Abstract
Melanoma has an unusual capacity to spread in early stage disease, prompting aggressive clinical intervention in very thin primary tumors. Despite these proactive efforts, patients with low-risk, low-stage disease can still develop metastasis, indicating the presence of permissive cues for distant spread. Here we show that constitutive activation of the small GTPase ARF6 (ARF6Q67L) is sufficient to accelerate metastasis in mice with BRAFV600E/Cdkn2aNULL melanoma at a similar incidence and severity to Pten loss, a major driver of PI3K activation and melanoma metastasis. ARF6Q67L promoted spontaneous metastasis from significantly smaller primary tumors than PTENNULL, implying an enhanced ability of ARF6-GTP to drive distant spread. ARF6 activation increased lung colonization from circulating melanoma cells, suggesting that the pro-metastatic function of ARF6 extends to late steps in metastasis. Unexpectedly, ARF6Q67L tumors showed upregulation of Pik3r1 expression, which encodes the p85 regulatory subunit of PI3 kinase (PI3K). Tumor cells expressing ARF6Q67L displayed increased PI3K protein levels and activity, enhanced PI3K distribution to cellular protrusions, and increased AKT activation in invadopodia. ARF6 is necessary and sufficient for activation of both PI3K and AKT, and PI3K and AKT are necessary for ARF6-mediated invasion. We provide evidence for aberrant ARF6 activation in human melanoma samples, which is associated with reduced survival. Our work reveals a previously unknown ARF6-PI3K-AKT pro-invasive pathway, it demonstrates a critical role for ARF6 in multiple steps of the metastatic cascade, and it illuminates how melanoma cells can acquire an early metastatic phenotype in patients.
Introduction
A hallmark of melanoma is early, aggressive spread of disease when primary tumors are as thin as 1 mm(1). The small GTPase adenosine diphosphate (ADP)-ribosylation factor 6 (ARF6) controls invasion of cutaneous melanoma(2–5) and other cancers(6). In its active, GTP-bound state, ARF6 promotes invasion whereas inactive, GDP-bound ARF6 reduces invasion(3–5). The critical role for ARF6 in cutaneous melanoma appears to be common, as knockdown of ARF6 uniformly inhibits invasion in a broad panel of human melanoma cell lines(2). Consistent with the in vitro phenotypes, our previous work suggest that ARF6 is necessary for metastasis. Specifically, pharmacologic inhibition of ARF6 activation reduced spontaneous metastasis in an orthotopic xenograft model of BRAF-mutant melanoma(2). In cutaneous melanoma, ARF6 is activated by WNT5A(2) and HGF(3), both of which result in increased invasion. In uveal melanoma, ARF6 is activated by mutant GNAQ to control tumor growth(7). Beyond melanoma, ARF6 is activated by and coordinates signaling from a variety of signals including EGFR(6), HER2/ERBB2(8), c-MET(9), VEGFR2(6), Frizzled(2,10), inflammatory receptors(6) and G-Protein Coupled Receptors(6). In addition, ARF6 has a critical role in the functional output of mutant p53(11) and RAS(12). Despite compelling evidence that ARF6 is necessary for invasion and metastasis, where ARF6 functions in the metastatic cascade and how it shapes the course of disease are unknown.
Aberrant PI3K pathway activation is a known driver of melanoma disease progression, with up to 70% of melanomas showing reduced PTEN expression or activation of the kinase AKT(13–16). Phosphorylated AKT (pAKT) levels increase significantly from benign nevi to primary melanoma, and from primary melanoma to metastasis. Furthermore, primary melanomas with increased pAKT levels have a worse prognosis(14). PTEN deletion or activation of PI3K or AKT1 accelerates metastasis in genetically engineered mouse models (GEMMs) of BRAF-mutant melanoma(17–19). Approximately 20% of BRAF mutant melanomas show genetic loss of PTEN or rare gain-of-function mutations in PIK3CA or AKT(20). Overall, genomic alterations predicted to activate the PI3K pathway have been detected in approximately 50% of all molecular subtypes of cutaneous melanoma(20). Importantly, activation of the PI3K pathway can also occur in the absence of mutation. The tumor microenvironment can be a rich source of stimulatory signals that promote disease progression and therapy resistance(21). Tumor stroma can generate growth factor ligand(s) that act in a paracrine fashion to stimulate MAPK/ERK and PI3K-AKT, causing resistance to BRAF inhibition in melanoma and other cancers(22). Thus, understanding mechanisms of PI3K activation in cancer may help improve treatment strategies.
In this study, we investigated the role of activated ARF6 in melanoma progression using clinically relevant, immunocompetent genetic models. Our work uncovered novel mechanisms of ARF6-mediated invasion via increased levels and activity of PI3 kinase, and support a role for activated ARF6 in multiple steps of metastasis. Further, we provide evidence for aberrant ARF6 activation in patient samples.
Materials and Methods
RCAS Virus Propagation and delivery in vivo
The RCAS-Cre ± ARF6Q67L plasmids were transfected into DF-1 cells and these cells were prepared for subcutaneous flank injection as previously described(17). HA-tagged ARF6Q67L expression was confirmed with anti-HA western blot.
Mouse Husbandry, Genotyping & Phenotyping
Animal studies were performed in accordance with a protocol approved by the University of Utah Institutional Animal Care and Use Committee. The DCT:TVA; BrafCA; Cdkn2af/f and DCT:TVA; BrafCA; Cdkn2af/f; Ptenf/f mice were previously described(17). Tumor growth was measured by caliper three times weekly. Mice were euthanized once the primary tumor reached a size of 2 cm in any direction, when skin ulceration occurred at the primary tumor site, or when the primary tumor caused symptoms due to deep local invasion. No mice necessitated sacrifice based on overall health. Metastases were evaluated by routine histology, performed by a board-certified pathologist (A.H.G.) who was blinded to genotypes/experimental group assignments. Metastatic volume in lungs was measured as cross-sectional tumor area (ImageJ pixels) from H&E stained slides. Metastatic tumors were imaged at 20x magnification for cross-sectional area. The cross-sectional area of spherical tumors larger than the 20x image capture field (6.5mm tumor diameter threshold), which occurred in Ptenf/f+Arf6Q67L mice, was derived from a diameter ratio, generating a radius (r) equivalent, where r = diameter of the large Ptenf/f+Arf6Q67L tumor / 6.5mm. The calculated cross sectional surface area = r2 x cross sectional area (in pixels of a 6.5mm tumor). Mice that failed to grow primary tumors were included in the tumor incidence calculations but were excluded from tumor growth, onset/latency, survival and the metastatic incidence analyses. pAKT and Ki67 immunohistochemical stains from primary tumors were evaluated in a blinded fashion.
Mouse tumor cell lines and experimental metastasis model
Early passage tumor cell lines from mice 6431, 6455, 7657, 5588, and 5523 were derived from primary melanomas removed from BrafCA;Cdkn2af/f mice that had been injected with DF-1 cells producing either Cre or Cre-ARF6Q67L virus. 6431, 6455, 7657, and 5523 melanoma cell lines were tested for mycoplasma by IDEXX, pre-treated with plasmocin (InvivoGen) as needed prior to injection into tail veins of NOD SCID mice at 5.0 × 105 cells per mouse. The mice were monitored for up to 4 weeks and sacrificed when the health of the animal necessitated euthanasia according to the guidelines of the University of Utah Institutional Animal Care and Use Committee. The lungs were harvested and fixed in 10% neutral buffered formalin overnight, transferred to 70% ethyl alcohol, and paraffin-embedded. 5 μm sections were mounted to glass slides and stained with H&E by the BMP research histology laboratory located at the Huntsman Cancer Institute, University of Utah.
Cell lines and compounds
LOX IMVI were acquired from the NCI-60 Human Cancer Cell Line Repository. SK-MEL-28, Yugen-8, and Yusac-2 cells were provided by D. Grossman (Huntsman Cancer Institute, University of Utah). DF-1 cells and A375-TVA (pDEST12.2-dsRED-LOXp eGFP) cells were provided by S. Holmen. SK-MEL-147, SK-MEL-5, SK-MEL-103, CACL, A375, HEY-T30, UACC.257, and UACC.62 were provided by M. VanBrocklin (Huntsman Cancer Institute, University of Utah). A2058, MeWo, SK-MEL-2, and WM266–4 cells were purchased from the American Type Culture Collection (ATCC). LOX IMVI, Yugen-8, and Yusac-2 cells were maintained in DMEM-high glucose media supplemented with 5% FBS (Invitrogen), pen/strep (Invitrogen), and maintained at 37°C, with 5% CO2. A375, A2058, MeWo, SK-MEL-2, and WM266–4 cells were maintained in DMEM-high glucose media supplemented with 10% FBS (Invitrogen) + pen/strep (Invitrogen), and incubated at 37°C, with 5% CO2. All other human melanoma cell lines were maintained in RPMI-1640-high glucose media supplemented with 10% FBS (ATCC) + pen/strep (Invitrogen), and maintained at 37°C, with 5% CO2. NIH3T3 cells were maintained in DMEM + sodium pyruvate + pen/strep (Invitrogen) and 10% calf serum (ATCC). DF-1 cells were grown in DMEM-high glucose media supplemented with 10% FBS (Invitrogen), gentamicin (Invitrogen), and maintained at 39°C, with 5% CO2. Mouse melanoma primary cells were cultured in DMEM with nutrient mixture F-12, supplemented with 10% FBS (ATCC), pen/strep (Invitrogen), 0.25ug/ml Amphotericin B (Invitrogen), and MEM Non-Essential Amino Acids Solution (Invitrogen). For Figure 5b, human melanoma cell lines were grown in RPMI-1640-high glucose media supplemented with 10% FBS (ATCC), pen/strep (Invitrogen), and maintained at 37°C, with 5% CO2. A375-TVA (pDEST12.2-dsRED-LOXp eGFP) cells were grown in DMEM-high glucose media supplemented with 10% FBS (Invitrogen), gentamicin (Invitrogen), and maintained at 37°C, with 5% CO2 and used to confirm expression of RCAS contructs. Human cell lines were authenticated using ATCC’s human STR identity profiling cell authentication service and the BRAF, NRAS, and KRAS mutation status was sequenced to confirm the expected (published) profile. GDC-0941 and MK2206 were purchased from Selleck Chemicals.
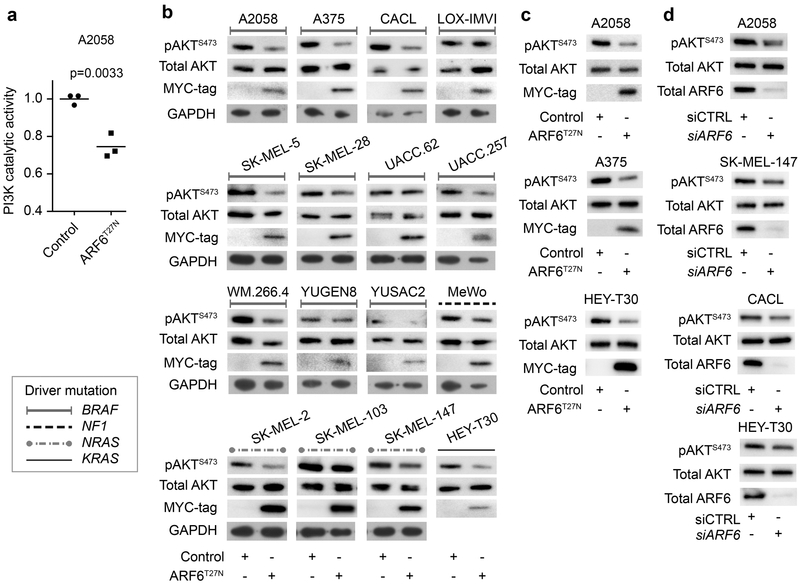
Figure 5. ARF6 is necessary for PI3K and AKT activation.
(a-c) Adenoviral delivery of MYC-tagged ARF6T27N, a GDP-bound, inactive form of ARF6. (a) Catalytic activity of endogenous PI3K in A2058 human melanoma cells. Unpaired t-test, two tailed. (b) 15 melanoma and one carcinoma (HEY-T30) cell lines were screened for AKT activation. 11/16 (68%) cell lines show reduced pAKTS473 with ARF6T27N expression. (c) Reproducible reduction in pAKT by ARF6T27N expression in A375 cells (n=4), A2058 cells (n=4), and HEY-T30 cells (n=5). (d) Reduced pAKT with siRNA knockdown of ARF6 in SK-MEL-147 (n=5), A2058 (n=4), and CACL (n=4) and HEY-T30 (n=6). See Figure S6 for quantitative data.
Proliferation Assay
Cell proliferation was measured by CyQUANT (Invitrogen) as previously described(7).
Apoptosis Assay
Apoptosis assays were performed using RealTime-Glo™ Annexin V Apoptosis Assay kit (Promega). 5,000 cells in 10% FBS or 0% FBS-containing medium were plated into each of four wells of duplicate 96-well plates. Annexin V luminescence and CyQUANT fluorescence (Invitrogen) were measured 24 hours later after cell attachment. Apoptosis index was calculated by Annexin V values normalized to cell numbers (CyQUANT values).
ARF6-GTP expression signature analysis
RNA was extracted from fresh-frozen mouse tumors using Qiagen RNAeasy Plus Universal Mini Kit after frozen section histologic confirmation of high tumor content. RNA-Sequencing was performed using Illumina TruSeq Stranded mRNA Library Preparation Kit with polyA selection followed by lllumina HiSeq 2500 125-cycle paired-end sequencing, performed by the High Throughput Genomics laboratory at the Huntsman Cancer Institute. The mouse GRCm38 FASTA and GTF files were downloaded from Ensembl release 90 and the reference database was created using STAR version 2.5.2b with splice junctions optimized for 125 base pair reads. Reads were trimmed of adapters using cutadapt version 1.16 and then aligned to the reference database using STAR in two pass mode to output a BAM file sorted by coordinates. Mapped reads were assigned to annotated genes in the GTF file using featureCounts version 1.5.1. The output files from cutadapt, FastQC, STAR and featureCounts were summarized using MultiQC to check for any sample outliers. Differentially expressed genes were identified using a 10% false discovery rate with DESeq2 version 1.16.0. Two BRAFV600E/Cdkn2ANULL control tumor samples were outliers in principle component analysis and were excluded from further analysis. Pathway analysis was performed with Illumina Base Space Correlation Engine. The RNAseq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE129392 (https://www.ncbi.nlm.gov/geo/query/acc.cgi?acc=GSExxx).
Proteomics
Protein extraction and reverse phase protein array of frozen mouse tumors was performed by the MD Anderson Cancer Center RPPA Core Facility. Analysis of human melanoma A2058 cells was accomplished with the Proteome Profiler Human Phospho-Kinase Array Kit, (R&D systems) according to the manufacturer’s instructions, using 600 ug of total cell lysates incubated with the phospho-kinase membrane array.
Western Blotting and antibodies
Cell lysates were prepared, protein was quantified and Western blots were performed as previously described(2). Antibodies dilutions for western blots include ARF6 (CST-5740) 1:1000, HA tag (CST-3724) 1:1000, MYC tag (CST-2276) 1:5000, AKT (CST-9272) 1:1000, pAKT S473 (CST-4060) 1:2000, p44/42 MAPK (Erk1/2) (CST-9107) 1:1000, phospho-p44/42 MAPK (Erk1/2 Thr202/Tyr204)(CST-4377) 1:2000. PI3K p85 (CST-4292) 1:1000, PI3K p110α (CST-4249), GAPDH (CST-5174) 1:5000.
Viral Vector Constructs
The retroviral vector system used for in vivo delivery of Cre and ARF6Q67L has been previously described(17). ARF6Q67L cDNA was provided in pCDNA3.1 by Zongzong Tong (Navigen, Inc.). RCAS-ARF6Q67L-HA-IRES-Cre was cloned by PCR amplification of ARF6Q67L using primers with EcoRI and NotI sites and then inserting the PCR product into a pENTR3c (Invitrogen) vector containing HA-IRES-Cre. This was followed by recombination into RCASBP(A) DV using LR Clonase Enzyme Mix (Invitrogen), per the manufacturer’s specifications. pENTR3c, Gateway reagents, and RCAS-Cre were provided by the S. Holmen lab (Huntsman Cancer Institute, University of Utah). The adenoviral constructs ARFQ67L and ARFT27N were provided by Zongzhong Tong (Navigen, Inc.) and then amplified by Vector Biolabs (Malvern, PA, USA). Ad-CMV-Null empty vector control was purchased directly from Vector Biolabs (Malvern, PA, USA).
Matrigel invasion assay
Twenty four hours after adenoviral transduction, invasion was performed as previously described(2) with minor modifications. 5 × 104 infected cells were seeded into chambers. Either vehicle (0.1% DMSO), MK2206 (1μM) or GDC-0941 (1μM) was added to both chambers. After 48 h incubation, media was replaced in the bottom chamber with Calcein AM (BD Biosciences) according to manufacturer’s instructions. Calcein-stained cells were visualized with a fluorescent microscope and quantified with plate reader at 485/530 nm.
Gelatin invasion assay
Invasion into gelatin labeled with Oregon Green 488 was performed as previously described(23,24), with minor modifications. In 8-well chambered coverglass, 25μl casts of gelatin were dried, fixed, quenched in PBS containing 0.5M glycine and then overlayed with 10μL Matrigel polymerized for 30–60 minutes at 37°C. The matrix was equilibrated in DMEM +10% FBS for 30 minutes at 37°C before adding cells. Prior to invasion, A2058 cells were transduced for 24 hours with 109 pfu/ml adenoviruses per 10cm2 dish. Cells were then treated with mitomycin C in preparation for invasion as described previously(2).
Immunofluorescence and confocal microscopy of invading cells
Prior to invasion, A2058 cells were cultured to 80% confluence and transduced for 24 hours with 109 pfu/ml adenoviruses per 10cm2 dish. Cells were then treated with mitomycin C in preparation for invasion as described previously(2). 24 hours prior to Matrigel invasion, Matrigel GFR (B-D) was thawed on ice overnight at 4°C. Transduced cells were harvested, washed three times in high glucose DMEM + Pyruvate + 0.1% FBS and resuspended to1.5 × 105/ml. During the cell preparation period, 40μl ice-cold Matrigel was pipetted to the center wells of an 8-well chambered coverglass (Thermo Fisher Scientific) on ice. 4μl ice-cold PBS was added to each 40μl dome of Matrigel. The domes were polymerized at 37°C /5% CO2 for 30 minutes. 350μl of A2058 cell suspension were added to each well and cells were incubated at 37°C / 5% CO2 for 48 hours. Media was gently aspirated and wells were rinsed once in 10% Neutral Buffered Formalin, then fixed for 30 minutes at room temperature in same, followed by three rinses in ‘PBSA’ (1xPBS +0.1% sodium azide.) Matrigels were permeablized for 40 minutes in PBSA+ 0.02%TritonX-100 at room temperature, followed by a blocking in PBSA + 3% v/v bovine serum albumin (BSA) for 60 minutes. Primary antibodies to pAKT S473 (CST-4060) and cortactin (Abcam 33333) were diluted 1:100 and 1:300, respectively, in PBSA + 3%BSA, and incubated on Matrigels overnight at 4°C. Antibodies were then aspirated and the wells were washed 4 × 20 minutes in PBSA. Signals were detected by fluorescently conjugated secondary antibodies; Donkey anti-Rabbit IgG Alexa 488, (pAKT) and Donkey anti-Mouse IgG, Alexa 594 (cortactin), diluted to 10μg/ml in PBSA+0.1% BSA and incubated for 50 minutes at room temperature in a darkened box. Wells were aspirated by gentle vacuum and washed 4 × 20 minutes at room temperature in PBSA. 5μg/ml DAPI in PBSA for added for 30 minutes, followed by 5 min washing in PBSA, and mounting in 40% w/v Polyvinyl pyrrolidone +4% v/v glycerol+0.1% sodium Azide dissolved in 1M Tris,pH 8.0. Images were collected on an Olympus Fluoview1000 scanning laser confocal microscope at 600x magnification and 1200x magnification. Invasion into gelatin labeled with Oregon Green 488 was performed as previously described(23,24), with minor modifications. In 8-well chambered coverglass, 25μl casts of gelatin were dried, fixed, quenched in PBS containing 0.5M glycine and then overlayed with 10μL Matrigel polymerized for 30–60 minutes at 37°C. The layered matrix was equilibrated in DMEM +10% FBS for 30 minutes at 37°C before adding cells. Staining of A2058 cells in gelatin followed 20 hours of invasion, fixation for 30 minutes at room temperature in freshly prepared 10% neutral buffered formalin, four washes in PBSA and storage, tightly wrapped, at 4°C. Wells were permeablized for 10 minutes in PBSA + 0.1% Triton X-100, followed by three washes in PBSA. Blocking, washing and primary and secondary antibodies were applied as described above. A glycerol-based antifade mount was applied. Images were collected with oil immersion at 1200x and 1800x magnification on an Olympus FV1000 laser scanning confocal microscope. For orthogonal views of invasion, 85 × 0.2μm, Z stacks were generated, (17μm depth) and 3-D animation images were produced in the FluoView software.
Tumor cells derived from BrafCA;Cdkn2af/f ± Arf6Q67L mice were plated to fibronectin-coated, chambered coverglass slides at a density of 6 × 104/ml or 5 ×103/well, and allowed to attach for 20 hours. Monolayers were fixed in 10% neutral buffered formalin x 20 minutes at room temperature, followed by washing x 3 in PBS + 0.1% NaZ. Wells were blocked and permeabilized x 45minutes in PBS + 0.1% NaZ + 0.01% MgCl2 +0.01% CaCl2 (PNMC solution) + 0.1% Saponin + 1%BSA, followed by incubation in primary antibodies. Rabbit anti-HA (CST-3724) 1:400. Mouse anti-Pi3Kinase: anti-p85 (Abcam ab86714) 1:100 or anti-p110 (B-D Biosciences 611398) 1:100, was diluted in PNMC solution and incubated overnight at 4°C. Slides were rinsed x 3 in PNMC solution. Secondary antibodies (Donkey-anti-Rabbit IgG Alexa 594 conjugate, Donkey anti-Mouse IgG Alexa 488 conjugate, Thermo Fisher Scientific) were applied at 10μg/ml each in blocking buffer x 2 hours in darkened chamber on ice. Slides were rinsed x 3 in PNMC solution and wells were mounted in a solution of 90% glycerol, 5μg/ml DAPI, PBS + 0.1% NaZ + 0.05% DABCO + 0.5% n-Propyl Gallate + 0.01% p-Phenylenediamine, with storage at 4°C in darkened chamber. Z-stacked images in 5 × 0.5μm slices were taken on an Olympus FV1000 scanning laser confocal microscope at 1200x magnification. Images were assessed by three independent, blinded investigators who evaluated a composite of random images from each condition. Quantification of p110α PI3K fluorescent signal on bulbo-spinous processes of mouse melanoma cells was performed according to standard protocols described in ImageJ tutorials (https://imagej.net/Image_Intensity_Processing#Getting_intensity_values_from_multiple_ROIs). Briefly, all regions of interest (ROI) were extracted from the RGB confocal images using the freehand selection tool. ROIs were uniformly thresholded and signal intensity was counted in ImageJ 64. Signal intensity was normalized to total pixels (surface area) in each ROI. All bulbo-spinous processes were quantified for all cells in eight distinct images per cell line.
RNA interference and viral transduction of cell lines
20 nM siRNA duplexes were transfected into cell lines using RNAiMax (Invitrogen) following the manufacturer’s recommendations. For A2058 cells, 70uL of RNAimax was used with 40nM siRNA. Forty-eight hours after transfection the cell lines were split 1:2, allowed to adhere for six hours, re-transfected, and allowed to grow for an additional 24 hours before harvesting. AllStars Negative Control siRNA (Qiagen) and ARF6-SMART pool (Dharmacon) siRNAs were used in these experiments. NIH3T3 cells were serum starved (0% FBS, DMEM) for six hours prior to adenoviral transduction and then incubated in virus-containing media for 18 hours prior to harvest of lysates. Transduction of A2058 cells was performed in whole media (10%FBS, DMEM) for 24 hours prior to harvest for invasion. Adenovirus was applied to A2058 and NIH3T3 at 1.0 × 107 pfu/10mL media.
Quantitative RT-PCR
RNA was extracted from fresh frozen mouse tumor using RNeasy Mini Kit with RNase Free DNase set (Qiagen). cDNA was synthesized from 5 ng of total RNA using either qScript cDNA Super mix (Quanta) or RETROscript reverse transcription kit (Invitrogen). Quantitative RT-PCR was performed with the Life Technologies QuantStudio 12K Flex instrument and QuantiTect SYBR Green PCR kit (Qiagen) with the following primers to ARF6-Q67L-HA: fw-GACAGGAACTGGTATGTGCA and rev-TCGTATGGGTAAGATTTGTAG. Primer amplification efficiency was validated and relative expression of Arf6Q67L-HA was determined by the ΔΔCT method. Samples were run in triplicate and normalized to Gapdh using Mm_Gapdh_3_SG Primer (Qiagen).
Histology
Mouse tissues were flash frozen or fixed in 10% neutral buffered formalin overnight, stored in 70% ethyl alcohol. Paraffin-embeddin and histologic sections were performed by the Research Histology Section of the Biorepository Molecular Pathology Laboratory in the Huntsman Cancer Institute, University of Utah.
Immunohistochemistry
Tissue sections were de-paraffinized and re-hydrated. Antigen retrieval was performed in citrate buffer (pH 6.0) at 120°C for 10 min. Sections were treated with 3% H2O2 and blocked with normal goat serum in 0.05% TBS-T for 60 min and washed x3 with 0.05% TBS-T. Primary antibodies were diluted in SignalStain Ab Diluent (Cell Signaling Technology). Sections were incubated overnight at 4°C with primary antibody, washed 5 min in 0.05% TBS-T and then probed with SignalStain Boost Detection Reagent-rabbit or mouse (Cell Signaling Technology) for 30 min at room temperature in a humidified chamber. Visualization was carried out with SignalStain DAB (Cell Signaling Technology). Sections were counterstained with hematoxylin. Antibodies against the following antigens were used: HA-tag (CST-3724) 1:500; phospho-AKT (Ser 473) (CST-4060) 1:100; Ki67 (MKI67) (UMAB107)1:300.
PI3K Catalytic Assays
PI3K activity was measured from endogenous PI3K complex, as previously described(25). Cell lysates were prepared with IP lysis buffer (Thermo Fisher Scientific). PI3 kinase complex from 1 mg of cell lysates was immunoprecipitated using protein A/G (Thermo Fisher Scientific) conjugated anti-p85 antibody (CST-4292) for 18hours at 4°C, and then washed three times in IP lysis buffer and one time in KBZ buffer (Echelon). After washing, PI3K activity was measured with PI3-Kinase Activity ELISA: Pico kit (Echelon).
Procurement of Patient Samples
Excess tumor and paired nontumor tissues from routine surgical resections at the Huntsman Cancer Institute were collected and flash frozen in liquid nitrogen, under University of Utah Institutional Review Board-approved protocol (#10924), by the Biorepository Molecular Pathology shared resource at the Huntsman Cancer Institute. Quality control (viability, tumor content) was performed by routine histology by a board-certified pathologist (A.H.G.).
ARF6-GTP pull-downs from patient samples
Human samples were prepared and ARF6-GTP pull-downs were performed and quantified as previously described(7).
TCGA melanoma survival analysis
ARF6 pathway genes (Table S1) for differential expression query were selected based on published literature. TCGA melanoma RNA-Seq data were extracted from all melanoma TCGA RNA-Seq data. Samples without survival data were excluded. Each gene was evaluated individually and those with statistical significance were then analyzed in the Stage III sub-cohort. Specifically, Stage III lymph node and skin metastases, as designated by TCGA, were selected for analysis. TCGA metadata were acquired from the GDC release 10.4, in which survival times were generated using days_to_death and days_to_last_follow_up data. Samples were stratified into ARF6-GAP-high (indicating higher overall ARF6-GAP gene expression) and ARF6-GAP low groups by median centering of expression levels for each gene. Survival p-value were calculated by log-rank (Mantel-Cox) test.
Statistical Analysis
For most experiments, statistical analyses were performed using GraphPad Prism 6.0f.
For statistical analyses of immunoblots from cell lines, the density of each band was normalized to an internal control protein and then the ratio of the normalized density of the band from the experimental treatment to the normalized density of the paired control treatment band was obtained. For patient samples, signals were normalized to total protein input as previously described(7). The 2×4 Fisher’s Exact test and the Linear by linear association test was performed with IBM SPSS Statistics software. For animal studies, sample size estimates were based on results from pilot studies and a power analysis was performed with G*Power software.
Results
ARF6-GTP accelerates melanoma metastasis and potentiates lung colonization in BrafCA;Cdkn2af/f mice
The impact of ARF6 on cancer progression in immunocompetent hosts is unknown, therefore we asked if expression of activated ARF6 (ARF6-GTP) alters tumor incidence, growth or metastasis in a clinically relevant genetically engineered mouse model (GEMM) of cutaneous melanoma(17). Invasion plays a role in multiple steps of the metastatic cascade, including early steps of primary tumor escape and intravasation into the lymphovascular system, and later extravasation. ARF6 is necessary for invasion in vitro, so we hypothesized that ARF6 activation would be sufficient to induce metastasis. To test our hypothesis, we utilized RCAS-mediated delivery of Cre recombinase specifically in the melanocytic lineage(17,26) to induce constitutive activation of BRAFV600E (BrafCA) and deletion of Cdkn2a (Cdkn2af/f). BrafCA;Cdkn2af/f mice display a high incidence of primary tumors. Although Cdkn2a loss is expected to cause invasion and metastases(27), metastasis has not been observed in BrafCA;Cdkn2af/f mice without additional alterations such as Pten loss or AKT activation(17).
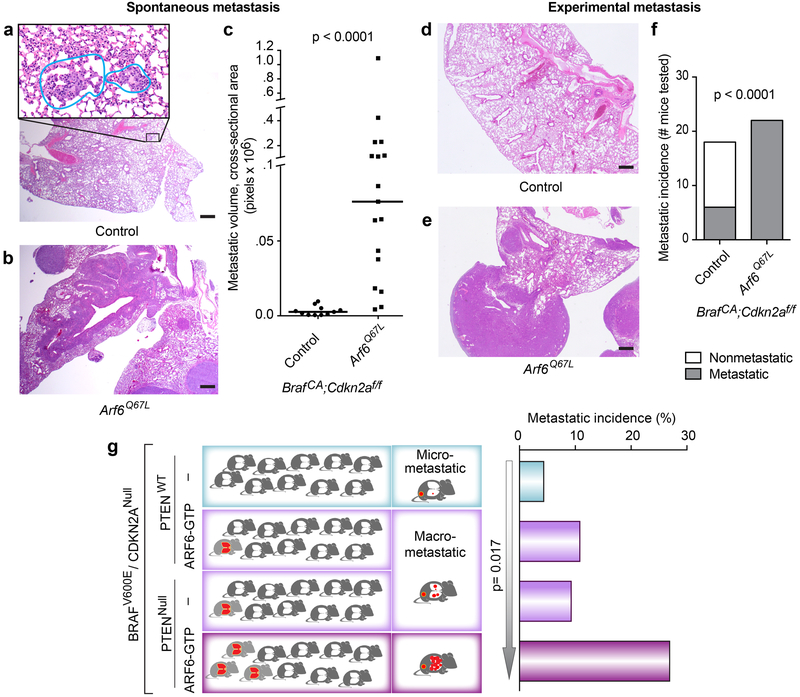
To model activated ARF6 (ARF6-GTP), we delivered a constitutively active mutant form, Arf6Q67L, along with Cre in RCAS, injected subcutaneously into the flank of Dct::TVA;BrafCA;Cdkn2af/f mice. We confirmed expression of HA-tagged ARF6Q67L by immunohistochemistry (IHC) (Figure S1a) and/or by qRT-PCR (Figure S1b). Tumor incidence, growth, and survival were similar between BrafCA;Cdkn2af/f (control) and BrafCA;Cdkn2af/f + Arf6Q67L mice (Figure S2a–c), indicating that ectopic ARF6-GTP does not alter primary tumorigenesis in this model. There was, however, an increase in spontaneous metastatic disease. Overall, the metastatic incidence increased from 4.3% to 11.1% in the ARF6Q67L cohort (Figure S2d), consistent with ARF6 driving metastasis. In mice with metastases, there was a dramatic increase in metastatic volume in the ARF6Q67L cohort (Figure 1a–c). Control mice showed negligible metastatic disease with infrequent and very early microscopic foci of tumor in the lungs (Figure 1a and c), whereas mice with ARF6Q67L demonstrated bulky metastatic disease (Figure 1b and c). Thus, ARF6-GTP was sufficient to expedite the metastatic process, leading to accelerated disease progression.
Figure 1. ARF6-GTP is sufficient to potentiate metastasis of BRAFV600E / Cdkn2aNULL tumors, and is equipotent as PTENNULL.
(a-c) Spontaneous pulmonary metastatic burden. (a) Micro-metastases (insert, outlined) from BrafCA;Cdkn2af/f (control) mouse. (b) Multiple, large, spontaneous metastases in BrafCA;Cdkn2af/f + Arf6Q67L mouse. (c) Metastatic volume in mice with metastases, Mann-Whitney test, two-tailed. (d-f) Lung colonization of tumor cells derived from BrafCA;Cdkn2af/f ± Arf6Q67L mice, injected into tail veins of NOD SCID mice. d) Benign lung, control mouse. e) Lung with tumor colonies. f) Number of mice with pulmonary tumor colonies by 4 weeks post-injection, Fisher’s exact test, two tailed. H&E images = 20x magnification (larger images, scale bar = 500μm) with 400x magnification of metastatic focus (panel a, insert, scale bar = 20μm). (g) Metastatic incidence and volume from BRAFV600E/CDKN2ANull tumors +/− ARF6-GTP or PTEN or both. Histogram (right): overall significance was evaluated by 2×4 Fisher’s exact test, p=0.042. 2×2 Fisher’s exact test, two-tailed, p=0.0498 for PTENNull vs. PTENNull+ ARF6-GTP. Arrow = Linear by Linear association test, based on the order shown. Diagram (left) illustrates the overall phenotypic patterns for each cohort, where the right column reflects the volume of disease burden (red) in metastatic mice and the left column reflects the overall incidence of metastatic mice (red). See also Figure S4e.
The metastatic cascade is a multistep process involving invasion and escape from the primary tumor, intravasation (invasion into lymphovascular system), survival in the circulation, extravasation (invasion into a distant organ), and survival and growth in distant organs(28). The pro-invasive role of ARF6 in cancer cells in vitro is well-established and consistent with our observation that ARF6 activation increases metastatic disease burden. A role for ARF6 in proliferation or survival is also possible. Although ARF6Q67L primary tumors did not show a growth advantage over controls, ARF6Q67L primary tumor cells showed increased proliferation and decreased apoptosis in vitro (Figure S2e and f), demonstrating context-dependent functions of activated ARF6. In contrast, ARF6-GTP had no impact on proliferative activity in primary tumors (Figure S2g). Together these data suggest that ARF6-GTP might convey a proliferative or survival advantage specifically to tumor cells en route to or colonizing distant organs. In addition to facilitating primary tumor escape of a metastatic cell, ARF6 may function later in the metastatic cascade. The increase in metastatic volume suggests a role for ARF6 after initial primary tumor escape.
To test the hypothesis that activated ARF6 promotes a late step in metastasis, we employed a tail vein-injection model of experimental metastasis. This approach monitors survival in the circulation, extravasation, invasion into distant organs, and/or end-organ colonization. We derived four mouse melanoma cell lines from BrafCA;Cdkn2af/f ± Arf6Q67L mouse tumors, delivered them separately to the circulation by tail vein injection into NOD SCID mice, and evaluated lung histology for metastases within 4 weeks post-injection. We found that ARF6-GTP dramatically enhanced metastatic lung colony formation in this model (Figure 1d–f). Specifically, circulating tumor cells expressing ARF6Q67L colonized the lungs in 100% of mice, whereas controls colonized the lungs in 33% of mice (Figure 1f). These data are consistent with previous work showing that expression of an inactive, GDP-bound form of ARF6 (T27N) in human LOX melanoma cells, injected into tail veins of Nude mice, reduces lung colonization(5). Thus, ARF6 activation provides a selective advantage for survival of circulating tumor cells, extravasation, end-organ invasion and/or establishment of metastatic colonies while inactivation of ARF6 curbs this pro-metastatic phenotype.
ARF6-GTP is sufficient to potentiate metastasis and exacerbates melanoma metastasis upon PTEN loss
In order to determine how ARF6 affects other known drivers of metastatic melanoma, we directly compared the BrafCA;Cdkn2af/f + Arf6Q67L mice to BrafCA;Cdkn2af/f;Ptenf/f mice(17). All mice engineered to express HA-tagged ARF6Q67L in tumors were confirmed by anti-HA immunostaining (see Figure S1a for examples) and/or by qRT-PCR (Figure S3). The overall incidence of primary tumor development was significantly higher and tumor onset (disease latency) was dramatically shorter in Ptenf/f mice (Figure S4a and b). Individual tumor growth rates were highly heterogeneous in Ptenf/f mice, causing erratic growth curves in the cohort, but trended higher than Arf6Q67L mice (Figure S4c). The difference in tumor growth was best represented in the survival data, where larger primary tumors in Ptenf/f mice translated into reduced survival after tumor onset (Figure S4d). No mice were sacrificed due to symptoms from metastasis. Rather, our experimental endpoint was limited by primary tumor growth and related symptoms that mandated animal sacrifice. These data suggest that PTEN loss accelerates tumorigenesis in BRAFV600E/Cdkn2aNull melanoma, in contrast to ectopic expression of ARF6-GTP.
Despite lagging behind in tumor incidence, disease latency and tumor growth, the Arf6Q67L mice demonstrated a metastatic incidence that is equivalent to the Ptenf/f mice (Figure 1g). Both cohorts showed an increase in overall metastatic incidence to 10–11% from about 4%, and both showed equivalent bulky metastatic disease (Figure S4e). No brain metastases were detected in the Ptenf/f mice, as previously reported(17), nor the Arf6Q67L mice. These phenotypes indicate that while PTEN loss incites aggressive primary tumor growth as well as progression to metastasis, ARF6-GTP function in this model specifically promotes metastasis. This is an important distinction, because it demonstrates that activation of ARF6 can promote a pro-metastatic state that is independent of tumor growth, possibly increasing the risk of distant spread in smaller tumors, a notorious clinical feature of melanoma.
We previously reported that PTEN loss cooperates with activated AKT1 in the BrafCA;Cdkn2af/f model to induce a more aggressive metastatic phenotype than each allele individually(17). Similarly, we found that combining PTENNull with ARF6-GTP dramatically and significantly increased the metastatic incidence and volume compared to each allele alone (Figures 1g and S4e). A linear-by-linear association test revealed a statistically significant increase in disease severity along an ordered spectrum. Thus, similar to AKT(17), activated ARF6 cooperates with PTEN loss to promote metastatic melanoma.
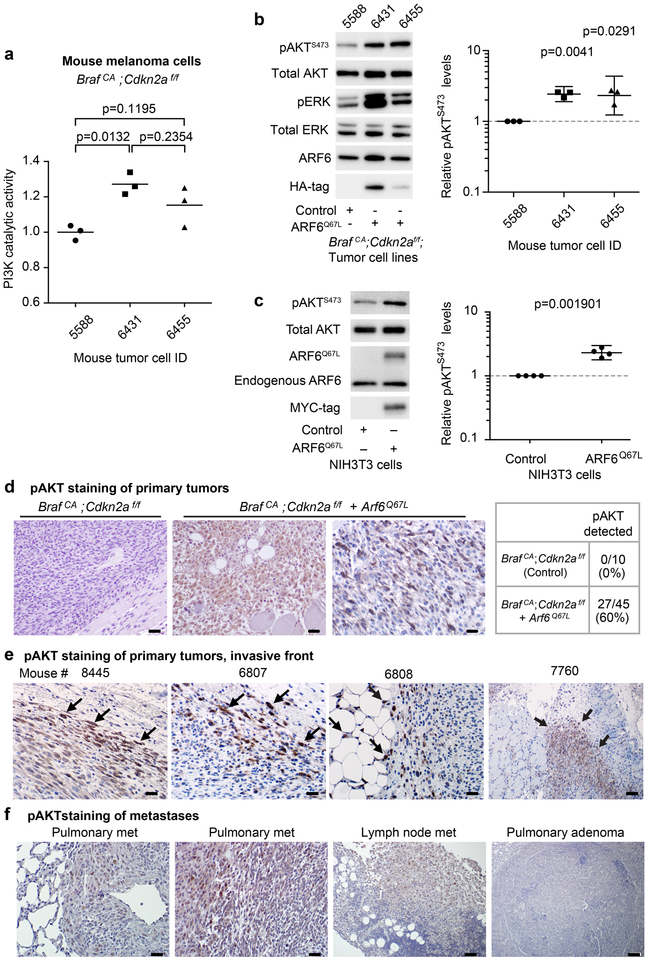
ARF6 activation leads to increased PI3 Kinase (PI3K) expression and AKT activation in BRAFV600E/CDKN2ANull tumors
ARF6 facilitates signaling of multiple proteins with direct and indirect transcriptional output, including ERK(3,7,10), PLC(7), Hedgehog(29), β-catenin(2,10) and YAP(7). These data suggest that ARF6 may impact global expression changes. To identify novel ARF6-regulated target genes and pathways, we analyzed transcriptomes from our BRAFV600E/CDKN2ANull ± ARF6Q67L mouse melanoma tumors. We detected a unique gene expression signature in the ARF6Q67L tumors (Figure 2a). This signature included upregulation of gene biogroups related to known ARF6-dependent cell functions in trafficking, cell adhesion, motility, providing an internal positive control that helps to validate the utility of RNA sequencing as a discovery tool for ARF6.
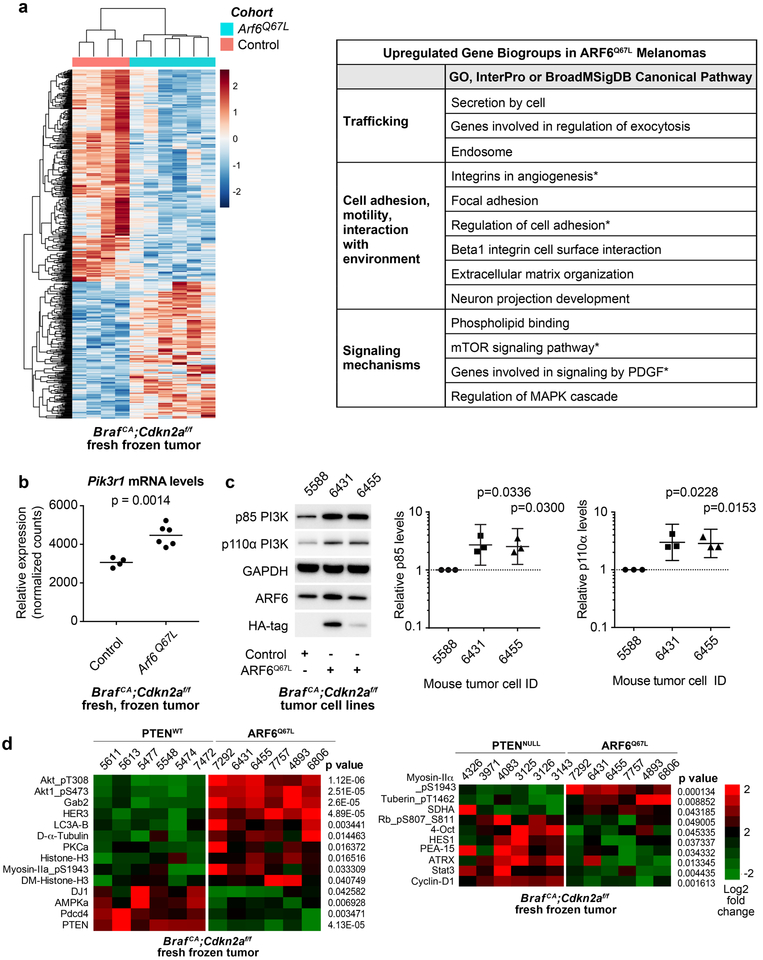
Figure 2: ARF6-GTP upregulates PI3K expression and AKT activation.
(a) RNA sequencing of mouse tumors reveals differential expression between melanomas from control (n=4) and Arf6Q67L mice (n=6). Left panel = heat map of relative expression shown, range of 2 to −2 (log2). Right panel = Upregulated gene biosets in ARF6Q67L tumors. * = gene sets that include Pik3r1. (b) ARF6Q67L induces 1.4503-fold increase in Pik3r1 expression in tumors. Unpaired t-test, two-tailed. (c) In mouse tumor cell lines derived from a distinct group of mice from those sequenced, ARF6Q67L increases both p85 and p110 PI3K protein levels. Ratio paired t-test, two-tailed. HA immunoblot detects ectopic HA-tagged ARF6Q67L. (d) Heatmap representing differences in signaling protein levels detected with reverse phase protein array. Columns represent individual mouse tumors. Unpaired t-test, two-tailed.
Interestingly, mTOR signaling components were upregulated in ARF6Q67L tumors (Figure 2a), including Pik3r1, which encodes the p85 regulatory subunit of PI3 kinase (PI3K). This finding is provocative because the PI3K pathway is strongly linked to melanoma disease progression and because our ARF6Q67L mice showed a comparable metastatic phenotype to mice with PTEN loss. Transcript levels of Pik3r1 were increased in ARF6Q67L tumors compared to BrafCA;Cdkn2af/f controls (Figure 2b). Likewise, p85 protein levels were significantly increased in primary tumor cell lines derived from ARF6Q67L mouse melanomas (Figure 2c). The p85 regulatory subunit facilitates PI3K signaling by stabilizing the p110 catalytic subunit of PI3K(30), and through recruitment of p110 to the plasma membrane (reviewed in(31)). Consistent with this function, we detected increased p110α protein levels in BrafCA;Cdkn2af/f + ARF6Q67L melanoma tumor cell lines (Figures 2c). Pik3ca mRNA levels were not altered in the ARF6Q67L tumors, suggesting that the increased p110 protein is secondary to ARF6-GTP – mediated increases in p85.
In order to broadly query for signaling proteins altered by ARF6 activation, we tested our mouse tumors with reverse phase protein array (RPPA). Compared to BRAFV600E/Cdkn2aNULL controls, ARF6Q67L tumors showed the most dramatic increase in phosphorylated AKT (pAKT) levels (Figure 2d). Other pathways upregulated include components of receptor tyrosine kinase signaling (HER3 and GAB2), calcium signaling (PKCa), autophagy (LC3A-B), dynamic chromatin structure (histone H3 and di-methyl histone H3), and cell motility (myosin IIa phosphorylation and α-tubulin (32–35)). Interestingly, myosin IIa upregulation distinguishes ARF6Q67L tumors from PTENNULL tumors (Figure 2d). ARF6Q67L tumors also show a stronger signal for tuberin T1462 phosphorylation than PTENNULL tumors. As Tuberin T1462 is a PI3K-regulated AKT substrate and constitutively phosphorylated in PTENNULL cells(36), these data suggest that ARF6-GTP is a potent activator of PI3K and/or AKT. Compared to ARF6Q67L, PTENNULL tumors show upregulation of cyclin-D1 and phosphorylation of RB, which could explain how the PTENNULL primary tumor growth rate exceeds ARF6Q67L tumors. Together, these findings help explain the overlapping but distinct phenotypes between PTENNULL and ARF6Q67L. ARF6-GTP may compensate for the lag in tumor growth, compared to PTEN loss, by activating cell motility pathways, ultimately reaching the same metastatic endpoint.
ARF6-GTP enhances PI3K distribution to cellular protrusions
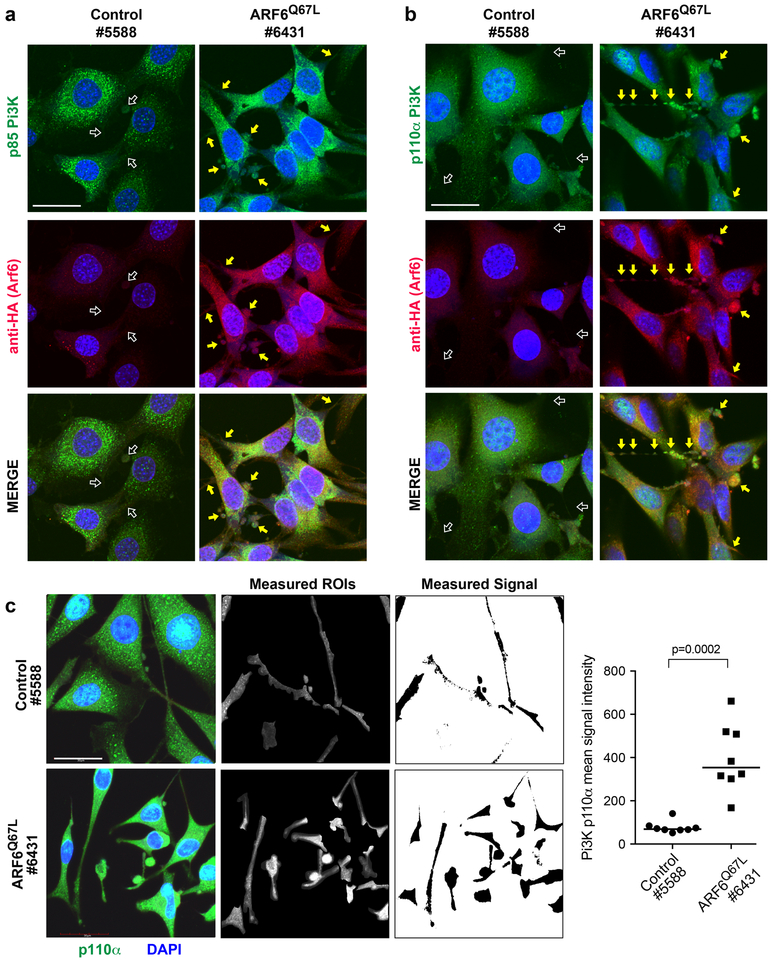
Because ARF6 is critical for intracellular trafficking, we next evaluated the localization of p85 and p110 in these tumor cells. BrafCA;Cdkn2af/f control mouse melanoma cells grown in vitro were polygonal shaped, whereas BrafCA;Cdkn2af/f+Arf6Q67L tumor cells were strikingly elongated/ spindle shaped (Figure 3 and S5). Both groups demonstrated prominent spinous/dendritic processes. Immunoflourescent staining confirmed that p85 and p110α levels increased in the presence of ARF6Q67L (Figure 3), consistent with transcriptome and Western blot analysis (Figure 2c and d). The localization pattern of endogenous p85 did not appear to change with ARF6 activation (Figure 3a), remaining mostly in the cell body. In contrast, endogenous p110α was more detectable in peripheral, protrusive structures of ARF6Q67L cells: in both bulbous and spinous processes (Figure 3b and c). These data suggest that ARF6-GTP is sufficient to control, either directly or indirectly, the subcellular localization of the catalytic subunit of PI3K to specific peripheral compartments within melanoma cells.
Figure 3: ARF6-GTP increases the distribution of p110α PI3K in cellular protrusions in melanoma.
Primary melanoma cells derived from mouse tumors, 1200x magnification, scale bars = 30μm. Left panels = BrafCA;Cdkn2af/f control (5588). Right panels = BrafCA;Cdkn2af/f + Arf6Q67L (6431), where arrows highlight bulbous and spinous cellular projections. (a) Green = anti-p85 PI3K. (b) Green = anti-p110a PI3K. (a-b) Red = anti-HA (HA-tagged ARF6Q67L). Blue = DAPI. (c) Quantification of p110α PI3K (green) in bulbs and spines (outlined regions, right panel). ROI=regions of interest selected for quantification of p110α signal intensity. All bulbo-spinous processes were quantified for all cells in eight distinct images per cell line Representative images shown, 1800x magnification, scale bars = 20μm. Mann-Whitney U test.
ARF6-GTP is sufficient for PI3K and AKT activation
Thus far, our data suggest that ARF6 activation upregulates expression and distribution of PI3K. Whether ARF6 also controls PI3K activation in melanoma, or any other cancer, is unknown. We asked whether ARF6-GTP is sufficient for activation of PI3K, or of its effector AKT, in our melanoma mouse model. We measured endogenous p110 PI3K catalytic activity in anti-p85 PI3K immunoprecipitates, as previously described(25). We found that the catalytic activity of endogenous PI3K increased in the presence of ARF6Q67L (Figure 4a). Further, we observed higher levels of activated AKT (pAKT) in ARF6Q67L mouse melanoma cells relative to controls (Figure 4b). Ectopic expression of ARF6Q67L was also sufficient to activate AKT in NIH3T3 cells (Figure 4c), suggesting a broader phenomenon.
Figure 4. ARF6-GTP is sufficient for PI3K and AKT activation.
(a) Catalytic activity of endogenous PI3K in mouse tumor cell lines, one–way ANOVA with Tukey’s multiple comparisons test (a). (b) Mouse melanoma cells BrafCA;Cdkn2af/f control (5588), BrafCA;Cdkn2af/f + Arf6Q67L (6431 and 6455). (c) Adenoviral delivery of MYC-tagged ARF6Q67L to serum-starved NIH3T3 cells. (b-c) Graphs show individual data points normalized to control along with geometric means, 95% C.I., ratio paired t test, two tailed. Phosphorylated AKTS473 detected by immunohistochemistry in (d) primary tumors from Arf6Q67L mice but not BrafCA;Cdkn2af/f controls. Scale bars = 20μm, 400x magnification. (e) pAKT stains tended to highlight tumor cells at the invasive front (arrows). Mouse tumors 8845, 6807 and 6808 scale bars = 20 μm, 400x magnification. In 6807 pAKT highlights tumor cells invading edematous subcutaneous tissue (top right of image). In 6808, pAKT highlights tumor cells invading adipose (left portion of image). Mouse 7760 shows pAKT highlighting tumor invading skeletal muscle, scale bar = 50 μm, 100x magnification. (f) Heterogeneous pAKT staining of metastatic tumors. Pulmonary metastasis (met), scale bars = 20 μm, 400x magnification. Lymph node met scale bars = 50 μm, 200x magnification. BrafCA;Cdkn2af/f mice are known to develop benign pulmonary adenomas (unrelated to melanoma) and these fail to stain with pAKT, providing a negative internal control (right panel, scale bar 100μm, 100x magnification).
In order to understand the role of ARF6-GTP in PI3K-AKT activation in vivo, we evaluated pAKT in histologic sections of our primary mouse tumors. Immunohistochemical staining revealed that pAKT is detectable in more than half of primary tumors from ARF6Q67L mice whereas pAKT was not detected in primary tumors from control mice (Figure 4d). Overall, pAKT staining was highly heterogeneous, detectable in only a fraction of the tumor cells (Figure 4d and e). Activated AKT was often limited to cells at the invasive front, at the interface with subcutaneous fibro-edematous connective tissue, skeletal muscle and/or adipose (Figure 4e). Activated AKT was also detected in metastatic tumors from BrafCA;Cdkn2af/ +Arf6Q67L mice (Figure 4f). Together these results demonstrate that ARF6-GTP is sufficient for activation of the PI3K pathway and suggest that ARF6-GTP may engage PI3K signaling in specific cell populations within a tumor.
ARF6 is necessary for PI3K and AKT activation in human cancer cells
In order to test if ARF6 is required for PI3K/AKT signaling and to extend our observations to human melanoma, we expressed a dominant negative ARF6 (ARF6T27N) in human melanoma A2058 cells. Ectopic expression of ARF6T27N significantly reduced PI3K catalytic activity in A2058 cells (Figure 5a). Further, ectopic expression of ARF6T27N reduced pAKT levels in 10 of 15 melanoma cell lines, independent of the primary driver gene mutation status (Figure 5b, c and S6a). ARF6T27N also reduced pAKT levels in a KRAS-mutant carcinoma cell line (HEY-T30), suggesting that ARF6 also regulates PI3K signaling in non-melanoma cancer cells (Figure 5b and S6a). Knockdown of ARF6 also reduced pAKT levels (Figure 5d and S6b). In a proteomic phosphokinase array, ARF6T27N incited the greatest impact on AKT and its substrate PRAS40, reducing phosphorylation of both (Figure S6c). Importantly, this proteomic survey corroborates our findings in mouse melanoma cells whereby AKT phosphorylation is one of the strongest signals in ARF6Q67L tumors (Figure 2e). Together, our in vitro and in vivo findings demonstrate that ARF6 is necessary and sufficient for PI3K activation and AKT signaling.
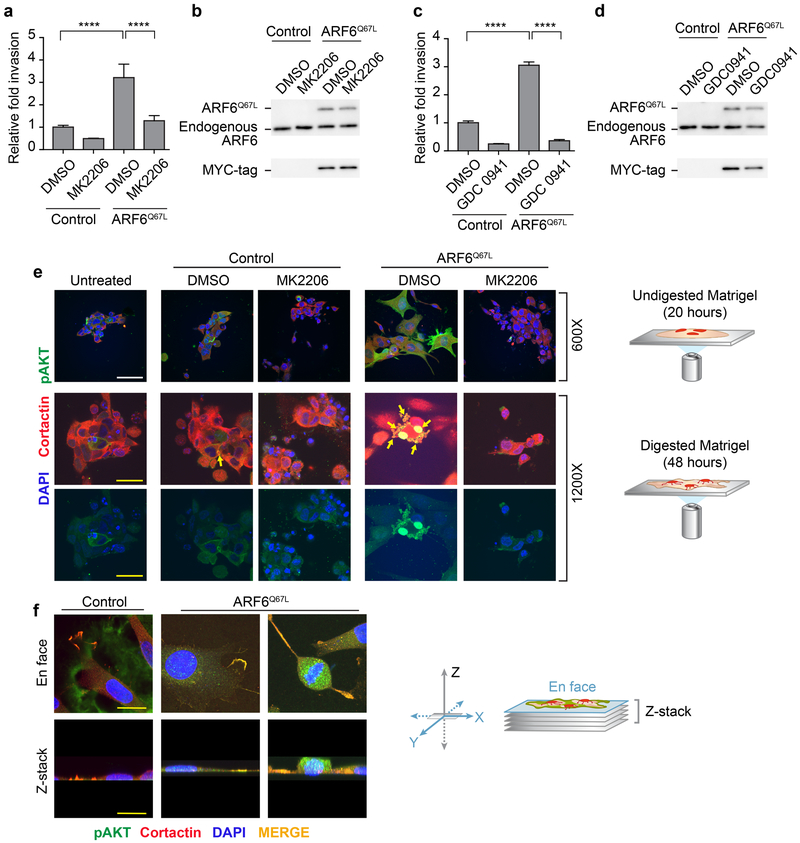
The PI3K/AKT pathway is necessary for ARF6-mediated invasion.
In the presence of ARF6-GTP, p110 PI3K localized to peripheral cellular protrusions (Figure 3b), As ARF6 has a prominent role in invadopodia function(3,5), our data suggest that ARF6 engages the PI3K-AKT pathway to facilitate invasion. The PI3K-AKT pathway has been implicated in cell motility/invasion(37), including in melanoma(17). We previously reported that knockdown of ARF6 or pharmacologic inhibition of ARF6 universally reduces invasion of human cutaneous melanoma cell lines in vitro(2). Muralidharan-Chari et al. showed that ectopic ARF6T27N inhibits invasion, whereas ARF6Q67L increases invasion of LOX melanoma cells(4). Consistent with these findings, ectopic ARF6Q67L increased Matrigel invasion of A2058 melanoma cells in vitro (Figure 6). With this system, we tested whether PI3K and AKT kinase activity are necessary for ARF6-mediated invasion. Indeed, the pan-AKT inhibitor MK2206 (Figure 6a and b) and the pan-Class I PI3K inhibitor GSK-0941 (Figure 6c and d) both abrogated ARF6Q67L-dependent Matrigel invasion, demonstrating that PI3K signaling is necessary for ARF6-mediated invasion.
Figure 6. PI3K and AKT are necessary for ARF6-GTP-dependent melanoma invasion.
(a,c) Matrigel invasion of A2058 cells is increased by ectopic expression of Myc-tagged ARF6Q67L (a-d). (a) The AKT inhibitor MK2206 (1μM) or (c) the PI3K inhibitor GDC0941 (1μM) eliminates ARF6Q67L-induced invasion. Error bars = S.D., ****p<0.0001, ANOVA with Tukey’s multiple comparisons test. (b, d) MYC-tagged ARF6Q67L expression relative to endogenous ARF6. (e) pAKT levels and localization in invading A2058 cells, plated on Matrigel dome, serum starved in PBS. Top row = 20 hours after plating, 600x magnification, scale bars = 60μM. Middle and bottom rows = 48 hours after plating, 1200x magnification, scale bars = 30μm. (f) ARF6Q67L–induced pAKT and cortactin colocalization in A2058 cells invading and degrading gelatin (labeled with Oregon Green 488).
In order to visualize the effect of ARF6-GTP on the PI3K-AKT pathway in invading melanoma cells, we plated A2058 cells on a dome of Matrigel and imaged cells penetrating and digesting the matrix. Cells were co-stained with anti-cortactin, a marker of invadopodia(38), and anti-pAKT (Figure 6e). Ectopic expression of ARF6Q67L triggered an increase in peripheral and cytosolic pAKT levels above background. The untreated and vector control cells showed a rounded morphology on undigested Matrigel whereas ARF6Q67L incited cell spreading into plump, polygonal shapes with extended cell processes (top row, Figure 6e). Treatment with the pan-AKT inhibitor MK2206 reduced pAKT staining and restored the rounded cell shape. During active Matrigel digestion, cellular protrusions were more readily visualized and endogenous pAKT localized with cortactin in ring-shaped structures (Figure 6e, middle and bottom rows, see arrows). This morphology is similar to the co-localization reported for cortactin and F-actin in invadopodia(38). ARF6Q67L expression led to an increase in these pAKT-rich invadopodia. ARF6-GTP induced condensation and colocalization of pAKT and cortactin in invadopodia and was also seen in melanoma cells invading and digesting gelatin matrix (Figure 6f). Our data are consistent with findings in breast cancer cells in which PI3K and AKT mediate invadopodia formation and function.(39)
Together these results demonstrate a previously unknown mechanism by which ARF6 facilitates invasion by engaging the PI3K-AKT pathway, which may help explain how ARF6 activation leads to metastasis. We propose a two-step mechanism of ARF6-PI3K-AKT signaling in invading melanoma cells. Activation of ARF6 induces 1) transcriptional upregulation of the p85 regulatory subunit of PI3K, causing stabilization of the p110 catalytic subunit, leading to 2) p110 transport to invadopodia, where AKT is activated.
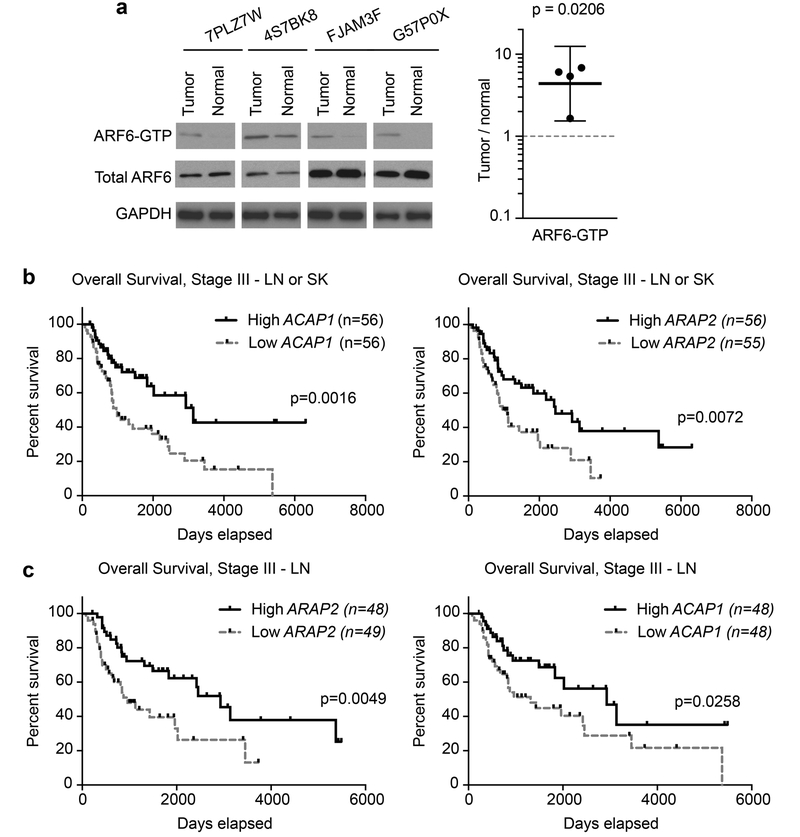
ARF6 is aberrantly activated in melanoma patient samples
We previously showed that pharmacologic inhibition of ARF6 limits metastasis, suggesting that ARF6-GTP is necessary for metastasis(17). Our current data reveal that ARF6-GTP is sufficient for metastasis and provide mechanistic insight. Yet, the clinical relevance of these findings remained uncertain because the activated, GTP-bound state of ARF6 in uncultured cancer specimens is unknown. Direct measurement of the activation state of small GTPases in patient samples is challenging due to the high instability of GTP binding and the significant input protein requirements for detection. Nevertheless, we asked whether ARF6-GTP levels are detectable and altered in tumors compared to nontumor tissue. We queried our frozen clinical biorepository for specimens of patient-matched melanoma and nontumor/normal tissue. Quality control analysis for tumor and tissue-type content was provided by H&E morphology evaluation. We limited our testing to samples with ample excess banked tissue and a high minimum viable tumor content of 70% for cancer samples. All nontumor, normal tissues were confirmed viable. With these inclusion criteria, we identified appropriate tissue pairs from 12 recent patients. Each pair was procured from a single surgical procedure (adjacent tissue in a wide resection) and processed simultaneously. GTP-bound ARF6 was detected in four of these pairs, including the normal tissue, reflecting the instability of the GTP-bound state and the variability in pre-analytic processing of the tissues, such as time to freezing. Among these four pairs, ARF6-GTP levels were higher in the tumors compared to matched normal tissues (Figure 7a). While these normal tissues do not contain a preponderance of the ideal control cell lineage, melanocytes, ARF6 activation has been reported in the predominate cell lineages of each of the matched normal tissues (uninvolved lymph node, fibroadipose, liver, and skin)(40) (41,42) (43,44) , as well as a variety of endothelial and epithelial cells (reviewed in(6)). These data support a biologic role for activated ARF6 specifically in tumor cell function.
Figure 7. Evidence for aberrant ARF6 activation in melanoma.
(a) Western blot of ARF6-GTP pull downs from paired patient samples with quantified ARF6-GTP levels per 500 mg of total protein input. Paired ratio t-test, two-tailed, bars = 95% c.i.. Each tumor sample is paired with its patient-matched, nontumor (normal) tissue type that was surgically resected and processed simultaneously. Normal tissues include lymph node, fibroadipose, liver, and skin (respectively, left to right). (b-c) Reduced ARF6-GAP expression in skin (SK) and lymph node (LN) metastasis correlates with a reduced overall survival in Stage III patients TCGA melanoma patients. The RNA expression ranks for ACAP1 and ARAP is shown. Higher rank indicates higher gene expression. Reduced ARAP2 and ACAP1 expression correlate with reduced overall survival from time of diagnosis. Log-rank (Mantel-Cox) test. See also Figure S7.
Biochemical measurement of small GTPase activation in patient samples is not scalable and, as demonstrated above, is limited by instability of the GTP-bound protein state and by significant tissue input requirements. These barriers to implementing biochemical interrogation of small GTPases in precious patient samples have limited our understanding of the contribution of many small GTPases to disease states. As an alternative to biochemical markers, we asked whether there is genomic evidence of ARF6 activation. ARF6 is expressed in a wide variety of cancer types, including melanoma(45), and high ARF6 expression correlates with reduced survival in pancreatic cancer(12). In prostate cancer ARF6 expression is increased in tumor cells compared to normal prostate and ARF6 staining tends to increase with Gleason grade(46).
ARF6 is activated by a variety of extracellular cues and by oncogenic mutations in vitro(6,7,12,29). Guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs) determine the active, GTP-bound state vs. the inactive, GDP-bound state of small GTPases, respectively. Upregulated ARF6 GEF expression is linked to cancer progression. For example, in breast cancer, expression of GEP100 increases with pathologic grade and with the transition from in situ to invasive cancer(47). In lung adenocarcinoma, increased HER2 and GEP100 correlate with lymph node metastasis.(8) In squamous cell carcinoma of the head and neck, high expression of AMAP1 correlates with reduced survival(48).
To investigate ARF6 pathway expression in a large melanoma cohort, we queried The Cancer Genome Atlas (TCGA) cutaneous melanoma RNAseq data(20) for alterations in the ARF6 pathway that correlate with overall survival from the time of diagnosis. We evaluated the expression levels of ARFs, ARF GEFs and GAPs, ARF6 effectors, and the ARF6-inhibiting ligand-receptor pair SLITs-ROBOs (Table S1). We found that a distinct pattern emerged: in contrast to the patterns reported for carcinomas, neither ARF6 nor ARF6 GEF expression was associated with survival in melanoma. Rather, the expression of ARF GAPs, which inactivate ARF6, was prognostic in cutaneous melanoma. Specifically, low ARF GAP expression significantly correlated with reduced overall survival compared to high ARF GAP expression (Figures 7b and S7). The six ARF GAP genes that are prognostic include four ARF6-specific GAPs (ACAP1(49), ADAP1(50), ARAP2(51), SMAP2(52)), an ARF1/ARF5 GAP (AGAP2(53)) and one predicted ARF GAP (AGFG2)(54,55). Low expression of each correlated with reduced overall survival in an individual gene analysis (Figure S7a). In lymph node and skin metastasis from Stage III TCGA patients, a subset of patients who are at significant risk of metastatic progression, ARAP2 and ACAP1 expression were strongly prognostic for reduced overall survival (Figure 7b and c). To a lesser extent, loss of ADAP1 and AGAP2 were associated with worse prognosis, although the association was not statistically significant (Figure S7b). Because the TCGA survival time begins at the date of diagnosis, this study is limited by a lack of primary tumors and may be biased by the lack of primary tumors that failed to metastasize. Nevertheless, downregulation of ARF6-specific GAPs (ARF6-suppressors) occurs during metastatic progression to Stage III and correlates with reduced overall survival in metastatic melanoma patients. These results suggest that ARF6 hyper-activation in melanoma is associated with metastatic progression and reveal the ARF6 pathway as a potential therapeutic target.
Discussion
In this study, we shed new light on the potency, mechanism of action, and potential clinical impact of ARF6 activation in cutaneous melanoma. In the active, GTP-bound state, ARF6 is not only sufficient to accelerate metastases, it confers equal metastatic capacity, from smaller tumors, compared to deletion of the tumor suppressor Pten. In essence, ARF6 activation renders BRAFV600E/Cdkn2aNULL tumors more competent for metastasis, acting as a catalyst for primary tumor escape and/or end-organ colonization. Early metastasis from very thin primary tumors is a long-standing clinical conundrum in melanoma. Our data suggests that activation of ARF6 may explain how this occurs. Furthermore, our work suggests that ARF6 functions in multiple stages of metastasis, including later stages such as survival and end-organ colonization of circulating tumor cells. These data are significant because targeting ARF6 may have therapeutic utility in patients with a significant risk of metastatic progression. Pharmacologic inhibitors of ARF6 are in preclinical development and studies show efficacy in several in vivo disease models, including melanoma(2,56–59). It will be important to test whether ARF6 pharmacologic inhibitors are effective in a variety of preclinical cancer model systems and to pair these studies with conditional genetic knockout models.
We have elucidated distinct roles for ARF6 in the metastatic cascade and uncovered a novel ARF6-PI3K-AKT pathway that is important for invasion. ARF6-GTP boosts expression of p85 PI3K, increasing p110 PI3K protein levels and enhancing localization of p110 PI3K to peripheral cellular compartments that are critical for melanoma invasion. This mechanism helps explain how ARF6 activation can synergize with Pten deletion in vivo to exacerbate metastasis. ARF6-induced PI3K expression and redistribution leads to increased pools of activated PI3K, while loss of PTEN eliminates the metabolism of its product PIP3, thus leading to increased PIP3 and activated AKT.
Our human data make a compelling case for the importance of ARF6 in cutaneous melanoma. ARF6-GTP is elevated in tumors compared to patient-matched, adjacent, non-tumor tissue. Furthermore, in a large TCGA melanoma cohort, transcriptome data implicates ARF6 activation, via ARF6 GAP loss, in reduced overall survival. These human data are provocative because they suggest aberrant activation of ARF6 in melanoma patient samples and a pathologic role for ARF6 in this disease. Overall, our data strongly suggest that activation of ARF6, via tumor-intrinsic or tumor microenvironmental signals, incites disease progression in melanoma patients at least in part by engaging the PI3K pathway.
Our in vivo model provides a unique paradigm for understanding trafficking-specialized small GTPases in cancer. ARFs and RABs are distinct among the RAS superfamily in that they are critical for endocytic and exocytic membrane trafficking. These proteins often show overlapping functions, such as promoting receptor tyrosine kinase and MAPK signaling and cancer cell invasion(6,60). Our work demonstrates that, similar to RAB35(25); and RAB25(61), ARF6 is also important for PI3K signaling. RAB35 appears to mediate late endosome-associated platelet-derived growth factor receptor activation of AKT(25). Ectopic RAB35Q67L associates with PI3K and increases pAKT levels in vitro. It is possible that ARF6 facilitates a similar RTK-PI3K endosomal signaling mechanism in melanoma. Using an in vivo approach, we discovered a distinct and unexpected mechanism of PI3K activation that begins with transcriptional upregulation of the p85 regulatory subunit of PI3K and ends with delivery of the p110 catalytic subunit of PI3K to invadopodia where PI3K and AKT are critical for ARF6-mediated invasion (graphical abstract).
While more work is needed to understand the role of ARF6 in fine-tuning the transcriptional output of signaling pathways, it is clear from our data and others that ARF6 can exert multiple levels of control over the PI3K pathway. A role for ARF6 in the PIP2 synthetic pathway was originally described by Honda et al.(62), who showed that synthesis of PIP2 by phosphatidylinositol 4-phosphate 5-kinaseα (PI4P5Kα) is stimulated by ARF proteins and phosphatidic acid. A more recent report shows that ARF6 controls PI4P5Kα -mediated PIP2 generation and AKT activation in benign hepatocytes(44). Unlike these previous studies, we show that ARF6 can control PI3K expression and, potentially, its catalytic activity. This is a unique role for ARF6 in PI3K-AKT signaling. In the future, it will be important to determine if targeting ARF6 can inhibit the PI3K pathway in metastatic progression, particularly in the presence of PTEN loss.
Disruption of the metastatic cascade, as a therapeutic approach for patients with local, regional or oligometastatic disease, remains an elusive but promising therapeutic goal in oncology(63). Reaching this goal begins with understanding the mechanistic underpinnings of metastasis and the development of preclinical metastatic models. ARF6 appears to be versatile in its capacity to facilitate distinct signaling pathways promoting cancer progression(6). In cutaneous melanoma, ARF6 has been linked to HGF-ERK mediated invasion(3) and WNT5A-β-catenin mediated invasion(2). Given that ARF6 is upstream of both MAPK/ERK(3,7,10,59,64) and now PI3K/AKT, it is possible that ARF6 regulates both of these pathways by exerting direct control over RAS. Regardless, the expanded reach of ARF6 in oncogenic signaling to both major arms of the RAS pathway has significant implications. RAS is an elusive pharmacologic target in cancer(65) while ARF6 inhibitors continue to show promise in preclinical models. In the future, it will be important to investigate a role for ARF6 in RAS-mediated oncogenesis and to test ARF6 pharmacologic inhibitors in cancers driven by unregulated RAS.
Supplementary Material
Significance.
These findings reveal a pro-metastatic role for ARF6 independent of tumor growth, which may help explain how melanoma spreads distantly from thin, early stage primary tumors.
Acknowledgments
This study was supported by the National Cancer Institute K08 CA188563–01A1, R01 CA121118, K99 CA230312, the Melanoma Research Alliance MRA 347651, a Harold J. Lloyd Charitable Trust Career Development Award, NCI R01CA202778, NIAMS R01AR064788, NHLBI R01HL130541, T32HL007576–31, U54CA209978. We thank several shared resources at the Huntsman Cancer Institute: High-Throughput Genomics, Biorepository Molecular Pathology, and Bioinformatics are supported by award P30CA042014 from the National Cancer Institute and the Preclinical Research Resource, which provided experimental metastasis modeling services. We also thank Health Science Cores at the University of Utah: Genomics Core Facility; DNA Sequencing Core Facility; Fluorescence Microscopy Core Facility, supported by NCRR Shared Equipment Grant # 1S10RR024761–01. We thank the MDACC RPPA Core facility, funded by NCI CA16672. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. We thank Drs. R. Andtbacka, J. Hyngstrom, and R.D. Kim for patient specimen procurement. We thank C. Stubben for bioinformatics support and Dr. K. Boucher for biostatistics input in the TCGA survival correlates. We thank Drs. D. Grossman and M. VanBrocklin for human cell lines, Dr. M. McMahon for scientific and technical input, Drs. J. Kaplan and A. Anderson for manuscript editing, Dr. ZZ Tong and Navigen, Inc. for ARF6 plasmid constructs and D. Lim for graphics preparation.
Footnotes
Competing interests: The University of Utah has filed intellectual property rights concerning ARF6 pathways. The University of Utah has licensed technology to Navigen Inc., a biotechnology company owned in part by the University of Utah Research Foundation. D.Y. Li is a cofounder of and consultant to Navigen Inc.
References
- 1.Wong SL, Balch CM, Hurley P, Agarwala SS, Akhurst TJ, Cochran A, et al. Sentinel lymph node biopsy for melanoma: American Society of Clinical Oncology and Society of Surgical Oncology joint clinical practice guideline. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2012;30:2912–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grossmann AH, Yoo JH, Clancy J, Sorensen LK, Sedgwick A, Tong Z, et al. The small GTPase ARF6 stimulates beta-catenin transcriptional activity during WNT5A-mediated melanoma invasion and metastasis. Science signaling 2013;6:ra14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tague SE, Muralidharan V, D’Souza-Schorey C. ADP-ribosylation factor 6 regulates tumor cell invasion through the activation of the MEK/ERK signaling pathway. Proceedings of the National Academy of Sciences of the United States of America 2004;101:9671–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, et al. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Current biology : CB 2009;19:1875–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muralidharan-Chari V, Hoover H, Clancy J, Schweitzer J, Suckow MA, Schroeder V, et al. ADP-ribosylation factor 6 regulates tumorigenic and invasive properties in vivo. Cancer research 2009;69:2201–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grossmann AH, Zhao H, Jenkins N, Zhu W, Richards JR, Yoo JH, et al. The small GTPase ARF6 regulates protein trafficking to control cellular function during development and in disease. Small GTPases 2016:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoo JH, Shi DS, Grossmann AH, Sorensen LK, Tong Z, Mleynek TM, et al. ARF6 Is an Actionable Node that Orchestrates Oncogenic GNAQ Signaling in Uveal Melanoma. Cancer cell 2016;29:889–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menju T, Hashimoto S, Hashimoto A, Otsuka Y, Handa H, Ogawa E, et al. Engagement of overexpressed Her2 with GEP100 induces autonomous invasive activities and provides a biomarker for metastases of lung adenocarcinoma. PloS one 2011;6:e25301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palacios F, Price L, Schweitzer J, Collard JG, D’Souza-Schorey C. An essential role for ARF6-regulated membrane traffic in adherens junction turnover and epithelial cell migration. EMBO J 2001;20:4973–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellon-Cardenas O, Clancy J, Uwimpuhwe H, D’Souza-Schorey C. ARF6-regulated endocytosis of growth factor receptors links cadherin-based adhesion to canonical Wnt signaling in epithelia. Molecular and cellular biology 2013;33:2963–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto A, Oikawa T, Hashimoto S, Sugino H, Yoshikawa A, Otsuka Y, et al. P53- and mevalonate pathway-driven malignancies require Arf6 for metastasis and drug resistance. J Cell Biol 2016;213:81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang C, Qin Y, Zhang B, Ji S, Shi S, Xu W, et al. ARF6, induced by mutant Kras, promotes proliferation and Warburg effect in pancreatic cancer. Cancer Lett 2017;388:303–11 [DOI] [PubMed] [Google Scholar]
- 13.Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer research 2004;64:7002–10 [DOI] [PubMed] [Google Scholar]
- 14.Dai DL, Martinka M, Li G. Prognostic significance of activated Akt expression in melanoma: a clinicopathologic study of 292 cases. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2005;23:1473–82 [DOI] [PubMed] [Google Scholar]
- 15.Davies MA, Stemke-Hale K, Lin E, Tellez C, Deng W, Gopal YN, et al. Integrated Molecular and Clinical Analysis of AKT Activation in Metastatic Melanoma. Clin Cancer Res 2009;15:7538–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bucheit AD, Chen G, Siroy A, Tetzlaff M, Broaddus R, Milton D, et al. Complete loss of PTEN protein expression correlates with shorter time to brain metastasis and survival in stage IIIB/C melanoma patients with BRAFV600 mutations. Clin Cancer Res 2014;20:5527–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho JH, Robinson JP, Arave RA, Burnett WJ, Kircher DA, Chen G, et al. AKT1 Activation Promotes Development of Melanoma Metastases. Cell Rep 2015;13:898–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr., et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nature genetics 2009;41:544–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh Durban V, Deuker MM, Bosenberg MW, Phillips W, McMahon M. Differential AKT dependency displayed by mouse models of BRAFV600E-initiated melanoma. J Clin Invest 2013;123:5104–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas N. Genomic Classification of Cutaneous Melanoma. Cell 2015;161:1681–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nature medicine 2013;19:1423–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012;487:500–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz B Invadopodia detection and gelatin degradation assay. Bio-protocol 2013;3:e997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz B, Yuen A, Iizuka S, Higashiyama S, Courtneidge SA. Notch increases the shedding of HB-EGF by ADAM12 to potentiate invadopodia formation in hypoxia. J Cell Biol 2013;201:279–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wheeler DB, Zoncu R, Root DE, Sabatini DM, Sawyers CL. Identification of an oncogenic RAB protein. Science 2015;350:211–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.VanBrocklin MW, Robinson JP, Lastwika KJ, Khoury JD, Holmen SL. Targeted delivery of NRASQ61R and Cre-recombinase to post-natal melanocytes induces melanoma in Ink4a/Arflox/lox mice. Pigment Cell Melanoma Res 2010;23:531–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng H, Jorapur A, Shain AH, Lang UE, Torres R, Zhang Y, et al. Bi-allelic Loss of CDKN2A Initiates Melanoma Invasion via BRN2 Activation. Cancer cell 2018;34:56–68 e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer 2003;3:453–8 [DOI] [PubMed] [Google Scholar]
- 29.Chabu C, Li DM, Xu T. EGFR/ARF6 regulation of Hh signalling stimulates oncogenic Ras tumour overgrowth. Nat Commun 2017;8:14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3’-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Molecular and cellular biology 1998;18:1379–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo J, Cantley LC. The negative regulation of phosphoinositide 3-kinase signaling by p85 and it’s implication in cancer. Cell Cycle 2005;4:1309–12 [DOI] [PubMed] [Google Scholar]
- 32.Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nature cell biology 2007;9:299–309 [DOI] [PubMed] [Google Scholar]
- 33.Chakrabarti KR, Hessler L, Bhandary L, Martin SS. Molecular Pathways: New Signaling Considerations When Targeting Cytoskeletal Balance to Reduce Tumor Growth. Clin Cancer Res 2015;21:5209–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Betapudi V, Gokulrangan G, Chance MR, Egelhoff TT. A proteomic study of myosin II motor proteins during tumor cell migration. J Mol Biol 2011;407:673–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norwood Toro LE, Wang Y, Condeelis JS, Jones JG, Backer JM, Bresnick AR. Myosin-IIA heavy chain phosphorylation on S1943 regulates tumor metastasis. Exp Cell Res 2018;370:273–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Molecular cell 2002;10:151–62 [DOI] [PubMed] [Google Scholar]
- 37.Xue G, Hemmings BA. PKB/Akt-dependent regulation of cell motility. J Natl Cancer Inst 2013;105:393–404 [DOI] [PubMed] [Google Scholar]
- 38.Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nature reviews Molecular cell biology 2011;12:413–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi H, Yoshida S, Muroi E, Yoshida N, Kawamura M, Kouchi Z, et al. Phosphoinositide 3-kinase signaling pathway mediated by p110alpha regulates invadopodia formation. J Cell Biol 2011;193:1275–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galandrini R, Micucci F, Tassi I, Cifone MG, Cinque B, Piccoli M, et al. Arf6: a new player in FcgammaRIIIA lymphocyte-mediated cytotoxicity. Blood 2005;106:577–83 [DOI] [PubMed] [Google Scholar]
- 41.Davies JC, Bain SC, Kanamarlapudi V. ADP-ribosylation factor 6 regulates endothelin-1-induced lipolysis in adipocytes. Biochem Pharmacol 2014;90:406–13 [DOI] [PubMed] [Google Scholar]
- 42.Davies JC, Tamaddon-Jahromi S, Jannoo R, Kanamarlapudi V. Cytohesin 2/ARF6 regulates preadipocyte migration through the activation of ERK1/2. Biochem Pharmacol 2014;92:651–60 [DOI] [PubMed] [Google Scholar]
- 43.Brooks R, Williamson R, Bass M. Syndecan-4 independently regulates multiple small GTPases to promote fibroblast migration during wound healing. Small GTPases 2012;3:73–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai MT, Katagiri N, Ohbayashi N, Iwasaki K, Ohkohchi N, Ding ST, et al. Regulation of HGF-induced hepatocyte proliferation by the small GTPase Arf6 through the PIP2-producing enzyme PIP5K1A. Sci Rep 2017;7:9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357. [DOI] [PubMed] [Google Scholar]
- 46.Morgan C, Lewis PD, Hopkins L, Burnell S, Kynaston H, Doak SH. Increased expression of ARF GTPases in prostate cancer tissue. Springerplus 2015;4:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morishige M, Hashimoto S, Ogawa E, Toda Y, Kotani H, Hirose M, et al. GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nature cell biology 2008;10:85–92 [DOI] [PubMed] [Google Scholar]
- 48.Sato H, Hatanaka KC, Hatanaka Y, Hatakeyama H, Hashimoto A, Matsuno Y, et al. High level expression of AMAP1 protein correlates with poor prognosis and survival after surgery of head and neck squamous cell carcinoma patients. Cell Commun Signal 2014;12:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson TR, Brown FD, Nie Z, Miura K, Foroni L, Sun J, et al. ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J Cell Biol 2000;151:627–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venkateswarlu K, Brandom KG, Lawrence JL. Centaurin-alpha1 is an in vivo phosphatidylinositol 3,4,5-trisphosphate-dependent GTPase-activating protein for ARF6 that is involved in actin cytoskeleton organization. The Journal of biological chemistry 2004;279:6205–8 [DOI] [PubMed] [Google Scholar]
- 51.Chen PW, Jian X, Yoon HY, Randazzo PA. ARAP2 signals through Arf6 and Rac1 to control focal adhesion morphology. The Journal of biological chemistry 2013;288:5849–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natsume W, Tanabe K, Kon S, Yoshida N, Watanabe T, Torii T, et al. SMAP2, a novel ARF GTPase-activating protein, interacts with clathrin and clathrin assembly protein and functions on the AP-1-positive early endosome/trans-Golgi network. Molecular biology of the cell 2006;17:2592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nie Z, Fei J, Premont RT, Randazzo PA. The Arf GAPs AGAP1 and AGAP2 distinguish between the adaptor protein complexes AP-1 and AP-3. Journal of cell science 2005;118:3555–66 [DOI] [PubMed] [Google Scholar]
- 54.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nature reviews Molecular cell biology 2011;12:362–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kahn RA, Bruford E, Inoue H, Logsdon JM Jr, Nie Z, Premont RT, et al. Consensus nomenclature for the human ArfGAP domain-containing proteins. J Cell Biol 2008;182:1039–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu W, London NR, Gibson CC, Davis CT, Tong Z, Sorensen LK, et al. Interleukin receptor activates a MYD88-ARNO-ARF6 cascade to disrupt vascular stability. Nature 2012;492:252–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hongu T, Funakoshi Y, Fukuhara S, Suzuki T, Sakimoto S, Takakura N, et al. Arf6 regulates tumour angiogenesis and growth through HGF-induced endothelial beta1 integrin recycling. Nat Commun 2015;6:7925. [DOI] [PubMed] [Google Scholar]
- 58.Davis CT, Zhu W, Gibson CC, Bowman-Kirigin JA, Sorensen L, Ling J, et al. ARF6 inhibition stabilizes the vasculature and enhances survival during endotoxic shock. J Immunol 2014;192:6045–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu W, Shi DS, Winter JM, Rich BE, Tong Z, Sorensen LK, et al. Small GTPase ARF6 controls VEGFR2 trafficking and signaling in diabetic retinopathy. J Clin Invest 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin X, Wang J, Wang X, Liu F, Jiang B, Zhang Y. Targeting Rabs as a novel therapeutic strategy for cancer therapy. Drug Discov Today 2017;22:1139–47 [DOI] [PubMed] [Google Scholar]
- 61.Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nature medicine 2004;10:1251–6 [DOI] [PubMed] [Google Scholar]
- 62.Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, et al. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 1999;99:521–32 [DOI] [PubMed] [Google Scholar]
- 63.Steeg PS. Targeting metastasis. Nat Rev Cancer 2016;16:201–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tushir JS, Clancy J, Warren A, Wrobel C, Brugge JS, D’Souza-Schorey C. Unregulated ARF6 activation in epithelial cysts generates hyperactive signaling endosomes and disrupts morphogenesis. Molecular biology of the cell 2010;21:2355–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCormick F KRAS as a Therapeutic Target. Clin Cancer Res 2015;21:1797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.