Abstract
TAF1/MRSX33 intellectual disability syndrome is an X-linked disorder caused by loss-of-function mutations in the TAF1 gene. How these mutations cause dysmorphology, hypotonia, intellectual and motor defects is unknown. Mouse models which have embryonically targeted TAF1 have failed, possibly due to TAF1 being essential for viability, preferentially expressed in early brain development, and intolerant of mutation. Novel animal models are valuable tools for understanding neuronal pathology. Here, we report the development and characterization of a novel animal model for TAF1 ID syndrome in which the TAF1 gene is deleted in embryonic rats using clustered regularly interspaced short palindromic repeats (CRISPR) associated protein 9 (Cas9) technology and somatic brain transgenesis mediated by lentiviral transduction. Rat pups, post-natal day 3, were subjected to intracerebroventricular (ICV) injection of either gRNA-control or gRNA-TAF1 vectors. Rats were subjected to a battery of behavioral tests followed by histopathological analyses of brains at post-natal day 14 and day 35. TAF1-edited rats exhibited behavioral deficits at both the neonatal and juvenile stages of development. Deletion of TAF1 lead to a hypoplasia and loss of the Purkinje cells. We also observed a decreased in GFAP positive astrocytes and an increase in Iba1 positive microglia within the granular layer of the cerebellum in TAF1-edited animals. Immunostaining revealed a reduction in the expression of the CaV3.1 T-type calcium channel. Abnormal motor symptoms in TAF1-edited rats were associated with irregular cerebellar output caused by changes in the intrinsic activity of the Purkinje cells due to loss of pre-synaptic CaV3.1. This animal model provides a powerful new tool for studies of neuronal dysfunction in conditions associated with TAF1 abnormalities and should prove useful for developing therapeutic strategies to treat TAF1 ID syndrome.
Keywords: TAF1, Intellectual disability syndrome, CRISPR/Cas9, X-linked disorder, cerebellum
Introduction
Mutations in TATA-box binding protein factor 1 (TAF1) protein are associated with a X-linked TAF1 intellectual disability (ID) syndrome (OMIM: 300966) (Hurst 2018; O'Rawe et al. 2015). Occurring in males, TAF1 ID syndrome (also known as mental retardation, X-linked, syndromic-33 disease, MRXS33) presents with abnormalities in global developmental (motor, cognitive, and speech), hypotonia, gait abnormalities, and cerebellar hypoplasia (Hurst 2018; O'Rawe et al. 2015). Exactly how mutations in TAF1 give rise to these neurological deficits remains unclear. TAF1 serves as a scaffold for the assembly of the transcription factor TFIID complex (Burley and Roeder 1996; Tora 2002). How the many functions of this protein complex are linked to the development of TAF1 ID syndrome is unknown. Before therapeutic options for the treatment of TAF ID syndrome can be evaluated, an animal model for this disease needs to be developed. Such a model should ideally recapitulate the major clinical features and disease progression reported in TAF1 ID patients.
We hypothesized that deletion of TAF1 by CRISPR/Cas9 editing, into the maturing brain during the post-natal days (Maries et al. 2003; Kornum et al. 2010; Gabery et al. 2012) using a lentiviral vector might replicate TAF1 ID symptoms. This mode of administration allows the vector diffusion and infection to occur before maturation of axon myelination and gliogenesis – two processes that restrict free diffusion in the brain as well as infection of the maturing cerebellum. Similar approaches have been utilized to develop a rat model of synucleinopathy (Aldrin-Kirk et al. 2014) and dystonia DYT1 (Fremont et al. 2017). We found that in neonatal rat pups, removal of TAF1 by CRISPR/Cas9 editing, results in defects in neonatal motor functions. Furthermore, the motor deficits were associated with loss of Purkinje cells in the cerebellum; the remaining Purkinje cells displayed abnormal firing frequencies. In addition, the motor defects persisted in TAF1-edited juvenile pups and were associated with morphologic abnormalities within the cerebellum.
Materials and Methods
Briefly, all biochemical, electrophysiology and behavior experiments were performed in a blinded fashion according to established protocols. Animal protocols were approved by the Institutional Animal Care and Use Committee of the College of Medicine at the University of Arizona and conducted in accordance with the Guide for Care and Use of Laboratory Animals published by the National Institutes of Health.
Animals
Pathogen-free, normal E18 pregnant Sprague-Dawley rats (Envigo Laboratories) were housed 1 per cage in temperature- (23 ± 3 °C) and light (12-h light/12-h dark cycle; lights on 07:00–19:00)-controlled rooms with standard rodent chow and water available ad libitum. The neonates are designated as post-natal day PD0 on the day of birth, the litter size range included in current study was 11-14 pups. For all behavior experiments, the experimenter was blinded to the treatments.
gRNA strategy for TAF1 gene targeting
Our strategy focused on targeting exon 1 of the TAF1 gene using a guide RNA (gRNA) as described previously (Moutal et al. 2017; Moutal, Cai, et al. 2018; Moutal, Sun, et al. 2018; Sandweiss et al. 2018). We identified the Taf1 gene at the position 66996430-67071877 of the Sprague dawley X chromosome (AC_000089.1). The exon 1 was located at the position 13..192 of this sequence and could also be found in the Taf1 mRNA sequence (NM_001191723.1). We targeted this exon to ensure total removal of the Taf1 protein. Using this approach, we expect minimal to none off-target activity of the Cas9 enzyme as we and others verified before (Moutal et al. 2017; Ma et al. 2017). The gRNA sequence (GTGTCTGACATGACGGCGGA, quality score 94, located at the position 65-84 within NM_001191723.1) was inserted into the restriction site of the pL-CRISPR.EFS.tRFP lentiplasmid (Cat#57819, Addgene, Cambridge, MA) (Heckl et al. 2014) a plasmid that allows for simultaneous expression of (i) the Cas9 enzyme; (ii) the gRNA; and (iii) a red fluorescent protein (tRFP) – to control for transduction efficiency. All plasmids were verified by Sanger sequencing (Eurofins, Louisville, KY). Lentiplasmids were packaged in lentiviruses by Viracore (USCF) at titers routinely above 107 infectious particles per ml.
Intracerebroventricular injections
All animal procedures were approved by the University of Arizona Institutional Animal Care and Use Committee and are in accordance with the NIH Guide for Care and Use of Laboratory Animals. Bilateral intracerebroventricular (i.c.v.) injections were performed as previously described in Sprague-Dawley (SD) rat pups on postnatal day 3 (Delenclos et al. 2017; Pang, Cai, and Rhodes 2003). Briefly, newborn SD rat puts were anesthetized by isoflurane. A 10 μl syringe (Hamilton Gastight Syringe, #1701) was used to pierce the skull (coordinates from bregma: −0.6 mm posterior, ± 1.75 mm lateral/medial, and −2.5 mm ventral), and 2.5 μl of CRISPR lentivirus (gRNA-control or gRNA-TAF1) was injected into each cerebral ventricle without opening the scalp. Neonatal rat pups were kept with parent until weaned.
Rats were sacrificed at set time points as follow: 14 days and 35 day of postnatal day. Of note, 14 day and 35 day groups were behaviorally assessed before being euthanized for electrophysiological and histological and protein expression analysis.
Cerebellar Slice Preparation
Ten to 14-day SD rat pups were used in the electrophysiological experiment. All animal procedures were carried out in accordance with the regulations of the Institutional Animal Care and Use Committee at the University of Arizona. The animals were decapitated after being deeply anesthetized with isoflurane, and their cerebellums were rapidly removed and placed in an ice-cold ACSF containing (in mM) 220 sucrose, 2.5 KCl, 1.25 Na2HPO4, 3.5 MgCl2, 0.5 CaCl2, 25 NaHCO3, and 20 D-glucose (with pH at 7.4 and osmolarity at 310 mOsm), bubbled with 95% O2 and 5% CO2. Parasagittal slices (320 μm thick) were cut using a VT 1200S vibratome (Leica, Germany). Slices were then incubated for at least 1 hour at 34°C in an oxygenated recording solution containing (in millimolar): 125 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 1.25 NaH2PO4, 26 NaHCO3, 25 D-glucose, with pH at 7.4 and osmolarity at 320 mOsm. The slices were then positioned in a recording chamber and continuously perfused with oxygenated recording solution at a rate of 1.5 to 3 mL/min before electrophysiological recordings at RT.
Whole Cell Patch-clamp Recording After incubation
Recordings were made from Purkinje Cells (PC) in lobules IV-VI, which were visually identified based on their location using infrared differential interference contrast video microscopy on an upright microscope (FN1; Nikon, Tokyo, Japan) equipped with a 3.40/0.80 water-immersion objective and a charge-coupled device camera. The pipettes were prepared by pulling glass capillaries on a model p-97 microelectrode puller (Sutter Instrument, Novato, USA). Patch pipettes had resistances of 3-5 MΩ. The internal solution was a K+-based solution containing (in mM): 120 potassium gluconate, 20 KCl, 2 MgCl 2, 2 Na2-ATP, 0.5 Na-GTP, 20 HEPES, 0.5 EGTA, with pH at 7.28 (with potassium hydroxide [KOH]) and osmolarity at 310 mOsm.
The whole-cell configuration was obtained in voltage-clamp mode. The membrane potential was held at −70 mV using PATCHMASTER software in combination with a patch clamp amplifier (EPC10; HEKA Elektronik, Lambrecht, Germany). To record spontaneous excitatory postsynaptic currents (sEPSCs), bicuculline methiodide (10 mM) was added to the recording solution to block γ-aminobutyric acid (GABA)-activated currents. Hyperpolarizing step pulses (5 mV in intensity, 50 milliseconds in duration) were periodically delivered to monitor the access resistance (15-25 MΩ), and recordings were discontinued if the access resistance changed by more than 20%. For each PC, sEPSCs were recorded for a total duration of 2 minutes. Currents were filtered at 3 kHz and digitized at 5 kHz. Data were further analyzed by the Mini-Analysis (Synatosoft Inc, NJ) and Clampfit 10.7 Program. When indicated 1μM TTA-P2 (T-type calcium channel blocker) (Choe et al. 2011) was added to the recording solution. The amplitude and frequency of sEPSCs were compared between neurons from the indicated groups. All recordings and analysis were done in a blinded fashion.
Neonatal Motor Test
At each testing day, the dams and the neonates were brought to the experimental room at the same time and left undisturbed for at least 1 h prior to testing.
Righting reflex test
This test evaluates the body’s tendency to regain its former body position after being displaced. The pups were placed on their backs on a flat surface (Yang et al. 2018). Their success or failure in repositioning themselves dorsal side up and the latency to falling off the platform were assessed over a 30 s period. For rats that failed to fall off, the time was recorded as 30 s. The measures were taken once every second day from PD5 to PD13.
Negative Geotaxis
This test evaluates motor coordination and vestibular sensitivity. Negative geotaxis was tested by placing the neonate on an inclined grid (at approximately 35μ inclination) with the head facing downward (Sukhanova et al. 2018). The grid had a rough surface to provide the neonate with a reasonable grip. We assessed the percentage of pups that successfully completed the test at each postnatal day and their latency to complete the geotaxis test in seconds (maximum 60 s). For those rats that failed to reorient themselves upward head, the time was recorded as 60 s. The measures were taken once every second day from PD5 to PD13.
Ambulation Test
Crawling is a behavior developed early in the rat pup between PND 0 - 5, at which point rats begin to transition to walking, from 5 - 10 days old (Feather-Schussler and Ferguson 2016). Ambulation can, however, be scored throughout the lifetime of a rat and can be determined at any age. As there is no potential for learning, the ambulation test can be repeated as many times as needed through the course of the experiment.
1. Place rats in a clear enclosure where rats are visible from the top as well as the side. Use gentle prodding by touching the pup's tail to motivate the pup to walk.
2. Score ambulation for 3 min using the following scale: 0 = no movement, 1 = crawling with asymmetric limb movement, 2 = slow crawling but symmetric limb movement, and 3 = fast crawling/walking.
Hind-limb Suspension Test
The hind-limb muscle strength, weakness, and fatigue in rat neonates was evaluated using the method previously described by EL-Khodor and co-workers (El-Khodor et al. 2008). This test also assesses general neuromuscular function and posture. In each trial, we placed the pups head-down, hanging by its hind limbs in a plastic 50 ml centrifuge tube with a cotton ball cushion at the bottom to protect the animal’s head upon its fall. The test was assayed over a 40 s period. The hind-limb score that assessed the positioning of the legs. The posture adopted by the pups was scored according to the following criteria: a score of ‘4’ indicated normal hind-limb separation; a score of ‘3’ indicated that the hind limbs were closer together than normal, without touching; a score of ‘2’ specified closer proximity of the hind limbs to each other, often touching; and a score of ‘1’ represented touching or clasping of the hind limbs. A score of 1 was given to a pup that failed to maintain its grasp of the tube. The score is an overall evaluation of the hind-limb position during the first 10 s of hanging onto the lip of the tube. The measures were taken once every second day, from PD5 to PD13.
Tail Suspension Test
The pups were suspended by their tails for 5-s and their hind-limb postures were scored 1–4, as defined above, using the modified method (Kim et al. 2017). The measurements were taken once every second day, from PD5 to PD13.
Juvenile Motor Test
After weaning the rat pups were separated from the dams and maintained 4 per cage. At each testing day, rats were brought to the experimental room at the same time and left undisturbed for at least 1 h prior to testing.
Open Field Test
The apparatus (W100 cm×D100 cm×H40 cm) is made of wood. The floor of this chamber was divided into 25 cm (5 × 5) squares. The rat pups were placed into one comer of an open field chamber and their behavior was observed for 5 min. Number of grooming; i.e. consisting of licking the fijr, washing face or scratching behavior.
Beam Walking Test
Animals were allowed to walk on a narrow flat stationary wooden beam (L100 cm×W2 cm) placed at a height of 100 cm from the floor to reach an enclosed escape platform. The time taken to cross the beam from one end to the other and beam walking score were also given as described previously (Rajasankar, Manivasagam, and Surendran 2009).
Hematoxylin & Eosin Staining
After dehydration of the tissue with 30% sucrose, twenty micron (20 μm) sections were cut, stained with hematoxylin and eosin (H&E) dye (Sigma), and mounted with Richard-Allan scientific mounting medium for microscopy.
Nissl Staining
After dehydration of the tissue with 30% sucrose, twenty-micron (20 μm) sections were cut, stained with cresyl violet dye (Sigma), and mounted with Richard-Allan scientific mounting medium for microscopy.
Immunohistochemical staining
Table 1 lists the antibody used in our studies. Rats were (n = 3–5 per genotype) were perfused with 4% formaldehyde in Phosphate buffer saline (PBS) at PD 14 day and PD 35 day of age, and the brains were extracted and post-fixed in 4% paraformaldehyde for 8 h at 4°C. Cerebellar sections were cut sagittally at 20 μm using a cryostat (Microm HM 505 E). After rinsing the sections in PBS for 5 min, the sections were incubated with a 0.1% H2O2 solution in PBS for 5 min, rinsed in PBS, and incubated for 30 min with 0.4% Triton X-100, rinsed in PBS and blocked with 8% goat serum, and 1% Triton X-100 in PBS. After blocking, the sections were incubated for overnight with anti-TAF1 (1:250, Millipore), anti-tRFP (1:250, Evrogen), anti-CaV3.1 (1:250 Alomone labs) antibody, anti-calbindin D-28 K polyclonal antibody (1:250, Millipore) and anti-β-III-tubulin (1:250, Millipore), anti-GFAP antibody rabbit polyclonal (1:250, Dako), Iba1 (1:250,Wako) diluted in 4% goat serum in PBS. The sections were washed in 1% goat serum in PBS, incubated with secondary antibody (anti-rabbit Alexa fluor 488 (TAF1, CaV3.1, GFAP, Iba1, and Calbindin) & anti-rabbit Alexa fluor 594 (tRFP)) in 4% goat serum for 2 h, washed, and incubated with DAPI for 5 mins. Sections were then washed and further air dried, and cover slipped with Richard Allan Scientific Mountant Medium (Thermo). All procedures were performed at room temperature. The expression of TAF1, tRFP, GFAP, and Iba1 were observed under a fluorescence microscope (LSM510, Carl Zeiss) using a 20x objective. In regards to TAF1, we conducted several immunofluorescence control tests. a) tissue sections were incubated with the secondary antibody only; b) tissue sections were incubated with antibody diluent with a nonimmunoglobulin of the same isotype (isotype control); c) samples were incubated with primary antibody only. These control experiments were all negative and demonstrated the specificity of the TAF1 antibody (Fig. S2). Similar control experiments were performed for the other antibodies used in this study (data not shown).
Table 1.
List of primary and secondary antibodies used in the study.
| Primary antibodies | ||||||
|---|---|---|---|---|---|---|
| Anti- | Abbreviation | Host | Company | Catalog number |
Dilutiona | |
| TATA-box binding protein factor 1 | TAF1 | Rabbit | Sigma Aldrich | AV32473 | 1:500 | |
| Calbindin-D28k | Calbindin | Rabbit | Sigma Aldrich | C2724 | 1:250 | |
| Low-Voltage-Activated CaV3.1 Calcium Channels | CaV3.1 | Rabbit | Almone labs | ACC-021 | 1:250 | |
| Glial fibrillary acidic protein | GFAP | Rabbit | Dako | Z0334 | 1:250 | |
| Ionizing calcium-binding adaptor molecule 1 | Iba1 | Rabbit | Wako Labs | 019-19741 | 1:250 | |
| Red fluorescent protein TagRFP | tRFP | Rabbit | Envitrogen | AB233 | 1:250 | |
| Tublin | Tublin | Mouse | Promega | G7121 | 1:250 | |
| Secondary antibodies | ||||||
| Anti- | Host | Label | Dilution | Company | ||
| Rabbit IgG | Goat | Alexa 488 | 1:250 | Life technologies | ||
| Rabbit IgG | Goat | Alexa 594 | 1:250 | Promega | ||
| Mouse IgG | Goat | Alexa 594 | 1:250 | Promega | ||
Dilutions were from original stocks supplied by the vendors
Immunofluorescence quantification
For TAF1 and CaV3.1, we quantified the fluorescence intensity. Briefly, the fluorescence intensity was measured in 9 different fields from 3 different animals in each experimental condition at 20X magnification. The relative fluorescence intensity was analyzed by Image J software (Pitzer et al. 2015;Guirado et al. 2018).
Morphometric analysis
Nucleated Purkinje cells in the anterior lobe (lobule I-III) of the cerebellar cortex in every fifth sections were counted using Icy software, and Purkinje cell linear density was calculated by dividing the number of cells by the linear distance of the Purkinje cell layer per section (Yamashita et al. 2011). We measured the thickness of the granular layer using Nissl stained slides and Icy software. In short, the mean granular layer thickness (mm) was determined in 9 fields from 3 different animal per experimental condition (El-Tahawy et al. 2018). We counted the number of GFAP+ and Iba1+ positive cells within the granular layer from 9 fields from 3 different animals per experimental condition and the mean number of GFAP+ and Iba1+ cells were enumerated as previously described (El-Tahawy et al. 2018).
Sanger Sequencing
The presence of mutation after CRISPR-Cas 9 editing was verified by Sanger Sequencing as previously described by our group (1). Briefly, a 508 bp fragment was amplified from genomic DNA using primers surrounding the PAM sequence in exon 1 of the TAF1 gene (Forward 5’-TCTGCCGCTCTTACCATAGC-3’ and Reverse 5’- AAACGCCCTCTATTACCGGC-3’). DNA sequencing was carried out on an Applied Biosystems 3730XL DNA Analyzer (Foster City, CA) in the UAGC Genomics Core Facility. The sequencing data was loaded into the ABI Sequence Scanner Software v1.0 for further analysis and genotype calling. Three different animals per each experimental condition were sequenced. All sequence traces were also manually reviewed to ensure the reliability of the data.
Statistics
All experiments were performed at least twice and in a blinded fashion. Data were analyzed with the software Prism 8 (GraphPad). Group differences were calculated using ANOVA followed by Tukey’s post hoc test as indicated in the figures. P values smaller than 0.05 were considered significant. Error bars in the graphs represent mean ± SEM. For TAF1 IHC quantification between naïve and gRNA-TAF1 edited animals, the data was analyzed by Student’s t-test with 0.05 considered significant.
Results
Design of CRISPR/Cas9 strategy for TAF1 gene editing in the CNS
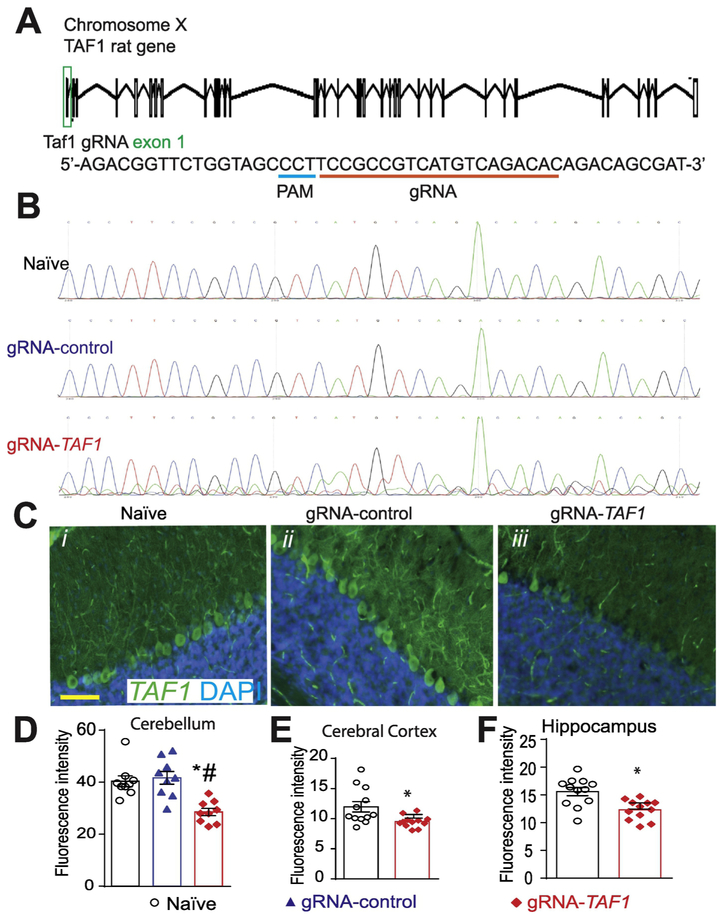
We designed a clustered regularly interspaced short palindromic repeats (CRISPR) associated protein-9 nuclease (Cas9) approach to induce a double stranded break in Exon 1 of the TAF1 gene (Fig. 1A). The primary reasons for selecting this region of the gene is that patients with TAF1 variants present with loss of function abnormalities (O'Rawe et al. 2015; Hurst 2018). Lentiviral particles were generated and were injected intracerebroventricularly (i.c.v.) into the brain of neonatal rats. Injection sites were verified with Evans blue dye (Fig. S1). We amplified a 508 bp fragment surrounding the PAM sequence within the rat TAF1 gene from brain tissue from 3 different animals in each treatment group. While there were no sequence abnormalities detected in the naïve and gRNA control groups, we found obvious evidence of mutations within the TAF1 gene near the PAM sequence in DNA isolated from TAF1 gRNA transduced animals (Fig. 1B). Using an antibody specific for TAF1, we observed reduced TAF1 protein expression in the cerebellum (Fig. 1C i-iii & 1D), cerebral cortex, and hippocampus (Fig. 1E & 1F);(see also Supplementary Fig. S2 & S3). These data indicate successful genome editing of the TAF1 gene resulting in reduced expression of TAF1 protein expression.
Figure 1. Scheme of guide RNA and PAM sequence targeting exon 1 in the rat TAF1 gene.
(A) Schematic showing exon/intron organization of the TAF1 gene in rat. To disrupt TAF1, we designed a gRNA targeting exon 1. We used a guide RNA (gRNA) targeting a non-coding region in exon 1 of the TAF1 gene. Both gRNAs are based on the reverse strand sequence and are indicated on the figure by the red line oriented from 5’ to 3’. The protospacer adjacent motif (PAM sequence) is indicated by a blue line. The gRNA pairs with its DNA target followed by a 5’NGG sequence (PAM sequence). Cas9 catalyzes a double-stranded cleavage on the genomic DNA 3 bp before the PAM sequence. Nucleotide positions are indicated based on the DNA sequence on the TAF1 gene. (B) Representative sequences (around the PAM sequence) of Exon 1 within the TAF1 gene in naïve, gRNA-control, and gRNA-TAF1 edited animals. Note the indel mutation(s) in the sequence of DNA isolated from the TAF1 edited animals. These experiments were performed with three different animals per each experimental condition. (C) Representative immunohistochemistry images of cerebellar Purkinje cells stained with an antibody against TAF1. Reduced TAF1 expression was observed in the TAF1-edited rat pups compared to control editing or naïve group of rat pups. Quantification of fluorescence intensity for the TAF1 immunohistochemistry in the (D) cerebellum, (E) cerebral cortex and (F) hippocampus. These experiments were performed with 4 male animals per each experimental condition in an investigator blinded manner. Scale bar = 200 μm. Data are shown as mean ± S.E.M. from 3 different fields from 4 animals per experimental condition (i.e. a total of twelve fields per group). *p<0.05 versus; naïve, #p<0.05 versus gRNA-control (ANOVA followed by Tukey’s test).
CRISPR/Cas9-mediated TAF1 deletion cause TAF1 ID-like motor deficits
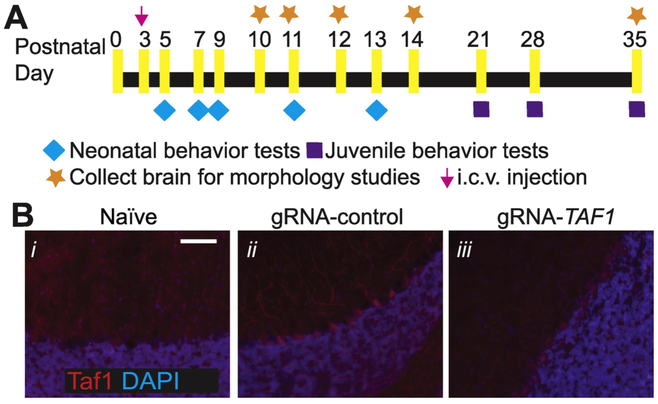
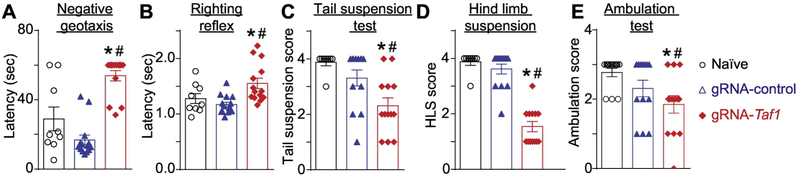
Motor dysfunction is a characteristic of TAF 1D syndrome (O'Rawe et al. 2015; Hurst 2018). Therefore, we performed a battery of neonatal motor tests to ascertain possible motor deficits between TAF-edited rat pups and control animals (Fig. 2A). Red fluorescent protein (RFP) staining indicated that cells within the cerebellum including Purkinje cells were transduced but not naïve animals (Fig. 2B i-iii). Negative geotaxis refers to the orienting response and movement expressed in opposition to cues of a gravitational vector (Feather-Schussler and Ferguson 2016). We observed that TAF1-edited pups displayed increased latency time in correcting their position relative to controls (Fig. 3A). The righting reflex evaluates the motor ability for a rat pup to flip onto its feet from a supine position (Feather-Schussler and Ferguson 2016). Similar to the negative geotaxis test, TAF1-edited pups showed an increased latency time to “right” their position (Fig. 3B). The tail suspension test is commonly used to measure stress and can also be used to assess motor alterations (i.e. motor coordination). In the tail suspension, the naïve and gRNA-control animals held their hind limbs wide apart from PD5 to PD13, whereas the TAF1-edited rats exhibited abnormal hind-limb clasping after PD7 (Fig. 3C). The hind-limb suspension test assesses the proximal hind limb muscle strength, weakness, and fatigue in the animal. The hind-limb suspension test also evaluates general neuromuscular function, body muscle strength, and posture. It is designed specifically for neonates and can be used on animals PD 2-14 (El-Khodor et al. 2008). TAF1-edited rat pups showed hind-limb weakness as demonstrated by a marked decrease in hanging score compared to control animals (i.e. gRNA-control and naïve rat pups at the same age) (Fig. 3D). Crawling is a behavior developed early in rodent pups between postnatal day (PD) 0-5, at which point pups start to transition to walking between 5-10 days old (Feather-Schussler and Ferguson 2016). Thus, the ambulation test takes advantage of this transitional time course. We noted significant differences in ambulation, at PD 14, between the TAF1-edited rat pups compared to gRNA-control and untreated (naïve) rat pups (Fig. 3E). Collectively, these data indicate motor dysfunction in CRISPR/Cas9- TAF1-edited treated rat pups.
Figure 2. Experimental design for these studies and localization of lentiviral transduction.
(A) Experimental design for neonatal and juvenile motor tests and histopathology studies. (B) Representative immunohistochemistry images of cerebellar Purkinje cells stained with an antibody against red fluorescent (RFP) protein. Images were obtained from slices of rat pups un-injected (naïve) or injected (with gRNA-control or gRNA-TAF1) at 35 days post-injection. These experiments were performed with 4 male animals per each experimental condition in an investigator blinded manner. Scale bar = 200 μm.
Figure 3. Behavioral assessment of neonatal motor functions.
The TAF1-edited rats showed increased latency time in negative geotaxis (A) and righting reflex test (B). Similarly, the TAF1-edited rat pups demonstrated decreased scores in the tail suspension (C), hind limb suspension (D), and ambulation (E) tests. Data are shown as mean ± S.E.M., n = 14 per condition. *p<0.05 versus; naïve, #p<0.05 versus gRNA-control (ANOVA followed by Tukey’s test). The experiments were conducted in an investigator blinded manner.
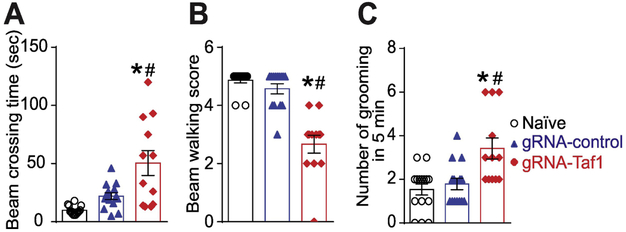
We continued our behavioral assessment past the weanling stage. We performed the beam walking test and open field test. The beam walking test can evaluate motor coordination and balance (Rajasankar, Manivasagam, and Surendran 2009). The beam crossing time (Fig. 4A) and number of foot slips errors were higher in the TAF1-edited animals at PD35 compared to controls (Fig. 4B). In the open field test, TAF1-edited animals displayed a higher level of anxiety as inferred from increased grooming frequency (Fig. 4C). Thus, TAF1-editing produces behavioral defects that appear to persist into the juvenile animals.
Figure 4. Behavioral assessment of juvenile motor functions.
The TAF1-edited animals showed behavior deficits compared to naïve and gRNA-control group animals. The TAF1-edited showed increased beam waking time (A) and had a decreased beaming score (B) in beaming walking tests relative to naïve and gRNA-control rats. (C) Grooming was increased in the TAF1-edited rats compared to the other groups. Data are shown as mean ± S.E.M., n = 5-8 per experimental condition. *p<0.05 versus; naïve, #p<0.05 versus gRNA-control (ANOVA followed by Tukey’s test). The experiments were conducted in an investigator blinded manner.
TAF1 deletion leads to cerebellar abnormalities
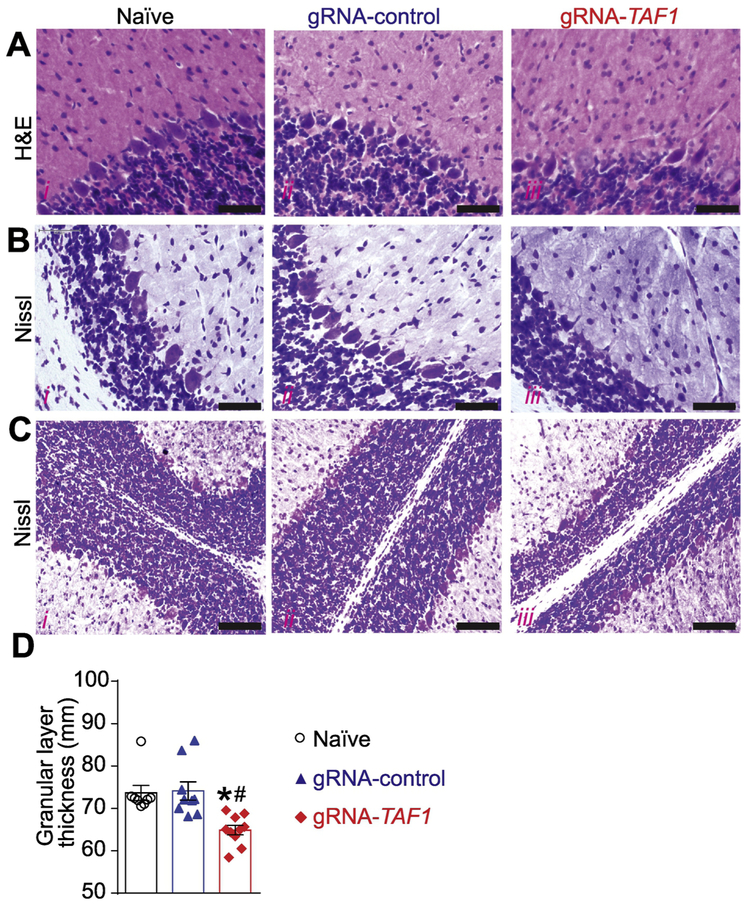
To examine a possible cellular basis for the behavioral defects, next we evaluated the morphology of the Purkinje cells by Hematoxylin and eosin (H&E) and Nissl staining. Our histopathology analysis showed that the architecture of the Purkinje cell layer was abnormal in TAF1-edited animals. In addition, the histopathology showed Purkinje cells were markedly hypoplastic in TAF1 treated animals relative to controls (Fig. 5A and 5B i-iii). Next, we assessed the thickness of the granular layer. We observed a decreased in the thickness of the granular layer in TAF-1 edited rats compared to control animals (Fig. 5C&D).
Figure 5. TAF1 gene editing results in morphologic changes in the cerebellum loss.
The morphology of the Purkinje cell layer was evaluated by Haemotoxylin and Eosin (A) and Nissl (B) Staining. Both H&E and Nissl staining showed abnormal architecture of the Purkinje cells as well as hypoplastic Purkinje cells. Scale bars: 500 μm. (C) The thickness of the Granular layer was evaluated by Nissl Staining. Nissl staining showed reduced granular layer thickness in gRNA-TAF1 group. Scale bars: 200 μm. (D) Summary of morphometric analysis of the Granular layer thickness. Data are shown as mean ± S.E.M., n = 9 fields per animal, 3 animals per experimental condition. *p<0.05 versus; naïve, #p<0.05 versus gRNA-control (ANOVA followed by Tukey’s test). The experiments were conducted in a blinded fashion.
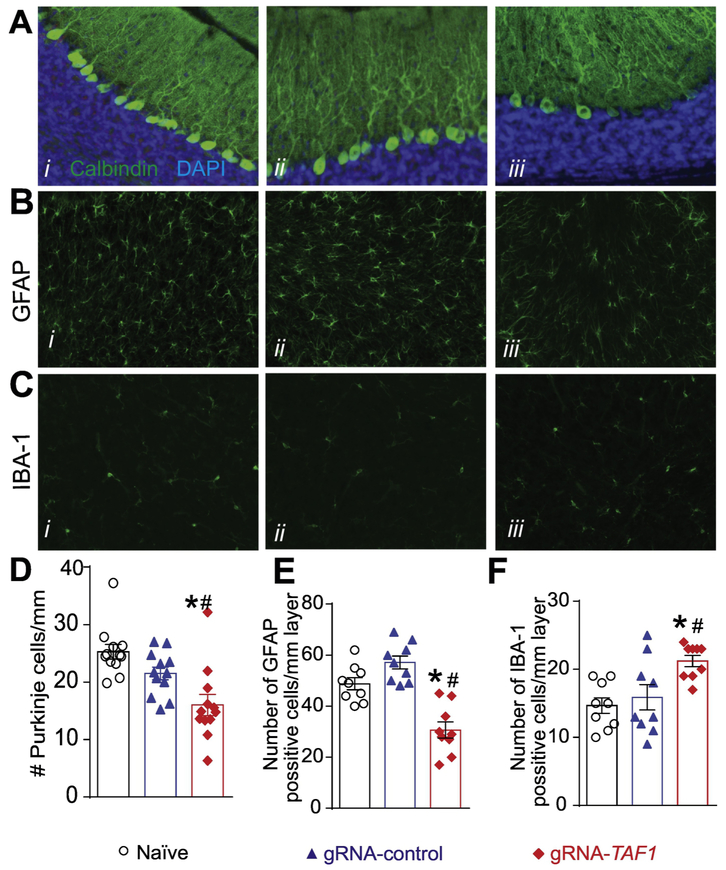
Purkinje cells can be identified by a Calbindin staining. Calbindin was abundant in the cell body, axon and dendritic tree of the naïve and gRNA-control animals, however some of the Purkinje cells in the TAF1-edited animals did not stain as positive as the control group animals (Fig. 6A i-iii). We also observed a decrease in the number of Purkinje cell in TAF1-edited animals compared to control animal (Fig. 6D).
Figure 6. TAF1 gene editing leads to changes in the granular layer of the cerebellum.
(A) Expression of calbindin was decreased in TAF1-edited animals as compared to naïve and CRISPR-control groups. Note also a decrease in the number of Calbindin positive Purkinje cells in TAF1-edited animals compared to control animals. B) Expression of GFAP (astrocytes marker) was decreased in TAF1-edited animals as compared to naïve and CRISPR-control groups. Note also a decrease in the number of GFAP positive cells in TAF1-edited animals compared to control animals. (C) Expression of Iba1 (microglia marker) was increased in TAF1-edited animals as compared to naïve and CRISPR-control groups. Note also increase in the number of Iba1 positive cells in TAF1-edited animals compared to control animals. (D) Summary of the number of Purkinje cells per linear density in each of the experimental conditions. Data are shown as mean ± S.E.M., n = 12 fields per animal, 4 animals per experimental condition. *p<0.05 versus; naïve, #p<0.05 versus gRNA-control (ANOVA followed by Tukey’s test). (E) and (F) Summary of the number of GFAP positive cells and Iba1 positive cells per linear density in each of the experimental conditions. Data are shown as mean ± S.E.M., n = 9 fields per animal, 3 animals per experimental condition. *p<0.05 versus; naïve, #p<0.05 versus gRNA-control (ANOVA followed by Tukey’s test). The experiments were conducted in an investigator blinded manner.
Glial cells, including astrocytes, oligodendrocytes, and microglia are by far the most abundant cells in the nervous system and located in the granular layer cerebellum (Domingues et al. 2016). Because the loss of Purkinje cells in TAF1 edited animals is not sufficient to explain the cerebellar hypoplasia found in patients, we examined whether TAF1 editing affected the thickness and cell content of the granular layer. We used glial fibrillary acidic protein (GFAP) as a marker for mature astrocytes in the granular layer. we found a reduced number of GFAP-positive cells in TAF1-edited animals compared to control animals (Fig. 6B &E). Next, we stained for Iba1, which is a marker for microglia. We detected an increases number of Iba1-positive cells in the granular layer of TAF1-edited rats compared to controls (Fig. 6C &F). Collectively, these findings imply that editing TAF1 alters the organization of the cerebellum during postnatal development.
TAF1 deletion leads to cerebral cortex abnormalities
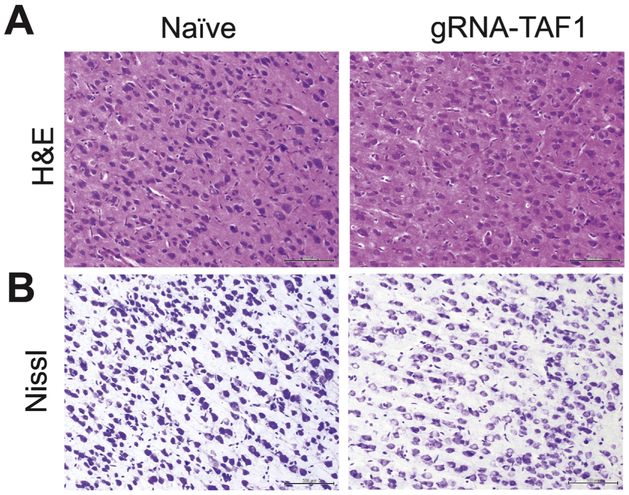
We also assessed other regions within the brain for morphologic abnormalities. There were not obvious morphologic differences between control and gRNA-TAF1 edited animals in other brain regions (i.e. frontal cortex, striatum, hippocampus, substantia nigra and pons);(see also Supplementary Fig. S4) with the exception of the cerebral cortex. In the cerebral cortex, neural cells in the control group were arranged tightly with regular shape and had an intact cell structure. The blue Nissl bodies in neural cells were visible and distinct. After TAF-1 editing, cerebral the morphology of the cells in the cortex clearly changed. The shape of the cells changed from triangular to rounded and a large number of cells were swollen. Also, the cells did not stain as distinctly as control cells with the H&E stain and Nissl dye (Fig. 7A&B).
Figure 7. TAF1 gene editing leads to abnormalities in the cerebral cortex.
The morphology of the neurons in the cerebral cortex was evaluated by Haemotoxylin and Eosin (A) and Nissl (B) Staining. Both H&E and Nissl staining showed the cell shape changed from triangular to rounded and a large number of cells were swollen in gRNA-TAF1 edited animals as compared to Naïve animals. Scale bars: 500 μm. The experiments were conducted in a blinded fashion.
CRISPR/Cas9-mediated TAF1 deletion is associated with reduced CaV3.1 protein expression
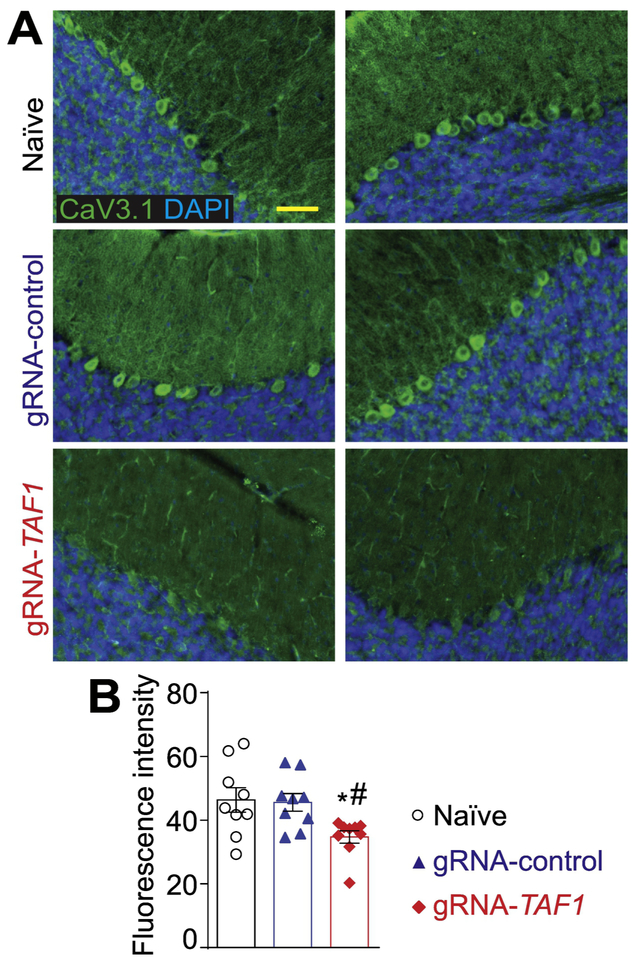
T-type calcium (Ca2+) channels play a key role in regulating membrane excitability in the brain (Aguado et al. 2016; Perez-Reyes 2003; Iftinca and Zamponi 2009). In the cerebellum the T-Type channel, CaV3.1, is expressed in Purkinje cells and appears to be involved in calcium influx (Asmara et al. 2017). For our immunofluorescence studies, we used a CaV3.1 antibody that had been previously utilized for CaV3.1 localization in other studies (Fernandez et al. 2015; Hurtado, Bub, and Herzlinger 2014). In addition, control adsorption experiments demonstrated the specificity of the CaV3.1 antibody on rat brain tissue sections (Supplementary Fig. S5C). Because we previously reported altered Ca2+ entry and reduced protein expression of CaV3.1 in TAF1 depleted SH-SY5Y cells (Hurst 2018), we investigated if CaV3.1 expression was altered after TAF1 editing in vivo. We observed comparable levels of expression of CaV3.1 in Purkinje cell slices from both naïve animals and gRNA-control injected animals, whereas CaV3.1 expression was markedly reduced in TAF1-edited animals at post-natal day 14 (data not shown) and at post-natal day 35 (Fig. 8A&B).
Figure 8. TAF1 gene editing leads to a downregulation of the CaV3.1 voltage-gated calcium channel.
(A) Representative photomicrographs of cerebellar slices from animals injected with (vehicle) or control or TAF1 gRNAs stained with an antibody against CaV3.1. Nuclei stained with DAPI. (B) Quantification of fluorescence intensity for the CaV3.1 immunohistochemistry. Scale bar = 200 μm. The experiments were performed in a blinded fashion. Data are shown as mean ± S.E.M. from 3 different fields from 3 animals per experimental condition (i.e. a total of nine fields per group). *p<0.05 versus; naïve, #p<0.05 versus gRNA-control (ANOVA followed by Tukey’s test). The experiments were conducted in an investigator blinded manner.
CRISPR/Cas9-mediated TAF1 deletion leads to Purkinje cells excitatory neurotransmission abnormalities
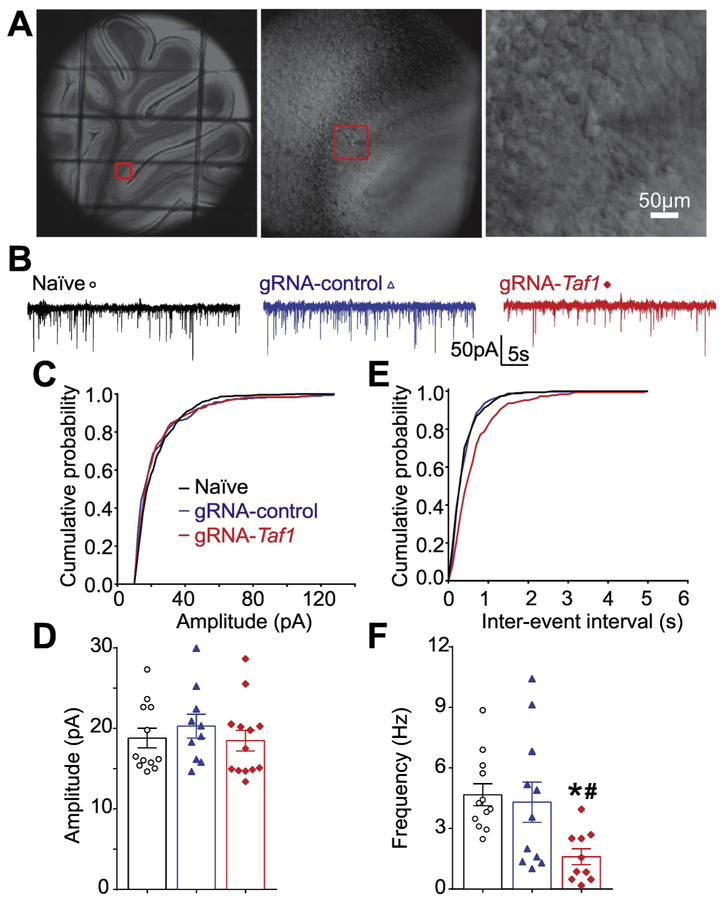
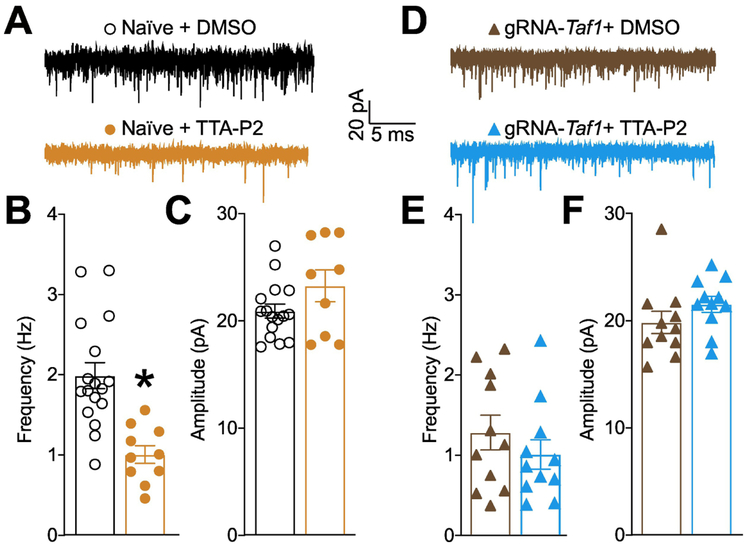
TAF1 editing decreased the levels of the pre-synaptic voltage gated calcium channel CaV3.1. In the cerebellum, CaV3.1 is expressed at the parallel fiber synapse and is involved in the delivery of the action potentials to the Purkinje neurons (Hildebrand et al. 2009; Yunker et al. 2003; Ly et al. 2013). To examine whether TAF1 editing can impair the neurotransmission at the level of the parallel fiber to purkinje synapse, we performed synaptic recordings of the Purkinje neurons in cerebellar slices (Fig. 9A) using whole-cell patch clamp, specifically measuring spontaneous excitatory postsynaptic currents (sEPSCs) (Fig. 9B). There was no change in amplitude of sEPSCs between the three groups (Fig. 9B-D). However, the frequency of sEPSCs was significantly decreased with TAF1-editing, compared with naïve or gRNA-control editing, respectively (Fig. 9F) (1.61 ± 0.40 Hz vs 4.67 ± 0.54 Hz, *P<0.05 vs Naïve; 1.61 ± 0.40 Hz vs 4.30 ± 1.00 Hz, #P<0.05 vs gRNA-control). To confirm that these effect were reflecting the loss of pre-synaptic CaV3.1 after Taf1 editing, we used the T-type calcium channel inhibitor TTA-P2 (Choe et al. 2011). In naïve rats, inhibition of CaV3.x channels (Fig. 10A) resulted in a decrease of the frequency (Fig. 10B) but not amplitude (Fig. 10C) of sEPSC thus showing that (i) T-type channels have an exclusive pre-synaptic function in neurotransmission to Purkinje cells and (ii) T-type channel inhibition recapitulates the phenotype observed after Taf1 editing (Fig. 9). Next, we used TTA-P2 on Taf1-edited rat cerebellum to measure whether the reduction of the frequency of sEPSC after Taf1 editing (Fig. 10D) was dependent on the loss of CaV3.1 function. TTA-P2 failed to further decrease the frequency of sEPSC in Taf1 edited rats (Fig. 10E) thus showing that all T-type calcium channel is non-functional in these animals. Similarly to naïve rats, TTA-P2 had no effect on sEPSC amplitude (Fig. 10F). These results confirm a presynaptic effect on excitatory neurotransmission on cerebellar Purkinje cells with TAF1-editing.
Figure 9. TAF1 gene editing leads to a decrease in frequency of spontaneous excitatory post synaptic current in cerebellar Purkinje cells.
Photomicrograph of cerebellar slice preparation with a progressive zoom with the rightmost panel showing positioning of the recording electrode to this region (A). Representative recording traces of cells from the indicated groups (B). The cumulative probability of amplitude (C) and inter-event interval (E). Summary of amplitudes (D) and frequencies (F) of sEPSCs for the indicated groups are shown. Data are shown as mean ± S.E.M., n = 12 Purkinje cells from at least 2 animals per experimental condition. *p<0.05 versus; naïve, #p<0.05 versus gRNA-control (ANOVA followed by Tukey’s test). The experiments were conducted in an investigator blinded manner.
Figure 10. T-type calcium channels control cerebellar sEPSC frequency and are inhibited after Taf1 editing.
(A) Representative recording traces of cells from naïve rats treated with 1μM TTA-P2 or 0.1% DMSO (vehicle). Bar graph with scatter plot showing the (B) frequency and (C) amplitude of sEPSCs for the indicated groups. Data are shown as mean ± S.E.M., n = 10-16 Purkinje cells from at least 3-4 animals per experimental condition. *p<0.05 versus; vehicle (Student’s t test). (D) Representative recording traces of cells from Taf1 edited rats treated with 1μM TTA-P2 or 0.1% DMSO (vehicle). Bar graph with scatter plot showing the (E) frequency and (F) amplitude of sEPSCs for the indicated groups. Data are shown as mean ± S.E.M., n = 11 Purkinje cells from at least 3-4 animals per experimental condition. *p<0.05 versus; vehicle (Mann-Whitney). The experiments were conducted in an investigator blinded manner.
Discussion
Mutations in TAF1, which is a component of the TFIID transcription factor complex, are implicated in an X-linked genetic syndrome that afflicts males (O'Rawe et al. 2015). Some of the clinical features associated with TAF1 ID syndrome include: delayed gross motor development, hypoplasia of the cerebellar vermis, and hypotonia (O'Rawe et al. 2015; Hurst 2018). By deleting the TAF1 gene in postnatal rats, we generated an animal model that replicates the clinical features of TAF1 ID syndrome. We observed motor deficits in both young rats and juveniles, morphological changes and a loss in Purkinje cells in the TAF1-edited animals. As in humans, we noted hypoplasia of the cerebellum, a pathological hallmark of dysfunction in TAF1 ID syndrome. Our model underscores the relevance of the cerebellum to this neurodegenerative disease and offers a powerful platform for examination of the cellular and molecular pathways underpinning TAF1 ID syndrome.
Our model of TAF1 ID allowed us to explore how cerebellar dysfunction contributes to disease. Two different pathways, parallel fibers (PF) from granule cells (GC) and climbing fibers (CF) from the cells in the inferior olive (I.O), project excitatory synaptic input to Purkinje neurons (Lee et al. 2018). The Purkinje neurons integrate these excitatory inputs and then send inhibitory GABAergic projections to the deep cerebellar nuclei (DCN) (D'Angelo 2018). In Purkinje cells, we found that the frequency of sEPSCs was significantly decreased after TAF1-editing, compared with naïve or control CRISPR. These results show that TAF1 editing impairs the presynaptic glutamatergic transmission on cerebellar Purkinje cells. This might lead to a disinhibition of the Purkinje neurons resulting in our observations of loss of motor activity and fine motor control. We found that the organization of Purkinje cell layer and morphology of Purkinje neurons are altered during postnatal development in TAF1-edited rats. We infer that the decrease of frequency of sEPSCs might result from the collapse of the cerebellar circuit. Additionally, the decreased density of Purkinje cells after TAF1-editing aggravates these effects and may be responsible for the decline in motor function.
T-type calcium channels are low voltage-activated calcium channels that transiently open to evoke tiny Ca2+ currents (reviewed in (Perez-Reyes 2003)). T-type calcium channels regulate calcium influx from the extracellular region by opening the calcium channel (Cazade et al. 2017), or activating calcium-induced calcium release from the internal calcium source (Kitchens, Burch, and Creazzo 2003; Coulon et al. 2009). These results suggest a critical role for T-type calcium channels in regulating intracellular calcium homeostasis and maintaining cellular function (Assandri et al. 1999; Chemin et al. 2000; Cazade et al. 2017). CaV3.1 T-type channels are abundant at the cerebellar synapse between parallel fibers and Purkinje cells where they contribute to synaptic depolarization (Ly et al. 2013). Consistent with our previous in vitro findings (Hurst 2018), we observed a decrease in CaV3.1 expression in Purkinje cells in the present study. Because CaV3.1 appears to be required for neuronal progenitor cell viability and the primary pathway by which Ca2+ enters into cerebellar neurons (Kim et al. 2018; Ly et al. 2013), our results suggests that dysregulated calcium signaling, due to decreased CaV3.1 protein levels caused by TAF1 gene knockout, could be important in the pathogenesis of TAF1 ID syndrome.
The granular layer of the cerebellum is composed of astrocytes, oligodendrocytes, and microglia (reviewed in (Domingues et al. 2016)). Collectively, these cells support the neurons within the cerebellum. Specifically, astrocytes provide physical support for neurons. Astrocytes also secrete gliotransmitters and growth factors like BDNF, required for synaptic plasticity, synaptogenesis, and neurogenesis (Henneberger et al. 2010; Ying et al. 2002). Thus, a reduced density of GFAP+ cells in the TAF1 edited animals may affect the survival and function of the Purkinje cells. The microglia are the primary immune cells within the CNS. In the present study, we observed an increase in Iba1 positive cells within the granular layer of the cerebellum in TAF1-edited animals. This suggests that the microglia may be responding to damage to the cerebellum induced by TAF1 editing and contributing to alterations in the integrity of the cerebellar circuit.
In addition to the cerebellum, we observed morphological changes in the cerebral cortex. The cerebral cortex and the cerebellum are both involved in movement and there are reciprocal interconnections by the cerebello-thalamo-cortical pathways which correlated with our studies (Mendoza and Merchant 2014). The motor cortex is the region within the cerebral cortex important for planning, control, and execution of voluntary movements. The abnormalities within the cerebral cortex could also contribute to the behavioral changes (i.e. impairment in beam walking) in gRNA-TAF1 edited animals observed in the our study.
In order for a gene to be transcribed, a transcriptional preinitiation complex (PIC) needs to assemble at its promoter. The first complex to bind the promoter is the general transcription factor, TFIID, consisting of the TATA-binding protein (TBP) and 13 different TBP-associated factors (TAFs) (Muller, Zaucker, and Tora 2010; Tora 2002). Mutations in several other human TFIID subunit coding genes (i.e. TBP, TAF2, TAF6, TAF8, and TAF13) have been implicated in human diseases including many with neurological outcomes and intellectual disability (Nakamura et al. 2001; Hellman-Aharony et al. 2013; Alazami et al. 2015; El-Saafin et al. 2018; Tawamie et al. 2017). The outcome of these mutations on TAFs has not been assessed in detail, due in part to the lack of the development of animal models for these disorders of transcription regulation as disruption of TFIID subunits can be embryonic lethal (Voss et al. 2000; El-Saafin et al. 2018). Our facile approach for assessing TAF1 function in vivo, using CRISPR/Cas9-based gene editing coupled lentiviral transduction, could be applied to these disorders of transcription regulation and provide insight into underlying mechanisms of disease. Although our work did not achieve germline deletion of TAF1, the approach used here provides an instructive example of how spatially and temporally restricted deletion may be sufficient to mimic hallmarks of these neurological disorders.
In conclusion, the present study shows that targeted knockdown of TAF1 can be achieved in the brain of rats with postnatal delivery of a CRISPR-Cas9 lentiviral vector. The current results demonstrate that this approach led to motor defects and morphological abnormalities within the cerebellum reminiscent of the human neurological disease-TAF1 ID syndrome. This model therefore provides an interesting opportunity for further studies investigating the effects of aberrant TAF1 on neuronal dysfunction and paves the way for testing of possible therapeutic strategies to treat TAF1 ID syndrome.
Supplementary Material
Highlights.
TAF-1 editing leads to behavioral defects at both the neonatal and juvenile stages of development.
Deletion of TAF-1 leads morphological abnormalities in both the cerebral cortex and the cerebellum.
The behavioral deficits seen in TAF1-edited animals is associated with irregular cerebellar outputs measured in Purkinje cells.
CaV3.1 accounts for the reduction in spontaneous excitatory post-synaptic currents in TAF1-edited animals.
Acknowledgements
We would like to thank Drs. Todd Vanderah and Tally Largent-Milnes for the i.c.v. injection facilities and behavior analysis. We would also like to thank Robert Hershoff for his help with the digital images. We also acknowledge the funding and support of the Senner Endowment for Precision Health, University of Arizona Health Sciences (M.N). This work was also supported by grants from the National Natural Science Foundation of China (81603088) and National Key Project of Research and Development of China (2018YFC1705501) to J.Y., and R01DA042852 from the National Institute on Drug Abuse to R.K.); and a Neurofibromatosis New Investigator Award from the Department of Defense Congressionally Directed Military Medical Research and Development Program (NF1000099) to R.K.
Footnotes
Conflict of interest
Mark A. Nelson, Ph.D. is Co-Founder and CEO of DesertDx, LLC
Rajesh Khanna, Ph.D. is Co-Founder and CSO of Regulonix, LLC
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguado C, Garcia-Madrona S, Gil-Minguez M, and Lujan R. 2016. 'Ontogenic Changes and Differential Localization of T-type Ca(2+) Channel Subunits Cav3.1 and Cav3.2 in Mouse Hippocampus and Cerebellum', Front Neuroanat, 10: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, Alhashem A, Bashiri FA, Al-Owain M, Kentab AY, Sogaty S, Al Tala S, Temsah MH, Tulbah M, Aljelaify RF, Alshahwan SA, Seidahmed MZ, Alhadid AA, Aldhalaan H, AlQallaf F, Kurdi W, Alfadhel M, Babay Z, Alsogheer M, Kaya N, Al-Hassnan ZN, Abdel-Salam GM, Al-Sannaa N, Al Mutairi F, El Khashab HY, Bohlega S, Jia X, Nguyen HC, Hammami R, Adly N, Mohamed JY, Abdulwahab F, Ibrahim N, Naim EA, Al-Younes B, Meyer BF, Hashem M, Shaheen R, Xiong Y, Abouelhoda M, Aldeeri AA, Monies DM, and Alkuraya FS. 2015. 'Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families', Cell Rep, 10: 148–61. [DOI] [PubMed] [Google Scholar]
- Aldrin-Kirk P, Davidsson M, Holmqvist S, Li JY, and Bjorklund T. 2014. 'Novel AAV-based rat model of forebrain synucleinopathy shows extensive pathologies and progressive loss of cholinergic interneurons', PLoS One, 9: e100869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmara H, Micu I, Rizwan AP, Sahu G, Simms BA, Zhang FX, Engbers JDT, Stys PK, Zamponi GW, and Turner RW. 2017. 'A T-type channel-calmodulin complex triggers alphaCaMKII activation', Mol Brain, 10: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assandri R, Egger M, Gassmann M, Niggli E, Bauer C, Forster I, and Gorlach A. 1999. 'Erythropoietin modulates intracellular calcium in a human neuroblastoma cell line', J Physiol, 516 (Pt 2): 343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, and Roeder RG. 1996. 'Biochemistry and structural biology of transcription factor IID (TFIID)', Annu Rev Biochem, 65: 769–99. [DOI] [PubMed] [Google Scholar]
- Cazade M, Bidaud I, Lory P, and Chemin J. 2017. 'Activity-dependent regulation of T-type calcium channels by submembrane calcium ions', Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemin J, Monteil A, Briquaire C, Richard S, Perez-Reyes E, Nargeot J, and Lory P. 2000. 'Overexpression of T-type calcium channels in HEK-293 cells increases intracellular calcium without affecting cellular proliferation', FEBS Lett, 478: 166–72. [DOI] [PubMed] [Google Scholar]
- Choe W, Messinger RB, Leach E, Eckle VS, Obradovic A, Salajegheh R, Jevtovic-Todorovic V, and Todorovic SM. 2011. 'TTA-P2 is a potent and selective blocker of T-type calcium channels in rat sensory neurons and a novel antinociceptive agent', Mol Pharmacol, 80: 900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon P, Herr D, Kanyshkova T, Meuth P, Budde T, and Pape HC. 2009. 'Burst discharges in neurons of the thalamic reticular nucleus are shaped by calcium-induced calcium release', Cell Calcium, 46: 333–46. [DOI] [PubMed] [Google Scholar]
- D'Angelo E 2018. 'Physiology of the cerebellum', Handb Clin Neurol, 154: 85–108. [DOI] [PubMed] [Google Scholar]
- Delenclos M, Faroqi AH, Yue M, Kurti A, Castanedes-Casey M, Rousseau L, Phillips V, Dickson DW, Fryer JD, and McLean PJ. 2017. 'Neonatal AAV delivery of alpha-synuclein induces pathology in the adult mouse brain', Acta Neuropathol Commun, 5: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues HS, Portugal CC, Socodato R, and Relvas JB. 2016. 'Oligodendrocyte, Astrocyte, and Microglia Crosstalk in Myelin Development, Damage, and Repair', Front Cell Dev Biol, 4: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khodor BF, Edgar N, Chen A, Winberg ML, Joyce C, Brunner D, Suarez-Farinas M, and Heyes MP. 2008. 'Identification of a battery of tests for drug candidate evaluation in the SMNDelta7 neonate model of spinal muscular atrophy', Exp Neurol, 212: 29–43. [DOI] [PubMed] [Google Scholar]
- El-Saafin F, Curry C, Ye T, Garnier JM, Kolb-Cheynel I, Stierle M, Downer NL, Dixon MP, Negroni L, Berger I, Thomas T, Voss AK, Dobyns W, Devys D, and Tora L. 2018. 'Homozygous TAF8 mutation in a patient with intellectual disability results in undetectable TAF8 protein, but preserved RNA polymerase II transcription', Hum Mol Genet, 27: 2171–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Tahawy NFG, Abdel Hafez SMN, Ramzy MM, Zenhom NM, and Abdel-Hamid HA. 2018. 'Effect of experimentally induced hypertension on cerebellum of postmenopausal rat', J Cell Physiol. [DOI] [PubMed] [Google Scholar]
- Feather-Schussler DN, and Ferguson TS. 2016. 'A Battery of Motor Tests in a Neonatal Mouse Model of Cerebral Palsy', J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez JA, McGahon MK, McGeown JG, and Curtis TM. 2015. 'CaV3.1 T-Type Ca2+ Channels Contribute to Myogenic Signaling in Rat Retinal Arterioles', Invest Ophthalmol Vis Sci, 56: 5125–32. [DOI] [PubMed] [Google Scholar]
- Fremont R, Tewari A, Angueyra C, and Khodakhah K. 2017. 'A role for cerebellum in the hereditary dystonia DYT1', Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabery S, Sajjad MU, Hult S, Soylu R, Kirik D, and Petersen A. 2012. 'Characterization of a rat model of Huntington's disease based on targeted expression of mutant huntingtin in the forebrain using adeno-associated viral vectors', Eur J Neurosci, 36: 2789–800. [DOI] [PubMed] [Google Scholar]
- Guirado R, Carceller H, Castillo-Gomez E, Castren E, and Nacher J. 2018. 'Automated analysis of images for molecular quantification in immunohistochemistry', Heliyon, 4: e00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, and Ebert BL. 2014. 'Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing', Nat Biotechnol, 32: 941–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman-Aharony S, Smirin-Yosef P, Halevy A, Pasmanik-Chor M, Yeheskel A, Har-Zahav A, Maya I, Straussberg R, Dahary D, Haviv A, Shohat M, and Basel-Vanagaite L. 2013. 'Microcephaly thin corpus callosum intellectual disability syndrome caused by mutated TAF2', Pediatr Neurol, 49: 411–16 e1. [DOI] [PubMed] [Google Scholar]
- Henneberger C, Papouin T, Oliet SH, and Rusakov DA. 2010. 'Long-term potentiation depends on release of D-serine from astrocytes', Nature, 463: 232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand ME, Isope P, Miyazaki T, Nakaya T, Garcia E, Feltz A, Schneider T, Hescheler J, Kano M, Sakimura K, Watanabe M, Dieudonne S, and Snutch TP. 2009. 'Functional coupling between mGluR1 and Cav3.1 T-type calcium channels contributes to parallel fiber-induced fast calcium signaling within Purkinje cell dendritic spines', J Neurosci, 29: 9668–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst SE, Liktor-Busa E, Moutal A, Parker S, Rice S, Szelinger S, Senner G, Hammer MF, Johnstone L, Ramsey K, Narayanan V, Perez-Miller S, Khanna M, Dahlin H, Lewis K, Craig D, Wang EH, Khanna R, Nelson MA. 2018. 'A novel variant in TAF1 affects gene expression and is associated with X-linked TAF1 intellectual disability syndrome', Neuronal Signalling, 2: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado R, Bub G, and Herzlinger D. 2014. 'A molecular signature of tissues with pacemaker activity in the heart and upper urinary tract involves coexpressed hyperpolarization-activated cation and T-type Ca2+ channels', FASEB J, 28: 730–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftinca MC, and Zamponi GW. 2009. 'Regulation of neuronal T-type calcium channels', Trends Pharmacol Sci, 30: 32–40. [DOI] [PubMed] [Google Scholar]
- Kim D, Bae CH, Jun YL, Jeon H, Koo S, and Kim S. 2017. 'Acupuncture alters pro-inflammatory cytokines in the plasma of maternally separated rat pups', Chin J Integr Med, 23: 943–47. [DOI] [PubMed] [Google Scholar]
- Kim JW, Oh HA, Lee SH, Kim KC, Eun PH, Ko MJ, Gonzales ELT, Seung H, Kim S, Bahn GH, and Shin CY. 2018. 'T-Type Calcium Channels Are Required to Maintain Viability of Neural Progenitor Cells', Biomol Ther (Seoul), 26: 439–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchens SA, Burch J, and Creazzo TL. 2003. 'T-type Ca2+ current contribution to Ca2+-induced Ca2+ release in developing myocardium', J Mol Cell Cardiol, 35: 515–23. [DOI] [PubMed] [Google Scholar]
- Kornum BR, Stott SR, Mattsson B, Wisman L, Ettrup A, Hermening S, Knudsen GM, and Kirik D. 2010. 'Adeno-associated viral vector serotypes 1 and 5 targeted to the neonatal rat and pig striatum induce widespread transgene expression in the forebrain', Exp Neurol, 222: 70–85. [DOI] [PubMed] [Google Scholar]
- Lee KY, Jang Y, Lee MH, Kim YI, Jung SC, Han SY, Kim SH, Park HS, and Kim DK. 2018. 'Intravenous Anesthetic, Propofol Affects Synaptic Responses in Cerebellar Purkinje Cells', Clin Psychopharmacol Neurosci, 16: 176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly R, Bouvier G, Schonewille M, Arabo A, Rondi-Reig L, Lena C, Casado M, De Zeeuw CI, and Feltz A. 2013. 'T-type channel blockade impairs long-term potentiation at the parallel fiber-Purkinje cell synapse and cerebellar learning', Proc Natl Acad Sci U S A, 110: 20302–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Marti-Gutierrez N, Park SW, Wu J, Lee Y, Suzuki K, Koski A, Ji D, Hayama T, Ahmed R, Darby H, Van Dyken C, Li Y, Kang E, Park AR, Kim D, Kim ST, Gong J, Gu Y, Xu X, Battaglia D, Krieg SA, Lee DM, Wu DH, Wolf DP, Heitner SB, Belmonte JCI, Amato P, Kim JS, Kaul S, and Mitalipov S. 2017. 'Correction of a pathogenic gene mutation in human embryos', Nature, 548: 413–19. [DOI] [PubMed] [Google Scholar]
- Maries E, Dass B, Collier TJ, Kordower JH, and Steece-Collier K. 2003. 'The role of alpha-synuclein in Parkinson’s disease: insights from animal models', Nat Rev Neurosci, 4: 727–38. [DOI] [PubMed] [Google Scholar]
- Mendoza G, and Merchant H. 2014. 'Motor system evolution and the emergence of high cognitive functions', Prog Neurobiol, 122: 73–93. [DOI] [PubMed] [Google Scholar]
- Moutal A, Cai S, Luo S, Voisin R, and Khanna R. 2018. 'CRMP2 is necessary for Neurofibromatosis type 1 related pain', Channels (Austin), 12: 47–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutal A, Sun L, Yang X, Li W, Cai S, Luo S, and Khanna R. 2018. 'CRMP2-Neurofibromin Interface Drives NF1-related Pain', Neuroscience, 381: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutal A, Yang X, Li W, Gilbraith KB, Luo S, Cai S, Francois-Moutal L, Chew LA, Yeon SK, Bellampalli SS, Qu C, Xie JY, Ibrahim MM, Khanna M, Park KD, Porreca F, and Khanna R. 2017. 'CRISPR/Cas9 editing of Nf1 gene identifies CRMP2 as a therapeutic target in neurofibromatosis type 1-related pain that is reversed by (S)-Lacosamide', Pain, 158: 2301–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Zaucker A, and Tora L. 2010. 'Developmental regulation of transcription initiation: more than just changing the actors', Curr Opin Genet Dev, 20: 533–40. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Jeong SY, Uchihara T, Anno M, Nagashima K, Nagashima T, Ikeda S, Tsuji S, and Kanazawa I. 2001. 'SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein', Hum Mol Genet, 10: 1441–8. [DOI] [PubMed] [Google Scholar]
- O'Rawe JA, Wu Y, Dorfel MJ, Rope AF, Au PY, Parboosingh JS, Moon S, Kousi M, Kosma K, Smith CS, Tzetis M, Schuette JL, Hufnagel RB, Prada CE, Martinez F, Orellana C, Crain J, Caro-Llopis A, Oltra S, Monfort S, Jimenez-Barron LT, Swensen J, Ellingwood S, Smith R, Fang H, Ospina S, Stegmann S, Den Hollander N, Mittelman D, Highnam G, Robison R, Yang E, Faivre L, Roubertie A, Riviere JB, Monaghan KG, Wang K, Davis EE, Katsanis N, Kalscheuer VM, Wang EH, Metcalfe K, Kleefstra T, Innes AM, Kitsiou-Tzeli S, Rosello M, Keegan CE, and Lyon GJ. 2015. 'TAF1 Variants Are Associated with Dysmorphic Features, Intellectual Disability, and Neurological Manifestations', Am J Hum Genet, 97: 922–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Cai Z, and Rhodes PG. 2003. 'Disturbance of oligodendrocyte development, hypomyelination and white matter injury in the neonatal rat brain after intracerebral injection of lipopolysaccharide', Brain Res Dev Brain Res, 140: 205–14. [DOI] [PubMed] [Google Scholar]
- Perez-Reyes E 2003. 'Molecular physiology of low-voltage-activated t-type calcium channels', Physiol Rev, 83:117–61. [DOI] [PubMed] [Google Scholar]
- Pitzer M, Lueras J, Warden A, Weber S, and McBride J. 2015. 'Viral vector mediated expression of mutant huntingtin in the dorsal raphe produces disease-related neuropathology but not depressive-like behaviors in wildtype mice', Brain Res, 1608: 177–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasankar S, Manivasagam T, and Surendran S. 2009. 'Ashwagandha leaf extract: a potential agent in treating oxidative damage and physiological abnormalities seen in a mouse model of Parkinson's disease', Neurosci Lett, 454: 11–5. [DOI] [PubMed] [Google Scholar]
- Sandweiss AJ, McIntosh MI, Moutal A, Davidson-Knapp R, Hu J, Giri AK, Yamamoto T, Hruby VJ, Khanna R, Largent-Milnes TM, and Vanderah TW. 2018. 'Genetic and pharmacological antagonism of NK1 receptor prevents opiate abuse potential', Mol Psychiatry, 23: 1745–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhanova IA, Sebentsova EA, Khukhareva DD, Manchenko DM, Glazova NY, Vishnyakova PA, Inozemtseva LS, Dolotov OV, Vysokikh MY, and Levitskaya NG. 2018. 'Gender-dependent changes in physical development, BDNF content and GSH redox system in a model of acute neonatal hypoxia in rats', Behav Brain Res, 350: 87–98. [DOI] [PubMed] [Google Scholar]
- Tawamie H, Martianov I, Wohlfahrt N, Buchert R, Mengus G, Uebe S, Janiri L, Hirsch FW, Schumacher J, Ferrazzi F, Sticht H, Reis A, Davidson I, Colombo R, and Abou Jamra R. 2017. 'Hypomorphic Pathogenic Variants in TAF13 Are Associated with Autosomal-Recessive Intellectual Disability and Microcephaly', Am J Hum Genet, 100: 555–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tora L 2002. 'A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription', Genes Dev, 16: 673–5. [DOI] [PubMed] [Google Scholar]
- Voss AK, Thomas T, Petrou P, Anastassiadis K, Scholer H, and Gruss P. 2000. 'Taube nuss is a novel gene essential for the survival of pluripotent cells of early mouse embryos', Development, 127: 5449–61. [DOI] [PubMed] [Google Scholar]
- Yamashita N, Mosinger B, Roy A, Miyazaki M, Ugajin K, Nakamura F, Sasaki Y, Yamaguchi K, Kolattukudy P, and Goshima Y. 2011. 'CRMP5 (collapsin response mediator protein 5) regulates dendritic development and synaptic plasticity in the cerebellar Purkinje cells', J Neurosci, 31: 1773–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Liu S, Zheng Y, Zhang M, Dang R, and Tang M. 2018. 'Maternal diet of polyunsaturated fatty acid influence the physical and neurobehaviour of rat offspring', Int J Dev Neurosci, 71: 156–62. [DOI] [PubMed] [Google Scholar]
- Ying SW, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TV, and Bramham CR. 2002. 'Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and upregulation of Arc synthesis', J Neurosci, 22: 1532–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunker AM, Sharp AH, Sundarraj S, Ranganathan V, Copeland TD, and McEnery MW. 2003. 'Immunological characterization of T-type voltage-dependent calcium channel CaV3.1 (alpha 1G) and CaV3.3 (alpha 1I) isoforms reveal differences in their localization, expression, and neural development', Neuroscience, 117: 321–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.