Abstract
Large-scale metagenomic and metatranscriptomic data analyses are often restricted by their gene-centric approach, limiting the ability to understand organismal and community biology. De novo assembly of large and mosaic eukaryotic genomes from complex meta-omics data remains a challenging task, especially in comparison with more straightforward bacterial and archaeal systems. Here, we use a transcriptome reconstruction method based on clustering co-abundant genes across a series of metagenomic samples. We investigated the co-abundance patterns of ∼37 million eukaryotic unigenes across 365 metagenomic samples collected during the Tara Oceans expeditions to assess the diversity and functional profiles of marine plankton. We identified ∼12,000 co-abundant gene groups (CAGs), encompassing ∼7 million unigenes, including 924 metagenomics-based transcriptomes (MGTs, CAGs larger than 500 unigenes). We demonstrated the biological validity of the MGT collection by comparing individual MGTs with available references. We identified several key eukaryotic organisms involved in dimethylsulfoniopropionate (DMSP) biosynthesis and catabolism in different oceanic provinces, thus demonstrating the potential of the MGT collection to provide functional insights on eukaryotic plankton. We established the ability of the MGT approach to capture interspecies associations through the analysis of a nitrogen-fixing haptophyte-cyanobacterial symbiotic association. This MGT collection provides a valuable resource for analyses of eukaryotic plankton in the open ocean by giving access to the genomic content and functional potential of many ecologically relevant eukaryotic species.
As an alternative to individual genome or transcriptome sequencing, environmental genomics has been used for many years to access the global genomic content of organisms from a given environment (Joly and Faure 2015). However, large-scale metagenomic and metatranscriptomic data analyses are often restricted by their gene-centric approach, limiting the ability to draw an integrative functional view of sampled organisms. Nevertheless, constructing gene catalogs from environmental samples provides a useful framework for a general description of the structure and functional capabilities of microbe-dominated communities (Venter et al. 2004; Qin et al. 2010; Brum et al. 2015; Sunagawa et al. 2015; Carradec et al. 2018). Gene-centric approaches allow deep and detailed exploration of communities of organisms, but they are usually undermined by limited contextual information for different genes, apart from taxonomic affiliation based on sequence similarity.
Several methods have been developed to shift the scientific paradigm from a gene-centric to an organism-centric view of environmental genomic and transcriptomic data. These methods use direct assemblies of metagenomic reads to generate contigs that encompass several genes. High recovery of bacterial and archaeal genomes has been achieved through traditional assembly strategies from both high- and low-diversity environmental samples (Dick et al. 2009; Albertsen et al. 2013; Tully et al. 2018). However, when dealing with complex communities of organisms, traditional genome assembly approaches are often impaired by the large amount of sequence data and the genome heterogeneity.
To circumvent these limitations, approaches based on reference genomes have been proposed (Caron et al. 2009; Pawlowski et al. 2012). Other approaches are based on binning of assembly contigs across a series of samples to extract information (Sharon et al. 2012; Albertsen et al. 2013). Significant results for bacteria and archaea have been achieved so far (Parks et al. 2017; Delmont et al. 2018; Nayfach et al. 2019; Pasolli et al. 2019). However, eukaryotic organisms have larger, more complex genomes that require significantly higher sequence coverage than bacteria and archaea. Even in cases where significantly long contigs can be obtained, the mosaic structure of eukaryotic genes and the difficulty of predicting them de novo from a genome sequence leads to poor gene recovery (Boeuf et al. 2019). Only eukaryotes possessing small genomes and a high proportion of mono-exonic genes are expected to provide results similar to those achieved for bacteria or archaea. Recently, a semisupervised method based on a model trained with a set of diverse references allowed eukaryotic genome reconstruction from complex natural environments (West et al. 2018; Olm et al. 2019).
Novel reference-independent clustering approaches that produce genomes from metagenomic data have recently been developed (Albertsen et al. 2013; Imelfort et al. 2014; Nielsen et al. 2014; Delmont et al. 2018; Kang et al. 2019) and successfully applied to prokaryote-dominated communities. One of these approaches efficiently delineated co-abundant gene groups (CAGs), the largest of which were termed metagenomic species (MGS), across a series of human gut microbiome samples (Nielsen et al. 2014). This method uses the metagenomic abundance profiles of a reference gene catalog, determined by stringent mapping of raw metagenomic reads onto sequences in this catalog, and defines clusters of genes showing similar variations of abundance profiles across a collection of samples.
The vast majority of the planktonic biomass in the global ocean consists of single-cell eukaryotes and multicellular zooplankton (Dortch and Packard 1989; Gasol et al. 1997). Globally, these organisms play an important role in shaping the biogeochemical cycles of the ocean and significantly impact food webs and climate. Despite recent advances in understanding their taxonomic and gene functional compositions (Venter et al. 2004; Joly and Faure 2015; Carradec et al. 2018), little is known about the biogeographical preferences and metabolic potential of many eukaryotic plankton species from an organism-centric perspective. Several collections of reference marine eukaryote organisms’ sequences have been created, the largest one being the Marine Microbial Eukaryotic Transcriptome Sequencing Project (MMETSP) collection (Keeling et al. 2014). However, the majority of fully sequenced marine eukaryotic genomes or transcriptomes are derived from cultured organisms. Due to the limited availability of cultured representatives of many dominant in the open ocean plankton, including zooplankton representatives, reference sequences represent only a small fraction of the natural biological diversity (De Vargas et al. 2015; Sibbald and Archibald 2017).
Here, we used the rationale of this reference-independent, gene co-abundance method (Nielsen et al. 2014) to delineate transcriptomes by mapping metagenomic sequencing data obtained from 365 metagenomic read sets generated from marine water samples collected from the global ocean during the Tara Oceans expedition onto the metatranscriptome-derived Marine Atlas of Tara Oceans Unigenes (MATOU-v1 catalog [Carradec et al. 2018]) obtained from the same set of Tara Oceans stations (Supplemental Fig. S1). The samples were collected from all the major oceanic provinces except the Arctic, typically from two photic zone depths (subsurface [SRF] and deep-chlorophyll maximum [DCM]) and across four size fractions (0.8–5 µm, 5–20 µm, 20–180 µm, and 180–2000 µm).
Results
Construction of the MGT collection
Of the 116,849,350 metatranscriptomic-based unigenes of the MATOU-v1 catalog, 37,381,609 (32%) were detected by metagenomic reads mapping in at least three different Tara Oceans samples and displayed no more than 90% of their total genomic occurrence signal in a single sample. The metagenomic RPKM-based abundance matrix of these unigenes was submitted to a canopy clustering process (see Methods) that regrouped unigenes based on the covariation of their genomic abundance across the samples. Of these unigenes, 7,254,163 (19.5%) were clustered into 11,846 co-abundance gene groups with sizes varying from 2 to 226,807 unigenes. Nine hundred twenty-four CAGs consisting of at least 500 unigenes were termed metagenomics-based transcriptomes (MGTs) as they may constitute a significant part of an organism's transcriptome and which encompass 6,946,068 unigenes (Supplemental Table S1). For subsequent analyses, we focused on these more complete 924 MGTs since they more accurately represent organisms’ transcriptomes (Supplemental Fig. S1). This MGT collection recruited a significant number of metagenomic reads across Tara Oceans stations with an average of 58.5% (up to 94.5% for some samples) of the reads (Supplemental Fig. S2). The average number of taxonomically assigned unigenes across the MGTs was 44.6% (up to 99.5%). We have detected 22 MGTs with completeness higher than 50% (average contamination 16%) and 58 MGTs with completeness higher than 20% (average contamination 10.5%). Contamination was computed using a set of 83 protistan-specific single-copy core genes (Simão et al. 2015) or a set of 139 bacterial-specific single-copy core genes (Campbell et al. 2013) within the Anvi'o package (ver 5.2) (Eren et al. 2015). Since unigenes often represent not full genes but their fragments, single-copy core genes may map to multiple unigenes representing the same gene, which may lead to artificially high levels of contamination. MGT completeness significantly improved after the CAP3 assembly step (see Supplemental Material), which resulted in 74 MGTs with completeness higher than 50% and 131 MGTs with completeness higher than 20% (Supplemental Table S1).
Taxonomic diversity of the MGT collection
We studied the distribution of taxonomically assigned unigenes for each MGT across major planktonic taxa. In several cases, we observed a homogeneous distribution of taxonomic affiliations, suggesting that the MGTs represented transcriptomes of individual organisms (Supplemental Table S1; Supplemental Fig. S11). The accuracy of the taxonomic affiliations of the unigenes varied throughout the samples and depended on (1) the conservation level of a given sequence across species and (2) the adequacy and robustness of a reference database in regard to a given taxonomic unit (Carradec et al. 2018).
For each MGT, global taxonomic affiliation was determined by the taxonomic node that covered at least 75% of the taxonomically assigned unigenes of that MGT (see Methods for more detail). The MGT collection mostly comprised eukaryotic representatives (728 MGTs: 78%, 6,380,849 unigenes), followed by bacteria (148 MGTs: 16%, 454,253 unigenes), archaea (2 MGTs: 0.2%, 2844 unigenes), and viruses (1 MGT: 0.1%, 877 unigenes). Presence of bacteria and archaea in the MATOU-v1 catalog, despite using polyadenylated RNA for the sequencing step, can be explained by (1) the true nonpolyadenylated nature of these transcripts or (2) the low level of eukaryotic annotations in regard to prokaryotes in reference databases (Carradec et al. 2018). In this study, we focused only on the MGTs from the domain Eukaryota.
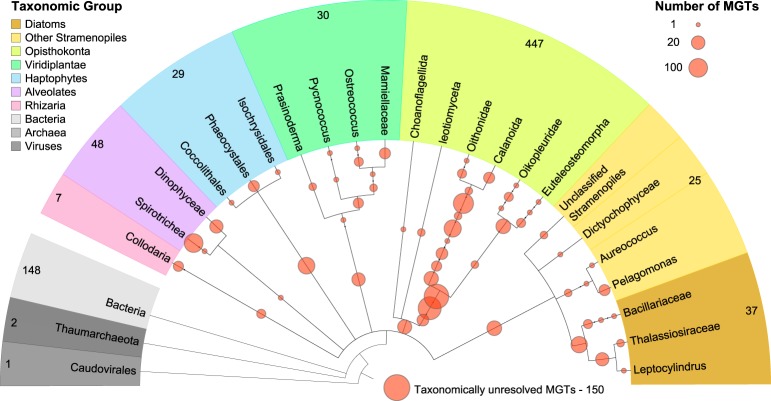
The overall taxonomic analysis of the MGT collection revealed that most of the major eukaryotic marine planktonic kingdoms (Worden et al. 2012) were covered, with the exception of Amoebozoa, Cryptophyta, and Rhodophyta (Fig. 1). Most of the MGTs with a low-resolution global taxonomic assignment (i.e., those for which the taxonomic affiliation could be assigned only at the kingdom level or higher) were related to the Opisthokonta group (447 out of 728 MGTs) or unclassified Eukaryota (105), whereas the well-defined eukaryotic MGTs (i.e., those for which the taxonomic affiliation could be assigned at the class level or deeper) belonged to unicellular algae: Stramenopiles (62), Alveolata (48), Viridiplantae (30), and Haptophyceae (29) lineages. This low taxonomic resolution of the MGTs could be due to (1) a low number of zooplankton organisms, including representatives from the Opisthokonta group, in the reference databases or (2) the presence of associations of several organisms in a given MGT. Overall, these observations suggest that the MGTs correspond to either organisms with available transcriptomes or those without sequenced representatives. Taxonomic diversity of the MGT collection differs significantly compared to the collections of reference transcriptomes derived from cultured organisms (Supplemental Fig. S3), including the MMETSP project (Keeling et al. 2014). Possible explanations for this include (1) the essentially coastal origin of cultured strains and (2) the absence of zooplankton in the MMETSP selected organisms.
Figure 1.
A taxonomic dendrogram representing the eukaryotic tree of life shows taxonomic positions of MGTs (orange circles) in relation to the major eukaryotic lineages. The size of the circles represents the number of MGTs positioned at a given taxonomic node. The total number of MGTs assigned to each taxonomic group is indicated on the outside of the tree.
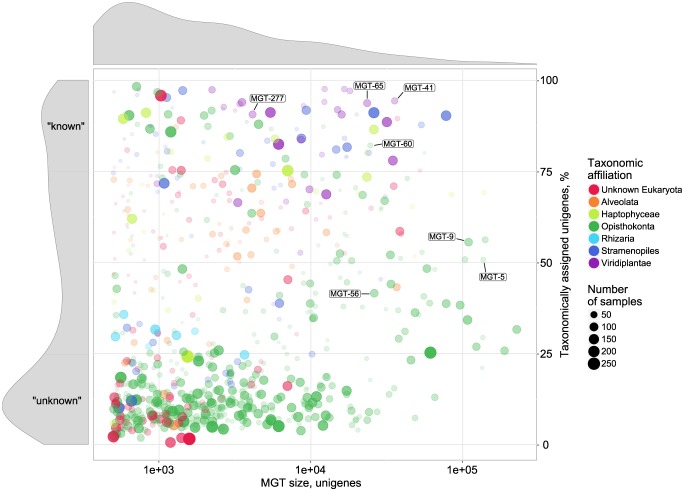
While 48.3% of the unigenes in the MATOU-v1 catalog and 47.3% of the unigenes captured in the MGTs were taxonomically assigned, we observed an uneven bimodal distribution of taxonomically assigned unigenes among the MGTs resulting in two distinct MGT groups (Fig. 2). The first group (228 MGTs, 24.7%) consisted of well-defined MGTs (>75% of their unigenes had a taxonomic assignment), representing “known” organisms, whereas the second group included 385 MGTs (41.7%) that were taxonomically poorly characterized (<25% of their unigenes were taxonomically assigned), which may represent currently undescribed genomes or mainly contain noncoding sequences and thus do not match with known proteins. The observed discontinuous distribution was not correlated with the MGT size or the number of samples in which an MGT was observed (Supplemental Fig. S4).
Figure 2.
A visual representation of the major MGT statistics including the MGT size (represented by the number of unigenes, x-axis) and the fraction of taxonomically assigned unigenes (y-axis). The circle size and its opacity represent the number of samples in which a given MGT was detected. Taxonomic affiliation of the MGTs to major eukaryotic lineages is color-coded. Size distribution of MGTs based on the number of unigenes is displayed on top of the main figure. Distribution of taxonomically assigned unigenes among MGTs is presented on the left-hand side panel of the figure. “Known” and “unknown” sections of the panel indicate the MGTs comprising >75% and <25% of taxonomically assigned unigenes, respectively. Highlighted MGTs were used for the biological validation of the MGT collection (see Results for more detail); Bathycoccus prasinos—MGT-41, MGT-65, MGT-277; Oithona nana—MGT-5, MGT-9, MGT-56, MGT-60.
Comparison with available transcriptomes
To assess the biological validity of the obtained MGTs, we investigated the distribution of unigenes from two marine planktonic reference organisms in the MGT collection. We analyzed reference transcriptomes from a single-celled microeukaryote Bathycoccus prasinos and a small multicellular zooplankton Oithona nana. The rationale for choosing these organisms as references was as follows: (1) They play an important role in the functioning of marine ecosystems; (2) their transcriptomes were publicly available (Keeling et al. 2014; Madoui et al. 2017); (3) they were hypothesized to be present in the data sets because of their high abundance in marine waters as demonstrated by an 18S rDNA survey from the same samples (De Vargas et al. 2015); and (4) they cover a range of organisms with substantially different transcriptome sizes (5.6–24 Mb), from phytoplankton and zooplankton groups, and from small and large size fractions of planktonic communities. We were able to recover an average of 68% (up to 77%) of the reference transcriptomes utilizing the MGT unigenes with at least 95% sequence identity over at least 50 amino acids (Fig. 2).
Segregation of the Bathycoccus ecotypes
To assess the potential of the MGT approach to segregate closely related biological entities, we focused on the MGTs highly similar to the reference transcriptomes of Bathycoccus prasinos. Bathycoccus is a genus of green algae from the order Mamiellales which is ecologically relevant because it is widely distributed in the global ocean and contributes significantly to primary production (Vannier et al. 2016; Limardo et al. 2017). Recent omics-based studies demonstrated the existence of at least two ecotypes of Bathycoccus (B1 and B2) which have identical 18S rRNA sequences but whose orthologous proteins share only 82 ± 6% nucleotide identity (Vaulot et al. 2012; Vannier et al. 2016).
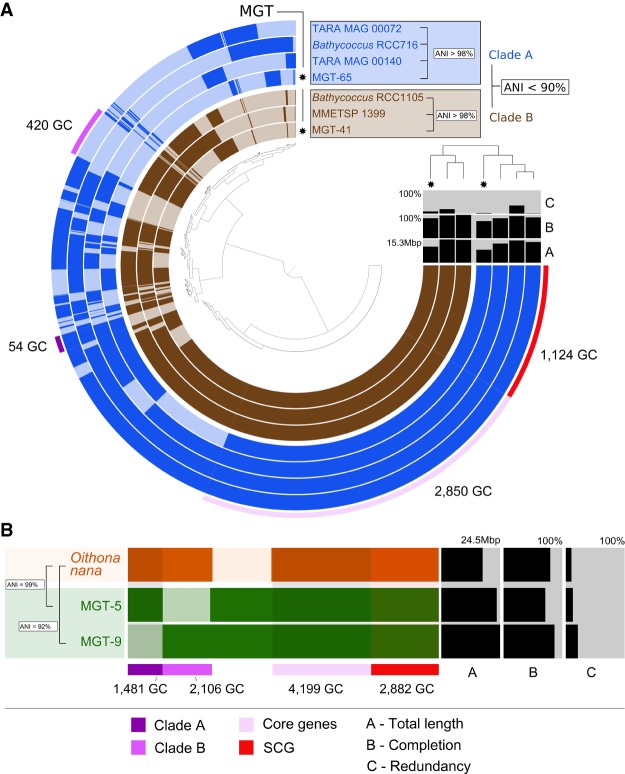
Both of these Bathycoccus ecotypes were detected in the MGT collection, and 99% of the total number of unigenes similar to B. prasinos were divided into three MGTs (MGT-41, MGT-65, MGT-277) (Supplemental Table S2). We focused on MGT-41 and MGT-65 because they comprised 95.2% of the signal in this group. Pangenomic analysis demonstrated a clear segregation between the two MGTs (Fig. 3A). The average nucleotide identity (ANI) analysis indicated <90% sequence similarity between them, further confirming their affiliation to different ecotypes. On the other hand, the ANI values between MGT-41 and its closest reference (isolate RCC1105) and MGT-65 and its closest reference (RCC716) were 98.2% and 98.8%, respectively. The estimated completeness of the assembled unigenes (computed based on a set of 83 protistan-specific single-copy core genes (Simão et al. 2015) was 85.5% for MGT-41 and 73.5% for MGT-65, suggesting a high level of the transcriptome recovery. The completeness estimation method applied here is based on the identification of a set of well-conserved single-copy core orthologs which usually demonstrate high levels of expression. Since not all genes from a given organism are always transcribed and thus some of them may not be captured in the MATOU-v1 catalog, this technique may overestimate the real completeness of our MGTs. However, to the best of our knowledge, this is currently the best available computational approach that estimates transcriptome completeness with a fairly high degree of confidence. MGT-41 and MGT-65 demonstrated different biogeographical preferences associated with environmental parameters, including temperature and oxygen concentrations (Supplemental Fig. S5). This observation supports previous findings about the differential biogeography of the Bathycoccus prasinos ecotypes B1 and B2 described in Vannier et al. (2016), leading us to assign MGT-41 to the ecotype B2 and MGT-65 to the ecotype B1. Together, these results further confirm the ability of the MGT analysis to segregate closely related eukaryotic plankton (even species with identical 18S rRNA gene sequences) in complex environmental samples.
Figure 3.
The pangenomes of (A) Bathycoccus prasinos and (B) Oithona nana compared to available sequenced references. Each layer represents an MGT, a reference genome, or transcriptome. Gene clusters are organized based on their distribution across samples. The dendrogram in the center organizes gene clusters based on their presence or absence in the samples. The top right dendrogram represents the hierarchical clustering of the samples based on the abundance of gene clusters. (ANI) Average nucleotide identity, (SCG) single-copy core genes, (GC) gene cluster.
Biogeography of the genus Oithona
Zooplankton, including cyclopoid copepods, also play an important role in marine ecosystems. They impact biogeochemical cycles and are key components of the oceanic food web (Roemmich and McGowan 1995; Keister et al. 2012; Steinberg and Landry 2017). However, only a few ecologically relevant references are currently available in public databases. Despite recent advances in the population genomics of the genus Oithona, one of the most abundant and widespread copepods in the pelagic ocean (Gallienne and Robins 2001; Madoui et al. 2017), more data are required to study the distribution of Oithona species and other copepods in the global ocean.
Ninety-eight percent of the Oithona-related unigenes were detected in four MGTs (MGT-5, MGT-9, MGT-56, and MGT-60), with MGT-5 and MGT-9 alone generating 87% of the signal (Supplemental Table S2). The estimated completeness of the assembled unigenes was 71% for MGT-5 and 87% for MGT-9 (Fig. 3B). MGT-5 demonstrated highly specific biogeographical preferences being observed at only 12 Tara Oceans stations, all from the Mediterranean Sea, which correlates well with the demonstrated biogeographic distribution of Oithona nana (Madoui et al. 2017). On the other hand, MGT-9 was detected at 41 stations, mostly located in the Pacific Ocean and the Mediterranean Sea but also in the Indian and Atlantic Oceans (Supplemental Fig. S12). These biogeographical preferences along with the significant difference in the ANI values relative to the reference transcriptome of O. nana (99% and 92% for MGT-5 and MGT-9, respectively) (Fig. 3B) suggest that MGT-9 may represent a different, yet genomically undescribed, species within the genus Oithona.
Functional insights from MGTs
After demonstrating the biological validity of the MGTs, we studied their potential to assess the functional state of the ecosystem through the analysis of ecologically relevant metabolic pathways and individual marine organisms. We analyzed the expression patterns, taxonomic affiliation, and geographical distribution of the genes coding for the key enzymes involved in the cycling of dimethylsulfoniopropionate (DMSP). We also investigated the interspecies relationship between an uncultivated unicellular cyanobacterium Candidatus Atelocyanobacterium thalassa (UCYN-A) and a haptophyte picoplankton alga of the class Prymnesiophyceae.
DMSP synthesis and degradation
Eukaryotic plankton, along with bacteria, are actively involved in the cycling of DMSP, an ecologically relevant organosulfur compound that can reach high concentrations in marine waters. DMSP is the precursor of the climate-active gas dimethyl sulfide (DMS), the largest natural source of sulfur to the atmosphere. This compound is also important from an organismal point of view because of its ability to act as an osmolyte, antioxidant, predator deterrent, and cryoprotectant in phytoplankton (Stefels et al. 2007; Bullock et al. 2017).
We investigated the expression patterns and geographical distribution of the genes involved in the production and degradation of DMSP across Tara Oceans stations. While the role of bacteria in DMSP cycling is well described (Reisch et al. 2011; Johnston et al. 2016; Bullock et al. 2017; Curson et al. 2017), recent identification of eukaryote counterpart genes (1) DSYB, coding for a methyl-thiohydroxybutyrate methyltransferase, a key enzyme involved in the DMSP synthesis in eukaryotes (Curson et al. 2018), and (2) Alma1 coding for dimethylsulfoniopropionate lyase 1, an enzyme responsible for the cleavage of DMSP into dimethyl sulfide and acrylate (Alcolombri et al. 2015; Johnson et al. 2016), allows investigation of the role of marine phytoplankton in the global DMSP cycle. Both of these genes were detected in the MGT collection, which allowed us to study their expression and biogeography from a genome-centric point of view (Supplemental Table S3).
Out of 1220 DSYB-related unigenes detected in the MATOU-v1 catalog, 1214 were taxonomically assigned to eukaryotes (Supplemental Fig. S6A). They were detected at all of the 66 sampling stations of the Tara Oceans cruise analyzed in this study and were mainly attributed to the pico-eukaryote size fraction (0.8–5 µm) (Supplemental Fig. S7). Forty-six DSYB-related unigenes were detected in 20 different MGTs, and their expression in 10 out of 20 MGTs represented more than 10% of the total DSYB expression in at least one sample. This further confirms the importance of the organisms related to these MGTs in the DMSP production (Fig. 4A). Two MGTs affiliated to the genus Phaeocystis (MGT-4 and MGT-13) were prevalent (up to 45% of the total DSYB expression signal) at the Southern Ocean stations (82–85), whereas a Haptophyte-related organism (MGT-29) was prevalent (from 10% to 35%, depending on the size fraction) at station 80 in the southern Atlantic. Phaeocystis-affiliated MGTs demonstrated high levels of DSYB expression both in the small size fraction (0.8–5 µm, corresponding to individual cells) and in large size fractions (20–180 and 180–2000 µm, corresponding to multicellular colonies which may form during P. antarctica blooms [Carlson et al. 1998; Smith et al. 1998; Wang et al. 2018]). A large portion of the total DSYB expression (from 10% to 70%) was also attributed to the Chloropicophyceae lineage (previously referred to as Prasinophytes clade VII) (Lopes Dos Santos et al. 2017a,b; Turmel et al. 2019): MGT-44 at the Pacific Ocean stations (93, 102, 109, 110, 122–125, 128, 136, 137, 139, and 144) and MGT-166 and MGT-179 at the Indian Ocean stations (36, 38, 39, and 41). In addition to these differences in their geographical distribution, MGT-166 and MGT-179 shared more genomic similarity between them (ANI = 95.23%), whereas MGT-44 was more distantly related (ANI = 86.86 and ANI = 85.97% with MGT-166 and MGT-179, respectively), suggesting that the former two may represent closely related organisms and the third one corresponds to a different transcriptome. However, given the small number of available Chloropicophyceae-related references in DNA databases, this hypothesis requires further investigation.
Figure 4.
Comparison of the DSYB (A) and Alma1 (B) relative gene expression per sample (x-axis) and per MGT (y-axis) across samples. Numbers near each point represent a Tara Oceans station. (SRF) Surface, (DCM) deep chlorophyll maximum. Y-axis (to the left of panel A) and size fractionation (to the right of panel B) are common for both figures. Tara Oceans provinces are applicable to both graphs and are specified on the right side of panel B. (ATLN) North Atlantic Ocean, (ATLS) South Atlantic Ocean, (SO) Southern Ocean, (INDIAN) Indian Ocean, (MEDIT) Mediterranean Sea, (PACN) North Pacific Ocean, and (PACS) South Pacific Ocean. (A) DSYB expression profiles across Tara Oceans stations and size fractions for 10 MGTs contributing significantly to the overall DSYB expression (at least 10% of the total DSYB expression in at least one sample). Red circles (MGT-4 and MGT-13) represent MGTs taxonomically assigned to the genus Phaeocystis; green circles (MGT-44, MGT-166, and MGT-179) represent Chloropicophyceae-affiliated MGTs; the rest of the circles represent other organisms. (B) Alma1 expression profiles across Tara Oceans stations and size fractions for nine MGTs contributing significantly to the overall Alma1 expression (at least 10% of the total Alma1 expression in at least one sample). Red circles (MGT-4, MGT-13, and MGT-67) represent MGTs taxonomically assigned to the genus Phaeocystis; yellow circles (MGT-178) represent Pelagomonas spp.-affiliated MGTs; and the rest of the circles represent other organisms.
It is well established that organisms from the classes Dinophyceae (dinoflagellates) and Prymnesiophyceae (coccolithophores) are major producers of DMSP, but only few studies are available that investigate the involvement of Chloropicophyceae in DMSP production (Keller 1989; Keller et al. 1989; Kiene et al. 1997). Our data suggest that there are at least two distinct representatives of Chloropicophyceae involved in DMSP biosynthesis and provide preliminary results on a possible pathway for DMSP production in this group.
We also investigated the expression of the Alma1 gene coding for a key enzyme of the DMSP degradation pathway (Alcolombri et al. 2015). We detected 1059 Alma1-related unigenes in the MATOU-v1 catalog (Supplemental Fig. S6B). The expression of these unigenes was detected mostly in the smallest size fraction (0.8–5 µm) at both depths (surface and DCM) at 66 sampled stations (Supplemental Fig. S8). No taxonomic affiliation was found for 153 Alma1 unigenes. The highest levels of Alma1 abundance and expression were detected at the Southern Ocean stations (Supplemental Fig. S8). Thirty-six Alma1-related unigenes were detected in 13 MGTs. Most of these Alma1 unigenes were taxonomically affiliated to the clade Alveolates (78%), followed by the family Haptophyceae (5%). However, 30 out of the 36 Alma1-related unigenes present in the MGT collection were concentrated in six MGTs taxonomically assigned to Haptophytes. Even though 48 MGTs possessed unigenes assigned to Alveolates, we did not detect any Alma1-containing MGTs affiliated to this group. In nine out of 13 MGTs containing Alma1 unigenes, Alma1 expression contributed more than 10% (in some cases, up to 40%) to the total Alma1 expression detected across 43 samples (Fig. 4B). MGTs demonstrating the highest levels of Alma1 expression were taxonomically assigned to Phaeocystis (MGT-4, MGT-13, and MGT-67) and Pelagomonas spp. (MGT-178).
Identification of interspecies associations
We investigated the interspecies relationship between an uncultivated diazotrophic unicellular cyanobacterium Candidatus Atelocyanobacterium thalassa (Zehr et al. 2008; Thompson et al. 2012) and a haptophyte picoplankton alga Braarudosphaera bigelowii (B. bigelowii) from the class Prymnesiophyceae. Both members of this association are abundant and widely distributed in the ocean and are ecologically relevant because of their ability to fix N2 (Zehr and Kudela 2011; Farnelid et al. 2016). Several UCYN-A genomes have been previously sequenced (Tripp et al. 2010; Bombar et al. 2014), whereas no genomic information is currently available for the algal host.
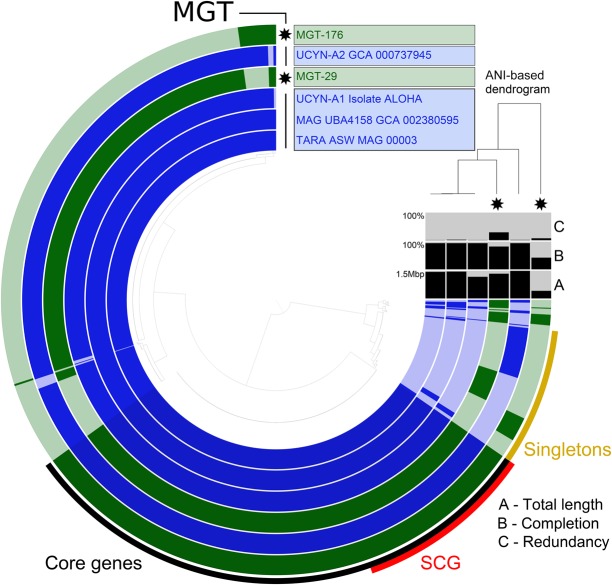
Detected in the MATOU-v1 catalog were 2616 UCYN-A-affiliated unigenes (see Methods). They were distributed among 41 Tara Oceans stations and were mostly present in the small size fraction (0.8–5 µm). The majority of the UCYN-A-affiliated unigenes (96%) were detected in two MGTs: 1742 unigenes in MGT-29 and 771 unigenes in MGT-176 (Supplemental Fig. S9). In addition to the unigenes affiliated with the diazotrophic cyanobacterium, MGT-29 also possessed ∼20,000 unigenes taxonomically assigned to the Haptophyte clade and possibly representing the eukaryotic host of this symbiosis, a Prymnesiophyte closely related to B. bigelowii. Together with the observation that the host's 18S rDNA V4 region was identified in the same samples as MGT-29, this suggests that the non-UCYN-A-affiliated genes of MGT-29 may be a part of the transcriptome of the host. Comparison of the MGTs comprising UCYN-A-related unigenes with reference genomes (Tripp et al. 2010; Bombar et al. 2014) and metagenome-assembled genomes (MAGs) (Parks et al. 2017; Delmont et al. 2018) demonstrated that MGT-29 unigenes covered 90.3% of the UCYN-A1 genome (Fig. 5). The estimated completeness of the UCYN-A genome computed based on a set of 139 bacterial-specific single-copy core genes (Campbell et al. 2013) was 82.7% for MGT-29 and 42.4% for MGT-176. The ANI value between MGT-29 and UCYN-A1 isolate ALOHA was 99.7%, which indicates a high genomic similarity. We hypothesize that MGT-176 may represent UCYN-A2 or another UCYN-A sublineage, because the ANI analysis demonstrated its higher genomic similarity with isolate SIO64986 (UCYN-A2) than isolate ALOHA (UCYN-A1) (97.1% and 94.3%, respectively).
Figure 5.
The pangenomes of MGT-29 and MGT-176 compared to available reference sequences of UCYN-A. Each layer represents an MGT, a reference genome, or a MAG. Gene clusters are organized based on their distribution across samples. The dendrogram in the center organizes gene clusters based on their presence or absence in the samples. The top right dendrogram represents the hierarchical clustering of the samples based on the ANI values. (ANI) Average nucleotide identity, (SCG) single-copy core genes.
In addition to the UCYN-A-related genes, MGT-29 also contained multiple core metabolism genes taxonomically assigned to the Haptophyte clade which may belong to the eukaryotic host of this symbiotic association, B. bigelowii (Supplemental Data Set S2). More specifically, we detected genes coding for enzymes driving major metabolic pathways in the haptophyte algae including glycolysis, the tricarboxylic acid (TCA) cycle, the pentose phosphate pathway, the GS-GOGAT cycle (ammonium assimilation through sequential actions of glutamine synthetase [GS] and glutamate synthase [GOGAT]), as well as multiple genes affiliated with the metabolism of fatty acids and amino acids. We also observed the presence of the gene bacA, coding for the ABC transporter involved in the transport of vitamin B12.
In addition to the UCYN-A symbiosis with a single-celled haptophyte, we detected other known microbial associations in the MGT collection (Supplemental Table S4). For example, MGT-738 partially captured an association between a diatom from the class Coscinodiscophyceae (47 unigenes) and the nitrogen-fixing heterocystous cyanobacteria, Richelia intracellularis (132 unigenes), an abundant organism in tropical and subtropical waters (Janson et al. 1999; Lyimo 2011). This MGT collection also detected an association between a pennate diatom from the family Bacillariaceae and a tintinnid ciliate, genes from both of which were detected in MGT-136 (243 and 192 unigenes affiliated to the diatom and tintinnid, respectively) (Vincent et al. 2018).
Discussion
Many eukaryotic lineages of ocean plankton remain largely undersampled, and as a result, there are few sequenced representatives for many ecologically important marine eukaryotic organisms. The MGT collection reported here represents a valuable resource for studying a range of eukaryotic planktonic organisms, including those that are largely unexplored using traditional omics techniques.
The gene clustering approach applied here provides an organism-centric view of the most abundant plankton populations. This approach allowed us to focus on the diversity and functional potential of marine eukaryotic organisms across major taxonomic lineages. The 924 MGTs generated from the Tara Oceans data sets contain an impressive diversity of taxa, allowing for comparative genomic studies in major eukaryotic groups, including Opisthokonta, Haptophytes, Stramenopiles, Alveolates, Archaeplastida, and Rhizaria (Fig. 1). Additionally, this collection of MGTs provides the first glimpse of the genomic content of a variety of organisms currently not available in culture, including copepods, one of the most prevalent zooplankton of the photic open ocean. Only a small number of MGTs have closely related cultured references, suggesting that many MGTs represent organisms which have only distant relatives within the publicly available collections of sequenced genomes or transcriptomes. These MGTs provide access to valuable genomic information currently not accessible through other DNA-based resources. For example, limited availability of full genomes or transcriptomes representing heterotrophic organisms prevents advances in studying their distribution, population structure, and functional potential. Access to the zooplankton transcriptomes through the MGT collection will increase our knowledge on their biogeography and complement the general lack of references for the copepods group. Alternatively, the analysis of the MGTs closely related to organisms which have sequenced representatives may lead to a re-evaluation of their genomic potential and provide additional information on their ecology.
DMSP biosynthesis and degradation by eukaryotes
We demonstrated the ability of the MGT approach to assess the contribution of eukaryotic plankton to ecologically important processes by focusing on the MGTs expressing genes coding for key enzymes involved in DMSP cycling. These genes included DSYB, coding for a methyl-thiohydroxybutyrate methyltransferase, a key enzyme of the eukaryotic DMSP synthesis pathway (Curson et al. 2018) and Alma1 coding for dimethylsulfoniopropionate lyase 1, an algal enzyme that cleaves DMSP into DMS and acrylate (Alcolombri et al. 2015). We revealed the importance of Phaeocystis spp. in the Southern Ocean as potential DMSP producers and degraders. Our data also indicated the involvement of at least two representatives from Chloropicophyceae in the biosynthesis of DMSP. One of them was primarily active at the oligotrophic equatorial Pacific stations, while the other one appeared to be restricted to the low oxygen stations (the Arabian basin and upwelling stations in the East Pacific). However, involvement of Chloropicophyceae in DMSP production is currently supported only by circumstantial evidence, mostly in early studies (Keller 1989; Keller et al. 1989; Kiene et al. 1997), which demonstrates a clear need for more sequenced references of this group of organisms. Our results also support recent findings stating that picoeukaryotes should be considered as important contributors to DMSP production through the DSYB pathway (Curson et al. 2018). These organisms may represent interesting targets for the experimental validation of their role in the global biogeochemical sulfur cycle and their impact on climate change as proposed in the CLAW hypothesis (Charlson et al. 1987; Ayers and Cainey 2007). Thus, the MGT approach allowed us to identify candidate organisms responsible for a large part of the eukaryotic DMSP biosynthesis and catabolism in the different regions of the open ocean.
Presence of different dominating groups of organisms involved in the DMSP cycling across oceanic regions suggests the importance of environmental conditions in shaping microbial community composition that defines the DMSP fate in the ocean. If environmental conditions change in a given ecological niche, we may expect that DMSP production and degradation rates would also change because of the transformations in the microbial community structure, which may lead to significant effects on climate change.
Interspecies associations
The importance of the MGT collection as a resource for studying marine interspecies interactions was demonstrated through detection of the ecologically relevant symbiosis between the metabolically streamlined nitrogen-fixing cyanobacterium UCYN-A and a single-celled haptophyte picoplankton alga. Initially, this symbiosis was discovered using a targeted approach that involved several culture-dependent and molecular techniques, proving it to be a challenging task (Zehr et al. 2017). Several UCYN-A sublineages have been defined, but limited information is currently available regarding their global distribution and, for some, the identity of the host (Thompson et al. 2014; Farnelid et al. 2016; Turk-Kubo et al. 2017). MGT-29 from our collection encompasses genes similar to those from UCYN-A1 strain ALOHA and genes taxonomically affiliated with the Haptophyte clade potentially representing the eukaryotic host. This suggests that MGT-29 may represent UCYN-A1 specifically associated with a closely related to B. bigelowii prymnesiophyte. More information is needed about the genomic content of the host cells for different UCYN-A sublineages to confidently state which symbiosis was detected.
No genomic information is available about these symbiotic hosts beyond 18S rRNA sequences (Hagino et al. 2013). As a result, many questions remain unanswered regarding the evolution of this symbiosis and the exact nature of the relationship between the two organisms. Through partial reconstruction of the host transcriptome, we provide the first glimpse of its genomic content, which will change the way in which this interspecies association can be studied. Better methods are needed for the accurate targeting of distinct UCYN-A/host associations, which will improve the understanding of the evolution and ecological characteristics of this symbiosis. Access to the genomic content of the host through the MGT collection, used in conjunction with the two closed UCYN-A genomes currently available in the databases (Tripp et al. 2010; Bombar et al. 2014), will provide a much-needed push in this direction.
Other ecologically important microbial associations were detected in the MGT collection. The diatom-cyanobacteria symbiotic populations captured in the MGT-738 which were previously observed in all major ocean basins (Foster and O'Mullan 2008) may encompass diatoms from several genera, including Hemiaulus, Rhizosolenia, and Chaetoceros. These associations may contribute as much new nitrogen (N) as the free-living diazotroph Trichodesmium, which is widely regarded as the most important player responsible for N2 fixation in the open ocean (Capone et al. 2008). It was reported that the contribution of the diatom symbioses to the global pool of N had been underestimated and that they should be included in global N models (Foster et al. 2011). In order to accurately do so, additional information on their genomic potential is required and can be accessed through the MGT collection.
Little is known about the nature of a diatom-tintinnid association detected in MGT-136. One hypothesis suggests a mutualistic symbiosis, where diatoms acquire increased motility and tintinnids benefit from silicification through increased protection (Vincent et al. 2018). Other data indicate that the tintinnid can be the only beneficiary of the association, whereas the diatom would play the role of the “victim” (Armbrecht et al. 2017). Recent studies suggest that diatom–tintinnid associations may be more common in the ocean than previously thought. However, their global ecological and biogeographical patterns remain poorly characterized (references within Vincent et al. 2018).
The MGT collection provides a valuable resource for the evaluation of these ecologically relevant associations by studying their distribution in major oceanic provinces and by exploring the expression patterns of key genes. These findings illustrate the ability of the MGT collection to depict more interspecies relationships in the ocean, thus potentially discovering previously unknown microbial associations (Supplemental Table S4), as well as to study their gene expression patterns.
Fragmentation of the MGTs
In addition to the MGTs, comprised of tens of thousands of unigenes, some of which cover eukaryotic reference transcriptomes with a high level of completeness, we also detected a number of smaller gene clusters which cannot reliably cover a full eukaryotic transcriptome. Several reasons may lead to the presence of these smaller MGTs representing partial eukaryotic transcriptomes. In some cases, not all of the genes from a specific organism can be detected in all of the samples where this organism is present—some genes may be missing or present at levels below the achieved sequence coverage. This may lead to the fragmentation of the MGTs, i.e., to the fact that genes from the same organism may be allocated to multiple CAGs and MGTs of various sizes (comprised of a different number of unigenes). Several possible scenarios exist: (1) Some accessory genes may be present and expressed in some subpopulations and missing in others; or (2) a sufficient sequencing depth was not achieved for some of the samples, resulting in only a partial genomic coverage. Thus, for organisms with sequencing depths below or near the limit of detection, some genes may lack corresponding reads, which would lead to incomplete coverage of the transcriptome by metagenomic reads. The situation when several CAGs of various sizes represent the same organism has been observed for the prokaryotic compartment of a human gut microbiome (Nielsen et al. 2014).
Limitations and advantages of the MGT method
General limitations relevant to interpreting the gene co-abundance data obtained using the MGT approach described here include its inability to incorporate the accessory genes in the analysis due to their inherent nature of not being present in all strains and its intrinsic inability to segregate organisms that form obligate symbioses because of their identical gene co-abundance profiles.
A recently developed computational tool may solve the former problem (Plaza Oñate et al. 2019), although further analyses on environmental data sets are needed to confirm its accuracy and efficiency. Alternatively, postprocessing of the MGT collection using methods based on differential sequence coverage of genes may be effective in cases where a significant bias in genome copy number of the associated organisms exists. Another caveat specific to our data sets is that genes expressed below the level of detection may be overlooked because the gene reconstruction has been performed using the metatranscriptomics data. However, the MGT approach has a number of advantages compared to other metagenome and metatranscriptome assembly methods. These include: (1) access to genomic content of organisms not available in culture (including zooplankton species) because of the culture-independent assembly and clustering of sequence data; and (2) de novo definition of gene clusters which allows for the reconstruction of transcriptomes with no need for references.
In this study, we applied a gene co-abundance clustering approach on a series of samples provided by the Tara Oceans expedition and demonstrated its efficiency for reconstructing high-quality eukaryotic transcriptomes. The resulting MGT collection provides a valuable resource for a comprehensive analysis of the eukaryotic plankton in the open sunlit ocean by providing access to biogeography, genomic content, and functional potential of many ecologically relevant eukaryotic species. This universal methodological framework can be implemented for transcriptome reconstruction of microscopic eukaryotic organisms in any environment provided that both metagenomic and metatranscriptomic data are available.
Methods
Sampling of eukaryotic plankton communities
The samples were collected during the 2009–2013 Tara Oceans expeditions from all the major oceanic provinces except the Arctic. For the majority of stations, samples were collected from two depths in the photic zone: subsurface and deep-chlorophyll maximum. Planktonic eukaryotic communities were collected in the 0.8- to 2000-µm range and divided into four size fractions (0.8–5 µm, 5–20 µm, 20–180 µm, and 180–2000 µm). A detailed description of the sampling strategies and protocols is available in the Supplemental Material and in Pesant et al. (2015). Biogeochemical data measured during the expedition are available in the Supplemental Material and in the Pangaea database (https://www.pangaea.de/).
DNA and RNA libraries were constructed and sequenced as detailed in Alberti et al. (2017) and processed as described in Carradec et al. (2018). Briefly, the raw data were filtered and cleaned to remove low-quality reads, adapters, primers, and ribosomal RNA-like reads. Resulting metatranscriptomic reads were assembled using Velvet v.1.2.07 (Zerbino et al. 2009) with a k-mer size of 63. Isoform detection was performed using Oases 0.2.08 (Schulz et al. 2012). Contigs smaller than 150 bp were removed from further analysis. The longest sequence from each cluster of contigs was kept as a reference for the gene catalog. The MATOU-v1 unigene catalog is accessible at https://www.genoscope.cns.fr/tara/.
Abundance computing and canopy clustering
The raw metagenomic (metaG) reads from 365 samples were mapped against the MATOU-v1 catalog as described in Carradec et al. (2018). Briefly, raw metagenomic reads from each sample were compared with the MATOU-v1 unigenes using the BWA tool (version 0.7.4) (Li and Durbin 2009), and those covering at least 80% of the read length with at least 95% of identity were retained for further analysis. In the case of several possible best matches, a random one was picked. For each unigene in each sample, the metagenomic abundance was determined in RPKM (reads per kilo base per million of mapped reads). To improve the clustering efficiency, we selected unigenes detected with metagenomic reads in at least three different samples and which had no more than 90% of their total genomic occurrence signal in a single sample. These two criteria are the default parameters of the canopy clustering tool (‐‐filter_min_obs 3 and ‐‐filter_max_top3_sample_contribution = 0.9). The metagenomic RPKM-based abundance matrix of these unigenes was submitted to the canopy clustering algorithm described in Nielsen et al. (2014) (the original code is available in Supplemental Code and at https://www.genoscope.cns.fr/tara/), which is a density-based clustering that does not take into account the sequence composition, as opposed to most binning tools. We used a max Pearson's correlation difference of 0.1 to define clusters, and then clusters were merged if canopy centroids’ distances were smaller than 0.05 (250 k iterations, default parameters).
A total of 7,254,163 unigenes were clustered into 11,846 co-abundant gene groups of at least two unigenes. CAGs with more than 500 unigenes are hereafter termed metagenomics-based transcriptomes. Nine hundred twenty-four MGTs were generated which encompassed 6,946,068 unigenes (95.8%). Since this method has never been applied to eukaryotic data, a smaller cutoff of 500 unigenes was used (compared to the original method applied to prokaryote-dominated communities [Nielsen et al. 2014]) to increase the number of resulting MGTs potentially representing individual organisms. For each sampling filter, we determined the fraction of metagenomics reads captured by the unigenes that compose the MGTs (Supplemental Fig. S2).
Taxonomic assignment
Taxonomic assignment of the unigenes is described in Carradec et al. (2018). Briefly, to determine a taxonomic affiliation for each of the unigenes, a reference database was built from UniRef90 (release of 2014–09–04) (Suzek et al. 2015), the MMETSP project (release of 2014–07–30) (Keeling et al. 2014), and Tara Oceans Single-cell Amplified Genomes (PRJEB6603). The database was supplemented with three Rhizaria transcriptomes (Collozoum, Phaeodaea, and Eucyrtidium), available through the European Nucleotide Archive under the reference PRJEB21821 (https://www.ebi.ac.uk/ena/data/view/PRJEB21821) and transcriptomes of Oithona nana (Madoui et al. 2017), available through the European Nucleotide Archive under the reference PRJEB18938 (https://www.ebi.ac.uk/ena/data/view/PRJEB18938). Sequence similarities between the gene catalog and the reference database were computed in protein space using DIAMOND (version 0.7.9) (Buchfink et al. 2015) with the following parameters: -e 1E-5 -k 500 -a 8 ‐‐more-sensitive. Taxonomic affiliation was performed using a weighted Lowest Common Ancestor approach. Subsequently, for each MGT, representative taxonomic level was determined by computing the deepest taxonomic node covering at least 75% of the taxonomically assigned unigenes of that MGT.
Completeness and contamination assessment
For each MGT, unigenes were further assembled using CAP3 (version date: 02/10/15) (Huang and Madan 1999). Assembled contigs and singletons were pooled, and completeness and contamination were computed using the Anvi'o package (ver 5.2) (Eren et al. 2015) with default parameters and a set of 83 protistan-specific single-copy core genes (Simão et al. 2015) for eukaryotes or a set of 139 bacterial-specific single-copy core genes (Campbell et al. 2013) for bacteria (Supplemental Table S1). Average nucleotide identity was computed using the dnadiff tool from the MUMmer package (ver 3.23) (Kurtz et al. 2004).
Functional characterization
DSYB-related unigenes were identified using Hidden Markov Models (HMMs) generated from 135 sequences extracted from Curson et al. (2018). These sequences were clustered using MMseqs2 (Steinegger and Söding 2017), and for each of the resulting 24 clusters, sequences were aligned using MUSCLE (Edgar 2004). HMM construction and unigenes catalog scanning were performed using HMMer (Wheeler and Eddy 2013). The DSYB HMM profile had significant matches (e-value ≤ 10−50) with 1220 unigenes in the MATOU-v1 catalog, 46 of which were found in the MGT collection (Supplemental Table S3).
Alma1-related unigenes were identified using HMMs generated from five sequences with demonstrated DMSP lyase activity, extracted from Alcolombri et al. (2015). These sequences were clustered using MMseqs2 (Steinegger and Söding 2017), and for each of the two resulting clusters, sequences were aligned using MAFFT v7.407 (Katoh and Standley 2013). HMM construction and unigenes catalog scanning were performed using HMMer (Wheeler and Eddy 2013). We identified 1069 positive unigenes (e ≤ 10−50) from the MATOU-v1 catalog; 36 of them were found in the MGT collection (Supplemental Table S3).
Unigene expression values were computed in RPKM. The expression of DSYB- and Alma1-related unigenes was normalized to the total number of reads mapped to a given MGT and to the total number of reads in a given sample (Fig. 4).
Comparison with reference transcriptomes
To assess the biological validity of the resulting MGT, the reference transcriptomes of Bathycoccus prasinos from the MMETSP database (release of 2014-07-30 [Keeling et al. 2014]) and the reference transcriptome of Oithona nana from Genoscope (Madoui et al. 2017) were used. Sequence similarities between the unigenes and the reference transcriptomes were computed in protein space using DIAMOND (version 0.7.9) (Buchfink et al. 2015) with the following parameters: -e 1Ee-5 -k 500 -a 8, and positive matches were defined as ≥95% identity over at least 50 amino acids.
Identification of potential interspecies interactions
We screened the MGT collection for potential interspecies associations by focusing on the MGTs that meet two criteria: (1) These MGTs must contain at least 10 unigenes from two different subkingdom taxonomic units; and (2) the number of unigenes associated with one of these taxonomic units must account for at least 5% of the number of unigenes associated with the other taxonomic unit. For example, MGT-29 contains 19,652 unigenes assigned to Haptophyceae and 1940 unigenes (9.9%) assigned to cyanobacteria. All the MGTs that met these criteria are listed in Supplemental Table S4. See Supplemental Material for more detail.
Statistical analysis
All statistical analyses and graphical representations were conducted in R (v 3.3.2) (R Core Team 2019) with the R package ggplot2 (v 2.2.1). The taxonomic dendrogram shown in Figure 1 was built using the phyloT and NCBI Taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy) toolkits of the Python ETE3 package and visualized using iTOL (Letunic and Bork 2016). The world maps were obtained using the R packages grid (v 3.3.2) and maps (v 3.2.0). Inkscape 0.92.3 was used to finalize the figures.
Data access
Sequencing data generated in this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under accession number PRJEB4352 for the metagenomics data and PRJEB6609 for the metatranscriptomics data. The unigene catalog generated in this study has been submitted to the ENA under accession number ERZ480625. The MGT collection data and environmental data are available in Supplemental Material in Supplemental Data Set S1, at https://www.genoscope.fr/tara/, and in the Pangaea database (https://www.pangaea.de/). MGT nucleic sequences in FASTA format and MGT post-assemblies generated through CAP3 are available at https://www.genoscope.fr/tara/. See Supplemental Material for more detail.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank the following people and sponsors whose commitment made this singular expedition possible: Centre National de la Recherche Scientifique (CNRS) (in particular, Groupement de Recherche GDR3280), the European Molecular Biology Laboratory (EMBL), Genoscope/CEA (Commissariat à l’Énergie Atomique et aux Énergies Alternatives), the French Government “Investissement d'Avenir” programs Oceanomics (ANR-11-BTBR-0008), and FRANCE GENOMIQUE (ANR-10-INBS-09), Fund for Scientific Research – Flanders, VIB, Stazione Zoologica Anton Dohrn, UNIMIB, agnès b., the Veolia Environment Foundation, Region Bretagne, World Courier, Illumina, Cap L'Orient, the EDF Foundation EDF Diversiterre, FRB, the Prince Albert II de Monaco Foundation, Etienne Bourgois, the Tara schooner and its captain and crew. Tara Oceans would not exist without continuous support from 23 institutes (https://oceans.taraexpeditions.org). We also thank C. Scarpelli for support in high-performance computing. Computations were performed using the platine, titane, and curie HPC machine provided through Grand Équipement National de Calcul Intensif (GENCI) grants (t2011076389, t2012076389, t2013036389, t2014036389, t2015036389, and t2016036389). This article is contribution number 104 of Tara Oceans.
Author contributions: A.V., P.W., and E.P. designed the research; A.V., Q.C., and E.P. generated the data; A.V., M.D., Q.C., A.A., T.O.D., and E.P. analyzed the data; and A.V. and E.P. wrote the paper, with assistance from all authors. All authors approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.253070.119.
References
- Alberti A, Poulain J, Engelen S, Labadie K, Romac S, Ferrera I, Albini G, Aury JM, Belser C, Bertrand A, et al. 2017. Viral to metazoan marine plankton nucleotide sequences from the Tara Oceans expedition. Sci Data 4: 170093 10.1038/sdata.2017.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. 10.1038/nbt.2579 [DOI] [PubMed] [Google Scholar]
- Alcolombri U, Ben-Dor S, Feldmesser E, Levin Y, Tawfik DS, Vardi A. 2015. Identification of the algal dimethyl sulfide-releasing enzyme: a missing link in the marine sulfur cycle. Science 348: 1466–1469. 10.1126/science.aab1586 [DOI] [PubMed] [Google Scholar]
- Armbrecht LH, Eriksen R, Leventer A, Armand LK. 2017. First observations of living sea-ice diatom agglomeration to tintinnid loricae in East Antarctica. J Plankton Res 39: 795–802. 10.1093/plankt/fbx036 [DOI] [Google Scholar]
- Ayers GP, Cainey JM. 2007. The CLAW hypothesis: a review of the major developments. Environ Chem 4: 366–374. 10.1071/EN07080 [DOI] [Google Scholar]
- Boeuf D, Edwards BR, Eppley JM, Hu SK, Poff KE, Romano AE, Caron DA, Karl DM, DeLong EF. 2019. Biological composition and microbial dynamics of sinking particulate organic matter at abyssal depths in the oligotrophic open ocean. Proc Natl Acad Sci 116: 11824–11832. 10.1073/pnas.1903080116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombar D, Heller P, Sanchez-Baracaldo P, Carter BJ, Zehr JP. 2014. Comparative genomics reveals surprising divergence of two closely related strains of uncultivated UCYN-A cyanobacteria. ISME J 8: 2530–2542. 10.1038/ismej.2014.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brum JR, Ignacio-Espinoza JC, Roux S, Doulcier G, Acinas SG, Alberti A, Chaffron S, Cruaud C, de Vargas C, Gasol JM, et al. 2015. Patterns and ecological drivers of ocean viral communities. Science 348: 1261498 10.1126/science.1261498 [DOI] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12: 59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- Bullock HA, Luo H, Whitman WB. 2017. Evolution of dimethylsulfoniopropionate metabolism in marine phytoplankton and bacteria. Front Microbiol 8: 637 10.3389/fmicb.2017.00637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JH, O'Donoghue P, Campbell AG, Schwientek P, Sczyrba A, Woyke T, Söll D, Podar M. 2013. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci 110: 5540–5545. 10.1073/pnas.1303090110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone DG, Bronk DA, Mulholland MR, Carpenter EJ. 2008. Nitrogen in the marine environment. Elsevier, Amsterdam. [Google Scholar]
- Carlson CA, Ducklow HW, Hansell DA, Smith WO. 1998. Organic carbon partitioning during spring phytoplankton blooms in the Ross Sea polynya and the Sargasso Sea. Limnol Oceanogr 43: 375–386. 10.4319/lo.1998.43.3.0375 [DOI] [Google Scholar]
- Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB. 2009. Protists are microbes too: a perspective. ISME J 3: 4–12. 10.1038/ismej.2008.101 [DOI] [PubMed] [Google Scholar]
- Carradec Q, Pelletier E, Da Silva C, Alberti A, Seeleuthner Y, Blanc-Mathieu R, Lima-Mendez G, Rocha F, Tirichine L, Labadie K, et al. 2018. A global ocean atlas of eukaryotic genes. Nat Commun 9: 373 10.1038/s41467-017-02342-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson RJ, Lovelock JE, Andreae MO, Warren SG. 1987. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 326: 655–661. 10.1038/326655a0 [DOI] [Google Scholar]
- Curson ARJ, Liu J, Bermejo Martínez A, Green RT, Chan Y, Carrión O, Williams BT, Zhang S-H, Yang G-P, Bulman Page PC, et al. 2017. Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat Microbiol 2: 17009 10.1038/nmicrobiol.2017.9 [DOI] [PubMed] [Google Scholar]
- Curson ARJ, Williams BT, Pinchbeck BJ, Sims LP, Martínez AB, Rivera PPL, Kumaresan D, Mercadé E, Spurgin LG, Carrión O, et al. 2018. DSYB catalyses the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nat Microbiol 3: 430–439. 10.1038/s41564-018-0119-5 [DOI] [PubMed] [Google Scholar]
- Delmont TO, Quince C, Shaiber A, Esen ÖC, Lee ST, Rappé MS, McLellan SL, Lücker S, Eren AM. 2018. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat Microbiol 3: 804–813. 10.1038/s41564-018-0176-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, Lara E, Berney C, Le Bescot N, Probert I, et al. 2015. Eukaryotic plankton diversity in the sunlit ocean. Science 348: 1261605 10.1126/science.1261605 [DOI] [PubMed] [Google Scholar]
- Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP, Banfield JF. 2009. Community-wide analysis of microbial genome sequence signatures. Genome Biol 10: R85 10.1186/gb-2009-10-8-r85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortch Q, Packard TT. 1989. Differences in biomass structure between oligotrophic and eutrophic marine ecosystems. Deep Sea Res Part Oceanogr Res Pap 36: 223–240. 10.1016/0198-0149(89)90135-0 [DOI] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi'o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3: e1319 10.7717/peerj.1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnelid H, Turk-Kubo K, Muñoz-Marín MdC, Zehr JP. 2016. New insights into the ecology of the globally significant uncultured nitrogen-fixing symbiont UCYN-A. Aquat Microb Ecol 77: 125–138. 10.3354/ame01794 [DOI] [Google Scholar]
- Foster RA, O'Mullan GD. 2008. Nitrogen-fixing and nitrifying symbioses in the marine environment. In Nitrogen in the marine environment, 2nd ed. (ed. Capone DG, et al. ), pp. 1197–1218. Academic Press, San Diego. [Google Scholar]
- Foster RA, Kuypers MMM, Vagner T, Paerl RW, Musat N, Zehr JP. 2011. Nitrogen fixation and transfer in open ocean diatom–cyanobacterial symbioses. ISME J 5: 1484–1493. 10.1038/ismej.2011.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallienne CP, Robins DB. 2001. Is Oithona the most important copepod in the world's oceans? J Plankton Res 23: 1421–1432. 10.1093/plankt/23.12.1421 [DOI] [Google Scholar]
- Gasol JM, del Giorgio PA, Duarte CM. 1997. Biomass distribution in marine planktonic communities. Limnol Oceanogr 42: 1353–1363. 10.4319/lo.1997.42.6.1353 [DOI] [Google Scholar]
- Hagino K, Onuma R, Kawachi M, Horiguchi T. 2013. Discovery of an endosymbiotic nitrogen-fixing cyanobacterium UCYN-A in Braarudosphaera bigelowii (Prymnesiophyceae). PLoS One 8: e81749 10.1371/journal.pone.0081749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res 9: 868–877. 10.1101/gr.9.9.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imelfort M, Parks D, Woodcroft BJ, Dennis P, Hugenholtz P, Tyson GW. 2014. GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ 2: e603 10.7717/peerj.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson S, Wouters J, Bergman B, Carpenter EJ. 1999. Host specificity in the Richelia–diatom symbiosis revealed by hetR gene sequence analysis. Environ Microbiol 1: 431–438. 10.1046/j.1462-2920.1999.00053.x [DOI] [PubMed] [Google Scholar]
- Johnson WM, Kido Soule MC, Kujawinski EB. 2016. Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J 10: 2304–2316. 10.1038/ismej.2016.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AW, Green RT, Todd JD. 2016. Enzymatic breakage of dimethylsulfoniopropionate—a signature molecule for life at sea. Curr Opin Chem Biol 31: 58–65. 10.1016/j.cbpa.2016.01.011 [DOI] [PubMed] [Google Scholar]
- Joly D, Faure D. 2015. Next-generation sequencing propels environmental genomics to the front line of research. Heredity (Edinb) 114: 429–430. 10.1038/hdy.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Li F, Kirton ES, Thomas A, Egan RS, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7: e7359 10.7717/peerj.7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, et al. 2014. The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol 12: e1001889 10.1371/journal.pbio.1001889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keister JE, Bonnet D, Chiba S, Johnson CL, Mackas DL, Escribano R. 2012. Zooplankton population connections, community dynamics, and climate variability. ICES J Mar Sci 69: 347–350. 10.1093/icesjms/fss034 [DOI] [Google Scholar]
- Keller MD. 1989. Dimethyl sulfide production and marine phytoplankton: the importance of species composition and cell size. Biol Oceanogr 6: 375–382. 10.1080/01965581.1988.10749540 [DOI] [Google Scholar]
- Keller MD, Bellows WK, Guillard RRL. 1989. Dimethyl sulfide production in marine-phytoplankton. Acs Symp Ser 393: 167–182. 10.1021/bk-1989-0393.ch011 [DOI] [Google Scholar]
- Kiene RP, Visscher PT, Keller MD, Kirst GO. 1997. Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum Press, New York. [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5: R12 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limardo AJ, Sudek S, Choi CJ, Poirier C, Rii YM, Blum M, Roth R, Goodenough U, Church MJ, Worden AZ. 2017. Quantitative biogeography of picoprasinophytes establishes ecotype distributions and significant contributions to marine phytoplankton. Environ Microbiol 19: 3219–3234. 10.1111/1462-2920.13812 [DOI] [PubMed] [Google Scholar]
- Lopes Dos Santos A, Gourvil P, Tragin M, Noël M-H, Decelle J, Romac S, Vaulot D. 2017a. Diversity and oceanic distribution of prasinophytes clade VII, the dominant group of green algae in oceanic waters. ISME J 11: 512–528. 10.1038/ismej.2016.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes Dos Santos A, Pollina T, Gourvil P, Corre E, Marie D, Garrido JL, Rodríguez F, Noël MH, Vaulot D, Eikrem W. 2017b. Chloropicophyceae, a new class of picophytoplanktonic prasinophytes. Sci Rep 7: 14019 10.1038/s41598-017-12412-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyimo T. 2011. Distribution and abundance of the cyanobacterium Richelia intracellularis in the coastal waters of Tanzania. J Ecol Nat Environ 3: 85–94. [Google Scholar]
- Madoui MA, Poulain J, Sugier K, Wessner M, Noel B, Berline L, Labadie K, Cornils A, Blanco-Bercial L, Stemmann L, et al. 2017. New insights into global biogeography, population structure and natural selection from the genome of the epipelagic copepod Oithona. Mol Ecol 26: 4467–4482. 10.1111/mec.14214 [DOI] [PubMed] [Google Scholar]
- Nayfach S, Shi ZJ, Seshadri R, Pollard KS, Kyrpides NC. 2019. New insights from uncultivated genomes of the global human gut microbiome. Nature 568: 505–510. 10.1038/s41586-019-1058-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, Plichta DR, Gautier L, Pedersen AG, Le Chatelier E, et al. 2014. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 32: 822–828. 10.1038/nbt.2939 [DOI] [PubMed] [Google Scholar]
- Olm MR, West PT, Brooks B, Firek BA, Baker R, Morowitz MJ, Banfield JF. 2019. Genome-resolved metagenomics of eukaryotic populations during early colonization of premature infants and in hospital rooms. Microbiome 7: 26 10.1186/s40168-019-0638-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW. 2017. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol 2: 1533–1542. 10.1038/s41564-017-0012-7 [DOI] [PubMed] [Google Scholar]
- Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, Armanini F, Beghini F, Manghi P, Tett A, Ghensi P, et al. 2019. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176: 649–662.e20. 10.1016/j.cell.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski J, Audic S, Adl S, Bass D, Belbahri L, Berney C, Bowser SS, Cepicka I, Decelle J, Dunthorn M, et al. 2012. CBOL Protist Working Group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol 10: e1001419 10.1371/journal.pbio.1001419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesant S, Not F, Picheral M, Kandels-Lewis S, Le Bescot N, Gorsky G, Iudicone D, Karsenti E, Speich S, Trouble R, et al. 2015. Open science resources for the discovery and analysis of Tara Oceans data. Sci Data 2: 150023 10.1038/sdata.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza Oñate F, Le Chatelier E, Almeida M, Cervino ACL, Gauthier F, Magoulès F, Ehrlich SD, Pichaud M. 2019. MSPminer: abundance-based reconstitution of microbial pan-genomes from shotgun metagenomic data. Bioinformatics 35: 1544–1552. 10.1093/bioinformatics/bty830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB. 2011. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473: 208–211. 10.1038/nature10078 [DOI] [PubMed] [Google Scholar]
- Roemmich D, McGowan J. 1995. Climatic warming and the decline of zooplankton in the California current. Science 267: 1324–1326. 10.1126/science.267.5202.1324 [DOI] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28: 1086–1092. 10.1093/bioinformatics/bts094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. 2012. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res 23: 111–120. 10.1101/gr.142315.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbald SJ, Archibald JM. 2017. More protist genomes needed. Nat Ecol Evol 1: 0145 10.1038/s41559-017-0145 [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Smith WO, Jr, Carlson CA, Ducklow HW, Hansell DA. 1998. Growth dynamics of Phaeocystis antarctica-dominated plankton assemblages from the Ross Sea. Mar Ecol Prog Ser 168: 229–244. 10.3354/meps168229 [DOI] [Google Scholar]
- Stefels J, Steinke M, Turner S, Malin G, Belviso S. 2007. Environmental constraints on the production and removal of the climatically active gas dimethylsulphide (DMS) and implications for ecosystem modelling. Biogeochemistry 83: 245–275. 10.1007/s10533-007-9091-5 [DOI] [Google Scholar]
- Steinberg DK, Landry MR. 2017. Zooplankton and the ocean carbon cycle. Annu Rev Mar Sci 9: 413–444. 10.1146/annurev-marine-010814-015924 [DOI] [PubMed] [Google Scholar]
- Steinegger M, Söding J. 2017. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 35: 1026–1028. 10.1038/nbt.3988 [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, Djahanschiri B, Zeller G, Mende DR, Alberti A, et al. 2015. Structure and function of the global ocean microbiome. Science 348: 1261359 10.1126/science.1261359 [DOI] [PubMed] [Google Scholar]
- Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH. 2015. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31: 926–932. 10.1093/bioinformatics/btu739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AW, Foster RA, Krupke A, Carter BJ, Musat N, Vaulot D, Kuypers MMM, Zehr JP. 2012. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337: 1546–1550. 10.1126/science.1222700 [DOI] [PubMed] [Google Scholar]
- Thompson A, Carter BJ, Turk-Kubo K, Malfatti F, Azam F, Zehr JP. 2014. Genetic diversity of the unicellular nitrogen-fixing cyanobacteria UCYN-A and its prymnesiophyte host. Environ Microbiol 16: 3238–3249. 10.1111/1462-2920.12490 [DOI] [PubMed] [Google Scholar]
- Tripp HJ, Bench SR, Turk KA, Foster RA, Desany BA, Niazi F, Affourtit JP, Zehr JP. 2010. Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature 464: 90–94. 10.1038/nature08786 [DOI] [PubMed] [Google Scholar]
- Tully BJ, Graham ED, Heidelberg JF. 2018. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci Data 5: 170203 10.1038/sdata.2017.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk-Kubo KA, Farnelid HM, Shilova IN, Henke B, Zehr JP. 2017. Distinct ecological niches of marine symbiotic N2-fixing cyanobacterium Candidatus Atelocyanobacterium thalassa sublineages. J Phycol 53: 451–461. 10.1111/jpy.12505 [DOI] [PubMed] [Google Scholar]
- Turmel M, Lopes dos Santos A, Otis C, Sergerie R, Lemieux C. 2019. Tracing the evolution of the plastome and mitogenome in the Chloropicophyceae uncovered convergent tRNA gene losses and a variant plastid genetic code. Genome Biol Evol 11: 1275–1292. 10.1093/gbe/evz074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier T, Leconte J, Seeleuthner Y, Mondy S, Pelletier E, Aury JM, de Vargas C, Sieracki M, Iudicone D, Vaulot D, et al. 2016. Survey of the green picoalga Bathycoccus genomes in the global ocean. Sci Rep 6: 37900 10.1038/srep37900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaulot D, Lepère C, Toulza E, la Iglesia RD, Poulain J, Gaboyer F, Moreau H, Vandepoele K, Ulloa O, Gavory F, et al. 2012. Metagenomes of the picoalga Bathycoccus from the Chile coastal upwelling. PLoS One 7: e39648 10.1371/journal.pone.0039648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74. 10.1126/science.1093857 [DOI] [PubMed] [Google Scholar]
- Vincent FJ, Colin S, Romac S, Scalco E, Bittner L, Garcia Y, Lopes RM, Dolan JR, Zingone A, de Vargas C, et al. 2018. The epibiotic life of the cosmopolitan diatom Fragilariopsis doliolus on heterotrophic ciliates in the open ocean. ISME J 12: 1094–1108. 10.1038/s41396-017-0029-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Maltrud ME, Burrows SM, Elliott SM, Cameron-Smith P. 2018. Impacts of shifts in phytoplankton community on clouds and climate via the sulfur cycle. Global Biogeochem Cycles 32: 1005–1026. 10.1029/2017GB005862 [DOI] [Google Scholar]
- West PT, Probst AJ, Grigoriev IV, Thomas BC, Banfield JF. 2018. Genome-reconstruction for eukaryotes from complex natural microbial communities. Genome Res 28: 569–580. 10.1101/gr.228429.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TJ, Eddy SR. 2013. nhmmer: DNA homology search with profile HMMs. Bioinformatics 29: 2487–2489. 10.1093/bioinformatics/btt403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, Janouskovec J, McRose D, Engman A, Welsh RM, Malfatti S, Tringe SG, Keeling PJ. 2012. Global distribution of a wild alga revealed by targeted metagenomics. Curr Biol 22: R675–R677. 10.1016/j.cub.2012.07.054 [DOI] [PubMed] [Google Scholar]
- Zehr JP, Kudela RM. 2011. Nitrogen cycle of the open ocean: from genes to ecosystems. Ann Rev Mar Sci 3: 197–225. 10.1146/annurev-marine-120709-142819 [DOI] [PubMed] [Google Scholar]
- Zehr JP, Bench SR, Carter BJ, Hewson I, Niazi F, Shi T, Tripp HJ, Affourtit JP. 2008. Globally distributed uncultivated oceanic N2-fixing cyanobacteria lack oxygenic photosystem II. Science 322: 1110–1112. 10.1126/science.1165340 [DOI] [PubMed] [Google Scholar]
- Zehr JP, Shilova IN, Farnelid HM, Muñoz-Marín MdC, Turk-Kubo KA. 2017. Unusual marine unicellular symbiosis with the nitrogen-fixing cyanobacterium UCYN-A. Nat Microbiol 2: 16214 10.1038/nmicrobiol.2016.214 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, McEwen GK, Margulies EH, Birney E. 2009. Pebble and Rock Band: heuristic resolution of repeats and scaffolding in the velvet short-read de novo assembler. PLoS One 4: e8407 10.1371/journal.pone.0008407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.