Abstract
Six candidate sHSP genes were identified from the Glyphodes pyloalis transcriptome. All sHSP genes included full-length open reading frames and shared high similarity with the sequences of other lepidopteran species. These sHSP genes encoded 175–191 amino acid residues, and the predicted proteins had a molecular weight from 19.5 to 21.8 kDa. All GpsHSPs were expressed at lower levels at larval stages. All GpsHSPs were expressed at higher levels at diapaused, prepupal, or pupal stages, suggesting that sHSPs may be involved in metamorphosis in G. pyloalis. In addition to the developmental stage, extreme temperatures can induce variations in the expression of sHSPs genes. All GpsHSPs were significantly upregulated in larvae following exposure to heat shock, except GpHSP21.4 which downregulated at 4 h following exposure to the cold shock treatment. Furthermore, Starvation influenced the expression patterns of GpsHSPs as a function of the duration of food deprivation. Four GpsHSPs increased their expression with time of starvation until reaching to the peak level at 6 d of starvation. Finally, parasitism by the endoparasitoid Aulacocentrum confusum He et van Achterberg (Hymenoptera: Braconidae)-induced fluctuations in the expression of all GpsHSPs, and the expression varied with time after parasitization. Our results from this study strongly suggest functional differentiation within the sHSPs subfamily in G. pyloalis. The present study would provide further insight into the roles of sHSPs in G. pyloalis and novel avenues for promoting integrated management of this pest.
Keywords: Glyphodes pyloalis, small heat shock protein, environmental stress, expression patterns, mulberry pest
Heat shock proteins (HSPs), known as stress proteins and highly conserved molecular chaperones, help organisms to response stress and protect organisms from damage to cell. (Kim et al. 1998, Bukau et al. 2006, Hartl et al. 2011). In insects, five major families of HSPs have been identified, including the ATP-independent small heat shock proteins (sHSPs) and the ATP-dependent HSP40, HSP60, HSP70, and HSP90 (Feder and Hofmann 1999, Huang et al. 2008, Gu et al. 2012). It has been known that HSPs have consequences on development, immunity, and other protective functions such as tolerance under high temperature, desiccation, anoxia/hypoxia, oxidative stress, and starvation (Sun and Macrae 2005, Sonoda et al. 2006, Nakamoto and Vígh 2007). The sHSP family is widely distributed across eukaryotes and exhibits diverse structures and functions in each compartment of the cell, mostly ranging in monomer size from 12 to 43 kDa (Huang et al. 2008, Gu et al. 2012). Compared with other families of HSPs, sHSPs are less conserved, even within a species (Brocchieri et al. 2008, Johnson 2012). sHSPs can prevent irreversible denaturation of substrate proteins and protect them from damage caused by abiotic stressors such as heavy metals, antibiotics, and radiation (Nakamoto and Vígh 2007). sHSPs can also defend against pathological stressors, including virus infection, inflammation, and parasitization (Nakatsue et al. 1998, Roelofs et al. 2006). In addition, sHSPs play a significant role in mediating endogenous physiological processes such as hormone secretion, cell differentiation, and histogeny (King and Macrae 2015). Moreover, sHSPs have been found to be involved in many developmental processes, including insect diapause, metamorphosis, embryo formation, apoptosis, and autophagy (Haslbeck et al. 2019, Rodríguez et al. 2019, Sudnitsyna and Gusev 2019).
The first insect sHSP was identified in and isolated from Drosophila melanogaster Meigen (Diptera: Drosophilidae) (Tissiéres et al. 1974). Thanks to the development of genomics, transcriptomics, and other sequencing technologies, an increasing number of sHSPs have been reported in insects, including Galeruca daurica Joannis (Coleoptera: Chrysomelidae), Antheraea pernyi Hübner (Lepidoptera: Saturniidae), Musca domestica Linnaeus (Diptera: Muscidae), Bactrocera dorsalis Hendel (Diptera: Tephritidae), Ericerus pela Chavannes (Homoptera: Coccidae), Plutella xylostella Linnaeus (Lepidoptera: Plutellidae) and Bombyx mori Linnaeus (Lepidoptera: Bombycidae). (Li et al. 2009; Liu et al. 2014, 2018; Chen and Zhang 2015; Dou et al. 2017; Tan et al. 2017; Tian et al. 2018).
The expression of sHSPs vary depending on insect developmental stage or environmental conditions insects live in (Morrow and Tanguay 2012). For example, four of the six sHSPs in the tobacco armyworm Spodoptera litura Fabricius (Lepidoptera: Noctuidae) are expressed at varying levels across developmental stages (Shen et al. 2011). In Apis cerana cerana Fabricius (Hymenoptera: Apidae), AccHSP22.6 was highly expressed at different developmental stages and was significantly upregulated by temperature (Zhang et al. 2014). In the parasitoid wasp, Pteromalus puparum Linnaeus (Hymenoptera: Pteromalidae), the expression level of PpHSP20 increases significantly after 6 h of starvation but decreases after 24 h (Wang et al. 2012). Rinehart et al. (2007) demonstrated that inhibiting the expression of ScHSP23 can effectively reduce cold tolerance in Sarcophaga crassipalpis Macquart (Diptera: Sarcophagidae).
Parasitism can be a stressor inducing sHSPs in hosts (Rinehart et al. 2002). For example, relative levels of the expression of sHSP genes in the host Pieris rapae Linnaeus (Lepidoptera: Pieridae) pupa are up-regulated in response to parasitism by P. puparum (Zhu et al. 2013). In contrast, sHSP23 is downregulated in Plodia interpunctella Hübner (Lepidoptera: Pyralidae) on the fourth day after envenomation by Bracon hebetor Say (Hymenoptera: Braconidae) (Shim et al. 2008).
The mulberry pyralid caterpillar, Glyphodes pyloalis Walker is a destructive pest of mulberry (Morus alba L.) trees in China and other Asian countries (Khosravi and Sendi 2010, Poltavsky et al. 2017). It can cause heavy losses to the sericultural industry by decreasing the production of mulberry leaves, which is the exclusive natural food for the silkworm B. mori. The population of G. pyloalis usually reaches a high level from mid-summer through to mid-autumn when temperature gets to as high as 42°C in eastern and southern China. High temperature has consequences on the expression of multiple genes in G. pyloalis (Liu et al. 2017). Yet, it remains unknown whether sHSPs are involved in these gene responses to heat waves. Aulacocentrum confusum (Hymenoptera: Braconidae) is a preponderant parasitoid wasp of G. pyloalis, and generally parasitizes G. pyloalis larvae during their second to fifth instar with the parasitism rate reaching up to 58.2% under field condition (Yu et al. 2003). Whether sHSPs play any role in eliminating or reducing the parasitization risk in G. pyloalis is also unknown.
The present study aimed to identify sHSPs from transcriptome database, to analyze their sequence characteristics, and to examine gene expressions of sHSPs in G. pyloalis exposed to extreme temperatures, starvation, or parasitism. The information obtained from this study can improve the understanding of the significance of physiological function of sHSP in G. pyloalis.
Materials and Methods
Insects
The laboratory stock of G. pyloalis was collected from the mulberry field at the campus of Jiangsu University of Science and Technology in the city of Zhenjiang (119°44′N 32°20′E), Jiangsu Province, China. The larvae were reared with fresh mulberry (variety ‘Yu71-1’) leaves in the insectary [26 ± 2°C, 60–80% relative humidity, and photoperiod of 14:10 (L: D) h]. Adult moths were provided as supplementary food with 10% honey solution while being kept with two males and one female in transparent glass jars (20 × 20 × 30 cm) for mating and oviposition. Fresh mulberry leaves were provided as the substrate for deposition of eggs.
The parasitoid A. confusum was collected from rearing parasitized G. pyloalis larvae attacking mulberry trees in the mulberry field. The parasitoid colony was established with G. pyloalis larvae as hosts in the insectary [26 ± 2°C, 60–80% relative humidity, and photoperiod of 14:10 (L: D) h]. A honey solution of 10% was supplied via cotton lines to adult parasitoids.
Sample Preparation
To determine the relative expression levels of the sHSPs at different life stages of G. pyloalis, samples of first-instar, third-instar, fifth-instar larvae, pupae, male, and female adult moths were collected and stored at −80°C until RNA extraction. To obtain diapaused larvae following Chen et al. (2015), the fifth-instar larvae were collected and reared at 16°C for 7–10 d. Once the larvae molted into the prepupal stage to enter into the diapause, they were collected and kept for half a month without pupation before the experiment.
In the thermal treatment, 3-d-old fourth-instar larvae were selected to undergo heat and cold shock. Three temperature levels, 40, 25, and 0°C, were imposed to the larvae (Yang et al. 2016, Liu et al. 2017). To evaluate the effect of exposure time on gene expression, two times of the exposure (1 and 4 h) were examined for their short- and long-term effects. In the starvation treatment, our preliminary trial showed that the fourth-instar larvae remained alive for 6–7 d without food while the first- to third-instar larvae were not be able to survive this period of starvation; beyond this period, the fifth-instar larvae molted into the prepupae stage when food was absent. So, we applied food deprivation treatment to the fourth-instar larvae at three levels: 2, 4, and 6 d. To avoid cannibalism, these larvae were individually reared in Petri dishes (9 cm in diameter). The survived larvae were collected and stored at −80°C until further analysis. The control larvae were fed ad arbitrium with mulberry leaves.
The parasitization experiment was conducted following the methods described by Zheng et al. (2017). Briefly, five of the fourth-instar G. pyloalis larvae with similar body weights were exposed to one 6-d-old mated and naive female wasp in a glass tube (2 × 2 × 10 cm). Once the parasitization by the wasp was finished, the parasitized larvae were removed and individually placed in Petri dishes (9 cm in diameter). Healthy fourth-instar larvae were used as the control. These larvae at 0, 2, 4, or 6 d after parasitization were stored at −80°C until RNA extraction.
RNA Isolation and Reverse Transcription
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instruction. RNase-Free DNase I was used to remove residual genomic DNA (Promega, Madison, WI, USA). The concentration of RNA was confirmed by the absorbance at 260 nm, and the purity was determined according to an OD260/280 ratio between 1.9 and 2.1 using a 2100 Bioanalyzer (Agilent Technologies). Agarose gel electrophoresis was used to ensure that the RNA was integrated. The PrimeScript RT reagent Kit (Takara, Dalian, China) was used to produce first-strand DNA, followed by reverse transcription following the manufacturer’s recommendations, and the synthesized cDNA was stored at −20°C.
Sequence Identification and Bioinformatics Analysis
cDNA sequences encoding sHSPs were retrieved from a previously constructed G. pyloalis transcriptome dataset (GenBank SRR9959773) using the TBLASTN algorithm (E-value < 1 × 10−5), and all candidate genes were manually checked using BLASTX (E-value < 1 × 10−5) against the GenBank nonredundant (nr) protein database at the National Center for Biotechnology Information (NCBI). Annotated sHSP protein sequences from other insect species (including Apis mellifera ligustica Linnaeus [Hymenoptera: Apidae], P. rapae, Sesamia inferens Walker [Lepidoptera: Noctuidae], Cnaphalocrocis medinalis Guenée [Lepidoptera: Pyralidae], B. mori, Lymantria dispar Linnaeus [Lepidoptera: Lymantriidae], and Pyrausta nubilalis Hubern [Lepidoptera: Pyralidae]) were used as queries. The open reading frames (ORFs) of the putative sHSPs were predicted using ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). The homologous sequences of the sHSPs were identified in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Motif scans (https://myhits.isb-sib.ch/cgi-bin/motif_scan) by choosing ‘PROSITE profiles’ as the mot_source were used to find all known motifs in the present sequences (Hulo et al. 2006). Then, we predicted the molecular weight and isoelectric point of the proteins with ExPASy (https://web.expasy.org/compute_pi/) by choosing ‘Average’ as the Resolution. Sequence alignments and homology analysis were performed using ClustalX 2.1 multiple alignment with default gap penalty parameters of gap opening 10 and extension 0.2. The neighbor-joining (NJ) tree is constructed based on the principle of minimum evolution and suggested to be high accuracy, fast computation and has been wildly and successfully used in phylogenetic analysis, especially in sHSPs (Gkouvitsas et al. 2008, Kriehuber et al. 2010, Dou et al. 2017). Therefore, a tree was constructed using MEGA 7.0 (Tamura et al. 2007) with a p-distance model and a pairwise deletion of gaps. The bootstrap support of tree branches was assessed by resampling amino acid positions from 1,000 replicates. The secondary structure predictions were performed with the online PHD software accessed via the NPS@Web server (http://npsa-pbil.ibcp.fr;Combet et al. 2000) by choosing ‘PHD’ as the Method and with the output width as 70. We named the sHSPs according to their molecular weight.
Real-Time Quantitative PCR
RT-qPCR was conducted in QuantStudio 6 Flex (Thermo Fisher Scientific) in a total reaction volume of 20 µl, including 10 µl of 2 × iQTM SYBR Green I (Takara-Bio, Dalian, China), each gene-specific primer (Supp Table 1 [online only]) at 10 µM and the cDNA template. A cDNA dilution series (1, 1/3, 1/9, 1/27, and 1/81) with the sample cDNA was used to construct the standard curve and calculate the efficiency of amplification. In addition, a dissociation curve analysis (60–95°C at the end of each RT-qPCR run) was used to verify amplification of a single product. The qPCR conditions were as follows: 95°C for 5 min and 40 cycles of 95°C for 15 s and 60°C for 31 s. Each sample was run in triplicate with three biological replicates, and the relative gene expression levels among the different samples were measured via the 2−ΔΔCt method (Livak and Schmittgen 2001). The G. pyloalis GAPDH gene (GenBank MK243490) was used as the housekeeping gene to normalize target gene expression since it had been evaluated as the most stable reference gene for the temporal and spatial distribution of G. pyloalis in our previous studies. Differences in the expression levels of each target gene among the treatments were compared by one-way ANOVA followed by the least significant difference (LSD) test using R 3.2.2 software (R Development Core Team 2015).
Results
Identification and Characterization of Six GpsHSPs
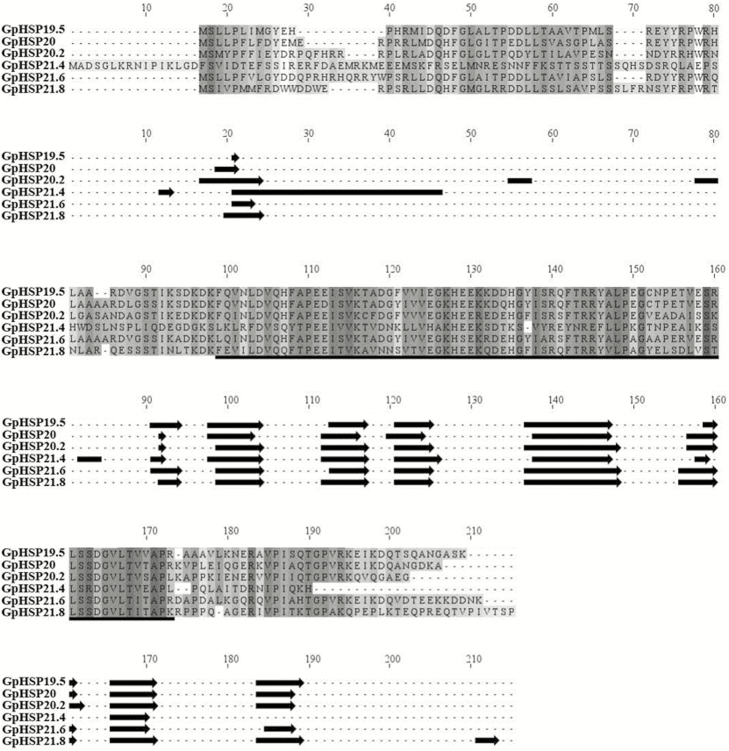
Six sHSP genes were identified from the G. pyloalis transcriptome and named according to their molecular weights (i.e., GpHSP19.5, 20, 20.2, 21.4, 21.6, and 21.8). The ORFs ranged from 525 to 573 bp and encoded 175 to 191 amino acids (Table 1). Their isoelectric points and predicted molecular weights are shown in Table 1. The deduced amino acid sequences of GpsHSPs contained a typical ɑ-crystalline domain consisting of six β-strands (Fig. 1). Unlike the conserved α-crystalline domain located in the C-terminal, their N-terminal is highly variable (Fig. 1).
Table 1.
Characteristics of sHSP mRNAs in Glyphodes Pyloalis
| Gene | GenBank accession number | Sequence length (bp) | Protein length (aa) | ORF | Molecular weight (kDa) | Isoelectric point | BLASTX best hit | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Description | Accession number | E-value | Identity(%) | |||||||
| GpHSP19.5 | MK225525 | 528 | 175 | complete | 19.5 | 6.30 | Bombyx mori | heat shock protein 22.6 | FJ602788.1 | 7E-41 | 78 |
| GpHSP20 | MK225526 | 534 | 177 | complete | 20.0 | 6.53 | Mamestra brassicae | heat shock protein 19.7 | AB251898.1 | 3E-104 | 80 |
| GpHSP20.2 | MK225527 | 537 | 178 | complete | 20.2 | 5.88 | Grapholita molesta | small heat shock protein 22.5 | KP843904.1 | 3E-10 | 83 |
| GpHSP21.4 | MK225528 | 564 | 187 | complete | 21.4 | 5.79 | Chilo suppressalis | heat shock protein 21.4 | JX491642.1 | 0 | 83 |
| GpHSP21.6 | MK225529 | 576 | 191 | complete | 21.6 | 7.03 | Helicoverpa armigera | heat shock protein 20.8 | KX845562.1 | 2E-100 | 80 |
| GpHSP21.8 | MK225530 | 576 | 191 | complete | 21.8 | 6.33 | Maruca vitrata | small heat shock protein 22.0 | KY565573.1 | 2E-157 | 85 |
Fig. 1.
Alignment of the deduced amino acid sequences of six GpsHSPs from Glyphodes pyloalis. Secondary structure elements are predicted for individual sequences. The conserved α-crystalline domain is underlined and is illustrated with cylinders (α-helices) and arrows (β-strands) below the alignment.
Phylogenetic Analysis of GpsHSPs
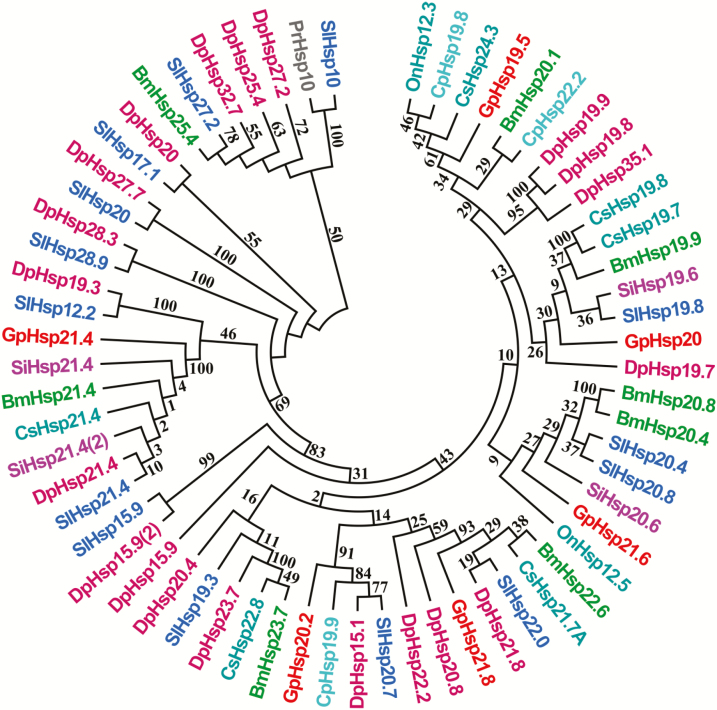
Results of the phylogenetic tree showed that insect sHSPs were divided into different clades (Fig. 2). Six G. pyloalis sHSPs were clustered into distinct branches, suggesting functional differentiation of GpsHSPs. GpHSP20 was grouped into a subbranch along with other insect sHSPs, which had similar molecular weight. By contrast, although GpHSP21.4, 21.6 and 21.8 had the similar molecular weight, they were divided into separate branches. GpHSP21.4 was clustered with HSP21.4 in S. inferens, B. mori, Chilo suppressalis Walker (Lepidoptera: Pyralidae), S. litura, and D. plexippus, suggesting that these sHSPs may be orthologs and undertake conserved functions. GpHSP21.4, GpHSP21.6, and GpHSP21.8 were grouped into a subbranch with other insect sHSPs but did not follow the similar molecular weight. GpHSP19.5 was assembled into a subbranch with C. suppressalis HSP24.3 and GpHSP20.2 fell into the subbranch with Cydia pomonella Linnaeus (Lepidoptera: Tortricidae) HSP19.9, D. plexippus HSP15.1, and S. litura HSP20.7 (Fig. 2).
Fig. 2.
Neighbor-joining phylogenetic tree of sHSP amino acid sequences from different insect species. The Glyphodes pyloalis sHSPs are labeled in red words. The full Latin names of all the species and the accession numbers of the genes from GenBank are presented with the corresponding abbreviations and listed in Supp Table 2 (online only). (Bm: Bombyx mori; Cp: Cydia pomonella; Cs: Chilo suppressalis; Dp: Danaus plexippus; On: Ostrinia nubilalis; Pr: Pieris rapae; Si: Sesamia inferens; Sl: Spodoptera litura)
Expression of GpsHSPs at Different Developmental Stages
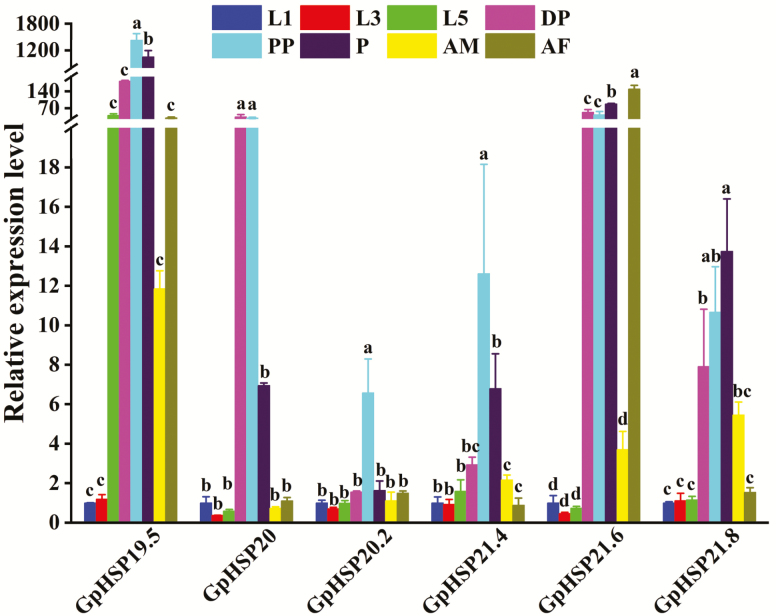
We used RT-qPCR to verify the expression levels of six GpsHSPs at different developmental stages (first-, third-, fifth-instar larvae, diapaused larvae, prepupae, pupae, female, and male adults). The results showed that all six sHSPs were expressed at relatively lower levels in first-, third-, and fifth-instar larvae than in other stages. GpHSP19.5, GpHSP20.2, and GpHSP21.4 reached their highest expression levels in the prepupae stage compared with the other stages. Furthermore, GpHSP20 showed the highest expression levels in the prepupae and diapause stages. Remarkably, GpHSP21.6 was highly transcribed in female adults but was expressed at extremely low levels in male adults. Meanwhile, it was expressed at high levels in pupae, diapause larvae and prepupae. GpHSP21.8 was expressed at the highest level at the pupal stage (Fig. 3).
Fig. 3.
Relative mRNA expression levels of six GpsHSPs at different developmental stages. first-instar larvae (L1), third-instar larvae (L3), fifth-instar larvae (L5), diapause larvae (DP), prepupae (PP), male adults (AM), and female adults (AF) of Glyphodes pyloalis. The relative expression levels of the GpsHSPs were determined by comparison with the first instar larvae. The different letters above the bars indicate significant differences (P < 0.05).
Expression of GpsHSPs at Different Temperatures
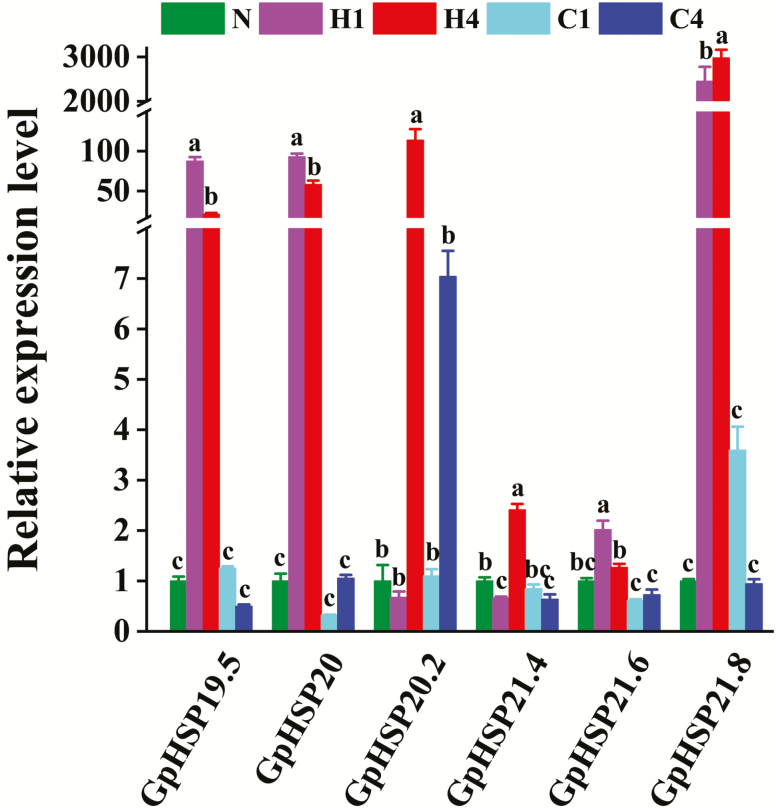
The temperature treatment significantly affected the expression of six GpsHSPs. The expression level of the sHSPs under the cold shock treatment did not significantly differ from that in the control, but GpHSP21.4 expression decreased after 4 h of the cold shock treatment. The expression levels of GpHSP19.5, 20, and 21.6 were highest after 1 h of heat treatment. Alternatively, the expression levels of GpHSP20.2, 21.4, and 21.8 were highest after 4 h of heat shock (Fig. 4).
Fig. 4.
Relative mRNA expression levels of six GpsHSPs at different temperatures. N: normal temperature control (25°C); H1: heat shock (40°C) for 1 h, H4: heat shock (40°C) for 4 h, C1: cold shock (0°C) for 1 h, C4: cold shock (0°C) for 4 h. Different lowercase letters indicate significant differences (P < 0.05).
Expression of GpsHSPs in Response to Starvation Stress
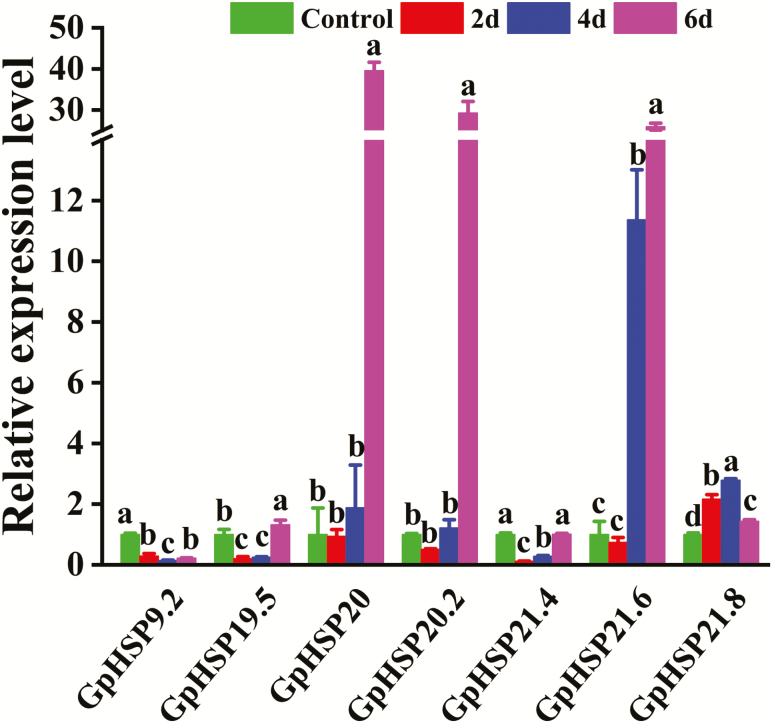
Starvation had a great impact on the expression levels of all six sHSPs, and this effect was dependent on the duration of food scarcity. The expression levels of GpHSP19.5, 20, 20.2, and 21.6 were highest until the sixth day of deprivation. In contrast, GpHSP21.8 was expressed most abundantly until the fourth day of starvation, while its expression level on the sixth day was lower than that on the second and fourth days. The expression level of GpHSP21.4 was comparable between the sixth day and the control but lower on the second and fourth day (Fig. 5).
Fig. 5.
Expression of GpsHSPs after different durations of starvation. Different lowercase letters indicate significant differences (P < 0.05).
Expression of GpsHSPs in Response to Parasitization Treatment
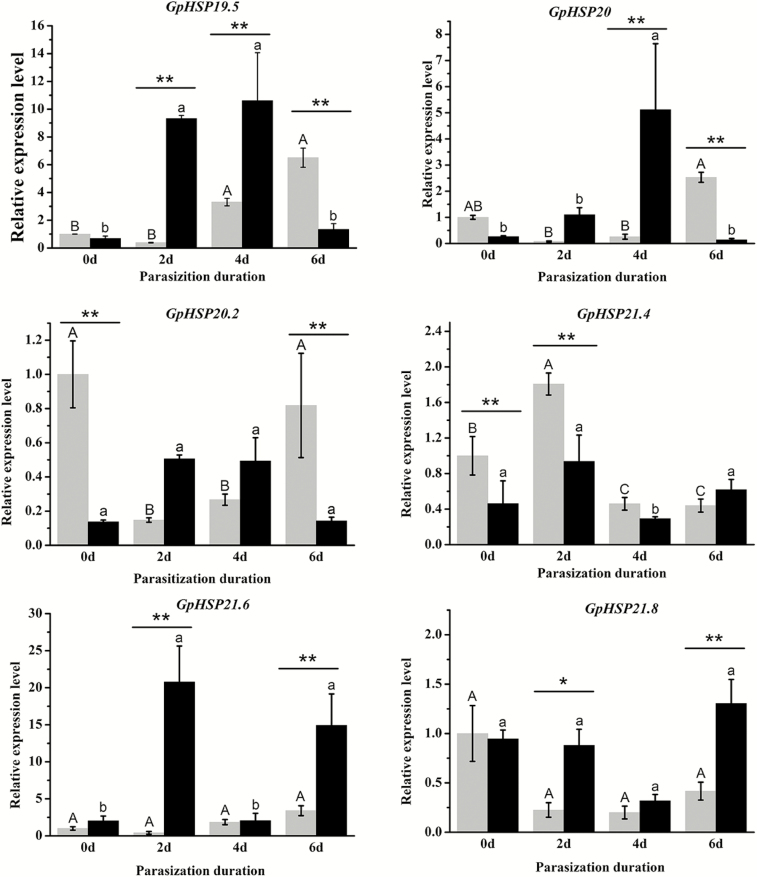
RT-qPCR validations were conducted to examine the mRNA transcription levels of six GpHSPs in G. pyloalis larvae parasitized by A. confusum. Parasitization significantly changed the expression levels of all candidate sHSPs in G. pyloalis larvae, and these differences exhibited temporal variability. When at parasitism (within one day after parasitization), GpHSP20.2 and 21.4 were expressed at lower levels in parasitized hosts than in healthy cohorts. The expression levels of GpHSP19.5, 21.6, and 21.8 were higher in the parasitoid-exposed groups than in unparasitized hosts, whereas GpHSP21.4 presented the opposite pattern 2 d after parasitization. After 4 d of development, the expression levels of GpHSP19.5 and 20.0 were higher in parasitized G. pyloalis larvae than in the control groups. The expression levels of GpHSP19.5, GpHSP20.0, and GpHSP20.2 were downregulated 6 d after parasitization compared with the healthy cohorts. In addition, GpHSP21.6 and GpHSP21.8 were expressed at higher levels in parasitized host larvae at this time point (Fig. 6).
Fig. 6.
Expression of GpsHSPs after parasitization by endoparasitoid, A. confusum for different periods. The gray bar represents the healthy groups and the black bar represents the parasitization groups. Different upper and lower case letters indicate significant differences between healthy and parasitization groups within the different days (P < 0.05). Single asterisks (P < 0.05) and double asterisks (P < 0.01) indicate significant differences between the healthy and parasitization groups within the same day.
Discussion
In the present study, a total of six sHSPs were identified from the G. pyloalis larval transcriptome dataset. In Grapholita molesta Busck (Lepidoptera: Tortricidae), fourteen new members from the sHSP families were identified by transcriptome sequencing (Zhang et al. 2015). The largest number of sHSP genes among insects has been reported in the silkworm B. mori until now. In contrast, in M. domestica, only four MdsHSPs have been identified (Tian et al. 2018). The variable numbers of sHSPs among insect species may result from gene duplication and a lack of data on these proteins. A full understanding of sHSP evolution could be achieved via genome-wide analysis in the future (Dou et al. 2017). Furthermore, apart from the conserved core domain existing in the members of the sHSP superfamily, which consists of the characteristic α-crystalline domain and its flanking sequences, variations in the N- and C-terminal regions contribute to the variability of the interspersed sequences of the protein and result in functional and structural differentiation of sHSPs (Kriehuber et al. 2010). In the present study, all GpsHSPs were predicted to contain the characteristic α-crystalline domains consisting of six β-strands. The further construction of a phylogenetic tree showed that all six GpsHSPs fell within homologous clades in terms of similar molecular weights, suggesting that the evolution of sHSPs is somewhat complex.
It has been shown that the expression of insect HSPs varies between developmental stages (Wang et al. 2017, Yang et al. 2019). The expression levels of PxHSP19.7 in P. xylostella (Sonoda et al. 2006) and LsHSP19.5, LsHSP20.8, and LsHSP21.7 in Liriomyza sativa Blanchard (Diptera: Agromyzidae) (Huang et al. 2009) is low at both larval and adult stages but highest at the pupal stage. Yet, SlHSP20.4 and SlHSP20.8 are expressed at a similar level across life stages in S. litura (Shen et al. 2011). In the present study, all sHSPs were expressed at higher levels in diapaused, prepupal, or pupal stages but lower at larval or adult stages. A previous study showed that the expression of HSP genes in S. crassipalpis increased upon entry into diapause (Yocum et al. 1998). In Choristoneura fumiferana Linnaeus (Lepidoptera: Tortricidae), most of the sHSP genes are involved in the diapause process, and they may play multiple and vital roles in different phases of this physiological process (Quan et al. 2018). Remarkably, the upregulation of sHSPs in diapaused insects is often explained in terms of increased cold tolerance and suppression of the development (Rinehart et al. 2007, King and Macrae 2015, Quan et al. 2018). So, we assume that the expression of sHSPs in diapaused larvae may be associated with overwintering in G. pyloalis. Alternatively, it may be correlated with metamorphosis as all GpHSPs were highly expressed at prepupal stage when the larvae terminated feeding and prepared for metamorphosis. It has been showed that sHSPs, as important chaperones, are markedly up-regulated during larval–pupal transformation, suggesting their essential roles in metamorphosis (Huang et al. 2009).
A crucial determinant of insect abundance and distribution is temperature, which at extremes can cause adaptive induction of HSP gene expression required for survival (King and Macrae 2015, Dou et al. 2017). A number of lepidopteran pests have been investigated to elucidate the relationship between temperature extremes and the expression level of sHSPs (Sun and Macrae 2005, Nakamoto and Vígh 2007, Haslbeck et al. 2019). sHSPs play important roles in insects upon exposure to thermal stresses (Gehring and Wehner 1995, Huang et al. 2009, Shen et al. 2011). In Harmonia axyridis Pallas (Coleoptera: Coccinellidae), the expression level of HaHSP16.25 and HaHSP21.00 increased initially and then decreased under the short-term cooling treatment, while the expression level of HaHSP10.87 and HaHSP21.56 increased during the entire process (Wang et al. 2017). In S. inferens, two sHSPs were significantly upregulated by a heat treatment, and three sHSPs were dramatically upregulated under low-temperature stresses (Sun et al. 2014). In C. pomonella, the relative expression level of sHSPs varied with different heat shock durations (Garczynski et al. 2011). In the present study, all sHSPs were highly expressed following exposure to heat stress as an increasing function of the duration of heat stress. The sHSPs in G. pyloalis reached their expression peaks after exposure to a short heat shock (1 h) but were notably down-regulated after exposure to a long heat shock (4 h), possibly because these HSPs responded more sensitively at the initial heat shock stage. In contrast, the higher expression level of GpsHSPs after exposure to a long heat shock may come as a result of an accumulation of expressed sHSPs with extension of heat shock (Gkouvitsas et al. 2008). Compared with heat shock associated with sHSPs, cold shock with them has received less attention until now. Recently, Tian et al. (2018) revealed that in M. domestica, one sHSP was expressed at a high level while another sHSP at a low level, following exposure to a cold treatment. In the present study, none of the GpsHSPs was expressed at a higher level following exposure to the cold shock than to normal temperature, yet even one sHSP was expressed at lower level under long cold treatment. Our previous study showed that one HSP70 in G. pyloalis was upregulated following 4-h exposure to a cold shock treatment (unpublished data). Therefore, it is assumed that the expression patterns of sHSPs associated with cold tolerance in insects are complicated or that sHSPs are less as active as other HSP classes under such stresses. More studies are required to unravel the mechanism behind the expression patterns of sHSPs in response to temperature extremes in G. pyloalis.
Starvation is a fatal stress in the wild and can interfere or destroy energy homeostasis in insects. In D. melanogaster, the function of DmHSP27 is associated with starvation stress (Hao et al. 2007). In starving M. domestica, the expression of MdHSP27 was downregulated as compared with the control (Tian et al. 2018). In a recent study, Xie et al. (2019) found that all adults of Tribolium castaneum Herbst (Coleoptera: Tenebrionidae) died within 18 d of starvation stress after the knockdown of TcHSP18.3, the longevity being much shorter than the control. These findings suggested that sHSPs can be sensitively responsive to starvation in insects. In the present study, four GpsHSPs were expressed at the highest level after 6 d following food deprivation, implying that these sHSPs could be activated specifically in response to long-term starvation. In contrast, two GpsHSPs showed a lower level of expression than the control at 2 or 4 d following the food deprivation, indicating that a shorter period of food starvation may suppress the expression of sHSPs. These sHSPs gene performances in G. pyloalis was similar to PpHSP20 in P. puparum (Wang et al. 2012). A possible explanation may lie that G. pyloalis cells can absorb nutrition through autophagy, which occurs in the comprehensive reorganization of cellular activities aimed at surviving low nutrient levels (Scott et al. 2004).
Parasitism causes severe stress on the host. Parasitoids inject at parasitization PDVs, venom or VLPs into their host bodies while depositing their eggs to guarantee a suitable environment for their offspring (Moreau and Asgari 2015). Our results of RT-qPCR showed that the expression level of the six GpsHSPs were significantly influenced by parasitism, and these effects depended on the developmental duration after parasitization. Similarly, in P. rapae, the PrHSP20 transcript level in pupae was higher at 12, 24, and 48 h after parasitization than in the healthy pupae (Zhu et al. 2013). Parasitism increases the relative expression level of sHSPs in P. interpunctella, indicating that sHSPs may be immunoregulatory agents that play important roles in the response to xenobiotic invasion (Shim et al. 2008). In contrast, GpHSP20.2 and GpHSP21.4 were downregulated immediately upon parasitization. A previous review showed that parasitism-associated factors can effectively regulate host development to create an environment favoring the development of parasitoids offspring (Beckage and Gelman 2004). Therefore, we assume that these GpsHSPs are inhibited following parasitism, especially during the initial stages.
Conclusion
In this study, we identified and characterized six sHSP genes in G. pyloalis. The structure of these sHSPs was predicted to contain conserved structural and functional domains and to incorporate typically variable C-terminal extensions. We determined the homology of GpsHSPs to sHSPs in other lepidopteran insect. Subsequently, we found that the six sHSPs exhibited unique expression patterns at different developmental stages and in response to various types of stresses. The information gained from the present study could provide a better understanding of the roles sHSPs play in G. pyloalis and thus helps in the integrative management of this pest.
Supplementary Material
Acknowledgments
We are grateful for the assistance of Dr. Sheng Qin at Jiangsu University of Science and Technology, Zhenjiang, China. This study was supported by the National Natural Science Foundation of China (31500312), the Key Research and Development program (Modern Agriculture) of Zhenjiang City (NY2019021), and the Special Fund for China Agriculture Research System (CARS-18).
References Cited
- Beckage N. E., and Gelman D. B.. . 2004. Wasp parasitoid disruption of host development: implications for new biologically based strategies for insect control. Annu. Rev. Entomol. 49: 299–330. [DOI] [PubMed] [Google Scholar]
- Brocchieri L., Conway de Macario E., and Macario A. J.. . 2008. hsp70 genes in the human genome: conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 8: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B., Weissman J., and Horwich A.. . 2006. Molecular chaperones and protein quality control. Cell. 125: 443–451. [DOI] [PubMed] [Google Scholar]
- Chen X. E., and Zhang Y.. . 2015. Identification of multiple small heat-shock protein genes in Plutella xylostella (L.) and their expression profiles in response to abiotic stresses. Cell Stress Chaperon. 20: 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. G., Yang Y. P., Zhang F., and Dai J. Z.. . 2015. Preliminary study on diapause termination and developmental duration of mulberry wintering larvae. Jiangsu Sericul. 37: 9–13. [Google Scholar]
- Combet C., Blanchet C., Geourjon C., and Deléage G.. . 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25: 147–150. [DOI] [PubMed] [Google Scholar]
- Dou W., Tian Y., Liu H., Shi Y., Smagghe G., and Wang J. J.. . 2017. Characteristics of six small heat shock protein genes from Bactrocera dorsalis: diverse expression under conditions of thermal stress and normal growth. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 213: 8–16. [DOI] [PubMed] [Google Scholar]
- Feder M. E., and Hofmann G. E.. . 1999. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61: 243–282. [DOI] [PubMed] [Google Scholar]
- Garczynski S. F., Unruh T. R., Guédot C., and Neven L. G.. . 2011. Characterization of three transcripts encoding small heat shock proteins expressed in the codling moth, Cydia pomonella (Lepidoptera: Tortricidae). Insect Sci. 18: 473–483. [Google Scholar]
- Gehring W. J., and Wehner R.. . 1995. Heat shock protein synthesis and thermotolerance in Cataglyphis, an ant from the Sahara desert. Proc. Natl. Acad. Sci. U. S. A. 92: 2994–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkouvitsas T., Kontogiannatos D., and Kourti A.. . 2008. Differential expression of two small Hsps during diapause in the corn stalk borer Sesamia nonagrioides (Lef.). J. Insect Physiol. 54: 1503–1510. [DOI] [PubMed] [Google Scholar]
- Gu J., Huang L. X., Shen Y., Huang L. H., and Feng Q. L.. . 2012. Hsp70 and small Hsps are the major heat shock protein members involved in midgut metamorphosis in the common cutworm, Spodoptera litura. Insect Mol. Biol. 21: 535–543. [DOI] [PubMed] [Google Scholar]
- Hao X., Zhang S., Timakov B., and Zhang P.. . 2007. The Hsp27 gene is not required for Drosophila development but its activity is associated with starvation resistance. Cell Stress Chaperones. 12: 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U., Bracher A., and Hayer-Hartl M.. . 2011. Molecular chaperones in protein folding and proteostasis. Nature. 475: 324–332. [DOI] [PubMed] [Google Scholar]
- Haslbeck M., Weinkauf S., and Buchner J.. . 2019. Small heat shock proteins: simplicity meets complexity. J. Biol. Chem. 294: 2121–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. H., Wang H. S., and Kang L.. . 2008. Different evolutionary lineages of large and small heat shock proteins in eukaryotes. Cell Res. 18: 1074–1076. [DOI] [PubMed] [Google Scholar]
- Huang L. H., Wang C. Z., and Kang L.. . 2009. Cloning and expression of five heat shock protein genes in relation to cold hardening and development in the leafminer, Liriomyza sativa. J. Insect Physiol. 55: 279–285. [DOI] [PubMed] [Google Scholar]
- Hulo N., Bairoch A., Bulliard V., Cerutti L., De Castro E., Langendijk-Genevaux P. S., Pagni M., and Sigrist C. J.. . 2006. The PROSITE database. Nucleic Acids Res. 34: D227–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L. 2012. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim. Biophys. Acta. 1823: 607–613. [DOI] [PubMed] [Google Scholar]
- Khosravi R., and Sendi J. J.. . 2010. Biology and demography of Glyphodes pyloalis Walker (Lepidoptera: Pyralidae) on mulberry. J. Asia-Pac. Entomol. 13: 273–276. [Google Scholar]
- Kim K. K., Kim R., and Kim S. H.. . 1998. Crystal structure of a small heat-shock protein. Nature. 394: 595–599. [DOI] [PubMed] [Google Scholar]
- King A. M., and MacRae T. H.. . 2015. Insect heat shock proteins during stress and diapause. Annu. Rev. Entomol. 60: 59–75. [DOI] [PubMed] [Google Scholar]
- Kriehuber T., Rattei T., Weinmaier T., Bepperling A., Haslbeck M., and Buchner J.. . 2010. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. Faseb J. 24: 3633–3642. [DOI] [PubMed] [Google Scholar]
- Li Z. W., Li X., Yu Q. Y., Xiang Z. H., Kishino H., and Zhang Z.. 2009. The small heat shock protein (sHSP) genes in the silkworm, Bombyx mori, and comparative analysis with other insect sHSP genes. BMC Evol. Biol. 9: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Su H., Li R., Li X., Xu Y., Dai X., Zhou Y., and Wang H.. . 2017. Comparative transcriptome analysis of Glyphodes pyloalis Walker (Lepidoptera: Pyralidae) reveals novel insights into heat stress tolerance in insects. BMC Genomics. 18: 974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. W., Yang P., Chen X. M., Xu D. L., and Hu Y. H.. . 2014. Cloning and expression analysis of four heat shock protein genes in Ericerus pela (Homoptera: Coccidae). J. Insect Sci. 14: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. N., Liu Y., Xin Z. Z., Zhu X. Y., Ge B. M., Li C. F., Wang D., Bian X. G., Yang L., Chen L., . et al. 2018. A small heat shock protein 21 (sHSP21) mediates immune responses in Chinese oak silkworm Antheraea pernyi. Int. J. Biol. Macromol. 111: 1027–1031. [DOI] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D.. . 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Meng E., Qiao T., Tang B., Hou Y., Yu W., and Chen Z.. . 2018. Effects of ovarian fluid, venom and egg surface characteristics of Tetrastichus brontispae (Hymenoptera: Eulophidae) on the immune response of Octodonta nipae (Coleoptera: Chrysomelidae). J. Insect Physiol. 109: 125–137. [DOI] [PubMed] [Google Scholar]
- Moreau S. J. M., and Asgari S.. . 2015. Venom proteins from parasitoid wasps and their biological functions. Toxins 7: 2385–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G., and Tanguay R. M.. . 2012. Small heat shock protein expression and functions during development. Int. J. Biochem. Cell Biol. 44: 1613–1621. [DOI] [PubMed] [Google Scholar]
- Nakamoto H., and Vígh L.. . 2007. The small heat shock proteins and their clients. Cell. Mol. Life Sci. 64: 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsue T., Katoh I., Nakamura S., Takahashi Y., Ikawa Y., and Yoshinaka Y.. . 1998. Acute infection of Sindbis virus induces phosphorylation and intracellular translocation of small heat shock protein HSP27 and activation of p38 MAP kinase signaling pathway. Biochem. Biophys. Res. Commun. 253: 59–64. [DOI] [PubMed] [Google Scholar]
- Poltavsky A. H., and Ilyina E. V.. . 2017. Glyphodes pyloalis Walker, 1859 (Lepidoptera, Crambidae) – a new species of tropical snout-moth for the fauna of Dagestan. Rus. J. Biol. Invasions 9: 101–112. [Google Scholar]
- Quan G., Duan J., Ladd T., and Krell P. J.. . 2018. Identification and expression analysis of multiple small heat shock protein genes in spruce budworm, Choristoneura fumiferana (L.). Cell Stress Chaperones. 23: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Developmental Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www. Rproject.org/.
- Rinehart J. P., Li A., Yocum G. D., Robich R. M., Hayward S. A., and Denlinger D. L.. . 2007. Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc. Natl. Acad. Sci. U. S. A. 104: 11130–11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinehart J. P., Denlinger D. L., and Rivers D. B.. . 2002. Upregulation of transcripts encoding select heat shock proteins in the flesh fly Sarcophaga crassipalpis in response to venom from the ectoparasitoid wasp Nasonia vitripennis. J. Invertebr. Pathol. 79: 62–63. [DOI] [PubMed] [Google Scholar]
- Rodríguez M. E., Arévalo D. E., Sanabria L. M., Carrión F. D. C., Fanelli M. A., and Rivarola V. A.. . 2019. Heat shock protein 27 modulates autophagy and promotes cell survival after photodynamic therapy. Photoch. Photobio. Sci., 18, 2. [DOI] [PubMed] [Google Scholar]
- Roelofs M. F., Boelens W. C., Joosten L. A., Abdollahi-Roodsaz S., Geurts J., Wunderink L. U., Schreurs B. W., van den Berg W. B., and Radstake T. R.. . 2006. Identification of small heat shock protein B8 (HSP22) as a novel TLR4 ligand and potential involvement in the pathogenesis of rheumatoid arthritis. J. Immunol. 176: 7021–7027. [DOI] [PubMed] [Google Scholar]
- Scott R. C., Schuldiner O., and Neufeld T. P.. . 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 7: 167–178. [DOI] [PubMed] [Google Scholar]
- Shen Y., Gu J., Huang L. H., Zheng S. C., Liu L., Xu W. H., Feng Q. L., and Kang L.. . 2011. Cloning and expression analysis of six small heat shock protein genes in the common cutworm, Spodoptera litura. J. Insect Physiol. 57: 908–914. [DOI] [PubMed] [Google Scholar]
- Shim J. K., Ha D. M., Nho S. K., Song K. S., and Lee K. Y.. . 2008. Upregulation of heat shock protein genes by envenomation of ectoparasitoid Bracon hebetor in larval host of Indian meal moth Plodia interpunctella. J. Invertebr. Pathol. 97: 306–309. [DOI] [PubMed] [Google Scholar]
- Sonoda S., Ashfaq M., and Tsumuki H.. . 2006. Cloning and nucleotide sequencing of three heat shock protein genes (hsp90, hsc70, and hsp19.5) from the diamondback moth, Plutella xylostella (L.) and their expression in relation to developmental stage and temperature. Arch. Insect Biochem. Physiol. 62: 80–90. [DOI] [PubMed] [Google Scholar]
- Sudnitsyna M. V., and Gusev N. B.. . 2019. Is the small heat shock protein HspB1 (Hsp27) a real and predominant target of methylglyoxal modification? Cell Stress Chaperones. 24: 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., and MacRae T. H.. . 2005. Small heat shock proteins: molecular structure and chaperone function. Cell. Mol. Life Sci. 62: 2460–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Lu M. X., Tang X. T., and Du Y. Z.. . 2014. Characterization and expression of genes encoding three small heat shock proteins in Sesamia inferens (Lepidoptera: Noctuidae). Int. J. Mol. Sci. 15: 23196–23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., and Kumar S.. . 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tan Y., Zhang Y., Huo Z. J., Zhou X. R., and Pang B. P.. . 2017. Molecular cloning of heat shock protein 10 (HSP10) and 60 (HSP60) cDNAs from Galeruca daurica (Coleoptera: Chrysomelidae) and their expression analysis. B. Entomol. Res. 108: 1–13. [DOI] [PubMed] [Google Scholar]
- Tian L., Wang X., Wang X., Lei C., and Zhu F.. . 2018. Starvation-, thermal- and heavy metal- associated expression of four small heat shock protein genes in Musca domestica. Gene. 642: 268–276. [DOI] [PubMed] [Google Scholar]
- Tissières A., Mitchell H. K., and Tracy U. M.. . 1974. Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J. Mol. Biol. 84: 389–398. [DOI] [PubMed] [Google Scholar]
- Wang H., Li K., Zhu J. Y., Fang Q., Ye G. Y., Wang H., Li K., and Zhu J. Y.. . 2012. Cloning and expression pattern of heat shock protein genes from the endoparasitoid wasp, Pteromalus puparum in response to environmental stresses. Arch. Insect Biochem. Physiol. 79: 247–263. [DOI] [PubMed] [Google Scholar]
- Wang H. J., Shi Z. K., Shen Q. D., Xu C. D., Wang B., Meng Z. J., Wang S. G., Tang B., and Wang S.. . 2017. Molecular cloning and induced expression of six small heat shock proteins mediating cold-hardiness in Harmonia axyridis (Coleoptera: Coccinellidae). Front. Physiol. 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Hu X. X., Zhai M. F., Yu X. J., Song X. W., Gao S. S., Wu W., and Li B.. . 2019. Characterization and functional analysis of hsp18.3 gene in the red flour beetle, Tribolium castaneum. Insect Sci. 26: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. J., Xu K. K., Cao Y., Meng Y. L., Liu Y., and Li C.. . 2019. Identification and expression analysis of four small heat shock protein genes in cigarette beetle, Lasioderma serricorne (Fabricius). Insects 10: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. Q., Zhang Y. L., Wang X. Q., Dong H., Gao P., and Jia L. Y.. . 2016. Characterization of multiple heat-shock protein transcripts from Cydia pomonella: their response to extreme temperature and insecticide exposure. J. Agric. Food Chem. 64: 4288–4298. [DOI] [PubMed] [Google Scholar]
- Yocum G. D., Joplin K. H., and Denlinger D. L.. . 1998. Upregulation of a 23 kDa small heat shock protein transcript during pupal diapause in the flesh fly, Sarcophaga, crassipalpis. Insect Biochem. Mol. Biol. 28: 677–682. [DOI] [PubMed] [Google Scholar]
- Yu H., and Zhou Q.. . 2003. Researches on parasitic bees of Diaphania pyloalis in Zhejiang Province. Sci. Sericul. 28: 330–334 [Google Scholar]
- Zheng Y., Liao C. W., Zhou Y., Zhang X. R., Wang J., Wu S. Y., Sheng S., and Wu F. A.. . 2017. Determination of body weight and nutrient content in haemolymph of Diaphania pyloalis larvae after parasitism by Aulacococentrum confusum. Sci. Sericul. 43: 744–749 [Google Scholar]
- Zhang Y. Y., Liu Y. L., Guo X. L., Li Y. L., Gao H. R., Guo X. Q., and Xu B. H.. . 2014. sHSP22.6, an intronless small heat shock protein gene, is involved in stress defence and development in Apis cerana cerana. Insect Biochem. Molec. 53: 1–12. [DOI] [PubMed] [Google Scholar]
- Zhang B., Zheng J. C., Peng Y., Liu X. X., Hoffmann A. A., and Ma C. S.. . 2015. Stress responses of small heat shock protein genes in Lepidoptera point to limited conservation of function across phylogeny. PLoS One 10: e132700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. Y., Wu G. X., Ye G. Y., and Hu C.. . 2013. Heat shock protein genes (hsp20, hsp75 and hsp90) from Pieris rapae: molecular cloning and transcription in response to parasitization by Pteromalus puparum. Insect Sci. 20: 183–193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.