Abstract
Fish and shellfish diseases are a constant threat to the sustainability and economic viability of aquaculture. Early diagnosis plays a vital role in management of fish and shellfish diseases. Traditionally, various biochemical and serological tests have been used for fish disease diagnosis. However, the time and expertise required for such diagnoses makes it difficult for aquaculturists to easily adopt them under production conditions. Polymerase chain reaction and probe‐based nucleic acid detection have become increasingly popular in fish and shellfish diagnostics. Recently, a novel technique called loop‐mediated isothermal amplification (LAMP) has been developed, which is highly sensitive and rapid. LAMP has been used for the detection of bacterial, viral, fungal and parasitic diseases in both animal and plants. In aquaculture, LAMP‐based detection of pathogens like Edwardsiella tarda, E. ictaluri, Nocardia seriolae, Tetracapsuloides bryosalmonae, white spot syndrome virus and infectious haematopoietic necrosis virus have been reported. In this review, the application of LAMP for the detection of aquaculture‐associated pathogens is discussed.
Keywords: bacteria, diagnostic method, fungi, loop‐mediated isothermal amplification, parasites, viruses
Introduction
Fish and shellfish culturists continually manage diseases that threaten the sustainability of aquaculture. The threat of newly emerging pathogens is also a concern for aquaculturists. The rapid diagnosis and prevention of diseases of fish and shellfish in culture systems is important in fish husbandry. Efficient management of disease usually begins by preventing the spread of disease. Disease diagnosis has been mainly based on clinical signs supported by isolation and identification of the aetiological agent. This requires a rapid and sensitive method to detect pathogens. On‐farm laboratories are not usually equipped with diagnostic equipment, thus small‐scale aquaculture operations depend on specialist fish disease laboratories for their diagnostic needs. However, laboratories routinely handling diagnostic cases are often very busy and take time to carry out diagnoses. In order for laboratories to handle large number of samples sensitive and rapid diagnostic kits are needed.
Traditional methods for identifying pathogens are mainly based on phenotypic characters. Common techniques used in diagnosis of fish and shellfish diseases include bacteriological analysis, virus isolation, histopathology and enzyme‐linked immunosorbent assay (ELISA)‐based techniques. These are often accurate and sensitive techniques although they require time and trained personnel. Nucleic acid‐based detection assays have also been used for the detection of pathogens. An excellent review on molecular diagnostics used to detect pathogens associated with fish and shellfish diseases has been published by Cunningham (2002). Various techniques, including restriction digestion of nucleic acid, nucleic acid hybridization and DNA amplification technology are currently being used. Polymerase chain reaction (PCR) and real‐time PCR have gained a wide popularity as a diagnostic tool. The advantage of PCR over other nucleic acid techniques is the ease of development of the detection system, its high sensitivity and rapidity.
In this paper, we review a novel method of DNA amplification known as loop‐mediated isothermal amplification (LAMP) of a target nucleic acid. This method has already been applied for the detection of several micro‐organisms (Table 1). The advantages and the potential application of this technique in detection of pathogens associated with aquaculture are reviewed.
Table 1.
List of pathogens detected by LAMP assays
| Species | Method | References |
|---|---|---|
| Bacteria | ||
| Legionella | LAMP | Annaka (2003) |
| MAP | LAMP | Enosawa et al. (2003) |
| Mycobacterium tuberculosis complex | LAMP | Iwamoto et al. (2003) |
| Escherichia coli O157:H7 | LAMP | Maruyama et al. (2003) |
| Edwardsiella tarda | LAMP | Savan et al. (2004) |
| Edwardsiella ictaluri | LAMP | Yeh et al. (2005) |
| Shigella and enteroinvasive E. coli | LAMP | Song et al. (2005) |
| Porphyromonas gingivalis | LAMP | Maeda et al. (2005) |
| Streptococcus pneumoniae | LAMP | Seki, Yamashita, Torigoe, Tsuda, Sato & Maeno (2005) |
| Viruses | ||
| Tomato yellow leaf curl virus | LAMP | Fukuta, Kato, Yoshida, Mizukami, Ishida, Ueda, Kanbe & Ishimoto (2003b) |
| Human herpes virus 6 | LAMP | Ihira et al. (2004) |
| WSSV | LAMP | Kono et al. (2004) |
| Varicella zoster virus | LAMP | Okamoto et al. (2004) |
| Adenoviral keratoconjunctivitis | LAMP | Wakabayashi et al. (2004) |
| Human herpes virus 7 | LAMP | Yoshikawa et al. (2004) |
| RSIV | LAMP | Caipang et al. (2004) |
| KHV | LAMP | Gunimaladevi et al. (2004) |
| Herpes simplex virus | LAMP | Enomoto et al. (2005) |
| Human influenza A viruses | LAMP | Poon et al. (2005) |
| NDV | LAMP | Pham, Nakajima, Ohashi & Onuma (2005) |
| JYMV | RT‐LAMP | Fukuta et al. (2003a) |
| Tomato spotted wilt virus | IC/RT‐LAMP | Fukuta et al. (2004) |
| IHNV | RT‐LAMP | Gunimaladevi et al. (2005) |
| Parasites | ||
| African trypanosomes | LAMP | Kuboki et al. 2003 |
| Babesia gibsoni infection | LAMP | Ikadai et al. (2004) |
| PKX | LAMP | El‐Matbouli & Soliman (2005) |
| Fungus | ||
| Paracoccidioidomycosis | LAMP | Endo et al. (2004) |
LAMP, loop‐mediated isothermal amplification; RT‐LAMP, reverse transcription LAMP; IC/RT‐LAMP, immunocapture/reverse transcription‐LAMP; PKX, Tetracapsuloides bryosalmonae; WSSV, white spot syndrome virus; KHV, koi herpes virus; RSIV, red sea bream iridovirus; IHNV, infectious haematopoietic necrosis virus; NDV, Newcastle disease virus; MAP, Mycobacterium avium subsp. paratuberculosis; JYMV, Japanese yam mosaic virus.
The LAMP method
Principle of LAMP
Loop‐mediated isothermal amplification is a sensitive strand displacement technique (Notomi, Okayama, Masubuchi, Yonekawa, Watanabe, Amino & Hase 2000). This method amplifies target DNA from a few copies to 109 copies in less than an hour under isothermal conditions. It is an offshoot of the basic strand displacement techniques which have been described thoroughly (Notomi et al. 2000). Briefly, four highly specific primers are constructed from the target DNA, one set of primers anneal to the target region one after the other on the same strand and the primer which anneals at the later stage displaces the strand formed by the first primer with the help of Bst DNA polymerase. The Bst polymerase has a strand displacement activity. This takes place on both strands and the primers are designed such that loops are formed. The reaction is carried out under isothermal conditions as denaturation of the strand takes place by strand displacement. The reactions produce a series of stem‐loop DNAs with various lengths. The four primers hybridize against six distinct sequences in the target DNA making it highly specific.
Design of primers
Designing primers for LAMP is a complex procedure compared with the normal PCR. A minimum of four primers are required for a LAMP reaction. The primers required are one pair of inner‐primers, which are usually over 40 mer in length, and shorter outer‐primers 25 mer in length. The technical specifications of each primer and the optimized annealing temperatures are described in detail by Notomi et al. (2000). The primers can be developed using Primer Explorer version 3 software (http://primerexplorer.jp/lamp3.0.0/) specifically designed to develop LAMP primers. By using additional sets of loop primers, the reaction time of LAMP can be further accelerated (Nagamine, Hase & Notomi 2002). Using loop primers the time required is reduced by half, making it a more efficient tool in disease diagnostic applications.
Requirements for LAMP reaction
The LAMP reaction is performed by the Bst DNA polymerase along with dNTPs and reaction buffer (Eiken Genome Co. Ltd, Tokyo, Japan). The reaction proceeds at isothermal conditions for 1 h or less depending on the efficiency of the developed primers and template DNA. The template DNA for LAMP is denatured for 5 min at 95 °C before setting up the LAMP reaction for isothermal amplification (Notomi et al. 2000). However, studies have shown that non‐denatured templates can also be used directly for LAMP‐mediated detection (Nagamine, Watanabe, Ohtsuka, Hase & Notomi 2001). The LAMP reaction is carried out at 60–65 °C for 45–60 min and the reaction terminated at 80 °C for 2 min (Fig. 1). The main advantage of the technique is that it does not need thermocyclers. As the amplification in made under isothermal conditions, only a water bath or heating block is needed to maintain the required temperature.
Figure 1.
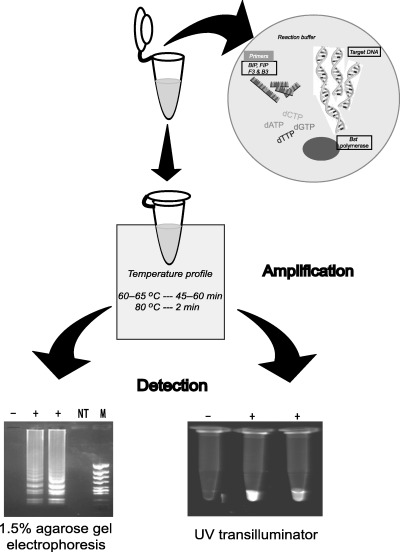
Schematic representation of the procedure used in detecting pathogens by loop‐mediated isothermal amplification. The reaction mixture and template is amplified at isothermal temperature and can be detected by staining the agarose gel or by incorporating the intercalating agents in the reaction tube and visualizing.
Visualization of amplified products
Several methods may be used to check for the LAMP reaction. The products are commonly visualized by agarose gel electrophoresis stained with an intercalating agent such as ethidium bromide or SYBR Green I. As the LAMP reaction produces products of various lengths of stem loop structures, the gel will show products from the minimum length of target DNA to the loading well, which appears as a smear and bands at the base of the gel. Because amplification of the target DNA is so high at the final stage it is vulnerable to contamination in subsequent amplifications. However, an advantage of the large amount of product that is generated is that it can be visualized on a UV‐transilluminator by incorporating intercalating agents directly into the LAMP‐amplified tubes (Notomi et al. 2000). Another method is by assessing the amount of white precipitate formed from magnesium pyrophosphate (Mori, Nagamine, Tomita & Notomi 2001). The DNA formed by amplification of prostate‐specific antigen by PCR for 35 cycles and LAMP for 60 min at 65 °C was 0.2 and 11.2 μg 25 μL−1, respectively (Mori et al. 2001). The huge amount of DNA formed by LAMP allows the detection of turbidity using a desktop centrifuge, as the precipitate accumulates at the bottom of the tube. Negative reactions neither show smearing nor turbidity in the reaction mixture.
Application of LAMP in the detection of micro‐organisms
Bacterial pathogens
There are several published reports regarding the use of LAMP for the detection of bacterial isolates from humans, fish and the aquatic environment. The first report of the application of LAMP to detect bacterial isolates was by Maruyama, Kenzaka, Yamaguchi, Tani & Nasu (2003). These authors developed a novel strategy using LAMP to detect Escherichia coli from the coastal environment, where the stx gene was detected by an in situ method. The amount of target DNA amplified is enormous and forms chains which cannot exit from the bacterial cell. The presence of huge amounts of target DNA facilitates detection using a nucleic acid probe. Song, Toma, Nakasone & Iwanaga (2005) developed a LAMP assay for Shigella and enteroinvasive E. coli by targeting the ipaH gene, with amplification within 2 h starting with as few as 8 CFU. Mycobacterium avium subsp. paratuberculosis (MAP) causing Johne's disease, a chronic progressive enteritis in ruminants, has a wide host range. LAMP primer sets were developed to detect the MAP IS900 fragment and were specific for MAP. Furthermore, the detection was simplified as the turbidity of the reaction allowed visual detection (Enosawa, Kageyama, Sawai, Watanabe, Notomi, Onoe, Mori & Yokomizo 2003). Iwamoto, Sonobe & Hayashi (2003) also developed an accurate detection assay for the M. tuberculosis complex, M. avium and M. intracellulare from sputum samples using LAMP. Detection of Legionella (Annaka 2003) and the periodontal pathogen Porphyromonas gingivalis (Maeda, Kokeguchi, Fujimoto, Tanimoto, Yoshizumi, Nishimura & Takashiba 2005) by LAMP was developed by targeting the 16sRNA gene.
Several reports on LAMP‐mediated diagnostic methods have been developed for bacterial pathogens associated with fish. The first use of LAMP for detection of an aquaculture pathogen was reported for edwardsiellosis (Savan, Igarashi, Matsuoka & Sakai 2004). Edwardsiella tarda was detected from infected Japanese flounder, Paralichthys olivaceus. LAMP primers were designed by targeting the haemolysin gene (Fig. 2). The specificity of LAMP was tested for five different E. tarda strains isolated from eel (E22), tilapia (E 381), flatfish (FPC 498), water from eel culture ponds (SU226) and sea bream (158). Non‐specific amplification was not seen in other enteric bacteria and those which produce enterotoxins. The optimum amplification regime for a clear visible banding pattern was at 65 °C for 45 min. Edwardsiella tarda could be detected from 10 and 103 CFU by LAMP and PCR, respectively. Thus, in this case, LAMP‐based detection was much more sensitive than PCR. Furthermore, LAMP could detect the pathogen from Japanese flounder pond water with an E. tarda count of 3.8 × 102 CFU. A LAMP‐based diagnostic system has been recently developed for detection of E. ictaluri from diseased catfish, Ictalurus punctatus (Yeh, Shoemaker & Klesius 2005). The LAMP primers were designed targeting the eip18 gene. The LAMP reaction amplified six different strains of E. ictaluri. Non‐specific amplification was not observed when tested with 12 other bacterial strains associated with channel catfish. The detection limit by LAMP was 20 CFU, more sensitive than real‐time PCR and traditional biochemical assays (Bilodeau, Waldbieser, Terhune, Wise & Wolters 2003). Edwardsiella ictaluri could be detected using genomic DNA from infected brain samples as template. The LAMP method has also been applied to the detection of nocardiosis (Kawakami, Kono & Sakai 2005). Nocardiosis is a serious pathogen affecting yellowtail, Seriola quinqueradiata, and amberjack, S. aureovittata. Nocardial diagnosis has been mainly achieved by isolation and biochemical identification. Recently, PCR detection has been possible for nocardiosis (Kono, Ooyama, Chen & Sakai 2001; Miyoshi & Suzuki 2002). In these studies, the detection limits by LAMP and PCR were 103 and 104 CFU, respectively. Compared with PCR a 10‐fold higher sensitivity is seen using LAMP. Furthermore, LAMP detection was superior to PCR when spleen DNA extracted from infected fish was used as template.
Figure 2.
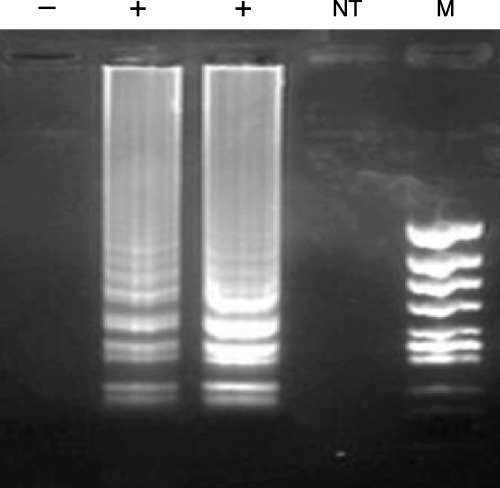
Loop‐mediated isothermal amplification reaction amplifying the haemolysin gene from Edwardsiella tarda isolates. The samples denoted as positive are E. tarda isolates. Amplification was not seen in the control (‐), the no template reaction (NT) and from the non‐specific DNA template. The products were run on a 1.5% agarose gel stained with ethidium bromide.
Detection of viruses
Detection and diagnosis of viruses is often difficult, and histopathology, virus isolation, Western blot and ELISA‐based detection techniques are often cumbersome, expensive, time consuming and require R & D support to develop viable methods. Advent of PCR has not only simplified the development of detection systems, but has also found various applications in virus diagnosis.
DNA viruses
There are a number of reports regarding the use of LAMP for diagnosis of infectious DNA viruses. LAMP assays have been developed for human herpes virus 6 (Ihira, Yoshikawa, Enomoto, Akimoto, Ohashi, Suga, Nishimura, Ozaki, Nishiyama, Notomi, Ohta & Asano 2004), herpes virus 7 (Yoshikawa, Ihira, Akimoto, Usui, Miyake, Suga, Enomoto, Suzuki, Nishiyama & Asano 2004) and herpes simplex virus (Enomoto, Yoshikawa, Ihira, Akimoto, Miyake, Usui, Suga, Suzuki, Kawana, Nishiyama & Asano 2005; Sugiyama, Yoshikawa, Ihira, Enomoto, Kawana & Asano 2005). The method was also used to detect human influenza viruses (H1 to H3) (Poon, Leung, Chan, Lee, Yuen, Guan & Peiris 2005).
Recently, detection of white spot syndrome virus (WSSV) infecting kuruma shrimp, Marsupenaeus japonicus, was reported from our laboratory (Kono, Savan, Sakai & Itami 2004). The detection limit of the viral DNA template was at the 10 fg level, whereas nested PCR‐mediated detection was limited to the 100 fg level. Detection by LAMP was superior to PCR in that it was faster and more sensitive. LAMP‐mediated detection for koi herpes virus (KHV) by targeting the thymidine kinase (tk) gene (AJ535112) has been developed. The detection limit was found to be similar to PCR primers targeting the tk gene from infected common carp, Cyprinus carpio (Gunimaladevi, Kono, Venugopal & Sakai 2004). Screening for KHV has become very important as trade of fancy carp is an easy route for geographical spread of the virus and a rapid and efficient system like LAMP is very suitable for this purpose. LAMP has also been used to detect red sea bream iridovirus (RSIV) (Caipang, Haraguchi, Ohira, Hirono & Aoki 2004). The RSIV DNA was amplified using DNA extracts obtained from spleen of infected red sea bream, Pagrus major. The protocol developed is 10 times more sensitive than conventional PCR for detecting RSIV. The authors also showed a correlation between the number of DNA copies of the virus and the corresponding turbidity, which can be used to quantify virus particles in infected fish.
RNA viruses
The use of reverse transcription (RT)‐PCR as a diagnostic tool for RNA viruses has been widely acknowledged; however, RT‐PCR suffers from constraints, the cost incurred per sample is high, the time required is high as each sample has to undergo RT and the efficiency is low. With the development of the LAMP method, an extended application of reverse transcription (RT)‐LAMP has been developed (Notomi et al. 2000). The ability of avian myeloblastosis virus (AMV) reverse transcriptase to withstand high temperatures, allows a single tube reaction of both RT and LAMP procedures.
The first report on the use of RT‐LAMP was for the detection of the Japanese yam mosaic potyvirus from infected leaves, propagules and roots of Japanese yam (Fukuta, Iida, Mizukami, Ishida, Ueda, Kanbe & Ishimoto 2003a). Recently, protocols for the detection of tomato spotted wilt virus (Fukuta, Ohishi, Yoshida, Mizukami, Ishida & Kanbe 2004), severe acute respiratory syndrome (SARS) coronavirus (Hong, Mai, Cuong, Parida, Minekawa, Notomi, Hasebe & Morita 2004; Poon, Leung, Tashiro, Chan, Wong, Yuen, Guan & Peiris 2004) and west Nile virus (Parida, Posadas, Inoue, Hasebe & Morita 2004) have been developed. Of 31 SARS samples only 64% could be detected as positive. LAMP was less sensitive than real‐time PCR conducted for SARS coronavirus (Poon et al. 2004). However, LAMP was superior to conventional PCR assay (Poon et al. 2004). In fish, RT‐LAMP has been reported for infectious haematopoietic necrosis virus (IHNV) (Gunimaladevi, Kono, Lapatra & Sakai 2005). This virus affects wild and hatchery‐reared salmonids causing widespread mortality. Several immunological and molecular methods to detect IHNV have been developed and RT‐PCR and real‐time PCR are commonly used for IHNV diagnosis. An RT‐LAMP protocol for detection of IHNV was developed targeting the G‐protein of the virus. A comparative analysis of RT‐LAMP, LAMP and nested PCR was conducted. LAMP and nested PCR require an additional 30–40 min as cDNA must be synthesized first. However, RT‐LAMP can directly use RNA as template, where the cDNA synthesis and target gene amplification is carried out in a single tube. In this study, LAMP was 10‐fold more sensitive than nested PCR. Although real‐time PCR is a superior method, RT‐LAMP might be a good alternative as the former can be expensive as a routine diagnostic tool.
Parasitic and fungal infections
Loop‐mediated isothermal amplification has been used for the detection of African trypanosomes, important protozoan parasites causing sleeping sickness in humans (Kuboki, Inoue, Sakurai, Di Cello, Grab, Suzuki, Sugimoto & Igarashi 2003). Two sets of LAMP primers were developed for detecting the Trypanosoma brucei group or T. congolense, targeting PFR‐A and ribosomal subunit P0 genes. The authors reported a 100 times higher sensitivity using the LAMP protocol compared with normal PCR in detecting trypanosome infection. Furthermore, detection of T. brucei from blood samples or cerebrospinal fluid by LAMP, shows that the system is not inactivated by tissue and blood‐derived inhibitors or genomic DNA as seen in PCR. Babesia gibsoni is a parasitic infection affecting dogs. Detection and analysis of B. gibsoni infection were performed with whole‐blood samples by PCR and LAMP targeting the 18S rDNA of the parasite (Ikadai, Tanaka, Shibahara, Matsuu, Uechi, Itoh, Oshiro, Kudo, Igarashi & Oyamada 2004).
Proliferative kidney disease (PKD) is a serious disease affecting salmonid fish (Hedrick, MacConnell & Kinkelin 1993). A myxozoan parasite, Tetracapsuloides bryosalmonae, is the causative agent of PKD. Apart from classical May–Grunwald–Giemsa staining to detect parasites from kidney imprints (Klontz & Chacko 1984), modern techniques like PCR targeting SSU‐rDNA (Kent, Khattra, Hervio & Devlin 1998), in situ hybridization (Morris & Adams 2002) and Mab (Adams, Richards & De Mateo 1992; Saulnier & De Kinkelin 1996) assays are available. Recently, El‐Matbouli & Soliman (2005) used LAMP for rapid diagnosis of PKD‐affected rainbow trout. Furthermore, a comparison of PKD‐LAMP with normal PCR was evaluated. Four sets of primers along with loop primers were designed targeting SSU‐rDNA of T. bryosalmonae. The loop primers were used for the acceleration of the LAMP reaction. The PKD‐LAMP was found to be 100‐fold more sensitive and low amounts of DNA as template could be amplified in 1 h.
Fungal infections
A LAMP assay was developed for Paracoccidiodes brasiliensis, an endemic fungus causing mycosis in Latin America, by targeting the gp43 gene (Endo, Komori, Ricci, Sano, Yokoyama, Ohori, Kamei, Franco, Miyaji & Nishimura 2004). DNA extracted from paraffin‐embedded tissue samples was used as template. The DNA extraction, LAMP assay and detection required 3 h, while nested PCR required 12 h. No reports exist on the use of LAMP for the detection of fungal infections in fish and shellfish. Presently, techniques like RAPD are used to detect fungi such as Aphanomyces (reviewed by Cunningham 2002). These techniques are technically demanding and have problems with reproducibility and LAMP could prove a suitable alternative.
Advantages and disadvantages of LAMP‐based diagnosis of pathogens
The LAMP reaction does not progress without the hybridization of six distinct sequences in the target DNA by four different highly specific primers, thus it is highly specific. Furthermore, the ability of the method to amplify from fewer copies of initial target DNA than PCR has been conclusively demonstrated (2004, 2005; Kono et al. 2004; Savan et al. 2004). The efficiency of LAMP does not seem to be affected by the presence of non‐target genomic DNA in the reaction mixture (Notomi et al. 2000), which is highly desirable in development of a diagnostic system. Detection of target DNA by LAMP compared with detection by two‐step nested‐PCR was at least equal or more sensitive (2004, 2005; Kono et al. 2004; Savan et al. 2004). The evidence suggests that LAMP is relatively more sensitive than conventional DNA‐based detection systems.
The high sensitivity of the LAMP system makes it susceptible to false positives because of carry‐over or cross‐contamination. Amplification and detection should therefore be carried out in separate working areas. As positive reactions are seen as a smear, together with some bands of low molecular weight, and are not seen as a single band as in PCR, the specificity of amplification should be thoroughly validated to ensure that the primers only amplify the target sequence of the specific pathogen. Alternatively, the specificity can be determined by cutting the amplified products using restriction enzymes specific to the target sequence. Detection of two or more pathogens in a single reaction, as in multiplex PCR or nested PCR, is not possible using LAMP.
Application of LAMP in aquaculture
Diagnosis of bacterial infections can be cumbersome because isolation and identification takes time and resources. Bacterial infection with mixed aetiology makes the diagnosis even more complex. Farm managers, together with their previous experience of the clinical signs of diseased fish, rely on diagnostic kits available in the market to detect bacterial agents. The available diagnostic kits are easy to use, do not require expensive equipment but take 12–24 h to obtain a result. PCR‐mediated detection has been developed for several diseases in aquaculture caused by bacterial or viral agents. The LAMP‐mediated diagnosis of edwardselliosis, WSSV, RSIV, IHNV and KHV clearly demonstrates the usefulness of the method as a diagnostic tool. The diagnosis can be based on specific DNA markers as target region for the bacteria. LAMP can also be used for the detection of bacterial resistance to antibiotics by targeting antimicrobial resistance genes. Using LAMP less time is required for detection, allowing more time to use management practices to minimize the spread of disease.
Viral diseases in fish and shellfish can cause mass mortalities and the use of specific pathogen‐free stocks is an excellent way to prevent the introduction of disease‐causing agents into culture systems. A major problem lies in the introduction of carriers from imported fish stocks, e.g. introduction of infectious pancreatic necrosis virus/IHNV infected fish into Japan (Yamazaki 1974). Along with robust legislation, a reliable and sensitive diagnostic technique is needed to detect known and emerging pathogens. The development of diagnostic kits based on nucleic acid detection is simpler and more cost‐effective than developing virus specific cell‐lines.
Nucleic acid‐based detection of protozoan and parasitic diseases in fish for efficient management practice in culture systems has been progressing in recent years. Many reports and reviews have been published on the detection of parasites using molecular methods in fish and shellfish (Gasser & Monti 1997; Gasser 1998; Cunningham 2002) including the use of ribosomal RNA genes and spacers by PCR. However, cross‐reaction with host tissues because of the presence of complementing conserved rDNA sequences has been suggested as a cause of false‐positive results (Perkins & Martin 1999). Using LAMP the specificity could be enhanced as the primers hybridize to six distinct sequences (El‐Matbouli & Soliman 2005).
References
- Adams A., Richards R.H. & De Mateo M.M. (1992) Development of monoclonal antibodies to PKX, the causative agent of proliferative kidney disease. Journal of Fish Diseases 15, 515–521. [Google Scholar]
- Annaka T. (2003) Rapid and simple detection of Legionella species by LAMP, a new DNA amplification method. Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi 14, 25–30. [PubMed] [Google Scholar]
- Bilodeau A.L., Waldbieser G.C., Terhune J.S., Wise D.J. & Wolters W.R. (2003) A real‐time polymerase chain reaction assay of the bacterium Edwardsiella ictaluri . Journal of Aquatic Animal Health 15, 80–86. [Google Scholar]
- Caipang C.M., Haraguchi I., Ohira T., Hirono I. & Aoki T. (2004) Rapid detection of a fish iridovirus using loop‐mediated isothermal amplification (LAMP). Journal of Virological Methods 121, 155–161. [DOI] [PubMed] [Google Scholar]
- Cunningham C. (2002) Molecular diagnosis of fish and shellfish diseases: present status and potential use in disease control. Aquaculture 206, 19–55. [Google Scholar]
- El‐Matbouli M. & Soliman H. (2005) Rapid diagnosis of Tetracapsuloides bryosalmonae, the causative agent of proliferative kidney disease (PKD) in salmonid fish by a novel DNA amplification method, loop‐mediated isothermal amplification (LAMP). Parasitology Research 96, 277–284. [DOI] [PubMed] [Google Scholar]
- Endo S., Komori T., Ricci G., Sano A., Yokoyama K., Ohori A., Kamei K., Franco M., Miyaji M. & Nishimura K. (2004) Detection of gp43 of Paracoccidioides brasiliensis by the loop‐mediated isothermal amplification (LAMP) method. FEMS Microbiology Letters 234, 93–97. [DOI] [PubMed] [Google Scholar]
- Enomoto Y., Yoshikawa T., Ihira M., Akimoto S., Miyake F., Usui C., Suga S., Suzuki K., Kawana T., Nishiyama Y. & Asano Y. (2005) Rapid diagnosis of herpes simplex virus infection by a loop‐mediated isothermal amplification method. Journal of Clinical Microbiology 43, 951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enosawa M., Kageyama S., Sawai K., Watanabe K., Notomi T., Onoe S., Mori Y. & Yokomizo Y. (2003) Use of loop‐mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis . Journal of Clinical Microbiology 41, 4359–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuta S., Iida T., Mizukami Y., Ishida A., Ueda J., Kanbe M. & Ishimoto Y. (2003a) Detection of Japanese yam mosaic virus by RT‐LAMP. Archives of Virology 148, 1713–1720. [DOI] [PubMed] [Google Scholar]
- Fukuta S., Kato S., Yoshida K., Mizukami Y., Ishida A., Ueda J., Kanbe M. & Ishimoto Y. (2003b) Detection of tomato yellow leaf curl virus by loop‐mediated isothermal amplification reaction. Journal of Virological Methods 112, 35–40. [DOI] [PubMed] [Google Scholar]
- Fukuta S., Ohishi K., Yoshida K., Mizukami Y., Ishida A. & Kanbe M. (2004) Development of immunocapture reverse transcription loop‐mediated isothermal amplification for the detection of tomato spotted wilt virus from chrysanthemum. Journal of Virological Methods 121, 49–55. [DOI] [PubMed] [Google Scholar]
- Gasser R.B. (1998) What's in that band? International Journal for Parasitology 28, 989–996. [DOI] [PubMed] [Google Scholar]
- Gasser R.B. & Monti J.R. (1997) Identification of parasitic nematodes by PCR‐SSCP of ITS‐2 rDNA. Molecular and Cellular Probes 11, 201–209. [DOI] [PubMed] [Google Scholar]
- Gunimaladevi I., Kono T., Venugopal M.N. & Sakai M. (2004) Detection of koi herpesvirus in common carp, Cyprinus carpio L., by loop‐mediated isothermal amplification. Journal of Fish Diseases 27, 583–589. [DOI] [PubMed] [Google Scholar]
- Gunimaladevi I., Kono T., Lapatra S.E. & Sakai M. (2005) A loop mediated isothermal amplification (LAMP) method for detection of infectious hematopoietic necrosis virus (IHNV) in rainbow trout (Oncorhynchus mykiss). Archives of Virology 150, 899–909. [DOI] [PubMed] [Google Scholar]
- Hedrick R.P., MacConnell E. & Kinkelin P. (1993) Proliferative kidney disease in salmonid fishes. Annual Review of Fish Diseases 3, 277–290. [Google Scholar]
- Hong T.C., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T., Hasebe F. & Morita K. (2004) Development and evaluation of a novel loop‐mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. Journal of Clinical Microbiology 42, 1956–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihira M., Yoshikawa T., Enomoto Y., Akimoto S., Ohashi M., Suga S., Nishimura N., Ozaki T., Nishiyama Y., Notomi T., Ohta Y. & Asano Y. (2004) Rapid diagnosis of human herpesvirus 6 infection by a novel DNA amplification method, loop‐mediated isothermal amplification. Journal of Clinical Microbiology 42, 140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikadai H., Tanaka H., Shibahara N., Matsuu A., Uechi M., Itoh N., Oshiro S., Kudo N., Igarashi I. & Oyamada T. (2004) Molecular evidence of infections with Babesia gibsoni parasites in Japan and evaluation of the diagnostic potential of a loop‐mediated isothermal amplification method. Journal of Clinical Microbiology 42, 2465–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto T., Sonobe T. & Hayashi K. (2003) Loop‐mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. Journal of Clinical Microbiology 41, 2616–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H., Kono T. & Sakai M. (2005) Detection of fish nocardiosis by loop‐mediated isothermal amplification. Journal of Applied Microbiology (in press). [DOI] [PubMed] [Google Scholar]
- Kent M.L., Khattra J., Hervio D.M. & Devlin R.H. (1998) Ribosomal DNA sequence analysis of the PKX myxosporean and their relationship to members of the genus Sphaerospora . Journal of Aquatic Animal Health 10, 21. [Google Scholar]
- Klontz G.W. & Chacko A.J. (1984) Methods to detect the organism causing proliferative kidney disease in salmonids. Bulletin of the European Association of Fish Pathologists 3, 33–36. [Google Scholar]
- Kono T., Ooyama T., Chen S.‐C. & Sakai M. (2001) Sequencing of 16S‐23S rRNA internal transcribed spacer and its application in the identification of Nocardia seriolae by polymerase chain reaction. Aquaculture Research 33, 1195–1197. [Google Scholar]
- Kono T., Savan R., Sakai M. & Itami T. (2004) Detection of white spot syndrome virus in shrimp by loop‐mediated isothermal amplification. Journal of Virological Methods 115, 59–65. [DOI] [PubMed] [Google Scholar]
- Kuboki N., Inoue N., Sakurai T., Di Cello F., Grab D.J., Suzuki H., Sugimoto C. & Igarashi I. (2003) Loop‐mediated isothermal amplification for detection of African trypanosomes. Journal of Clinical Microbiology 41, 5517–5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Kokeguchi S., Fujimoto C., Tanimoto I., Yoshizumi W., Nishimura F. & Takashiba S. (2005) Detection of periodontal pathogen Porphyromonas gingivalis by loop‐mediated isothermal amplification method. FEMS Immunology and Medical Microbiology 43, 233–239. [DOI] [PubMed] [Google Scholar]
- Maruyama F., Kenzaka T., Yamaguchi N., Tani K. & Nasu M. (2003) Detection of bacteria carrying the stx2 gene by in situ loop‐mediated isothermal amplification. Applied and Environmental Microbiology 69, 5023–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi Y. & Suzuki S. (2002) A PCR method to detect Nocardia seriolae in fish sample. Fish Pathology 38, 93–97. [Google Scholar]
- Mori Y., Nagamine K., Tomita N. & Notomi T. (2001) Detection of loop‐mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochemical and Biophysical Research Communications 289, 150–154. [DOI] [PubMed] [Google Scholar]
- Morris D.J. & Adams A. (2002) PCR and in situ hybridization of Tetracapsula bryosalmonae (PKX), the causative agent of proliferative kidney disease In: Molecular Diagnosis of Salmonid Diseases (ed. by Cunningham C.), pp. 299–313. Kluwer Academic Publishers, Dordecht. [Google Scholar]
- Nagamine K., Watanabe K., Ohtsuka K., Hase T. & Notomi T. (2001) Loop‐mediated isothermal amplification reaction using a nondenatured template. Clinical Chemistry 47, 1742–1743. [PubMed] [Google Scholar]
- Nagamine K., Hase T. & Notomi T. (2002) Accelerated reaction by loop‐mediated isothermal amplification using loop primers. Molecular and Cellular Probes 16, 223–229. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N. & Hase T. (2000) Loop‐mediated isothermal amplification of DNA. Nucleic Acids Research 28, E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Posadas G., Inoue S., Hasebe F. & Morita K. (2004) Real‐time reverse transcription loop‐mediated isothermal amplification for rapid detection of West Nile virus. Journal of Clinical Microbiology 42, 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins S.L. & Martin J.M. (1999) Conserved polymerase chain reaction primers fail in diagnosis of parasitic infections. Journal of Parasitology 85, 982–984. [PubMed] [Google Scholar]
- Pham H.M., Nakajima C., Ohashi K. & Onuma M. (2005) Loop‐mediated isothermal amplification for rapid detection of Newcastle disease virus. Journal of Clinical Microbiology 43, 1646–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Leung C.S., Tashiro M., Chan K.H., Wong B.W., Yuen K.Y., Guan Y. & Peiris J.S. (2004) Rapid detection of the severe acute respiratory syndrome (SARS) coronavirus by a loop‐mediated isothermal amplification assay. Clinical Chemistry 50, 1050–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Leung C.S., Chan K.H., Lee J.H., Yuen K.Y., Guan Y. & Peiris J.S. (2005) Detection of human influenza A viruses by loop‐mediated isothermal amplification. Journal of Clinical Microbiology 43, 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier D. & De Kinkelin P. (1996) Antigenic and biochemical study of PKX, the myxosporean causative of proliferative kidney disease of salmonid fish. Diseases of Aquatic Organisms 27, 103–114. [Google Scholar]
- Savan R., Igarashi A., Matsuoka S. & Sakai M. (2004) Sensitive and rapid detection of edwardsiellosis in fish by a loop‐mediated isothermal amplification method. Applied and Environmental Microbiology 70, 621–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Yamashita Y., Torigoe H., Tsuda H., Sato S. & Maeno M. (2005) Loop‐mediated isothermal amplification method targeting the lytA gene for detection of Streptococcus pneumoniae . Journal of Clinical Microbiology 43, 1581–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T., Toma C., Nakasone N. & Iwanaga M. (2005) Sensitive and rapid detection of Shigella and enteroinvasive Escherichia coli by a loop‐mediated isothermal amplification method. FEMS Microbiology Letters 243, 259–263. [DOI] [PubMed] [Google Scholar]
- Sugiyama H., Yoshikawa T., Ihira M., Enomoto Y., Kawana T. & Asano Y. (2005) Comparison of loop‐mediated isothermal amplification, real‐time PCR, and virus isolation for the detection of herpes simplex virus in genital lesions. Journal of Medical Virology 75, 583–587. [DOI] [PubMed] [Google Scholar]
- Yamazaki T. (1974) Infectious pancreatic necrosis of rainbow trout. Fish Culture 11, 36–40. [Google Scholar]
- Yeh H.‐Y., Shoemaker C.A. & Klesius P.H. (2005) Evaluation of a loop‐mediated isothermal amplification method for rapid detection of channel catfish Ictalurus punctatus important bacterial pathogen Edwardsiella ictaluri . Journal of Microbiological Methods 63, 36–44. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T., Ihira M., Akimoto S., Usui C., Miyake F., Suga S., Enomoto Y., Suzuki R., Nishiyama Y. & Asano Y. (2004) Detection of human herpesvirus 7 DNA by loop‐mediated isothermal amplification. Journal of Clinical Microbiology 42, 1348–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]


