Abstract
Aims: To determine the suitability of murine norovirus (MNV) as a surrogate for human norovirus (HuNoV) in heat inactivation studies.
Methods and Results: MNV, hepatitis A virus (HAV) and HuNoV genogroup I and II (GI and GII) specific real‐time quantitative reverse transcription (qRT)‐PCR assays were used to determine the effects of heat exposure (63 and 72°C) for up to 10 min in water and milk. Using culture assays, MNV and HAV showed similar reductions in infectivity over time. Both HuNoV GI and GII showed lower log reductions in qRT‐PCR titre following heat exposure than either MNV or HAV. No significant protective effect of milk was observed for any virus.
Conclusions: MNV is as suitable a surrogate for HuNoV as HAV. In heat inactivation studies at 63 and 72°C, qRT‐PCR results indicate that HuNoV is less susceptible to heat than either HAV or MNV and so neither virus may be an appropriate surrogate for HuNoV.
Significance and Impact of the Study: Caution should be used when extrapolating surrogate virus data for HuNoV. Although not conclusive, our results suggest that HuNoV may be more resistant to heat than either HAV or MNV.
Keywords: heat inactivation, hepatitis A virus, human norovirus, murine norovirus, real‐time quantitative reverse transcription PCR, virus surrogate
Introduction
Human noroviruses (HuNoV), belonging to the genus Norovirus in the Caliciviridae family, are associated with person‐to‐person, food and waterborne outbreaks of gastroenteritis. Efforts to fully characterize their properties are hampered because HuNoV has yet to be grown readily in cell culture and there is no animal model (Duizer et al. 2004a; Straub et al. 2007). Because of these limitations, various surrogate viruses have been used to predict HuNoV behaviour following inactivation treatments by heat, high pressure and disinfection. Previously, feline calicivirus (FeCV) (Doultree et al. 1999; 2003a, 2003b, 2005a, 2005b; Duizer et al. 2004b; Bae and Schwab 2008; Buckow et al. 2008), canine calicivirus (CaCV) (De Roda Husman et al. 2004; Duizer et al. 2004b), hepatitis A virus (HAV) (Bidawid et al. 2000; Mariam and Cliver 2000; Kingsley et al. 2002; Hewitt and Greening 2004), poliovirus (Mariam and Cliver 2000; Strazynski et al. 2002; Bae and Schwab 2008) and bacteriophages (Mariam and Cliver 2000; Dawson et al. 2005; Bae and Schwab 2008) have been used as surrogates for HuNoV. Some of these viruses have demonstrated their unsuitability as HuNoV surrogates. For example, FeCV is readily inactivated by heat (Slomka and Appleton 1998) and loses significant viability when subjected to low pH (Hewitt and Greening 2004), in contrast to HuNoV which is resistant at pH 2·7 for 3 h at room temperature (Dolin et al.1972).
In 2004, murine norovirus (MNV), a genogroup V culturable norovirus that infects mice, was reported and characterized (Karst et al. 2003). The virus is readily cultured on the mouse dendritic cell line, RAW 264.7 and can be quantified easily using a plaque assay. Because of its taxonomic relationship to HuNoV, MNV is considered a potential surrogate for increasing knowledge of HuNoV properties and mode of replication (2004, 2006; Ward et al. 2007). Recently, MNV has been used as a surrogate for HuNoV in studies of high pressure processing (Kingsley et al. 2007; Buckow et al. 2008), UV, pH, temperature, disinfection (Cannon et al. 2006; Belliot et al. 2008; Lee et al. 2008) and in survival studies in water (Bae and Schwab 2008) and shellfish (Kingsley et al. 2007). However, it is not certain that any results obtained for MNV are truly representative of HuNoV. HAV is also regarded as a potential surrogate for HuNoV as both viruses share similar transmission pathways and are known to be environmentally stable (Carter 2005; Boone and Gerba 2007).
We determined the effects of heat on HAV, MNV and two strains of HuNoV using two common food processing temperatures (63 and 72°C) over different time periods. Inactivation experiments were carried out in both water and a protein‐rich matrix (milk) to determine whether the protein protected the viruses from the effects of heat. The results for all viruses were compared using real‐time quantitative reverse transcription PCR (qRT‐PCR) and culture where applicable.
Materials and methods
Viruses and cells
HuNoV suspensions (10% w/v) were prepared from faecal specimens collected from cases of gastroenteritis cases as previously described (Greening et al. 2001). Two HuNoV strains of known genotypes (GI.2 and GII.4) were used. HAV (cytopathic strain HM‐175) and foetal rhesus monkey kidney (FRhK‐4) cells were kindly provided by Professor M.D. Sobsey (University of North Carolina, Chapel Hill, NC, USA). MNV (MNV‐1 strain) was kindly provided by Professor H. Virgin (Washington University School of Medicine, MO, USA) and RAW 264.7 cells from the Department of Microbiology and Immunology, University of Otago. Virus titres were determined by real‐time qRT‐PCR titration of 10‐fold dilutions of each virus stock to the respective endpoint, which was designated as 1 RT‐PCR unit (RT‐PCRU). The titres of HuNoV GI and GII were 2 × 108 RT‐PCRU ml−1, and titres of HAV and MNV were 108 RT‐PCRU ml−1. MNV (plaque forming units, PFU) and HAV (infectious units, IU) titres were determined by culture assays as described below.
Heat inactivation
Water or milk was seeded with stock viruses (HuNoV GI, HuNoV GII, HAV and MNV), diluted 1/10 (1/200 for HuNoV) in cell growth medium (GM). Dilution of viruses was necessary to give the required concentrations and to dilute any possible effects from the different matrices (faecal material for HuNoV and cell culture medium for MNV and HAV). Final concentrations were 107 RT‐PCRU ml−1 (3 × 105 PFU ml−1) for MNV, 107 RT‐PCRU ml−1 (5 × 105 IU ml−1) for HM175 HAV, and 5 × 106 RT‐PCRU ml−1 for both HuNoV GI and GII.
Samples were dispensed in 100 μl volumes in to 0·2 ml PCR tubes and heated at 63 or 72°C for 0, 1, 2, 5 and 10 min. A PCR machine (GeneAmp PCR System 9700) was used as a heating block to ensure good temperature transfer. At each time point, samples were removed from the heating block and immediately cooled on ice. Each experiment was performed in triplicate.
Culture assays for MNV and HAV
For MNV, RAW 264.7 cells were cultured in GM consisting of minimum essential medium (MEM; Gibco®; Invitrogen Corp, Carlsbad, CA, USA) containing 10% (v/v) low endotoxin foetal bovine serum (FCS) and 100 units penicillin G sulphate and 100 μg ml−1 streptomycin sulphate (PS; Gibco). RAW 264.7 cells were plated out in 6‐well plates (3·5 cm diameter) at a density of 3 × 106 cells per well in 3 ml of GM and incubated at 37°C with 5% CO2 for 24 h. Following heat treatment, all samples were diluted in GM to give the desired MNV PFU number (10–50 PFU per well) and the volume made up to 500 μl per well with GM prior to inoculation on the confluent cells. Plates were incubated at 37°C for 1 h in a shaking incubator (60 rev min−1). The inoculum was then removed and 2 ml of 2% (w/v) low melting point agarose (Lonza, Rockland, ME, USA) in 2× complete MEM media added to each well and incubated at 37°C with 5% CO2. After 48 h, a second agar overlay with 1% (w/v) neutral red (BDH, Poole, UK) was added to each well and plaques counted after 6 h. Plates with between 10–50 PFU per well were used to determine the virus titres. Results were expressed as PFU ml−1. The amount of undiluted inoculums (water and milk) that could be added to the cells without toxicity occurring was also determined.
For HAV, 104 FRhK‐4 cells (100 μl) per well were plated in a 96 microtitre well plate in 2% (v/v) FCS in MEM (Gibco) supplemented with 0·1 mmol l−1 nonessential amino acids (Gibco) and PS (Gibco). This was followed immediately by the addition of serial dilutions of virus (100 μl) per well, with at least eight replicates per dilution. Plates were incubated for up to 21 days at 37°C in a humidified 5% CO2 atmosphere and examined for cytopathic effects (CPE) characteristic of HAV. Results were expressed as infectious units (IU) ml−1. For MNV and HAV, each experimental replicate (n = 3) was assayed in duplicate.
Real‐time qRT‐PCR assays
Samples (200 μl) were extracted using the Roche High Pure Viral Nucleic Acid Kit (Roche Molecular Biochemicals Ltd, Mannheim, Germany) and eluted in 50 μl elution buffer as per manufacturer’s instructions. For all viruses, one‐step real‐time RT‐PCR assays using the Platinum Quantitative RT‐PCR ThermoScript One Step System (Invitrogen) were performed using primers and probes specific for each virus. The primers for HAV and HuNoV have been previously described (Kageyama et al. 2003; Hewitt and Greening 2004). MNV specific primers and probes were designed using Genscript primer design software (http://www.genscript.com) (Table 1).
Table 1.
Primers and probe used for the detection of murine norovirus (MNV) by real‐time qRT‐PCR
| Name | Sequence (5′–3′) | Location |
|---|---|---|
| MNV probe* | CCTTCCCGACCGATGGCATC | 6578–6597 |
| MNV for primer | TGCAAGCTCTACAACGAAGG | 6520–6539 |
| MNV rev primer | CACAGAGGCCAATTGGTAAA | 6626–6645 |
*Probe is FAM‐BHQ1 labelled at the 5′ and 3′ ends respectively.
Real‐time qRT‐PCR assays were carried out using the Rotor‐Gene™ 3000 or 6000 real‐time rotary analyzer (Corbett Research Ltd, Sydney, Australia). Raw data were analysed using Rotor‐Gene™ software. Data were transformed to RT‐PCRU using a standard curve generated from serial dilutions of virus stock preparations. For HuNoV GI/GII, MNV and HAV, each experimental replicate (n = 3) was assayed in duplicate as for the culture assays.
Data analysis
The average virus titre from the duplicate assays for each experiment was determined, then the average from the three replicate experiments calculated. Log10 reductions, −log10 N t/N 0 ± SD, were determined, where N t is the virus titre detected at each time (t) point and N 0 is the initial titre. Statistical significance of the log reductions of qRT‐PCR titres for each virus was determined for each condition using t‐test analysis (Microsoft Excel; Microsoft, Redmond, WA, USA). Results with P‐values of <0·05 were considered significant.
The time required to reduce the virus titres by 1 log (D‐values) was determined by regression analysis (Excel) and calculated as the negative reciprocal of the slope of log titres (culture) of each virus following each treatment vs time. The D‐value calculations assume that inactivation was constant for each virus and matrix in the first minute.
Results
The maximum detectable reduction in infectivity was 3·5 logs for both MNV and HAV. Similar log decreases in infectivity and D‐values were observed for MNV and HAV in both milk and water (2, 3). The D‐values ranged from 0·6–1·1 min at 63°C and ≤0·5 min at 72°C, which represents a 3‐log reduction over the first minute (Table 2).
Table 2.
Inactivation times (D‐values) of murine norovirus (MNV), and hepatitis A virus (HAV) in water and milk at 63 and 72°C
| Matrix | Temperature | D‐value (min) | |
|---|---|---|---|
| MNV | HAV | ||
| Water | 63°C | 0·9 | 0·6 |
| 72°C | ≤0·3 | ≤0·3 | |
| Milk | 63°C | 0·7 | 1·1 |
| 72°C | 0·5 | ≤0·3 | |
Table 3.
Comparison of log reductions (±SD) in infectivity titres and RT‐PCR titres for murine norovirus (MNV), and hepatitis A virus (HAV) in water and milk at 63 and 72°C
| Time (min) | 63°C | 72°C | ||||||
|---|---|---|---|---|---|---|---|---|
| Log reduction in infectivity | Log reduction in RT‐PCR titre | Log reduction in infectivity | Log reduction in RT‐PCR titre | |||||
| MNV | HAV | MNV | HAV | MNV | HAV | MNV | HAV | |
| (a) Water | ||||||||
| 0 | 0·00 ± 0·04 | 0·00 ± 0·05 | 0·00 ± 0·13 | 0·00 ± 0·25 | 0·00 ± 0·01 | 0·00 ± 0·20 | 0·00 ± 0·21 | 0·00 ± 0·02 |
| 1 | 1·09 ± 0·04 | 1·26 ± 0·19 | −0·01 ± 0·06 | 0·39 ± 0·04 | ≥3·5 | ≥3·5 | 0·21 ± 0·06 | 0·46 ± 0·02 |
| 2 | 3·43 ± 0·30 | 2·34 ± 0·22 | 0·18 ± 0·10 | 0·49 ± 0·15 | ≥3·5 | ≥3·5 | 0·44 ± 0·20 | 0·57 ± 0·03 |
| 5 | 3·13 ± 0·78 | ≥3·5 | 0·41 ± 0·05 | 0·45 ± 0·02 | ≥3·5 | ≥3·5 | 0·88 ± 0·04 | 0·67 ± 0·01 |
| 10 | 3·28 ± 0·04 | ≥3·5 | 0·99 ± 0·11 | 0·50 ± 0·03 | ≥3·5 | ≥3·5 | 1·02 ± 0·11 | 1·00 ± 0·09 |
| (b) Milk | ||||||||
| 0 | 0·00 ± 0·02 | 0·00 ± 0·19 | 0·00 ± 0·03 | 0·00 ± 0·02 | 0·00 ± 0·02 | 0·00 ± 0·42 | 0·00 ± 0·04 | 0·00 ± 0·10 |
| 1 | 1·35 ± 0·34 | 1·60 ± 0·39 | 0·47 ± 0·15 | 0·54 ± 0·09 | ≥3·5 | 2·22 ± 0·06 | 0·35 ± 0·20 | 0·47 ± 0·05 |
| 2 | 1·85 ± 0·27 | 2·72 ± 0·03 | 0·52 ± 0·12 | 0·60 ± 0·06 | ≥3·5 | ≥3·5 | 0·67 ± 0·04 | 0·50 ± 0·05 |
| 5 | ≥3·5 | 3·35 ± 0·20 | 0·38 ± 0·07 | 0·65 ± 0·04 | ≥3·5 | ≥3·5 | 0·82 ± 0·06 | 0·57 ± 0·03 |
| 10 | ≥3·5 | ≥3·5 | 1·03 ± 0·07 | 0·65 ± 0·06 | ≥3·5 | ≥3·5 | 0·91 ± 0·02 | 0·72 ± 0·02 |
At 63°C for 1 min, 1·09 ± 0·04 and 1·35 ± 0·34‐log reductions in infectivity were observed for MNV in water and milk respectively, and 1·26 ± 0·19 and 1·60 ± 0·39‐log reductions for HAV (Table 3). These log reductions were not significantly different between HAV and MNV or between water and milk (P > 0·05). After 5 and 10 min exposure at 63°C, both MNV and HAV showed reductions in infectivity titre of more than 3·0 logs.
At 72°C, a rapid decrease in infectivity of MNV and HAV in both matrices with increased time was observed (Table 3). There was at least a 3·5‐log reduction in infectivity observed after 2 min at 72°C for both viruses in each matrix (Table 2).
The qRT‐PCR titre decreased for all viruses over time in both matrices at 63 and 72°C, although the maximum decrease for any virus was 1·0 log, observed for both MNV and HAV in water heated to 72°C for 10 min. For MNV and HAV, the log reductions in qRT‐PCR titre observed were much lower than those observed by culture, particularly at 72°C where the maximum measurable 3·5‐log reduction in infectivity titre was observed for both viruses (Table 3). By comparison, a maximum 1·0‐log reduction in qRT‐PCR titre was observed for both HAV and MNV at 72°C. HuNoV GI and GII strains showed lower reductions in qRT‐PCR titre over time compared with MNV and HAV for both temperatures and matrices. This was particularly evident at 63°C, where the decrease in HuNoV titres was less than 0·25 log over the time period (Figure 1).
Figure 1.
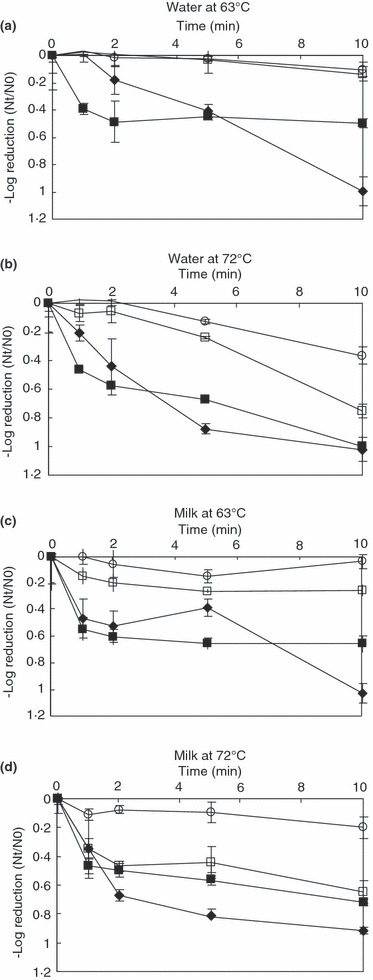
Effect of heat on qRT‐PCR titre of HuNoV GI (○), HuNoV GII (□), MNV (♦) and HAV () in (a) water at 63°C, (b) water at 72°C, (c) milk at 63°C and (d) milk at 72°C.
Interestingly, in both matrices there was a rapid 0·5 log reduction in HAV qRT‐PCR titre at both 63°C and 72°C within the first minute of heating, followed by a slower reduction in qRT‐PCR titre thereafter. MNV showed a similar inactivation profile, except in water at 63°C, where no reduction was seen. This effect was not observed for the HuNoV strains, both of which showed little or no reduction in qRT‐PCR titre (≤0·25 log) over 10 min at 63°C, and showed a gradual reduction over time at 72°C (Fig. 1).
Significant differences (P < 0·05) in log reductions of qRT‐PCR titres were observed between both HuNoV GI and HAV at both temperatures in both matrices, indicating that, by qRT‐PCR, HuNoV GI and HAV do not share the same heat sensitivity properties (Table 4). Similar results were observed for NoV GII and HAV, except in the milk matrix at 72°C, where no significant differences were seen at any time point.
Table 4.
P‐values for the effect of heat on human norovirus genogroup I and II (GI and GII), murine norovirus (MNV), and hepatitis A virus (HAV) at 63 and 72°C in water and milk over time. The conditions where no significance difference was observed are shown in bold (P > 0·05)
| Time (min) | GI and MNV | GII and MNV | GI and HAV | GII and HAV | GI and GII |
|---|---|---|---|---|---|
| (a) Water at 63°C | |||||
| 1 | 0·66 | 0·72 | <0·05 | <0·05 | 0·92 |
| 5 | <0·05 | <0·05 | <0·05 | <0·05 | 0·88 |
| 10 | <0·05 | <0·05 | <0·05 | <0·05 | 0·48 |
| (b) Water at 72°C | |||||
| 1 | <0·05 | <0·05 | <0·05 | <0·05 | 0·08 |
| 5 | <0·05 | <0·05 | <0·05 | <0·05 | <0·05 |
| 10 | <0·05 | <0·05 | <0·05 | <0·05 | <0·05 |
| (c) Milk at 63°C | |||||
| 1 | <0·05 | <0·05 | <0·05 | <0·05 | <0·05 |
| 5 | <0·05 | 0·08 | <0·05 | <0·05 | 0·05 |
| 10 | <0·05 | <0·05 | <0·05 | <0·05 | <0·05 |
| (d) Milk at 72°C | |||||
| 1 | 0·18 | 0·97 | <0·05 | 0·08 | <0·05 |
| 5 | <0·05 | <0·05 | <0·05 | 0·19 | <0·05 |
| 10 | <0·05 | <0·05 | <0·05 | 0·26 | <0·05 |
Significant differences in log reductions of MNV qRT‐PCR titres compared with both HuNoV GI and GII titres were observed at most time points, especially with increased heating time at 63°C in water and milk (Table 4). Significant differences in log reductions of qRT‐PCR titre were observed between HuNoV GI and GII under all conditions except in water at 63°C. In all cases, HuNoV GI was more resistant to heat inactivation than HuNoV GII.
Discussion
The aim of this study was to investigate the suitability of MNV as a surrogate for HuNoV in heat inactivation studies and to compare MNV properties with HAV. As HuNoV cannot be quantified by culture methods, we used qRT‐PCR to compare the HuNoV response with MNV and HAV.
The qRT‐PCR results suggest that neither HAV nor MNV reflect the response of the selected HuNoV strains (GI.2 and GII.4) to heat inactivation at the temperatures evaluated in this study. The apparent differences in the reductions of HuNoV qRT‐PCR titre compared with MNV and HAV may be explained by differences in their susceptibility to heat at 63 and 72°C. On heating, changes in the virus capsid structure occur, resulting in loss of infectivity. It is possible that in some viruses RNA may also be destroyed which will result in a lower qRT‐PCR titre. For HuNoV, we observed that the reduction in qRT‐PCR titre was greater at 72°C than at 63°C, since presumably more damage was occurring at 72°C. Although qRT‐PCR results do not fully reflect the degree of damage on the infectivity status (Richards 1999), they may provide some indication of the extent of capsid damage. These results are in agreement with our previous studies (Hewitt and Greening 2006) and that of other researchers who also found heat inactivation affected the infectivity more than the integrity of the viral genome (Baert et al. 2008). From the qRT‐PCR results, we can hypothesize that loss of HuNoV infectivity may be lower than for HAV and MNV, although currently this cannot be confirmed by culture. Significant differences in log reduction were observed between HuNoV GI and GII, with GI being less affected by heat than HuNoV, MNV and HAV.
Our results show that MNV and HAV exhibited similar log reductions in infectivity with heat. Both viruses showed inactivation of greater than 3·5 logs after 2 min at 72°C, and approximately 1–2 log reduction in infectivity at 63°C over the first minute. Cannon et al. (2006) and Lee et al. (2008) also report significant reductions in MNV titres at similar temperatures.
We conducted our experiments in water and milk to determine whether milk would have a protective effect. Other researchers have demonstrated the protective effects of milk on viruses such as HAV (Parry and Mortimer 1984; Bidawid et al. 2000) and poliovirus (Strazynski et al. 2002). We did not demonstrate any detectable protection from inactivation for any of the viruses in the milk matrix over the time periods studied, similar to the findings of Mariam and Cliver (2000).
For MNV, the reduction in qRT‐PCR titre mirrored the decrease in infectivity, albeit by a lesser amount. However the reduction in qRT‐PCR titre for HAV, compared with the other viruses used in this study, was interesting. The initial decrease of HAV qRT‐PCR titre in the first minute with little further decrease thereafter was similar to the effect reported for HAV infectivity by Bidawid et al. (2000) who noted that rapid inactivation with the first minute was followed by a slower inactivation rate (‘plateau’ effect). It was postulated that this may have been due to the effect of the presence of nonaggregated virus particles that would be inactivated quickly and aggregates that would take longer to be inactivated. If this is correct and the qRT‐PCR titre does give some indication of capsid damage, then it does not explain why MNV and HuNoV did not follow the same trend, as it is reasonable to assume that aggregates and nonaggregated virus particles would also be present in MNV and HuNoV preparations.
In summary, caution should be used when extrapolating surrogate virus data for HuNoV. Although not conclusive, our results suggest that HuNoV may be more resistant to heat than either HAV or MNV. In this study, MNV has been shown to have similar heat inactivation properties to HAV and may be suitable as HAV as a surrogate for other studies of enteric virus survival. However, neither HAV nor MNV may be appropriate surrogates for HuNoV.
Acknowledgements
This work was supported by the New Zealand Food Safety Authority as part of the ESR contract for scientific services. We thank Dr Christiane Wobus (Department of Microbiology and Immunology, University of Michigan Medical School, MI, USA) for advice on MNV culture. We appreciate the contributions of Dr Andrew Hudson, Dr Sandro Wolf and Dr Phil Carter for critical review of the manuscript and Dr Catherine McLeod for her scientific input during the experimental design of the project.
References
- Bae, J. and Schwab, K.J. (2008) Evaluation of murine norovirus, feline calicivirus, poliovirus, and MS2 as surrogates for human norovirus in a model of viral persistence in surface water and groundwater. Appl Environ Microbiol 74, 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baert, L. , Wobus, C.E. , Van Coillie, E. , Thackray, L.B. , Debevere, J. and Uyttendaele, M. (2008) Detection of murine norovirus 1 by using plaque assay, transfection assay, and real‐time reverse transcription‐PCR before and after heat exposure. Appl Environ Microbiol 74, 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliot, G. , Lavaux, A. , Souihel, D. , Agnello, D. and Pothier, P. (2008) Use of murine norovirus as a surrogate to evaluate resistance of human norovirus to disinfectants. Appl Environ Microbiol 74, 3315–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidawid, S. , Farber, J.M. , Sattar, S.A. and Hayward, S. (2000) Heat inactivation of hepatitis A virus in dairy foods. J Food Prot 63, 522–528. [DOI] [PubMed] [Google Scholar]
- Boone, S.A. and Gerba, C.P. (2007) Significance of fomites in the spread of respiratory and enteric viral disease. Appl Environ Microbiol 73, 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckow, R. , Isbarn, S. , Knorr, D. , Heinz, V. and Lehmacher, A. (2008) Predictive model for inactivation of feline calicivirus, a norovirus surrogate, by heat and high hydrostatic pressure. Appl Environ Microbiol 74, 1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, J.L. , Papafragkou, E. , Park, G.W. , Osborne, J. , Jaykus, L.A. and Vinje, J. (2006) Surrogates for the study of norovirus stability and inactivation in the environment: A comparison of murine norovirus and feline calicivirus. J Food Prot 69, 2761–2765. [DOI] [PubMed] [Google Scholar]
- Carter, M.J. (2005) Enterically infecting viruses: pathogenicity, transmission and significance for food and waterborne infection. J Appl Microbiol 98, 1354–1380. [DOI] [PubMed] [Google Scholar]
- Dawson, D.J. , Paish, A. , Staffell, L.M. , Seymour, I.J. and Appleton, H. (2005) Survival of viruses on fresh produce, using MS2 as a surrogate for norovirus. J Appl Microbiol 98, 203–209. [DOI] [PubMed] [Google Scholar]
- De Roda Husman, A.M. , Bijkerk, P. , Lodder, W. , Van Den Berg, H. , Pribil, W. , Cabai, A. , Gehringer, P. , Sommer, R. et al. (2004) Calicivirus inactivation by nonionizing (253.7‐nanometer‐wavelength [UV]) and ionizing (gamma) radiation. Appl Environ Microbiol 70, 5089–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolin, R. , Blacklow, N.R. , DuPont, H. , Buscho, R.F. , Wyatt, R.G. , Kasel, J.A. , Hornick, R. and Chanock, R.M. (1972) Biological properties of Norwalk agent of acute infectious nonbacterial gastroenteritis. Proc Soc Exp Biol Med 140, 578–583. [DOI] [PubMed] [Google Scholar]
- Doultree, J.C. , Druce, J.D. , Birch, C.J. , Bowden, D.S. and Marshall, J.A. (1999) Inactivation of feline calicivirus, a Norwalk virus surrogate. J Hosp Infect 41, 51–57. [DOI] [PubMed] [Google Scholar]
- Duizer, E. , Schwab, K.J. , Neill, F.H. , Atmar, R.L. , Koopmans, M.P. and Estes, M.K. (2004a) Laboratory efforts to culture noroviruses. J Gen Virol 85, 79–87. [DOI] [PubMed] [Google Scholar]
- Duizer, E. , Bijkerk, P. , Rockx, B. , De Groot, A. , Twisk, F. and Koopmans, M. (2004b) Inactivation of caliciviruses. Appl Environ Microbiol 70, 4538–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening, G.E. , Mirams, M. and Berke, T. (2001) Molecular epidemiology of ‘Norwalk‐like viruses’ associated with gastroenteritis outbreaks in New Zealand. J Med Virol 64, 58–66. [DOI] [PubMed] [Google Scholar]
- Hewitt, J. and Greening, G.E. (2004) Survival and persistence of norovirus, hepatitis A virus, and feline calicivirus in marinated mussels. J Food Prot 67, 1743–1750. [DOI] [PubMed] [Google Scholar]
- Hewitt, J. and Greening, G.E. (2006) Effect of heat treatment on hepatitis A virus and norovirus in New Zealand greenshell mussels (Perna canaliculus) by quantitative real‐time reverse transcription PCR and cell culture. J Food Prot 69, 2217–2223. [DOI] [PubMed] [Google Scholar]
- Kageyama, T. , Kojima, S. , Shinohara, M. , Uchida, K. , Fukushi, S. , Hoshino, F.B. , Takeda, N. and Katayama, K. (2003) Broadly reactive and highly sensitive assay for Norwalk‐like viruses based on real‐time quantitative reverse transcription‐PCR. J Clin Microbiol 41, 1548–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst, S.M. , Wobus, C.E. , Lay, M. , Davidson, J. and Virgin, H.W. (2003) STAT1‐dependent innate immunity to a Norwalk‐like virus. Science 299, 1575–1578. [DOI] [PubMed] [Google Scholar]
- Kingsley, D.H. , Hoover, D.G. , Papafragkou, E. and Richards, G.P. (2002) Inactivation of hepatitis A and a calicivirus by high hydrostatic pressure. J. Food Prot 65, 1605–1609. [DOI] [PubMed] [Google Scholar]
- Kingsley, D.H. , Holliman, D.R. , Calci, K.R. , Chen, H. and Flick, G.J. (2007) Inactivation of a norovirus by high‐pressure processing. Appl Environ Microbiol 73, 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Zoh, K. and Ko, G. (2008) Inactivation and UV disinfection of murine norovirus with TiO2 under various environmental conditions. Appl Environ Microbiol 74, 2111–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariam, T.W. and Cliver, D.O. (2000) Small round coliphages as surrogates for human viruses in process assessment. Dairy, Food and Environ Sanitation 20, 684–689. [Google Scholar]
- Parry, J.V. and Mortimer, P.P. (1984) The heat sensitivity of hepatitis A virus determined by a simple tissue culture method. J Med Virol 14, 277–283. [DOI] [PubMed] [Google Scholar]
- Richards, G.P. (1999) Limitations of molecular biological techniques for assessing the virological safety of foods. J Food Prot 62, 691–697. [DOI] [PubMed] [Google Scholar]
- Slomka, M.J. and Appleton, H. (1998) Feline calicivirus as a model for heat inactivation studies of small round structured viruses in shellfish. Epidemiol Infect 121, 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub, T.M. , Honer zu Bentrup, K. , Orosz‐Coghlan, P. , Dohnalkova, A. , Mayer, B.K. , Bartholomew, R.A. , Valdez, C.O. , Bruckner‐Lea, C. et al. (2007) In vitro cell culture infectivity assay for human noroviruses. Emerg Infect Dis 13, 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazynski, M. , Kramer, J. and Becker, B. (2002) Thermal inactivation of poliovirus type 1 in water, milk and yoghurt. Int J Food Microbiol 74, 73–78. [DOI] [PubMed] [Google Scholar]
- Thurston‐Enriquez, J.A. , Haas, C.N. , Jacangelo, J. and Gerba, C.P. (2003a) Chlorine inactivation of adenovirus type 40 and feline calicivirus. Appl Environ Microbiol 69, 3979–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston‐Enriquez, J.A. , Haas, C.N. , Jacangelo, J. , Riley, K. and Gerba, C.P. (2003b) Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Appl Environ Microbiol 69, 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston‐Enriquez, J.A. , Haas, C.N. , Jacangelo, J. and Gerba, C.P. (2005a) Inactivation of enteric adenovirus and feline calicivirus by ozone. Water Res 39, 3650–3656. [DOI] [PubMed] [Google Scholar]
- Thurston‐Enriquez, J.A. , Haas, C.N. , Jacangelo, J. and Gerba, C.P. (2005b) Inactivation of enteric adenovirus and feline calicivirus by chlorine dioxide. Appl Environ Microbiol 71, 3100–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, V.K. , McCormick, C.J. , Clarke, I.N. , Salim, O. , Wobus, C.E. , Thackray, L.B. , Virgin, H.W., IV and Lambden, P.R. (2007) Recovery of infectious murine norovirus using pol II‐driven expression of full‐length cDNA. Proc Natl Acad Sci USA 26, 11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus, C.E. , Karst, S.M. , Thackray, L.B. , Chang, K.O. , Sosnovtsev, S.V. , Belliot, G. , Krug, A. , Mackenzie, J.M. et al. (2004) Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol 2, e432. doi: DOI: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus, C.E. , Thackray, L.B. and Virgin, H.W. (2006) Murine norovirus: a model system to study norovirus biology and pathogenesis. J Virol 80, 5104–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]


