; Supplemental Figs. S2, S3) were performed using GraphPad Prism 6 software. Life span data are presented as Kaplan-Meier survival plots with a log-rank Mantel-Cox test for statistical comparison of survival plots. For box and whisker plots, the boxes extend from the 25th to 75th percentiles, the horizontal line represents the median, and the error bars represent the minimum and maximum data points. Statistical comparisons were performed using a two-sample t-test with a Welch's correction for unequal variance (Figs. 1C, 2A–D,I, 4; Supplemental Fig. S2A,B,D,E), or a one-way ANOVA followed by Tukey's multiple comparisons test (Figs. 2J,K, 5A,B; Supplemental Fig. S2F,G). Statistical analysis of RNA-seq and ChIP-seq data is described above.
Supplementary Material
Acknowledgments
We thank members of the Thummel laboratory, W. Barry, and M. Metzstein for helpful discussions; J. Tennessen for discussions and his contributions to the list of Drosophila metabolism genes in Supplemental Table S2; G. Storelli for comments on the manuscript; B. Edgar for stocks; the Bloomington Stock Center for providing stocks; and FlyBase for critical information that made these studies possible. The Huntsman Cancer Institute High-Throughput Genomics and Bioinformatic Analysis Shared Resource at the University of Utah is supported by the National Cancer Institute (P30CA042014). Metabolomics analysis was performed at the Metabolomics Core Facility at the University of Utah, which is supported by the National Institutes of Health (NIH; S10 OD016232-01, S10 OD021505-01, and U54 DK110858-01). This research was supported by NIH grants F32DK116487 and 5T32DK091317 to K.B., and 5R01DK075607 to C.S.T.
Author Contributions: C.S.T. supervised the work. K.B. designed and interpreted experiments with input from M.M.R. and E.J.H.; K.B., M.M.R., G.L., and M.A.H. performed the experiments. K.B. and C.S.T. wrote and revised the paper.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.335281.119.
References
- Aguila JR, Suszko J, Gibbs AG, Hoshizaki DK. 2007. The role of larval fat cells in adult Drosophila melanogaster. J Exp Biol 210: 956–963. 10.1242/jeb.001586 [DOI] [PubMed] [Google Scholar]
- Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguere V, et al. 2007. ERRγ directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab 6: 13–24. 10.1016/j.cmet.2007.06.007 [DOI] [PubMed] [Google Scholar]
- Barry WE, Thummel CS. 2016. The Drosophila HNF4 nuclear receptor promotes glucose-stimulated insulin secretion and mitochondrial function in adults. Elife 5: e11183 10.7554/eLife.11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- B'Chir W, Dufour CR, Ouellet C, Yan M, Tam IS, Andrzejewski S, Xia H, Nabata K, St-Pierre J, Giguère V. 2018. Divergent role of estrogen-related receptor α in lipid- and fasting-induced hepatic steatosis in mice. Endocrinology 159: 2153–2164. 10.1210/en.2018-00115 [DOI] [PubMed] [Google Scholar]
- Bodine JH, Orr PR. 1925. Respiratory metabolism - physiological studies on respiratory metabolism. Biological Bulletin 48: 1–14. 10.2307/1536588 [DOI] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. 2006. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126: 789–799. 10.1016/j.cell.2006.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EL, Hazen BC, Eury E, Wattez JS, Gantner ML, Albert V, Chau S, Sanchez-Alavez M, Conti B, Kralli A. 2018. Estrogen-related receptors mediate the adaptive response of brown adipose tissue to adrenergic stimulation. iScience 2: 221–237. 10.1016/j.isci.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byerly MS, Al Salayta M, Swanson RD, Kwon K, Peterson JM, Wei Z, Aja S, Moran TH, Blackshaw S, Wong GW. 2013. Estrogen-related receptor β deletion modulates whole-body energy balance via estrogen-related receptor γ and attenuates neuropeptide Y gene expression. Eur J Neurosci 37: 1033–1047. 10.1111/ejn.12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest-Marcotte A, Dufour CR, Wilson BJ, Tremblay AM, Eichner LJ, Arlow DH, Mootha VK, Giguere V. 2010. The homeobox protein Prox1 is a negative modulator of ERRα/PGC-1α bioenergetic functions. Genes Dev 24: 537–542. 10.1101/gad.1871610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough JR. 1985. Energy metabolism during mammalian embryogenesis. Biochem Soc Trans 13: 77–79. 10.1042/bst0130077 [DOI] [PubMed] [Google Scholar]
- Deblois G, Giguère V. 2013. Oestrogen-related receptors in breast cancer: control of cellular metabolism and beyond. Nat Rev Cancer 13: 27–36. 10.1038/nrc3396 [DOI] [PubMed] [Google Scholar]
- DiAngelo JR, Birnbaum MJ. 2009. Regulation of fat cell mass by insulin in Drosophila melanogaster. Mol Cell Biol 29: 6341–6352. 10.1128/MCB.00675-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguère V. 2007. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRα and γ. Cell Metab 5: 345–356. 10.1016/j.cmet.2007.03.007 [DOI] [PubMed] [Google Scholar]
- Gándara L, Wappner P. 2018. Metabo-devo: a metabolic perspective of development. Mech Dev 154: 12–23. 10.1016/j.mod.2018.02.004 [DOI] [PubMed] [Google Scholar]
- Gargano JW, Martin I, Bhandari P, Grotewiel MS. 2005. Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Exp Gerontol 40: 386–395. 10.1016/j.exger.2005.02.005 [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O'Connor-Giles KM. 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss JM, Garbacz WG, Xie W. 2015. Constitutive activities of estrogen-related receptors: transcriptional regulation of metabolism by the ERR pathways in health and disease. Biochim Biophys Acta 1852: 1912–1927. 10.1016/j.bbadis.2015.06.016 [DOI] [PubMed] [Google Scholar]
- Kim DK, Gang GT, Ryu D, Koh M, Kim YN, Kim SS, Park J, Kim YH, Sim T, Lee IK, et al. 2013. Inverse agonist of nuclear receptor ERRγ mediates antidiabetic effect through inhibition of hepatic gluconeogenesis. Diabetes 62: 3093–3102. 10.2337/db12-0946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko EV, Mazina MY, Krasnov AN, Vorobyeva NE. 2019. The Drosophila nuclear receptors EcR and ERR jointly regulate the expression of genes involved in carbohydrate metabolism. Insect Biochem Mol Biol 112: 103184 10.1016/j.ibmb.2019.103184 [DOI] [PubMed] [Google Scholar]
- Krejci A, Tennessen JM. 2017. Metabolism in time and space - exploring the frontier of developmental biology. Development 144: 3193–3198. 10.1242/dev.150573 [DOI] [PubMed] [Google Scholar]
- Kumar L, Futschik ME. 2007. Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2: 5–7. 10.6026/97320630002005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman JJ, Kelly DP. 2002. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol 29: 339–345. 10.1046/j.1440-1681.2002.03655.x [DOI] [PubMed] [Google Scholar]
- Lehmann FO, Dickinson MH. 1997. The changes in power requirements and muscle efficiency during elevated force production in the fruit fly Drosophila melanogaster. J Exp Biol 200: 1133–1143. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chawla G, Hurlburt AJ, Sterrett MC, Zaslaver O, Cox J, Karty JA, Rosebrock AP, Caudy AA, Tennessen JM. 2017. Drosophila larvae synthesize the putative oncometabolite L-2-hydroxyglutarate during normal developmental growth. Proc Natl Acad Sci 114: 1353–1358. 10.1073/pnas.1614102114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Zhang Z, Teng CT. 2005. Estrogen-related receptor-γ and peroxisome proliferator-activated receptor-γ coactivator-1α regulate estrogen-related receptor-α gene expression via a conserved multi-hormone response element. J Mol Endocrinol 34: 473–487. 10.1677/jme.1.01586 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguère V. 2003. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor α. Mol Cell Biol 23: 7947–7956. 10.1128/MCB.23.22.7947-7956.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila J, Hietakangas V. 2017. Regulation of carbohydrate energy metabolism in Drosophila melanogaster. Genetics 207: 1231–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkey AB, Wong CK, Hoshizaki DK, Gibbs AG. 2011. Energetics of metamorphosis in Drosophila melanogaster. J Insect Physiol 57: 1437–1445. 10.1016/j.jinsphys.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Misra J, Kim DK, Choi HS. 2017. ERRγ: a junior orphan with a senior role in metabolism. Trends Endocrinol Metab 28: 261–272. 10.1016/j.tem.2016.12.005 [DOI] [PubMed] [Google Scholar]
- Miyazawa H, Aulehla A. 2018. Revisiting the role of metabolism during development. Development 145: dev131110 10.1242/dev.131110 [DOI] [PubMed] [Google Scholar]
- Miyazawa H, Yamaguchi Y, Sugiura Y, Honda K, Kondo K, Matsuda F, Yamamoto T, Suematsu M, Miura M. 2017. Rewiring of embryonic glucose metabolism via suppression of PFK-1 and aldolase during mouse chorioallantoic branching. Development 144: 63–73. 10.1242/dev.138545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman LP, Fink JL, Narzinski K, Ramachandran PV, Hathiramani SS, Cagan RL, Baranski TJ. 2011. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis Model Mech 4: 842–849. 10.1242/dmm.007948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman LP, Fink JL, Ramachandran PV, Patterson BW, Okunade AL, Maier E, Brent MR, Turk J, Baranski TJ. 2013. Role of fat body lipogenesis in protection against the effects of caloric overload in Drosophila. J Biol Chem 288: 8028–8042. 10.1074/jbc.M112.371047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj R, Sharpley MS, Chi F, Braas D, Zhou Y, Kim R, Clark AT, Banerjee U. 2017. Nuclear localization of mitochondrial TCA cycle enzymes as a critical step in mammalian zygotic genome activation. Cell 168: 210–223.e11. 10.1016/j.cell.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Sampaio JL, Brankatschk M, Carvalho M, Mahmoud A, Shevchenko A, Eaton S. 2012. Lipoproteins in Drosophila melanogaster—assembly, function, and influence on tissue lipid composition. PLoS Genet 8: e1002828 10.1371/journal.pgen.1002828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Stephens M. 2017. False discovery rates: a new deal. Biostatistics 18: 275–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storelli G, Nam HJ, Simcox J, Villanueva CJ, Thummel CS. 2019. Drosophila HNF4 directs a switch in lipid metabolism that supports the transition to adulthood. Dev Cell 48: 200–214.e6. 10.1016/j.devcel.2018.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AA, Thummel CS. 2003. Temporal profiles of nuclear receptor gene expression reveal coordinate transcriptional responses during Drosophila development. Mol Endocrinol 17: 2125–2137. 10.1210/me.2002-0430 [DOI] [PubMed] [Google Scholar]
- Tennessen JM, Baker KD, Lam G, Evans J, Thummel CS. 2011. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab 13: 139–148. 10.1016/j.cmet.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JM, Barry WE, Cox J, Thummel CS. 2014a. Methods for studying metabolism in Drosophila. Methods 68: 105–115. 10.1016/j.ymeth.2014.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JM, Bertagnolli NM, Evans J, Sieber MH, Cox J, Thummel CS. 2014b. Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 4: 839–850. 10.1534/g3.114.010652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, DeBerardinis RJ. 2017. Understanding the intersections between metabolism and cancer biology. Cell 168: 657-669. 10.1016/j.cell.2016.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods 6: 431–434. 10.1038/nmeth.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KP, Rifkin SA, Hurban P, Hogness DS. 1999. Microarray analysis of Drosophila development during metamorphosis. Science 286: 2179–2184. 10.1126/science.286.5447.2179 [DOI] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer International Publishing, New York, NY. [Google Scholar]
- Wigglesworth VB. 1949. The utilization of reserve substances in Drosophila during flight. J Exp Biol 26: 150–163. [DOI] [PubMed] [Google Scholar]
- Xia H, Dufour CR, Giguère V. 2019. ERRα as a bridge between transcription and function: role in liver metabolism and disease. Front Endocrinol 10: 206 10.3389/fendo.2019.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.