Abstract
Background
Remdesivir has received significant attention for its potential application in the treatment of COVID-19, caused by SARS-CoV-2. Remdesivir has already been tested for Ebola virus disease treatment and found to have activity against SARS and MERS coronaviruses. The remdesivir core contains GS-441524, which interferes with RNA-dependent RNA polymerases alone. In non-human primates, following IV administration, remdesivir is rapidly distributed into PBMCs and converted within 2 h to the active nucleoside triphosphate form, while GS-441524 is detectable in plasma for up to 24 h. Nevertheless, remdesivir pharmacokinetics and pharmacodynamics in humans are still unexplored, highlighting the need for a precise analytical method for remdesivir and GS-441524 quantification.
Objectives
The validation of a reliable UHPLC-MS/MS method for remdesivir and GS-441524 quantification in human plasma.
Methods
Remdesivir and GS-441524 standards and quality controls were prepared in plasma from healthy donors. Sample preparation consisted of protein precipitation, followed by dilution and injection into the QSight 220 UHPLC-MS/MS system. Chromatographic separation was obtained through an Acquity HSS T3 1.8 μm, 2.1 × 50 mm column, with a gradient of water and acetonitrile with 0.05% formic acid. The method was validated using EMA and FDA guidelines.
Results
Analyte stability has been evaluated and described in detail. The method successfully fulfilled the validation process and it was demonstrated that, when possible, sample thermal inactivation could be a good choice in order to improve biosafety.
Conclusions
This method represents a useful tool for studying remdesivir and GS-441524 clinical pharmacokinetics, particularly during the current COVID-19 outbreak.
Introduction
Remdesivir has received significant attention for its potential application in the treatment of COVID-19, caused by SARS-CoV-2, a zoonotic pathogen that emerged in 2019. Remdesivir is a monophosphoramidate prodrug of an adenine nucleotide analogue. Its core is a 1′-cyano-substituted adenine C-nucleoside ribose analogue (GS-441524) linked to another small molecule through an ester bond; this linkage confers better penetration into cells.1,2 From 2015, remdesivir was tested in healthy volunteers during Phase I clinical trials and it then entered Phase II studies in the context of Ebola virus disease.3,4 It soon revealed activity against SARS and MERS coronaviruses (CoVs).5,6 CoVs are positive-sense, single-stranded RNA viruses that infect a wide range of animal hosts. In humans they are known to cause upper respiratory tract infections and pneumonia.6 Due to the effectiveness of GS-441524 in interfering with the activity of viral RNA-dependent RNA polymerases and inhibiting viral RNA synthesis, remdesivir is being developed for the treatment of COVID-19, with encouraging preliminary data.7–9 Concerning its metabolism, Warren et al.1 demonstrated that, upon IV administration of a 10 mg/kg dose in rhesus monkeys, remdesivir exhibited a short plasma half-life (t½ = 0.39 h) with rapid systemic elimination followed by the appearance of transient systemic levels of a key intracellular intermediate alanine metabolite and more persistent levels of GS-441524 (detectable for over 24 h in plasma). Thereafter, remdesivir was rapidly distributed into PBMCs and converted within 2 h into the active nucleoside triphosphate form.1
However, information about remdesivir pharmacokinetics (PK) and pharmacodynamics in humans is inadequate and no therapeutic or toxic ranges have been reported; this is partially due to the small number of patients treated with remdesivir. Therapeutic drug monitoring (TDM), consisting of the measurement of drug concentrations in biological fluids in order to optimize drug posology and avoid toxic effects or therapeutic failures, is already well established in several areas, such as in HIV treatment, and may be useful in the context of COVID-19 therapy.10–12 Therefore, both for PK studies and for possible future TDM, there is the emerging need for a reliable analytical method for the quantification of remdesivir and its metabolite GS-441524 in human plasma. Here we present the first UHPLC method coupled with tandem MS (UHPLC-MS/MS), validated according to FDA and EMA guidelines, for both remdesivir and GS-441524 determination.
Materials and methods
Laboratory certification
The Laboratory of Clinical Pharmacology and Pharmacogenetics (Amedeo di Savoia Hospital, University of Turin) is certified for ‘Design, development and application of determination methods for clinical analytes and drugs. Pharmacogenetic analyses.’ (ISO 9001:2015; certificate no. 18960/08/S), ‘Design and production of diagnostic medical devices in vitro’ (EN ISO 13485:2012; certificate no. DM/17/154/S) and Phase I trials (AIFA; certificate no. IT-64386). See www.tdm-torino.org .
Chemicals
HPLC-grade acetonitrile (ACN) and methanol (MetOH) were purchased from VWR Chemicals (Radnor, PA, USA); MS-grade H2O (MilliQ) was produced with a Milli-DI system coupled with a Synergy 185 system by Millipore (Milan, Italy); DMSO and 6,7-dimethyl-2,3-di(2-pyridyl)quinoxaline [QX; purity 98.5%, used as internal standard (IS)] were purchased from Sigma–Aldrich Corporation (Milan, Italy). Blank plasma from healthy donors was supplied by the Blood Bank of Città della Salute e della Scienza of Turin (Italy).
Remdesivir (purity 98.3%) and its metabolite GS-441524 (purity 98%) were kindly donated by CoQua Lab (Turin, Italy). All powders were stored at −20°C in the dark, in order to prevent any possible degradation.
Stock solutions, IS, standards (STDs) and quality controls (QCs)
Remdesivir and GS-441524 stock solutions (1 mg/mL) were prepared in a mixture of DMSO:MetOH 50:50 (v:v) and QX stock solution (1 mg/mL) in a mixture of H2O:MetOH 5:95 (v:v).
Remdesivir and GS-441524 stock solutions were stored at −80°C in the dark until use (with expiry date 6 months later), while QX stock solution was stored at 4°C (with expiry date 1 year later).
Series of aliquots of the highest STD sample of the calibration curve, STD 9, and QCs were prepared by independently spiking blank plasma with stock solutions and then stored at −80°C. The same calibration ranges and QC concentrations were used both for remdesivir and GS-441524, in accordance with the little information reported in the literature: STD 9, 1000 ng/mL; QC H (high), 800 ng/mL; QC M (medium), 100 ng/mL; QC L (low), 10 ng/mL; and STD 1 (the lowest point of the calibration curve), 3.91 ng/mL.13 An overview of all the concentrations is reported in Table S1 (available as Supplementary data at JAC Online).
STDs 1–8 of the calibration curve were prepared by serial 1:1 dilutions of STD 9.
UHPLC-MS/MS analysis
A Perkin Elmer LX-50® UHPLC system coupled with a Triple Quadrupole QSight 220® (Perkin Elmer, Milan, Italy) was used for the chromatographic analysis. Chromatographic separation was obtained on an Acquity® HSS T3 1.8 μm, 2.1 × 50 mm column (Waters, Milan, Italy), protected by a physical filter (‘Frit’, 0.2 μm, 2.1 mm; Waters, Milan, Italy) precolumn, at 40°C using a column thermostat. The gradient elution was obtained by using two different mobile phases: Phase A (H2O + formic acid 0.05%) and Phase B (ACN + formic acid 0.05%) (Table 1). Positive electrospray ionization (ESI+) was used for all the analytes. Multiple reaction monitoring (MRM) traces (m/z) were quantified as: 603.15 > 200 for remdesivir, 292 > 163 for GS-441524 and 313.2 > 78.05 for QX. All instruments settings are detailed in Tables 2 and 3.
Table 1.
Chromatographic gradient
| Time (min) | % Phase A | % Phase B | Flow (mL/min) |
|---|---|---|---|
| 0.00 | 95 | 5 | 0.4 |
| 0.30 | 95 | 5 | 0.4 |
| 0.35 | 70 | 30 | 0.4 |
| 1.50 | 30 | 70 | 0.4 |
| 1.80 | 10 | 90 | 0.4 |
| 2.80 | 10 | 90 | 0.4 |
| 2.90 | 95 | 5 | 0.4 |
| 4.00 | 95 | 5 | 0.4 |
Phase A: H2O + 0.05% formic acid; Phase B: ACN + 0.05% formic acid.
Table 2.
General instrument settings
| Variables | Setting |
|---|---|
| Drying gas temperature (°C) | 130 |
| HSID temperature (°C) | 270 |
| Nebulizer gas (L/h) | 350 |
| Electrospray V1 positive (V) | 5000 |
| Source temperature (°C) | 350 |
| Multipole 1 RF (V) | 370 |
| Collision pressure (AU) | 410 |
HSID, hot-surface induced desolvation; RF, radio frequency; V, volts; AU, arbitrary units.
Table 3.
Analyte-specific parameters
| Variable | Remdesivir | GS-441524 | QX (IS) |
|---|---|---|---|
| Quantification trace (m/z) | 603.15>200 | 292>163 | 313.20>78.05 |
| collision energy (V) | −53 | −32 | −50 |
| entrance voltage (V) | 15 | 43 | 30 |
| collision cell lens 2 (V) | −116 | −64 | −80 |
| Secondary ion trace (m/z) | 603.15>318 | 292>147 | 313.20>246.15 |
| collision energy (V) | −28 | −50 | −50 |
| entrance voltage (V) | 12 | 2 | 30 |
| collision cell lens 2 (V) | −104 | −80 | −80 |
| Ionization | ESI+ | ESI+ | ESI+ |
Sample extraction protocol
The extraction procedure consisted of a low-cost and rapid protein precipitation: briefly, 100 μL of IS working solution [H2O:MetOH 70:30 (v:v) with QX added to a concentration of 100 ng/mL] and 600 μL of precipitant solution, consisting of a mixture of MetOH:ACN 50:50 (v:v), were added to a volume of 50 μL of plasma sample/calibration STD/QC. After being vortexed for 30 s, samples underwent centrifugation (21 000 g for 10 min at 4°C); 300 μL of the supernatant was then diluted with 600 μL of pure water, mixed and injected (8 μL) into the UHPLC system.
Specificity, selectivity, accuracy, precision and limit of quantification/detection
Six interday validation sessions were performed, as stipulated by FDA and EMA guidelines.14–16 Accuracy and interday imprecision were evaluated, performing quantification of the three different QC samples in duplicate during each validation session; intraday imprecision was evaluated in five intraday replicates. Interday and intraday imprecision were expressed as the relative standard deviation (RSD) at each QC concentration. Integration was performed, considering peak areas for each analyte.
Specificity and selectivity were evaluated using six individual sources of the blank plasma matrix, individually analysed and evaluated for interference. Also, the extent of any interference caused by possible coadministered medications was investigated: briefly, an aliquot of blank plasma was spiked with fourteen antiretroviral drugs currently used for the treatment of HIV (amprenavir, atazanavir, cobicistat, darunavir, dolutegravir, efavirenz, elvitegravir, etravirine, lopinavir, maraviroc, nevirapine, raltegravir, rilpivirine and ritonavir) and analysed.15 The absence of detectable interfering peaks at the analyte retention times was considered as lack of interference.
The upper limit of quantification (ULOQ) corresponded to STD 9, the highest calibration STD, for both the analytes; the lower limit of quantification (LLOQ) for each analyte was the lowest concentration of analyte in a sample that could be quantified reliably, with a deviation from the nominal concentration (measure of accuracy) and RSD (measure of precision) lower than 20% and with a signal-to-noise ratio higher than five.15 On the other hand, the limit of detection (LOD) was considered as the lowest dilution of LLOQ that yielded a signal-to-noise ratio higher than three.
In order to ensure good coverage, even in the case of therapeutic regimens differing from those adopted against Ebola virus (such as in the case of CoVs), the defined calibration range was used to quantify a STD higher than the ULOQ, spiked at a concentration of 3000 ng/mL for both analytes.
Recovery (REC) and extraction efficiency (EE)
REC was evaluated during six validation sessions at high, medium and low concentrations by comparing peak areas from extracted QCs (pre-spiked) with those obtained by the direct injection of a chemical mix containing both the drugs and the IS at the same concentrations as the QCs (rec H, rec M and rec L).14 The EE was measured by comparing the areas of peaks of pre- and post-spiked samples.
Matrix effect (ME) and IS-normalized ME (IS-nME)
Separate plasma samples from six healthy donors were used for the preparation of STDs and for the evaluation of ME. The ME was calculated by comparing the signal from the analysis of post-extraction spiked samples (post-spiked) at high, medium and low QC levels with those from direct injection of the same concentration of analytes without matrix, as described by Taylor17 and in FDA guidelines (post-extraction addition method).14
The IS-nME effect was calculated as described by De Nicolò et al.18–20
Stability and impact of thermal inactivation
As a preliminary experiment, the photostability of the analytes was tested: three concentrations of rec were considered (rec H, rec M and rec L) and analysed by keeping two aliquots for each level, one in the dark and the other under the light on the benchtop for 4 h (in excess of the maximum time requested by the extraction protocol).
Stability was assessed by maintaining single aliquots of the QCs in the following conditions: 24 h benchtop at room temperature (RT), 24 h at 37°C, 24 h at 4°C, 24 h at −20°C and 1, 2, 4, 5 and 7 months at −80°C. Three freeze–thaw cycles were monitored. Furthermore, in order to measure the processed sample stability, extracted samples were maintained for 24 h and 7 days in the autosampler at 10°C.15 All the abovementioned tested conditions were compared with freshly extracted QCs, which had been stored at −80°C since preparation.
Finally, in the context of biosafety, the effect of thermal inactivation on analytes was studied. Based on the study by Rabenau et al.21 concerning SARS-CoV (which demonstrated that heat treatment at 56°C for 30 min reduced the virus titre to below the detection limit), three aliquots of QCs, coming from −80°C storage, were directly placed and maintained at 58°C for 38 min. Considering also the importance of disulphide bonds for the maintenance of enzyme conformation (considering in this case plasma esterases) and the impact of heat on disulphide bonds, three aliquots of QCs that underwent thermal inactivation were then kept for 24 h on the benchtop at RT and finally compared with freshly extracted QCs.22,23
Results
Specificity and selectivity
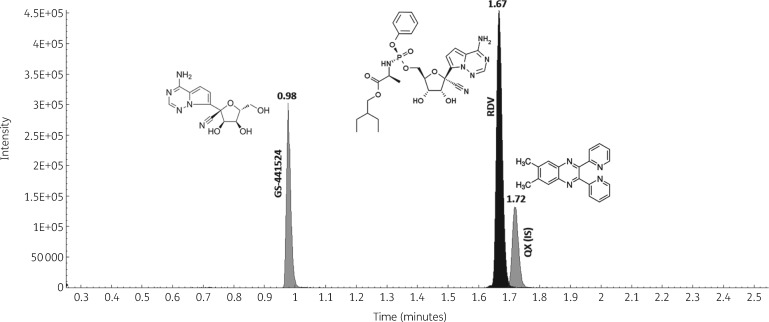
Mean retention times for the considered analytes were 0.98 min for GS-441524, 1.67 min for remdesivir and 1.72 min for QX, the IS (Figure 1). Blank plasma, alone and spiked with antiretroviral drugs, presented no interfering peaks at the analyte retention times (Figure S1).
Figure 1.
Overlaid chromatograms of GS-441524, remdesivir (RDV) and QX (the IS), with respective retention times, obtained from the injection of STD 9, the highest point of the calibration curve.
Accuracy, imprecision, ULOQs, LLOQs and LODs
Accuracy and imprecision values for each analyte at the three QC levels are summarized in Table 4. All these parameters satisfied the FDA and EMA guidelines. The ULOQ coincided with STD 9 for both remdesivir and GS-441524 (1000 ng/mL), the LLOQ value for both the analytes was 0.98 ng/mL while the LOD values were 0.24 ng/mL for remdesivir and 0.98 ng/mL for GS-441524 (Figure S2).
Table 4.
Overview of method validation parameters
| Imprecision (RSD), % |
||||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy, % | intraday | interday | Mean REC, % (RSD, %) | Mean EE, % (RSD, %) | Mean ME, % (RSD, %) | Mean IS-nME, % (RSD, %) | ||
| Remdesivir | High QC level | 104 | 2 | 6 | 67 (6) | 66 (7) | 2 (2) | −10 (1) |
| Medium QC level | 100 | 1 | 6 | 67 (8) | 67 (11) | −1 (3) | −6.9 (3) | |
| Low QC level | 87 | 5 | 6 | 78 (4) | 67 (9) | 16 (7) | 3 (7) | |
| LLOQ | 118 | 10 | 12 | |||||
| Mean (RSD) | 102 | 4.5 | 7.5 | 71 (6) | 67 (9) | 6 (4) | −5 (4) | |
| GS-441524 | High QC level | 96 | 2 | 3 | 104 (6) | 99 (5) | 5 (4) | 1 (4) |
| Medium QC level | 102 | 6 | 4 | 99 (5) | 105 (17) | −3 (21) | −9.2 (10) | |
| Low QC level | 92 | 9 | 11 | 104 (10) | 112 (9) | −7 (10) | −10 (10) | |
| LLOQ | 81 | 9 | 14 | |||||
| Mean (RSD) | 93 | 6 | 8 | 102 (7) | 105 (10) | −2 (12) | −6 (8) | |
RSD, relative standard deviation; REC, recovery; EE, extraction efficiency; ME, matrix effect; IS-nME, internal standard-normalized matrix effect.
Calibration curves had a good fit with ‘linear through zero’ regression models, with a 1/x weighting factor, to ensure high accuracy at low concentrations. Determination coefficients (r2) of calibration curves were all above 0.998.
The defined calibration range revealed the ability to quantify the highest STD (3000 ng/mL for both remdesivir and GS-441524), without requiring a pre-dilution step, and with a deviation from the nominal concentration lower than 20%.
REC, EE, ME and IS-nME
All the parameters satisfied the FDA and EMA guidelines and are detailed in Table 4. Mean values were as follows: REC was 71% (RSD 6%) for remdesivir and 102% (RSD 7%) for GS-441524; EE was 67% (RSD 9%) for remdesivir and 105% (RSD 10%) for GS-441524; ME was 6% (RSD 4%) for remdesivir and −2% (RSD 12%) for GS-441524; IS-nME was −5% (RSD 4%) for remdesivir and −6% (RSD 8%) for GS-441524.
Stability and impact of thermal inactivation
No photodegradation was observed for remdesivir or GS-441524. All results obtained from the stability tests are reported in Tables 5 and 6. Both remdesivir and GS-441524 remained stable in QCs conserved at −80°C for over 4 months; moreover, remdesivir was shown to be stable in the stock solution for at least 10 months (GS-441524 stock solution had not been tested yet). Nevertheless, remdesivir, when dissolved in plasma, was found to be unstable at RT and 4°C, even for 24 h; in contrast, in extracted plasma samples, remdesivir was stable for up to 7 days in the autosampler (10°C).
Table 5.
Degradation (%) of remdesivir and GS-441524 in different conditions: aliquots of QCs maintained in different conditions and ‘stressed’
| After 24 h at |
Freeze and thaw |
Storage at −80°C, months |
Thermal inactivationa |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QC standard | RT | 37°C | 4°C | −20°C | 2°C | 3°C | 1 | 2 | 4 | 5 | 7 | standard | standard + 24 h RT |
| Remdesivir | |||||||||||||
| high | 93 | 99 | 17 | 0 | 6 | 6 | 0 | 4 | 0 | 0 | 0 | 9 | 26 |
| medium | 95 | 100 | 17 | 0 | 10 | 4 | 8 | 0 | 0 | 7 | 0 | 5 | 24 |
| low | 100 | 100 | 22 | 2 | 5 | 8 | 16 | 2 | 0 | 0 | 0 | 4 | 27 |
| GS-441524 | |||||||||||||
| high | 0 | 0 | 1 | 0 | 1 | 0 | NA | NA | 0 | ongoing | ongoing | 4 | 0 |
| medium | 5 | 5 | 1 | 0 | 8 | 0 | NA | NA | 0 | ongoing | ongoing | 0 | 1 |
| low | 0 | 0 | 0 | 0 | 4 | 0 | NA | NA | 0 | ongoing | ongoing | 0 | 0 |
NA, not available.
Standard thermal inactivation was 58°C for 38 min.
Table 6.
Degradation (%) of remdesivir and GS-441524 in different conditions: processed samples and the stock solution stabilities
| QC standard | Processed sample stability [(10°C) post-extraction] |
10 months in stock solution | |
|---|---|---|---|
| 24 h autosampler | 7 days autosampler | ||
| Remdesivir | |||
| high | 0 | 0 | 0 |
| medium | 4 | 0 | 0 |
| low | 7 | 0 | 0 |
| GS-441524 | |||
| high | 0 | 0 | ongoing |
| medium | 0 | 0 | ongoing |
| low | 0 | 0 | ongoing |
Finally, the inactivation of three QC levels by maintaining them at 58°C for 38 min did not have a significant impact on analyte concentrations (mean degradation observed was 7% for remdesivir and none for GS-441524); interestingly, the QCs that were previously stressed by heat and then kept at RT for 24 h showed only a mean 26% degradation for remdesivir (no degradation was observed for GS-441524).
Discussion
We report the first, to the best of our knowledge, published method for remdesivir and GS-441524 quantification using a highly precise quantitative technology, UHPLC-MS/MS. The validation procedure here reported suggests that it is feasible to perform TDM for remdesivir and GS-441524, which could then be applied to identify therapeutic and/or toxic ranges, in order to individualize dosing, avoid toxicity and minimize the risk of therapeutic failures. This assay is important because it could be applied to clinical research, not only for COVID-19, but also for Ebola virus disease. In 2019, interim analysis of the PALM clinical trial comparing four therapeutic agents for the Ebola virus (Zaire) outbreak found lower mortality rates for two monoclonal antibody products (mab114, REGN-EB3) and these drugs have been prioritized over remdesivir.24 However, remdesivir remains relevant as an investigational therapeutic agent for other Ebola strains (Sudan and Bundibugyo) and for Marburg virus disease, where therapies are currently lacking.
The marked remdesivir degradation, observed only in the presence of unextracted plasma, may be due to intense residual activity of esterases, probably inhibited by the low temperature when stored at −20°C and −80°C. This phenomenon may explain the relatively low REC and EE (around 70%); in pre-spiked samples, remdesivir is possibly degraded by plasma esterases, as already demonstrated for artesunate in the context of malaria, whereas this reaction does not occur in post-spiked samples, where the plasma does not contain the proteins anymore, and degradation does not occur in chemical mixtures spiked with the drug.25 Another confirmation of this phenomenon comes from the observation that after thermal inactivation QCs are more stable if kept at RT for 24 h, probably because plasma residual esterases lose their original conformation when excessively stressed by heat. These findings have implications for sample collection, transportation, storage and biosafety when processing for TDM or PK evaluation of remdesivir in tropical countries, where ambient temperatures may be high and access to a cold chain for sample transportation and storage may be limited. In this study, we observed that if samples undergo thermal inactivation immediately after withdrawal and they are then stored in a freezer (−20°C might be cold enough), a large proportion of degradation can be avoided.
Importantly, in PBMCs, nucleoside triphosphate represents the predominant metabolite and it tends to accumulate (with a t½ of 14 h). Consequently, the development of a parallel method for the intracellular quantification of remdesivir and of the triphosphate active form in the near future is guaranteed, following an already tested protocol.26–28
Conclusions
Although this method was not applied to real-life samples (due to a couple of factors: remdesivir is in Phase II evaluation and the number of treated patients is still low, to date, in Italy), it represents the first step in order to ensure a useful tool for the study of remdesivir PK.
Supplementary Material
Acknowledgements
With special thanks to Dr Massimo Tempestilli (Spallanzani Hospital, Rome) for his kind support. We thank the EU for their support through the EDCTP2 programme.
Funding
This research is part of the EDCTP2 programme supported by the European Union (RIA2018EF-083).
Transparency declarations
None to declare.
Supplementary data
Table S1 and Figures S1 and S2 are available as Supplementary data at JAC Online.
References
- 1. Warren TK, Jordan R, Lo MK. et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016; 531: 381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siegel D, Hui HC, Doerffler E. et al. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J Med Chem 2017; 60: 1648–61. [DOI] [PubMed] [Google Scholar]
- 3. Bixler SL, Duplantier AJ, Bavari S.. Discovering drugs for the treatment of Ebola virus. Curr Treat Options Infect Dis 2017; 9: 299–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.EMA. Medicinal products under development for the treatment of Ebola. 2016. https://www.ema.europa.eu/en/documents/referral/assessment-report-article-53-procedure-medicinal-products-under-development-treatment-ebola_en.pdf.
- 5. Sheahan TP, Sims AC, Graham RL. et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 2017; 9: eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agostini ML, Andres EL, Sims AC. et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 2018; 9: e00221–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends 2020; 14: 69–71. [DOI] [PubMed] [Google Scholar]
- 8. Liu W, Morse JS, Lalonde T. et al. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem 2020; 21: 730–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang M, Cao R, Zhang L. et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020; 30: 269–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu X, Ma Q, Zhang F.. Therapeutic drug monitoring in highly active antiretroviral therapy. Expert Opin Drug Saf 2010; 9: 743–58. [DOI] [PubMed] [Google Scholar]
- 11. Aarnoutse RE, Schapiro JM, Boucher CA. et al. Therapeutic drug monitoring: an aid to optimising response to antiretroviral drugs? Drugs 2003; 63: 741–53. [DOI] [PubMed] [Google Scholar]
- 12. Punyawudho B, Singkham N, Thammajaruk N. et al. Therapeutic drug monitoring of antiretroviral drugs in HIV-infected patients. Expert Rev Clin Pharmacol 2016; 9: 1583–95. [DOI] [PubMed] [Google Scholar]
- 13.WHO. Appendix 4: Ad-hoc Expert Consultation on clinical trials for Ebola Therapeutics. 2018. https://www.who.int/ebola/drc-2018/treatments-approved-for-compassionate-use-update/en/.
- 14.FDA. Guidance for Industry: Bioanalytical Method Validation. 2013. https://www.regulations.gov/document?D=FDA-2013-D-1020-0002
- 15.EMA. Guideline on bioanalytical method validation. 2011. https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-bioanalytical-method-validation_en.pdf
- 16. Lynch KL. CLSI C62-A: a new standard for clinical mass spectrometry. Clin Chem 2016; 62: 24–9. [DOI] [PubMed] [Google Scholar]
- 17. Taylor PJ. Matrix effects: the Achilles heel of quantitative high-performance liquid chromatography-electrospray-tandem mass spectrometry. Clin Biochem 2005; 38: 328–34. [DOI] [PubMed] [Google Scholar]
- 18. De Nicolò A, Cantu M, D’Avolio A.. Matrix effect management in liquid chromatography mass spectrometry: the internal standard normalized matrix effect. Bioanalysis 2017; 9: 1093–105. [DOI] [PubMed] [Google Scholar]
- 19. De Nicolò A, Avataneo V, Rabbia F. et al. UHPLC-MS/MS method with sample dilution to test therapeutic adherence through quantification of ten antihypertensive drugs in urine samples. J Pharm Biomed Anal 2017; 142: 279–85. [DOI] [PubMed] [Google Scholar]
- 20. Avataneo V, D’Avolio A, Cusato J. et al. LC-MS application for therapeutic drug monitoring in alternative matrices. J Pharm Biomed Anal 2019; 166: 40–51. [DOI] [PubMed] [Google Scholar]
- 21. Rabenau HF, Cinatl J, Morgenstern B. et al. Stability and inactivation of SARS coronavirus. Med Microbiol Immunol 2005; 194: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yin X, Hu D, Li JF. et al. Contribution of disulfide bridges to the thermostability of a type A feruloyl esterase from Aspergillus usamii. PLoS One 2015; 10: e0126864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Futami J, Miyamoto A, Hagimoto A. et al. Evaluation of irreversible protein thermal inactivation caused by breakage of disulphide bonds using methanethiosulphonate. Sci Rep 2017; 7: 12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mulangu S, Dodd LE, Davey RT Jr. et al. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med 2019; 381: 2293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindegardh N, Hanpithakpong W, Kamanikom B. et al. Major pitfalls in the measurement of artemisinin derivatives in plasma in clinical studies. J Chromatogr B Analyt Technol Biomed Life Sci 2008; 876: 54–60. [DOI] [PubMed] [Google Scholar]
- 26. D’Avolio A, De Nicolo A, Simiele M. et al. Development and validation of a useful HPLC-UV method for quantification of total and phosphorylated-ribavirin in blood and erythrocytes of HCV+ patients. J Pharm Biomed Anal 2012; 66: 376–80. [DOI] [PubMed] [Google Scholar]
- 27. Agnesod D, De Nicolo A, Simiele M. et al. Development and validation of a useful UPLC-MS/MS method for quantification of total and phosphorylated-ribavirin in peripheral blood mononuclear cells of HCV+ patients. J Pharm Biomed Anal 2014; 90: 119–26. [DOI] [PubMed] [Google Scholar]
- 28. De Nicolo A, Bonifacio G, Boglione L. et al. UHPLC-MS/MS method with automated on-line solid phase extraction for the quantification of entecavir in peripheral blood mononuclear cells of HBV+ patients. J Pharm Biomed Anal 2016; 118: 64–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.