Abstract
Miniproteins are a diverse group of protein scaffolds characterized by small (1–10 kDa) size, stability, and versatility in drug-like roles. Coming largely from native sources, they have been widely adopted into drug development pipelines. While their structures and capabilities are diverse, the approaches to their utilization share more similarities to each other than to more widely used modalities (e.g. antibodies or small molecules). This review highlights recent advances in miniprotein-based approaches to otherwise poorly addressed clinical needs, including structure-based and functional characterization. We also summarize their unique screening strategies and pharmacology considerations. Through a greater understanding of the unique properties that make them attractive for drug design, miniproteins can be effectively utilized against targets that are intractable by other approaches.
Keywords: Protein therapeutics, peptide screening, protein pharmacology, cystine-dense peptides, affibodies, stapled peptides
Molecular Therapeutic Modalities are Diverse in Structure and Capabilities
Pharmacological interventions can broadly be segregated into two categories with implications to their capabilities, development strategies, and regulatory hurdles. Small molecule drugs (typically < 1000 Da) are the historical gold standard for drug development, typically targeting a small cleft in an enzyme or macromolecule and generally of limited immunogenicity. Their liabilities are often a forced tradeoff between specificity and potency, particularly when targeting a protein:protein interaction (PPI). Also, in oncology, the limited interface surface area makes each target protein contact crucially important and particularly susceptible to mutational resistance selection. The second category is biologic drugs (see Glossary), currently dominated by antibody therapeutics but also including modalities such as peptides, antisense RNA, gene therapy, and cell therapy (Box 1). Antibodies accounted for 7 of the top 15 sellers in the US drug market in 2018 [https://www.genengnews.com/wp-content/uploads/2019/03/GEN_Apr19_AList_Table.jpg], and methods for their screening and general development are well established. Antibodies excel at target binding and PPI disruption with potency and specificity. However, they carry certain liabilities, including inability to access intracellular targets [1], poor tissue penetration [2,3], complex manufacturing and storage [4], immunogenic potential [5], and limitations to targets of sufficiently poor homology in host animals for B-cell reactivity. Many high-value targets like Ras, Myc, and amyloidogenic inflammatory molecules are considered “undruggable” because they are found in locations antibodies cannot easily access (e.g. solid tumor cores, brain, gut, cytosol) or lack pockets amenable to potent and selective small molecules [6].
Box 1. Generalities of molecular therapeutic modalities. Effective therapeutics come from many sources, but some high-level differences between categories can guide discovery campaigns. Genetic or cell-based modalities (antisense RNA, gene therapy, and cell therapy) are beyond the scope of this review. Of the discrete, targeted molecular modalities, we favor categorization into large proteins, peptides, small molecules, and miniproteins; the latter occupy a distinct class that straddles the size range of peptides and proteins but offers a unique combination of molecular properties, which we highlight compared to properties most typically occurring in the other categories in Table 1. Large proteins are almost universally generated from whole or fragments of native sources; antibody-based therapeutics are exemplary of this. Existing functional and manufacturing knowledge facilitates their design and development, but required control of posttranslational modifications, folding, and process-related impurities necessitates much more extensive development pathways than for the other classes [4]. Miniproteins are smaller, rigid scaffolds that can be produced recombinantly or synthetically. Their small size typically results in short serum half-life, but they can bind with high affinity, and the small, robust scaffolds may act in tissues and compartments that are unavailable to larger proteins. Typical peptides lack the rigidity of miniproteins, hampering both stability and binding affinity, but can be screened rapidly in high diversity (e.g. phage display) and are often produced in high quantities through facile synthetic means [19]. The shorter sequences of miniproteins and peptides are also more amenable to defined drug conjugation. Finally, small molecules are excellent at engaging targets in binding pockets inaccessible to large molecules and can achieve great potency [80]. They are amenable to oral dosing in tablet form, which is most favorable for patients, and they lack the immunogenic potential of proteinaceous drugs. Their weakness is in engaging a target on a flat interface, where potency and specificity are limited.
Choosing a category best equipped for the task at hand can facilitate efficient resource engagement. An extracellular target in the bloodstream with a flat interface is a candidate for a large protein therapeutic. A miniprotein approach would be favored for a flat protein interface in a location of poor accessibility to antibodies (e.g. intracellular) or a tissue of high proteolytic activity (such as the GI tract). Peptide screening could be used to mimic native peptide:target interactions based on a minimal protein interface. Finally, enzymatic activity disruption is often best accomplished with small molecules. Knowledge of the target characteristics and biodistribution will guide this decision prior to screen initiation.
Miniproteins, the topic of this review, are biologics of smaller molecular weight (1–10 kDa) which may retain the potency and specificity advantages of antibodies while avoiding some of their liabilities. The boundaries herein were based on two criteria: first, the existence of a rigid tertiary structure within an independent stable folding unit; second, being of sufficiently small size (less than ~100 amino acids) that synthetic production, complete characterization, and regulation as a small molecule drug rather than as a biological product are possible. While small size is a hallmark of miniproteins, their shared properties confer functional utility and pharmacology that warrant their identification as a distinct class that encompasses molecules that may fall within the size range of peptides or proteins (Box 1). Miniproteins contain stable tertiary structures that facilitate specificity and potency in addition to resistance to proteolysis, reduction, and denaturation. Their size also facilitates computational design and SAR as well as favorable penetration through tissue and manufacturing flexibility. Thus they offer the potential to address difficult biological compartments and difficult targets.
Subcategories and Classes of Miniproteins
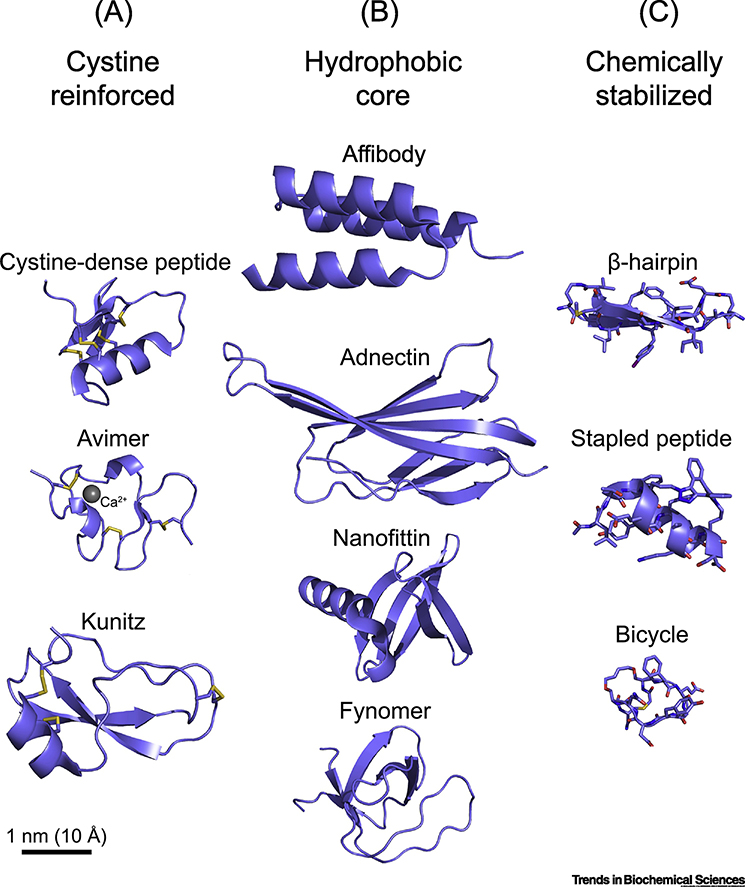
Miniproteins can themselves be segregated into three general subcategories, based on biophysical properties that impact their pharmacologic capabilities and biosynthetic production strategies (Figure 1 and Table 1).
Figure 1.
Miniprotein classes illustrated. Examples of miniprotein scaffolds within the three general subcategories of cystine-reinforced, hydrophobic core, and chemically-stabilized are shown. See Table 1 for the RCSB PDB ID of each chosen scaffold. Images were scaled to demonstrate relative size differences.
Table 1.
Typical characteristics and considerations for miniproteins with comparison to other modalities
| Molecular properties | ||||||||||||
| Category | Subcategory | Class | Common screening methods | Size (kDa) | Description | Specificity | PPI disruption | Enzymatic activity Inhibition | Product homogeneity | |||
| Miniproteins | Cystine reinforced | Avimers | Phage display | 4 | Loop-rich A-domains from human cell surface receptors, stabilized by cystines and a coordinated calcium ion. | High | Excellent | Moderate as novel function, excellent as native function. | Moderate to High | |||
| Kunitz | Yeast display | 7 | Protease inhibitor domain with a hydrophobic core and cystine stabilization. | |||||||||
| CDP | Yeast display, mammalian display | 3–7 | Diverse structure and taxonomic lineage, folding and stability driven primarily by cystine dense core. | |||||||||
| Hydrophobic core | Affibody | Phage display, bacterial display | 7 | Three-helix bundle from FcR. | High | Excellent | Moderate | Moderate to High | ||||
| Adnectin + Centyrins | mRNA display, CIS display | 10 | Fibronectin type 3 (10Fn3) fragment. Highly sheet rich, similar to VH. | |||||||||
| Nanofittins + Affitins | Ribosome display | 7 | 7–kDa DNA binding protein family, such as Sulfolobus acidocaldarius Sac7d; mini β-barrel capped by an α-helix. | |||||||||
| Fynomer | Phage display, OBOC | 7 | SH3 domain of Fyn kinase. Some β-sheet elements with surface-exposed loops. | |||||||||
| Chemically stabilized | β-hairpins | Phage display | 1–2 | Pair of short β-sheets joined by small turn, often cyclized. | High | Moderate to excellent | High | High | ||||
| Stapled peptides | Phage display, OBOC | 1–2 | Single α-helix with chemical staple to stabilize helical fold and impart additional function. | |||||||||
| Bicycles | Phage display, OBOC | 1–2 | Short peptide loop with three reactive residues cross-linked to form bicyclic compound. | |||||||||
| Typical Large proteins | Rational design, mammal immunization for antibodies | > 10 | Whole or modular domain of a protein, applying native function as the therapeutic with minimal structural modifications. | High | Excellent | Poor | Poor | |||||
| Typical (unstructured) Peptides | Phage display, OBOC, rational design | 0.2–4 | Short, flexible amino acid polymer. Minimal, if any, secondary structure. | Moderate to High | Moderate | High | High to Excellent | |||||
| Typical Small molecules | Single-well cell-based assays | < 1 | Synthetic organic molecules of non-protein origin. Many are from natural sources. | Low to Moderate | Poor | Excellent | Excellent | |||||
| Pharmacologic Properties | Manufacturing | |||||||||||
| Category | Subcategory | Class | Tissue/cell penetration | Viable compartments | Serum half-life | Route | Production | Characterization | Cost of goods | Typical storage | PDB used in Fig. 1 | Ref |
| Miniproteins | Cystine reinforced | Avimers | Moderate to high | Extracellular, intracellular, proteolytic | Typically short, can be engineered | Parenteral, some oral | Synthetic or recombinant (eukaryotic or prokaryotic) | Straightforward | Low to moderate | Refrigerated or room temperature | 1AJJ | [17] |
| Kunitz | 5C67 | [87] | ||||||||||
| CDP | 6ATW | [67] | ||||||||||
| Hydrophobic core | Affibody | Moderate to high | Extracellular | Typically short, can be engineered | Parenteral | Synthetic or recombinant (eukaryotic or prokaryotic) | Straightforward | Low to moderate | Refrigerated or room temperature | 3MZW | [88] | |
| Adnectin + Centyrins | 3QWQ | [7] | ||||||||||
| Nanofittins + Affitins | 2XIW | [10] | ||||||||||
| Fynomer | 4AFQ | [8] | ||||||||||
| Chemically stabilized | β-hairpins | Moderate to High | Extracellular, intracellular, proteolytic | Typically short, can be engineered | Parenteral, some oral | Synthetic or recombinant (prokaryotic) for primary screening, synthetic for manufacture | Straightforward | Low to moderate | Refrigerated or room temperature | 5V63 | [89] | |
| Stapled peptides | 5AFG | [90] | ||||||||||
| Bicycles | 5EEL | [91] | ||||||||||
| Typical large proteins | Poor | Extracellular | Antibodies: extended. Others: variable |
Parenteral | Recombinant (eukaryotic) | Complex | High | Refrigerated | [92] | |||
| Typical (unstructured) peptides | Moderate to high | Extracellular, proteolytic with non-natural amino acids | Typically short, can be engineered | Parenteral, some intranasal, oral | Synthetic or recombinant (eukaryotic or prokaryotic) | Straightforward | Low to moderate | Refrigerated or room temperature | [19] | |||
| Typical small molecules | High to excellent | Extracellular, intracellular, proteolytic | Typically short | Oral, parenteral, other | Synthetic | Facile | Low | Room temperature | [80] | |||
Hydrophobic core miniproteins are rich in helices and sheets
The largest miniproteins are the hydrophobic core miniproteins, whose structure and stability are driven by rigid secondary structural elements (α-helix and β-sheet) arranged around a hydrophobic core whose solvent exclusion drives its folding. Several such classes have found use in preclinical and clinical testing. For example, adnectins and centyrins are based on fibronectin type 3 (10Fn3) domains [7] and are reminiscent of antibody VH domains with prominent loops extending out from a quasi-β-barrel structure. Similarly, fynomers are adapted from the SH3 domain of Fyn kinase [8] and are primarily β-barrels with exposed loops for binding. Affibodies are simple three-helix domains based on the antibody-engaging Z-domain of Protein A [9]. Further, nanofittins and affitins are a relatively recent addition to this class and possess extraordinary thermal stability as they are adapted from various 7-kDa DNA binding proteins from thermophilic archea [10]. All four classes are of 7–10 kDa in size and have no need for chemical or oxidative stapling; as such, they can be produced in bacteria in large quantities [7,8]. While their structures are diverse, their binding is mainly driven by similar strategies as in antibody Fv domains: randomized sequences within loops or helices attached to a rigid superstructure. Their thermostability vary (melting temperatures in the 40’s to 80’s °C) [11] and are generally utilized when one wants the pharmacologic function of an antibody on a small, globular scaffold.
Cystine-reinforced miniproteins use disulfides to drive folding and provide rigidity With their folding and stability dependent on Cys-Cys disulfides (cystines), cystine-reinforced miniproteins often require that screening efforts utilize eukaryotes (yeast or mammalian cells) to facilitate cystine formation in the oxidative secretory pathway [12,13]. The largest class consists of cystine-dense peptides (CDPs), including many spider venoms and plant defense peptides. They are of tremendous taxonomic diversity representing a wealth of structurally diverse scaffolds that nevertheless have similar biophysical properties (suggesting convergent evolution of drug-like capabilities in rigid, cystine-stabilized scaffolds) [12]. Several native CDPs are naturally cell-penetrant [14] or blood-brain barrier-penetrant [15], adding versatility to their potential pharmacologic utility. Further, the striking protease resistance provided by the cystine knot can enable activity in aggressive environments such as the gastrointestinal (GI) tract [16]. Kunitz domains have similarities to CDPs but contain a hydrophobic core and are specialized for protease inhibition; hundreds of such proteins exist in nature, such as the popular engraftment scaffold human APPI [13].
Lastly, avimers are based on the loop-rich A-domains of human cell surface receptors, and, unlike CDPs or Kunitz domains, require coordination of a calcium ion. Avimers have been shown to be highly amenable to multimerization [17]. While smaller (4–7 kDa) than hydrophobic core miniproteins, the screening strategies for cystine-reinforced miniproteins are similar to that of larger scaffolds. Binding is imparted by identifying surface exposed regions to engraft known binder motifs or randomized sequences, or by whole-protein (sparing cysteines) mutational screening.
Chemically stabilized miniproteins are rigidified versions of peptides
Most similar to small molecule drugs in both size (1–2 kDa) and functionality are the chemically stabilized miniproteins. They have unique properties but can roughly be considered as miniature, rigidified versions of β-sheets (β-hairpins), α-helices (stapled peptides), or loops (bicycles) found at native protein-protein interfaces. All rely on a chemical crosslinking step to stabilize what would otherwise be unstable or unstructured peptide sequences [18]; we consider them to be miniproteins rather than simple peptides because of the rigidity, functionality, and quasi-tertiary structure imparted by the constraining bonds. Non-stabilized linear peptides can occasionally approach such structural characteristics through selection or rational design, but the addition of chemical staples serves to further improve thermostability and affinity (due to a lower entropic penalty upon binding) [19]. Chemically stabilized miniproteins lack the potential surface area of binding found in the larger miniprotein scaffolds, and as such, potency can be lacking when attempting to use them in conventional interface disruption roles; however, chemically-versatile staples and non-natural amino acids can impart such desirable properties as thermal and protease stability [20], oral bioavailability [21], and cell penetration [22].
Pharmacologic Capabilities of Miniproteins
Miniprotein functions are as diverse as those of native proteins. Broadly, there are five pharmacologic capabilities for which miniproteins have been utilized: agonist, antagonist, steric blocker, ferry, and joiner. This characterization can drive strategy for leveraging a particular miniprotein’s strengths against a target of interest and its disease-associated activity.
Agonist miniproteins augment target activity
Agonists are generally either based on mimicking the native ligand’s target engagement mechanism or incorporation of the native ligand itself in a scaffold for better PK/PD (pharmacokinetic and pharmacodynamic, describing drug distribution/elimination and observed effect, respectively) parameters. For example, a melanocortin receptor agonist was designed by engraftment of the MSH pharmacophore onto the cyclic CDP kalata B1 as a candidate obesity treatment; the engrafted variant was less potent than the native ligand but had high specificity and extreme stability to proteolytic degradation [23]. Similarly, antidepressant stapled peptides, based upon the relaxin-3 neuropeptide and acting as agonists for the relaxin family peptide receptor 3, are also in development and have shown promising activity in rat models of depression and anxiety [24].
Indirect agonism, in the form of T-cell reactivity and immune tolerance induction, is also feasible by CDP engraftment. One case concerns multiple sclerosis, where self-peptides from myelin oligodendrocyte glycoprotein (MOG) are subject to auto-immune targeting, but administration of exogenous peptides are unlikely to elicit immune tolerance due to poor stability. This group engrafted a fragment of MOG into a stable kalata B1 scaffold; when administered with a potent adjuvant, it rendered mice resistant to a subsequent inflammatory stimulus used to model multiple sclerosis [25].
Antagonist miniproteins suppress or eliminate target function
Antagonists serve to interfere with a specific function of the protein. Amongst hydrophobic core miniproteins, CD4- and gp41-binding adnectins were identified that specifically and potently (< 10 nM EC50) inhibit HIV virion fusion with T-cell membranes [26,27]. Cystine-reinforced miniprotein inhibitors are also in development on account of their native inhibitory or agonistic activities: potent and selective native CDP inhibitors have been found against targets like ASIC1a, whose native activity can potentiate neuronal injury after stroke [28], and Kv1.3, whose inhibition can convert inflammatory microglia into a less active, repair-oriented state [29]. Furthermore, tumor cells produce proteases that aid migration, and as Kunitz domain miniproteins are specialized protease inhibitors, they are drawing interest in oncology; for example, a randomly screened canine tapeworm Kunitz domain protein EgKl-1 showed moderately potent (1–5 μM) ability to inhibit cancer cell growth and migration [30]. This highlights the utility of screening for diverse functions in native miniproteins, as their drug-like properties can yield surprising capabilities that would not rationally be ascribed to evolutionary pressure.
The smaller size of chemically stabilized miniproteins lends itself well to precise, mechanistic target inhibition. In recent years, stapled peptides [31] and bicycles [32] were developed to inhibit the oncogenic function of β-catenin, a target in the Wnt pathway and cell-cell adhesion [33]. Similarly, stapled peptides targeting the EGFR/HER2/HER3 juxtamembrane interface were developed that impair dimerization and cross-phosphorylation, reducing tumor burden in murine models of lung and breast cancer [34]. The peptides were more effective than tyrosine kinase inhibitors or monoclonal antibodies, with the ability to avoid resistance mechanisms and access an important intracellular domain. In general, a detailed structural knowledge of the target and its binding/response to native inhibitors will provide the greatest payoff in miniprotein antagonist design, as small motifs capable of potent inhibition can be inserted into a stable scaffold making them into useful drugs.
Steric blocker miniproteins interfere with disease-associated PPIs
Steric blockers of PPIs are the most populous category of known miniproteins in clinical and preclinical development, and the only category in which all three miniprotein subcategories have made substantial progress. In oncology, there have been promising examples of integrin-blocking knottin CDPs [35], VEGFR-blocking adnectins [36], TEAD-blocking CDPs [12], RAS-blocking bicycles [37], and MCL-1-blocking stapled peptides [38]. Beyond oncology, a sub-nM affinity adnectin blocks PCSK9 from participating in LDLR recycling, aiding in normalization of cholesterol levels [39], while administration of an affibody against amyloid β peptide improved cognitive function in Alzheimer’s model mice [40]. In some cases, a native ligand is used for computational or rational design, while in others, the target is randomly screened to find any binder and then counterscreened to confirm disruption of the interaction of interest.
Ferry miniproteins alter the pharmacokinetics and/or biodistribution of other drugs
Vehicles for drug delivery are an increasingly popular role for miniproteins. The favorable stability and varied biodistribution properties of miniproteins have been shown in some instances to synergize with existing, potent small molecules to append the targeting specificity of a biologic while maintaining the tissue penetration and clearance of small molecule pharmacology. Utilizing tumor homing miniproteins for imaging modalities is prevalent; a few examples include a native-derived knottin CDP with tumor accumulation in mice and human patients [41,42], an affibody raised against PD-L1 [43], and an EGFR-binding nanofittin [10]. All are intended to aid in diagnosis (positron emission tomography) or tumor imaging and surgical resection (near-IR fluorescence) with high signal-to-background ratios.
Rapid peripheral clearance and high tissue penetration of miniproteins are an inherent advantage in such applications. A recent work with a bicycle targeting MTI-MMP demonstrated much higher levels of injected dose accumulated in tumors than with an equipotent monoclonal antibody [44]. Cytotoxic molecules can also be targeted to neoplasms, though care must be taken that the usual organs of miniprotein excretion (kidney and liver) are not exposed to excessive levels of the agent. One example is HER2-targeting affibodies that deliver aurostatin E [45]. Meanwhile, loop grafting a knottin CDP scaffold to generate integrin binders can improve gemcitabine potency in glioma cells through mechanisms like insensitivity to nucleoside transporter overexpression [46].
Joiner miniproteins bring two entities into close proximity to induce activity
Two distinct targets can be brought into close proximity (joined) via exposure to a chemical or genetic fusion of miniproteins against one or both targets. This is useful for providing artificially-high concentrations of membrane components, binding partners, or substrates to targets of interest. Chimeric antigen receptor (CAR) T-cells are a prominent example of this strategy. They typically use surface-tethered antibody domains that recognize markers on target cells, linking the cells together spatially while initiating a signaling cascade that activates a cytotoxic response, but an EGFR-binding adnectin can also be used in this capacity [47]. As for soluble heterodimeric reagents, one group has fused a centyrin against FcγRIIB with anti-OX40 antibody constructs, creating a potent T-cell activator [48].
Strategies for Primary Screening of Miniprotein Target Binders
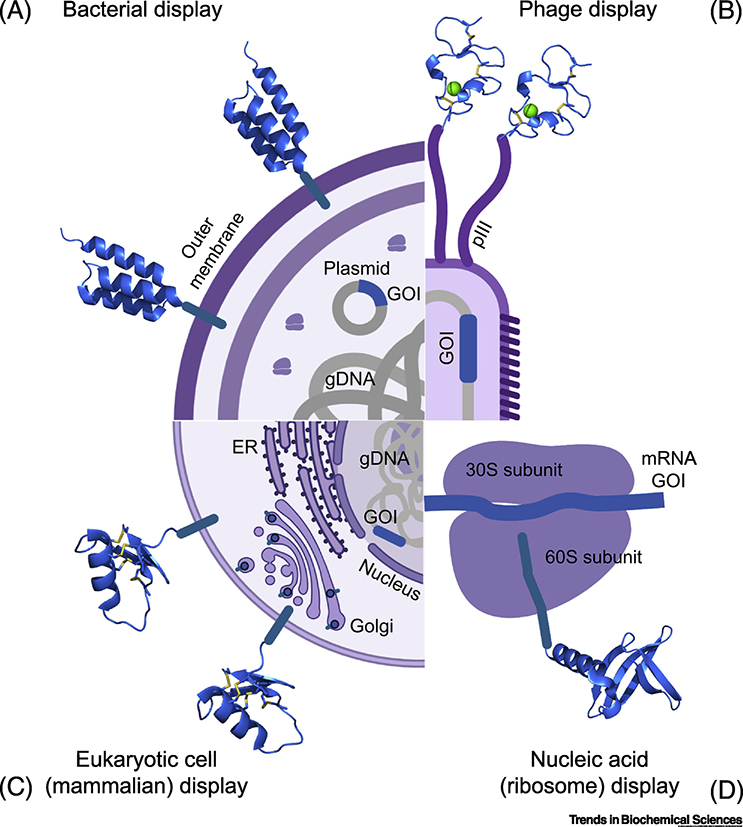
As with other targeted interventions, identifying a miniprotein that binds to a target of interest is the first step in their development. Most miniproteins are screened using some variation of display technology (Figure 2) followed by affinity selection (chromatography or flow cytometry), causing the coding sequence to travel with the protein for later deconvolution. With that in mind, the choice of display technology is mainly driven by the folding properties of the miniprotein in question. Hydrophobic core miniproteins robustly fold without requirements for chemistry steps or oxidation. Hence, they are often produced using common techniques such as phage display [49], bacterial display [50], or direct nucleic acid display (e.g. mRNA display [26], CIS display [48], or ribosome display [10]). These methods require little more than a bacterial incubator or thermocycler, though the isolated particles may require either host cells (phage) or exogenous enzymatic processes (nucleic acid display) for gene amplification. Conversely, cystine-reinforced miniproteins often require an oxidative environment to promote accurate cystine formation. This is most commonly accomplished by display on eukaryotic cells, primarily yeast but occasionally mammalian [12,51]. Eukaryotic (especially mammalian) display can increase the initial screening timeline, but may speed development by producing molecules with the correct disulfide topologies that translate authentically to soluble leads with good developability. Some CDPs can be screened in bacteria or phage [52], but these are pre-selected for prokaryotic folding compatibility, limiting scaffold diversity. Finally, chemically stabilized miniproteins can be screened with high diversity phage display libraries [53–55]; in situ stapling on the phage surface is often accomplished by incorporating cysteines and using redox reactive linkers. The necessity of chemical conjugations adds complexity to the screening protocol, but methods for streamlining this process exist (particularly in a synthetic production setting). Whatever platform is used, affinity maturation of primary screen hits can be performed in the same setting. It is usually approached with either random (e.g. error-prone PCR) or saturating (e.g. discrete variant pools) mutagenesis, depending on economical constraints and the desired affinity characteristics.
Figure 2.
Surface display screening strategies. Four techniques are highlighted: phage display (top right; M13 phage protein pIII fused to an avimer), bacterial display (top left; E. coli displaying an affibody from its outer membrane), eukaryotic cell display (bottom left; mammalian cell displaying a CDP from its plasma membrane), and nucleic acid display (bottom right; ribosome display with a nanofittin). All are variations on a theme: the miniprotein (blue ribbon structure) is tethered to an entity that contains the miniprotein’s encoding sequence (blue line labeled “GOI” for gene of interest). The miniprotein is exposed to a target protein, which is either labeled or immobilized, followed by specific collection of the complex of target, miniprotein, and its tethered entity (bringing with it the GOI). For phage and nucleic acid display, this is often done with bead-based affinity methods. For cell display, flow cytometry is commonly used via cell staining with fluorescently labeled target protein. Multiple rounds of binding, enrichment, and growth/amplification are typical. After final collection, the GOI is then sequenced, identifying the candidate target-binding miniprotein. Note: the bottom left mammalian cell is illustrated with an ER and Golgi apparatus to highlight the importance of the eukaryotic secretory pathway for proper CDP folding. gDNA: genomic DNA.
For those who need high diversity screening incorporating non-natural amino acids (NNAAs), one bead one compound (OBOC) screening can be considered [37,56]. OBOC utilizes sequential mixing and separation of a pool of beads for each round of single amino acid peptide extension, ensuring that a given bead only contains a single miniprotein or peptide species. Beads are collected through similar affinity-driven methods to phage or cell display, followed by mass spectrometry for identification. Alternatively, NNAAs can be incorporated using techniques like RaPID, wherein engineered aminoacyl-tRNA synthetases charge tRNAs with NNAAs, followed by mRNA display [57].
Computer-aided or manual rational design is also plausible with miniproteins for which a confident target:ligand co-crystal structure exists, producing candidates predicted to bind the target per simulated thermodynamic modeling or simply by mimicking the known binding element. This method is most often applied to scaffolds rich in rigid secondary structure elements (α-helices and β-sheets) [12,58,59], as their range of motion is more limited and binding simulations are more accurate at predicting favorable energetics. These scaffolds can be further stabilized by insertion of cystine staples [58], producing molecules of similar stability to native cystine-reinforced miniproteins. Box 2 provides further elaboration on computational screening strategies; while powerful, they require training and computational resources that are not always accessible. In that case, if the interface is simple enough (e.g. a single helix in a groove [60] or a loop engraftment [61]), candidates based on rational sequence substitution can be produced and tested with low diversity screening methods in mind.
Box 2. Computational design of miniprotein binders. Using tools like Rosetta protein design software, it is possible to screen in silico for target engagement using miniprotein scaffolds. This is of particular utility in ligand mimicking, when the goal is to identify a miniprotein that binds not just in the same site, but with the same amino acid interactions as a known binder. This can sometimes be accomplished simply through rational design based on studying the known binder at the interface, but oftentimes the interface is much larger than can reasonably be transferred as a single pharmacophore. In this case, protein design software can take an element of the known interaction to design a novel thermodynamically favorable interface around it. In its most basic form, this design process is performed in three steps: grafting, docking, and design [81]. First, a user-supplied fragment of an existing binder is tested for engraftment capability onto a proposed miniprotein scaffold, then tested for docking at the native binding site. If the fit here is favorable (sufficient contact area with no steric overlap), the amino acids at the interface can be mutated to design a novel binder candidate; for example, inserting basic amino acids in close proximity to an acidic residue on the target to create a salt bridge.
For this purpose, it is important to distinguish whether one is using existing, native scaffold crystal structures (e.g. RCSB PDB files), or de novo generated virtual scaffolds. Existing miniprotein scaffold crystal structures are limited in number and diversity (particularly because high resolution is required for a high success rate) but have the advantage that their stability can be better predicted based on the behavior of the parental protein. Alternatively, one can use scaffolds generated in silico in great numbers and structural diversity, typically rich in rigid secondary structural elements with minimal loops. This strategy increases your shots on goal for grafting/docking/design, but at the cost of using scaffolds that have not been evolved in organisms for effective folding and function. Either strategy is viable, and one should consult with experts at computational protein design for advice, but the stability and rigidity of miniprotein scaffolds makes them excellent candidates for this approach. This is because software can better predict the stability and thermodynamics of an interface dominated by rigid elements (a characteristic antibody CDR regions lack), allowing candidate binder generation with higher confidence for real-world performance.
Optimizing the Pharmacology of Miniproteins
A novel, potent target binder is not automatically a high-value therapeutic candidate. While miniprotein scaffolds have inherent drug-like properties, optimization may be required to meet the desired biological outcome in the complex system of the human body. Common parameters for improvement may include pharamacokinetics, resistance to enzymes and reduction, and immunogenicity. Furthermore, access to some targets requires cell penetration (see Box 3). For a review containing a list of advanced miniprotein clinical candidates, we recommend Simeon et al. 2018 [62].
Box 3. Cell penetration of miniproteins for cytosolic targets. Many compelling targets for miniprotein drug development are intracellular, necessitating delivery across one or more biological membranes. While cell penetration has long been investigated, a more accurate approach requires consideration of the exact subcellular localization of the relevant molecules. Beyond cellular uptake, endosomal escape may be important to effect and must be assessed properly by experimental design [82,83]. Some venom-derived CDPs or fragments thereof act on intracellular targets or traffic to specific intracellular locations that may be useful to realize targeted delivery [14,84,85]. Studying the mechanisms through which they natively access the cytosol could be fruitful for general miniprotein cell penetration strategies. Locking ɑ-helical conformation with introduction of a hydrophobic staple can significantly increase cellular uptake of some peptides, especially those possessing a moderate positive charge [22], while other approaches utilize a concentrated patch of arginine residues on helical miniproteins [86]. However, the mechanisms of uptake are still under investigation [83] and some peptides have decreased uptake after stapling. Indeed, common features in many instances of cellular uptake are a cationic domain and a covalent locking of configuration. Nevertheless, there is not yet a common mechanistic understanding of the rules for imparting cytosolic delivery.
The miniprotein’s pharmacokinetics is often targeted for optimization, depending on the desired dosing interval and tissue exposure profile. It is particularly important to consider target engagement at the site of action in addition to conventional serum half-life characterization [63]. Extended local exposure due to binding or uptake accompanied by more rapid serum clearance has the potential to abrogate off-target toxicities and can be a pharmacological advantage of miniproteins. The target profile may also be a function of acute versus chronic treatment, the kinetics of the pharmacodynamic response, and the treatment setting. Miniproteins typically have fairly short serum half-lives, but several approaches are clinically validated to increase the serum half-life if desired. Linkage to a polymer, such as polyethylene glycol (PEG), is a common technique to extend the half-lives of proteins. Specifically, PEGylation was successfully used to increase the half-life of a cystine-reinforced helical miniprotein designed to block IL-6 in inflammatory disease [52]. Harnessing the long serum retention of albumin, either directly or indirectly, is another established approach. Albumin fusion has been used to extend the half-life of avimers and affibodies [64,65], and fatty acid binding to albumin was used to extend the half-life of a bicycle miniprotein [44]. Alternatively, FcRn-mediated recycling can also increase the half-life of miniproteins via fusion to Fc or full-length IgG [66]. However, increasing miniprotein size to reduce clearance could also restrict tissue penetration, a tradeoff that should be considered when employing this strategy. Formulation and route also play a role; for example, PLGA (a biodegradable copolymer of lactic and glycolic acid) encapsulation has been used to extend exposure of miniproteins by providing a mechanism for prolonged release [https://rapharma.com/wp-content/uploads/2019/04/Peptides-Congress-Presentation-April-2019vFINAL_042419.pdf].
Resistance to proteolytic degradation and disulfide reduction is often necessary for the desired target exposure, especially in aggressive compartments such as the GI tract, the cytosol, and topical locations. A higher density of covalent crosslinks may contribute to the tremendous stability of some miniprotein scaffolds versus proteins such as antibodies. For instance, a panel of 44 diverse CDPs contained members with striking resistance to pepsin (77%), 100°C (45%), trypsin (5%), and even dithiothreitol (9%) [67]. These contained 3–4 disulfide bonds within ~40 residues, whereas IgG1 contains 16 disulfide bonds within ~1350 amino acids. Further, of 5,311 de novo miniprotein scaffolds with hydrophobic cores that were designed to bind botulinum neurotoxin B, only those containing cystines demonstrated protease resistance [58]. Adding extra disulfide bonds to various Kunitz domain miniproteins substantially increased resistance to mesotrypsin [13]. Chemical crosslinks (e.g. staples) have also been shown to increase resistance to proteases. By locking the secondary or tertiary structure of a miniprotein, stabilization of the scaffold can provide increased conformational specificity as well as rigidity, both of which improve affinity [12,68]. Screens for improved stability may be performed in model fluids with proteases, glutathione, and pH relevant to the target compartments or in actual biological fluids in vitro or in vivo. Finally, resistance to proteolysis can be introduced by altering the backbone chemistry, such as incorporating D-amino acids [69] or β amino acids [70].
Lastly, immunogenicity is a potential risk for any proteinaceous therapeutic. Such a response can neutralize the activity of the therapeutic or cause dangerous responses to the therapeutic or endogenous proteins [71]. However, the relatively short sequences of miniproteins renders them amenable to sequence optimization to minimize potential T-cell epitopes, and the extreme protease resistance of some miniproteins should render them resistant to the proteolytic processing required for MHC class II presentation. The former can be approached through in silico modeling, in vitro T-cell activation assays, or in vivo immunogenicity assessments, and the use of D-amino acids has also been considered [72]. However, the predictive power of these approaches are limited by a lack of clinical validation [73]. Miniproteins including CDPs, fynomers, avimers, and β-hairpins have shown low immunogenicity as assessed by antibody formation or maintained exposure after repeat dosing [17,59,74–76]. However, anti-drug antibodies have been detected in humans dosed with some Kunitz or adnectin miniproteins [77,78]. Some of these antibodies recognized engineered loops added to the scaffolds while others recognized the Pichia manufacturing organism, but in both cases, they did not affect efficacy and/or exposure; association with rare anaphylaxis was unclear. Immunogenicity can be a function of primary or tertiary structure, but also of factors beyond initial drug design such as impurities, aggregation, dosing route/schedule, formulation, and patient population. As such, immunogenicity must be mitigated by a network of approaches that includes clinical assessment.
Concluding remarks
Miniprotein scaffolds have enormous potential to impact disease intervention by approaching the countless targets that are inaccessible to antibodies and lack pockets that can be modulated by small molecules. Rather than supplanting existing modalities, we believe miniproteins should be employed when their size, stability, and specificity fulfill a need wanting in other approaches. Extensive structural and mechanistic knowledge of the target is recommended before sophisticated miniprotein intervention strategies are tested. However, tools for achieving this detailed understanding at the atomic level, combined with increasingly powerful and accurate protein design software and established methods for optimizing drug-like properties, permit creative approaches to unmet medical needs.
Further work to advance miniprotein therapeutics includes detailed consideration of the pharmacology of disease targets, as well as technologies that facilitate development (see Outstanding Questions). For a given disease pathway, efficacious target engagement [63,79] requires adequate drug exposure to the tissue and/or cellular compartment where the target is located, necessitating understanding of drug and target biodistribution and of duration of effect upon target engagement. Advancing our understanding of the pharmacologic properties of miniproteins, as compared to conventional modalities, will allow researchers to determine which targets could be favorably approached with miniproteins. Additionally, demonstration of the ability of miniproteins to effectively modulate targets that are “undruggable” by antibodies or small molecules will increase confidence in committing programs to these approaches; where possible, a side-by-side comparison with a conventional approach is ideal. Continuing development of technologies that make intracellular delivery and non-parenteral delivery routes more off-the-shelf will also facilitate utilization of miniproteins, as will establishment of platform manufacturing and regulatory approaches. Similarly, improvements in computational software that increase accessibility with limited training will enable more researchers to use these tools. Being an unconventional modality targeting unmet clinical needs, many of these efforts are well-suited for academia.
Outstanding Questions.
While the protein and peptide screening landscape is vast, not all small proteins contain the drug-like properties of miniproteins. Can we develop computational algorithms or economical in vitro assays to aid in the identification of novel miniprotein scaffolds with bona fide drug-like properties, as opposed to the current practice of hypothesis-driven research?
Can the regulatory agencies establish a clear framework of CMC (Chemistry, Manufacturing, and Controls) expectations for miniproteins, similar to existing guidance for small molecule drugs and large protein (e.g. antibody-based) biological products?
Can clinical datasets be correlated sufficiently to allow prediction of minimally immunogenic miniproteins, taking into account unique characteristics like a scaffold’s protease resistance or chemical staples?
Is there a generalizable strategy to design and evaluate biodistribution to tissues and targets to achieve the desired tissue exposure profile?
Can we adapt miniprotein pharmacology assays (e.g. protease resistance, serum half life, biodistribution, immunogenicity) to pooled candidate libraries for more rapid selection of candidates and scaffolds with favorable drug-like qualities?
Can computational interface design tools be adapted to using scaffolds of substantial rigidity but minimal ɑ-helical or β-sheet secondary structural elements?
By specifically focusing on so-called “undruggable” protein targets in poorly-accessible locations, can miniprotein therapeutics carve out a niche as a primary modality for targets or conditions that have been intransigent to conventional small molecule or antibody approaches?
With small molecule-like pharmacology and protein-like utility, miniproteins are increasingly being utilized as a favorable approach to intransigent disease pathways; we are excited to see the field continue exploring the capabilities of miniprotein clinical candidates.
Highlights.
Therapeutics based on miniprotein scaffolds can have antibody-like affinity and functionality while avoiding some of the liabilities of larger scaffolds such as poor tissue or cell penetration, protease and reduction sensitivity, complex manufacturing and characterization, or (for antibodies) reliance on targets of poor host homology.
Amongst miniproteins, three categories (hydrophobic core, cystine-reinforced, and chemically-stabilized) emerge with distinct biochemical and biophysical characteristics but similar utility.
Most screening campaigns use a variant of surface display, though chemical conjugation can be utilized if necessary.
Miniprotein candidates that do not naturally possess favorable serum half-life, cell penetration, or protease resistance properties can often be engineered through evolution, genetic fusion, or chemical modification for these characteristics.
Acknowledgements
We wish to thank our collaborators and the members of our laboratories, but in particular Roland Strong and Claudia Jochheim, for helpful conversations and fact-checking during manuscript preparation. This work was funded by NIH Grants NCI/5 R01 CA223674-02 (J.M.O.) and Project Violet (Z.R.C. and J.M.O.). Figure 2 was created with BioRender.
Glossary
- Agonist
molecule that stimulates a target protein or complex’s native activity
- Biologic drugs
here encompassing therapeutics derived from biological sources, including cells, proteins, nucleic acids, sugars, or complex combinations thereof. Note, certain peptides and proteins may be regulated as biological products or as small molecule drugs by the US FDA
- CIS display
nucleic acid display wherein the displayed miniprotein is fused to the RepA initiation protein, attaching it to its own template DNA
- Cystine-dense peptide
One of a structurally and taxonomically diverse class of cystine-reinforced miniproteins, including knottins and defensins, whose folding and stability are entirely dependent on a core of cystine disulfides (3 or more) without a hydrophobic core
- Cystine knot
a disulfide topology common to CDPs containing a two-cystine macrocycle through which a third cystine passes
- Disulfide topology
the specific pairing pattern of cysteines when multiple cystines are present
- Hydrophobic core
the interior of a protein or miniprotein containing aromatic or aliphatic (non-polar) side chains, sequestered from solvent
- In situ stapling
synthetic chemical stapling (typically thiol-reactive) applied to intact phage displaying proteins with reactive amino acid side chains
- Kunitz domain
a family of globular, disulfide-stabilized miniproteins with a hydrophobic core that canonically serve as protease inhibitors, particularly in animals
- Preclinical
vertebrate animal models meant to approximate patient pharmacology
- Rational design
protein design based on incorporating known target:ligand interactions derived from biophysical characterization
- SAR
structure-activity relationship, or how the structural and chemical nature of the molecule impact biological function
- Scaffold
a protein tertiary fold template, usually of modular folding capability
- Stapling
intramolecular covalent bonds that serve to stabilize the scaffold, imparting rigidity, thermostability, and/or protease resistance
Footnotes
Competing Interests
Z.R.C. has a paid consulting relationship with Blaze Bioscience, which develops miniprotein therapeutics. N.W.N. is an employee of Blaze Bioscience, and J.M.O. is a founder and shareholder of Blaze Bioscience.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Trenevska I et al. (2017) Therapeutic Antibodies against Intracellular Tumor Antigens. Front. Immunol 8, 1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cruz E and Kayser V (2019) Monoclonal antibody therapy of solid tumors: Clinical limitations and novel strategies to enhance treatment efficacy. Biol. Targets Ther 13, 33–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thurber GM et al. (2008) Antibody tumor penetration: transport opposed by systemic and antigen-mediated clearance. Adv. Drug Deliv. Rev 60, 1421–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shukla AA et al. (2017) Evolving trends in mAb production processes. Bioeng. Transl. Med 2, 58–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Almagro JC et al. (2017) Progress and Challenges in the Design and Clinical Development of Antibodies for Cancer Therapy. Front. Immunol 8, 1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verdine GL and Walensky LD (2007) The challenge of drugging undruggable targets in cancer: Lessons learned from targeting BCL-2 family members. Clin. Cancer Res 13, 7264–7270 [DOI] [PubMed] [Google Scholar]
- 7.Sachdev E et al. (2015) Adnectin-Targeted Inhibitors: Rationale and Results. Curr. Oncol. Rep 17, 1–6 [DOI] [PubMed] [Google Scholar]
- 8.Schlatter D et al. (2012) Generation, characterization and structural data of chymase binding proteins based on the human Fyn kinase SH3 domain. MAbs 4, 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ståhl S et al. (2017) Affibody Molecules in Biotechnological and Medical Applications. Trends Biotechnol. 35, 691–712 [DOI] [PubMed] [Google Scholar]
- 10.Goux M et al. (2017) Nanofitin as a New Molecular-Imaging Agent for the Diagnosis of Epidermal Growth Factor Receptor Over-Expressing Tumors. Bioconjug. Chem 28, 2361–2371 [DOI] [PubMed] [Google Scholar]
- 11.Daunert S et al. (2017) Beyond Antibodies as Binding Partners: The Role of Antibody Mimetics in Bioanalysis. Annu. Rev. Anal. Chem 10, 293–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crook ZR et al. (2017) Mammalian display screening of diverse cystine-dense peptides for difficult to drug targets. Nat. Commun 8, 2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen I et al. (2019) Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin. J. Biol. Chem 294, 5105–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mabrouk K et al. (2007) Critical amino acid residues of maurocalcine involved in pharmacology, lipid interaction and cell penetration. Biochim. Biophys. Acta - Biomembr. 1768, 2528–2540 [DOI] [PubMed] [Google Scholar]
- 15.Veiseh M et al. (2007) Tumor paint: A chlorotoxin:Cy5.5 bioconjugate for intraoperative visualization of cancer foci. Cancer Res. 67, 6882–6888 [DOI] [PubMed] [Google Scholar]
- 16.Busby RW et al. (2013) Pharmacologic properties, metabolism, and disposition of linaclotide, a novel therapeutic peptide approved for the treatment of irritable bowel syndrome with constipation and chronic idiopathic constipation. J. Pharmacol. Exp. Ther 344, 196–206 [DOI] [PubMed] [Google Scholar]
- 17.Silverman J et al. (2005) Multivalent avimer proteins evolved by exon shuffling of a family of human receptor domains. Nat. Biotechnol 23, 1556–1561 [DOI] [PubMed] [Google Scholar]
- 18.Tsomaia N (2015) Peptide therapeutics: Targeting the undruggable space. Eur. J. Med. Chem 94, 459–470 [DOI] [PubMed] [Google Scholar]
- 19.Henninot A et al. (2018) The Current State of Peptide Drug Discovery: Back to the Future? J. Med. Chem 61, 1382–1414 [DOI] [PubMed] [Google Scholar]
- 20.Angelini A et al. (2012) Bicyclization and tethering to albumin yields long-acting peptide antagonists. J. Med. Chem 55, 10187–10197 [DOI] [PubMed] [Google Scholar]
- 21.Nielsen DS et al. (2017) Orally Absorbed Cyclic Peptides. Chem. Rev 117, 8094–8128 [DOI] [PubMed] [Google Scholar]
- 22.Chu Q et al. (2015) Towards understanding cell penetration by stapled peptides. Medchemcomm 6, 111–119 [Google Scholar]
- 23.Eliasen R et al. (2012) Design, synthesis, structural and functional characterization of novel melanocortin agonists based on the cyclotide kalata B1. J. Biol. Chem 287, 40493–40501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marwari S et al. (2019) Intranasal administration of a stapled relaxin-3 mimetic has anxiolytic- and antidepressant-like activity in rats. Br. J. Pharmacol DOI: 10.1111/bph.14774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang CK et al. (2014) Molecular grafting onto a stable framework yields novel cyclic peptides for the treatment of multiple sclerosis. ACS Chem. Biol 9, 156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wensel D et al. (2017) Discovery and Characterization of a Novel CD4-Binding Adnectin with Potent Anti-HIV Activity. Antimicrob. Agents Chemother 61, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wensel D et al. (2018) A Novel gp41-Binding Adnectin with Potent Anti-HIV Activity Is Highly Synergistic when Linked to a CD4-Binding Adnectin. J. Virol 92, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chassagnon IR et al. (2017) Potent neuroprotection after stroke afforded by a double-knot spider-venom peptide that inhibits acid-sensing ion channel 1a. Proc. Natl. Acad. Sci 114, 3750–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y et al. (2015) Toxins targeting the Kv1.3 Channel: Potential immunomodulators for autoimmune diseases. Toxins (Basel). 7, 1749–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranasinghe SL et al. (2018) Kunitz type protease inhibitor EgKI-1 from the canine tapeworm Echinococcus granulosus as a promising therapeutic against breast cancer. PLoS One 13, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossmann TN et al. (2012) Inhibition of oncogenic Wnt signaling through direct targeting of β-catenin. Proc. Natl. Acad. Sci. U. S. A 109, 17942–17947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertoldo D et al. (2016) Phage Selection of Peptide Macrocycles against β-Catenin to Interfere with Wnt Signaling. ChemMedChem 11, 834–839 [DOI] [PubMed] [Google Scholar]
- 33.Clevers H and Nusse R (2012) Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 [DOI] [PubMed] [Google Scholar]
- 34.Maisel SA et al. (2019) Stapled EGFR peptide reduces inflammatory breast cancer and inhibits additional HER-driven models of cancer. J. Transl. Med 17, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JW et al. (2015) A chemically cross-linked knottin dimer binds integrins with picomolar affinity and inhibits tumor cell migration and proliferation. J. Am. Chem. Soc 137, 6–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aghaabdollahian S et al. (2019) Enhancing bioactivity, physicochemical, and pharmacokinetic properties of a nano-sized, anti-VEGFR2 Adnectin, through PASylation technology. Sci. Rep 9, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trinh TB et al. (2016) Discovery of a Direct Ras Inhibitor by Screening a Combinatorial Library of Cell-Permeable Bicyclic Peptides. ACS Comb. Sci 18, 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rezaei Araghi R et al. (2018) Iterative optimization yields Mcl-1-targeting stapled peptides with selective cytotoxicity to Mcl-1-dependent cancer cells. Proc. Natl. Acad. Sci. U. S. A 115, E886–E895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell T et al. (2014) Pharmacologic Profile of the Adnectin BMS-962476, a Small Protein Biologic Alternative to PCSK9 Antibodies for Low-Density Lipoprotein Lowering. J. Pharmacol. Exp. Ther 350, 412–424 [DOI] [PubMed] [Google Scholar]
- 40.Boutajangout A et al. (2019) Affibody-Mediated Sequestration of Amyloid β Demonstrates Preventive Efficacy in a Transgenic Alzheimer’s Disease Mouse Model. Front. Aging Neurosci 11, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baik FM et al. (2016) Fluorescence Identification of Head and Neck Squamous Cell Carcinoma and High-Risk Oral Dysplasia With BLZ-100, a Chlorotoxin-Indocyanine Green Conjugate. JAMA Otolaryngol. Neck Surg 142, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patil CG et al. (2019) Phase 1 Safety, Pharmacokinetics, and Fluorescence Imaging Study of Tozuleristide (BLZ-100) in Adults With Newly Diagnosed or Recurrent Gliomas. Neurosurgery 0, 1–9 [DOI] [PubMed] [Google Scholar]
- 43.González Trotter DE et al. (2017) In Vivo Imaging of the Programmed Death Ligand 1 by 18 F PET. J. Nucl. Med 58, 1852–1857 [DOI] [PubMed] [Google Scholar]
- 44.Eder M et al. (2019) Bicyclic peptides as a new modality for imaging and targeting of proteins overexpressed by tumors. Cancer Res. 79, 841–852 [DOI] [PubMed] [Google Scholar]
- 45.Serwotka-Suszczak AM et al. (2017) A conjugate based on anti-HER2 diaffibody and auristatin e targets HER2-positive cancer cells. Int. J. Mol. Sci 18, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox N et al. (2016) Integrin-Targeting Knottin Peptide–Drug Conjugates Are Potent Inhibitors of Tumor Cell Proliferation. Angew. Chemie - Int. Ed 55, 9894–9897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han X et al. (2017) Adnectin-Based Design of Chimeric Antigen Receptor for T Cell Engineering. Mol. Ther 25, 2466–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang D et al. (2018) FcγRII-binding Centyrins mediate agonism and antibody-dependent cellular phagocytosis when fused to an anti-OX40 antibody. MAbs 10, 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hulme JT et al. (2018) Novel protein therapeutic joint retention strategy based on collagenbinding Avimers. J. Orthop. Res 36, 1238–1247 [DOI] [PubMed] [Google Scholar]
- 50.Fleetwood F et al. (2015) Simultaneous targeting of two ligand-binding sites on VEGFR2 using biparatopic Affibody molecules results in dramatically improved affinity. Sci. Rep 4, 7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore SJ and Cochran JR (2012) Engineering Knottins as Novel Binding Agents In Protein Engineering for Therapeutics Part B (1st edn), 503pp. 223–251, Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- 52.Ranganath S et al. (2015) Discovery and characterization of a potent interleukin-6 binding peptide with neutralizing activity in Vivo. PLoS One 10, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barkan DT et al. (2016) Clustering of disulfide-rich peptides provides scaffolds for hit discovery by phage display: application to interleukin-23. BMC Bioinformatics 17, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watson GM et al. (2017) Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target. J. Med. Chem 60, 9349–9359 [DOI] [PubMed] [Google Scholar]
- 55.Corbi-Verge C et al. (2017) Strategies to Develop Inhibitors of Motif-Mediated Protein-Protein Interactions as Drug Leads. Annu. Rev. Pharmacol. Toxicol 57, 39–60 [DOI] [PubMed] [Google Scholar]
- 56.Gates ZP et al. (2018) Xenoprotein engineering via synthetic libraries. Proc. Natl. Acad. Sci 115, E5298–E5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bashiruddin NK and Suga H (2015) Construction and screening of vast libraries of natural product-like macrocyclic peptides using in vitro display technologies. Curr. Opin. Chem. Biol 24, 131–138 [DOI] [PubMed] [Google Scholar]
- 58.Chevalier A et al. (2017) Massively parallel de novo protein design for targeted therapeutics. Nature 550, 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhardwaj G et al. (2016) Accurate de novo design of hyperstable constrained peptides. Nature 538, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muppidi A et al. (2012) Rational design of proteolytically stable, cell-permeable peptide-based selective Mcl-1 inhibitors. J. Am. Chem. Soc 134, 14734–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moore SJ et al. (2013) Engineering agatoxin, a cystine-knot peptide from spider venom, as a molecular probe for in vivo tumor imaging. PLoS One 8, e60498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simeon R and Chen Z (2018) In vitro-engineered non-antibody protein therapeutics. Protein Cell 9, 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durham TB and Blanco MJ (2015) Target Engagement in Lead Generation. Bioorganic Med. Chem. Lett 25, 998–1008 [DOI] [PubMed] [Google Scholar]
- 64.Smith R et al. (2013) FGF21 Can Be Mimicked In Vitro and In Vivo by a Novel Anti-FGFR1c/βKlotho Bispecific Protein. PLoS One 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong D et al. (2016) Human Serum Albumin and HER2-Binding Affibody Fusion Proteins for Targeted Delivery of Fatty Acid-Modified Molecules and Therapy. Mol. Pharm. 13, 3370–3380 [DOI] [PubMed] [Google Scholar]
- 66.Klupsch K et al. (2019) COVA4231, a potent CD3/CD33 bispecific FynomAb with IgG-like pharmacokinetics for the treatment of acute myeloid leukemia. Leukemia 33, 805–808 [DOI] [PubMed] [Google Scholar]
- 67.Correnti CE et al. (2018) Screening, large-scale production and structure-based classification of cystine-dense peptides. Nat. Struct. Mol. Biol DOI: 10.1038/s41594-018-0033-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bird GH et al. (2010) Hydrocarbon double-stapling remedies the proteolytic instability of a lengthy peptide therapeutic. Proc. Natl. Acad. Sci 107, 14093–14098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng Z and Xu B (2016) Inspiration from the mirror: D-amino acid containing peptides in biomedical approaches. Biomol. Concepts 7, 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Checco JW et al. (2015) Targeting diverse protein-protein interaction interfaces with α/βpeptides derived from the Z-domain scaffold. Proc. Natl. Acad. Sci. U. S. A 112, 4552–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Services, U.S.D. of H. and H. et al. (2014) Guidance for Industry: Immunogenicity Assessment for Therapeutic Protein Products. http://www.fda.gov/Drugs/DevelopmentApprovalProcess/HowDrugsareDevelopedandApproved/ApprovalApplications/TherapeuticBiologicApplications/ucm080536.htm at <http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm338856.pdf>
- 72.Mandal K et al. (2012) Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography. Proc. Natl. Acad. Sci. U. S. A 109, 14779–14784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brinks V et al. (2013) Preclinical models used for immunogenicity prediction of therapeutic proteins. Pharm. Res 30, 1719–1728 [DOI] [PubMed] [Google Scholar]
- 74.Grabulovski D et al. (2007) A novel, non-immunogenic Fyn SH3-derived binding protein with tumor vascular targeting properties. J. Biol. Chem 282, 3196–3204 [DOI] [PubMed] [Google Scholar]
- 75.Wach A et al. (2018) Pharmacokinetics and Safety of Intravenous Murepavadin Infusion in Healthy Adult Subjects Administered Single and Multiple Ascending Doses. Antimicrob. Agents Chemother 62, e02355–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tarcha EJ et al. (2017) Safety and pharmacodynamics of dalazatide, a Kv1.3 channel inhibitor, in the treatment of plaque psoriasis: A randomized phase 1b trial. PLoS One 12, e0180762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Craig TJ et al. (2015) Characterization of Anaphylaxis After Ecallantide Treatment of Hereditary Angioedema Attacks. J. Allergy Clin. Immunol. Pract 3, 206–212.e4 [DOI] [PubMed] [Google Scholar]
- 78.Tolcher AW et al. (2011) Phase I and pharmacokinetic study of CT-322 (BMS-844203), a targeted adnectin inhibitor of VEGFR-2 based on a domain of human fibronectin. Clin. Cancer Res. 17, 363–371 [DOI] [PubMed] [Google Scholar]
- 79.Cook D et al. (2014) Lessons learned from the fate of AstraZeneca’s drug pipeline: A fivedimensional framework. Nat. Rev. Drug Discov 13, 419–431 [DOI] [PubMed] [Google Scholar]
- 80.Stocks M (2013) The small molecule drug discovery process – from target selection to candidate selection In Introduction to Biological and Small Molecule Drug Research and Development pp. 81–126, Elsevier [Google Scholar]
- 81.Silva D-A et al. (2016) Motif-Driven Design of Protein–Protein Interfaces. In Methods in Molecular Biology 1414pp. 285–304 [DOI] [PubMed] [Google Scholar]
- 82.Deprey K et al. (2019) Trapped! A Critical Evaluation of Methods for Measuring Total Cellular Uptake versus Cytosolic Localization. Bioconjug. Chem 30, 1006–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pei D and Buyanova M (2019) Overcoming Endosomal Entrapment in Drug Delivery. Bioconjug. Chem 30, 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wiranowska M et al. (2011) Clathrin-mediated entry and cellular localization of chlorotoxin in human glioma. Cancer Cell Int. 11, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gurrola GB et al. (2010) Imperatoxin a, a cell-penetrating peptide from scorpion venom, as a probe of Ca2+-release channels/ryanodine receptors. Pharmaceuticals 3, 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Appelbaum JS et al. (2012) Arginine Topology Controls Escape of Minimally Cationic Proteins from Early Endosomes to the Cytoplasm. Chem. Biol 19, 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scott CJ and Taggart CC (2010) Biologic protease inhibitors as novel therapeutic agents. Biochimie 92, 1681–1688 [DOI] [PubMed] [Google Scholar]
- 88.Frejd FY and Kim KT (2017) Affibody molecules as engineered protein drugs. Exp. Mol. Med 49, e306–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Obrecht D et al. (2012) β-Hairpin protein epitope mimetic technology in drug discovery. Drug Discov. Today Technol 9, e63–e69 [DOI] [PubMed] [Google Scholar]
- 90.Walensky LD and Bird GH (2014) Hydrocarbon-stapled peptides: Principles, practice, and progress. J. Med. Chem 57, 6275–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heinis C et al. (2009) Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat. Chem. Biol 5, 502–507 [DOI] [PubMed] [Google Scholar]
- 92.Zhao L et al. (2012) Clinical pharmacology considerations in biologics development. Acta Pharmacol. Sin 33, 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]