Abstract
Purpose
Legionella pneumophila pneumonia is a life-threatening, environmentally acquired infection identifiable via Legionella urine antigen tests (LUAT). We aimed to identify cumulative incidence, demographic distribution, and undetected disease outbreaks of Legionella pneumonia via positive LUAT in a single eastern Wisconsin health system, with a focus on urban Milwaukee County.
Methods
A multilevel descriptive ecologic study was conducted utilizing electronic medical record data from a large integrated health care system of patients who underwent LUAT from 2013 to 2017. A random sample inclusive of all positive tests was reviewed to investigate geodemographic differences among patients testing positive versus negative. Statistical comparisons used chi-squared or 2-sample t-tests; stepwise regression followed by binary logistic regression was used for multivariable analysis. Positive cases identified by LUAT were mapped to locate hotspots; positive cases versus total tests performed also were mapped by zip code.
Results
Of all LUAT performed (n=21,599), 0.68% were positive. Among those in the random sample (n=11,652), positive cases by LUAT were more prevalent in the June–November time period (86.2%) and younger patients (59.4 vs 67.7 years) and were disproportionately male (70.3% vs 29.7%) (P<0.0001 for each). Cumulative incidence was higher among nonwhite race/ethnicity (1.91% vs 1.01%, P<0.0001) but did not remain significant on multivariable analysis. Overall, 5507 tests were performed in Milwaukee County zip codes, yielding 82 positive cases by LUAT (60.7% of all positive cases in the random sample). A potential small 2016 outbreak was identified.
Conclusions
Cumulative incidence of a positive LUAT was less than 1%. LUAT testing, if done in real time by cooperative health systems, may complement public health detection of Legionella pneumonia outbreaks.
Keywords: Legionella; antigens, bacterial; pneumonia, bacterial; disease outbreaks
The genus Legionella consists of 60 species and 3 subspecies.1,2 All Legionella species are gram-negative bacteria, most of which are considered pathogenic for humans.1,2 Legionella pneumophila contains 16 different serogroups. Of these, Legionella pneumophila serogroup 1 was the cause of the 1976 Philadelphia outbreak of Legionella pneumonia (ie, Legionnaires’ disease) as well as the cause of approximately 84% of cases worldwide.1,3,4
All Legionella species have been detected in natural aqueous environments, including river systems, hot springs, and moist soils.1–3,5–12 As a facultative intracellular parasite of free-living amoeba, Legionella also exists within biofilms, which can be disrupted, leading to bacterial aerosolization followed by inhalation and infection.1,5,8,11 Importantly, Legionella has been associated with built environmental features, such as cooling towers and potable water, as well as household, medical, and business sources, including hot water heaters, showers, mist and ice machines, and decorative fountains.4,9–11,13–23 In addition, Legionella has been found in mobile equipment such as street cleaners and cruise ships.9,10,24–26
Following inhalation, Legionella undergoes phagocytosis by, and reproduction within, alveolar macrophages.1,8,11 This process kills the macrophages and continues until the immune system initiates a severe inflammatory response.1 After 2–10 days of incubation, the host will typically show signs of pneumonia, including fever, cough, and chest pain.1,2,8,9 Patients with chronic obstructive pulmonary disease, a history of smoking for 10 years or more, use of immunosuppressant drugs, a high prevalence of comorbid conditions, or who are more than 50 years old are at increased risk of developing Legionella pneumonia.2,7,9,27,28
The incidence of Legionella pneumonia in the United States has increased since 1976 and, more recently, by 192% (from 3.9 cases/million-persons in 2000 to 11.5 cases/million-persons in 2009).2,9,29 The increase in reported incidence rates may be due to increased awareness and diagnostic testing. Legionella pneumonia is classified as community-acquired, travel-related, or nosocomial.30,31 Identification of Legionella pneumonia disease incidence and prevalence is difficult due to reliance on “a passive surveillance system dependent on health care providers and laboratories actually testing for and reporting cases.”29 In the United States, from 2005 to 2009, only 24% of domestic Legionella pneumonia cases were considered travel-related,29 suggesting that home addresses or zip codes may be potentially useful in identifying regional “hotspots” of Legionella pneumonia infection on an occurrence or recurrence basis.
Legionella urine antigen testing (LUAT) is the most commonly used diagnostic test for identification of Legionella pneumonia.9,32,33 A positive test is diagnostic for Legionella pneumonia.34 LUAT is generally specific for Legionella pneumophila serogroup 1, the most common serogroup to cause pneumonia, although cross-reactivity with other Legionella serogroups exists. LUAT has 70%–80% sensitivity and up to 99% specificity for Legionella pneumonia; sensitivity may exceed 95% for Legionella pneumophila serogroup 1 infection, specifically.29,33–36 In order to presumptively link a particular source to a patient, a similar positive Legionella culture or PCR would need to be obtained from the source and the patient. Despite variable sensitivity, epidemiologic surveillance utilizing LUAT is being used in parts of the United States. In New York City, case reports predominately based on LUAT, as well as linked home and work zip codes, are utilized to identify Legionella outbreaks and possible environmental sources of infection.31
Spatial data is continuously increasing in value for infectious disease surveillance and outbreak recognition.37 Despite the limitations of only including patients who have sought care within a particular health care system,37 large integrated medical systems’ electronic medical record (EMR) data may be a source of discovery of occult disease epidemics. For example, our recent report on investigating the use of Blastomyces antigen for the diagnosis of blastomycosis revealed disease outbreak data complementary to health department data.38
There is limited literature regarding the epidemiology of Legionella pneumonia infections in Wisconsin. Most notable are outbreak reports in the state, including one linked to a decorative hospital fountain22 and one linked to multiple sources in Milwaukee.32,39 A recent report from the Centers for Disease Control and Prevention (CDC) identified 78% of Wisconsin Legionella pneumonia cases to be located within Milwaukee alone.39 Consequently, we investigated the cumulative incidence and geodemographic distribution of positive cases by LUAT within a large integrated health care system serving eastern Wisconsin, with emphasis on urban Milwaukee County. Moreover, this study was conducted in order to determine the feasibility of using large health system data to enhance public health surveillance and source identification, leading to targeted clinician, patient, and public education on a geographic basis.
METHODS
Study Population, Design, Data Sources
This study included inpatients and outpatients of all ages, genders, and ethnicities in a large integrated health care system who had LUAT (by enzyme immunoassay) performed from January 1, 2013, through December 31, 2017. This health system (15 hospitals and 159 outpatient clinics) spans eastern Wisconsin and part of northeastern Illinois. LUAT results on specimens were obtained from our affiliated laboratory system and further reviewed through the EMR for missing or additional variables of interest. If available, home address, work address, and employment information (missing from most subjects) was collected from individuals with positive LUAT.
Overall, 21,599 LUAT results were initially identified. If a patient tested positive more than once, only the encounter in which they had their first positive test was included in the analyses. For all other patients, only the encounter in which they had their first negative test was included. Given data saturation, a random sample of subjects with negative LUAT were examined until analysis revealed that more than 90% of all Milwaukee County zip codes were represented by more than 50 subjects (>60% were represented by >100 subjects; median number of subjects per zip code was 152). This reduced the total number of examined subjects, including all with positive LUAT and the random sample of those with negative LUAT, to 11,652. Positive and negative cases by LUAT were compared to determine associations of positive cases regarding the following variables: age, gender, race/ethnicity, year and month of LUAT, and home zip code at time of LUAT.
Comparison of Health System Demographics to U.S. Census Data
To allow the reader to compare the demographic makeup of the health system investigated in this study to those of all individuals in the catchment area (412 total zip codes), several comparisons were made. U.S. Census data was obtained from the American Community Survey 5-Year Summary Data for 2017 for the zip code tabulation areas that comprised the health system catchment area. Included were counts of that population by race/ethnicity for all ages and counts by age/gender category. Using the health system patient database, patient zip codes as of January 1, 2018, were used to determine whether the patient lived within the catchment area at that time. Catchment area health system data on race/ethnicity for all ages, and age/gender data for adult age groups (only 4 of 11,652 study subjects were less than 18 years old), were similarly compared to the Census data.
Statistical Analysis
Basic descriptive, and summary statistics were calculated using Minitab® statistical software (Version 13, State College, PA). Categorical and continuous variables were analyzed using chi-squared tests and 2-sample t-tests (assuming normal distribution), respectively (Mann-Whitney test for median age). Stepwise regression (for significantly associated predictors) followed by binary logistic regression was used for multivariable analysis to determine odds ratios (an appropriate statistic for this type of retrospective analysis40). P-values less than 0.05 were deemed significant.
Geographic patterns were mapped using GIS mapping software (ArcGIS Desktop 10.6, Esri, Redlands, CA) and SAS statistical software (Version 9.4, SAS Institute Inc, Cary, NC). Positive cases by LUAT with zip codes within Wisconsin were mapped to locate hotspots specifically within Milwaukee County, which consists of 35 zip codes as identified in the Milwaukee Health Report.41 We also examined the proportion of positive LUAT among all tests by zip code and, for Milwaukee County, by a 5-tiered socioeconomic status (SES) zip code grouping based on the combination of an income index and an education index as used by the Center for Urban Population Health (Milwaukee, WI).41
RESULTS
Of all LUAT results identified in the original data set of the affiliated laboratory, 146 (0.68%) were positive. On further chart review of the random sample inclusive of all positive tests, we identified a total of 142 individuals with positive tests, as there was no data available for 4 individuals. Additionally, 4 patients had more than 1 positive test, leaving a total of 138 individuals found positive by LUAT in the final analyzed data set, 3 of whom had zip codes outside of Wisconsin.
The study population is outlined in Table 1. Overall, patients of nonwhite race/ethnicity were significantly younger than white patients in our study sample (57.0 vs 70.0 years; P<0.0001). Among those in the random sample (n=11,652), positive cases by LUAT were younger (59.4 vs 67.7 years) and disproportionately male (70.3% vs 29.7% female); P<0.0001 for both. The proportion of positive cases by LUAT was higher among nonwhite race/ethnicity (1.91% vs 1.01%; P<0.0001). Nonwhite race/ethnicity, however, did not remain significant in multivariable analysis of all subjects (Table 1).
Table 1.
Patient Geodemographic Predictors Associated With Negative vs Positive LUAT
| Predictor | Overall (n=11,652) | Negative LUAT (n=11,514) | Positive LUAT (n=138) | Univariable P (Positives vs Negatives) | Multivariable P (Positives vs Negatives) | Multivariable Odds Ratio (95% CI) |
|---|---|---|---|---|---|---|
| Race/Ethnicitya | ||||||
| White, % (n) | 81.4 (9384) | 81.5 (9289) | 69.9 (95) | reference | reference | reference |
| Black, % (n) | 12.3 (1415) | 12.1 (1381) | 25.0 (34) | <0.0001 | 0.019 | 1.67 (1.09–2.56) |
| AN/AI, % (n) | 0.4 (47) | 0.4 (47) | -- | |||
| Asian, % (n) | 0.9 (103) | 0.9 (103) | -- | |||
| Hispanic, % (n) | 4.9 (562) | 4.9 (555) | 5.2 (7) | 0.751 | 0.866 | 0.93 (0.42–2.06) |
| 2 or more, % (n) | 0.1 (13) | 0.1 (13) | -- | |||
| PI/Hawaiian, % (n) | 0.03 (4) | 0.04 (4) | -- | |||
| White vs nonwhitea | ||||||
| White, % (n) | 81.4 (9384) | 81.5 (9289) | 69.9 (95) | <0.0001 | 0.123 | 1.37 (0.92–2.04) |
| Nonwhite, % (n) | 18.6 (2144) | 18.5 (2103) | 30.2 (41) | |||
| Gender | ||||||
| Male, % (n) | 48.9 (5700) | 48.7 (5603) | 70.3 (97) | <0.0001 | <0.0001 | 2.38 (1.64–3.45) |
| Female, % (n) | 51.1 (5952) | 51.3 (5911) | 29.7 (41) | |||
| Mean age when tested, years | 67.6 | 67.7 | 59.4 | <0.0001 | <0.0001 | 0.98 (0.97–0.99) |
| Median age when tested, years | 69.0 | 69.0 | 59.0 | <0.0001 | ||
| Year | ||||||
| 2013, % (n) | 20.9 (2438) | 20.8 (2392) | 33.3 (46) | 0.0007 | reference | reference |
| 2014, % (n) | 20.0 (2335) | 20.1 (2319) | 11.6 (16) | 0.001 | 0.38 (0.21–0.68) | |
| 2015, % (n) | 20.0 (2332) | 20.1 (2310) | 15.9 (22) | 0.035 | 0.57 (0.34–0.96) | |
| 2016, % (n) | 18.9 (2201) | 18.9 (2179) | 15.9 (22) | 0.049 | 0.59 (0.35–1.00) | |
| 2017, % (n) | 20.1 (2346) | 20.1 (2314) | 23.2 (32) | 0.286 | 0.78 (0.49–1.24) | |
| Time of year | ||||||
| January, % (n) | 10.5 (1228) | 10.6 (1225) | 2.2 (3) | <0.0001 | reference | reference |
| February, % (n) | 8.1 (944) | 8.2 (944) | -- | 0.996 | N/A | |
| March, % (n) | 10.2 (1184) | 10.3 (1182) | 1.5 (2) | 0.677 | 0.68 (0.11–4.10) | |
| April, % (n) | 8.7 (1015) | 8.8 (1014) | 0.7 (1) | 0.430 | 0.40 (0.04–3.87) | |
| May, % (n) | 7.9 (922) | 8.0 (917) | 3.6 (5) | 0.287 | 2.18 (0.52–9.17) | |
| June, % (n) | 7.0 (819) | 7.0 (808) | 8.0 (11) | 0.020 | 4.66 (1.28–17.04) | |
| July, % (n) | 7.2 (837) | 7.0 (806) | 22.5 (31) | 0.001 | 14.13 (4.29–46.56) | |
| August, % (n) | 7.3 (853) | 7.2 (827) | 18.8 (26) | 0.001 | 11.33 (3.41–37.64) | |
| September, % (n) | 7.7 (900) | 7.6 (877) | 16.7 (23) | 0.001 | 10.24 (3.06–34.28) | |
| October, % (n) | 7.9 (919) | 7.9 (905) | 10.1 (14) | 0.006 | 5.76 (1.65–20.14) | |
| November, % (n) | 7.9 (919) | 7.9 (905) | 10.1 (14) | 0.007 | 5.58 (1.59–19.52) | |
| December, % (n) | 9.5 (1112) | 9.6 (1104) | 5.8 (8) | 0.126 | 2.83 (0.75–10.70) | |
AN/AI, Alaskan Native/American Indian; LUAT, Legionella urine antigen test; N/A, not available; PI, Pacific Islander.
Race/Ethnicity was unknown for 124 subjects.
Positive cases by LUAT were disproportionately diagnosed within the year 2013 versus 2014–2017 (33.3% vs 20% expected; P=0.0007). During the months of June–November there was an increase in positive cases (86.2% vs 50.1% expected; P<0.0001), peaking in the months July–September (58.0% of cases vs 25.2% expected). Gender, age, and year and month of test remained significant predictors in the final multivariable model (Table 1).
In models for the random sample of which only Wisconsin zip codes wherein 100 or more patients were tested, nonwhite race/ethnicity did not remain significant in multivariable analysis, which included age and gender, with or without year and month of test, and 3-digit zip code group (eg, 532XX) — all the variables that were found positively associated in univariable analysis.
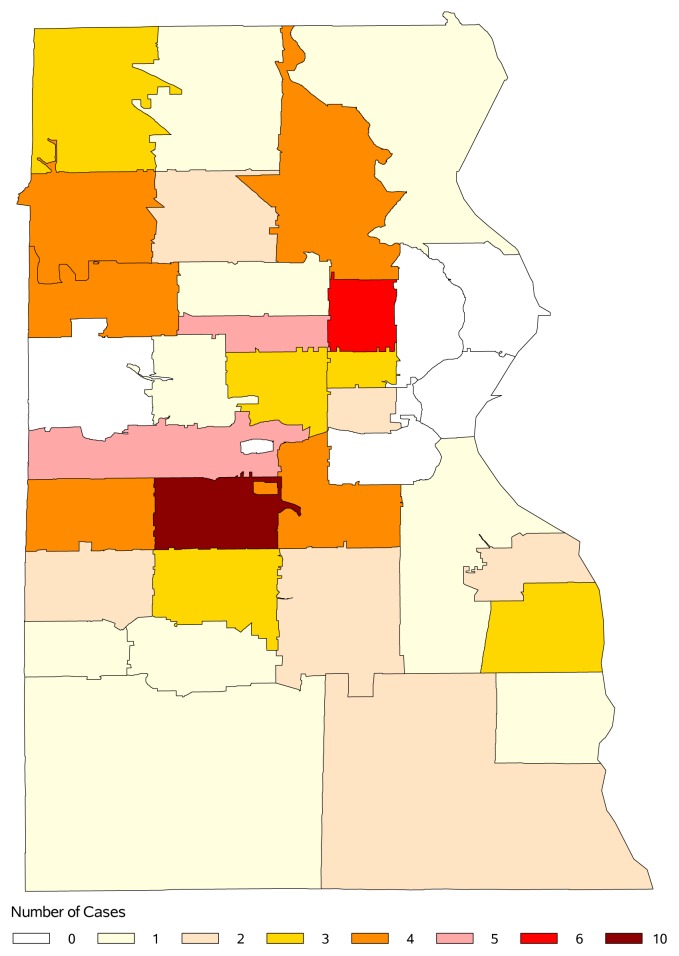
Geographically, most tests were performed in the southeastern region of Wisconsin, specifically within Milwaukee County. Among zip code group 532XX, which includes the city of Milwaukee and surrounding cities, there was a higher cumulative incidence by LUAT compared to the remainder of the study regions (54.1% vs 45.9%, Table 2). The predicted portion of positive cases (based on a proportionally even distribution of cases) for 532XX was 39.6% (P=0.006). Positive cases in and around Milwaukee County were further mapped (Figure 1). Three zip codes were further explored based on having 5 or more positive cases. The first two of these zips consistently had a few positive cases each year — one being a Milwaukee neighborhood (6 cases) and another where the cities of Milwaukee, West Milwaukee, West Allis, and Greenfield intersected (10 cases). The third zip code identified was in West Allis, with a temporospatial cluster of all 5 positive cases in 2016. This may represent a small outbreak.
Table 2.
Positive Legionella Pneumonia Cases Each Year by Zip Code Group
| Zip Code Group* | 2013 | 2014 | 2015 | 2016 | 2017 | Total | Total % |
|---|---|---|---|---|---|---|---|
| 530XX (n=14) | 5 | 4 | 4 | 2 | 3 | 18 | 13.3% |
| 531XX (n=22) | 15 | 0 | 8 | 2 | 5 | 30 | 22.2% |
| 532XX (n=23) | 23 | 11 | 7 | 13 | 19 | 73 | 54.1% |
| 534XX (n=2) | 0 | 1 | 0 | 0 | 2 | 3 | 2.2% |
| 539XX (n=1) | 0 | 0 | 0 | 1 | 0 | 1 | 0.7% |
| 541XX (n=2) | 1 | 0 | 1 | 1 | 0 | 3 | 2.2% |
| 542XX (n=3) | 0 | 0 | 0 | 2 | 1 | 3 | 2.2% |
| 543XX (n=1) | 0 | 0 | 0 | 0 | 1 | 1 | 0.7% |
| 549XX (n=3) | 1 | 0 | 1 | 0 | 1 | 3 | 2.2% |
|
| |||||||
| Total number | 45 | 16 | 21 | 21 | 32 | 135 | |
|
| |||||||
| Total percentage | 33.3% | 11.9% | 15.6% | 15.6% | 23.7% | ||
n reflects the number of zip codes within a designated zip code group.
Figure 1.
Map of positive Legionella urine antigen tests (after duplicate tests removed), by zip code, in Milwaukee County, Wisconsin, 2013–2017.
The ratio of positive LUAT results to all LUAT tests within a given zip code was mapped (Figure 2). Overall, within Milwaukee County, there were 4 zip codes in which 4% or more of the 32–107 patients tested had a positive test.
Figure 2.
Map of proportion of positive Legionella urine antigen test (LUAT) results among all LUAT tests within a given zip code in eastern Wisconsin. Within each percentage group, the larger the circle, the more patients who were tested in a given zip. The majority of tested patients resided in zip codes near the greater Milwaukee area.
Based on the updated Milwaukee Health Report SES breakdown,41 of the 5507 LUAT performed in Milwaukee County zip codes, 82 cases tested positive for Legionella. Of the positive tests, 3.7% were from a high SES zip, 14.6% from medium-high, 23.2% from medium, 32.9% from medium-low, and 25.6% from a low SES zip. When compared to expected positive test distribution — 8.2%, 20.5%, 29.0%, 21.7%, and 20.6%, respectively — the observed positive test distribution was significantly different (P=0.038, Table 3).
Table 3.
Observed vs Expected Distribution of 82 LUAT-Positive Subjects per Zip Code SES Group, Milwaukee County, Wisconsin
| Zip SES Status41 | Total Tested Subjects | Observed Positive Cases, n (%) | Expected Positive Cases, n (%) |
|---|---|---|---|
| High | 450 | 3 (3.7%) | 6.7 (8.2%) |
| Medium-high | 1126 | 12 (14.6%) | 16.8 (20.5%) |
| Medium | 1599 | 19 (23.2%) | 23.8 (29.0%) |
| Medium-low | 1197 | 27 (32.9%) | 17.8 (21.7%) |
| Low | 1135 | 21 (25.6%) | 16.9 (20.6%) |
|
| |||
| TOTAL | 5507 | 82 (100%) | 82.0 (100%) |
χ2(4 degrees of freedom): 10.133; P=0.038.
LUAT, Legionella urine antigen test; SES, socioeconomic status.
Regarding the generalizability of our results, Table 4 compares the known race/ethnicity of our health system patients, by category, to the expected number (and percentage of total), based on Census data for all individuals in our catchment area (412 total zip codes). As only 4 of our study subjects were children, we were able and chose to compare subject age/gender, by category, to the expected number/percentages predicted by Census data, restricted to adults only. This data is presented in Table 5. Note that all statistical comparisons in Tables 4 and 5 are significant due, in part, to the extremely large sample sizes. Overall, for adult patients, our health system included 1,905,632 of 3,341,806 (57.0%) of the total Census population for the catchment area and was 53.0% female compared to 51.0% female in the Census population.
Table 4.
Race/Ethnicity Comparisons: Health System Population vs Predicted According to U.S. Census Data
| Predictor | Health System Population (n=2,057,014) | Predicted (n=2,057,014) | P |
|---|---|---|---|
| Race/Ethnicitya | <0.0001 | ||
| White, % (n) | 76.8% (1,580,183) | 73.2% (1,505,734) | |
| Black, % (n) | 11.7% (240,975) | 8.2% (168,264) | |
| Alaskan Native/American Indian, % (n) | 0.4% (9084) | 0.5% (10,285) | |
| Asian, % (n) | 2.3% (47,602) | 3.3% (67,264) | |
| Hispanic, % (n) | 7.9% (163,125) | 11.1% (228,946) | |
| 2 or more, % (n) | 0.7% (13,885) | 3.7% (76,110) | |
| Pacific Islander/Hawaiian, % (n) | 0.1% (2160) | 0.02% (411) | |
| White vs nonwhitea | <0.0001 | ||
| White, % (n) | 76.8% (1,580,183) | 73.2% (1,505,734) | |
| Nonwhite, % (n) | 23.2% (476,831) | 26.8% (551,280) |
All ages included; unknown race/ethnicity was omitted.
Table 5.
Adult Age/Gender Category Comparisons: Health System Population vs Predicted According to U.S. Census Data
| Category (Gender/Age Group) | Health System Population | Predicted | P |
|---|---|---|---|
| Female (n=1,009,949)a | <0.0001 | ||
| 18–29 years, % (n) | 19.5% (196,461) | 19.2% (194,213) | |
| 30–39 years, % (n) | 16.9% (170,771) | 16.0% (161,390) | |
| 40–49 years, % (n) | 14.5% (146,239) | 16.6% (167,752) | |
| 50–59 years, % (n) | 16.6% (167,907) | 19.1% (193,506) | |
| 60–69 years, % (n) | 14.0% (141,675) | 14.6% (146,948) | |
| 70–79 years, % (n) | 8.7% (87,365) | 8.1% (82,210) | |
| ≥80 years, % (n) | 9.9% (99,531) | 6.3% (63,930) | |
| Male (n=894,212)a | <0.0001 | ||
| 18–29 years, % (n) | 19.7% (175,726) | 21.2% (189,841) | |
| 30–39 years, % (n) | 17.3% (154,415) | 16.7% (148,976) | |
| 40–49 years, % (n) | 15.3% (137,079) | 17.1% (153,536) | |
| 50–59 years, % (n) | 17.4% (155,819) | 19.5% (174,640) | |
| 60–69 years, % (n) | 14.7% (130,990) | 14.4% (129,124) | |
| 70–79 years, % (n) | 8.3% (74,315) | 7.1% (63,131) | |
| ≥80 years, % (n) | 7.4%% (65,868) | 3.9% (34,964) |
Unknown age/gender subjects omitted.
DISCUSSION
As evidenced by a recent Legionella pneumonia outbreak in Flint, Michigan,23 the leading types of infection from drinking water in the United States have shifted from gastrointestinal to respiratory.18 Annually, waterborne diseases cost approximately $1 billion, with Legionella being the most expensive due to long length of hospital stay.19 Prompt diagnosis of legionellosis using LUAT and other modalities may limit patient morbidity, mortality, and cost.9,10
In our integrated health system, LUAT appears to be commonly used as means of legionellosis identification (an average of 4320 tests per year in a health system with 30,000 billed diagnoses of all pneumonia types each year). Overall, less than 1% tested positive, indicative of the relative infrequency of this potentially life-threatening disease. This is considering an overall sensitivity of LUAT in detecting Legionella pneumonia of ≤80%, due in part to only detecting Legionella pneumophila serogroup 1.29,33,35
Our study revealed positive cases by LUAT to be disproportionately male and of nonwhite race/ethnicity, which reinforces and is consistent with current literature.2,9,13,29,42,43 However, we identified positive cases by LUAT to be relatively younger (59.4 vs 67.7 years), which differs from previous studies.2,9,27 This may be due to our study sample including significant numbers of elderly, debilitated patients with various pneumonia symptoms having a panel of organism-specific diagnostic tests done upon hospital admittance that might otherwise not be needed (ie, overtesting of elderly population). A contributing factor to our hypothesis is the recommendation of the American Thoracic Society and the Infectious Diseases Society of America that LUAT be performed for patients that do not respond to antibiotic therapy, have severe pneumonia with intensive care, are immunocompromised, have a history of excessive alcohol use, have traveled within the past 2 weeks, or are older than 50 years of age.2 Additionally, in our study, the increased proportion of Legionella pneumonia cases in those of nonwhite race/ethnicity appeared to be confounded by the relatively younger age of the nonwhite population tested.
Our study also revealed that positive cases by LUAT disproportionately occurred within the year 2013, for unknown reasons. Whether the extreme heat events in the summer of 2012 or other weather factors contributed to this finding is not clear from our study. Additional research is needed on the effects of heat and humidity on Legionella ecology and infection in Wisconsin.44,45 Pathogen survival is correlated with warm (15°C to 27°C) and humid (≥80.0%) months, with previous associations of infection with both precipitation and increased humidity.12,46,47 Local seasonal fluctuations, even of a stable climate, may be a factor in the temporal variation of Legionella pneumonia incidence rates if within a humid environment as opposed to dry.9 In our study, most positive cases were identified in the months of June–November, which is consistent with previous reports describing seasonality, specifically within summer to autumn, as a major epidemiologic factor within the Mid-Atlantic United States, England, Wales, and the Netherlands.9,12,47 According to the CDC, 62% of Legionella pneumonia cases within the United States from 2000 to 2009 occurred during the months of June–October.29 Our study further reinforces this seasonality pattern. However, it may become a less apparent factor with increased age (>65 years).47
We have identified a correlation between SES of a particular zip-coded region and positive findings of Legionella pneumonia infection. Within Milwaukee County, the majority of positive cases by LUAT came from patients of medium-low to low SES. High levels of poverty have been found to increase one’s risk for Legionella.43 Unlike other pneumonias, Legionella is, in part, an opportunistic premise plumbing pathogen (OPPP), which is disinfectant-resistant.16 Some 30% of the human population may be exposed to OPPPs through shared habitats (ie, showers, dishwashers),15,16 as well as vacant housing, renter-occupied housing, and homes built pre-1970.16,43 The inability to renew these old water systems perhaps contributes to outbreaks within areas of lower SES.8 The presence of nearby industrial structures, large buildings with cooling towers, or other sources of potable water should be investigated as potential causes of increased Legionella pneumonia cases within lower SES neighborhoods.21 Air pollutants within the ozone layer have been associated with higher temperatures and may contribute to the increased vulnerability to heat within highly populated areas.4,45 This could be an additional contributing factor to the increased proportion of positive cases within Milwaukee County.
Currently, positive Legionella pneumonia cases are reported to the CDC through the National Notifiable Disease Surveillance System and a Supplemental Legionnaires Disease Surveillance System. The CDC has investigated Legionella pneumonia cases as a means of enhancing general public health surveillance and suggests targeted clinician and patient education on a geographic basis.39 Our study illustrates the benefits of using large health care system data to identify potential Legionella pneumonia outbreaks or hotspots through real-time EMR abstraction and health department follow-up,32 a structure similar to that implemented by New York City’s health department.31,37
Limitations
There are several limitations of our study, including the potential for ecologic inference fallacy (deducing inferences about individuals based on inferences regarding the group that includes those individuals). As this is retrospective data, the information within each individual patient’s chart was dependent on the accuracy and detail in which the information was entered. Limitations in data collection for positive cases included incomplete occupational details at time of diagnosis. Final diagnoses were not known for LUAT-negative subjects. Underlying medical conditions and extent of additional testing were not captured for study subjects, and this limits interpretation of demographic factors that increase the likelihood of positive LUAT. Additionally, due to lack of follow-up questioning associated with positive LUAT, there can be no individual postulated sites of exposure.
Moreover, there is a lack of social and travel history for patients.9,17,48–50 The identification of a source of exposure within built environments was limited by the possibility of mobile sources such as street sweepers, vehicle cooling units, and asphalt paving machines.10,25,26 Our study was limited because highly immunosuppressed patients may not produce the substrate for which LUAT tests,2 and we did not capture whether patients were immunocompromised and if there were differences in detection among those testing positive or negative. LUAT-positive subjects in this study presumably represented the <5% of persons who were exposed to the Legionella organisms in the environment.34 Thus, surveillance based on such data may miss many potential environmental sources.
Lastly, while our data was obtained from a large multihospital health system (a potential strength), and included negative as well as positive LUAT data, it represents a single health system within a single geographic region, not from a broad, comprehensive community survey. This could have limited both generalizability and identification of case clusters, although our study identified clusters within Milwaukee County. Even so, our health system represents the majority of persons residing in our catchment area, based on Census comparison data, supporting the representativeness of our sample. Our study sample was generally similar in median age (69 vs 68 years) and sex (48.9% vs 46.2% male) but lower in proportion of black race (12.3% vs 19.8%), as compared to a recent large university study of patients hospitalized with pneumonia.51 Perhaps with communitywide data, including from other health systems, and/or an even greater population density, we might recognize “rolling epidemics” of legionellosis in Milwaukee County similar to that described for the cities of New York and Sydney.14 Obviously, this would require resources for cooperative health system laboratories and information technology departments to develop processes for monitoring and reporting of continuous readouts of LUAT results.
CONCLUSIONS
The general demographic and seasonal features of patients with positive LUAT in this large eastern Wisconsin health system were similar to those reported in the literature. A correlation between positive LUAT and lower SES was identified; thus, focused study on built environments near clustered cases in these areas may reveal additional sources of infection. We hope that our report might prompt a formal investigation of the utility of combined health department and EMR surveillance. LUAT surveillance of cooperating health system electronic medical records, if done in real time, may complement public health detection of Legionella pneumonia outbreaks and high-risk areas.
Patient-Friendly Recap.
The bacterium Legionella pneumophila is the major cause of Legionnaires’ disease (a type of pneumonia). This disease can be diagnosed from a urine test that detects Legionella antigens (LUAT).
The authors found that 0.68% of the LUATs performed in their health system over the study period tested positive. Average age of positive cases (~60 years) was younger than that of negative (ie, other pneumonia) cases.
A small unrecognized outbreak may have occurred during the time of the study.
If surveilled in real time using electronic records, patient data and interview after a positive LUAT result could aid public health efforts to identify where in the environment these infections come from.
Acknowledgments
We would like to thank Julie Prabucki for initial laboratory data acquisition. We also thank Andrew Marek for his acquisition of comparison health system data and U.S. Census data from the American Community Survey as well as his contribution to the GIS mapping associated with this project.
Footnotes
Author Contributions
Study design: Kram, Baumgardner. Data acquisition or analysis: all authors. Manuscript drafting: Toberna, William, Kram, Baumgardner. Critical revision: all authors.
Conflicts of Interest
None.
References
- 1.Edelstein PH, Roy CR. Legionnaires’ disease and Pontiac fever. In: Bennett JE, Dolin R, Blaser MJ, editors. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, Eighth Edition. Philadelphia, PA: Elsevier Saunders; 2015. pp. 2633–44.e6. [Google Scholar]
- 2.Cunha BA, Burillo A, Bouza E. Legionnaires’ disease. Lancet. 2016;387:376–85. doi: 10.1016/S0140-6736(15)60078-2. [DOI] [PubMed] [Google Scholar]
- 3.Valavane A, Chaudhry R. The summer of seventy-six – Legionella pneumophila monologue. Emerg Infect Dis. 2017;23:1202–3. doi: 10.3201/eid2307.160546. [DOI] [Google Scholar]
- 4.Chaudhry R, Sreenath K, Arvind V, Vinayaraj EV, Tanu S. Legionella pneumophila serogroup 1 in the water facilities of a tertiary healthcare center, India. Emerg Infect Dis. 2017;23:1924–25. doi: 10.3201/eid2311.171071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassell K, Gacek P, Warren JL, Raymond PA, Cartter M, Weinberger DM. Association between sporadic legionellosis and river systems in Connecticut. J Infect Dis. 2018;217:179–87. doi: 10.1093/infdis/jix531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumgardner DJ. Soil-related bacterial and fungal infections. J Am Board Fam Med. 2012;25:734–44. doi: 10.3122/jabfm.2012.05.110226. [DOI] [PubMed] [Google Scholar]
- 7.Kenagy E, Priest PC, Cameron CM, et al. Risk factors for Legionella longbeachae Legionnaires’ disease, New Zealand. Emerg Infect Dis. 2017;23:1148–54. doi: 10.3201/eid2307.161429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506–26. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phin N, Parry-Ford F, Harrison T, et al. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect Dis. 2014;14:1011–21. doi: 10.1016/S1473-3099(14)70713-3. [DOI] [PubMed] [Google Scholar]
- 10.Orkis LT, Harrison LH, Mertz KJ, Brooks MM, Bibby KJ, Stout JE. Environmental sources of community-acquired Legionnaires’ disease: a review. Int J Hyg Environ Health. 2018;221:764–74. doi: 10.1016/j.ijheh.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Bartley PB, Ben Zakour NL, Stanton-Cook M, et al. Hospital-wide eradication of a nosocomial Legionella pneumophila serogroup 1 outbreak. Clin Infect Dis. 2016;62:273–9. doi: 10.1093/cid/civ870. [DOI] [PubMed] [Google Scholar]
- 12.Simmering JE, Polgreen LA, Hornick DB, Sewell DK, Polgreen PM. Weather-dependent risk for Legionnaires’ disease, United States. Emerg Infect Dis. 2017;23:1843–51. doi: 10.3201/eid2311.170137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez JL, Polyak CR, Kolavic SA, Brokaw JK, Birkmire SE, Valcik JA. Investigation of a cluster of Legionella pneumophila infections among staff at a federal research facility. Mil Med. 2001;166:753–8. doi: 10.1093/milmed/166.9.753. [DOI] [PubMed] [Google Scholar]
- 14.MacIntyre CR, Dyda A, Bui CM, Chughtai AA. Rolling epidemic of Legionnaires’ disease outbreaks in small geographic areas. Emerg Microbes Infect. 2018;7(1):36. doi: 10.1038/s41426-018-0051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida M, Furuya N, Hosokawa N, et al. Legionella pneumophila contamination of hospital dishwashers. Am J Infect Control. 2018;46:943–5. doi: 10.1016/j.ajic.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 16.Falkinham JO, 3rd, Hilborn ED, Arduino MJ, Pruden A, Edwards MA. Epidemiology and ecology of opportunistic premise plumbing pathogens: Legionella pneumophila, Mycobacterium avium, and Pseudomonas aeruginosa. Environ Health Perspect. 2015;123:749–58. doi: 10.1289/ehp.1408692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chochlakis D, Sandalakis V, Deramarou M, Tselentis Y, Psaroulaki A. Legionellosis: a walk-through to identification of the source of infection. Cent Eur J Public Health. 2017;25:235–9. doi: 10.21101/cejph.a4660. [DOI] [PubMed] [Google Scholar]
- 18.Beer KD, Gargano JW, Roberts VA, et al. Surveillance for waterborne disease outbreaks associated with drinking water – United States, 2011–2012. MMWR Morb Mortal Wkly Rep. 2015;64:842–8. doi: 10.15585/mmwr.mm6431a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collier SA, Stockman LJ, Hicks LA, Garrison LE, Zhou FJ, Beach MJ. Direct healthcare costs of selected diseases primarily or partially transmitted by water. Epidemiol Infect. 2012;140:2003–13. doi: 10.1017/S0950268811002858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pepper IL, Gerba CP. Risk of infection from Legionella associated with spray irrigation of reclaimed water. Water Res. 2018;139:101–7. doi: 10.1016/j.watres.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 21.De Filippis P, Mozzetti C, Messina A, D’Aló GL. Prevalence of Legionella in retirement homes and group homes water distribution systems. Sci Total Environ. 2018;643:715–24. doi: 10.1016/j.scitotenv.2018.06.216. [DOI] [PubMed] [Google Scholar]
- 22.Patrick M ABC News. Hospital fountain linked to disease outbreak in Wisconsin. [Accessed November 7, 2017]. Published 2012 Jul. 11. https://abcnews.go.com/Health/hospital-fountain-linked-disease-outbreak-wisconsin/story?id=15339838.
- 23.Zahran S, McElmurry SP, Kilgore PE, et al. Assessment of the Legionnaires’ disease outbreak in Flint, Michigan. Proc Natl Acad Sci U S A. 2018;115:E1730–9. doi: 10.1073/pnas.1718679115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inglis TJJ, Spittle C, Carmichael H, et al. Legionnaires’ disease outbreak on a merchant vessel, Indian Ocean, Australia, 2015. Emerg Infect Dis. 2018;24:1345–8. doi: 10.3201/eid2407.171978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valero N, de Simón M, Gallés P, et al. Street cleaning trucks as potential sources of Legionella pneumophila. Emer Infect Dis. 2017;23:1880–2. doi: 10.3201/eid2311.161390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coscollá M, Fenollar J, Escribano I, González-Candelas F. Legionellosis outbreak associated with asphalt paving machine, Spain, 2009. Emerg Infect Dis. 2010;16:1381–7. doi: 10.3201/eid1609.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berkelman RL, Pruden A. Prevention of Legionnaires’ disease in the 21st century by advancing science and public health practice. Emerg Infect Dis. 2017;23:1905–7. doi: 10.3201/eid2311.171429. [DOI] [Google Scholar]
- 28.Wickramasekaran RN, Sorvillo F, Kuo T. Legionnaires’ disease and associated comorbid conditions as causes of death in the U.S., 2000–2010. Public Health Rep. 2015;130:222–9. doi: 10.1177/003335491513000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hicks LA, Garrison LE, Nelson GE, Hampton LM Centers for Disease Control and Prevention. Legionellosis – United States, 2000–2009. MMWR Morb Mortal Wkly Rep. 2011;60:1083–7. [Google Scholar]
- 30.Lapierre P, Nazarian E, Zhu Y, et al. Legionnaires’ disease outbreak caused by endemic strain of Legionella pneumophila, New York, New York, USA, 2015. Emerg Infect Dis. 2017;23:1784–91. doi: 10.3201/eid2311.170308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzhenry R, Weiss D, Cimini D, et al. Legionnaires’ disease outbreaks and cooling towers, New York City, New York, USA. Emerg Infect Dis. 2017;23:1769–76. doi: 10.3201/eid2311.161584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wisconsin Department of Health Services, Division of Public Health. Increased reports of laboratory-confirmed cases of legionellosis (Legionnaires’ disease) [Accessed March 20, 2020]. Issued 2018 Jul. 9. https://www.dhs.wisconsin.gov/dph/memos/communicable-diseases/index.htm.
- 33.Shimada T, Noguchi Y, Jackson JL, et al. Systematic review and metaanalysis: urinary antigen tests for Legionellosis. Chest. 2009;136:1576–85. doi: 10.1378/chest.08-2602. [DOI] [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention. Legionella (Legionnaires’ disease and Pontiac fever) Diagnosis, treatment, and prevention. [Accessed March 20, 2020]. Last reviewed 2020 Jan 15. https://www.cdc.gov/legionella/clinicians/diagnostic-testing.html.
- 35.Helbig JH, Uldum SA, Lück PC, Harrison TG. Detection of Legionella pneumophila antigen in urine samples by the BinaxNOW immunochromatographic assay and comparison with both Binax Legionella Urinary Enzyme Immunoassay (EIA) and Biotest Legionella Urine Antigen EIA. J Med Microbiol. 2001;50:509–16. doi: 10.1099/0022-1317-50-6-509. [DOI] [PubMed] [Google Scholar]
- 36.Badoux P, Euser SM, Bruin JP, Mulder PPG, Yzerman EPF. Evaluation of the bioNexia Legionella test, including impact of incubation time extension, for detection of Legionella pneumophila serogroup 1 antigen in urine. J Clin Microbiol. 2017;55:1733–7. doi: 10.1128/JCM.02448-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee EC, Asher JM, Goldlust S, Kraemer JD, Lawson AB, Bansal S. Mind the scales: harnessing spatial big data for infectious disease surveillance and inference. J Infect Dis. 2016;214(suppl 4):S409–13. doi: 10.1093/infdis/jiw344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baumgardner DJ. Use of urine antigen testing for Blastomyces in an integrated health system. J Patient Cent Res Rev. 2018;5:176–82. doi: 10.17294/2330-0698.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biedrzycki PA, Stanley M, Radmer F, Lauf S Centers for Disease Control and Prevention. Notes from the field: increase in reported legionellosis – Milwaukee, Wisconsin, June–September 2013. MMWR Morb Mortal Wkly Rep. 2014;63(3):63. [PMC free article] [PubMed] [Google Scholar]
- 40.Lang TA, Secic M. How to Report Statistics in Medicine, Second Edition. Washington, DC: American College of Physicians; 2006. [Google Scholar]
- 41.Health Compass Milwaukee. Milwaukee Health Report: socioeconomic status and health. [Accessed May 30, 2019]. http://www.healthcompassmilwaukee.org/tiles/index/display?id=146057311458936237.
- 42.Falcó V, Fernándex de Sevilla T, Alegre J, Ferrer A, Martínez Vázquez JM. Legionella pneumophila: a cause of severe community-acquired pneumonia. Chest. 1991;100:1007–11. doi: 10.1378/chest.100.4.1007. [DOI] [PubMed] [Google Scholar]
- 43.Gleason JA, Ross KM, Greeley RD. Analysis of population-level determinants of legionellosis: spatial and geovisual methods for enhancing classification of high-risk areas. Int J Health Geogr. 2017;16(1):45. doi: 10.1186/s12942-017-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christenson M, Geiger SD, Phillips J, Anderson B, Losurdo G, Anderson HA. Heat vulnerability index mapping for Milwaukee and Wisconsin. J Public Health Manag Pract. 2017;23:396–403. doi: 10.1097/PHH.0000000000000352. [DOI] [PubMed] [Google Scholar]
- 45.Schatz J, Kucharik CJ. Seasonality of the urban heat island effect in Madison, Wisconsin. J Appl Meteorol Climatol. 2014;53:2371–86. doi: 10.1175/JAMC-D-14-0107.1. [DOI] [Google Scholar]
- 46.Fisman DN, Lim S, Wellenius GA, et al. It’s not the heat, it’s the humidity: wet weather increases legionellosis risk in the greater Philadelphia metropolitan area. J Infect Dis. 2005;192:2066–73. doi: 10.1086/498248. [DOI] [PubMed] [Google Scholar]
- 47.Cilloniz C, Ewig S, Gabarrus A, et al. Seasonality of pathogens causing community-acquired pneumonia. Respirology. 2017;22:778–85. doi: 10.1111/resp.12978. [DOI] [PubMed] [Google Scholar]
- 48.Snowman WJ, Holtzhauer FJ, Halpin TJ, Correa-Villasenor A. The role of indoor and outdoor occupations in the seroepidemiology of Legionella pneumophila. J Infect Dis. 1982;145:275. doi: 10.1093/infdis/145.2.275. [DOI] [PubMed] [Google Scholar]
- 49.Fraser DW, Tsai TR, Orenstein W, et al. Legionnaires’ disease: description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–97. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 50.Beauté J, Zucs P, de Jong B. Risk for travel-associated Legionnaires’ disease, Europe, 2009. Emerg Infect Dis. 2012;18:1811–6. doi: 10.3201/eid1811.120496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramirez JA, Wiemken TL, Peyrani P, et al. Adults hospitalized with pneumonia in the United States: incidence, epidemiology, and mortality. Clin Infect Dis. 2017;65:1806–12. doi: 10.1093/cid/cix647. [DOI] [PubMed] [Google Scholar]