Abstract
Stylosanthes (Papilionoideae, Leguminosae) is a predominantly Neotropical genus with ~48 species that include worldwide important forage species. This study presents the chromosome number and morphology of eight species of the genus Stylosanthes (S. acuminata, S. gracilis, S. grandifolia, S. guianensis, S. hippocampoides, S. pilosa, S. macrocephala, and S. ruellioides). In addition, staining with CMA and DAPI, in situ hybridization with 5S and 35S rDNA probes, and estimation of DNA content were performed. The interpretation of Stylosanthes chromosome diversification was anchored by a comparison with the sister genus Arachis and a dated molecular phylogeny based on nuclear and plastid loci. Stylosanthes species showed 2n = 20, with low cytomolecular diversification regarding 5S rDNA, 35S rDNA, and genome size. Arachis has a more ancient diversification (~7 Mya in the Pliocene) than the relatively recent Stylosanthes (~2 Mya in the Pleistocene), and it seems more diverse than its sister lineage. Our data support the idea that the cytomolecular stability of Stylosanthes in relation to Arachis could be a result of its recent origin. The recent diversification of Stylosanthes could also be related to the low morphological differentiation among species, and to the recurrent formation of allopolyploid complexes.
Keywords: Arachis, cytogenetics, evolution, Leguminosae, Stylosanthes
Introduction
Chromosomal evolution, including polyploidization (auto or allopolyploidy), chromosomal rearrangements, and heterochromatin polymorphisms are known to be important mechanisms promoting speciation in plants (Raskina et al., 2008; Soltis and Soltis, 2009; Reis et al., 2014; Pinto et al., 2016). However, chromosome changes may be neutral, with no direct impact on the adaptability or reproductive viability of the individuals carrying the polymorphism (Levin, 2002), and will not necessarily result in speciation. For example, Pimentel et al. (2017) investigated patterns of diversification and chromosomal evolution in Pooideae (Poaceae) in the light of past environmental changes. In this group, the haploid basic chromosome number has remained stable, with no direct association of chromosome transitions with diversification shifts.
One aspect that would influence the accumulation of cytomolecular diversity in plant lineages over time is the age of the group (Levin, 2002; Soltis and Soltis, 2009; Susek et al., 2016). Therefore, it would be expected that groups of recent diversification would have more stable and uniform karyotypes, while those with older divergences would have time to accumulate more cytomolecular differences, resulting in higher karyotypical diversity. However, it is not clear if the timing of diversification of a given lineage reflects variation at the cytogenetic level. For example, there are cases of recently evolved lineages with highly variable karyotypes (e.g., genus Nothoscordum [Amaryllidaceae] Souza et al., 2012), and ancient groups showing high karyotype stability (e.g., subfamily Bombacoideae [Malvaceae] Costa et al., 2017).
In recent years, different studies reported new insights on the phylogeny and karyotype evolution of legumes, making this group an interesting model to investigate diversification and karyotype evolution. The period between the origin and diversification of Leguminosae was estimated to be short, with the divergence of the main lineages around 60 million years ago (Mya) (Lavin et al., 2005). Within the Leguminosae, subfamily Papilionoideae comprises one of the most diverse and ecologically successful plant radiations, presenting a diversification history stemming from the early Cenozoic (Lavin et al., 2005; Cardoso et al., 2012; LPWG - Legume Phylogeny Working Group, 2017). Wong et al. (2017) suggested that a single whole genome duplication (WGD) event near to the base of Fabales was associated with the onset of Leguminosae diversification and that subsequent chromosome number reductions contributed to the success and diversification of Papilionoideae.
Within the papilionoid Pterocarpus clade (Cardoso et al., 2013), the sister genera Arachis and Stylosanthes represent a useful model for analyzing the evolution of karyotypic diversity in closely related lineages of similar size. The South American genus Arachis comprises approximately 82 species arranged in nine sections that were grouped according to the geographic distribution, morphology, and cross-compatibility (Krapovickas and Gregory 1994; Valls and Simpson, 2005), and includes widely cultivated crops (peanut) and forages. Stylosanthes is a predominantly neotropical genus with nearly 48 species with low inter and intraspecific morphological variation (Stace and Cameron, 1984; Costa, 2006), including important species cultivated as forage in Africa and Australia (Costa, 2006). Due to their adaptability to low fertility soils and nitrogen fixing capacity, some species of the genus are grown to recover degraded soils (Sheltom et al., 2005; Starr et al., 2013; Liu et al., 2016).
The species of Arachis are mostly autogamous and have the basic chromosome number x = 10 (except x = 9 in A. decora Krapov., W.C.Greg. & Valls, A. porphyrocalyx Valls & C.E.Simpson, A. palustris Krapov., W.C.Greg. & Valls, and A. praecox Krapov., W.C.Greg. &Valls). There are diploid (2n = 2x = 20 or 2n = 2x = 18) and tetraploids species (2n = 4x = 40) with six different genomes (A, B, D, F, G, and K) (Fernández and Krapovickas, 1994; Peñaloza and Valls, 2005; Silvestri et al., 2015; Ortiz et al., 2017). DNA contents are known for 23 species of Arachis, with an average of 2C = 2.83 pg (http://data.kew.org/cvalues). On the other hand, Stylosanthes karyotype data are extremely scarce, with most data coming from chromosome counting, and few species with genome size and molecular cytogenetics were investigated (Vieira et al., 1993; Chandra and Kaushal, 2009; Lira, 2015; Marques et al., 2018). Most of the species of the genus are diploids (2n = 20) but few polyploid species (2n = 40, 60) were reported (Cameron, 1967; Karia, 2008). Their chromosomes are small and with similar morphology, making it difficult to recognize chromosome pairs and to interpret the karyotype evolution (Vieira et al., 1993). A recent study based on cytogenetic and genomic data of the allopolyploidy S. scabra (2n = 40) revealed its origin from two diploid progenitors (Marques et al., 2018).
Phylogenetic and cytogenetic analyses are useful to investigate the time and mode of genome evolution, as well as to examine the impact of chromosome changes in plant diversification (e.g., Escudero et al., 2012). The combination of fluorescent in situ hybridization (FISH) and/or fluorochrome banding with phylogenetic comparative methods is a powerful tool for reconstructing detailed karyotype evolution (Guerra, 2012; Van-Lume et al., 2017). These techniques are useful to display chromosome morphological features, heterochromatin distribution, and physical locations of repetitive DNA in plants. Such combination of methods is particularly interesting in revealing relationships among species and their genomic organizations, helping to understand the evolutionary history of plant groups (Reis et al., 2014; Ortiz et al., 2017; Samoluk et al., 2017). Here, we analyzed chromosome numbers and morphology of eight species of Stylosanthes, including chromomycin A3 (CMA) and 4’-6-diamidino-2-phenylindole (DAPI) chromosome banding, FISH for 5S and 35S ribosomal DNA (rDNA), as well as genome size estimate by flow cytometry. These data were used to investigate the relationship between timing of diversification and karyotypic diversity, using for comparison the sister genus Arachis, which has been extensively characterized in terms of cytology. The interpretation of chromosome variation within genera was anchored by a dated molecular phylogeny based on nuclear and plastid sequences.
Materials and Methods
Plant material
The present study was based on material obtained from seeds of 12 accessions of eight Stylosanthes species [S. acuminata, S. gracilis, S. grandifolia, S. guianensis, S. hippocampoides, S. macrocephala, S. pilosa, and S. ruellioides]. Five accessions of S. guianensis (1480, 4171, 1463, LC2538, and cv. Mineirão) were also investigated (see Table S1 (69.5KB, pdf) ).
For cytogenetic analysis, root tips obtained from seeds or seedlings were pretreated with 8-hydroxyquinoline (0.003 M) for 7 h at room temperature, fixed in ethanol:acetic acid (3:1; v/v) for 24 h at room temperature, and then stored at -20 °C. Fixed root tips were washed in distilled water and digested in a solution of 2% (w/v) cellulase /20% (v/v) pectinase (Onozuka) at 37 °C for 5 h. The slides were prepared according to Carvalho and Saraiva (1993, 1997).
Chromosome banding
Chromosome banding was performed according to Schweizer (1976) with few modifications. Slides were stained with CMA (0.5 mg/mL) for 1h, dystamicyn (0.1 mg/mL) for 30 min and DAPI (2 μg/mL) for 30 min. The slides were mounted in glycerol:McIlvaine buffer pH 7.0 (1:1 v/v), and examined using an epifluorescence microscope (Olympus BX51).
Fluorescent in situ hybridization (FISH)
Fluorescent in situ hybridization (FISH) was performed using a 18S (located in a repetitive unit known as 45S rDNA, Roa and Guerra, 2015; or 35S rDNA, Garcia et al., 2017) spacer rDNA probes from Triticum aestivum L. (Gerlach and Bedbrook, 1979) and 5S rDNA probes from Zea mays L. (courtesy of Koo and J. Jiang). Each probe was labeled with digoxigenin by nick translation using DIG-Nick Translation Mix (Roche), and then hybridized according to Jiang et al. (1995) with some modifications. The hybridization mixture was denatured at 90 °C for 10 min and immediately transferred to an icebox. The slides were denatured at 85 °C for 1 min and treated with a series of alcohol washes (70% ice cold, 90% and 100% at room temperature for 5 min each). The hybridization mixture was then added to the slides and the chromosomes allowed to hybridize at 37 °C for 24 h. Post hybridization washes were carried out using 2 SSC buffer (0.3 mol/L sodium citrate, 0.03 mol/L sodium chloride, pH 7) and 1 PBS buffer (0.136 mol/L sodium chloride, 0.27 mol/L potassium chloride, 0.1 mol/L dibasic sodium phosphate, 0.2 mol/L monobasic potassium phosphate, pH 7.4). Probes were detected with anti-DIG conjugate with rhodamine (Sigma) and post detection washes were performed using 1 TNT buffer (0.1 mol/L Tris, 0.15 mol/L sodium chloride, 0.05% Tween-20) and 1X PBS at room temperature. Metaphases were counter-stained with 2 μg/mL of DAPI (Sigma). The slides were mounted in Vectashield (Vector, Burlingame, California, USA), and samples were rehybridized. Images with 5S (red) and 18S (green pseudocolor) signals were merged using Adobe Photoshop CS5.
Flow cytometry
Nuclear DNA content estimation was performed according to Galbraith et al. (1983) using a FacsCanto II Flow Cytometer. Approximately 20-30 mg of young and fresh leaves and the same amount of tissue of standard references (Pisum sativum L. 9.09 pg) were chopped with 1 mL of WPB lysis buffer (Loureiro et al., 2007). The histograms were analyzed with Flowing Software 2.5.
DNA nuclear content (pg) of each sample was estimated by the relative fluorescence intensity of the sample and the internal reference standard. Three samples of each species/accessions were measured according to Dolezel et al. (2003):
were PIFI is the fluorescence intensity of propidium iodide in G1 cells. DNA content among S. guianensis accessions was analyzed by ANOVA using Genes computational software (Cruz, 2013).
Phylogenetic sampling
The phylogenetic analysis of Stylosanthes and its sister genus Arachis was based on ITS nuclear region (ITS1-5.8S-ITS2) and matK plastid gene. ITS and matK sequences of 25 species of Arachis were retrieved from previous studies (see GenBank references in Table 1; Lavin et al., 2001; Bechara et al., 2010). However, only four matK sequences were available for Arachis species. New ITS and matK sequences for ten accessions of Stylosanthes were generated (Genbank accession numbers MH223599 - MH223618). The related species Chapmannia floridana Torr. and A. Gray (Cardoso et al., 2013) was included as an outgroup (Genbank accession numbers KJ772648 and AF203562).
Table 1. Species analyzed with their respective chromosome numbers, DNA content (average and coefficient of variation), and GenBank accession numbers.
| Genus / Species | 2n | 2C-value (pg) | CV (%) | nº of rDNA sites | GenBank No. | References | |||
|---|---|---|---|---|---|---|---|---|---|
| 5S | 35S | ITS | matK | C-values | rDNA | ||||
| Arachis L. | |||||||||
| A. batizocoi Krapov. & W.C.Greg. | 20 | 2.83 | 3.65 | 1 | 3 | AY615256.1 | - | Samoluk et al., 2014 | Robledo and Seijo 2010 |
| A. cardenasii Krapov. & W.C.Greg. | 20 | 3.01 | 4.15 | 1 | 4 | AY615236.1 | - | Samoluk et al., 2014 | Robledo et al., 2009 |
| A. correntina (Burkart) Krapov. & W.C.Greg. | 20 | 2.85 | 3.55 | 1 | 2 | AF203554.1 | - | Samoluk et al., 2014 | Seijo et al., 2004 |
| A. cruziana Krapov., W.C. Greg. & C.E. Simpson | 20 | 2.59 | 4.33 | 3 | 3 | AY615259.1 | - | Samoluk et al., 2014 | Robledo and Seijo, 2010 |
| A. duranensis Krapov. & W.C.Greg. | 20 | 2.55 | 3.76 | 1 | 2 | AY615240.1 | - | Samoluk et al., 2014 | Seijo et al., 2004 |
| A. helodes Krapov. & Rigoni | 20 | 2.81 | 2.59 | 1 | 3 | AY615241.1 | - | Samoluk et al., 2014 | Robledo et al., 2009 |
| A. hypogaea L. | 40 | 2.80 | 2.80 | 2 | 5 | AF156675.2 | KX257487.1 | Samoluk et al., 2014 | Robledo and Seijo, 2010 |
| A. ipaensis Krapov. | 20 | 3.19 | 3.71 | 1 | 3 | AY615257.1 | - | Samoluk et al., 2014 | Robledo and Seijo, 2010 |
| A. kretschmeri Krapov. & W.C.Greg. | 20 | - | - | 1 | 1 | AY615220.1 | - | - | - |
| A. kuhlmannii Krapov. & W.C.Greg. | 20 | 3.09 | 4.11 | 1 | 3 | AY615219.1 | - | Samoluk et al., 2014 | Robledo et al., 2009 |
| A. magna Krapov., W.C.Greg. & C.E.Simpson | 20 | 3.22 | 3.70 | 1 | 4 | AF203555.1 | - | Samoluk et al., 2014 | Robledo and Seijo, 2010 |
| A. major Krapov. & W.C.Greg. | 20 | - | - | - | - | AY615229.1 | AF203597.1 | - | - |
| A. monticola Krapov. & Rigoni | 40 | 2.85 | 2.76 | 2 | 5 | AY615239.1 | - | Samoluk et al., 2014 | Robledo and Seijo, 2010 |
| A. pintoi Krapov. & W.C.Greg. | 20 | 5.95 | - | 1 | 2 | AJ320397.1 | AF203596.1 | Singh et al., 1996 | Lavia et al., 2011 |
| A. schininii Krapov. Valls & C.E.Simpson | 20 | 3.18 | 4.11 | 1 | 2 | AY615248.1 | - | Samoluk et al., 2014 | Robledo et al., 2009 |
| A. simpsonii Krapov. & W.C.Greg. | 20 | 3.08 | 3.94 | 1 | 3 | AY615247.1 | - | Samoluk et al., 2014 | Robledo et al., 2009 |
| A. stenosperma Krapov. | 20 | 2.96 | 3.36 | 1 | 3 | AY615252.1 | - | Samoluk et al., 2014 | Robledo et al., 2009 |
| A. triseminata Krapov. & W.C.Greg. | 20 | 3.05 | - | 1 | 2 | AF204233.1 | AF203599.1 | Singh et al., 1996 | Raina et al., 1999 |
| A. valida Krapov. & W.C.Greg. | 20 | 3.16 | 4.12 | 1 | 4 | AY615244.1 | - | Samoluk et al., 2014 | Robledo and Seijo, 2010 |
| A. villosa Benth. | 20 | 3.04 | 2.94 | 1 | 2 | AF203558.1 | - | Samoluk et al., 2014 | Robledo et al., 2009 |
| A. williamsii Krapov. & W.C.Greg. | 20 | 3.2 | 3.68 | 1 | 1 | AY615255.1 | - | Samoluk et al., 2014 | Seijo et al., 2004 |
| Stylosanthes Sw. | |||||||||
| S. acuminata M.B.Ferreira & Sousa Costa | 20 | 2.2 | 4.19 | 1 | 2 | MH223618 | MH223603 | This study | This study |
| S. gracilis Kunth | 20 | 2.5 | 2.5 | 1 | 1 | MH223617 | MH223604 | his study | This study |
| S. grandifolia M.B.Ferreira & Sousa Costa | 20 | 2.5 | 4.21 | 1 | 1 | MH223609 | MH223599 | This study | This study |
| S. guianensis (Aubl.) Sw. 1463 | 20 | 2.81 | 4.66 | 1 | 1 | MH223611 | MH223606 | This study | This study |
| S. guianensis (Aubl.) Sw. 1480 | 20 | 3.02 | 3.48 | 1 | 1 | MH223613 | MH223602 | This study | This study |
| S. guianensis (Aubl.) Sw. 4171 | 20 | 3.07 | 4.11 | 1 | 1 | MH223612 | MH223607 | This study | This study |
| S. guianensis (Aubl.) Sw. LC2538 | 20 | - | - | 1 | 1 | - | - | - | - |
| S. guianensis (Aubl.) Sw. Mineirão | 20 | 2.80 | 3.06 | 1 | 1 | MH223610 | MH223605 | This study | This study |
| S. hamata (L.) Taub. | 20 | 1.80 | - | 1 | 1 | - | - | Marques et al., 2018 | Marques et al., 2018 |
| S. hippocampoides Mohlenbr | 20 | 2.53 | 3.85 | 1 | 1 | MH223614 | MH223608 | This study | This study |
| S. macrocephala M.B.Ferreira & Sousa Costa | 20 | 2.05 | 3.16 | 1 | 1 | MH223616 | MH223601 | This study | This study |
| S. ruellioides Mart. ex Benth. | 20 | - | - | 1 | 1 | MH223615 | MH223600 | - | This study |
| S. scabra Vogel | 40 | 2.86 | - | 2 | 1 | - | - | Marques et al., 2018 | Marques et al., 2018 |
| 20 | - | - | - | - | - | - | - | - | |
| S. pilosa M.B.Ferreira & Sousa Costa | |||||||||
| S. viscosa (L.) Sw. | 20 | 1.36 | - | 1 | 1 | - | - | Marques et al., 2018 | Marques et al., 2018 |
DNA extraction, polymerase chain reaction (PCR), and sequencing
DNA extraction was performed according to Doyle and Doyle (1987) using seed cotyledon/embryo tissue (30 mg per species). DNA quantification was done in a NanoDrop 2000c spectrophotometer (Thermo Scientific). The complete ITS region (ITS1-5.8S-ITS2), was amplified with universal primers ITS 4 and ITS 5 (White et al., 1990). The maturase K (matK) gene was also amplified using universal primers 1RKIM and 3FKIM (Bremer et al., 2002). Amplifications were performed in 50 μL of reaction volume containing 200 ng of genomic DNA and final concentrations of 1 buffer, 0.3 mM MgCl, 0.2 mM DNTP, 0.1 μM of each specific primers, 1.25 U Taq DNA polymerase, 1 TBT and MiliQ water to complete the volume. ITS PCRs were incubated at 95 ºC for 5 minutes, followed by 35 cycles of 1 min at 95 ºC, 1 min at 55 ºC, 1 min at 72 ºC, and finally, 1 min at 72 ºC in a thermocycler (Applied Biosystems). matK reactions were run using the same program with few adjustments: annealing temperature (56 ºC) and extension time (10 min. After visualizing the PCR products in agarose 1% gel, the products were purified by precipitation. Purified PCR products were sequenced in a 3500 Genetic Analyzer (Applied Biosystems).
Phylogenetic analysis and character reconstruction
DNA sequences were analyzed, edited, and aligned using Geneious software (version 7.1.9). An incongruence length difference test (ILD) was made in PAUP* (40.b10) (Swofford, 2002) to determine the statistical significance of incongruence between the data partitions. Bayesian inference search was performed with the ITS1-5.8S-ITS2 + matK concatenated alignment. Since matK sequences were missing for most Arachis species (see Table 1) we also performed a phylogenetic analysis with ITS only to compare with the topology of the concatenated alignment (Figure S1 (122KB, pdf) ). The Bayesian inference search was performed using CIPRES Science Gateway (Miller et al., 2010) using MrBayes 3.2.1 on XSEDE (Ronquist and Huelsenbeck, 2003) plugin, with 10,000,000 generations.
To interpret the evolution of cytomolecular diversification of the genus Stylosanthes, a molecular clock analysis was performed using BEAST v.1.8.0 (Drummond et al., 2012a). We used the Aikake Criterion in Jmodeltest2 (Darriba et al., 2012) to select the best evolutionary model, which identified the GTR+I+G model for both partitions. Analyses were run using an uncorrelated log normal relaxed clock and a Yule Process speciation model. Two independent runs of 20,000,000 generations each were performed, sampling every 1,000 generations. In order to verify the effective sampling of all parameters and assess convergence of independent chains, we examined their posterior distributions in Tracer v.1.6. (Rambaut et al., 2014) and the MCMC sampling was considered sufficient at effective sampling sizes (ESS) higher than 200. After removing 25% of samples as burn-in, the independent runs were combined and a maximum clade credibility (MCC) tree was constructed using TreeAnnotator v.1.8.2. (Drummond et al., 2012a). Divergence time between Arachis and Stylosanthes estimated by a previous study (13.8 ± 1.7 Mya; Lavin et al., 2004) was used as a secondary calibration point using a normal prior.
The interpretation of karyotype data evolution of Stylosanthes and Arachis was done by plotting 5S rDNA, 35S rDNA, and genome size data on the dated phylogeny obtained. For this, the ancestral state reconstruction of cytomolecular characters (number of 5S and 35S rDNA) were performed in Mesquite v. 3.51 (Maddison and Maddison, 2018). The trace character history function was used with the 50% majority-rule consensus tree from the Bayesian inference analyses. The ancestral state was inferred using maximum parsimony, in which all changes are equally probable. The number of 5S and 35S rDNA sites was assumed as continuous data. Chromosome data of species were taken from the literature (Marques et al., 2018) or from this work, and they were treated as an unordered, multistate character.
Results
All species of Stylosanthes here investigated were diploids (2n = 20) with predominantly metacentric chromosomes, measuring on average 2.7 μm (Figures 1 -4). Most of the species (S. hippocampoides, S. gracilis, S. macrocephala, S. pilosa, S. ruellioides, and S. guianensis accessions 1480, 1463, 4171, and LC2538), showed two CMA+/DAPI- signals in the short arms of the smaller chromosomal pair. In addition to those bands, in S. ruellioides CMA-/DAPI+ proximal bands were also observed in all chromosomes (Figure 1). On the other hand, S. acuminata, S. grandifolia, and S. guianensis cv. Mineirão showed four CMA+ bands, two of them in the short arms of the smaller chromosome pair and the other two in the proximal region of a large chromosome pair (Figure 1d, g, l).
Figure 1. DAPI/CMA banding profile in Stylosanthes species. (a) S. guianensis 1480, (b) S. guianensis 4171, (c) S. guianensis 1463, (d) S. acuminata, (e) S. guianensis LC2538, (f) S. pilosa, (g) S.guianensis cv. Mineirão, (h) S. ruellioides, (i) S. gracilis, (j) S. macrocephala, (k) S. hippocampoides, (l) S. grandifolia. Bar = 10μm.
Figure 2. 5S (red) and 35S (green) probes mapped in Stylosanthes species by fluorescent in situ hybridization. (a) S. hippocampoides, (b) S. gracilis, (c) S. grandifolia, (d) S. macrocephala, (e) S. acuminata, (f) S. ruellioides, (g) S. guianensis LC2538, two extra points are distended satellites, (h) S. guianensis cv. Mineirão, (i) S. guianensis 4171, two extra points are distended satellites, (j) S. guianensis 1480 Bar = 10μm.
Figure 3. Idiograms of Stylosanthes species showing chromosome length (L), arm ratio (AR), 5S (red), CMA bands colocalized with 35S r DNA sites (green), and CMA bands colocalized with 5S (yellow).
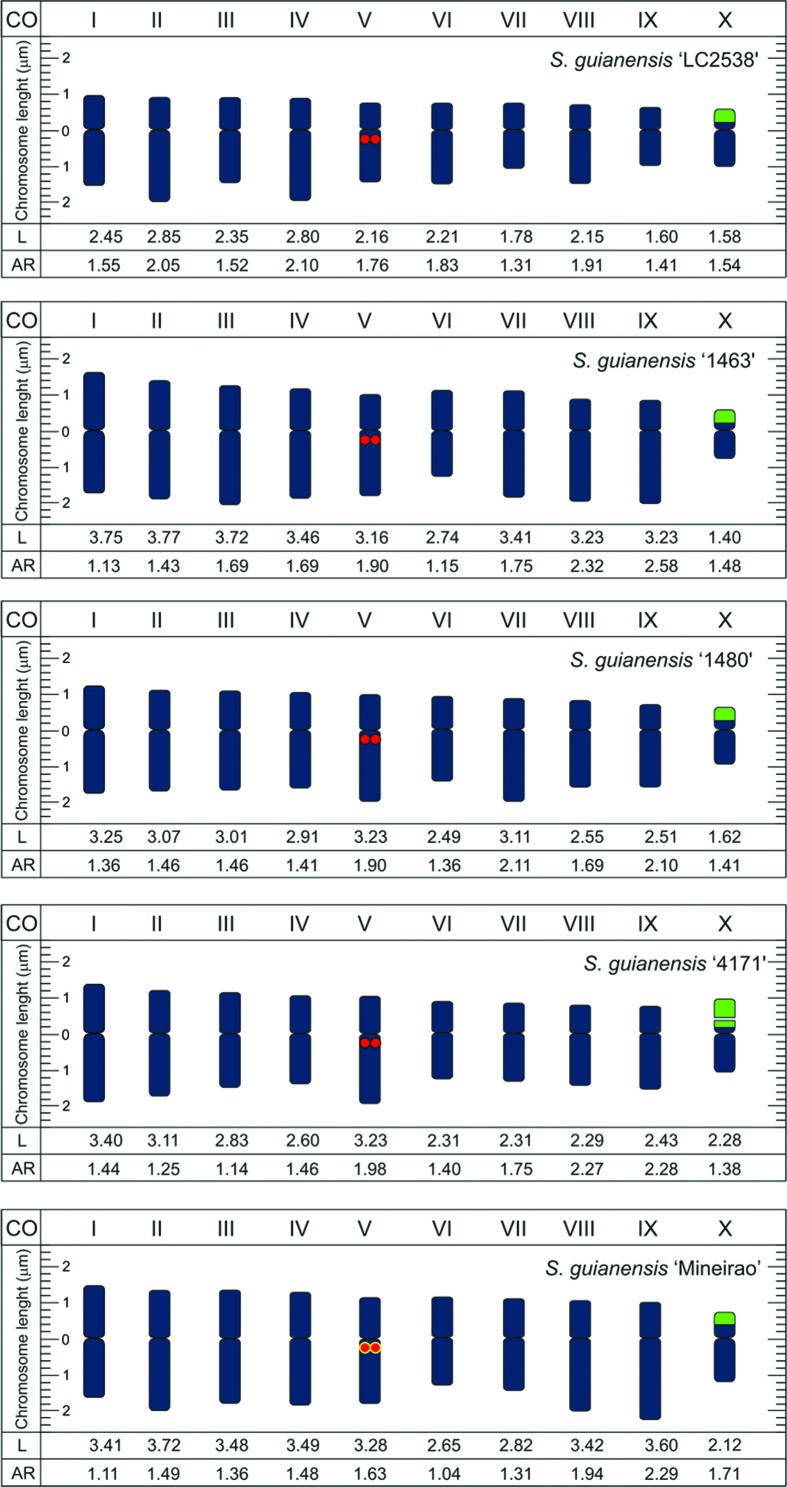
Figure 4. Idiograms of S. guianensis accessions showing chromosome length (L), arm ratio (AR), 5S (red), CMA bands co-localized with 35S r DNA sites (green), and CMA bands co-localized with 5S (yellow).
FISH analyses revealed 35S rDNA sites co-localized with the CMA+ bands in the short arms of the smaller acrocentric pair, as it was observed in S. gracilis, S. grandifolia, S. hippocampoides, S. macrocephala, and S. ruellioides (Figure 2a-d, f and Figure 3). The 5S rDNA sites were localized in the proximal region of one chromosome pair (Figure 2a-j and Figure 3). In S. acuminata we observed four 35S rDNA sites (Figure 2e). No variation in the number of rDNA sites was observed among S. guianensis accessions, being possible to map one pair of 5S rDNA and one pair of 35S rDNA (Figure 2g-j and Figure 4).
The DNA content among Stylosanthes species ranged from 2.05 to 3.07 pg (Table 1), with S. macrocephala showing the lowest value (2.05 pg) and S. guianensis accessions the highest (2.8 to 3.07). These values were analyzed by ANOVA and there were no significant differences between accessions (p=10.6194).
Molecular phylogenetic analysis including Stylosanthes (eight spp.) and Arachis (21 spp.) were performed. ILD test revealed no significant incongruence (p < 0.05) between matK and ITS datasets. Thus, Bayesian inference was conducted with the concatenated alignment matK + ITS since this alignment produced better resolved relationships. The concatenated alignment contains 1,180 base pairs, where the matK region seems to be more conserved (identity = 98.3%) in both groups when compared to the ITS region (identity = 91.5%). The genera Arachis and Stylosanthes were recovered as monophyletic (Figure 5), with the diversification of Arachis starting on the upper Miocene (7.10 Mya; credibility interval 7.80–4.16 Mya), followed by a more recent origin of Stylosanthes at (2.90 Mya; CI 3.25–2.60 Mya). The clade represented by S. acuminata, S. gracilis, S.grandifolia, S. hippocampoides, and the four accessions of S. guianensis showed more recent diversification in the Pleistocene (~3 Mya).
Figure 5. Chronogram of Arachis and Stylosanthes species based on BEAST analysis using the plastid matK and nuclear ITS combined datasets. Blue bars indicate 95% highest posterior density intervals. a, comparative DNA content evolution, with symbols next to accessions proportional to genome size. b, ancestral character estimation of number of rDNA sites along the branches and nodes of the phylogeny. The color of edges in the tree represents observed and reconstructed values for chromosome number on the tree. Red colors correspond to relatively high numbers of rDNA sites; whereas dark blue colors represent low number of observed and reconstructed rDNA sites.
Available cytogenetic data and genome size were also considered to interpret the molecular phylogenetic of the genera Stylosanthes and Arachis (Table 1, Figure 5). We categorized DNA contents into smaller (2C = 1.36 to 2.83 pg) and larger (2C = 2.84 to 3.28 pg). Most Stylosanthes fell into the 2C = 1.36–2.83 pg range size, except for two S. guianensis accessions (2C = 3.02 and 2C =3.07 pg). On the other hand, most Arachis species presented 2C values from 2C =2.87 to 3.28 pg, except for A. duranensis, A. batizocoi, A. cruziana, and A. helodes (2C = 2.55, 2.83, 2.59 and 2.81 pg, respectively) (Figure 5a). The number of 5S rDNA sites ranged from one to three in Arachis. All Stylosanthes species showed one pair of the rDNA 5S site. Regarding the 35S rDNA sites, it was possible to identify higher variation in Arachis (one to five pairs). For Stylosanthes, every species presented only one pair of the same site (except S. acuminata) (Figure 5b).
Discussion
Karyotype stability in Stylosanthes
Although most of the species of the genus Stylosanthes are diploids, few polyploid species, such as S. hamata and S. scabra with 2n= 40, and S. erecta with 2n=60 were reported (Cameron, 1967; Karia, 2008; Polido et al., 2015). The diploid chromosome number (2n = 2x = 20) here observed is in agreement with previous report for the genus Stylosanthes (Vieira et al., 1993; Lira, 2015; Marques et al., 2018). Morphometric analysis of the Stylosanthes karyotypes revealed that the chromosomal profile was similar for all diploid species previously investigated (Vieira et al., 1993), corroborating our findings. The predominance of metacentric chromosomes suggests a symmetrical karyotype, which is generally observed in other members of Leguminosae (Bandel, 1974; Kumari and Bir, 1989; Vieira et al., 1993; Pinto et al., 2016; Van-Lume et al., 2017).
The stability or variability in legume karyotypes can be associated with the timing of diversification, reproduction strategies, and other factors related to the evolutionary history of each group (Soltis and Soltis, 2009; Gu et al., 2016). Genome comparisons have showed conserved syntenic blocks between papilionoid genomes, especially among phylogenetically closely related species (Young and Bharti, 2012). In the genus Phaseolus (Papilionoideae), for example, a karyotypic stability of chromosomal numbers (except for dysploidy in the clade Leptostachyus) revealed by C-banding and fluorochrome staining (Zheng et al., 1993; Almeida and Pedrosa-Harand, 2013) was observed and associated with a recent diversification over the last 5 Mya (Delgado-Salinas et al., 2006). In this genus, the use of BAC-FISH mapping indicated a high level of macro-collinearity among homologous chromosomes (Almeida and Pedrosa-Harand, 2013; Fonsêca and Pedrosa-Harand, 2017). However, other legumes with recent diversification such as Vigna may present polymorphic karyotypes (Delgado-Salinas et al., 2011; She et al., 2014), indicating that time alone would not be the only factor responsible for the accumulation of karyotypic variability.
Genome size in Stylosanthes
The small variation of DNA content (1.4 fold) in the Stylosanthes species analyzed here corroborates a scenario of karyotypic stability. Although the average of DNA estimation is in agreement with other estimations for the genus, we observed different C-values for two diploid species (S. seabrana B.L.Maass & ‘t Mannetje and S. viscosa) for which 5.45 pg were reported in the literature (Chandra and Kaushal, 2009). Variation of DNA content within a genus may be due to different factors such as recombination, deletion and retrotransposition (Piegu et al., 2006; Baziz et al., 2014). Among closely related species, other mechanisms appear to have some impact on genome size variations, such as expansion of tandem repeated DNA sequences, variation in intron size, and transfer of organellar DNA to the nucleus (Deutsch and Long, 1999; Morgante et al., 2002; Adams and Palmer, 2003; Baziz et al., 2014). Compared to other Leguminosae, the amount of DNA in Stylosanthes seems to be relatively high. Leucaena macrophylla Benth. has 2C value = 0.62 pg, Trifolium arvense L. was described with 2C = 0.78 pg and Lotus coimbrensis Willd. with 2C = 0.90 pg, although, Lathyrus latifolius L. (2C = 21.76 pg) and Vicia faba L. (2C = 54.8 pg) have the highest DNA content within the family (Cheng and Grant, 1973; Bennett et al., 1982; Hartman et al., 2000; Vizintin et al., 2006). The reason why the DNA contents of eukaryotic genomes vary independently remains a matter of speculation. The same is true for the questions of whether there is a general tendency for increase or decrease of genome size and whether genome size and/or chromosome number have an adaptive value. Some authors hypothesized that three processes of genome evolution (shrinkage, expansion, and equilibrium) might be involved in achieving the optimal balance between genomic stability and plasticity (Gregory, 2001; Schubert and Giang, 2016).
Heterochromatin pattern in Stylosanthes
The co-localization of all 35S rDNA sites with CMA+ bands as found in Stylosanthes species is commonly reported in plants (Siljak-Yakolav et al., 2003). Interestingly, the co-localization of 5S rDNA with heterochromatic bands, as observed in S. guianensis cv. Mineirão and S. grandifolia, is a rare condition in other higher plants (Cabral et al., 2006; Vasconcelos et al., 2010; Bernardes et al., 2013). Karyotype variability in Papilionoideae has been characterized by comparative cytogenetic studies of heterochromatin bands and rDNA sites distribution. In the genus Crotolaria, different heterochromatin types were observed, suggesting the occurrence of replacement of repetitive DNA families during the genus diversification (Mondin et al., 2007; Mondin and Aguiar-Perencin, 2011; Morales et al., 2011). In Astragalus, it was reported variation in number, intensity, and position of CMA bands along the chromosomes, likewise 5S rDNA sites (Baziz et al., 2014), while cultivated species of Canavalia showed variations in the rDNA positions (She et al., 2017). In Lotus japonicus (Regel) K. Larsen and L. filicaulis Durieu, for which three 35S loci have been identified, initially no polymorphism of this type was observed (Pedrosa et al., 2002). Nevertheless, a comparative cytogenetic map built for Lotus uliginosus (L.) Schkuhr (2n = 12) revealed intra and interspecific chromosomal rearrangements in L. japonicus, L. filicaulis, and L. burttii Borsos. Changes in the number, size, and position of rDNA sites were observed, as well as an intraspecific heteromorphism of the 5S rDNA site in L. uliginosus (Ferreira et al., 2012). Comparative analyses have demonstrated variations in the position of 5S and 45S rDNA sites in Medicago L. species, giving evidence of new rearrangements throughout the evolutionary history of the genus (Yu et al., 2017). Meanwhile, the genus Lens showed variations mainly in 5S sites (Balyan et al., 2002; Fernandez et al., 2005). Although wide cytogenetic variation related to the repetitive fraction of the genome (heterochromatic bands and rDNA sites) has been reported in different Papilionoid genera, there is little information on the degree of intergeneric variability of whole genomes (Schubert and Giang, 2016).
Variation in heterochromatin composition was also observed in Stylosanthes. However, only two species showed different number and positions in CMA+ bands. Thus, comparing with Arachis, this variability of heterochromatin can still be considered low (Silvestri et al., 2015). Interestingly, DAPI+ proximal bands were observed only in S. ruellioides. This species belongs to the same clade of Stylosanthes scabra complex (S. viscosa, S. hamata, and S. scabra) (Marques et al., 2018), that also showed DAPI+ bands in a (peri)centromeric location, suggesting that these species are closely related.
Evolutionary trends in Stylosanthes genome
The knowledge about the karyotype organization in Stylosanthes contributes to a better understanding of its evolutionary history, which is so far exemplified by the general stability in genome size and chromosome numbers. Here we evaluated the evolution of karyotypic diversity in Stylosanthes by comparing the variation in the number of rDNA sites and DNA content between its sister clade. In general, cytogenetic data give evidence of cytomolecular stability of Stylosanthes in relation to Arachis (Vieira et al., 1993; Silvestri et al., 2015), suggesting that the age of the group could be an important factor to be considered in karyotype diversification. However, a more representative sampling within Stylosanthes and Arachis in the dating analysis would be required to confirm the age estimates of major lineages within these genera. Low morphological differentiation, phylogenetic similarity of plastid and nuclear DNA sequences, as well as crosses between species (allopolyploidization) from different clades support the cytomolecular stability in the recently evolved Stylosanthes (Vander Stappen et al., 2003). In contrast, Arachis presents a high variability of the CMA/DAPI banding and rDNA sites distribution (5S and 18S), being possible to identify six genomes (A, B, D, F, G, and K) (Fernández and Krapovickas, 1994; Peñaloza and Valls, 2005; Silvestri et al., 2015; Ortiz et al., 2017). Smartt et al. (1978) initially established the A and B genomes based on the two different chromosome complements in the allotetraploid A. hypogaea (AABB). The “A chromosomes” have a differential condensation pattern during prometaphase (Fernández and Krapovickas, 1994) and have a large heterochromatic band DAPI+ in the centromeric region (Seijo et al., 2004). The remaining species with symmetric karyotypes, but without “A chromosomes”, have been assigned to one single genome group named B genome or non-A genome (Seijo et al., 2004; Robledo and Seijo, 2010). In addition, Arachis presents a greater diversity of species and morphological variability compared to Stylosanthes.
Although rDNA occupies a large fraction of the nuclear genome, it is also an unstable genomic region and the reasons for this instability are not fully understood (Kobayashi, 2008; Totta et al., 2017). Recent studies have explored the dynamics of rDNA loci under an explicit phylogenetic framework. Totta et al. (2017) inferred that a stasis in 45S rDNA site number occurred during most of the evolutionary history of Cistus (Cistaceae) and allied genera. The authors suggested that most of the multiple shifts involving changes in the number of the rDNA loci likely occur since the Middle Pleistocene in this group, and that rDNA stasis in site number may have been underestimated in diploids. The terminal positions of 35 rDNA loci might also facilitate higher frequencies of interlocus homogenization than is found in interstitial or pericentromeric loci (Garcia et al., 2017)
Analysis of the evolutionary rates of DNA sequence data in the rainforest tree genus Swartzia (Papilionoideae) indicates that it diversified rapidly after its origin, probably during the Miocene (Torke, 2006). Such information, coupled with cytogenetic features (small chromosomes, two 5S, and 45S rDNA sites in metaphases as well as two positive CMA bands) are preliminary evidences of strongly conserved karyotypes in a recently evolved species-rich lineage (Pinto et al., 2016). The stability observed in some groups of recent divergence was also identified in other papilionoid genera such as Phaseolus (Fonsêca and Pedrosa-Harand, 2013, 2017). The lupin genus (Lupinus) encompasses about 270 annual and perennial species distributed in both the Old World (generally around the Mediterranean basin) and the New World (primarily North and South America) (Drummond et al., 2012b). The Old World lineage originated between 17–20 Mya while New World species evolved around 2–5 Mya. The Old Word lupins show a high level of genomic diversification characterized by variation in chromosome numbers (2n = 32–52), basic chromosome numbers (x = 5–7, 9, 13), nuclear genome size, patterns of chromatin modifications at the chromosomal level, and 5S/45S rDNA sites number (Naganowska et al., 2002, 2003; Hajdera et al., 2003; Susek et al., 2016, 2017). On the other hand, New World lupins have more uniform karyotypes in terms of their genome structure, and have chromosome numbers of either 2n = 36 or 2n = 48 with a fixed basic chromosome number x = 6 (Naganowska et al., 2006; Susek et al., 2016). This reinforces the view that within the same group, lineages with older diversification tends to show high karyotype diversity compared to more recent lineages, as was reported here for the sister genera Arachis and Stylosanthes.
Acknowledgments
We thank the financial support received from Coordenação de Aperfeiçoamento Pessoal de Nível Superior (CAPES – Finance Code 001), National Council for Scientific and Technological Development (CNPq), Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG) and Fundação de Amparo à Pesquisa e Tecnologia do Estado de Pernambuco (FACEPE; APQ-0970-2.03/2015).
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Marcelo Guerra
Conflict of interest
The authors declare that there is no conflict of interest.
Author contributions
GS, LFV and SMS conceived and designed the study; ALF, AF and LMP conducted the experiments; ALF, GS and LFV wrote the manuscript. MAC, MFS and SMS have contributed with biological material and manuscript writing. All authors read and approved the final version of the manuscript.
References
- Adams KL, Palmer JD. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- Almeida C, Pedrosa-Harand A. High macro-collinearity between lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.) as revealed by comparative cytogenetic mapping. Theor Appl Genet. 2013;126:1909–1916. doi: 10.1007/s00122-013-2106-9. [DOI] [PubMed] [Google Scholar]
- Balyan HS, Houben A, Ahne R. Karyotype analysis and physical mapping of 18S-5.8S-25S and 5S ribosomal RNA loci in species of genus Lens Miller (Fabaceae) Caryologia. 2002;55:121–128. [Google Scholar]
- Baziz K, Benamara-Bellagha M, Pustahija F, Brown SC, Siljak-Yakovlev S, Khalfallah N. First karyotype analysis, physical rDNA mapping, and genome size assessment in 4 North African Astragalus taxa (Fabaceae) Turk J Bot. 2014;38:1248–1258. [Google Scholar]
- Bennett MD, Smith JB, Heslop-Harrison JS. Nuclear DNA amounts in angiosperms. Proc R Soc Lond B. 1982;216:179–199. [Google Scholar]
- Bandel G. Chromosome numbers and evolution in the Leguminosae. Caryologia. 1974;27:17–32. [Google Scholar]
- Bechara MD, Moretzsohn MC, Palmieri DA, Monteiro JP, Bacci M, Martins J, Valls JFM, Lopes CR, Gimenes MA. Phylogenetic relationships in genus Arachis based on ITS and 5.8 S rDNA sequences. BMC Plant Biol. 2010;10:255. doi: 10.1186/1471-2229-10-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes ECS, Benko-Iseppon AM, Vasconcelos S, Carvalho R, Brasileiro-Vidal AC. Intra and interespecific chromosome polymorphisms in cultivated Cichorium L. species (Asteraceae) Genet Mol Biol. 2013;36:357–363. doi: 10.1590/S1415-47572013005000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer B, Bremer K, Heidari N, Erixon P, Olmstead RG, Anderberg AA, Källersjö M, Barkhordarian E. Phylogenetics of asterids based on 3 coding and 3 non-coding chloroplast DNA markers and the utility of non-coding DNA at higher taxonomic levels. Mol Phylogenet Evol. 2002;24:274–301. doi: 10.1016/s1055-7903(02)00240-3. [DOI] [PubMed] [Google Scholar]
- Cabral JS, Felix LP, Guerra M. Heterochromatin diversity and its co-localization with 5S and 45S rDNA sites in chromosomes of four Maxillaria species (Orchidaceae) Genet Mol Biol. 2006;29:659–664. [Google Scholar]
- Cameron DF. Chromosome number and morphology of some introduced Stylosanthes species. Aust J Agric Res. 1967;18:375–379. [Google Scholar]
- Cardoso D, Queiroz LP, Pennington RT, Lima HC, Fonty E, Wojciechowski MF, Lavin M. Revisiting the phylogeny of Papilionoid legumes: New insights from comprehensively sampled early-branching lineages. Am J Bot. 2012;99:1991–2013. doi: 10.3732/ajb.1200380. [DOI] [PubMed] [Google Scholar]
- Cardoso D, Pennington RT, De Queiroz LP, Boatwright JS, Van Wyk BE, Wojciechowski MF, Lavin M. Reconstructing the deep-branching relationships of the papilionoid legumes. S Afr J Bot. 2013;89:58–75. [Google Scholar]
- Carvalho CR, Saraiva LS. A new heterochromatin banding pattern revealed by modified HKG banding technique in maize chromosomes. Heredity. 1993;70:515–519. [Google Scholar]
- Carvalho CR, Saraiva LS. High-resolution HKG-banding in maize mitotic chromosomes. J Plant Res. 1997;110:417–420. [Google Scholar]
- Chandra A, Kaushal P. Identification of diploid Stylosanthes seabrana Aaccessions from existing germplasm of S. scabra utilizing genome-specific STS. Mol Biotechnol. 2009;42:282–291. doi: 10.1007/s12033-009-9154-z. [DOI] [PubMed] [Google Scholar]
- Cheng RIJ, Grant WF. Species relationships in the Lotus corniculatus group as determined by karyotype and cytophotometric analyses. Can J Genet Cytol. 1973;15:101–115. [Google Scholar]
- Costa N. Revisão do gênero Stylosanthes. D. Sc. Thesis. Universidade Técnica de Lisboa; Lisboa: 2006. p. 470. [Google Scholar]
- Costa L, Oliveira A, Carvalho-Sobrinho J, Souza G. Comparative cytomolecular analyses reveal karyotype variability related to biogeographic and species richness patterns in Bombacoideae (Malvaceae) Plant Syst Evol. 2017;303:1131–1144. [Google Scholar]
- Cruz CD. GENES - a software package for analysis in experimental statistics and quantitative genetics. Acta Sci Agron. 2013;35:271–276. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Salinas A, Bibler R, Lavin M. Phylogeny of the genus Phaseolus (Leguminosae): A recent diversification in an ancient landscape. Syst Bot. 2006;31:779–791. [Google Scholar]
- Delgado-Salinas A, Thulin M, Pasquet R, Weeden N, Lavin M. Vigna (Leguminosae) sensu lato: The names and identities of the American segregate genera. Am J Bot. 2011;98:1694–1715. doi: 10.3732/ajb.1100069. [DOI] [PubMed] [Google Scholar]
- Deutsch M, Long M. Intron-exon structure of eukaryotic model organisms. Nucleic Acids Res. 1999;27:3219–3228. doi: 10.1093/nar/27.15.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel J, Bartos J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry A. 2003;51:127–128. doi: 10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond CS, Eastwood RJ, Miotto ST, Hughes CE. Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): Testing for key innovation with incomplete taxon sampling. Syst Biol. 2012;61:443–460. doi: 10.1093/sysbio/syr126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero M, Hipp AL, Waterway MJ, Valente LM. Diversification rates and chromosome evolution in the most diverse angiosperm genus of the temperate zone (Carex, Cyperaceae) Mol Phylogenet Evol. 2012;63:650–655. doi: 10.1016/j.ympev.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Ferreira J, Mendes S, Dall’Agnol M, Sandal N, Sato S. Comparative analyses in Lotus: The cytogenetic map of Lotus uliginosus Schkuhr. Cytogenet Genome Res. 2012;37:42–49. doi: 10.1159/000339617. [DOI] [PubMed] [Google Scholar]
- Fernández A, Krapovickas A. Cromosomas y evolución em Arachis (Leguminosae) Bonplandia. 1994;8:187–220. [Google Scholar]
- Fernandez M, Ruiz ML, Linares C, Fominaya A, Pérez de la Vega M. 5S rDNA genome regions of Lens species. Genome. 2005;48:937–942. doi: 10.1139/g05-052. [DOI] [PubMed] [Google Scholar]
- Fonsêca A, Pedrosa-Harand A. Karyotype stability in the genus Phaseolus evidenced by the comparative mapping of the wild species Phaseolus microcarpus. . Genome. 2013;56:335–343. doi: 10.1139/gen-2013-0025. [DOI] [PubMed] [Google Scholar]
- Fonsêca A, Pedrosa-Harand A. Cytogenetics and comparative analysis of Phaseolus species. In: Vega P, Santalla M, Marsolais M, editors. The Common Bean Genome. Springer; Cham: 2017. pp. 57–68. [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the cell-cycle in intact plant-tissues. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Garcia S, Kovarík A, Leitch AR, Garnatje T. Cytogenetic features of rRNA genes across land plants: analysis of the Plant rDNA database. Plant J. 2017;89:1020–1030. doi: 10.1111/tpj.13442. [DOI] [PubMed] [Google Scholar]
- Gerlach WL, Bedbrook JR. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol Rev Camb Philos Soc. 2001;76:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- Gu Y, Xing S, He C. Genome-wide analysis indicates lineage-specific gene loss during Papilionoideae evolution. Genome Biol Evol. 2016;8:635–648. doi: 10.1093/gbe/evw021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra M. Cytotaxonomy: the end of childhood. Plant Biosyst. 2012;146:703–710. [Google Scholar]
- Hajdera I, Siwinska D, Hasterok R, Maluszynska J. Molecular cytogenetic analysis of genome structure in Lupinus angustifolius and Lupinus cosentinii . Theor Appl Genet. 2003;107:988–996. doi: 10.1007/s00122-003-1303-3. [DOI] [PubMed] [Google Scholar]
- Hartman TPV, Jones J, Blackhall NW, Power JB, Cocking EC, Davey MR. Cytogenetics, molecular cytogenetics and genome size in Leucaena. In: Guttenberger H, Borzan Z, Schlarbaum SC, Hartman TPV, editors. Cytogenetic Studies of Forest Trees and Shrubs - review, present status and outlook on the future. Arbora Publishers; Zvolen: 2000. pp. 57–70. [Google Scholar]
- Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC. Metaphase and interphase fluorescence in situ hybridization mapping of the rice with bacterial artificial chromosomes. Proc Natl Acad Sci U S A. 1995;92:4487–4491. doi: 10.1073/pnas.92.10.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karia CT. Caracterização genética e morfoagronômica de germoplasma de Stylosanthes guianensis (Aubl.) D. Sc. Thesis. Universidade Federal de Goiás; Goiânia: 2008. p. 140. [Google Scholar]
- Kobayashi T. A new role of the rDNA and nucleolus in the nucleus – rDNA instability maintains genome integrity. Bioessays. 2008;30:267–272. doi: 10.1002/bies.20723. [DOI] [PubMed] [Google Scholar]
- Krapovickas A, Gregory WC. Taxonomia del Genero “Arachis (Leguminosae)”. Bonplandia. 1994;8:1–186. [Google Scholar]
- Kumari S, Bir SS. Karyomorphological evolution in Caesalpiniaceae. J Cytol Genet. 1989;24:149–163. [Google Scholar]
- Lavia GI, Ortiz AM, Robledo G, Fernández A, Seijo G. Origin of triploid Arachis pintoi (Leguminosae) by autopolyploidy evidenced by FISH and meiotic behaviour. Ann Bot. 2011;108:103–111. doi: 10.1093/aob/mcr108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Pennington RT, Klitgaard BB, Sprent JI, Lima HC, Gasson PE. The dalbergioid legumes (Fabaceae): Delimitation of a pantropical monophyletic clade. Am J Bot. 2001;88:503–533. [PubMed] [Google Scholar]
- Lavin M, Schrire BP, Lewis G, Pennington RT, Delgado-Salinas A, Thulin M, Hughes CE, Matos AB, Wojciechowski MF. Metacommunity process rather than continental tectonic history better explains geographically structured phylogenies in legumes. Philos Trans R Soc Lond B Biol Sci. 2004;359:1509–1522. doi: 10.1098/rstb.2004.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of leguminosae implicates a rapid classification of lineages during the Tertiary. Syst Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Levin DA. The role of chromosomal change in plant evolution. Oxford University Press; New York: 2002. p. 230. [Google Scholar]
- Lira ICSA. Caracterização citogenética e morfoagronômica de acessos de Stylosanthes spp. (Fabaceae-Papilionoideae) coletados no nordeste brasileiro. M. Sc. Thesis. Universidade estadual de Feira de Santana; Feira de Santana: 2015. p. 60. [Google Scholar]
- Liu PD, Xue YB, Chen ZJC, Liu GD, Tian J. Characterization of purple acid phosphatases involved in extracellular dNTP utilization in Stylosanthes. . J Exp Bot. 2016;67:4141–4154. doi: 10.1093/jxb/erw190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro J, Rodriguez E, Dolezel J, Santos C. Two new nuclear isolation buffers for plant DNA flow cytometry: A test with 37 species. Ann Bot. 2007;100:875–888. doi: 10.1093/aob/mcm152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LPWG - Legume Phylogeny Working Group A new subfamily classification of the leguminosae based on a taxonomically comprehensive phylogeny. Taxon. 2017;66:44–47. [Google Scholar]
- Marques A, Moraes L, Santos MA, Costa I, Costa L, Nunes T, Melo N, Simon MF, Leitch AR, Almeida C, et al. Origin and parental genome characterization of the allotetraploid Stylosanthes scabra Vogel (Papilionoideae, Leguminoseae), an important legume pasture crop. Ann Bot. 2018;122:1143–1159. doi: 10.1093/aob/mcy113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Proceedings of the Gateway Computing Environments Workshop (GCE) New Orleans: 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- Mondin M, Aguiar-Perencin M. Heterochromatin patterns and ribosomal DNA loci distribution in diploid and polyploid Crotalaria species (Leguminosae, Papilionoideae), and inferences on karyotype evolution. Genome. 2011;54:718–726. doi: 10.1139/g11-034. [DOI] [PubMed] [Google Scholar]
- Mondin M, Santos-Serejo J, Aguiar-Perencin MLR. Karyotype characterization of Crotalaria juncea (L.) by chromosome banding and physical mapping of 18S-5.8S-26S and 5S rRNA gene sites. Genet Mol Biol. 2007;30:65–72. [Google Scholar]
- Morales AG, Aguiar-Perecin MLR, Mondin M. Karyotype characterization reveals an up and down of 45S and 5S rDNA sites in Crotalaria (Leguminosae-Papilionoideae) species of the section Hedriocarpae subsection Macrostachyae. Genet Resour Crop Evol. 2011;59:277–288. [Google Scholar]
- Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with non-repetitive DNA in plant genomes. Nat Genet. 2002;30:194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- Naganowska B, Zielinska A. Physical mapping of 18S-25S rDNA and 5S rDNA in Lupinus via fluorescent in situ hybridization. Cell Mol Biol Lett. 2002;7:665–670. [PubMed] [Google Scholar]
- Naganowska B, Wolko B, Sliwinska E, Kaczmarek Z. Nuclear DNA content variation and species relationships in the genus Lupinus (Fabaceae) Ann Bot. 2003;92:349–355. doi: 10.1093/aob/mcg145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganowska B, Wolko B, Sliwinska E, Kaczmarek Z, Schifino-Wittmann M. 2C DNA variation and relationships among New World species of the genus Lupinus (Fabaceae) Plant Syst Evol. 2006;256:147–157. [Google Scholar]
- Ortiz AM, Robledo G, Seijo G, Valls JFM, Lavia GI. Cytogenetic evidences on the evolutionary relationships between the tetraploids of the section Rhizomatosae and related diploid species (Arachis, Leguminosae) J Plant Res. 2017;130:791–807. doi: 10.1007/s10265-017-0949-x. [DOI] [PubMed] [Google Scholar]
- Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A. Chromosomal map of the model legume Lotus japonicus . Genetics. 2002;161:1661–1672. doi: 10.1093/genetics/161.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñaloza A, Valls JFM. Chromosome number and satellite chromosome morphology of eleven species of Arachis (Leguminosae) Bonplandia. 2005;14:65–72. [Google Scholar]
- Piegu B, Guyot R, Picault N, Roulin A, Sanyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, et al. Doubling genome size without polyploidization: Dynamics of retrotransposition driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel M, Escudero M, Sahuquillo E, Minaya MA, Catalán P. Are diversification rates and chromosome evolution in the temperate grasses (Pooideae) associated with major environmental changes in the Oligocene-Miocene? PeerJ. 2017;5:e3815. doi: 10.7717/peerj.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto RB, Mansano VF, Torke BM, Forni-Martins ER. Evidence for a conserved karyotype in Swartzia (Fabaceae, Papilionoideae): Implications for the taxonomy and evolutionary diversification of a species-rich neotropical tree genus. Brittonia. 2016;68:93–101. [Google Scholar]
- Polido CA, Moraes AP, Forni- Martins ER. IAPT/IOPB Chromosome data 20. Taxon. 2015;64:1347. [Google Scholar]
- Raina SN, Mukai Y. Detection of a variable number of 18S-5.8 S-26S and 5S ribosomal DNA loci by fluorescent in situ hybridization in diploid and tetraploid Arachis species. Genome. 1999;42:52–59. [Google Scholar]
- Rambaut A, Suchard M, Xie W, Drummond A. Tracer v. 1.6. 2014. http://beast.community/tracer.
- Raskina O, Barber JC, Nevo E, Belyayev A. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res. 2008;120:351–357. doi: 10.1159/000121084. [DOI] [PubMed] [Google Scholar]
- Reis AC, Sousa SM, Vale AA, Pierre PMO, Franco AL, Campos JMS, Vieira RF, Viccini LF. Lippia alba (Verbenaceae): A new tropical autopolyploid complex? Am J Bot. 2014;101:1002–1012. doi: 10.3732/ajb.1400149. [DOI] [PubMed] [Google Scholar]
- Roa F, Guerra M. Non-random distribution of 5S rDNA sites and its association with 45s rDNA in plant chromosomes. Cytogenet Genome Res. 2015;146:243–249. doi: 10.1159/000440930. [DOI] [PubMed] [Google Scholar]
- Robledo G, Seijo G. Species relationships among the wild B genome of Arachis species (section Arachis) based on FISH mapping of rDNA loci and heterochromatin detection: a new proposal for genome arrangement? Theor Appl Genet. 2010;121:1033–1046. doi: 10.1007/s00122-010-1369-7. [DOI] [PubMed] [Google Scholar]
- Robledo G, Lavia GI, Seijo G. Species relations among wild Arachis species with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin detection. Theor Appl Genet. 2009;118:1295–1307. doi: 10.1007/s00122-009-0981-x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Samoluk SS, Chalup L, Robledo G, Seijo JG. Genome sizes in diploid and allopolyploid Arachis L. species (section Arachis) Genet Resour Crop Evol. 2014;62:747–763. [Google Scholar]
- Samoluk SS, Robledo G, Bertioli D, Seijo JG. Evolutionary dynamics of an at-rich satellite DNA and its contribution to karyotype differentiation in wild diploid Arachis species. Mol Genet Genomics. 2017;292:283–296. doi: 10.1007/s00438-016-1271-3. [DOI] [PubMed] [Google Scholar]
- Schubert I, Giang THV. Genome stability and evolution: Attempting a holistc view. Trends Plant Sci. 2016;21:749–757. doi: 10.1016/j.tplants.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Schweizer D. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma. 1976;58:307–324. doi: 10.1007/BF00292840. [DOI] [PubMed] [Google Scholar]
- Seijo JG, Lavia GI, Fernández A, Krapovickas A, Ducasse D, Moscone EA. Physical mapping of the 5S and 18S–25S rRNA genes by FISH as evidence that Arachis duranensis and A. ipaensis are the wild diploid progenitors of A. hypogaea (Leguminosae) Am J Bot. 2004;91:1294–1303. doi: 10.3732/ajb.91.9.1294. [DOI] [PubMed] [Google Scholar]
- She CW, Jiang XH, Ou LJ, Liu J, Long KL, Zhang LH, Duan WT, Zhao W, Hu JC. Molecular cytogenetic characterisation and phylogenetic analysis of the seven cultivated Vigna species (Fabaceae) Plant Biol. 2014;17:268–280. doi: 10.1111/plb.12174. [DOI] [PubMed] [Google Scholar]
- She CW, Wei L, Jiang XH. Molecular cytogenetic characterization and comparison of the two cultivated Canavalia species (Fabaceae) Comp Cytogen. 2017;11:579–600. doi: 10.3897/CompCytogen.v11i4.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltom HM, Franzel S, Peters M. Adoption of tropical legume technology around the world: Analysis of success. In: McGilloway DA, editor. Grassland: A Global Resource. Wageningen Academic Publishers; Wageningen: 2005. pp. 149–166. [Google Scholar]
- Silvestri MC, Ortiz AM, Lavia GI. rDNA loci and heterochromatin positions support a distinct genome type for `x = 9 species’ of section Arachis (Arachis, Leguminosae) Plant Syst Evol. 2015;301:555–562. [Google Scholar]
- Siljak-Yakovlev S, Peccenini S, Muratovic E, Zoldos V, Robin O, Vallès J. Chromosomal differentiation and genome size in three European mountain Lilium species. Plant Syst Evol. 2003;236:165–173. [Google Scholar]
- Singh KP, Raina SN, Singh AK. Variation in chromosomal DNA associated with the evolution of Arachis species. Genome. 1996;39:890–897. doi: 10.1139/g96-112. [DOI] [PubMed] [Google Scholar]
- Smartt J, Gregory WC, Gregory MP. The genomes of Arachis hypogaea. 1. Cytogenetic studies of putative genome donors. Euphytica. 1978;27:665–675. [Google Scholar]
- Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annu Rev Plant Biol. 2009;60:561–88. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- Souza LGR, Crosa O, Speranza P, Guerra M. Cytogenetic and molecular evidence suggest multiple origins and geographical parthenogenesis in Nothoscordum gracile (Alliaceae) Ann Bot. 2012;109:987–999. doi: 10.1093/aob/mcs020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stace HM, Cameron D. Cytogenetics and the evolution of Stylosanthes. In: Stace HM, Edye LA, editors. The Biology and Agronomy of Stylosanthes. Academic Press Australia; Sydney: 1984. pp. 49–72. [Google Scholar]
- Starr CR, Corrêa RS, Filgueiras TS, Hay JDV, Santos PF. Plant colonization in a gravel mine revegetated with Stylosanthes spp. in a Neotropical savanna. Landsc Ecol Eng. 2013;9:189–201. [Google Scholar]
- Susek K, Bielski WK, Hasterook R, Naganowska B, Wolko B. A first glimpse of wild lupin karyotype variation as revealed by comparative cytogenetic mapping. Front Plant Sci. 2016;7:1152. doi: 10.3389/fpls.2016.01152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek K, Braszewska-Zalewska A, Bewick AJ, Hasterok R, Schmitz RJ, Naganowska B. Epigenomic diversification within the genus Lupinus . PLoS One. 2017;12:e0179821. doi: 10.1371/journal.pone.0179821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torke BM. Systematics and evolutionary diversification in the species-rich neotropical tree genus Swartzia (Leguminosae - Papilionoideae). PhD Thesis. Washington University; Missouri: 2006. [Google Scholar]
- Totta C, Rosato M, Ferrer-Galego P, Lucchese F, Rosselló JA. Temporal frames of 45S rDNA site-number variation in diploid plant lineages: lessons from the rock rose genus Cistus (Cistaceae) Biol J Linnean Soc. 2017;120:626–636. [Google Scholar]
- Valls JFM, Simpson CE. New species of Arachis (Leguminosae) from Brazil, Paraguay and Bolivia. Bonplandia. 2005;14:35–64. [Google Scholar]
- Van-Lume B, Esposito T, Diniz-Filho JAF, Gagnon E, Lewis GP, Souza G. Heterochromatic and cytomolecular diversification in the Caesalpinia group (Leguminosae): Relationships between phylogenetic and cytogeographical data. Perspect Plant Ecol Syst. 2017;29:51–63. [Google Scholar]
- Vander Stappen J, Marant S, Volckaert G. Molecular characterization and phylogenetic utility of the rDNA external transcribed spacer region in Stylosanthes (Fabaceae) Theor Appl Genet. 2003;107:291–298. doi: 10.1007/s00122-003-1245-9. [DOI] [PubMed] [Google Scholar]
- Vasconcelos S, Souza AA, Gusmão CLS, Milani M, Benko-Iseppon AM, Brasileiro-Vidal AC. Heterochromatin and rDNA 5S and 45S sites as reliable cytogenetic markers for castor bean (Ricinus communis, Euphorbiaceae) Micron. 2010;41:746–753. doi: 10.1016/j.micron.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Vieira MLC, Aguiar-Perencin MLR, Martins PS. A cytotaxonomic study in twelve taxa of Stylosanthes Sw., Leguminosae. Cytologia. 1993;58:305–311. [Google Scholar]
- Vizintin L, Javornik B, Bohanec B. Genetic characterization of selected Trifolium species as revealed by nuclear DNA content and ITS rDNA region analysis. Plant Sci. 2006;170:859–866. [Google Scholar]
- White TJ, Bruns T, Lee SJWT, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; New York: 1990. pp. 315–322. [Google Scholar]
- Wong MM, Vaillancourt RE, Freeman JS, Hudson CJ, Bakker FT, Cannon CH, Ratnam W. Novel insights into karyotype evolution and whole genome duplications in legumes. bioRxiv:099044 2017 [Google Scholar]
- Young ND, Bharti AK. Genome-enabled insights into legume biology. Annu Rev Plant Biol. 2012;63:283–305. doi: 10.1146/annurev-arplant-042110-103754. [DOI] [PubMed] [Google Scholar]
- Yu F, Wang H, Zhao Y, Liu R, Dou Q, Dong J, Wang T. Karyotypic evolution of the Medicago complex: sativa-caerulea-falcata inferred from comparative cytogenetic analysis. BMC Evol Biol. 2017;17:104. doi: 10.1186/s12862-017-0951-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Nakata M, Irifune K, Tanaka R, Morikawa H. Fluorescent banding pattern analysis of eight taxa of Phaseolus and Vigna in relation to their phylogenetic relationships. Theor Appl Genet. 1993;87:38–43. doi: 10.1007/BF00223741. [DOI] [PubMed] [Google Scholar]
Internet Resources
- Flowing Software 2.5. http://www.uskonaskel.fi/flowingsoftware/
- Kew Royal Botanic Gardens. [5 April 2017]. http://data.kew.org/cvalues/CvalServlet?querytype=2.
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis, version 3.51. 2018. http://www.mesquiteproject.org.
- Swofford DL. PAUP* (* Phylogenetic Analysis Using PAUP) 2002. https://paup.phylosolutions.com/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.