Abstract
Aims: To determine the detection limit of diagnostic negative staining electron microscopy for the diagnosis of pathogens that could be used for bioterrorism.
Methods and Results: Suspensions of vaccinia poxvirus and endospores of Bacillus subtilis were used at defined concentrations as a model for poxviruses and spores of anthrax (Bacillus anthracis), both of which are pathogens that could be used for bioterrorist attacks. Negative staining electron microscopy was performed directly or after sedimentation of these suspensions on to the sample supports using airfuge ultracentrifugation. For both virus and spores, the detection limit using direct adsorption of a 10‐μl sample volume onto the sample support was 106 particles per ml. Using airfuge ultracentrifugation with a sample volume of 80 μl, the detection limit could be reduced to 105 particles per ml for spores and to 5 × 104 particles per ml for poxviruses. The influence on particle detection of incubation time, washing and adsorption procedures was investigated.
Conclusions: The reproducibility and sensitivity of the method were acceptable, particularly considering the small sample volume and low particle number applied onto the sample support.
Significance and Impact of the Study: Diagnostic negative staining electron microscopy is used for the diagnosis of pathogens in emergency situations because it allows a rapid examination of all particulate matter down to the nanometre scale. This study provides precise detection limit for the method, an important factor for the validation and improvement of the technique.
Keywords: airfuge, bioterrorism, detection limit, diagnosis, negative staining, particle enrichment, poxvirus, spore
Introduction
Terrorist attacks using human or animal pathogens could have a serious impact on society, mainly because a single local distribution of a pathogen, depending on its transmissibility and virulence, can affect a large number of people or animals (Inglesby et al. 2002). Moreover, panic and uncertainty can easily be initiated using harmless mock substances in copycat attacks. Recent examples demonstrating the profound impact and consequences of even limited bioterrorist attacks include the letters containing anthrax spores that were sent shortly after the events of September 2001 (Lane et al. 2001) and the distribution of anthrax spores by the Aum cult in Japan in 1995 (Olson 1999).
In response to these attacks, authorities worldwide increased their preparedness for dealing with bioterrorism (e.g. Nolte et al. 2004). One major focus was to set up and improve procedures for the rapid and reliable diagnosis (or exclusion) of relevant pathogens in samples suspected to contain such organisms (Sapsford et al. 2008). Because of a number of unique advantages, electron microscopy formed an integral part of these procedures (Hazelton and Gelderblom 2003; Miller 2003, 2004). Electron microscopy allows the direct imaging in a sample of all particles down to a size of a fraction of a nanometre and hence the instant recognition of different morphological groups of pathogens (Hazelton and Gelderblom 2003; Curry et al. 2006). The method provides basic information on the particulate composition of a sample and therefore gives a degree of direction and control for more precise techniques (e.g. nucleic acid amplification, immunological assays) that use specific probes to detect a pathogen but which can sometimes fail if the pathogen has mutated or if detection is impaired by inhibitors. Indeed, diagnostic electron microscopy can reveal and describe new versions of a pathogen and thereby facilitate their precise identification. The importance of this approach was clearly demonstrated by the collaborative identification of the pathogen causing SARS in which electron microscopy gave the first clue that a coronavirus was responsible (World Health Organisation 2003).
The fastest method for diagnostic electron microscopy uses negative staining, a technique introduced by Brenner and Horne (1959). Samples, usually in the form of a suspension, are adsorbed onto the surface of a thin transparent plastic film supported by a metal grid, stained with heavy metals for stabilization and contrast (Miller 1986; Harris and Horne 1991) and then inspected using a transmission electron microscope. This whole procedure can be carried out within only a few minutes. Virus particles can usually be assigned to a particular virus family, while bacteria or fungi can only be recognized to a more general morphological group that provides direction for a more focused diagnosis (Gelderblom et al. 1991; Curry et al. 2006). Most importantly, as well as being a valuable quick and simple diagnostic technique, there is a significant amount of reference data available in the literature concerning negative staining electron microscopy (Biel and Gelderblom 1999a). However, despite having been used for many decades, quantitative data on the detection limits of the method are limited (Hammond et al. 1981; Biel and Gelderblom 1999a; Biel et al. 2004; Nitsche et al. 2007).
The purpose of our study was therefore to determine and to evaluate the detection limit of the negative staining technique for the diagnosis of bioterrorism‐relevant pathogens. As test models we used suspensions of the poxvirus vaccinia and the endospores of Bacillus subtilis (Fig. 1) to represent at least two classes of micro‐organisms relevant in bioterrorism (list of possible bioterrorism agents by the CDC (Centers for Disease, Control and Prevention), USA; http://emergency.cdc.gov/agent/agentlist-category.asp). In addition, the influence of variables during preparation (e.g. incubation time and washing) on the efficiency of detection was evaluated. Knowledge of the detection limit and a comparison with results published for negative staining of, for instance clinical specimens, is important because samples and preparations are handled differently. Samples from a suspected bioterrorist attack are usually first inactivated by chemicals or by radiation, which may interfere with adsorption to the support (see discussion in Madeley and Biel 2006; Gelderblom et al. 2007). Moreover, a diagnosis should include fungal spores and bacteria, particles that are usually denser and bigger than viruses. Adsorption and washing conditions may, according to the morphological class of particles (size, shape, density), differentially affect the number of particles bound to the grid. Finally, the results of this study are also relevant for accredidation and certification procedures carried out in diagnostic laboratories during the establishment of quality management systems aimed at improving the overall quality of diagnosis.
Figure 1.
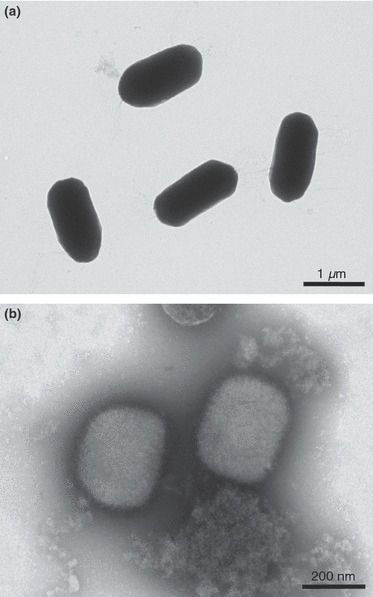
Negative staining of spore and virus particles. (a) Spores of Bacillus subtilis are oval particles with a high density that prevents visualization of internal details. (b) Vaccinia poxvirus shows the typical brick‐shape morphology and surface structure of orthopoxvirus, a subgroup of the poxvirus family.
Materials and methods
Test suspensions
Bacterial endospores of B. subtilis (JH 642) were generated by cultivation in sporulation medium (SM‐Medium; Sterlini and Mandelstam 1969). The degree of sporulation was followed by light microscopy until the spore number was sufficiently high. Spores were harvested and washed twice in 0·05 M HEPES buffer and stored in buffer at 2–10°C. The quality of the spore preparation was evaluated by thin section electron microscopy (Laue et al. 2007). The criteria for a suitable preparation were a regular spore ultrastructure and the absence of intact vegetative bacteria. Spore suspensions were centrifuged, and sedimented spores were fixed with a mixture of 10% paraformaldehyde and 0·05% glutaraldehyde in 0·05 M HEPES buffer, pH 7·2. Spore concentrations were determined using a Neubauer counting chamber.
Vaccina virus (VR 1530) was propagated in Vero cells. The culture medium was collected after development of cytopathic effect and fixed with 2% paraformaldehyde in 0·05 mol l−1 HEPES (final concentration). Virus particle concentration was determined according to Geister and Peters (1963) using a reference suspension with a defined concentration of plastic beads mixed with the test suspension. Three 1 ml samples of a suspension of 200‐nm polystyrene beads (Duke Scientific Corp.) were dried in a 37°C oven for several days. The particle concentration was calculated from the dry weight and the weight of a single particle (calculated from the bead density as provided by the producer and our own size measurements using electron microscopy). The bead suspension and the virus suspension were then mixed at a similar concentration (as determined from initial quality control experiments using negative staining electron microscopy). After adsorption onto a support for negative staining, particles were counted in the electron microscope (40 regions on four grids) and the concentration of the unknown suspension was calculated from the mean relation between particle number of known and unknown suspension determined for the different regions, taking into account all preceding dilutions.
Negative staining
The protocol for negative staining was standardized as far as possible, particularly with regard to manual procedures. Four hundred mesh grids (Agar Scientific) coated with a formvar film and reinforced by a thick carbon layer were used as a support. All incubations were carried out using droplets (30 μl unless otherwise stated) on a sheet of parafilm. To increase the hydrophilicity and stickiness of the surface, grids were placed on a drop of alcian blue (Fluka, Sigma‐Aldrich, Munich, Germany; 1% in 1% acetic acid in water) for 10 min (Lang et al. 1981). Grids were then washed over four droplets of double‐distilled water, and the bulk of the water was removed from the grid surface using filter paper. The test suspension was brought into contact with the grid surface either by placing the grid, with the hydrophilic surface facing down, on a drop of the suspension (grid‐on‐drop procedure) or by adding 10 μl of the suspension to the hydrophilic surface of the grid (drop‐on‐grid procedure). After incubation for 10 min, grid surface was washed on four drops of double‐distilled water for 5 s each. Excess water was removed from the grid surface using a filter paper, and the grid was placed on a drop of either 0·5% uranyl acetate (spores) or 1% phosphotungstic acid at neutral pH (virus) for 5 s. Finally, grids were dried using a filter paper. To ensure that the forces applied to the adsorbed particles were reproducible, grids were held perpendicular to the surface of the filter paper for all such drying steps.
Pilot experiments using different staining solutions (uranyl acetate or phosphotungstic acid) for negative staining of viruses yielded similar results (data not shown) but because of a lower variability among grids and better visibility of particles, phosphotungstic acid was selected for the experiments using the virus suspensions.
Airfuge ultracentrifugation
Ultracentrifugation of particles directly onto the grid was performed using an airfuge (Beckman) equipped with a fixed angle rotor (A100‐18, 18°). Grids were placed in adaptors (Type S/N 833101329, Beckman; No. 11093, Laborgeraete Beranek, Weinheim, Germany), which maintain the grid surface parallel to the axis of the rotor (Gelderblom 2006). Centrifuge tubes were filled with 80 μl of test suspension, and adaptors with grids in place were added. The period between filling the tubes with the test suspension and commencing centrifugation was between 7 and 10 min (for six tubes). Centrifugation was carried out for 10 min at a pressure of 20 psi which corresponds to a centrifugal force of about 120 000 g (clearing factor is approx. 15; Beckman). These values were chosen based on a calculation of the time to fully sediment virus particles of the lowest known density [i.e. 100 Svedberg; Liebermann 1978; time (h) = clearing factor/density (Svedberg)]. After adaptors were removed from the centrifuge tubes, grids were released by inverting the adapters and adding 80 μl of double‐distilled water to form a hanging drop. Grids were washed on two drops of double‐distilled water, and excess water removed using a filter paper before placing them on a drop of negative staining solution as described earlier.
Electron microscopy and sampling
Grids were examined using a transmission electron microscope (Tecnai12 BioTwin, FEI) operated at 120 kV. Micrographs were recorded with a CCD camera (Megaview III, OSIS) at a resolution of 1376 × 1024 pixels. The microscope’s standard sample holder allows approx. 970 squares of the grid with a mesh size of about 46 × 46 μm (visible area) to be viewed. Particle numbers were estimated using a systematic sampling strategy in which 22 grid squares, distributed evenly over the grid, were selected for counting. This procedure is similar to the routine sampling strategy used to inspect diagnostic samples. For samples with low counts, the number of grid squares was increased by a factor of 4 (i.e. 88 grid squares in total). Results were processed using Microsoft Excel, and diagrams were generated using the Origin software.
The similarity (homogeneity) of particle number distributions was analysed statistically at the 95% confidence level according to Kolmogoroff–Smirnoff (Sachs 1992). This test does not require values to have a standardized normal distribution, which is particularly important with low particle counts.
Results
Reproducibility of particle detection
The reproducibility of the negative staining method with regard to the number of particles detected was evaluated in several independent experiments. The shelf life of the alcian blue used to increase the hydrophilicity and ‘stickiness’ of the grids was tested by coating grids with alcian blue at different time points (0, 1, 3, 6 months) after preparing the solution and adding 10 μl of a spore suspension (108 particles ml−1). There was no statistical difference in the resulting particle numbers on the grids (Table 1, No. 1–4). There was also no statistical difference between grids treated with different batches of alcian blue solution (compare Table 1, No. 5 with No. 6).
Table 1.
Spore number on grids in independent experiments (1–6) using a spore suspension of 108 spores per ml. In experiments 1–4, the shelf life of the alcian blue solution was tested at defined intervals of storage (d = days). In experiments 5 and 6, different batches of alcian blue solution were used than in experiments 1–4
| Experiment | Particles/square | Interquartile range | No. Grids |
|---|---|---|---|
| 1 (d 0) | 11 | 2 | 3 |
| 2 (d 30) | 12 | 1 | 3 |
| 3 (d 90) | 12 | 1 | 3 |
| 4 (d 180) | 10 | 0.5 | 3 |
| 5 | 10 | 0 | 3 |
| 6 | 10 | 0.5 | 3 |
Moreover, a comparison of experiments in which different suspensions of B. subtilis spores (different strain, different preparation and estimated particle number) were used also yielded a similar particle number at a given spore concentration (not shown). Negative staining experiments using virus suspensions also gave no significant inter‐experiment differences (e.g. one particle/grid square vs two particles/grid square for 107 particles per ml suspension; n = 3 grids per measurement; compare also with Table 5). The reproducibility of the negative staining method in terms of particle number is therefore appropriate.
Table 5.
Adsorption of spores or virus particles on grids in relation to particle concentration of the sample suspension applied
| Concentration (part per ml) | Particles/22 squares | Interquartile range | Particles/square* | Interquartile range | No. Grids |
|---|---|---|---|---|---|
| Spores | |||||
| 108 | 244 | 22 | 11 | 1.0 | 3 |
| 107 | 28 | 8 | 1 | 0.4 | 3 |
| 106 | 11 | 2 | 0.5 | 0.1 | 3 |
| 105 | 1 | 0.4 | 0.05 | 0.02 | 3 |
| Virus | |||||
| 108 | 336 | 137 | 15 | 6 | 3 |
| 107 | 33 | 10 | 2 | 0.5 | 3 |
| 106 | 3 | 2 | 0.1 | 0.07 | 3 |
| 105 | 1 | 0.5 | 0.05 | 0.02 | 3 |
*Value for each grid was calculated from respective number counted for 22 grid squares.
Particle adsorption and influence of incubation time and washing steps
Particles in suspension can be adsorbed on the surface of a grid by either adding a drop of the suspension on the alcian blue‐coated carbon‐plastic film (drop‐on‐grid) or by placing the grid, with the coated film facing down, onto a drop of the suspension (grid‐on‐drop). With the drop‐on‐grid procedure, dense particles are sedimented onto the grid by gravity while with the grid‐on‐drop procedure particles can be adsorbed that do not sediment significantly at normal gravity but which concentrate at the liquid‐air interface (Johnson and Gregory 1993; Cyrklaff et al. 1994). The performances of the two adsorption procedures for suspensions of spores and vaccinia virus were therefore compared. In all experiments, the drop‐on‐grid procedure gave a higher particle number than the grid‐on‐drop procedure (Table 2). Even with the virus suspension, the difference between the two procedures was statistically significant although the relative difference between the two procedures is smaller with virus than with spores (Table 2).
Table 2.
Comparison of different particle adsorption procedures (drop‐on‐grid vs grid‐on‐drop)
| Suspension | Adsorption | Particles/square | Interquartile range | No. Grids |
|---|---|---|---|---|
| Spore, 108 part per ml | drop‐on‐grid | 9 | 0 | 3 |
| grid‐on‐drop | 0 | 0 | 3 | |
| Spore, 5 × 108 part per ml | drop‐on‐grid | 52 | 8 | 3 |
| grid‐on‐drop | 1 | 2 | 3 | |
| Virus, 108 part per ml | drop‐on‐grid | 10 | 2 | 3 |
| grid‐on‐drop | 4 | 3 | 3 |
Washing grids after adsorption of particles and before negative staining is useful to remove low‐molecular weight components (Harris and Horne 1991) that might mask the structure of relevant particles or that interact with the staining solution (e.g. phosphate salts form precipitates with uranyl acetate). However, relevant particles are lost during washing in addition to unwanted components. The loss of particles during washing was quantified by successively increasing the number of washing steps from zero to six in twofold steps. With both spore and virus suspensions, a significant loss of particles during washing with double‐distilled water was seen (Table 3). The loss was slightly pronounced with spores than with virus suspensions (compare relative differences between the different washing conditions, Table 3). The relative loss between no washing and the standard four step washing procedure was 42% for spores and 34% for virus suspensions.
Table 3.
Loss of particles during washing. Absolute numbers represent the median of particles per 22 grid squares of three grids (virus) or six grids (two experiments; spores). Relative values (in per cent) represent difference (Δ) in particle number between two succeeding washing procedures
| Suspension | without washing | 2 washing steps | Δ (%) | 4 washing steps | Δ (%) | 6 washing steps | Δ (%) |
|---|---|---|---|---|---|---|---|
| Spore, 106 part per ml | 19 | 15 | 24 | 11 | 24 | 6 | 45 |
| Virus, 107 part per ml | 50 | 42 | 16 | 33 | 21 | 22 | 33 |
To evaluate the influence of sample incubation time on the resulting particle number on the grid, both test suspensions were incubated for 1, 5, 10 and 20 mins using the drop‐on‐grid method. The particle number for spores increased rapidly by a factor of 10 from 1 to 5 min but began to level off thereafter (Table 4, Fig. 2a). In contrast, with the virus suspension, particle numbers increased only slowly from 1 to 10 min but at a higher rate between 10 and 20 min, approx. following an exponential slope (Table 4, Fig. 2b). Remarkably, the absolute number of virus particles on the grid after 1 min of incubation was about 10 times higher than with spores (Table 4).
Table 4.
Particle number in relation to incubation time. Absolute numbers represent the median of particles per 22 grid squares of three grids. Difference (Δ) in particle number between two succeeding incubation times is expressed as the quotient of absolute values.
| Suspension | 1 min | 5 min | Δ (1/5 min) | 10 min | Δ (5/10 min) | 20 min | Δ (10/20 min) |
|---|---|---|---|---|---|---|---|
| Spore, 107 part per ml | 1 | 10 | 10 | 19 | 1.9 | 26 | 1.4 |
| Virus, 107 part per ml | 12 | 25 | 2 | 42 | 1.7 | 101 | 2.4 |
Figure 2.
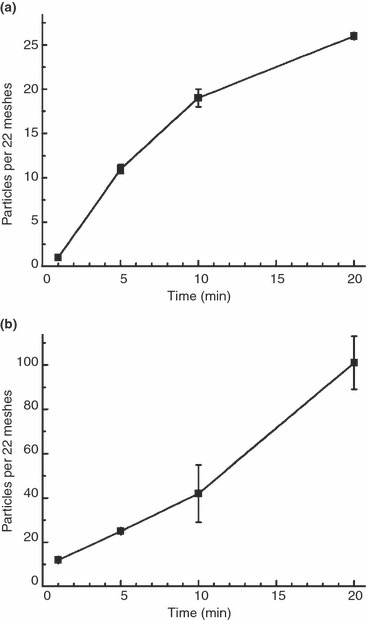
Particle number in relation to incubation time. (a) Spore suspension. (b) Virus suspension.
Detection limit of the negative staining method
To estimate the detection limit of the drop‐on‐grid method, the number of adsorbed particles on the grid after 10 min of incubation was determined in relation to the concentration in the suspensions applied. With both virus and spores, the number of particles adsorbed on the grid increased almost linearly with increasing suspension concentration (Table 5, Fig. 3). The critical concentration at which detection of particles would be difficult using the systematic sampling approach defined in this study (i.e. sampling 22 grid squares distributed evenly over the grid) was 105 particles per ml. At this concentration, a median value of only one particle/22 grid squares could be detected for both suspensions of virus and of spores. Using the interquartile range as a measure of variability, it was clear that the number of particles detected can drop below 1 per 22 grid squares, i.e. no detection at all (Table 5). In contrast, at 106 particles per ml, the number of particles detected in 22 grid squares systematically distributed over the grid should be sufficient (Table 5).
Figure 3.
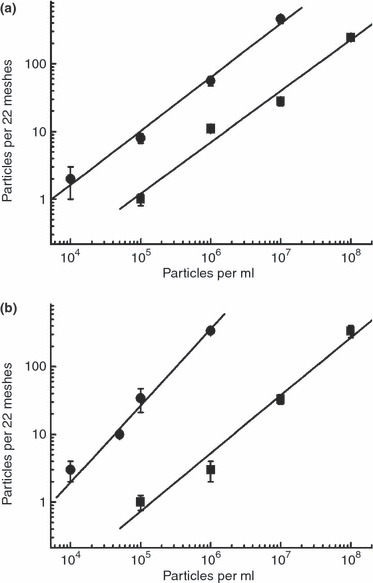
Particle number on the grid in relation to concentration of the test suspension. Squares represent values of the drop‐on‐grid procedure. Circles represent values of the airfuge enrichment. (a) Spore suspension. (b) Virus suspension.(•) Airfuge and (▪) Drop‐on‐grid.
To check the accuracy of the 106 particles per ml detection limit, several independent control experiments were performed using suspensions of either 105 or 106 particles per ml. Detection was not always possible with grids treated with 105 particles per ml (4 of 10 grids were positive for spores; 1 of 10 grids were positive for virus particles) whereas a suspension of 106 particles per ml always resulted in particle detection (n = 10 grids).
Detection limit of the negative staining method using airfuge sedimentation onto the grid
Airfuge concentration of spores or virus particles onto the grid resulted in a higher particle number than adsorption of particles by the drop‐on‐grid procedure (compare Table 5 with Table 6 and Fig. 3). However, the calculated enrichment factors were different for spores and virus suspensions. Virus particles were far more enriched by airfuge sedimentation than were spores (Table 7). Moreover, while enrichment factors were almost the same for different concentrations of spores they varied for different concentrations of virus (Table 7).
Table 6.
Airfuge concentration of spores or virus particles on grids in relation to particle concentration of the sample suspension applied
| Concentration (part per ml) | Particles/22 squares | Interquartile range | Particles/square* | Interquartile range | No. Grids |
|---|---|---|---|---|---|
| Spores | |||||
| 107 | 459 | 125 | 21 | 6 | 2 |
| 106 | 56 | 18 | 3 | 0.8 | 3 |
| 105 | 8 | 3 | 0.4 | 0.1 | 3 |
| 5 × 104 | 4 | 0.5 | 0.2 | 0.02 | 3 |
| 104 | 2 | 2 | 0.07 | 0.08 | 6 |
| Virus | |||||
| 106 | 340 | 26 | 15 | 1 | 3 |
| 105 | 34 | 26 | 2 | 1 | 3 |
| 5 × 104 | 10 | 2 | 0.5 | 0.07 | 3 |
| 104 | 3 | 2 | 0.1 | 0.09 | 3 |
*Value for each grid was calculated from respective number counted for 22 grid squares.
Table 7.
Enrichment factor of the airfuge concentration procedure
| Concentration (part per ml) | Airfuge* | Drop‐on‐grid* | Enrichment factor |
|---|---|---|---|
| Spores | |||
| 107 | 459 | 28 | 16 |
| 106 | 56 | 11 | 5 |
| 105 | 8 | 1 | 8 |
| Virus | |||
| 106 | 340 | 3 | 113 |
| 105 | 34 | 1 | 34 |
*numbers in particles/22 grid squares.
The detection limit for spores and virus is 104 particles per ml according to the particle number determined at different concentrations (Table 6). However, at this concentration, the variability (expressed as the interquartile range) is relatively high (Table 6) implying that detection of particles may not be possible on every grid investigated. To check the reliability of the detection limit, several control experiments were conducted in which suspensions at concentrations of 105 or 104 particles per ml were used for airfuge sedimentation onto grids. While detection of virus or spores was possible for all grids tested at 105 particles per ml (spores, n = 10 grids; virus, n = 6 grids), at the lower concentration of 104 particles per ml, some grids revealed no virus or spore particles in a systematical screening of 22 grid squares (five of six grids were positive for spores; four of six grids were positive for virus particles).
Because the detection limit appears to be close to 104 particles per ml (Table 6) and because the likelihood of finding a particle at this concentration was high (but not 100%), an intermediate concentration of 5 × 104 particle per ml was tested. The resulting median particle number on the grids, together with a small interquartile range, imply successful detection of spores or virus on all grids treated with 5 × 104 particles per ml (corresponding to 4000 particles in the sample volume of 80 μl) using airfuge adsorption (Table 6). Testing grids (n = 11) with a virus suspension of 5 × 104 particles per ml supported this reliable detection while a similar test using spore suspension failed (2 of 11 grids were negative for spores).
Discussion
Reproducibility and reliability are important criteria for a diagnostic method. Our results demonstrate that negative staining of test suspensions of bacterial endospores or poxviruses is reproducible with regard to the number of particles detected on the grid. Alcian blue pretreatment of grids seems to be a reliable procedure because it did not add significantly to the variability of the results, possibly because the treatment is applied directly before adding the sample to the grid. Moreover, alcian blue solution can be stored for at least 6 months (at 2–10°C) without any significant loss of efficiency, reducing the risk of impaired reproducibility in practice. Pretreatment of grids by glow discharge is very popular because it renders them hydrophilic without adding any chemicals to the surface (Reissig and Orrell 1970). However, the efficiency of binding particles to the surface is dependent on several factors of the discharge procedure (e.g. duration) and the time postdischarge (Harris 1997; Shibata and Miyazaki 2002). It is still unclear what influence these parameters have on the detection of particles. Clearly, more experiments are needed to investigate the influence of different chemical or physical grid pretreatments on the efficiency of particle adsorption.
The detection limit, i.e. the minimal detectable concentration of particles in the test suspension, was similar for both poxviruses and bacterial endospores. Without airfuge sedimentation at least 106 particles per ml and applying the suspension onto the grid (drop‐on‐grid procedure) were necessary for reliable detection. With both poxviruses and spores, the application of a drop of the test suspension onto the grid was more efficient than placing the grid onto a drop of suspension (grid‐on‐drop procedure) indicating that particle density is high enough for a sufficient sedimentation by gravity. The detection limit determined here is in the range given by other studies using virus suspensions (Katz et al. 1984; Biel and Gelderblom 1999a; Johnsen et al. 2006). However, detection is not only influenced by the concentration of the relevant particles but also by the overall composition of the suspension. Constituents of a suspension can interfere with adhesion of the particles to the grid or can cover the particles (Kurth et al. 2008) thereby reducing the likelihood of detection. The detection limit proposed in this study should therefore be examined in further experiments using mixtures of test suspensions with environmental test samples (e.g. dust, soil, drying powder) and/or clinical test samples. Moreover, the detection limit for particles possessing a density lower than that of poxviruses should be determined. Experiments using clinical samples and particle estimations based on the measurement of genomic units suggest that the detection limit under these conditions may be higher (Biel et al. 2004; Nitsche et al. 2007).
Enrichment by airfuge sedimentation onto grids placed in centrifuge adaptors (Gelderblom 2006) increased the detection sensitivity by a factor of 10 for spores (i.e. 105 particles per ml) and by a factor of 50 for poxviruses (i.e. 5 × 104 particles per ml). The enrichment factor for poxviruses is similar to that given by other authors using the airfuge (Müller et al. 1981; Biel and Gelderblom 1999a), although it is difficult to make direct comparisons because the airfuge parameters were not identical and, moreover, enrichment factors were not determined at the detection limit. Higher enrichment factors might be achieved using the particle counting EM‐90 Beckman rotor (Hammond et al. 1981) although a direct comparison of the two rotor systems, including a critical evaluation of practical handiness, has yet to be published.
Differences between the detection limit of spores and virus using airfuge enrichment may be explained by their different sizes and therefore, different susceptibilities to the mechanical forces exerted on the particles. Spores may be lost from the grid during manipulation after centrifugation, i.e. removal of the adaptor from the centrifugation tube and removal of the grid from the adaptor. Enhanced susceptibility of spores to mechanical forces is supported by the higher loss of spores (compared with virus) during washing.
Several factors influence the detection limit of the negative staining method. In this study, we studied the impact of three factors: (i) procedure for applying the suspension (drop‐on‐grid versus grid‐on‐drop); (ii) incubation time and (iii) number of washing steps. In our hands, drop‐on‐grid always resulted in higher particle numbers than grid‐on‐drop. The difference between the two procedures was more pronounced with the spores than with the poxvirus test suspension, possibly because of the higher density of the spores. In this respect, it would be interesting to know at which particle density the drop‐on grid procedure becomes less efficient than grid‐on‐drop for a given incubation time. Because both the sedimentation and the enrichment of particles at the interface between the liquid and air or the grid surface are time‐dependent, it would be also helpful to know more about the kinetics of both processes.
The impact of incubation time on adsorbed particle number was investigated using the drop‐on‐grid procedure. Particle numbers increased with time for both viruses and spores. However, while with spores the particle adsorption followed a saturation kinetic during the 20‐min observation time, with the virus an almost exponential increase in particle number was observed. This result can be explained by the different densities of the particles, leading to a faster sedimentation of spores than viruses. It is, however, remarkable that the virus suspension gave comparatively high particle numbers at short incubation times (i.e. 1 and 5 min). This difference may be because of a higher susceptibility of spores to removal from the grid by washing. As the detection limit is dependent on incubation time, a compromise between sensitivity and overall processing time must be found for each experiment or diagnostic test. Based on our experiments, a slight increase in the sensitivity of detection (i.e. by a factor of 1·4 for spores and 2·4 for virus) is likely if the incubation time is increased to 20 min.
Although washing the grids directly after particle adsorption is helpful because it eliminates solutes or other contaminants that may interfere with detection (Harris and Horne 1991; Biel and Gelderblom 1999b), it may also reduce the actual number of particles by desorption (Müller et al. 1980). We showed a significant loss of particles only after two washing steps and after four washing steps (the standard used in this study) the loss was about 40% for spores and about 30% for virus particles. This difference may be related to the larger size of spores, making them more prone to removal than the smaller virus particles. Washing should therefore be reduced to a minimum to avoid a reduction in detection sensitivity and further testing should establish an optimal compromise between the visibility of particles and their retention on the grid.
The time available for observation also has an impact on the detection limit: the sensitivity of detection increases with observation. However, in practice, observation time is limited by the situation, especially on the time available for a diagnosis. Screening 20 grid squares systematically distributed over the grid already takes approx. 10–15 min depending on various parameters such as staining quality, particle number, size and distribution. A possible moderate gain in detection sensitivity should therefore be carefully weighed against the loss in detection speed. Moreover, while scanning across the grid in a more or less even sampling pattern, it is possible to concentrate on certain regions of the grid that show an interesting particle distribution. Open view negative staining electron microscopy may therefore find particles that are present at a low number on the grid, somewhat lowering the detection limit in practice. However, diagnostic quality is also dependent on the competence of the electron microscopist and overall diagnostic performance must therefore be regularly evaluated by blinded testing and maintained or improved by continuous practice (Biel and Gelderblom 1999a).
The detection limit may also be improved by optimizing the pretreatment of grids. No comparative data are available on the efficiency of the different pretreatment procedures such as, for instance, glow discharge or coating with the different chemicals (e.g. poly‐l‐lysine, bacitracin). Preliminary experiments using virus suspensions suggest no significant difference between grids that have been treated with alcian blue or by glow discharge whereas with bacteria and endospores alcian blue‐treated grids performed better (M. Laue, unpublished data). However, as only a few pretreatment parameters have so far been tested systematically, there are many options to further improve efficiency. For example, increasing the concentration of chemicals applied to the grid surface or using alternative strategies to modify the surface properties of the grid (e.g. by advanced surface chemistry) may be beneficial.
One other strategy to improve the detection limit is to increase the number of particles applied to the grid by either sample concentration (e.g. by ultracentrifugation) or by directly applying a larger volume to the grid. The latter may be possible for the airfuge procedure because adaptors containing deeper recesses are available (No. 11094; Laborgeraete Beranek). Preliminary tests using these adaptors have resulted in increased particle numbers on the grid compared to the use of adaptors with a flat recess (M. Laue, unpublished data). Other studies have used tubes and/or adaptors in conventional ultracentrifuges to apply larger volumes than can usually be achieved with the airfuge (Müller 1969; Mathews and Buthala 1970). It may be worth evaluating modern table‐top ultracentrifuges able to process larger volumes for their ability, by simply inserting the grids into the centrifuge tube, to deposit particles directly onto a grid. Such a direct approach has already been successfully used with the airfuge (Alain et al. 1987), resulting in almost the same particle number achieved using conventional adaptors (M. Laue, unpublished data).
Despite the low detection efficiency compared to other methods (e.g. nucleic acid amplification methods), negative staining of bioterrorism‐relevant pathogens is important because it is completely independent of a need for specific probes (such as antibodies or nucleic acids) and reveals all particulate nonsolute matter in a sample down to a sub‐nanometre size. It therefore serves as an independent control for other methods applied in parallel to identify possible bioterrorism‐related pathogens. Our experiments demonstrate that bacterial endospores and poxviruses, two candidate pathogens for bioterrorism, can be reliably detected up to a defined detection limit under ideal conditions, even if they are modified by inactivating chemicals. As contaminants in the samples can mask particles and may therefore reduce the detection efficiency (Kurth et al. 2008), the detection limit must be tested in further experiments using more realistic samples, such as mixtures of test organisms and particles mimicking environmental samples.
Acknowledgements
The authors thank Marianne Hohoff for the preparation of spores, and Mandy Büchner, Pia Henselmann, Maria Martin, Silvie Muschter and Janett Piesker for conducting the many negative staining experiments and for their patient counting at the microscope. In addition, we thank the two referees for their helpful suggestions and Dr Steve Norley for polishing the English.
References
- Alain, R. , Nadon, F. , Séguin, C. , Payment, P. and Trudel, M. (1987) Rapid virus subunit visualization by direct sedimentation of samples on electron microscope grids. J Virol Methods 16, 209–216. [DOI] [PubMed] [Google Scholar]
- Biel, S.S. and Gelderblom, H.R. (1999a) Diagnostic electron microscopy is still a timely and rewarding method. J Clin Virol 13, 105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel, S.S. and Gelderblom, H.R. (1999b) Electron microscopy of viruses In Virus Cell Culture ed. Cann A. pp. 111–147. Oxford: Oxford University Press. [Google Scholar]
- Biel, S.S. , Nitsche, A. , Kurth, A. , Siegert, W. , Özel, M. and Gelderblom, H.R. (2004) Detection of human polyomaviruses in urine from bone marrow transplant patients: comparison of electron microscopy with PCR. Clin Chem 50, 306–312. [DOI] [PubMed] [Google Scholar]
- Brenner, S. and Horne, R.W. (1959) A negative staining method for high resolution electron microscopy of viruses. Biochim Biophys Acta 34, 103–110. [DOI] [PubMed] [Google Scholar]
- Curry, A. , Appleton, H. and Dowsett, B. (2006) Application of transmission electron microscopy to the clinical study of viral and bacterial infections: present and future. Micron 37, 91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyrklaff, M. , Roos, N. , Gross, H. and Dubochet, J. (1994) Particle‐surface interaction in thin vitrified films for cryo‐electron microscopy. J Microsc-Oxford 175, 135–142. [Google Scholar]
- Geister, R. and Peters, D. (1963) Ein vereinfachtes direktes Zählverfahren für Virussupensionen ab 105 Partikel/ml. Z Naturforsch C 18b, 266–267. [Google Scholar]
- Gelderblom, H.R. (2006) Virus enrichment using the Airfuge for rapid diagnostic EM in infectious diseases. Rotor 4, 4–5. [Google Scholar]
- Gelderblom, H.R. , Renz, H. and Özel, M. (1991) Negative staining in diagnostic virology. Micron Microsc Acta 22, 435–447. [Google Scholar]
- Gelderblom, H. , Bannert, N. and Pauli, G. (2007) Arguments pro disinfection in diagnostic electron microscopy: a response to Madeley and Biel. J Infect 54, 307–308. [DOI] [PubMed] [Google Scholar]
- Hammond, G.W. , Hazelton, P.R. , Chuang, I. and Klisko, B. (1981) Improved detection of viruses by electron microscopy after direct ultracentrifuge preparation of specimens. J Clin Microbiol 14, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, J.R. (1997) Negative staining and cryoelectron microscopy: the thin film techniques Royal Society Handbooks, Vol. 35. Oxford, UK: BIOS Scientific. [Google Scholar]
- Harris, R. and Horne, R. (1991) Negative staining In Electron Microscopy in Biology: A Practical Approach ed. Harris J.R. and Horne R. pp. 203–232. Oxford: IRL Press. [Google Scholar]
- Hazelton, P.R. and Gelderblom, H.R. (2003) Electron microscopy for rapid diagnosis of infectious agents in emergent situations. Emerg Infect Dis 9, 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglesby, T.V. , O’Toole, T. , Henderson, D.A. , Bartlett, J.G. , Ascher, M.S. , Eitzen, E. , Friedlaender, A.M. , Gerberding, J. et al. (2002) Anthrax as a biological weapon, 2002. Updated recommendations for management. J Am Med Assoc 287, 2236–2252. [DOI] [PubMed] [Google Scholar]
- Johnsen, C.K. , Böttiger, B. and Blom, J. (2006) Confirmation of electron microscopy results by direct testing of viruses adhered to grids using nucleic acid amplification techniques. J Virol Methods 134, 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.P.C. and Gregory, D.W. (1993) Viruses accumulate spontaneously near droplet surfaces: a method to concentrate viruses for electron microscopy. J Microsc-Oxford 171, 125–136. [DOI] [PubMed] [Google Scholar]
- Katz, D. , Straussman, Y. and Shahar, A. (1984) A simplified microwell pseudoreplica for the detection of viruses by electron microscopy and immunoelectron microscopy. J Virol Methods 9, 185–192. [DOI] [PubMed] [Google Scholar]
- Kurth, A. , Achenbach, J. , Miller, L. , Mackay, I.M. , Pauli, G. and Nitsche, A. (2008) Orthopoxvirus detection in environmental specimens during suspected bioterror attacks: inhibitory influences of common household products. Appl Environ Microbiol 1, 32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, H.C. , Montagne, J.L. and Fauci, A.S. (2001) Bioterrorism: a clear and present danger. Nat Med 7, 1271–1273. [DOI] [PubMed] [Google Scholar]
- Lang, R.D.A. , Nermut, M.V. and Williams, L.D. (1981) Ultrastructure of sheep erythrocyte plasma membrane and cytoskeletons bound to solid supports. J Cell Sci 49, 383–399. [DOI] [PubMed] [Google Scholar]
- Laue, M. , Niederwöhrmeier, B. and Bannert, N. (2007) Rapid diagnostic thin section electron microscopy of bacterial endospores. J Microbiol Meth 70, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermann, H. (1978) Die Ultrazentrifugation in der Virusdiagnostik. Arch Exp Vet 32, 427–434. [PubMed] [Google Scholar]
- Madeley, C.R. and Biel, S.S. (2006) For debate: is disinfection of specimens, which may contain unknown or bio‐terrorist organisms, essential before electron microscopic examination. J Infect 53, 70–74. [DOI] [PubMed] [Google Scholar]
- Mathews, J. and Buthala, D.A. (1970) Centrifugal sedimentation of virus particles for electron microscopic counting. J Virol 5, 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.E. (1986) Detection and identification of viruses by electron microscopy. J Electron Microsc Tech 4, 265–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.E. (2003) Bioterrorism and electron microscopy differentiation of poxviruses from herpesviruses: does and don′ts. Ultrastruct Pathol 27, 133–140. [DOI] [PubMed] [Google Scholar]
- Miller, S.E. (2004) Surveillance of bioterrorism agents: considerations for EM laboratories. Micros Today 4, 56–59. [Google Scholar]
- Müller, G. (1969) Elektronenmikroskopische Partikelzählung in der Virologie. I. Zentrifugierröhrchen zur direkten Sedimentation von Viren auf Netzträger. Arch Gesamte Virusforsch 27, 339–351. [PubMed] [Google Scholar]
- Müller, G. , Nielsen, G. and Baigent, C.L. (1980) Electron microscopical particle counting in virology. III. Parameters governing the efficiency of sedimentation techniques and its improvement with the application of poly‐L‐lysine. Arch Virol 64, 311–318. [DOI] [PubMed] [Google Scholar]
- Müller, G. , Gelderblom, H. and Nielsen, G. (1981) Zur Effektivität von Airfuge und Poly‐L‐Lysin in der elektronenmikroskopischen Präparation von Virussuspensionen. GIT Fachzeitschrift Labor Suppl 25, 7–11. [Google Scholar]
- Nitsche, A. , Gelderblom, H.R. , Eisendle, K. , Romani, N. and Pauli, G. (2007) Pitfalls in diagnosing human poxvirus infections. J Clin Virol 38, 165–168. [DOI] [PubMed] [Google Scholar]
- Nolte, K.B. , Hanzlick, R.L. , Payne, D.C. , Kroger, A.T. , Oliver, W.R. , Baker, A.M. , McGowan, D.E. , DeJong, J.L. et al. (2004) Medical examiners, coroners and biological terrorism. A guidebook for surveillance and case management. Morb Mortal Wkly Rep 53, 1–36. [PubMed] [Google Scholar]
- Olson, K.B. (1999) Aum Shinrikyo: once and future threat? Emerg Infect Dis 5, 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissig, M. and Orrell, S.A. (1970) A technique for the electron microscopy of protein‐free particle suspensions by the negative staining method. J Ultrastr Res 32, 107–117. [DOI] [PubMed] [Google Scholar]
- Sachs, L. (1992) Angewandte Statistik. Berlin: Springer. [Google Scholar]
- Sapsford, K.E. , Bradburne, C. , Delehanty, J.B. and Mednitz, I.L. (2008) Sensors for detecting biological agents. Mater Today 11, 38–49. [Google Scholar]
- Shibata, Y. and Miyazaki, T. (2002) Anode glow discharge plasma treatment enhances calcium phosphate adsorption plates. J Dent Res 81, 841–844. [DOI] [PubMed] [Google Scholar]
- Sterlini, J.M. and Mandelstam, J. (1969) Commitment to sporulation in Bacillus subtilis and its relationship to development of Actinomycin resistance. Biochem J 113, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organisation (2003) A multicentre collaboration to investigate the cause of severe acute respiratory syndrome. Lancet 361, 1730–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]


