Abstract
In Arabidopsis, LTR retrotransposons are activated by mutations in the chromatin gene DECREASE in DNA METHYLATION 1 (DDM1), giving rise to 21- to 22-nt epigenetically activated siRNA (easiRNA) that depend on RNA DEPENDENT RNA POLYMERASE 6 (RDR6). We purified virus-like particles (VLPs) from ddm1 and ddm1rdr6 mutants in which genomic RNA is reverse transcribed into complementary DNA. High-throughput short-read and long-read sequencing of VLP DNA (VLP DNA-seq) revealed a comprehensive catalog of active LTR retrotransposons without the need for mapping transposition, as well as independent of genomic copy number. Linear replication intermediates of the functionally intact COPIA element EVADE revealed multiple central polypurine tracts (cPPTs), a feature shared with HIV in which cPPTs promote nuclear localization. For one member of the ATCOPIA52 subfamily (SISYPHUS), cPPT intermediates were not observed, but abundant circular DNA indicated transposon “suicide” by auto-integration within the VLP. easiRNA targeted EVADE genomic RNA, polysome association of GYPSY (ATHILA) subgenomic RNA, and transcription via histone H3 lysine-9 dimethylation. VLP DNA-seq provides a comprehensive landscape of LTR retrotransposons and their control at transcriptional, post-transcriptional, and reverse transcriptional levels.
Long terminal repeat (LTR) retrotransposons and endogenous retroviruses (ERVs) are a major component of the large genomes of most animal and plant species (Huang et al. 2012; Wang et al. 2014). The mouse genome, for example, contains more than 1 million ERVs, of which only a handful are autonomous elements (Huang et al. 2012; Gagnier et al. 2019). In Arabidopsis thaliana, ancient Ty3/gypsy-type ATHILA elements comprise ∼3% of the genome, mostly in pericentromeric regions, and relatively young Ty1/copia ATCOPIA elements (∼1% of the genome) are often found in euchromatic regions (Pereira 2004; Marco and Marín 2008). Inhibition of retrotransposons by small RNA has been reported in metazoans and plants, as well as in fission yeast, and occurs at the transcriptional and post-transcriptional levels. In Drosophila, piwi-interacting RNA (piRNA) trigger transcriptional silencing of transposons in the germline (Czech et al. 2018), resembling fission yeast in this respect (Volpe et al. 2002). In contrast, Ago2 and Dcr2 lie in the small interfering RNA pathway, and their mutation results in increased somatic retrotransposition (Xie et al. 2013). In mammalian embryos, 3′ tRNA fragments (3′-tRF) control transposition of LTR retrotransposons both after transcription and by direct inhibition of reverse transcription (Schorn et al. 2017). In Arabidopsis, transcriptional activation of some Ty1/copia retrotransposons is triggered by stress, and their retrotransposition rates are enhanced by loss of the 24-nt small RNA pathway (Mirouze et al. 2009; Ito et al. 2011). In contrast, in ddm1 mutants and wild-type (WT) pollen, most transposons are transcriptionally active and their RNA transcripts are processed into 21- to 22-nt easiRNA (Lippman et al. 2004; Slotkin et al. 2009). In ddm1 mutants, epigenetically activated siRNA (easiRNA) are generated by RDR6 and diverse miRNA (Nuthikattu et al. 2013; Creasey et al. 2014) from the nonfunctional ATHILA2 and ATHILA6 Ty3/gypsy retrotransposons but also from the functional Ty1/copia element EVADE. In the WT, retroelements generate easiRNA only in pollen, where they are targeted at the primer binding site (PBS) by miR845, and biogenesis occurs via a noncanonical pathway (Borges et al. 2018).
Ty1/copia elements in plants have a single ORF that encodes both the capsid protein (GAG), and the polyprotein (POL) composed of reverse transcriptase, RNase H, and integrase. These proteins coassemble with their genomic RNA (gRNA) into virus-like particles (VLPs), the cytoplasmic compartments encapsulated by GAG in which retrotransposon cDNA intermediates are produced (Supplemental Fig. S1; Peterson-Burch and Voytas 2002; Sabot and Schulman 2006; Finnegan 2012; Pachulska-Wieczorek et al. 2016). Ty3/gypsy elements also have a single GAG-POL ORF, although the POL proteins are in a different order. In both Drosophila and plants, the Ty1/copia GAG protein is translated from an abundant, alternatively spliced subgenomic RNA (Yoshioka et al. 1990; Chang et al. 2013). In Arabidopsis, Ty1/copia elements, the subgenomic GAG RNA is more efficiently translated than unspliced GAG-POL transcripts, and blocking splicing leads to significant reduction of GAG protein translation (Oberlin et al. 2017).
After VLP formation in the cytoplasm, LTR retrotransposons proliferate through tRNA-primed reverse transcription of gRNA, followed by nuclear import of double-stranded cDNA and integration into new loci (Supplemental Fig. S1; Chapman et al. 1992; Schorn and Martienssen 2018). In yeast and Arabidopsis, tRNA-iMet initiates reverse transcription of the LTR from the PBS to the 5′ end of the R region making minus-strand strong-stop DNA (Chapman et al. 1992; Mules et al. 1998; Griffiths et al. 2018; Schorn and Martienssen 2018). RNase H degrades the template RNA upstream of the PBS, and minus-strand strong-stop DNA is transferred to the 3′ LTR to prime minus-strand cDNA synthesis toward the PBS (Supplemental Fig. S1). During the extension of minus-strand cDNA synthesis, the template RNA is degraded except for an RNase H–resistant polypurine tract (PPT) near the 3′ LTR (Wilhelm et al. 2001). This PPT RNA fragment primes plus-strand strong-stop DNA synthesis up to U5 and the PBS sequence from the translocated minus-strand (Supplemental Fig. S1B). Then, the plus-strand cDNA is transferred to the 5′ end to prime full-length double-stranded DNA. Additional central PPT (cPPT) can also initiate plus-strand synthesis, which is displaced by the 3′ end of plus-strand DNA causing DNA flaps to form during Ty1 replication (Garfinkel et al. 2006). cPPT and DNA flaps have been found in the lentivirus HIV-1, where they play roles in nuclear import and in preventing mutagenesis by the cytidine deaminase APOBEC (Zennou et al. 2000; VandenDriessche et al. 2002; Wurtzer et al. 2006; Hu et al. 2010).
The capacity for specific retrotransposons to proliferate by integration into new genomic loci can only be confirmed by observing these insertions using transposition assays, by whole-genome sequencing in an active background, or by comparing insertion sites among individuals within a population. However, a number of methods including mobilome-seq (Lanciano et al. 2017) and ALE-seq (Cho et al. 2019) have been developed to catalog elements that may be transpositionally competent by sequencing transposition intermediates before integration. Short reads from mobilome-seq cover specifically circular extrachromosomal DNA (ecDNA), which arise from recombination of linear ecDNA. ALE-seq generates reads covering only the 5′-LTRs of linear ecDNA and depends on the use of reverse transcription primers specific to the PBS of the elements being queried. These linear intermediates are more directly predictive of transposition competence, as circular products do not undergo chromosomal integration (Sloan and Wainberg 2011; Lanciano et al. 2017). We have developed an alternative strategy that enables comprehensive sequencing of products from isolated VLPs. VLPs have been isolated in yeast and Drosophila (Eichinger and Boeke 1988; Kenna et al. 1998; Bachmann et al. 2004) as well as in plants (Jaaskelainen et al. 1999; Bachmair et al. 2004), but sequencing of their cDNA contents has not been reported. Our method captures linear products as well as abortive linear and circular intermediates missed by other approaches. By sequencing these intermediates from different genetic backgrounds, insights can be gained into mechanisms of genetic and epigenetic regulation.
In this study, we examined multiple layers of LTR retrotransposon control in Arabidopsis, using ddm1 mutants in which transposons are transcriptionally active and ddm1rdr6 mutants deficient in easiRNA (Vongs et al. 1993; Lippman et al. 2004; Creasey et al. 2014). We sought to determine whether easiRNA act to silence retroelements at transcriptional and post-transcriptional levels using genome-wide polysomal RNA (translatome), chromatin immunoprecipitation (ChIP), and small RNA sequencing. Furthermore, we performed VLP DNA sequencing to capture the full complement of cDNA intermediates generated during retrotransposition for the first time and to evaluate whether their sequence features are diagnostic of functional potential.
Results
Characterization of functional LTR retrotransposons by VLP DNA sequencing
Functional LTR retrotransposons form VLPs assembled from GAG proteins (Supplemental Fig. S1A; Sabot and Schulman 2006). Reverse transcription occurs inside the VLPs, and double-stranded cDNA products are subsequently imported into the nucleus bound to the integrase protein. After integration into new genomic loci, these insertions transcribe additional gRNA. We purified VLPs after treatment with DNase I (Methods), and sequenced cDNA products from WT, ddm1, and ddm1rdr6 using both Illumina short-read and Oxford Nanopore Technologies (ONT) long-read sequencing platforms (Supplemental Fig. S2). The two most productive elements, EVADE and the ATCOPIA52 subfamily element AT3TE76225, constituted 1.2% and 3.4% of mapped bases, respectively, across ddm1 short-read libraries. Despite its outsized representation in the mutant libraries, AT3TE76225 has not been observed to reintegrate and, as we show later, accumulates nonproductive circular intermediates. Consequently, we named this element SISYPHUS. EVADE is one of two full-length elements of the ATCOPIA93 family in A. thaliana Col-0, whereas the other element, ATTRAPE, is transcriptionally nonfunctional (Mirouze et al. 2009; Marí-Ordóñez et al. 2013). When EVADE is transcriptionally activated, it is the most successful retroelement by far in terms of copy number increases, although transposition of elements of the ATGP3, ATCOPIA13, ATCOPIA21, and ATCOPIA51 subfamilies has also been detected under nonstressed conditions (Tsukahara et al. 2009; Quadrana et al. 2019). Differential analysis of uniquely mapped VLP DNA short sequencing reads with replicates from WT, ddm1, and ddm1rdr6 mutants revealed enrichment from all of these subfamilies as well as ATGP10, ATCOPIA48, ATCOPIA52, and ATCOPIA76 elements, consistent with active reverse transcription (Fig. 1; Supplemental Figs. S2, S3; Supplemental Table S1). The proportion of elements in each subfamily significantly enriched in VLP DNA-seq in each mutant is shown in Supplemental Figure S2B. Long-read coverage of EVADE and other active COPIA elements spanned the entire element and was increased in ddm1rdr6 (Fig. 1B; Supplemental Fig. S2B; Supplemental Table S1). Furthermore, linear ecDNA was increased in ddm1rdr6 by Southern blot (Fig. 2A). Only small numbers of linear near full-length ONT reads were found for SISYPHUS, EVADE, ATCOPIA51, and ATGP3 elements, suggesting the double-stranded cDNA is exported immediately after completion of reverse transcription or otherwise turned into circular DNA. In contrast, VLP DNA from ATHILA families were more enriched in ddm1rdr6, indicating regulation by easiRNA, but comprised only small fragments derived mostly from LTRs, likely reflecting abortive retrotransposition intermediates from these nonfunctional elements (Supplemental Fig. S3A; Marco and Marín 2008).
Figure 1.
VLP DNA-seq data of LTR retrotransposons in ddm1 and ddm1rdr6. (A) Differential analysis of paired-end sequencing of VLP DNA using Illumina short-read platform. The statistical significance of three comparisons of wild type (WT), ddm1, and ddm1rdr6 is shown with |log2(fold-change)| ≥ 2 and FDR threshold at 5%. Each point corresponds to an annotated transposable element. Multiple ATHILA subfamilies were combined and labeled as “ATHILA.” (B) Coverage of short- and long-read VLP DNA-seq at representative LTR retrotransposon loci (EVADE, AT5TE20395; ATGP3, AT1TE45315; ATCOPIA51, AT1TE36035; SISYPHUS, AT3TE76225) were plotted for ddm1 and ddm1rdr6. Mean read counts per million mapped reads and 95% confidence intervals of biological replicates are shown for WT (yellow; n = 3), ddm1 (blue; n = 2), and ddm1rdr6 (orange, n = 3) short-read libraries. VLP DNA replicate samples were pooled for each genotype and sequenced in aggregate by ONT long-read sequencing. In the LTR retrotransposon annotation, abbreviations for conserved protein domains within the GAG-POL ORF are as follows: (AP) amino peptidase, (INT) integrase, (RT) reverse transcriptase, and (RH) RNase H. Blue and red lines indicate primer binding sites (PBSs) and polypurine tracts (PPTs). The 21- to 22-nt small RNA (sRNA) data were obtained from a previous study (Creasey et al. 2014). Target positions of miRNAs are indicated as arrows (for details, see Supplemental Table S4). Central PPT (cPPT) positions are indicated as dashed lines. Elevated coverage at the edges of strong-stop intermediate and flap DNA is shown as asterisks above ddm1 short-read data.
Figure 2.
Extrachromosomal DNA of LTR retrotransposons in ddm1 and ddm1rdr6. (A) Southern blotting using an EVADE probe was performed with undigested genomic DNA of F1 and F2 plants from the same parental lines. Integrated DNA copies (IC) and extrachromosomal DNA copies (EC) are indicated. Ethidium bromide (EtBr) staining was used for loading control. (B) Discordant short-read alignments from SISYPHUS (AT3TE76225) and EVADE in ddm1. Read pair orientations (forward or reverse for the first and second mate): RR and FF reads align in the same direction to the reference, indicating inversions, whereas RF reads face outward, indicating circular templates. LTR regions are indicated as blue bars. (C) Inverse PCR with genomic DNA to detect circular extrachromosomal DNA from ATCOPIA51, SISYPHUS, EVADE, and ATGP3 in ddm1 plants. (D) Inverse PCR with VLP DNA and reverse-forward (RF) outward reading primers for SISYPHUS and EVADE. (C,D) PCR primers are listed in Supplemental Table S6.
cDNA can exist in both linear and circular forms, and circular forms were previously reported for EVADE in Arabidopsis and a few other LTR retrotransposons in rice (Reinders et al. 2013; Lanciano et al. 2017). Outward-facing paired-end read alignments from Illumina VLP-seq reads consistent with junction-crossing reads from circular templates were absent from the vast majority of elements but were observed in ddm1 and ddm1rdr6 samples for ATCOPIA52, ATCOPIA93, ATCOPIA51, ATGP3, and ATHILA subfamily elements (Fig. 2B). SISYPHUS was exceptional, with >2% of read pairs mapped nonconcordantly in both mutants, whereas the proportion in EVADE was just 0.2% (Supplemental Table S1). Circular ecDNA formation was confirmed by inverse PCR (Supplemental Fig. S1D) whose products corresponded to one-LTR in size (Fig. 2C,D), and SISYPHUS was by far the most abundant. Double-stranded one-LTR circular products are thought to be generated by integrase-mediated autointegration in VLP, or as gapped intermediates in cDNA synthesis (Garfinkel et al. 2006; Sloan and Wainberg 2011; Munir et al. 2013). In contrast, two-LTR (tandem) circular DNA with junction nucleotides is formed in the nucleus by nonhomologous end joining and enhanced when integrase is nonfunctional (Garfinkel et al. 2006; Sloan and Wainberg 2011). The inverse PCR products of SISYPHUS were one-LTR in size, suggesting the circular DNA was either a gapped double-stranded circular intermediate or else a double-stranded product of autointegration into same strands or opposite strands (Supplemental Fig. S1), which results in deletion circles or inversion circles, respectively (Garfinkel et al. 2006; Sloan and Wainberg 2011; Munir et al. 2013). Both inversion and deletion circles were detected in large numbers based on outward facing reverse-forward and forward-forward paired-end reads, respectively, indicating auto-integration was the major source of these circles (Fig. 2B).
In yeast, auto-integration occurs near the cPPT, taking advantage of a DNA flap structure (Garfinkel et al. 2006). There was no strong indication of a DNA flap based on polypurine sequences and read alignment in SISYPHUS. We mapped individual long reads to investigate the integration sites (Fig. 3; Supplemental Figs. S1C, S3B). Deletion circles are predicted to have either the 5′ or the 3′ LTR, as well as a deleted portion of the full-length cDNA, up to the integration site, whereas inversion circles have an inverted portion separating the two LTRs (Garfinkel et al. 2006). Many of the SISYPHUS ONT reads fell into these categories, comprising either the 5′ or the 3′ LTR contiguous with a truncated or inverted portion of the retrotransposon (Supplemental Fig. S1C). These structural variants indicated the presence of circularly permuted reads, which were presumably arbitrarily sheared during library preparation. Among all the COPIA and GYPSY elements examined, only SISYPHUS gave rise to large numbers of these structural variants (Supplemental Table S1). The inversions spanned diverse regions of the element, consistent with inversion circles. The deleted portions terminated at inferred autointegration sites, which were distributed throughout the length of the element, consistent with the lack of a cPPT flap in SISYPHUS. One possibility is that nuclear import of double-stranded cDNA is not efficient for SISYPHUS, leading to elevated autointegration inside the VLP. This could be owing to mutations in nuclear localization (Kenna et al. 1998) or else to reduced translation of the integrase gene (see below), although read distributions were comparable for ddm1 and ddm1rdr6, so easiRNA likely did not play a major role.
Figure 3.
Alignments of ONT long reads from ddm1 VLP DNA. The central polypurine tract (cPPT), PBS, and PPT positions are indicated as dashed lines relative to full and LTR annotation of SISYPHUS (AT3TE76225), EVADE (AT5TE20395), and ATGP3 (AT1TE45315). Gaps in individual reads are indicated with black horizontal lines, and sequence mismatches are shown as colored dots in the read alignments. Pileups of linear intermediates are observed for EVADE, whereas a continuous distribution of fragment lengths is observed in SISYPHUS.
In sharp contrast, in EVADE we observed discontinuous regions of read alignments flanked by multiple cPPT, defined as 15- to 19-nt polypurine sequences (Figs. 1B, 3; Supplemental Fig. S3B). These regions represent active replication intermediates, generated by both minus-strand and plus-strand strong stop DNA, as well as extension products that terminate at cPPT and DNA flaps. The numbers of these intermediates, as well as their abundance, were significantly elevated in long-read sequencing data from ddm1rdr6 double mutants (Figs. 1B, 3; Supplemental Fig. S3B). ATGP3 also had elevated levels of strong stop intermediates but had few if any cPPT and no circular reads.
21- to 22-nt easiRNA control retrotransposition
In a previous study, dcl2/4 mutants lacking 21- to 22-nt small RNA were shown to accumulate 24-nt small RNA from an EVADE transgene driven by an ectopic promoter, leading to transcriptional silencing (Marí-Ordóñez et al. 2013). In contrast, rdr6 had no effect on the EVADE transgene, which can be interpreted as evidence that easiRNA might not inhibit transposition in WT cells. We tested whether easiRNA contribute to control of endogenous EVADE in ddm1 and ddm1rdr6 mutants by analyzing DNA copy numbers and RNA levels using quantitative PCR (qPCR) and RT-qPCR (Methods). Both ddm1 and ddm1rdr6 contained higher copy numbers of EVADE than the WT, implying high rates of transposition, whereas copy numbers of ATGP3 and SISYPHUS remained constant. By using qPCR, we detected an increase from two copies of EVADE in WT to 12 copies in ddm1 to 40 copies in ddm1rdr6 F2 siblings (Fig. 4A). Similar increases were observed in F2 and F3 rdr6 progeny from a parental +/rdr6 F1 with active EVADE elements (Fig. 4C) inherited epigenetically (Marí-Ordóñez et al. 2013). We detected parallel increases in gRNA levels, reflecting these increases in copy number (Fig. 4B,D). Consistent with gRNA levels, extrachromosomal EVADE copies were also more abundant in ddm1rdr6 than in ddm1 (Fig. 2A). RNase H cleavage products just upstream of the PBS, which are a hallmark of active transposition (Schorn et al. 2017), were readily detected for EVADE in both ddm1 and ddm1rdr6 (Supplemental Fig. S4A,B). We conclude that easiRNA actually inhibit EVADE retrotransposition in ddm1 mutants by down-regulating RNA levels.
Figure 4.
DNA and RNA levels of LTR retrotransposons in ddm1 and rdr6 mutants. (A) DNA copy numbers of EVADE, ATGP3, and SISYPHUS in ddm1 and ddm1rdr6 were normalized with a single copy gene (AT5G13440). (B) RT-qPCR data of EVADE elements using POL primers; y-axis indicates relative levels of EVADE genomic RNA to WT after normalization to ACT2. (C,D) EVADE DNA copy number and genomic RNA levels were analyzed in F2 and F3 progenies of F1 plants carrying active EVADE epigenetically inherited from parental rdr6/+ (Epi) crossed with WT pollen. Error bars, SD (n = 3).
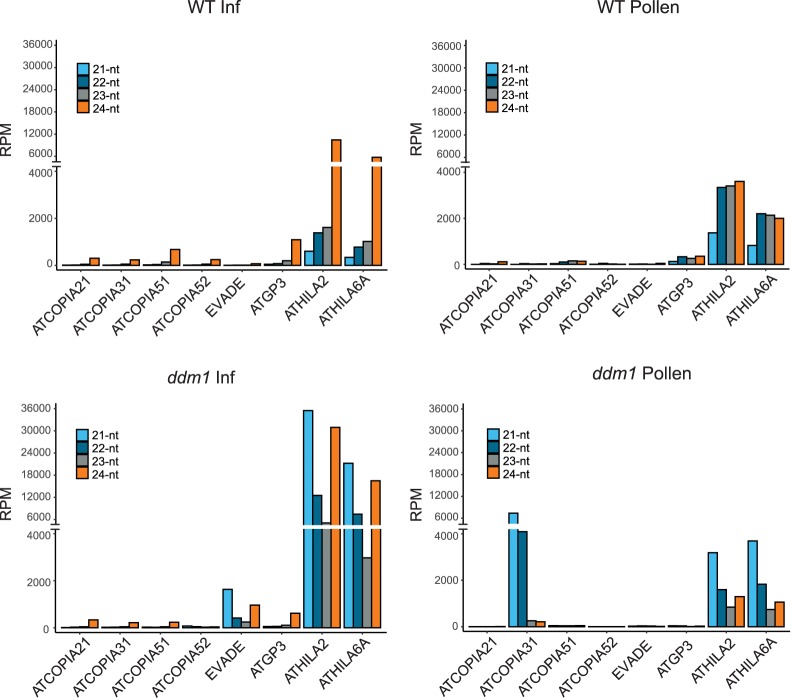
In backcrosses to WT plants, EVADE activity is inherited epigenetically, but copy number increases are thought to be limited by a switch from 21-nt to 24-nt siRNA accompanied by remethylation and silencing (Mirouze et al. 2009; Marí-Ordóñez et al. 2013; Reinders et al. 2013). Active EVADE elements can be resilenced through the female gametophyte but not through the male gametophyte (Reinders et al. 2013), in which easiRNA normally accumulate (Slotkin et al. 2009; Borges et al. 2018). We sequenced small RNA from WT and ddm1 flower buds and pollen and found that 21- to 22-nt easiRNA from EVADE were abundant in ddm1 inflorescence tissues but absent from pollen (Fig. 5). In contrast, ATHILA2 and ATHILA6A easiRNA were present in WT pollen (Slotkin et al. 2009), whereas ATCOPIA31 21- to 22-nt easiRNA were strongly up-regulated in ddm1 pollen. Thus, the absence of EVADE easiRNA in pollen must be because of transcriptional repression independent of DDM1 and likely accounts for the lack of paternal resilencing (Reinders et al. 2013).
Figure 5.
Small RNA profiles of representative LTR retrotransposons; 21-, 22-, 23-, and 24-nt small RNA levels in inflorescence tissues and pollen of WT and ddm1. Reads per million (RPM) were calculated from entire elements, including LTR and coding sequences.
Post-transcriptional suppression by easiRNA
Because easiRNA in ddm1 mutants depend on AGO1 associated with 21- to 22-nt small RNA (Nuthikattu et al. 2013) and because AGO1 represses translation of target mRNA (Li et al. 2013), we tested whether easiRNA can affect translation efficiency of transposon transcripts. Translating ribosome affinity immunopurification (TRAP) RNA-seq has been shown to estimate polysomal occupancy and translation efficiency in plants (Juntawong et al. 2014). Furthermore, microsome-polysomal fractionation has revealed that microRNA-dependent translational control takes place on the endoplasmic reticulum (Li et al. 2013). We generated TRAP lines of 35S:FLAG-RPL18 in ddm1 and ddm1rdr6 mutant backgrounds and performed total RNA-seq, total-polysomal RNA-seq, and microsome-polysomal RNA-seq. The polysomal RNA occupancy (polysomal RNA / total RNA) was obtained for 3903 transposable elements defined as ORFs from TAIR10 annotation (see Methods). As for the comparison between ddm1 and ddm1rdr6, we could detect the effect of the rdr6 mutation in microsome-polysomal RNA-seq data for known targets of RDR6, such as ARF4 (Marin et al. 2010), and for a handful of transposons (Fig. 6A; Supplemental Fig. S5; Supplemental Tables S2, S3). Among 31 up-regulated transposons in ddm1rdr6 relative to ddm1, 26 elements belonged to ATHILA LTR retrotransposon families (Supplemental Table S3), which are a major source of RDR6-dependent easiRNA. Although ATHILA elements in A. thaliana cannot transpose, a subgenomic mRNA encoding ORF2 (the “env” gene) is spliced from the full-length mRNA (Wright and Voytas 2002; Havecker et al. 2004) and was enriched on polysomes (Supplemental Fig. S5; Supplemental Table S3). This subgenomic RNA is targeted extensively by miRNA, which trigger easiRNA production (Creasey et al. 2014). The other three elements were ATENSPM3, LINE1_6, and VANDAL3, all of which have been identified as active elements in ddm1 mutants or in population level studies of transposon variants (Stuart et al. 2016). These non-LTR and DNA transposons are also targets of miRNA and generate RDR6-dependent easiRNA (Creasey et al. 2014). EVADE easiRNA are generated from the GAG subgenomic RNA (Marí-Ordóñez et al. 2013), but polysomal occupancy was not increased in ddm1rdr6 (Fig. 6B). GAG subgenomic mRNA from SISYPHUS was highly enriched in polysomes, consistent with previous studies (Oberlin et al. 2017), whereas the relative abundance of EVADE POL transcripts on polysomes indicates higher translation rates of integrase and reverse transcriptase (Oberlin et al. 2017). Unlike for ATHILA, polysome association of COPIA transcripts was unaffected by RDR6.
Figure 6.
Translatome profiles of ddm1 and ddm1rdr6. (A) Differential analysis of polysomal RNA-seq data between ddm1 and ddm1rdr6. Polysomal RNA-seq values were normalized by total RNA seq values to reflect polysomal enrichment (Methods). Red dots indicate significantly regulated genes or transposable elements (TEs) by cut-off values of |log2(fold-change)| > 0.5 and P-values < 0.01 which include ARF4 as an internal control. Significantly regulated ATHILA family elements are labeled with blue dots. (B) Total RNA and microsome-polysomal RNA (M poly) levels are shown for EVADE (AT5TE20395) and SISYPHUS (AT3TE76225). Mean RPM mapped reads and 95% confidence intervals of three biological replicates are shown for ddm1 (blue) and ddm1rdr6 (orange). Conserved protein domains, PBS and PPT, small RNA profiles, and miRNA target sites are indicated as in Figure 1.
easiRNA require miRNA triggers that target these transcripts (Creasey et al. 2014), and SISYPHUS LTRs were targeted by a single miRNA in the R region of the LTR. Consistent with this miRNA acting as a trigger, easiRNA accumulated along the length of the mRNA between the LTRs (Fig. 6B; Supplemental Table S4). In the case of EVADE, four miRNA were predicted to target the gRNA somewhere along its length. miR2938 was predicted to target the start codon of the GAG gene immediately 5′ of the easiRNA cluster, whereas miR5648-5p targets the 3′ end of the easiRNA cluster (Supplemental Fig. S4C,D; Supplemental Table S4). The 21- to 22-nt easiRNA were significantly reduced in ddm1dcl1 compared with ddm1, suggesting that miRNA were involved (Creasey et al. 2014). The single mutant dcl1 does not accumulate as much LTR retrotransposon RNA and easiRNA as ddm1. EVADE easiRNAs were also down-regulated in ddm1dcl1 (Supplemental Fig. S4E). miR2938 and miR5648-5p expression were reported in pollen and root cells (Grant-Downton et al. 2009; Breakfield et al. 2012). We did not detect miRNA-mediated cleavage by PARE-seq (Creasey et al. 2014) or by RACE-PCR in inflorescence tissues, but secondary siRNA, such as easiRNA, do not require cleavage so long as miRNA recognition recruits RdRP (Axtell et al. 2006; de Felippes et al. 2017). Although 21- to 22-nt easiRNA is mainly produced by RDR6, in somatic cells, RDR2 is involved in the 24-nt siRNA pathway. RDR2 and RDR6 are the major RdRP in Arabidopsis (Borges and Martienssen 2015). Consistent with induction without cleavage, EVADE easiRNA were not phased (Arribas-Hernandez et al. 2016). miR5663 was detected in inflorescence tissues (Supplemental Fig. S4F) and targets the EVADE intron near the splice acceptor site (Supplemental Fig. S4C). The level of unspliced RNA was increased in ddm1dcl1 mutants (Supplemental Fig. S4G), indicating that miR5663 might target unspliced gRNA and promote the accumulation of spliced GAG RNA, but further experiments would be required to show this requirement. Negative regulation of P-element splicing by piRNA has been reported in Drosophila (Teixeira et al. 2017). ATCOPIA21 and ATCOPIA51 had no strongly predicted miRNA targets, and easiRNA were barely detected in somatic tissues (Fig. 1B; Supplemental Fig. S3A; Oberlin et al. 2017), accounting for lack of regulation by RDR6. In contrast, significant levels were detected in pollen (Fig. 5; Borges et al. 2018), in which most gypsy and copia class retrotransposons are targeted by miR845, a pollen-specific miRNA that targets the PBS (Borges et al. 2018).
Transcriptional repression by easiRNA
In plants, both 21-/22-nt easiRNA and 24-nt siRNA species have the capacity to direct DNA methylation via the RNA directed DNA methylation (RdDM) pathway (Borges and Martienssen 2015). As such, we sought to more comprehensively define the impact of RDR6-dependent easiRNA on transposable elements at the transcriptional level. Genome-wide analysis of bisulfite sequencing revealed only minimal differences in DNA methylation between ddm1 and ddm1rdr6 (Creasey et al. 2014). However, transcriptional repression can also be achieved via repressive histone modifications, such as histone H3 lysine-9 dimethylation (H3K9me2), which in many organisms is known to be guided by small RNA (Volpe et al. 2002; Fagegaltier et al. 2009; Gu et al. 2012; Martienssen and Moazed 2015). We therefore performed H3K9me2 ChIP-seq in WT, ddm1, and ddm1rdr6 plants. As expected, ddm1 mutants showed genome-wide loss of H3K9me2 relative to WT (Supplemental Fig. S6; Supplemental Table S5). We identified a subset of regions that, in contrast to the rest of the genome, maintained high levels of H3K9me2 in ddm1. These loci were composed almost entirely of ATHILA family elements and lost H3K9me2 in ddm1rdr6 mutants (Fig. 7; Supplemental Fig. S6). The RDR6-dependent accumulation of H3K9me2 at these loci was significantly correlated with levels of 21- to 22-nt easiRNA accumulation (Fisher's exact test, P = 3−10), and the subset of ATHILA elements regulated by RDR6-dependent H3K9me2 had higher levels of 21- to 22-nt and 24-nt sRNAs than those that were unaffected (Supplemental Fig. S6C; Supplemental Code). In contrast, COPIA elements were not associated with RDR6-dependent H3K9me2 (Fisher's exact test, P = 0.98), showing only modest increases in H3K9me2 and 24-nt siRNAs in ddm1rdr6 (Supplemental Fig. S7). Taken together, these results strongly imply an as-yet-unexplored role for easiRNA in transcriptional control of transposable elements via targeting of the repressive histone modification H3K9me2.
Figure 7.
ATHILA family elements gain RDR6-dependent H3K9me2 in ddm1. H3K9me2 signal at transposable elements from multiple ATHILA families was analyzed in WT, ddm1, and ddm1rdr6 genotypes and correlated with previously published small RNA data (Creasey et al. 2014). RDR6-dependent gains in H3K9me2 colocalize with increased 21- to 22-nt siRNAs in ddm1. Plots depict transposable elements annotations scaled to 5 kb, as well as 2 kb upstream of and downstream from each feature. H3K9me2 ChIP data were normalized by H3, and small RNA data were normalized by counts per million.
Discussion
Sequencing of VLP DNA detected all known functional LTR retrotransposons in Arabidopsis, as well as some nonfunctional ones. Full-length VLP DNA read coverage from ATCOPIA and ATGP families (Fig. 1B; Supplemental Fig. S3A) corresponded to relatively young and low-copy elements known to transpose. Ancient ATHILA elements did not make full-length VLP DNA, confirming these gypsy retrotransposons are nonfunctional (Havecker et al. 2004; Marco and Marín 2008), but short products matching the LTR appeared to correspond to aborted strong stop replication intermediates (Supplemental Fig. S3A). Similar LTR fragments from ATHILA2 comprise an important family of dispersed centromeric satellite repeats known as 106B (May et al. 2005; Thompson et al. 1996), and retrotransposition might account for their origin. Thus, functional and nonfunctional retrotransposons could be readily distinguished even though nonfunctional ATHILA elements are present in copy numbers three to four orders of magnitude higher than functional ATCOPIA and ATGP elements. As for SISYPHUS, nonproductive one-LTR circular DNA, corresponding to autointegration “suicide” products, markedly accumulated in the VLP at levels far higher than productive retrotransposons such as EVADE. In contrast, two-LTR circular products of SISYPHUS were very rare, whereas small amounts of EVADE two-LTR products were present as previously described (Reinders et al. 2013), presumably owing to recombination of nonintegrated copies by host DNA repair enzymes in the nucleus. Both short-read and long-read sequencing revealed that these auto-integration products in SISYPHUS VLP led to nonfunctional deletion and inversion circles, accounting for lack of transposition.
We found that some retrotransposons are regulated post-transcriptionally by RNA interference, whereas others are regulated at the transcriptional level by histone H3 lysine-9 methylation guided by small RNA. RDR6-dependent easiRNA inhibit retrotransposition by post-transcriptional silencing of gRNA, by translational suppression of subgenomic RNA, and by controlling transcription via histone modification. ATHILA elements are no longer functional, but they are the primary source of easiRNA that arise by miRNA targeting of a spliced subgenomic RNA encoding the “ENV” protein (Creasey et al. 2014). These easiRNA inhibit polysome association of this subgenomic RNA and also inhibit transcript levels by guiding histone H3K9me2. This transcriptional silencing occurred in the absence of DNA methylation in ddm1 mutants. In plants, RNAi-dependent histone modification is thought to depend on RNA-dependent DNA methylation, found in asymmetric CHH contexts. As CHH methylation stays more or less the same in ddm1 whereas H3K9me2 is increased (Fig. 7), this might indicate the existence of a novel pathway for RNA-guided histone methylation, resembling that found in Drosophila, Caenorhabditis elegans, and fission yeast, which lack DNA methylation. Further investigation will be required to establish if such a pathway exists.
In contrast to ATHILA, linear extrachromosomal copies of EVADE accumulated in ddm1 and were further enriched by mutations in RDR6. Like ATHILA, EVADE is targeted by three or four miRNA that likely trigger easiRNA from the subgenomic GAG gene transcript, which is found associated with polysomes (Oberlin et al. 2017). However, association of the EVADE GAG mRNA with polysomes was unaffected in ddm1rdr6 mutants. Instead, levels of gRNA increased threefold, suggesting that EVADE easiRNA act post-transcriptionally to target gRNA directly. SISYPHUS easiRNA arose from full-length gRNA between the two LTR. Polysomal association of full-length EVADE GAG-POL is far more abundant than SISYPHUS GAG-POL, although both were unchanged in the absence of RDR6 (Fig. 6). As the integrase protein is translated from this transcript, this could contribute to lack of nuclear integration of SISYPHUS relative to EVADE. Thus, although easiRNA have a significant impact on COPIA gRNA accumulation and so inhibit increases in copy number, they have only limited impact on translation. The 22-nt tRNA-derived small RNA fragments (3′CCA-tRFs) were recently shown to inhibit ERVs in mammalian cells by targeting the PBS by RNA interference (Schorn et al. 2017), and it is possible that EVADE easiRNA may have a similar function in plants.
In conclusion, long-read and short-read sequencing of VLP DNA has revealed features that distinguish functional and nonfunctional replication intermediates and provides a powerful tool for identifying active transposons from complex genomes and for investigating molecular signatures of LTR retrotransposons. One such feature is the cPPT, which is present in EVADE but absent in SISYPHUS. cPPTs are hallmarks of the most active retrotransposons, including Ty1 in yeast, as well as HIV and other lentiviruses, in which cPPT is thought to be important for nuclear import of double-stranded cDNA (Zennou et al. 2000; VandenDriessche et al. 2002). Our work shows that these features may play a significant role in the activity of EVADE, the most active retrotransposon in Arabidopsis, and that their absence may account for the lack of nuclear integration of SISYPHUS and for high levels of “suicide” by autointegration. By comparing VLP sequencing, transcriptome sequencing and translatome sequencing, we have been able to establish the multiple levels at which easiRNA regulate the Arabidopsis LTR retrotransposons. Our methods are widely applicable to other plant and animal models and to human cells, especially those with genomes that contain very large numbers of nonfunctional LTR retrotransposons. By leveraging long-read sequencing, complete sequences of active transposon intermediates can be studied even when no reference genome is available.
Methods
Plant materials
All genotypes in this study are of Col-0 background, including WT, dcl1-11, ddm1-2, and rdr6-15. Genotyping primers are listed in Supplemental Table S6. Homozygous plants of ddm1-2 and ddm1-2 rdr6-15 were generated from heterozygous ddm1-2 backcrossed five times with Col-0 (ddm1-2 BC5), and their second generation was used for VLP DNA-seq experiments. For polysomal RNA-seq experiments, inbred ddm1-2 was independently crossed to 35S:FLAG-RPL18 and to rdr6-15 35S:FLAG-RPL18. The F3 plants were used for polysomal RNA purification.
gDNA extraction and DNA analyses
Whole inflorescence stems of 4-wk-old Arabidopsis plants were frozen and ground in liquid nitrogen. Total gDNA was isolated using a Nucleon PhytoPure kit (GE healthcare). EVADE DNA copy number was quantified using qPCR with EVADE qPCR primers and single copy gene primers as a reference (the primers are listed in Supplemental Table S6). Southern blotting was performed using EVADE Probe B as previously described (Mirouze et al. 2009).
ChIP
ChIP was performed with two biological replicates of 10-d-old seedlings using H3K9me2 (Abcam ab1220) and H3 (Abcam ab1791) antibodies, following a previously described protocol (Ingouff et al. 2017). Sequencing libraries were prepared using NEBNext Ultra II DNA library prep kit for Illumina (New England Biolabs) with size-selection for ∼200-bp insert DNA. The ChIP-seq libraries were sequenced using Illumina NextSeq high output SR 76 with 76-cycle single reads. Two biological replicates were prepared and sequenced for each genotype of interest. Before alignment, adapter trimming was performed using Trimmomatic (Bolger et al. 2014), and read quality was assessed with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned to the TAIR10 reference genome using BWA-MEM (Li 2013) with default parameters. Only primary alignments were retained, and optical and PCR duplicates were removed using SAMtools (Li et al. 2009). Peak calling was performed using MACS2 (Zhang et al. 2008) broad peak calling with a Q-value cutoff of 0.05 and normalization by signal per million reads. Peaks that were differentially regulated across genotypes were identified using MAnorm (Shao et al. 2012) and confirmed between biological replicates. Annotation of these differentially regulated peaks was performed using a combination of BEDOPS (Neph et al. 2012) tools and custom scripts (Supplemental Code). deepTools (Ramírez et al. 2014) was used to visualize the data.
RNA extraction and RT-qPCR
Total RNA was isolated from the same tissues used for gDNA extraction with Direct-zol RNA miniprep plus (Zymo Research). DNase I was treated on column. cDNA was synthesized with SuperScript VILO master mix (Thermo Fisher Scientific). qPCR was performed using iQ SYBR Green supermix. Primers are listed in Supplemental Table S6.
Polysomal RNA-seq
Total polysome was isolated using ribosome immunopurification as described previously (Mustroph et al. 2009, 2013). Briefly, inflorescence tissues of FLAG-RPL18 lines were ground in liquid nitrogen and transferred to polysome extraction buffer (PEB). Cell debris was removed by centrifugation and filtering with Miracloth. The supernatant was taken and transferred to prewashed EZview anti-FLAG agarose beads (Sigma-Aldrich) for 2 h at 4 °C. The agarose beads containing polysomes were washed once with PEB and three times with washing buffer. Polysomes were eluted using 3× FLAG peptide (Sigma-Aldrich) and used for RNA extraction with Direct-zol RNA miniprep kit (Zymo Research) including DNase I treatment. Ribosomal RNA (rRNA) in the samples was depleted by a Ribo-Zero magnetic kit (plant leaf) (Epicentre). Then, rRNA free samples were used for RNA-seq library preparation using a ScriptSeq v2 RNA-seq library preparation kit (EPicentre). Microsome-polysomal RNA was obtained using a previously described method with some modifications (Li et al. 2013). Briefly, 2 g of frozen tissues was suspended to 7 mL microsome extraction buffer (MEB). After removing cell debris by filtration with Miracloth and centrifugation at 10,000g for 15 min at 4 °C, the supernatant was transferred on the top of 1.7 M/0.6 M sucrose cushions and applied to ultracentrifugation using swing rotor at 140,000g for 1 h at 4 °C. The microsome fraction of the 1.7 M/0.6 M layer interface was harvested and diluted 10 times by MEB and centrifuged at 140,000g for 0.5 h at 4 °C to obtain microsome pellet. The pellet was resuspended with 8 mL PEB and used for ribosome immunopurification and RNA-seq library preparation as described above. The PE 101 sequencing data were obtained using Illumina HiSeq 2000 platform. The paired-end reads were mapped to Arabidopsis TAIR10 genome using TopHat (Trapnell et al. 2009), and the polysome occupancy (polysomal RNA / total RNA) was calculated using the systemPipeR package (Backman and Girke 2016) (Supplemental Code) with raw count data obtained by Cuffnorm.
VLP DNA-seq
Virus-like particles were purified using modified method reported previously (Bachmair et al. 2004). Four grams of 4-wk-old whole inflorescence stems was ground with 10 mL of ice-cold VLP extraction buffer and 10 mL of sea sand on ice. Ten milliliters of the extraction buffer and Triton X-100 was added and mixed. The slurry was transferred to a 50-mL tube and centrifuged for 5 min at 180g and 4°C. The supernatant was carefully transferred onto 5 mL of prechilled 15% sucrose and 10 mM potassium phosphate buffer (pH 7.2) and ultracentrifuged for 1.5 h at 109,000g and 4°C using a fixed angle rotor. The pellet was washed with the 15% sucrose buffer and resuspended with 4 mL particle suspension buffer to obtain VLP fractions. To remove non-VLP DNA, 0.5 mL of the VLP sample was treated with 5 μL of 1 mg/mL DNase I for 10 min at 37°C. Twenty microliters of 0.25 M EDTA, 50 μL of 10% SDS, 25 μL of 10 mg/mL proteinase were added and incubated for 10 min at 65°C. VLP DNA was purified by 0.5 mL of equilibrated (pH 8.0) phenol:chloroform:IAA (25:24:1) mixture three times and with 0.5 mL chloroform:IAA (24:1) once. The last aqueous fraction was transferred into a new 1.5-mL tube and used for 100% ethanol precipitation with 40 μL 3 M sodium acetate (pH 7.0). The DNA pellet was washed with 70% ethanol, dried, and resuspended with 100 μL TE buffer. One microliter of RNase A (10 mg/mL) was added to the VLP DNA sample and incubated 10 min. The treated DNA sample was purified using DNA Clean & Concentrator (Zymo Research). The DNA was sheared to 650 bp using Covaris S220 and subsequently used for DNA-seq library preparation with NEBNext ultra DNA library prep kit (New England Biolabs). The paired-end sequencing data sets with 101-nt read length (PE101) were obtained by Illumina HiSeq 2000. Adapters were trimmed from raw reads with Skewer (Jiang et al. 2014) in paired-end mode, and read pairs with both mates >25 nt were retained. Reads were aligned to the TAIR10 genome with STAR (Dobin et al. 2013) in two-pass mode to improve spliced alignment at unannotated introns. Intact bacteria copurified with VLP, as indicated by large numbers of reads mapping to bacterial genomes (up to 95% in WT), and these were discarded. Reads mapping equally well to multiple locations were randomly assigned, and chimeric/split read alignments were output separately from concordant alignments. Optical and PCR duplicates were removed from the alignments with the Picard toolkit (https://broadinstitute.github.io/picard/). Counts of reads mapping to the TAIR10 transposon annotations were computed with featureCounts (Liao et al. 2014). Pairwise differential expression at TAIR10 transposon loci was tested across three WT, two ddm1, and three ddm1rdr6 replicates using quasi-likelihood F-tests in edgeR (Robinson et al. 2010), controlling FDR at 5% and a log2(fold-change) threshold of two (Supplemental Code).
ONT long-read libraries were prepared as follows: 10 ng per genotype of purified VLP DNA extract was pooled from the replicate samples and initially amplified following the conditions in the “1D low-input genomic DNA with PCR” (SQK-LSK108) protocol with reagents. End-repair, dA-tailing, and PCR adapter ligation were performed, followed by 16 cycles of PCR amplification. PCR products were purified and concentrated with Ampure XP beads (Agencourt), and 300 ng of eluate per sample was carried through to library preparation following the “1D genomic DNA by ligation” protocol with SKQ-LSK109 reagents. Libraries were loaded onto r9.4 (FLO-MIN106) flow cells and sequenced on a GridION X5. Base-calling was performed offline with Guppy v2.3.1 using the default r9.4.1 model. By using Porechop (https://github.com/rrwick/Porechop), ONT sequencing adapters were trimmed from 5′ ends, 3′ ends, and the middle of raw reads. Reads with middle adapters were split. Remaining reads >100 bp were aligned to the TAIR10 reference with minimap2 (Li 2018) for coverage and read alignment plots. Structural variants were called on NGMLR (Sedlazeck et al. 2018) alignments using Sniffles (Sedlazeck et al. 2018) with default parameters, except minimum read support was reduced to three. Ribbon was used to visualize complex structural variants (Nattestad et al. 2016).
5′ RACE PCR
5′ RACE PCR was performed using a FirstChoice RLM-RACE kit (Thermo Fisher Scientific) without the treatments of calf intestine alkaline phosphatase and tobacco acid pyrophosphatase. A gene-specific primer was used for cDNA synthesis after adapter ligation (Supplemental Table S6). First and second nested PCR was performed with the primers are listed.
Small RNA-seq data
Small RNA-seq libraries from inflorescence and pollen for comparisons of 21-, 22-, and 24-nt small RNA between WT and ddm1 were prepared as previously described (Borges et al. 2018). WT pollen sample was previously deposited in the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) database (GSM2829912). Briefly, small RNAs were purified by running total RNA from pollen and inflorescence tissues on acrylamide gels (15% polyacrylamide, 7 M urea) with size-selection of 18-to-30-nt regions. Small RNAs were extracted from the gel bands using TRIzol LS (Thermo Fisher Scientific) and Direct-zol columns (Zymo Research). Libraries were prepared with the TruSeq small RNA sample preparation kit (Illumina) and sequenced in Illumina MiSeq platform. Data analysis was performed as previously reported (Borges et al. 2018). We obtained 21- to 22-nt small RNA data sets from inflorescence (Creasey et al. 2014) from NCBI GEO accession GSE52951. After adapter trimming with Skewer, reads were quality filtered with fastp (Chen et al. 2018) and aligned to the TAIR10 genome with ShortStack (Axtell 2013) with default parameters except “‐‐bowtie_m 1000 ‐‐ranmax 50.”
LTR retrotransposon annotation
GenomeTools was used to structurally annotate retrotransposons across the TAIR10 genome. First, LTRharvest (Ellinghaus et al. 2008) was run to detect LTR sequences with at least 85% similarity separated by 1–15 kbp flanked by target site duplications and the TGCA motif. Then, LTRdigest (Steinbiss et al. 2009) was run to annotate internal transposon features including the PBS, PPT, and GAG and POL protein homology.
Genome Browser Figures
Genome-wide read coverage for VLP DNA, small RNA, total, and polysomal RNA libraries was calculated with bamCoverage from deepTools (Ramírez et al. 2014) and normalized to reads per nucleotide per million mapped reads and plotted across the genome with Gviz (Hahne and Ivanek 2016) or Integrative Genomics Viewer (IGV) (Thorvaldsdottir et al. 2013).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE128932.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Vincent Colot, Leandro Quadrana, Tetsuji Kakutani, Matthias Benoit, and all members of the Martienssen laboratory for discussions. We also thank Julia Bailey-Serres for helpful discussion and providing 35S:FLAG-RPL18 seeds. Research in the Martienssen laboratory is supported by the U.S. National Institutes of Health (NIH) grant R01 GM067014, the National Science Foundation Plant Genome Research Program, and the Howard Hughes Medical Institute. The authors acknowledge assistance from the Cold Spring Harbor Laboratory Shared Resources, which are funded in part by a Cancer Center Support grant (5PP30CA045508).
Author contributions: S.C.L., E.E., and R.A.M. designed the study; S.C.L., E.E., F.B., J.-S.P., and P.L. performed the experiments; S.C.L., E.E., B.B., F.B., and A.S. analyzed the data and their significance; S.C.L., E.E., B.B., and R.A.M. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.259044.119.
References
- Arribas-Hernandez L, Marchais A, Poulsen C, Haase B, Hauptmann J, Benes V, Meister G, Brodersen P. 2016. The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell 28: 1563–1580. 10.1105/tpc.16.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. 2013. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19: 740–751. 10.1261/rna.035279.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. 2006. A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577. 10.1016/j.cell.2006.09.032 [DOI] [PubMed] [Google Scholar]
- Bachmair A, Garber K, Takeda S, Sugimoto K, Kakutani T, Hirochika H. 2004. Biochemical analysis of long terminal repeat retrotransposons. Methods Mol Biol 260: 73–82. 10.1385/1-59259-755-6:073 [DOI] [PubMed] [Google Scholar]
- Bachmann AS, Corpuz G, Hareld WP, Wang G, Coller BA. 2004. A simple method for the rapid purification of copia virus-like particles from Drosophila Schneider 2 cells. J Virol Methods 115: 159–165. 10.1016/j.jviromet.2003.09.025 [DOI] [PubMed] [Google Scholar]
- Backman T, Girke T. 2016. systemPipeR: NGS workflow and report generation environment. BMC Bioinformatics 17: 388 10.1186/s12859-016-1241-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Martienssen RA. 2015. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16: 727–741. 10.1038/nrm4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Parent JS, van Ex F, Wolff P, Martínez G, Köhler C, Martienssen RA. 2018. Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat Genet 50: 186–192. 10.1038/s41588-017-0032-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakfield NW, Corcoran DL, Petricka JJ, Shen J, Sae-Seaw J, Rubio-Somoza I, Weigel D, Ohler U, Benfey PN. 2012. High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Res 22: 163–176. 10.1101/gr.123547.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Jääskeläinen M, Li SP, Schulman AH. 2013. BARE retrotransposons are translated and replicated via distinct RNA pools. PLoS One 8: e72270 10.1371/journal.pone.0072270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KB, Byström AS, Boeke JD. 1992. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc Natl Acad Sci 89: 3236–3240. 10.1073/pnas.89.8.3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Benoit M, Catoni M, Drost HG, Brestovitsky A, Oosterbeek M, Paszkowski J. 2019. Sensitive detection of pre-integration intermediates of long terminal repeat retrotransposons in crop plants. Nat Plant 5: 26–33. 10.1038/s41477-018-0320-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey KM, Zhai J, Borges F, Van Ex F, Regulski M, Meyers BC, Martienssen RA. 2014. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. 10.1038/nature13069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Munafò M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, Hannon GJ. 2018. piRNA-guided genome defense: from biogenesis to silencing. Annu Rev Genet 52: 131–157. 10.1146/annurev-genet-120417-031441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felippes FF, Marchais A, Sarazin A, Oberlin S, Voinnet O. 2017. A single miR390 targeting event is sufficient for triggering TAS3-tasiRNA biogenesis in Arabidopsis. Nucleic Acids Res 45: 5539–5554. 10.1093/nar/gkx119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger DJ, Boeke JD. 1988. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell 54: 955–966. 10.1016/0092-8674(88)90110-9 [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9: 18 10.1186/1471-2105-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D, Bougé AL, Berry B, Poisot E, Sismeiro O, Coppée JY, Théodore L, Voinnet O, Antoniewski C. 2009. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc Natl Acad Sci 106: 21258–21263. 10.1073/pnas.0809208105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan DJ. 2012. Retrotransposons. Curr Biol 22: R432–R437. 10.1016/j.cub.2012.04.025 [DOI] [PubMed] [Google Scholar]
- Gagnier L, Belancio VP, Mager DL. 2019. Mouse germ line mutations due to retrotransposon insertions. Mob DNA 10: 15 10.1186/s13100-019-0157-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel DJ, Stefanisko KM, Nyswaner KM, Moore SP, Oh J, Hughes SH. 2006. Retrotransposon suicide: formation of Ty1 circles and autointegration via a central DNA flap. J Virol 80: 11920–11934. 10.1128/JVI.01483-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant-Downton R, Le Trionnaire G, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, Twell D, Dickinson H. 2009. MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics 10: 643 10.1186/1471-2164-10-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Catoni M, Iwasaki M, Paszkowski J. 2018. Sequence-independent identification of active LTR retrotransposons in Arabidopsis. Mol Plant 11: 508–511. 10.1016/j.molp.2017.10.012 [DOI] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. 2012. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet 44: 157–164. 10.1038/ng.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne F, Ivanek R. 2016. Visualizing genomic data using Gviz and Bioconductor. Methods Mol Biol 1418: 335–351. 10.1007/978-1-4939-3578-9_16 [DOI] [PubMed] [Google Scholar]
- Havecker ER, Gao X, Voytas DF. 2004. The diversity of LTR retrotransposons. Genome Biol 5: 225 10.1186/gb-2004-5-6-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Saenz DT, Fadel HJ, Walker W, Peretz M, Poeschla EM. 2010. The HIV-1 central polypurine tract functions as a second line of defense against APOBEC3G/F. J Virol 84: 11981–11993. 10.1128/JVI.00723-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CR, Burns KH, Boeke JD. 2012. Active transposition in genomes. Annu Rev Genet 46: 651–675. 10.1146/annurev-genet-110711-155616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Selles B, Michaud C, Vu TM, Berger F, Schorn AJ, Autran D, Van Durme M, Nowack MK, Martienssen RA, et al. 2017. Live-cell analysis of DNA methylation during sexual reproduction in Arabidopsis reveals context and sex-specific dynamics controlled by noncanonical RdDM. Genes Dev 31: 72–83. 10.1101/gad.289397.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J. 2011. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472: 115–119. 10.1038/nature09861 [DOI] [PubMed] [Google Scholar]
- Jaaskelainen M, Mykkanen AH, Arna T, Vicient CM, Suoniemi A, Kalendar R, Savilahti H, Schulman AH. 1999. Retrotransposon BARE-1: expression of encoded proteins and formation of virus-like particles in barley cells. Plant J 20: 413–422. 10.1046/j.1365-313x.1999.00616.x [DOI] [PubMed] [Google Scholar]
- Jiang H, Lei R, Ding SW, Zhu S. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15: 182 10.1186/1471-2105-15-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J. 2014. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci 111: E203–E212. 10.1073/pnas.1317811111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna MA, Brachmann CB, Devine SE, Boeke JD. 1998. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol 18: 1115–1124. 10.1128/MCB.18.2.1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciano S, Carpentier MC, Llauro C, Jobet E, Robakowska-Hyzorek D, Lasserre E, Ghesquiere A, Panaud O, Mirouze M. 2017. Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLoS Genet 13: e1006630 10.1371/journal.pgen.1006630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN].
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, et al. 2013. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574. 10.1016/j.cell.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. 2004. Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476. 10.1038/nature02651 [DOI] [PubMed] [Google Scholar]
- Marco A, Marín I. 2008. How Athila retrotransposons survive in the Arabidopsis genome. BMC Genomics 9: 219 10.1186/1471-2164-9-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. 2010. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117. 10.1105/tpc.109.072553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A, Marchais A, Etcheverry M, Martin A, Colot V, Voinnet O. 2013. Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet 45: 1029–1039. 10.1038/ng.2703 [DOI] [PubMed] [Google Scholar]
- Martienssen R, Moazed D. 2015. RNAi and heterochromatin assembly. Cold Spring Harb Perspect Biol 7: a019323 10.1101/cshperspect.a019323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA. 2005. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet 1: e79 10.1371/journal.pgen.0010079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, Ossowski S, Cao J, Weigel D, Paszkowski J, Mathieu O. 2009. Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461: 427–430. 10.1038/nature08328 [DOI] [PubMed] [Google Scholar]
- Mules EH, Uzun O, Gabriel A. 1998. In vivo Ty1 reverse transcription can generate replication intermediates with untidy ends. J Virol 72: 6490–6503. 10.1128/JVI.72.8.6490-6503.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir S, Thierry S, Subra F, Deprez E, Delelis O. 2013. Quantitative analysis of the time-course of viral DNA forms during the HIV-1 life cycle. Retrovirology 10: 87 10.1186/1742-4690-10-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Juntawong P, Bailey-Serres J. 2009. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol 553: 109–126. 10.1007/978-1-60327-563-7_6 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Girke T, Bailey-Serres J. 2013. Isolation and analysis of mRNAs from specific cell types of plants by ribosome immunopurification. Methods Mol Biol 959: 277–302. 10.1007/978-1-62703-221-6_19 [DOI] [PubMed] [Google Scholar]
- Nattestad M, Chin C-S, Schatz MC. 2016. Ribbon : visualizing complex genome alignments and structural variation. bioRxiv 10.1101/082123 [DOI]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, et al. 2012. BEDOPS: high-performance genomic feature operations. Bioinformatics 28: 1919–1920. 10.1093/bioinformatics/bts277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S, McCue AD, Panda K, Fultz D, DeFraia C, Thomas EN, Slotkin RK. 2013. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol 162: 116–131. 10.1104/pp.113.216481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlin S, Sarazin A, Chevalier C, Voinnet O, Mari-Ordonez A. 2017. A genome-wide transcriptome and translatome analysis of Arabidopsis transposons identifies a unique and conserved genome expression strategy for Ty1/Copia retroelements. Genome Res 27: 1549–1562. 10.1101/gr.220723.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachulska-Wieczorek K, Le Grice SF, Purzycka KJ. 2016. Determinants of genomic RNA encapsidation in the Saccharomyces cerevisiae long terminal repeat retrotransposons Ty1 and Ty3. Viruses 8: 193 10.3390/v8070193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira V. 2004. Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol 5: R79 10.1186/gb-2004-5-10-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson-Burch BD, Voytas DF. 2002. Genes of the Pseudoviridae (Ty1/copia retrotransposons). Mol Biol Evol 19: 1832–1845. 10.1093/oxfordjournals.molbev.a004008 [DOI] [PubMed] [Google Scholar]
- Quadrana L, Etcheverry M, Gilly A, Caillieux E, Madoui MA, Guy J, Bortolini Silveira A, Engelen S, Baillet V, Wincker P, et al. 2019. Transposition favors the generation of large effect mutations that may facilitate rapid adaption. Nat Commun 10: 3421 10.1038/s41467-019-11385-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Mirouze M, Nicolet J, Paszkowski J. 2013. Parent-of-origin control of transgenerational retrotransposon proliferation in Arabidopsis. EMBO Rep 14: 823–828. 10.1038/embor.2013.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabot F, Schulman AH. 2006. Parasitism and the retrotransposon life cycle in plants: a hitchhiker's guide to the genome. Heredity (Edinb) 97: 381–388. 10.1038/sj.hdy.6800903 [DOI] [PubMed] [Google Scholar]
- Schorn AJ, Martienssen R. 2018. Tie-break: host and retrotransposons play tRNA. Trends Cell Biol 28: 793–806. 10.1016/j.tcb.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. 2017. LTR-retrotransposon control by tRNA-derived small RNAs. Cell 170: 61–71.e11. 10.1016/j.cell.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, Schatz MC. 2018. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods 15: 461–468. 10.1038/s41592-018-0001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. 2012. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol 13: R16 10.1186/gb-2012-13-3-r16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan RD, Wainberg MA. 2011. The role of unintegrated DNA in HIV infection. Retrovirology 8: 52 10.1186/1742-4690-8-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdžić M, Becker JD, Feijó JA, Martienssen RA. 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. 10.1016/j.cell.2008.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbiss S, Willhoeft U, Gremme G, Kurtz S. 2009. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res 37: 7002–7013. 10.1093/nar/gkp759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Eichten SR, Cahn J, Karpievitch YV, Borevitz JO, Lister R. 2016. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. eLife 5: e20777 10.7554/eLife.20777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Okuniewska M, Malone CD, Coux R-X, Rio DC, Lehmann R. 2017. piRNA-mediated regulation of transposon alternative splicing in soma and germ line. Nature 552: 268–272. 10.1038/nature25018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson HL, Schmidt R, Dean C. 1996. Identification and distribution of seven classes of middle-repetitive DNA in the Arabidopsis thaliana genome. Nucleic Acids Res 24: 3017–3022. 10.1093/nar/24.15.3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, Kakutani T. 2009. Bursts of retrotransposition reproduced in Arabidopsis. Nature 461: 423–426. 10.1038/nature08351 [DOI] [PubMed] [Google Scholar]
- VandenDriessche T, Thorrez L, Naldini L, Follenzi A, Moons L, Berneman Z, Collen D, Chuah MK. 2002. Lentiviral vectors containing the human immunodeficiency virus type-1 central polypurine tract can efficiently transduce nondividing hepatocytes and antigen-presenting cells in vivo. Blood 100: 813–822. 10.1182/blood.V100.3.813 [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. 1993. Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928. 10.1126/science.8316832 [DOI] [PubMed] [Google Scholar]
- Wang W, Haberer G, Gundlach H, Gläßer C, Nussbaumer T, Luo MC, Lomsadze A, Borodovsky M, Kerstetter RA, Shanklin J, et al. 2014. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat Commun 5: 3311 10.1038/ncomms4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Uzun O, Mules EH, Gabriel A, Wilhelm FX. 2001. Polypurine tract formation by Ty1 RNase H. J Biol Chem 276: 47695–47701. 10.1074/jbc.M106067200 [DOI] [PubMed] [Google Scholar]
- Wright DA, Voytas DF. 2002. Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res 12: 122–131. 10.1101/gr.196001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S, Goubard A, Mammano F, Saragosti S, Lecossier D, Hance AJ, Clavel F. 2006. Functional central polypurine tract provides downstream protection of the human immunodeficiency virus type 1 genome from editing by APOBEC3G and APOBEC3B. J Virol 80: 3679–3683. 10.1128/JVI.80.7.3679-3683.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Donohue RC, Birchler JA. 2013. Quantitatively increased somatic transposition of transposable elements in Drosophila strains compromised for RNAi. PLoS One 8: e72163 10.1371/journal.pone.0072163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Honma H, Zushi M, Kondo S, Togashi S, Miyake T, Shiba T. 1990. Virus-like particle formation of Drosophila copia through autocatalytic processing. EMBO J 9: 535–541. 10.1002/j.1460-2075.1990.tb08140.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101: 173–185. 10.1016/S0092-8674(00)80828-4 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.