Abstract
The common laboratory zebrafish can regenerate functional cardiac muscle after cataclysmic damage or loss, by activating programs that direct the division of spared cardiomyocytes. Heart regeneration is not a linear series of molecular steps and synchronized cellular progressions, but rather an imperfect, relentless process that proceeds in an advantaged competition with scarring until recovery of the lost heart function. In this review, we summarize recent advances in our understanding of signaling events that have formative roles in injury-induced cardiomyocyte proliferation in zebrafish, and we forecast advances in the field that are needed to decipher heart regeneration.
Introduction
Zebrafish have become a powerful model system to study key biological events like embryogenesis, disease, behavior, and regeneration. Their external fertilization, rapid early development, and transparency of embryos benefit the study of fundamental developmental mechanisms, advances in ecotoxicology, and new drug discovery [1–3]. The ability of zebrafish to regenerate its fins was first reported in the late 1980’s, although the phenomenon of fin regeneration in teleost fish had been recognized two centuries prior [4, 5]. Since then, many other tissues of zebrafish have been reported to regenerate after injury [6, 7].
Heart regeneration in zebrafish was first described in 2002 as a process in which a new wall of muscle is built through the division of cardiomyocytes (CMs), after surgical removal of a fifth of the cardiac ventricle [8]. While one of the early models invoked a progenitor cell precursor to these proliferating cardiomyocytes [9], the use of genetic fate-mapping approaches has unambiguously identified the source of muscle to be pre-existing muscle cells, stimulated by injury to divide [10, 11]. By contrast, and reviewed extensively elsewhere [12, 13], adult mammals have a limited ability to provoke cardiomyocyte division upon injury, and the injured mammalian heart heals a major injury like myocardial infarction predominantly by scarring and tissue remodeling. This is considered a process of repair, not regeneration, and, while a scarred human heart can and often does perform at a sufficient level for decades, an initial injury/repair event can leave the heart vulnerable to further injury and failure.
Heart regeneration is a complex process that involves communication between multiple cell types. As opposed to morphogenesis in embryos, where many tissues grow and mature in synchrony to increase size and improve physiological function, heart muscle regeneration is more of a solo performance and does not adhere to a strict time window to perform its chief goal – to outdo the competing process of scarring. Research over the past 17 years has significantly advanced what we know of the signals exchanged during regeneration, although there remains much to learn. Whereas findings in other model systems like mice have provided many insights into the regenerative capacity of the heart, we limit this brief review to what is known of the signaling mechanisms that direct cardiomyocyte proliferation during zebrafish heart regeneration.
The cellular players in heart regeneration
To regenerate efficiently, CMs need intrinsic programs that are competent for division, as well as the ability to receive extrinsic proliferative cues. Without question, CMs are built to work. They are massive cells that rhythmically contract and are filled with sarcomeres and energy-supplying mitochondria for that purpose. Mammalian hearts have a high percentage (>50%) of polyploid CMs, with multiple diploid nuclei or single polyploid nuclei, or both, with the proportions depending on species [14]. Higher DNA content possibly enables the generation of more contractile and energizing machinery on a per-cell basis. In turn, it is just as likely that the highly differentiated structure of CMs restricts their ability to complete cytokinesis during organ growth or in response to injury. As most or all teleost and amphibian vertebrates that have been examined have a high proportion (>90%) of mononuclear, diploid CMs, the association of ploidy and regeneration is strong, and it has been fortified in several recent studies [15–17]. One of these reports examined zebrafish and described experiments inducing the expression of a dominant-negative form of the Ect2 guanine nucleotide exchange factor in CMs transiently during heart development, to yield adults with a ~50% complement of polyploid CMs [15]. The authors found that these genetically manipulated CMs participated less vigorously in regeneration than did predominantly diploid CMs. The key manipulation in this study was elegant and precisely controlled, though it is conceivable that some event in addition to multinucleation occurred under Ect2 inhibition that might contribute to the regenerative defects. Zebrafish CMs also undergo some level of dedifferentiation, a dedicated program that includes induction of cardiac transcription factors, tempering of the contractile machinery, and acquisition of a more glycolytic program [10, 11, 18], events that require further elucidation and are expected to further improve the competency of CMs to divide.
The heart is not pure, uncaged muscle, but instead has a diverse cast of supporting cells. The wall of the zebrafish ventricle is, like all vertebrate hearts, lined by an outer mesothelial lining called the epicardium and an inner endothelial lining called the endocardium (Fig. 1). The wall is vascularized and innervated, and there is a minor fibroblast component. When the heart is in an acute phase of injury, the first responders are inflammatory cells like neutrophils, macrophages, and T-cells [19–22]. Importantly, many of these non-myocardial tissues have been formally examined for their requirements during regeneration, by genetic cell ablation or genetic inactivation of a key regulator. For instance, induced genetic depletion of the epicardium and its derivatives disrupts CM proliferation and muscle regeneration, after which the process recovers as the epicardium itself regenerates from survivors [23]. Blocking rapid vascularization of the injury area by inhibiting angiogenic communication, or blocking vascularization that occurs with later cardiogenesis, also impacts muscle regeneration [21, 24]. Recently, the lympathic system has been implicated in clearance of collagen and fibrin during cardiac repair [25, 26]. Genetic ablation of regulatory T-cells, which circulate and home to the injury site, impairs heart regeneration and other examples of tissue regeneration in zebrafish [27]. As a final example, Mahmoud and O’Meara et al. found that the presence of nerves is a positive factor for cardiomyocyte proliferation and heart regeneration in both zebrafish and neonatal murine contexts [28].
Figure 1. Schematic representation of the zebrafish heart and key cell types.
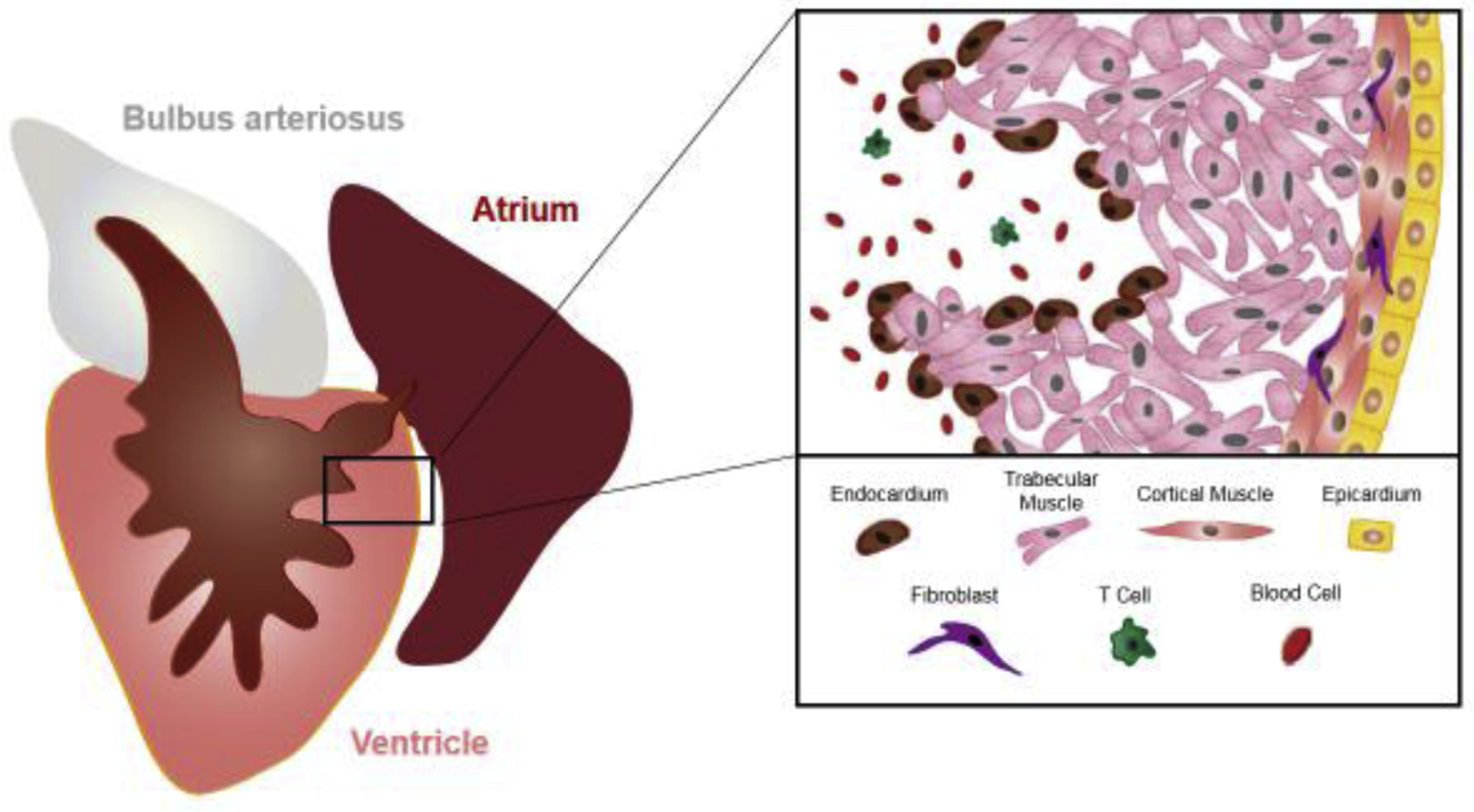
The zebrafish ventricle retains a heavily trabeculated anatomy, with a thin outer wall of cortical muscle. Several cardiac cell types and their general location within the heart are shown. “All cardiac cell types (e.g. vascular tissue, nerves) are not depicted in this simplified cartoon”.
These cell populations collectively have the potential to influence regeneration in many ways. This could be as a structural scaffolding: a heart with a injury ablating half of all CMs will regenerate much more quickly than a heart injured by resection or cryoinjury where only 20–25% of tissue is lost, most likely due to the spared architecture of the non-muscle linings [29, 30]. Also, likely to be key are the classic functions of the various cells, e.g. extracellular matrix deposition and vascular support for epicardial-derived cells [31–35], nutrient provision by vascular tissue, or debris clearance by macrophages and neutrophils [19, 36]. Finally, these cells can directly act as a source of signals conducive to CM proliferation.
Instructive cardiac mitogens
A simple mechanism for muscle regeneration would be this: injury-induced release of a potent signal promotes CM proliferation. In theory, one should be able to experimentally uncouple the mitogenic signal from the injury, evidence for which would be that overexpression of a factor(s) on its own induces CM proliferation when introduced to uninjured, adult cardiac tissue. Such factors can be referred to as “instructive” in this regard, following the language of developmental biology. In recent years, three diffusible or secreted factors have been found to provoke CM proliferation when overexpressed in the absence of injury. One is the extracellular factor extracellular factor Neuregulin 1 (Nrg1), which binds to the receptor tyrosine kinase ErbB4 and induces heterodimerization with ErbB2 [37]. Based on an earlier study implicating Nrg1 as a stimulant for CM proliferation in mice [38], Gemberling et al. found that induced overexpression of Nrg1 in adult zebrafish sharply increases CM proliferation after injury [39], ultimately causing cardiomegaly through hyperplasia. Blocking ErbB2 function pharmacologically with the drug AG1478 reduced indicators of CM proliferation after injury. Key questions remain from this study, most notably which specific ligands and receptors from the array of possibilities actually participate, given that no specific genetic mutations were employed. Nrg1 has been reported to be synthesized in epicardial cells as well as in T-regulatory cells [27, 39].
Vascular endothelial growth factor a (Vegfa) is a well-known inducer of endothelial cell proliferation, and angiogenesis logically tracks tissue regeneration [21, 24]. An initial study by Marin-Juez et al. reported that induced expression of an inhibitory form of Vegfa can block regenerative angiogenesis [24]. A later study found that induced cardiac expression of vegfaa hypervascularized the adult heart, but also led, unexpectedly, to CM hyperplasia and thickening of the muscular wall in the absence of injury [40]. As had been observed with ectopic expression of Nrg1, dedifferentiation programs such as expression of the cardiogenic transcription factor GATA binding protein 4 (gata4) were activated. Upon injury, vegfaa-overexpression in cardiomyocytes impaired cardiac repair at the injury site, even in the presence of ectopic growth away from the wound [40]. This suggested that the location and/or amount of signal impacts the response. Moving forward, it will be critical to define how the signaling pathway leading to cardiogenesis differs from that leading to angiogenesis, and whether, for instance, the hyperplastic effect in zebrafish is analogous to the hypertrophic effect that has been assigned to Vegf ligands when delivered to cultured mammalian CMs [41].
Vitamin D is a third instructive signal for heart regeneration, having been initially implicated to promote CM division by a Fluorescence Ubiquitin Cell Cycle Indicator (FUCCI)-based in vivo screen in transgenic zebrafish embryos treated with FDA-approved drugs [42]. Treatment with the vitamin D analog Alfacalcidol was sufficient to sharply boost in vivo CM proliferation in embryonic and adult CMs, and induced expression of an activated vitamin D receptor led to profound cardiomegaly in juvenile animals. A dominant-negative vitamin D receptor blocked heart regeneration when it was experimentally induced in adult CMs [42]. Interestingly, vitamin D treatment elevated the cardiac gene expression of many factors associated with ErbB2 signaling; plus, its effects on CM proliferation were disrupted by treatment with the pharmacological ErbB2 inhibitor AG1478. Vitamin D signaling, though heavily studied, is a complex pathway, and it remains unclear how signals might be regulated through processing enzymes or downstream mediators to guide regeneration. Moreover, vitamin D has been a target of several clinical research trials to define effects, if any, on cardiovascular disease, without clear evidence of specific benefits [43, 44].
These instructive factors each represent a basis for the important idea that a single factor on its own, whether an encoded protein or a hydrophobic drug, can drive the crucial event in heart regeneration. Each has the ability to (directly or indirectly) coincidentally increase proliferation indices or other biological responses in multiple cardiac cell types, thereby enabling coordinated growth of key tissues for the organ. In zebrafish, it will be essential to compare and contrast the effects these instructive factors have on molecular machinery, and to learn how these highly potent developmental factors are regulated – individually, and in shared networks - for the function of innate heart regeneration.
Permissive cardiogenic influences
Many ligands, receptors, and transcription factors have been shown to be required for zebrafish heart regeneration, although these molecules on their own have not been shown to promote CM proliferation in the absence of injury. Such mitogenic influences can be considered “permissive”, to contrast their effects from the features of instructive CM mitogens.
Intrinsic factors acting in CMs
Several transcription factors that regulate cardiac development are re-expressed after injury (Fig. 2). Regulatory sequences of the embryonic cardiogenesis genes gata4, nk2 homeobox 2.5 (nkx2.5), hand2, t-box 5 (tbx5), and t-box 10 (tbx20) are activated in CMs (and in some cases other cardiac cell types) upon injury [9, 10, 45, 46]. Induced expression of a dominant-negative Gata4 in CMs impaired proliferation and heart regeneration [46], while hand2 augmentation could increase CM proliferation after injury [45]. The others have yet to be interrogated functionally. A handful of reliable Cre-based transgenic strains are available for inducible recombination of floxed alleles in zebrafish CMs; however, there is a dire paucity of strains with loxP sequence-flanked gene sequences. Conditional gene deletion is a methodology that the field must advance in the coming years.
Figure 2. Schematic of pathways implicated in zebrafish heart regeneration.
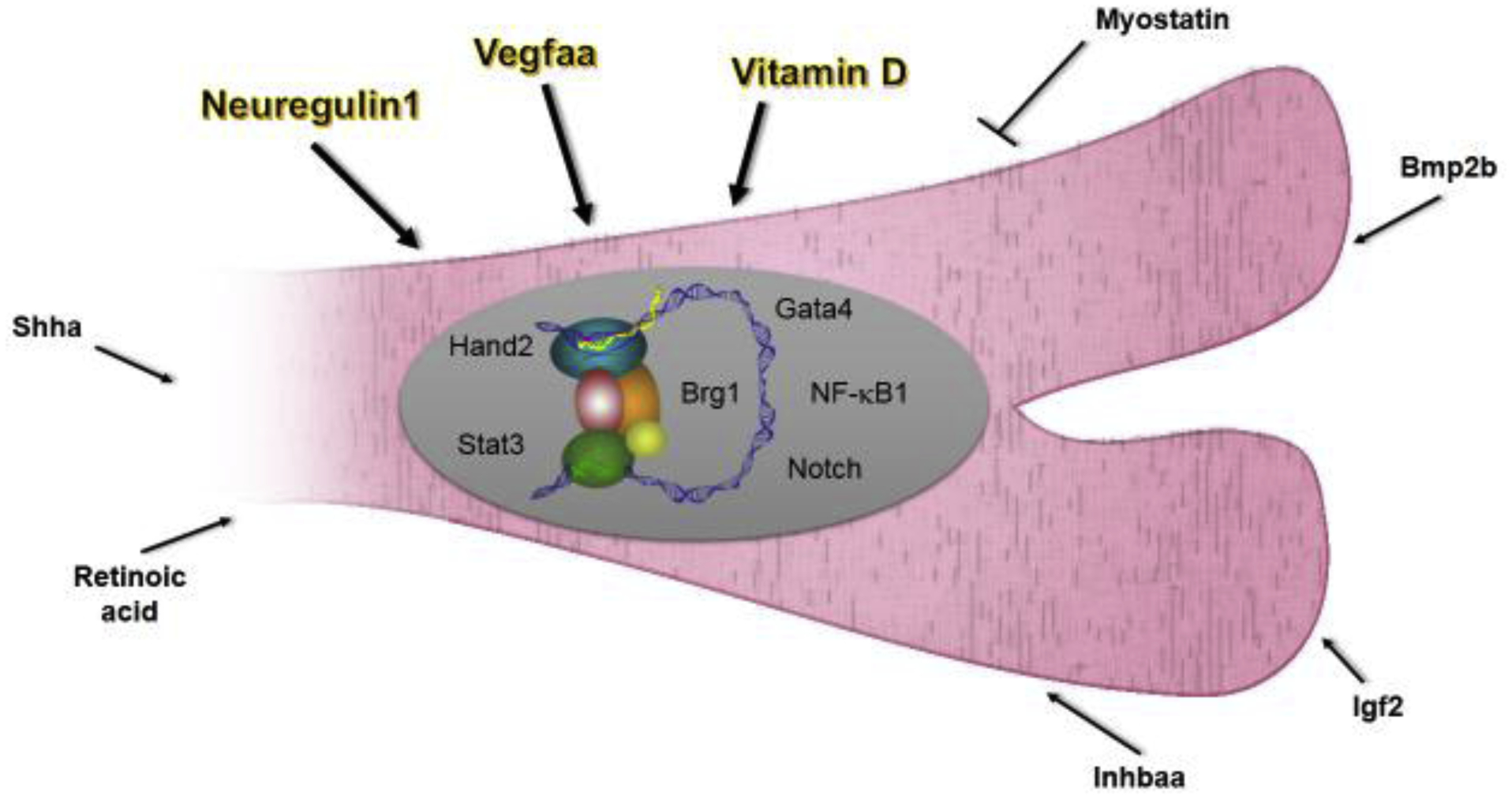
Pathways described here are shown as either intrinsic or extrinsic effectors. Instructive factors are highlighted and have bolded arrows. Putative cellular sources of extrinsic factors: Retinoic acid - endocardium, epicardium; Shha - epicardium; Nrg1 - epicardium, T-cells; Vegfaa – endocardium, epicardium; Bmp2b - epicardium; Igf2 - epicardium, endocardium.
Jak/Stat3 signaling within cells can be activated by dozens of ligands including Interleukin 11a (Il11a), Interleukin 11b (il11b), and Leukemia inhibitory factor (Lif). Transcription of the il11a ligand gene, the co-receptor interleukin 6 signal transducer (Il6st), the feedback regulator suppressor of cytokine signaling 3b (socs3b), and the transcription factor mediator signal transducer and activator of transcription 3 (stat3) are induced after cardiac injury, and induced expression of a dominant-negative Stat3 cassette in CMs blocked regeneration [47]. Stat3 ostensibly acts at least in part by regulating the secreted protein Relaxin 3a (Rln3a), a known transcriptional target of Stat3. The authors reported that rln3a is upregulated upon cardiac injury, and that systemic delivery of human recombinant RLN3 increased CM proliferation after injury [47]. Another pathway activated in CMs during regeneration involves the NF-κB transcription factor complex, which, like Jak/Stat signaling, is also essential for the mammalian hypertrophy response [48, 49]. In the presence of a dominant-negative IκBSR, which retains NfκB transcription factors in the cytoplasm, disassembly of sarcomeres, proliferation and induction of gata4 regulatory sequences were each disrupted after injury [50]. These inhibitory effects could not be rescued by gata4 overexpression, which implies that the downstream network of NfκB is complex; indeed, the initiating ligand has not been elucidated.
In addition to transcription factors themselves, epigenetic regulation, like chromatin remodeling or histone modification, have been implicated in zebrafish heart regeneration [51]. Transgenic myocardial inhibition of Brahma-related gene-1 (Brg1), a component of the ATP-dependent chromatin remodeling complex SWI/SNF, inhibited injury-induced CM proliferation, potentially due to increased expression of cyclin-dependent kinase inhibitors cdkn1a and cdkn1c [51]. RNAseq and ChiP-seq screens using purified gata4-expressing CMs revealed that Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit (Ezh2), a component of the polycomb repressor complex 2, suppresses expression of structural genes involved in sarcomere formation by H3K27-tri-methlation [52]. When a transgenic histone H3 with a mutated methylation site was ectopically expressed, CMs at the wound site retained a mature state – with intact sarcomere structure and reduced expression of embryonic cardiac myosin heavy chain. This study combines with others that have identified broad changes in histone regulation during zebrafish heart regeneration [53, 54]. The initial signals that trigger large-scale and local changes in chromatin structure during heart regeneration await elucidation.
Extrinsic factors from neighboring cells
An injured zebrafish heart engages a host of secreted signals from neighboring non-myocardial cells (Fig. 2). Retinoic acid, its production controlled by the enzyme Retinaldehyde dehydrogenase 2 (Raldh2), is synthesized by the epicardium and endocardium within hours of injury [55]. Broad transgenic inhibition of retinoic acid receptors (RARs) impairs CM proliferation, although requirements for RARs have yet to be attributed to a specific cell type. Signaling by release of the membrane-bound transcription factor Notch has also been implicated in communication between the myocardium and epicardium or endocardium [56, 57]. Although Notch is dispensable for activation of injury markers in endocardial cells, blocking Notch signaling via the Notch inhibitor Dominant negative mastermind-like (DN-MAML) in endothelial cells (including the endocardium) was reported to decrease CM proliferation [57]. Transcriptome analysis of injured DN-MAML-expressing hearts identified Wnt antagonists Wnt inhibitory factor 1 (Wif1) and Notum1b as likely Notch targets. Notably, pharmacological suppression of Wnt by the small-molecule antagonist IWR-1-endo partially rescued the phenotype caused by conditional loss of Notch, and Wnt inhibition at injury sites was proposed to be required for normal CM proliferation [57].
Myostatin and activin Inhibin subunit beta aa (Inhbaa) are TGFβ ligands that are antagonistically regulated during tissue repair [58, 59]. While myostatin expression is reduced in the ventricular wall, inhbaa is upregulated in cells at the wound after cryoinjury [58]. Interestingly, either transgenic myostatin overexpression (OE) or inhbaa knock-out resulted in decreased CM proliferation, whereas knock-out of myostatin or inhbaa-OE caused hyperplasia and hypertrabeculation with late stage pericardial edema, respectively. The authors reported that Myostatin and Inhbaa bind to two distinct Activin receptors respectively, and these in turn lead to the activation of distinct Smads [58]. inhbaa-OE caused an increase in CM proliferation independently of Nrg1-Erbb2-signaling; thus, it would be interesting to determine how heart regeneration is impacted by coincident overexpression of both inhbaa and Nrg1. Wu et al investigated potential roles for Bone morphogenetic protein (BMP) signaling during regeneration, after observations that bone morphogenetic protein 2b (bmp2b) RNA levels increased in the wound border zone and epicardium after cryoinjury [60]. Induced global overexpression of bmp2b decreased the wound size, while overexpression of BMP-inhibitor noggin3 delayed muscle repair. Transgenic bmp2b increased CM proliferation only slightly in injured hearts, and had no effect on CM proliferation in uninjured hearts; however, Noggin inhibition of BMP signaling limited CM de-differentiation and cell cycle entry. Signaling pathways like Insulin-like growth factor (Igf) and Sonic Hedgehog (Shh) have also been implicated in CM proliferation by various loss-of-function studies, with the epicardium in each case reported as a key ligand source [61, 62]. Notably Sugimoto et al. used inducible Cre-based techniques to disrupt the shha gene specifically in epicardial and epicardial-derived cells, providing elegant genetic evidence that this tissue is a significant source of Hedgehog ligand during heart regeneration [63].
Outlook
It is now evident that experimental disruption of many candidate factors individually can have no apparent effect on heart regeneration – the authors know this firsthand, and such results do not make their way easily into publications. Genetic redundancy is a generally fascinating but still confounding issue, and mechanisms of compensation are likely in play with a subset of these factors [64]. Evolution of the genetic toolset for adult zebrafish can address this, as well as the challenges of tying results like those summarized in this review into coherent regulatory networks. When groups use the same injury models and methods of genetic manipulation, it is more straightforward for them to reproduce findings and perform tests of epistasis.
We emphasize that a key area to pursue more deeply is how signals, especially potent, instructive signals, are induced and restricted at the level of chromatin structure, gene regulatory elements, and transcription factor-DNA complexes. This is reviewed more extensively elsewhere [65]. As a method to identify signals, high-resolution proteomes of heart regeneration would be of great interest, but defining them effectively has challenges, such as the limited amount of tissue and dominance of profiles by contractile and mitochondrial proteins. Recently developed technologies might help unveil the regeneration proteome at new detail, including large-scale assessment of protein modification and protein-protein interaction dynamics [66–70]. Before a clear equation for regeneration is derived, it in fact is likely that sufficient nuggets will have been mined from the study of heart regeneration in laboratory models to already initiate effective therapies. The discovery of instructive influences in zebrafish, and recent similar discoveries in mice, suggests that such interventions employing potent triggers are on the horizon [14, 71–74].
Acknowledgements
We thank Nutishia Lee for artwork, and Juan Manuel Gonzales-Rosa for comments on the manuscript. M.I.P is funded by the Life Science Research Foundation (LSRF) – Fellowship – sponsored by Astellas Pharma. K.D.P. acknowledges support from the American Heart Association, Foundation Leducq, and NIH (R01 HL081674, R01 HL131319, and R01 HL136182).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
- 1.Bambino K, and Chu J (2017). Zebrafish in Toxicology and Environmental Health. Curr Top Dev Biol 124, 331–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saleem S, and Kannan RR (2018). Zebrafish: an emerging real-time model system to study Alzheimer’s disease and neurospecific drug discovery. Cell Death Discov 4, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lele Z, and Krone PH (1996). The zebrafish as a model system in developmental, toxicological and transgenic research. Biotechnol Adv 14, 57–72. [DOI] [PubMed] [Google Scholar]
- 4.Broussonet M (1786). Observations sur la régénérations de quelques parties du corps des poissons. Hist. d. l’Acad. Roy. des Sciences [Google Scholar]
- 5.Pfefferli C, and Jazwinska A (2015). The art of fin regeneration in zebrafish. Regeneration (Oxf) 2, 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gemberling M, Bailey TJ, Hyde DR, and Poss KD (2013). The zebrafish as a model for complex tissue regeneration. Trends Genet 29, 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marques IJ, Lupi E, and Mercader N (2019). Model systems for regeneration: zebrafish. Development 146. [DOI] [PubMed] [Google Scholar]
- 8.Poss KD, Wilson LG, and Keating MT (2002). Heart regeneration in zebrafish. Science 298, 2188–2190. [DOI] [PubMed] [Google Scholar]
- 9.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, and Poss KD (2006). A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607–619. [DOI] [PubMed] [Google Scholar]
- 10.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, and Poss KD (2010). Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464, 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jopling C, Sleep E, Raya M, Marti M, Raya A, and Izpisua Belmonte JC (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doppler SA, Deutsch MA, Serpooshan V, Li G, Dzilic E, Lange R, Krane M, and Wu SM (2017). Mammalian Heart Regeneration: The Race to the Finish Line. Circ Res 120, 630–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzahor E, and Poss KD (2017). Cardiac regeneration strategies: Staying young at heart. Science 356, 1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirose K, Payumo AY, Cutie S, Hoang A, Zhang H, Guyot R, Lunn D, Bigley RB, Yu H, Wang J, et al. (2019). Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science 364, 184–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **15.Gonzalez-Rosa JM, Sharpe M, Field D, Soonpaa MH, Field LJ, Burns CE, and Burns CG (2018). Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish. Dev Cell 44, 433–446 e437. [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence that experimentally induced polyploidy can inhibit cardiomyocyte proliferation and heart regeneration.
- 16.Alkass K, Panula J, Westman M, Wu TD, Guerquin-Kern JL, and Bergmann O (2015). No Evidence for Cardiomyocyte Number Expansion in Preadolescent Mice. Cell 163, 1026–1036. [DOI] [PubMed] [Google Scholar]
- 17.Patterson M, Barske L, Van Handel B, Rau CD, Gan P, Sharma A, Parikh S, Denholtz M, Huang Y, Yamaguchi Y, et al. (2017). Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat Genet 49, 1346–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honkoop H, de Bakker DE, Aharonov A, Kruse F, Shakked A, Nguyen PD, de Heus C, Garric L, Muraro MJ, Shoffner A, et al. (2019). Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang WC, Yang CC, Chen IH, Liu YM, Chang SJ, and Chuang YJ (2013). Treatment of Glucocorticoids Inhibited Early Immune Responses and Impaired Cardiac Repair in Adult Zebrafish. PLoS One 8, e66613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Preux Charles AS, Bise T, Baier F, Marro J, and Jazwinska A (2016). Distinct effects of inflammation on preconditioning and regeneration of the adult zebrafish heart. Open Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison MR, Bussmann J, Huang Y, Zhao L, Osorio A, Burns CG, Burns CE, Sucov HM, Siekmann AF, and Lien CL (2015). Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Developmental cell 33, 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itou J, Oishi I, Kawakami H, Glass TJ, Richter J, Johnson A, Lund TC, and Kawakami Y (2012). Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development 139, 4133–4142. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Cao J, Dickson AL, and Poss KD (2015). Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 522, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marin-Juez R, Marass M, Gauvrit S, Rossi A, Lai SL, Materna SC, Black BL, and Stainier DY (2016). Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc Natl Acad Sci U S A 113, 11237–11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gancz D, Raftrey BC, Perlmoter G, Marin-Juez R, Semo J, Matsuoka RL, Karra R, Raviv H, Moshe N, Addadi Y, et al. (2019). Distinct origins and molecular mechanisms contribute to lymphatic formation during cardiac growth and regeneration. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison MR, Feng X, Mo G, Aguayo A, Villafuerte J, Yoshida T, Pearson CA, Schulte-Merker S, and Lien CL (2019). Late developing cardiac lymphatic vasculature supports adult zebrafish heart function and regeneration. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *27.Hui SP, Sheng DZ, Sugimoto K, Gonzalez-Rajal A, Nakagawa S, Hesselson D, and Kikuchi K (2017). Zebrafish Regulatory T Cells Mediate Organ-Specific Regenerative Programs. Dev Cell 43, 659–672 e655. [DOI] [PubMed] [Google Scholar]; T regulatory cells play a key role in zebrafish heart regeneration, at least in part through the production of context-specific factors.
- 28.Mahmoud AI, O’Meara CC, Gemberling M, Zhao L, Bryant DM, Zheng R, Gannon JB, Cai L, Choi WY, Egnaczyk GF, et al. (2015). Nerves Regulate Cardiomyocyte Proliferation and Heart Regeneration. Dev Cell 34, 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, et al. (2011). The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 138, 3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chablais F, Veit J, Rainer G, and Jazwinska A (2011). The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol 11, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, and Poss KD (2011). tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 138, 2895–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore-Morris T, Guimaraes-Camboa N, Banerjee I, Zambon AC, Kisseleva T, Velayoudon A, Stallcup WB, Gu Y, Dalton ND, Cedenilla M, et al. (2014). Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J Clin Invest 124, 2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snider P, Standley KN, Wang J, Azhar M, Doetschman T, and Conway SJ (2009). Origin of cardiac fibroblasts and the role of periostin. Circ Res 105, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Puig A, Mosquera JL, Jimenez-Delgado S, Garcia-Pastor C, Jorba I, Navajas D, Canals F, and Raya A (2019). Proteomics analysis of extracellular matrix remodeling during zebrafish heart regeneration. Mol Cell Proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez-Iranzo H, Galardi-Castilla M, Sanz-Morejon A, Gonzalez-Rosa JM, Costa R, Ernst A, Sainz de Aja J, Langa X, and Mercader N (2018). Transient fibrosis resolves via fibroblast inactivation in the regenerating zebrafish heart. Proc Natl Acad Sci U S A 115, 4188–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans MA, Smart N, Dube KN, Bollini S, Clark JE, Evans HG, Taams LS, Richardson R, Levesque M, Martin P, et al. (2013). Thymosin beta4-sulfoxide attenuates inflammatory cell infiltration and promotes cardiac wound healing. Nat Commun 4, 2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wadugu B, and Kuhn B (2012). The role of neuregulin/ErbB2/ErbB4 signaling in the heart with special focus on effects on cardiomyocyte proliferation. Am J Physiol Heart Circ Physiol 302, H2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bersell K, Arab S, Haring B, and Kuhn B (2009). Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138, 257–270. [DOI] [PubMed] [Google Scholar]
- 39.Gemberling M, Karra R, Dickson AL, and Poss KD (2015). Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife 4 **Nrg1 overexpression leads to overt cardiomyocyte proliferation in the absence of injury – the first instructive cardiomyocyte mitogen in adult zebrafish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *40.Karra R, Foglia MJ, Choi WY, Belliveau C, DeBenedittis P, and Poss KD (2018). Vegfaa instructs cardiac muscle hyperplasia in adult zebrafish. Proc Natl Acad Sci U S A 115, 8805–8810. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vegfaa is shown to have mitogenic effects on adult zebrafish cardiomyocytes.
- 41.Zentilin L, Puligadda U, Lionetti V, Zacchigna S, Collesi C, Pattarini L, Ruozi G, Camporesi S, Sinagra G, Pepe M, et al. (2010). Cardiomyocyte VEGFR-1 activation by VEGF-B induces compensatory hypertrophy and preserves cardiac function after myocardial infarction. FASEB J 24, 1467–1478. [DOI] [PubMed] [Google Scholar]
- **42.Han Y, Chen A, Umansky KB, Oonk KA, Choi WY, Dickson AL, Ou J, Cigliola V, Yifa O, Cao J, et al. (2019). Vitamin D Stimulates Cardiomyocyte Proliferation and Controls Organ Size and Regeneration in Zebrafish. Dev Cell 48, 853–863 e855. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vitamin D has mitogenic effects on cardiomyocytes at many different contexts and stages.
- 43.Pilz S, Verheyen N, Grubler MR, Tomaschitz A, and Marz W (2016). Vitamin D and cardiovascular disease prevention. Nat Rev Cardiol 13, 404–417. [DOI] [PubMed] [Google Scholar]
- 44.Tebben PJ, Singh RJ, and Kumar R (2016). Vitamin D-Mediated Hypercalcemia: Mechanisms, Diagnosis, and Treatment. Endocr Rev 37, 521–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schindler YL, Garske KM, Wang J, Firulli BA, Firulli AB, Poss KD, and Yelon D (2014). Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Development 141, 3112–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta V, Gemberling M, Karra R, Rosenfeld GE, Evans T, and Poss KD (2013). An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr Biol 23, 1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang Y, Gupta V, Karra R, Holdway JE, Kikuchi K, and Poss KD (2013). Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc Natl Acad Sci U S A 110, 13416–13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purcell NH, Tang G, Yu C, Mercurio F, DiDonato JA, and Lin A (2001). Activation of NF-kappa B is required for hypertrophic growth of primary rat neonatal ventricular cardiomyocytes. Proc Natl Acad Sci U S A 98, 6668–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freund C, Schmidt-Ullrich R, Baurand A, Dunger S, Schneider W, Loser P, El-Jamali A, Dietz R, Scheidereit C, and Bergmann MW (2005). Requirement of nuclear factor-kappaB in angiotensin II- and isoproterenol-induced cardiac hypertrophy in vivo. Circulation 111, 2319–2325. [DOI] [PubMed] [Google Scholar]
- 50.Karra R, Knecht AK, Kikuchi K, and Poss KD (2015). Myocardial NF-kappaB activation is essential for zebrafish heart regeneration. Proc Natl Acad Sci U S A 112, 13255–13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *51.Xiao C, Gao L, Hou Y, Xu C, Chang N, Wang F, Hu K, He A, Luo Y, Wang J, et al. (2016). Chromatin-remodelling factor Brg1 regulates myocardial proliferation and regeneration in zebrafish. Nat Commun 7, 13787. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chromatin structure as regulated by Brg1 can influence heart regeneration in zebrafish
- 52.Ben-Yair R, Butty VL, Busby M, Qiu Y, Levine SS, Goren A, Boyer LA, Burns CG, and Burns CE (2019). H3K27me3-mediated silencing of structural genes is required for zebrafish heart regeneration. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, Gemberling M, Goldman JA, Black BL, and Poss KD (2016). Modulation of tissue repair by regeneration enhancer elements. Nature 532, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldman JA, Kuzu G, Lee N, Karasik J, Gemberling M, Foglia MJ, Karra R, Dickson AL, Sun F, Tolstorukov MY, et al. (2017). Resolving Heart Regeneration by Replacement Histone Profiling. Dev Cell 40, 392–404 e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, and Poss KD (2011). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell 20, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao L, Borikova AL, Ben-Yair R, Guner-Ataman B, MacRae CA, Lee RT, Burns CG, and Burns CE (2014). Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc Natl Acad Sci U S A 111, 1403–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.*.Zhao L, Ben-Yair R, Burns CE, and Burns CG (2019). Endocardial Notch Signaling Promotes Cardiomyocyte Proliferation in the Regenerating Zebrafish Heart through Wnt Pathway Antagonism. Cell Rep 26, 546–554 e545. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tissue specific knock down of Notch signaling in the endocardium impacts zebrafish heart regeneration.
- 58.Dogra D, Ahuja S, Kim HT, Rasouli SJ, Stainier DYR, and Reischauer S (2017). Opposite effects of Activin type 2 receptor ligands on cardiomyocyte proliferation during development and repair. Nat Commun 8, 1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chablais F, and Jazwinska A (2012). The regenerative capacity of the zebrafish heart is dependent on TGFbeta signaling. Development 139, 1921–1930. [DOI] [PubMed] [Google Scholar]
- *60.Wu CC, Kruse F, Vasudevarao MD, Junker JP, Zebrowski DC, Fischer K, Noel ES, Grun D, Berezikov E, Engel FB, et al. (2016). Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration. Dev Cell 36, 36–49. [DOI] [PubMed] [Google Scholar]; A new profiling method reveals local signaling by BMPs as important for zebrafish heart regeneration
- 61.Choi WY, Gemberling M, Wang J, Holdway JE, Shen MC, Karlstrom RO, and Poss KD (2013). In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 140, 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang Y, Harrison MR, Osorio A, Kim J, Baugh A, Duan C, Sucov HM, and Lien CL (2013). Igf Signaling is Required for Cardiomyocyte Proliferation during Zebrafish Heart Development and Regeneration. PLoS One 8, e67266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sugimoto K, Hui SP, Sheng DZ, and Kikuchi K (2017). Dissection of zebrafish shha function using site-specific targeting with a Cre-dependent genetic switch. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Gunther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai SL, et al. (2019). Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duncan EM, and Alvarado AS (2019). Regulation of Genomic Output and (Pluri)potency in Regeneration. Annu Rev Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, and Ting AY (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, and Ting AY (2015). Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods 12, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. (2006). Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643. [DOI] [PubMed] [Google Scholar]
- 69.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, and Hamon C (2003). Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75, 1895–1904. [DOI] [PubMed] [Google Scholar]
- 70.Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, and Roux KJ (2016). An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell 27, 1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mohamed TMA, Ang YS, Radzinsky E, Zhou P, Huang Y, Elfenbein A, Foley A, Magnitsky S, and Srivastava D (2018). Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell 173, 104–116 e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leach JP, Heallen T, Zhang M, Rahmani M, Morikawa Y, Hill MC, Segura A, Willerson JT, and Martin JF (2017). Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 550, 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, Kain D, Rajchman D, Leach J, Riabov Bassat D, et al. (2017). The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 547, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakada Y, Canseco DC, Thet S, Abdisalaam S, Asaithamby A, Santos CX, Shah AM, Zhang H, Faber JE, Kinter MT, et al. (2017). Hypoxia induces heart regeneration in adult mice. Nature 541, 222–227. [DOI] [PubMed] [Google Scholar]


