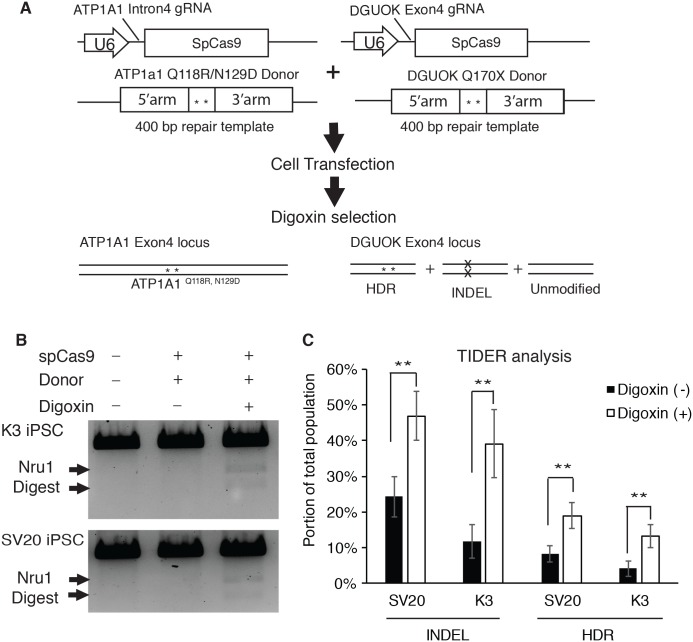
Figure 4. Selecting for mutation of ATP1A1 by digoxin enriches HDR and INDEL-mediated mutagenesis at the DGUOK gene.
(A) Diagram of CRISPR co-targeting appraoch. Two CRISPR plasmids PX459 containing guide RNAs (ATP1A1 Intron 4 and DGUOK exon 4), and two linearized repair templates were transfected into two different iPSC lines (K3 and SV20). The DGUOK repair template introduced a NruI restriction enzyme cutting sequence to facilitate RFLP analysis. Cell selection was demonstrated in Fig. 1B. (B) Cells with or without culture in 1 µM digoxin were collected and genomic DNA was extracted from the whole population. RFLP analysis was performed to determine the level of enrichment of HDR-driven mutagenesis (for uncropped image see Fig. S6). (C) Out-out PCR amplicons from (B) were sequenced and subjected to TIDER analysis (for raw data see Data S2). The genome editing experiment was performed in five replicates (n = 5). Data are shown as mean ± SD. Statistical differences between each two means (Digoxin-treated V.S. non-treated) were determined by student T-test (*p < .05, **p < .01) (SV20 Indel: p = 5.1 × 10−4, K3 Indel: p = 3.34 × 10−4, SV20 HDR: p = 1.33 × 10−3, K3 HDR: p = 3.64 × 10−4).