Abstract
Mitochondrial DNA (mtDNA) deletions are a common cause of human mitochondrial diseases. Mutations in the genes encoding components of the mitochondrial replisome, such as DNA polymerase gamma (Pol γ) and the mtDNA helicase Twinkle, have been associated with the accumulation of such deletions and the development of pathological conditions in humans. Recently, we demonstrated that changes in the level of wild-type Twinkle promote mtDNA deletions, which implies that not only mutations in, but also dysregulation of the stoichiometry between the replisome components is potentially pathogenic. The mechanism(s) by which alterations to the replisome function generate mtDNA deletions is(are) currently under debate. It is commonly accepted that stalling of the replication fork at sites likely to form secondary structures precedes the deletion formation. The secondary structural elements can be bypassed by the replication-slippage mechanism. Otherwise, stalling of the replication fork can generate single- and double-strand breaks, which can be repaired through recombination leading to the elimination of segments between the recombination sites. Here, we discuss aberrances of the replisome in the context of the two debated outcomes, and suggest new mechanistic explanations based on replication restart and template switching that could account for all the deletion types reported for patients.
Keywords: Mitochondria, DNA replication, human diseases, Pol γ, Twinkle
Introduction
Most animal mitochondrial DNA (mtDNA) is a compact, circular double-stranded molecule of approximately 16 kb, composed of 37 genes. Thirteen of these genes encode essential subunits of the mitochondrial respiratory chain, which in turn is responsible for the bulk of cellular ATP production via the oxidative phosphorylation (OXPHOS) process (Zeviani and Di Donato, 2004; McKinney and Oliveira, 2013). The number of mtDNA copies and the amount of mitochondria inside a cell type/tissue may vary dynamically to accommodate the cellular metabolic needs (Taylor and Turnbull, 2005). Considering the direct relationship between mtDNA copy number and the synthesis of respiratory chain subunits, the mtDNA replicative machinery, the so-called replisome, is one of the most important factors for proper maintenance of this genome and appropriate OXPHOS function.
The minimum mitochondrial replisome is composed of a set of three nuclear genome-encoded proteins: the replicative mtDNA helicase Twinkle, DNA polymerase γ (Pol γ), and the mitochondrial single-stranded DNA-binding protein (mtSSB) (Figure 1) (Korhonen et al., 2004; McKinney and Oliveira, 2013; Ciesielski et al., 2016). During replication fork progression, the homohexameric/heptameric, ring-shaped Twinkle translocates on one DNA strand in the 5’-3’ direction, hydrolyzing nucleotide tri-phosphate and promoting the unwinding of the parental double-stranded DNA (dsDNA) (reviewed in Kaguni and Oliveira, 2016). Using the resulting single-stranded DNA (ssDNA) as template, the heterotrimeric Pol γ synthesizes a new mtDNA strand also in the 5’-3’ fashion and proofreads it using its 3’-5’ exonuclease activity. The catalytic subunit (Pol γ-α) is responsible for such activities, which are highly stimulated by its accessory Pol γ-β subunit (reviewed in Kaguni, 2004). The parental ssDNA exposed at the replication fork is protected from nucleolysis through the binding of the homotetrameric mtSSB, which also further stimulates the dsDNA unwinding by Twinkle and DNA synthesis/proofreading by Pol γ, most likely coordinating their enzymatic functions during mtDNA replication (Korhonen et al., 2004; Oliveira and Kaguni, 2011).
Figure 1. The mitochondrial replisome at a replication fork during mtDNA heavy-strand synthesis. The nuclear-encoded proteins that form the replisome are represented by the crystal structure of Pol γ (PDB: 4ZTZ), and the models of mtSSB (Oliveira et al., 2011) and Twinkle (Kaguni and Oliveira, 2016). The dimeric accessory subunit of Pol γ is depicted in two tons of gray. The software Pymol (www.pymol.org) was used to analyze the structures and models and to create the figure. The scheme is not meant to detail structural and/or functional aspects of the replisome components; please see Ciesielski et al. (2016) for such information.
Here, we provide a substantial review of the literature, highlighting the importance of the mitochondrial replisome functions for the mechanistic interpretations of mtDNA deletion formation, one of the most common causes of human mtDNA diseases. We discuss the clinical features of these diseases, with the support of research data from a wide range of cell culture and animal models, and reconstituted in vitro systems. We also provide an overview of the pathogenesis of mtDNA disorders and the molecular features of mtDNA deletions, describing previously proposed mechanisms for their formation inside mitochondria. Moreover, we present novel hypotheses that could be tested experimentally, to improve our understanding of the most abundant form of mutation in the human mitochondrial genome.
Pathogenesis of mtDNA disorders
mtDNA diseases are metabolic disorders with an occurrence of ~1 in 5000 human individuals (Chinnery et al., 2000; Elliott et al., 2008). These are generally classified as primary, when arising from mutations in the mtDNA itself, or secondary, if mutations or alterations in the expression levels of nuclear genes encoding factors important for mtDNA metabolism are detected (Schon et al., 2012; Alston et al., 2017). The most common causes of primary mtDNA diseases are single large-scale deletions and point mutations, whereas the secondary class may arise from mtDNA depletion and multiple mtDNA deletions. Given that multiple copies of mtDNA are present in a cell, a mix of aberrant and wild-type molecules may be found in varying proportions in different tissues. This is called a heteroplasmic state, which typically does not manifest as a disease condition unless the number of mutated genomes exceed a threshold of approximately 60%. This phenomenon is tissue-dependent and some tissues may withstand higher loads of aberrant molecules (Wong, 2007; Tuppen et al., 2010).
An epidemiological survey indicated that 1 in ~200 human individuals are carriers of a pathogenic mtDNA point mutation (Elliott et al., 2008). Carriers of a primary, pathogenic mtDNA point mutation may remain asymptomatic for generations, up to the point in which the mutated mtDNA molecules reach the threshold level. The difference between the relatively high frequency of carriers and low frequency of diseased individuals can be explained by a balance between a genetic bottleneck and negative selection during female germ line development (for alternative possibilities, see Tuppen et al., 2010; Otten et al., 2018). mtDNA copy number undergoes a radical decrease in early stages of oogenesis, followed by a significant increase towards the end of the process, enabling the generation of eggs with levels of mutant mtDNA molecules higher (or lower) than in the mother’s somatic tissues. When the mutation is too severe, the eggs carrying high levels of such mtDNA are usually unviable and the mutation is often negatively selected (Fan et al., 2008; Wai et al., 2008). As a result, the offspring of a mother carrying a relatively mild, but yet pathogenic mutation may exhibit various levels of heteroplasmy, ranging from a virtual wild-type homoplasmy, to a predominantly aberrant haplotype with symptoms of the related mitochondrial disease (Wilson et al., 2016; Alston et al., 2017; Otten et al., 2018). Because there is no applicable way to assess the mtDNA mutation load in maternal oocytes, a prediction of reoccurrence risk is almost impossible, although up to ~25% of pathogenic mtDNA point mutations may occur de novo at early developmental stages (Sallevelt et al., 2017).
The primary, single large-scale deletions were the first mtDNA defects described (Holt et al., 1988) and remain the most common sporadic mutations on mtDNA, accounting for approximately a quarter of all mitochondrial disorders in the human population (Schaefer et al., 2008; Pitceathly et al., 2012; Grady et al., 2014; Gorman et al., 2015). In contrast to point mutations, pathogenic mtDNA deletions typically arise de novo and, with rare exceptions (Chinnery et al., 2004), are not inherited by the offspring (Ng and Turnbull, 2016; Kauppila et al., 2017). Phenotypically, primary deletions manifest as the often-fatal Pearson syndrome in infancy, Kearns-Sayre syndrome (KSS) in childhood and adolescence, or late onset progressive external ophthalmoplegia (PEO) (Rocha et al., 2018; Russell et al., 2018). In the clinical scope, the mtDNA mutation load found in sporadic KSS, PEO and Pearson’s syndrome is extremely high (>80% in affected tissues) (Moraes et al., 1995). This ‘clonal expansion’ of the deletion-bearing mtDNA molecules (ΔmtDNA) in affected individuals can hardly be explained by the sporadic character of the pathogenesis. In fact, a recent comparative study of the expansion of various aberrant mtDNA molecules in dividing induced pluripotent stem cells demonstrated that ΔmtDNA are preferentially replicated compared to controls or those bearing point mutations (Russell et al., 2018). This finding is in line with other analyses implying that point mutation-bearing mtDNA molecules do not exhibit advantageous replication and clonal expansion (Pallotti et al., 1996; Wilson et al., 2016), and supports the idea of preferential expansion of ΔmtDNA (Picard et al., 2016). The mechanism of clonal expansion of ΔmtDNA remains unknown (Picard et al., 2016; Kowald and Kirkwood, 2018). Interestingly, the accumulation of primary ΔmtDNA has also been associated with aging of healthy human individuals (Cortopassi et al., 1992; Bua et al., 2006). These deletions arise often in post-mitotic tissues, such as heart, brain and skeletal muscles. However, in these cases, even though the levels of ΔmtDNA in individual cells can be very high, the overall amount of mtDNA deletions in the tissue is low, compared to pathological conditions (Cortopassi et al., 1992; Schon et al., 2012; Kauppila et al., 2017).
Secondary mtDNA diseases result from mutations in nuclear genes and their inheritance follows the Mendelian pattern, often in an autosomal dominant fashion (Ng and Turnbull, 2016; Alston et al., 2017). The major causes of secondary mtDNA diseases identified to date involve defects in the mitochondrial replisome: mutations in the POLG1 and TWNK genes, which encode respectively Pol γ-α and Twinkle (Van Goethem et al., 2001; Copeland, 2014; Nurminen et al., 2017; Rahman and Copeland, 2019). Pathogenic mutations in genes related to other mitochondrial processes, such as OXPHOS, fusion and fission, intra-mitochondrial translation, etc., have also been found and are reviewed elsewhere (Ng and Turnbull, 2016; Alston et al., 2017). To date, mutations in >250 nuclear genes encoding mitochondrial proteins have been related to mitochondrial disorders in general (Mayr et al., 2015).
The most common consequence of defects in the mitochondrial replisome is the accumulation of multiple large-scale deletions in mtDNA (Rahman and Copeland, 2019), which are formatively similar to the primary single large-scale deletions described above (Reeve et al., 2008; Schon et al., 2012). Furthermore, multiple large-scale deletions generated by an aberrant mitochondrial replisome are most commonly associated with myopathies and PEO in adult patients, which may also be manifest in the primary syndromes such as PEO and KSS (Van Goethem et al., 2001; Agostino et al., 2003; Di Fonzo et al., 2003; Filosto et al., 2003; Wanrooij et al., 2004; González-Vioque et al., 2006; Lehmann et al., 2016; Grier et al., 2018). These similarities may suggest that both primary and secondary mtDNA deletions may emerge upon similar shortcomings of the mtDNA replication machinery. In addition, a recent comparative study demonstrated that ΔmtDNA molecules generated in patients with POLG1 mutations undergo clonal expansion similarly to the single large deletions sporadically generated in patients. In contrast, ΔmtDNA generated in patients with a mutation in OPA1, which encodes a protein involved in mitochondrial fusion, did not expand clonally (Trifunov et al., 2018). In both POLG1-related and sporadic deletions (but not in OPA1-related deletions), the authors observed an increase in the overall mtDNA copy number, which may link the phenomenon of clonal expansion to a compensatory upregulation of replication as a response to inefficient OXPHOS.
In contrast to primary mtDNA diseases, defects of the mitochondrial replisome may lead to different, frequently coexisting, mtDNA aberrances: point mutations are often found along with mtDNA deletions in the same patient/tissue. A mouse line expressing a proofreading-deficient Pol γ-α variant, the so-called mutator mouse, may serve as an important model for the understanding of this phenomenon, as the accumulation of multiple large-scale deletions in its mtDNA has been observed (Trifunovic et al., 2004; Fuke et al., 2014), in addition to the expected accumulation of point mutations (Vanderstraeten et al., 1998; Spelbrink et al., 2000). Despite a previously considered possibility that the accumulation of point mutations may predispose mtDNA molecules to deletions, no correlation has been found between the distribution of point mutations and the deletion breakpoints (Wanrooij et al., 2004; Hudson and Chinnery, 2006). Furthermore, heterozygous mice encoding the proofreading-deficient Pol γ-α on a single allele do not accumulate point mutations (they may happen, but are likely corrected by the product of the wild-type POLG1 allele), but still develop large-scale deletions (Fuke et al., 2014). The parallel occurrence of mtDNA point mutations and deletions may suggest that the proofreading activity might not be the most critical function of the exonuclease domain of Pol γ-α, whose active site was abolished in the mutator mouse (Nurminen et al., 2017). In support, some mutations in the exonuclease domain result in decreased nucleotide polymerization rather than defects in proofreading (Szczepanowska and Foury, 2010), and the exonucleolytic activity by Pol γ is necessary for the in vitro production of ligatable 5’ ends for proper mtDNA replication (Macao et al., 2015).
mtDNA depletion is relatively rare and typically causes early-onset mitochondrial diseases (Moraes et al., 1991; Suomalainen and Isohanni, 2010). Although the inefficient replication of mtDNA is an obvious cause, the exact mechanism(s) remain(s) unexplained. Intriguingly, POLG1 mutations that cause point mutations and deletions in the mtDNA can also cause mtDNA depletion when the individual carries two mutated, pathogenic alleles. Moreover, the same POLG1 mutations can cause early onset encephalopathy with severe mtDNA depletion or late-onset PEO with ataxia (Horvath et al., 2006; Nguyen et al., 2006; Tzoulis et al., 2006). Therefore, mtDNA depletion syndromes appear to result from similar insufficiencies of the mitochondrial replisome as other late-onset secondary mitochondrial diseases. However, more studies are necessary to fully understand the pathogenesis of the depletion syndromes.
Formation of mtDNA deletions
Since KSS was first reported by Kearns and Sayre (1958), there have been many reports describing deletions of different sizes and at different mtDNA positions among patients (Holt et al., 1988; Nelson et al., 1989; Montiel-Sosa et al., 2013; Damas et al., 2014; Saldaña-Martínez et al., 2019). The most common pathogenic large-scale deletion of 4977 bp (the so-called `common deletion’) is precisely flanked by perfect direct repeats (DR) of 13 bp, at nucleotide positions 13,447–13,459 (within the ND5 gene) at the 5’ end, and at positions 8470–8482 (within the ATPase8 gene) at the 3’ end (Schon et al., 1989; Samuels et al., 2004). As a result, the genes for two complex V subunits, one complex IV subunit, four complex I subunits and five tRNAs are lost. Approximately 60% of mtDNA deletions reported to date are similarly flanked by perfect DR sequences; these are called class I deletions. Of the remaining, 30% are flanked by imperfect repeats (class II deletions) and 10% have no flanking repeats (class III deletions) (Mita et al., 1990; Samuels et al., 2004; Reeve et al., 2008; Chen et al., 2011). The secondary, multiple large-scale deletions generated due to mutated POLG1 or TWNK bear characteristics of the class II deletions because of the imperfect flanking repeats observed (Zeviani et al., 1989; Wanrooij et al., 2004).
Notably, the vast majority of all deletions occurs between the mtDNA sites OH and OL (Figure 2) (Reeve et al., 2008; Damas et al., 2014; Belmonte et al., 2016), recognized as origins of the heavy- and the light-strand replication, respectively (vertebrate mtDNA strands are denoted as heavy and light due to their distinct nucleotide composition; for details, see Ciesielski et al., 2016). This striking accumulation of deletions between the two replication origins suggests that their formation mechanism is related to the replication process (Tuppen et al., 2010). According to the strand-displacement model of mtDNA replication, synthesis of the new heavy- (leading) strand initiates at OH and proceeds using the light-strand as template. The displaced parental heavy-strand remains single-stranded, coated by mtSSB or hybridized to RNA. After synthesis reaches approximately two-thirds of the mtDNA circumference, the replisome unveils OL on the parental heavy-strand, enabling replication of the new light- (lagging) strand to initiate at this site by a new replisome and proceed in the opposite direction (see the extensive reviews of current models of mtDNA replication by Holt and Jacobs, 2014; Ciesielski et al., 2016). The first model of mtDNA deletion formation was proposed upon the analysis of the common deletion (class I) and is based on the replication-slippage mechanism (Shoffner et al., 1989). In this scenario, incidental breaks of the displaced parental heavy-strand between the 3’ and 5’ DR enable annealing of the 3’ DR from that strand to the distant complimentary 5’ DR on the parental light-strand, brought to proximity by structural rearrangements during synthesis of the nascent heavy-strand (Shoffner et al., 1989; Mita et al., 1990). Subsequent synthesis from OL would omit the stretch between the break point and the paired repeats, generating a deletion (Figure 3A). This mechanism has been found especially appealing in early reports indicating that the mtDNA molecules bearing the common deletion retain the 3’ DR but not the 5’ DR, which indeed fits the model (Shoffner et al., 1989; Degoul et al., 1991; Chen et al., 2011). In support, a recent elegant report using mito-TALENS in human cultured cells determined that nicks in the heavy-strand, in the vicinity of the 5’ DR are sufficient and necessary to yield the common deletion (Phillips et al., 2017). In vivo, reactive oxygen species (ROS) could pose as the potential source of damage that triggers the DR mispairing. Replicating mtDNA molecules are associated with the inner mitochondrial membrane, frequently in close proximity to the OXPHOS complexes (Rajala et al., 2014). Hence, mtDNA can be permanently exposed to ROS, with well-established detrimental effects (Shokolenko et al., 2009).
Figure 2. Diagram of the distribution of selected deletions within the human mitochondrial genome. The location of the deletions corresponds to the distribution of lines inside the mtDNA ideogram. Each line represents a single deletion. The location of 152 reported single mtDNA deletions is presented as black lines. The location of 86 reported multiple mtDNA deletions associated with progressive external ophthalmoplegia (PEO), including those generated upon mutations in POLG1 and TWNK, is presented as light blue lines. The location of the common deletion is presented as the red line. Origins of replication OH and OL, as well as the termination associated sequence (TAS) are labeled. Protein-coding genes are colored in green; ribosomal RNAs are colored in brown; transfer RNAs are colored in grey; the non-coding region (NCR) is colored in light grey. The diagram was generated with and modified from the mitochondrial DNA breakpoints database, MitoBreak (Damas et al., 2014).
Figure 3. Schematic representation of the debated mechanisms of mitochondrial DNA deletion formation. (A) In the replication-slippage (i.e. slipped mispairing) mechanism, a break of the displaced heavy DNA strand (indicated with a red X), generated during an asynchronous replication process, enables pairing of the 3’ flanking region of that heavy-strand (grey box) with the 5’ flanking region of the light-strand (black box), ahead of the leading DNA synthesis (dashed arrow). The resulting unpaired DNA flap (dotted line) is then degraded and ligated (solid arrow) with the remainder of the heavy-strand. Replication initiated from OL would yield a daughter mtDNA molecule carrying the deletion of a segment ahead of the 3’ flanking region. The scheme represents the model proposed by Shoffner et al. (1989), and it was modified to incorporate the location of the heavy-strand breaks according to the results reported in Phillips et al. (2017). The position of the flanking regions corresponds to the 3’ and 5’ direct repeats (DRs) of the common deletion. (B) The formation of deletions in the homologous recombination- or microhomology-mediated end-joining-based mechanism (upper panel) is initiated by a double strand break (indicated with a red X), which occurs predominantly within the region between the two replication origins. Excision of both heavy and light-strands in 3’-5’ direction (dotted lines) enables pairing of the unveiled complementary flanking regions (grey and black boxes). Next, flap excision and subsequent ligation would generate a daughter mtDNA molecule carrying the deletion and retaining one flanking region. The scheme represents the model proposed by Krishnan et al. (2008). Class III mtDNA deletions have been suggested to form via nonhomologous end-joining of linear molecules (lower panel), which to date remain uncharacterized. The scheme has been modified from Chen et al. (2011).
Even though the replication-slippage mechanism could explain the class I deletions, it hardly explains the formation of class II and III deletions, which constitute approximately one third of the reported cases (Mita et al., 1990; Degoul et al., 1991). An alternative was proposed by Schon and colleagues, who based on combined analyses of cases of class I (including the common deletion), II and III deletions, suggested that recombination may underlie their formation (Schon et al., 1989; Mita et al., 1990). This idea was further developed into a model that assumed that cross-pairing of DR in the formation of class I deletions, or imperfect repeats in the formation of class II deletions, would facilitate efficient homologous recombination or microhomology-mediated end-joining (MMEJ) events, which, similarly to the replication-slippage mechanism, would yield a copy of a single flanking sequence in the daughter ΔmtDNA (Krishnan et al., 2008; Tadi et al., 2016) (Figure 3B). A later study on multiple large-scale deletions in patients with mutated POLG1 or TWNK indicated that double-stranded breaks (DSBs) could be involved in the formation of ΔmtDNA (Wanrooij et al., 2004). Homologous recombination is one of the fundamental DSB repair mechanisms in a wide spectrum of genetic systems, which made the recombination-mediated mechanism of ΔmtDNA formation plausible, although historically lack of recombination has been erroneously associated with animal mtDNA (Harrison, 1989; Avise, 2000). The model was further supported by circumstantial evidence for recombination in mitochondria, including the concurrent accumulation of linear and ΔmtDNA forms, the observation of four-way junctions, catenates and other recombination intermediates, the identification of specific nuclear recombination factors in mitochondria, and others (Srivastava and Moraes, 2005; Bacman et al., 2009; de Souza-Pinto et al., 2009; Fukui and Moraes, 2009; Pohjoismäki et al., 2009, 2011; Sage et al., 2010; Ciesielski et al., 2018). Perhaps the most compelling evidence comes from a study that demonstrated that mitochondrial protein extracts from distinct rat tissues and from HeLa cells were able to mediate joining of DNA substrates bearing microhomologies between 5 and 22-nt, which are similar to the flanking regions of the class I and II deletions (Tadi et al., 2016). Additionally, nonhomologous end-joining (Figure 3B) (Lieber, 2010), which would account for the formation of the least frequent class III deletions (Srivastava and Moraes, 2005; Fukui and Moraes, 2009; Nissanka et al., 2018), has been clearly reported for the yeast Saccharomyces cerevisiae (Kalifa et al., 2012), but substantial evidence from mitochondria of humans or animal models is yet to be delivered (Fishel et al., 2007; Tadi et al., 2016; Nissanka et al., 2018).
A shortcoming of the proposed DSB repair-mediated mechanism of deletion formation, however, lies in recently published data demonstrating that linearized mtDNA molecules have a very short half-life in cells, being rapidly degraded by unknown nucleases, with no evidence for any robust DSB repair process (Moretton et al., 2017). Efficient elimination of linearized mtDNA molecules, as a result of DSBs induced by targeted enzymatic cleavage of mutant mtDNA in patient-derived cells, has been previously shown to reduce heteroplasmy by allowing repopulation of the organelles with the undigested wild-type mtDNA molecules (Bacman et al., 2013; Gammage et al., 2014). Moraes and colleagues have recently discovered that this rapid degradation of linear mtDNA molecules depends on the exonucleolytic activity of Pol γ (but not on its polymerase activity) and that large deletions of mtDNA are accumulated in the exonuclease-deficient Pol γ mice (Nissanka et al., 2018), which is in line with the earlier reports on the mutator mouse (Trifunovic et al., 2004; Fuke et al., 2014). These results imply that the DSB-induced degradation of linearized molecules and recombinational repair are actually competing/complementing processes, with a significant advantage for the former. Earlier reports did in fact indicate low levels of DSB repair in mitochondria (Bacman et al., 2009; Fukui and Moraes, 2009). Overall, the correlation between the frequency of DSBs and deletion formation in mitochondria appears to be well documented (Fukui and Moraes, 2009; Ciesielski et al., 2018; Nissanka et al., 2018), and in the light of these reports, including our own studies, they are very likely related. Perhaps, the coexistence of the rapid degradation of linearized mtDNA and the low efficiency of DSB repair could actually explain the clonal expansion phenomenon, as the rapid degradation of many cleaved molecules (which could arise in abundance, for example, from uncoupling of the mitochondrial replisome, as discussed in the next section) would give advantage to the replication of those few repaired ΔmtDNA, which consequently would repopulate mitochondria.
In the putative DSB repair mechanism, the resulting free ends are susceptible to being resected completely by 3’-5’ exonucleases, allowing the homologous (or micro-homologous) repeats to pair. The resulting overhangs are exposed to degradation, so subsequent ligation of the nicks on both strands would generate circular ΔmtDNA capable of being replicated (Figure 3B) (Krishnan et al., 2008). Pol γ is the only 3’-5’ exonuclease in mitochondria, hence it is a likely candidate to promote the putative resection of DSB-afflicted mtDNA molecules. Notably, Kunz and coworkers have recently proposed an appealing model for linear mtDNA degradation, in which Pol γ acts in concert with another mitochondrial exonuclease, MGME1, to degrade both DNA strands simultaneously, according to their substrate preferences, i.e. 3’-5’ for Pol γ, and 5’-3’ for MGME1 (Kornblum et al., 2013; Peeva et al., 2018). However, other studies indicate that the role of MGME1 is secondary, if not dispensable (Moretton et al., 2017; Nissanka et al., 2018). Perhaps, MGME1 action fluctuates depending on the physiological context: when functional, MGME1 and Pol γ would degrade linear mtDNA fragments; if MGME1 is unfunctional or absent, Pol γ alone would generate overhangs, which could later hybridize and serve as material for deletion formation. Markedly, MGME1 knockout results in the accumulation of mtDNA deletions in mice (Matic et al., 2018), which is consistent with the idea of higher frequency of overhangs in the absence of MGME1. Other enzymes necessary for the putative maturation of DSB-repaired mtDNA are also present in mitochondria. Fen1 and Dna2 are nucleases specific to the processing of 5’-DNA flaps (Zheng et al., 2008; Kazak et al., 2013). Recently, the activity of mitochondrial ligase III, Lig3, has been demonstrated to be important for the MMEJ in mitochondrial extracts (Tadi et al., 2016), and along with MGME1 and Dna2, it appears to be necessary for the formation of the common deletion in cultured cells (Phillips et al., 2017). However, the same study also implicated the mitochondrial replisome components (Pol γ, Twinkle, and mtSSB) in the formation of the common deletion, which underscores the role of the replication process in deletion formation or indicates new roles for the replication proteins in DSB repair that are yet to be described. Nevertheless, it seems that the suite of proteins necessary to mediate DSB repair by recombination is present in human mitochondria.
It has been suggested in several reports that both mechanisms, the replication-slippage and the DSB repair, may coexist and facilitate the formation of distinct deletion classes; e.g., replication-slippage would account for the class I deletions, whereas DSB repair would lead to classes II and III, or, alternatively, that replication-slippage may be specific to sporadic deletions, and DSB repair to the secondary deletions (Degoul et al., 1991; Mita et al., 1990; Wanrooij et al., 2004; Tuppen et al., 2010; Chen et al., 2011; Ciesielski et al., 2018). However, comparative studies of all three classes of deletions revealed remarkable similarities in the distribution of deletion breakpoints, which were independent of the presence of repeat sequences (Samuels et al., 2004; Reeve et al., 2008; Guo et al., 2010). These analyses imply that classifying the deletions in three distinct groups on the basis of sequence homology may be somewhat misleading, and that the same principal mechanisms operate in the formation of almost all mtDNA deletions.
The role of the replisome in the formation of mtDNA deletions
One major difference between the two models of deletion formation lies in the role of the mitochondrial replisome proteins. According to the early replication-slippage mechanism, deletions are technically formed during the replication process, hence the activity of the replisome is critical (Shoffner et al., 1989). In the DSB repair-related model, deletions are formed in an independent process of recombination/recircularization, by a yet uncharacterized machinery that facilitates one of the proposed recombination types (Krishnan et al., 2008; Chen, 2013). Importantly, the latter scenario received significant recognition upon observations that DSBs and the formation of deletions correlate with the stalling of the replication fork (Wanrooij et al., 2004,2007; Pohjoismäki et al., 2011). Correspondingly, some studies indicate that, while the distribution of the 5’ deletion breakpoints is more variable, the 3’ breakpoints predominantly localize in the vicinity of the termination associated sequence (TAS) site at the end of the D-loop structure (Zeviani et al., 1989; Samuels et al., 2004; Wanrooij et al., 2004; Damas et al., 2014; Jemt et al., 2015) (Figure 2). TAS is the prominent replication arrest site even under normal physiological conditions: ~90% of all replication events initiated at OH terminate there, yielding the 7S DNA and the characteristic triplex structure of the D-loop region (Doda et al., 1981; Bowmaker et al., 2003; Ciesielski et al., 2016). This co-localization of the prominent replication arrest site and the 3’ breakpoints of deletions indicates that replication failure precludes the formation of deletions.
In agreement with these observations, studies of the biochemical properties of pathogenic mitochondrial replisome components linked their dysfunctions with replication stalling. The most commonly reported pathogenic mutation of POLG1, the A467T replacement (Chan and Copeland, 2009; Nurminen et al., 2017), compromises the Pol γ holoenzyme formation (i.e., the interactions between Pol γ-α and the accessory Pol γ-β) and results in reduction of the polymerase activity and stalling during DNA synthesis in vitro (Chan et al., 2005). Affected patients accumulate deletions but manifest variable clinical symptoms (Rajakulendran et al., 2016; Nurminen et al., 2017). In addition, all known pathogenic mutations in the POLG2 gene (which encodes Pol γ-β) affect Pol γ holoenzyme assembly and stability, which as for POLG1 A467T, results in the accumulation of ΔmtDNA (Young et al., 2011; Copeland, 2014). In vivo and in vitro studies of error-prone Pol γ variants, such as POLG1 Y995C or the earlier discussed exonuclease-deficient variant, indicated that these also stall frequently during DNA synthesis and lead to the accumulation of multiple deletions (Wanrooij et al., 2004, 2007; Szczepanowska and Foury, 2010; Song et al., 2011). Notably, it is currently considered that the lower fidelity of these Pol γ variants results in mechanistic shortcomings, which could also underlie the formation of deletions (Chan and Copeland, 2009; Szczepanowska and Foury, 2010; Nurminen et al., 2017).
Mutations in TWNK that alter the biochemical activities of Twinkle result in the accumulation of multiple large-scale deletions in mtDNA and the development of PEO (Suomalainen et al., 1997; Spelbrink et al., 2001; Tyynismaa et al., 2005; Copeland, 2014). The characterization of mtDNA replication intermediates (RIs) using bidimensional-agarose gel electrophoresis (for the detailed experimental procedure see Reyes et al., 2007, 2009) in cells expressing pathogenic helicase variants indicated multiple replication stalling events and frequent DSBs (Wanrooij et al., 2007; Goffart et al., 2009; Pohjoismäki et al., 2011). Intriguingly, overexpression of defective Twinkle in mice (creating the so-called “deletor mouse” line) resulted in less severe symptoms as compared to the mutator mouse (Tyynismaa et al., 2005), perhaps because the endogenous wild-type TWKN locus is still intact in the deletor mouse. Even though this mouse model accumulates multiple deletions, it exhibits relatively good physical performance and no traits of accelerated aging, which are the hallmarks of the mutator mouse (Trifunovic et al., 2004). The major difference between the two mouse models is the accumulation of point mutations by the mutator mouse. Interestingly, a heterozygous mouse, expressing the proofreading-deficient Pol γ from a single allele, does not exhibit point mutation accumulation, as mentioned earlier, nor the progeroid phenotype, but like the deletor mouse, bears deletions and symptoms typical of PEO (Fuke et al., 2014). These results indicate that mtDNA deletions (including the primary single large-scale deletions) result in similar phenotypic effects, which implies a uniform mechanism for their formation. In the case of Pol γ defects, however, the co-accumulation of point mutations may further alter/complicate clinical symptoms, which could explain the larger clinical spectrum of POLG1-disorders (Rahman and Copeland, 2019). In the case of Twinkle dysfunctions, mtDNA replication stalling in cultured human cells and in tissues of six-week-old deletor mice is evident, although the former never accumulates deletions, and the latter only shows ΔmtDNA much later in life (Goffart et al., 2009). These findings indicate that the events that lead to mtDNA deletion in post-mitotic tissues can be modelled in proliferating cells, again pointing to a common mechanism of deletion formation irrespective of tissue or cell type.
Stalling of the replication fork may directly result from mtDNA secondary structures, which are resolved during normal replication, but pose hurdles for a defective replisome. Such idea has been proposed to explain class I deletions, as their flanking regions have high potential to form hairpin-like secondary structures due to sequence complementarity (Mita et al., 1990; Damas et al., 2012). The difficulty of applying this explanation to class II and III deletions, which share micro- to no complementary flanking regions, drove scientists to consider other recombination-based scenarios (Mita et al., 1990; Krishnan et al., 2008). However, computational analyses of mtDNA sequences indicated that the general distribution of the deletion breakpoints overlaps with the distribution of sites with high potential to form guanine-quadruplex (GQ) structures (Oliveira et al., 2013; Bharti et al., 2014; Dong et al., 2014). These are non-B DNA secondary structures characterized by planar stacks of guanines interacting by nonconventional Hoogsteen hydrogen bonds (Hou and Wei, 1996; Bharti et al., 2014; Dong et al., 2014). In the nucleus, GQs have been demonstrated to hinder DNA replication and cause genome instability (Ribeyre et al., 2009; Lopes et al., 2011). Furthermore, sequences with GQ-forming potential have been associated with deletions and duplications in the genomic DNA of cancer cells (Ribeyre et al., 2009; De and Michor, 2011). Recently, GQs within human mtDNA have been observed directly using fluorescence microscopy (Huang et al., 2015), and computational analyses have associated GQs with the formation of sporadic and secondary mtDNA deletions (Dong et al., 2014). Regarding the common deletion, both the 5’ and the 3’ 13 bp repeats have the potential to form GQs in three overlapping configurations (Oliveira et al., 2013; Dong et al., 2014). Markedly, a single nucleotide polymorphism within the 5’ repeat, 8472C>T, abolishes the guanine run shared by all three possible GQs and, consequently, the formation of the common deletion is significantly reduced in the haplogroup bearing such substitution (whereas the distribution of other deletions seems to be unaltered) (Guo et al., 2010). The TAS region has earlier been predicted to form stem-loop structures (Doda et al., 1981; Brown et al., 1986; Pereira et al., 2008), however, it also has the potential to form a GQ structure (Dong et al., 2014). Importantly, the GQ-forming potential does not exclude the formation of other secondary structures, such as hairpin-, cruciform- and cloverleaf-like (Damas et al., 2012). In fact, in their comprehensive study on the GQ-forming potential of mtDNA, Kaufman and coworkers have also confirmed the potential for the formation of other secondary structures facilitated by direct repeats (Dong et al., 2014). Perhaps an additive potential for the formation of GQs and other secondary structures may explain the higher frequency of deletions facilitated by direct repeats (class I). Direct evidence for the relevance of mtDNA secondary structures for ΔmtDNA formation comes from studies on the Pif1 helicase, which catalyzes the resolution of GQs and other secondary structures in the nucleus and in mitochondria (Paeschke et al., 2013; Mendoza et al., 2016). Pif1 ablation in mice increases the mtDNA deletion load in skeletal muscles (Bannwarth et al., 2016), and its depletion in human cell lines results in a 2-fold increase in the common deletion formation in response to TALEN-induced breaks of the mtDNA heavy-strand (Phillips et al., 2017).
Nearly all putative GQs are predicted to form on the heavy-strand, which is consistent with its high G-content (Oliveira et al., 2013; Dong et al., 2014). Considering the strand-displacement replication model, secondary structures on the heavy-strand would primarily affect the synthesis of the nascent light-strand. Notably, studies on mtDNA RIs indicated that, in comparison to the heavy-strand synthesis, the light-strand synthesis is significantly delayed and exhibits higher susceptibility to halting in the presence of dideoxynucleotides (Wanrooij et al., 2007). Moreover, Falkenberg and coworkers demonstrated directly that subnormal levels of Pol γ-α cause inefficient replication of the nascent light-strand (with unaltered replication of the nascent heavy-strand), and result in mtDNA deletions and PEO in patients (Roos et al., 2013). Stalling of Pol γ on the parental heavy-strand would generate breaks consistent with the replication-slippage model (see above, Figure 4A). Also, secondary structures inherent to the heavy-strand (formed before another replication round) would pose a hurdle for the helicase, which operates on such a strand (Figure 4B). In fact, Brosh and coworkers demonstrated that in vitro the wild-type Twinkle is incapable of resolving intra-strand GQs (Bharti et al., 2014). Additionally, analysis of mtDNA RIs in cells expressing defective Twinkle variants indicated that replication is abolished due to arrest in the vicinity of TAS (Wanrooij et al., 2007), which is consistent with the observed mtDNA copy number decrease in associated cases. Hence, the replicative helicase is highly vulnerable to secondary structures formed on the heavy-strand, to the extent of halting of the processive replication.
Figure 4. Schematic representation of potential stalling of a mitochondrial replisome at G-quadruplex structures (GQs) formed on the heavy-strand template (denoted as H). GQs (shown as three rhomboids) pose as a potential structural obstacle for the lagging-strand synthesis by Pol γ on the heavy-strand template (A), or for the Twinkle helicase during leading-strand synthesis on the light-strand template (denoted as L) (B). The red line represents the RNA primer generated at OL. Alternative priming sites generated by PrimPol on the heavy-strand template (not depicted here), permit passing the obstacles and would explain the “rescue replication”, as suggested by Torregrosa-Munumer et al. (2017). See text for more details.
In striking consistency with the accumulation of GQs on the heavy-strand, Sfeir and coworkers recently reported that not only double-strand breaks, but “just a nick” in the heavy-strand in proximity to the 5’ breakpoint is sufficient to generate the common deletion in mtDNA molecules of human cells in culture (Phillips et al., 2017). Furthermore, the authors of this study demonstrated that the formation of the common deletion requires the presence of Pol γ, Twinkle and mtSSB, as their depletion resulted in a dramatic drop in ΔmtDNA upon induced cleavage. This study advocates that the replisome components are involved in the formation of deletions and/or that replication and deletion formation are somehow linked. This is somewhat contradictory to the fact that a dysfunction of the replisome (either by the knockdown of one of its component or by the presence of a mutated constituent) may also result in deletions of all classes (Reeve et al., 2008). In our opinion, this inconsistency could be explained by the existence of a threshold effect for replisome activity, above which defective replisomes retain competence for genome replication (but may also facilitate the formation of replication-related deletions); below this threshold, replication halts resulting in mtDNA depletion (which in fact occurs in early-onset syndromes). In support of such a threshold hypothesis, a decrease in the levels of Pol γ-α transcripts by half in a heterozygous POLG1 knockout mouse does not affect its development, whereas the homozygous POLG1 knockout is lethal at the early embryonic stage (Hance et al., 2005). This lack of an ‘intermediate’ phenotype implies significant tolerance to the limited level/activity of Pol γ. Also, the phenomenon of clonal expansion seems to be consistent with the concept of threshold activity, as the lack of replication capacity would also exclude any genome expansion, and yet it has recently been documented for deletions resulting from defective Pol γ (Trifunov et al., 2018). Therefore, each individual who develops beyond the embryonic stage must possess the capacity to replicate mtDNA. A direct implication of this hypothesis is that deletion formation would not be dictated by the replication capacity, but rather by the frequency of which the deletion-forming mechanism is “triggered”, i.e. the frequency of heavy-strand breaks, which would also determine the severity of the resulting condition. A defective replisome could induce such breaks in every replication round explaining why mutations in TWNK and POLG1 are the most common causes of mtDNA diseases. Interestingly, in contrast to a defective Pol γ, the RIs resulting from a defective Twinkle that stalls before reaching OL, appear to be fully dsDNA (Wanrooij et al., 2007). This might suggest that cells expressing defective Twinkle could have a lower frequency of single-strand breaks in the heavy-strand, as these would be prevented in vivo by annealing to a second strand of DNA (e.g., a nascent strand, product of replication) or of RNA (as product of the RITOLS/bootlace process). On the other hand, the observation that fully double-stranded RIs appear before the fork reaches OL implies that the stalling of Twinkle may initiate light-strand synthesis independently of OL. This finds support in earlier reports that suggested the existence of alternative light-strand replication initiation sites (Brown et al., 2005), as well as in a recent report demonstrating that Twinkle may promote the initiation of replication at sites distinct from the two origins (Cluett et al., 2018). Pohjoismaki and coworkers recently demonstrated that such advantageous replication mechanism does in fact exist and can rescue the stalled mtDNA replication (Torregrosa-Muñumer et al., 2017). This `rescue replication’ involves the cooperative action of Pol γ and PrimPol, which is an RNA/DNA polymerase also found in the nucleus, capable of laying primers on the mtDNA to be utilized by Pol γ (García-Gómez et al., 2013; Stojkovic et al., 2016; Xu et al., 2019). The activity of such an alternative replication mechanism could explain the less deleterious symptoms associated with the deletor mouse, specifically the lack of motor function impairment, which is present in the exonuclease-deficient Pol γ mice.
Stalling of the replication fork appears to be a common consequence of various defects to the replisome components that precludes heavy-stand breaks, which directly triggers the formation of deletions. However, in the case of primary deletions, the source of heavy-strand breaks remains puzzling, as all the replisome components are supposedly functional. As mentioned earlier, Sfeir and coworkers proposed that the breaks could result from ROS activity. However, this view is currently debated as there is no clear association of elevated ROS and accumulation of ΔmtDNA (Nissanka et al., 2018), and neither the mutator nor the deletor mice exhibit elevated ROS (Trifunovic et al., 2005; Tyynismaa et al., 2005; Edgar et al., 2009). Another possibility considered by Sfeir and coworkers was an uncontrolled activity of DNA metabolic enzymes. In agreement with this idea, in our recent study on mtDNA replication in Drosophila melanogaster, we observed that an elevated expression of wild-type Twinkle resulted in the accumulation of ΔmtDNA, which expanded clonally to the extent that no wild-type mtDNA was detected by Southern-blotting (Ciesielski et al., 2018). Deletions have also been reported for mice overexpressing the wild-type helicase (Ylikallio et al., 2010). Although there are structural and functional differences between the fly and mammalian Twinkle and Pol γ homologues (Stiban et al., 2014; Ciesielski et al., 2015; Oliveira et al., 2015), these observations prompted us to speculate that not only mutated replisome components but also dysregulation of their levels, in a way that would change their stoichiometry at the replication fork, is deleterious and may result in mtDNA deletion formation. Importantly, overexpression of Twinkle in our study, as well as in the recent study by Holt and coworkers (Cluett et al., 2018), resulted in the accumulation of RIs consistent with replisome stalling. This observation strongly suggests that stalling of the replication fork is a universal step in the formation of deletions in both primary and secondary syndromes.
Markedly, the effects of overexpressing Twinkle in fly cells resemble the effects of halting Pol γ in human cells with dideoxynucleotides, to which the mitochondrial replicase is highly sensitive. In both cases, RIs are consistent with replication stalling and exhibit long stretches of ssDNA, as well as substantial incorporation of RNA (Wanrooij et al., 2007; Ciesielski et al., 2018). These features are consistent with desynchronization between unwinding of the template DNA and DNA synthesis per se. The milestone report on the human minimal mitochondrial replisome implied that effective DNA synthesis in the “mini-circle” assays requires functional interaction between both enzymes (Korhonen et al., 2004). In support, it has recently been demonstrated that a specific pathogenic mutation in human POLG1 does not alter the biochemical properties of the enzyme, but interrupts its functional interaction with Twinkle (Qian et al., 2015). Therefore, it is highly likely (although still speculative) that during processive DNA synthesis, the mitochondrial replisome, similarly to many other replication systems, acts as a complex containing the two enzymes and, perhaps mtSSB (Lee and Richardson, 2011), at least for the mtDNA heavy-strand synthesis. It seems likely that uncoupling the activity of both enzymes (e.g. upon overactivity of the helicase or halting of Pol γ) results in the generation of single-stranded stretches ahead of the polymerase, vulnerable to breaks. In fact, we observed that Twinkle overexpression in fly cells results in a 50% increase in the abundance of linear mtDNA molecules (Ciesielski et al., 2018). Interestingly, dideoxynucleotide-restriction of Pol γ activity generates the same pattern of RIs as defective Pol γ variants bearing human mutations, an observation that is supportive of the presumed uniform mechanism of deletion formation (Wanrooij et al., 2007). Considering the possibility of mitochondrial replisome uncoupling, one would need to address the etiology of such an event in healthy tissues before trying to understand disease and aging. In this notion, certain xenobiotics, such as berberine, a plant alkaloid used in the treatment of diabetes and other conditions (Kong et al., 2004; Cicero and Baggioni, 2016), possess GQ-stabilizing properties and accumulate in mitochondria (Pereira et al., 2007; Bazzicalupi et al., 2013). Accumulation of such agents could potentially stall the activity of a fully functional mitochondrial replisome (Naeem et al., 2018), which poses as an appealing circumstantial cause for the formation of primary mtDNA deletions, especially those accumulating with aging. This possibility warrants further investigation.
Stalling of the replication fork often results in single- or double-strand breaks (reviewed in Mirkin and Mirkin, 2007; Cannan and Pederson, 2016). The mechanism by which a stalled replication fork generates breaks on mtDNA remains elusive. Generally, single-stranded DNA is more vulnerable to breaks, and double-strand breaks are a consequence of unrepaired single-strand breaks in a double-stranded DNA molecule. The heavy-strand displaced during mtDNA replication is therefore the “weak side”, as the light-strand remains double-stranded. This is consistent with the idea that heavy-strand breaks is the major trigger for mtDNA deletion formation, as discussed above. Oxidation inflicted by ROS is often considered a direct break-inducing damage, however, as mentioned earlier, this possibility is currently under debate. Alternatively, breaks could result from the tension imposed on mtDNA by the progressing replisome, which usually results in positive supercoiling ahead of the fork. If unresolved, superhelical strains may be relieved by inter-winding of strands behind the fork, which in the case of a single-stranded molecule, could result in breaks in a phosphodiester bond. We find this possibility especially interesting, as it could also result in the formation of mtDNA catenanes (if no break occurs), which is another hallmark of Twinkle overexpression that we and others have observed (Pohjoismäki et al., 2009; Ciesielski et al., 2018). Nevertheless, the link between replication fork stalling and mtDNA breaks warrants further experimental evidence.
What exactly happens after the break?
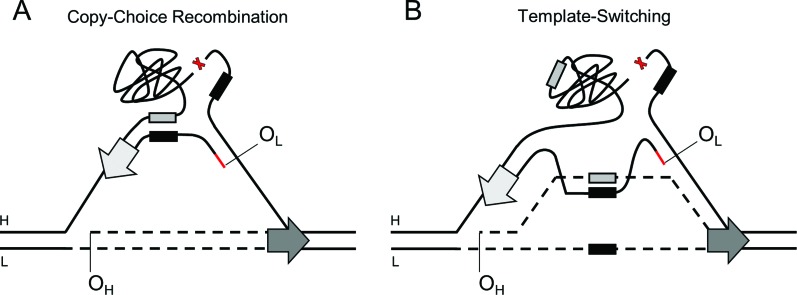
Although replication stalling is acknowledged as an important culprit behind pathological mtDNA deletions, almost nothing is known about the fate of stalled replication intermediates, such as their subsequent processing, and which enzymatic players are involved. The replication-slippage model proposed earlier is based in essence on the concept of ‘slipped mispairing’ of direct repeats between both DNA strands (Shoffner et al., 1989). The shortcoming of this scenario is that for the slipped mispairing to occur, the heavy-strand would need to be displaced ahead of the fork during leading-strand synthesis (Figure 3A), for which there is no evidence during the regular replication process. Interestingly, such RIs would be generated upon the hypothesized replisome uncoupling. However, taking the evidence that replication stalling during mtDNA light-strand synthesis underlies the formation of deletions, another possibility needs to be considered: that the mispairing does not have to occur between the two parental strands, but may in fact take place between the parental heavy-strand and the nascent light-strand. In such a case, a replisome stalled on, for example, a GQ site during light-strand synthesis, downstream of the 5’ DR (of the common deletion), would generate a break like those induced with mito-TALENS. The nascent light-strand containing a copy of the 5’ DR could potentially anneal to the 3’ DR of the parental heavy-strand (Figure 5A). This would create a new primed template sufficient for replication restart. The daughter, deletion-bearing mtDNA molecules would be consistent with those observed in class I deletions (and others), i.e., retaining the 3’ flanking region. Such mechanism of the replication-slippage model (known specifically as copy-choice recombination) has been proposed upon intensive studies on prokaryotic and viral replication systems (Viguera et al., 2001, and references therein). Notably, both Pol γ and Twinkle share viral ancestry (Oliveira et al., 2015; Kaguni and Oliveira, 2016), and the copy-choice recombination mechanism agrees with major findings regarding the formation of mtDNA deletions: i) this mechanism depends on replication stalling on the heavy-strand template; ii) the mispairing of flanking sites could occur independently of the heavy-strand replication, which resolves the issue of the annealing of parental strands; iii) repair of the nicked parental heavy-strand would require exonuclease and ligase activities, consistent with the reported importance of MGME1, Dna2 and Lig3 for the formation of the common deletion (Phillips et al., 2017); and iv) no specialized recombination machinery is required, consistent again with the report by Sfeir and coworkers. Remarkably, as this work was under review, a report was published providing strong in vitro evidence for the copy-choice recombination mechanism (Persson et al., 2019), which occurs in a way very much similar to our predictions (Figure 5A).
Figure 5. Schematic representation of the mechanisms of mtDNA deletion formation via copy-choice recombination (A) or template-switching (B). In both scenarios, DNA synthesis of the light-strand initiated at OL, using the heavy-strand template (denoted as H), is stalled after the 5’ deletion’s flanking region (black box), which promotes template breaking (indicated with a red X) between the deletion flanking regions. The 5’ flanking region of the nascent light-strand may pair with the complementary 3’ flanking region of the parental heavy-strand (gray box), which would enable replication restart (A). Alternatively, the nascent light-strand may invade the nascent heavy-strand (which remains paired with the parental light-strand, denoted as L; both strands represented as dashed lines), and pair with its complementary 3’ flanking region (gray box). Such a duplex structure would serve as a transient primed-template for DNA synthesis, until the damage site is bypassed and replication can resume on the parental heavy-strand (B). The block arrows represent the replisome progressing in the indicated directions during heavy- (dark gray) and light- (light gray) strand syntheses. The red line represents an RNA primer generated at OL.
Interestingly, Pol γ, like other polymerases, is tolerant to a certain level of primer-template mismatch, as long as the 3’ end is complementary (Bebenek and Kunkel, 2000; Longley et al., 2001). Perhaps, this could explain the class II and III deletions based on imperfect repeats and micro- to no homologies, and the higher frequency of the class I deletions, which would be based on the high complementarity of the flanking regions. Additionally, utilization of a mismatched primer-template substrate by Pol γ could potentially give rise to point mutations, which as mentioned above, are frequently found concomitantly to deletions, although no correlation between the distributions of deletion breakpoints and point mutations in the mtDNA has been detected to date (Wanrooij et al., 2004).
Alternatively, replication stalling on the heavy-strand template would also enable the nascent light-strand to invade and pair with the nascent heavy-strand, serving as a “bypass” template to overcome a stalling site. This possibility is consistent with the strand-displacement model in which the nascent heavy-strand would be laid on the light-strand template before the initiation of DNA synthesis at the OL. After bypassing the structural hurdle, replication would continue back on the heavy-strand template. This so-called template-switching mechanism (Figure 5B) is well documented in various organisms (Giannattasio et al., 2014; Lovett, 2017), and could take place in mitochondria by the recently reported strand-exchange activity of Twinkle (Sen et al., 2016). Template-switching typically results in the accumulation of catenated circular DNA molecules (Giannattasio et al., 2014), which was in fact observed along with mtDNA deletions in our study on the effects of Twinkle overexpression in fly cells (Ciesielski et al., 2018). A similar phenotype was observed in human heart muscle, as well as upon Twinkle overexpression in mice heart, brain, and skeletal muscle (Pohjoismäki et al., 2009). Since mtDNA replication and deletion formation could be dependent on the mitochondrial nucleoid context, which still needs to be better understood especially in different post-mitotic tissues, it is possible that both copy-choice recombination and template-switching play a role in vivo, although this is highly speculative at this point.
In our opinion, the results of the studies presented in this review strongly support the replication-based models of mtDNA deletion formation, which is initiated upon replication stalling on and possibly breaks of the heavy-strand template. This promotes mispairing of the nascent light-strand with complementary sites on another accessible DNA strand. Resolution of the slipped-template may require a simple replication restart or a template-switching mechanism. We believe that the proposed mechanism accommodates all the major findings in the field. Notably, this replication-mediated recombination model does not exclude the existence of independent DSB-repair systems in mitochondria, such as homologous recombination or non-homologous end-joining. However, further studies are warranted to provide strong biochemical evidence for such processes, including the complete identification of their protein players and the associated enzymatic activities.
Acknowledgments
GLC and MTO would like to dedicate this article to Dr. Laurie S. Kaguni in gratitude for years of mentorship. We also wish to thank Dr. Mike Gerards and Ana Paula C. Rodrigues for their critical comments on the manuscript, and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (grant numbers 2014/02253-6, 2017/04372-0 and 306974/2017-7 to MTO) for financial support.
Footnotes
Associate Editor: Nadja Souza-Pinto
Conflict of interest
Authors declare no conflict of interest.
Author contributions
GLC and CBP drafted the initial version of the manuscript, MTO critically edited, revised and assembled the final version.
References
- Agostino A, Valletta L, Chinnery PF, Ferrari G, Carrara F, Taylor R, Schaefer AM, Turnbull DM, Tiranti V, Zeviani M. Mutations of ANT1, Twinkle, and POLG1 in sporadic progressive external ophthalmoplegia (PEO) Neurology. 2003;60:1354–1356. doi: 10.1212/01.wnl.0000056088.09408.3c. [DOI] [PubMed] [Google Scholar]
- Alston CL, Rocha MC, Lax NZ, Turnbull DM, Taylor RW. The genetics and pathology of mitochondrial disease. J Pathol. 2017;241:236–250. doi: 10.1002/path.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC. Phylogeography: The History and Formation of Species. Harvard University Press; Cambridge: 2000. p. 464. [Google Scholar]
- Bacman SR, Williams SL, Moraes CT. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res. 2009;37:4218–4226. doi: 10.1093/nar/gkp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Pinto M, Peralta S, Moraes CT. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat Med. 2013;19:1111–1113. doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannwarth S, Berg-Alonso L, Augé G, Fragaki K, Kolesar JE, Lespinasse F, Lacas-Gervais S, Burel-Vandenbos F, Villa E, Belmonte F, et al. Inactivation of Pif1 helicase causes a mitochondrial myopathy in mice. Mitochondrion. 2016;30:126–137. doi: 10.1016/j.mito.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzicalupi C, Ferraroni M, Bilia AR, Scheggi F, Gratteri P. The crystal structure of human telomeric DNA complexed with berberine: an interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res. 2013;41:632–638. doi: 10.1093/nar/gks1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K, Kunkel TA. Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harb Symp Quant Biol. 2000;65:81–91. doi: 10.1101/sqb.2000.65.81. [DOI] [PubMed] [Google Scholar]
- Belmonte FR, Martin JL, Frescura K, Damas J, Pereira F, Tarnopolsky MA, Kaufman BA. Digital PCR methods improve detection sensitivity and measurement precision of low abundance mtDNA deletions. Sci Rep. 2016;6:25186. doi: 10.1038/srep25186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti SK, Sommers JA, Zhou J, Kaplan DL, Spelbrink JN, Mergny JL, Brosh RM. DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative Twinkle helicase. J Biol Chem. 2014;289:29975–29993. doi: 10.1074/jbc.M114.567073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowmaker M, Yang MY, Yasukawa T, Reyes A, Jacobs HT, Huberman JA, Holt IJ. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J Biol Chem. 2003;278:50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- Brown GG, Gadaleta G, Pepe G, Saccone C, Sbisà E. Structural conservation and variation in the D-loop-containing region of vertebrate mitochondrial DNA. J Mol Biol. 1986;192:503–511. doi: 10.1016/0022-2836(86)90272-x. [DOI] [PubMed] [Google Scholar]
- Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19:2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am J Hum Genet. 2006;79:469–480. doi: 10.1086/507132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannan WJ, Pederson DS. Mechanisms and consequences of double-strand DNA break formation in chromatin. J Cell Physiol. 2016;231:3–14. doi: 10.1002/jcp.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SS, Copeland WC. DNA polymerase gamma and mitochondrial disease: understanding the consequence of POLG mutations. Biochim Biophys Acta. 2009;1787:312–319. doi: 10.1016/j.bbabio.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SS, Longley MJ, Copeland WC. The common A467T mutation in the human mitochondrial DNA polymerase (POLG) compromises catalytic efficiency and interaction with the accessory subunit. J Biol Chem. 2005;280:31341–31346. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]
- Chen T, He J, Huang Y, Zhao W. The generation of mitochondrial DNA large-scale deletions in human cells. J Hum Genet. 2011;56:689–694. doi: 10.1038/jhg.2011.97. [DOI] [PubMed] [Google Scholar]
- Chen XJ. Mechanism of homologous recombination and implications for aging-related deletions in mitochondrial DNA. Microbiol Mol Biol Rev. 2013;77:476–496. doi: 10.1128/MMBR.00007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery PF, Johnson MA, Wardell TM, Singh-Kler R, Hayes C, Brown DT, Taylor RW, Bindoff LA, Turnbull DM. The epidemiology of pathogenic mitochondrial DNA mutations. Ann Neurol. 2000;48:188–193. [PubMed] [Google Scholar]
- Chinnery PF, DiMauro S, Shanske S, Schon EA, Zeviani M, Mariotti C, Carrara F, Lombes A, Laforet P, Ogier H, et al. Risk of developing a mitochondrial DNA deletion disorder. Lancet. 2004;364:592–596. doi: 10.1016/S0140-6736(04)16851-7. [DOI] [PubMed] [Google Scholar]
- Cicero AF, Baggioni A. Berberine and its role in chronic disease. Adv Exp Med Biol. 2016;928:27–45. doi: 10.1007/978-3-319-41334-1_2. [DOI] [PubMed] [Google Scholar]
- Ciesielski GL, Bermek O, Rosado-Ruiz FA, Hovde SL, Neitzke OJ, Griffith JD, Kaguni LS. Mitochondrial single-stranded DNA-binding proteins stimulate the activity of DNA polymerase γ by organization of the template DNA. J Biol Chem. 2015;290:28697–28707. doi: 10.1074/jbc.M115.673707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesielski GL, Oliveira MT, Kaguni LS. Animal mitochondrial DNA replication. Enzymes. 2016;39:255–292. doi: 10.1016/bs.enz.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesielski GL, Nadalutti CA, Oliveira MT, Jacobs HT, Griffith JD, Kaguni LS. Structural rearrangements in the mitochondrial genome of Drosophila melanogaster induced by elevated levels of the replicative DNA helicase. Nucleic Acids Res. 2018;46:3034–3046. doi: 10.1093/nar/gky094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluett TJ, Akman G, Reyes A, Kazak L, Mitchell A, Wood SR, Spinazzola A, Spelbrink JN, Holt IJ. Transcript availability dictates the balance between strand-asynchronous and strand-coupled mitochondrial DNA replication. Nucleic Acids Res. 2018;46:10771–10781. doi: 10.1093/nar/gky852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland WC. Defects of mitochondrial DNA replication. J Child Neurol. 2014;29:1216–1224. doi: 10.1177/0883073814537380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortopassi GA, Shibata D, Soong NW, Arnheim N. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc Natl Acad Sci U S A. 1992;89:7370–7374. doi: 10.1073/pnas.89.16.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J, Carneiro J, Gonçalves J, Stewart JB, Samuels DC, Amorim A, Pereira F. Mitochondrial DNA deletions are associated with non-B DNA conformations. Nucleic Acids Res. 2012;40:7606–7621. doi: 10.1093/nar/gks500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J, Carneiro J, Amorim A, Pereira F. MitoBreak: tThe mitochondrial DNA breakpoints database. Nucleic Acids Res. 2014;42:D1261–1268. doi: 10.1093/nar/gkt982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Maynard S, Hashiguchi K, Hu J, Muftuoglu M, Bohr VA. The recombination protein RAD52 cooperates with the excision repair protein OGG1 for the repair of oxidative lesions in mammalian cells. Mol Cell Biol. 2009;29:4441–4454. doi: 10.1128/MCB.00265-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degoul F, Nelson I, Amselem S, Romero N, Obermaier-Kusser B, Ponsot G, Marsac C, Lestienne P. Different mechanisms inferred from sequences of human mitochondrial DNA deletions in ocular myopathies. Nucleic Acids Res. 1991;19:493–496. doi: 10.1093/nar/19.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fonzo A, Bordoni A, Crimi M, Sara G, Del Bo R, Bresolin N, Comi GP. POLG mutations in sporadic mitochondrial disorders with multiple mtDNA deletions. Hum Mutat. 2003;22:498–499. doi: 10.1002/humu.9203. [DOI] [PubMed] [Google Scholar]
- Doda JN, Wright CT, Clayton DA. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc Natl Acad Sci U S A. 1981;78:6116–6120. doi: 10.1073/pnas.78.10.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong DW, Pereira F, Barrett SP, Kolesar JE, Cao K, Damas J, Yatsunyk LA, Johnson FB, Kaufman BA. Association of G-quadruplex forming sequences with human mtDNA deletion breakpoints. BMC Genomics. 2014;15:677. doi: 10.1186/1471-2164-15-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar D, Shabalina I, Camara Y, Wredenberg A, Calvaruso MA, Nijtmans L, Nedergaard J, Cannon B, Larsson NG, Trifunovic A. Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metab. 2009;10:131–138. doi: 10.1016/j.cmet.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008;83:254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, MacGregor GR, Wallace DC. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filosto M, Mancuso M, Nishigaki Y, Pancrudo J, Harati Y, Gooch C, Mankodi A, Bayne L, Bonilla E, Shanske S, et al. Clinical and genetic heterogeneity in progressive external ophthalmoplegia due to mutations in polymerase gamma. Arch Neurol. 2003;60:1279–1284. doi: 10.1001/archneur.60.9.1279. [DOI] [PubMed] [Google Scholar]
- Fishel ML, Vasko MR, Kelley MR. DNA repair in neurons: So if they don’t divide what’s to repair? Mutat Res. 2007;614:24–36. doi: 10.1016/j.mrfmmm.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Fuke S, Kametani M, Yamada K, Kasahara T, Kubota-Sakashita M, Kujoth GC, Prolla TA, Hitoshi S, Kato T. Heterozygous Polg mutation causes motor dysfunction due to mtDNA deletions. Ann Clin Transl Neurol. 2014;1:909–920. doi: 10.1002/acn3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet. 2009;18:1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammage PA, Rorbach J, Vincent AI, Rebar EJ, Minczuk M. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol Med. 2014;6:458–466. doi: 10.1002/emmm.201303672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gómez S, Reyes A, Martínez-Jiménez MI, Chocrón ES, Mourón S, Terrados G, Powell C, Salido E, Méndez J, Holt IJ, et al. PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell. 2013;52:541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Zwicky K, Follonier C, Foiani M, Lopes M, Branzei D. Visualization of recombination-mediated damage bypass by template switching. Nat Struct Mol Biol. 2014;21:884–892. doi: 10.1038/nsmb.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A, Spelbrink JN. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum Mol Genet. 2009;18:328–340. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Vioque E, Blázquez A, Fernández-Moreira D, Bornstein B, Bautista J, Arpa J, Navarro C, Campos Y, Fernández-Moreno MA, Garesse R, et al. Association of novel POLG mutations and multiple mitochondrial DNA deletions with variable clinical phenotypes in a Spanish population. Arch Neurol. 2006;63:107–111. doi: 10.1001/archneur.63.1.107. [DOI] [PubMed] [Google Scholar]
- Gorman GS, Schaefer AM, Ng Y, Gomez N, Blakely EL, Alston CL, Feeney C, Horvath R, Yu-Wai-Man P, Chinnery PF, et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol. 2015;77:753–759. doi: 10.1002/ana.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady JP, Campbell G, Ratnaike T, Blakely EL, Falkous G, Nesbitt V, Schaefer AM, McNally RJ, Gorman GS, Taylor RW, et al. Disease progression in patients with single, large-scale mitochondrial DNA deletions. Brain. 2014;137:323–334. doi: 10.1093/brain/awt321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier J, Hirano M, Karaa A, Shepard E, Thompson JLP. Diagnostic odyssey of patients with mitochondrial disease: Results of a survey. Neurol Genet. 2018;4:e230. doi: 10.1212/NXG.0000000000000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Popadin KY, Markuzon N, Orlov YL, Kraytsberg Y, Krishnan KJ, Zsurka G, Turnbull DM, Kunz WS, Khrapko K. Repeats, longevity and the sources of mtDNA deletions: Evidence from ‘deletional spectra’. Trends Genet. 2010;26:340–343. doi: 10.1016/j.tig.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum Mol Genet. 2005;14:1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- Harrison RG. Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trends Ecol Evol. 1989;4:6–11. doi: 10.1016/0169-5347(89)90006-2. [DOI] [PubMed] [Google Scholar]
- Holt IJ, Jacobs HT. Unique features of DNA replication in mitochondria: a functional and evolutionary perspective. Bioessays. 2014;36:1024–1031. doi: 10.1002/bies.201400052. [DOI] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- Horvath R, Hudson G, Ferrari G, Fütterer N, Ahola S, Lamantea E, Prokisch H, Lochmüller H, McFarland R, Ramesh V, et al. Phenotypic spectrum associated with mutations of the mitochondrial polymerase gamma gene. Brain. 2006;129:1674–1684. doi: 10.1093/brain/awl088. [DOI] [PubMed] [Google Scholar]
- Hou JH, Wei YH. The unusual structures of the hot-regions flanking large-scale deletions in human mitochondrial DNA. Biochem J. 1996;318:1065–1070. doi: 10.1042/bj3181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WC, Tseng TY, Chen YT, Chang CC, Wang ZF, Wang CL, Hsu TN, Li PT, Chen CT, Lin JJ, et al. Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res. 2015;43:10102–10113. doi: 10.1093/nar/gkv1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G, Chinnery PF. Mitochondrial DNA polymerase-gamma and human disease. Hum Mol Genet. 2006;15:R244–252. doi: 10.1093/hmg/ddl233. [DOI] [PubMed] [Google Scholar]
- Jemt E, Persson Ö, Shi Y, Mehmedovic M, Uhler JP, Dávila López M, Freyer C, Gustafsson CM, Samuelsson T, Falkenberg M. Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 2015;43:9262–9275. doi: 10.1093/nar/gkv804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- Kaguni LS, Oliveira MT. Structure, function and evolution of the animal mitochondrial replicative DNA helicase. Crit Rev Biochem Mol Biol. 2016;51:53–64. doi: 10.3109/10409238.2015.1117056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa L, Quintana DF, Schiraldi LK, Phadnis N, Coles GL, Sia RA, Sia EA. Mitochondrial genome maintenance: Roles for nuclear nonhomologous end-joining proteins in Saccharomyces cerevisiae. Genetics. 2012;190:951–964. doi: 10.1534/genetics.111.138214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppila TES, Kauppila JHK, Larsson NG. Mammalian mitochondria and aging: An update. Cell Metab. 2017;25:57–71. doi: 10.1016/j.cmet.2016.09.017. [DOI] [PubMed] [Google Scholar]
- Kazak L, Reyes A, He J, Wood SR, Brea-Calvo G, Holen TT, Holt IJ. A cryptic targeting signal creates a mitochondrial FEN1 isoform with tailed R-Loop binding properties. PLoS One. 2013;8:e62340. doi: 10.1371/journal.pone.0062340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns TP, Sayre GP. Retinitis pigmentosa, external ophthalmophegia, and complete heart block: unusual syndrome with histologic study in one of two cases. AMA Arch Ophthalmol. 1958;60:280–289. [PubMed] [Google Scholar]
- Kong W, Wei J, Abidi P, Lin M, Inaba S, Li C, Wang Y, Wang Z, Si S, Pan H, et al. Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat Med. 2004;10:1344–1351. doi: 10.1038/nm1135. [DOI] [PubMed] [Google Scholar]
- Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum C, Nicholls TJ, Haack TB, Schöler S, Peeva V, Danhauser K, Hallmann K, Zsurka G, Rorbach J, Ius A, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet. 2013;45:214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowald A, Kirkwood TBL. Resolving the enigma of the clonal expansion of mtDNA deletions. Genes (Basel) 2018;9:E126. doi: 10.3390/genes9030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan KJ, Reeve AK, Samuels DC, Chinnery PF, Blackwood JK, Taylor RW, Wanrooij S, Spelbrink JN, Lightowlers RN, Turnbull DM. What causes mitochondrial DNA deletions in human cells? Nat Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Richardson CC. Choreography of bacteriophage T7 DNA replication. Curr Opin Chem Biol. 2011;15:580–586. doi: 10.1016/j.cbpa.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann D, Kornhuber ME, Clajus C, Alston CL, Wienke A, Deschauer M, Taylor RW, Zierz S. Peripheral neuropathy in patients with CPEO associated with single and multiple mtDNA deletions. Neurol Genet. 2016;2:e113. doi: 10.1212/NXG.0000000000000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. J Biol Chem. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- Lopes J, Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou MP, Foiani M, Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett ST. Template-switching during replication fork repair in bacteria. DNA Repair (Amst) 2017;56:118–128. doi: 10.1016/j.dnarep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macao B, Uhler JP, Siibak T, Zhu X, Shi Y, Sheng W, Olsson M, Stewart JB, Gustafsson CM, Falkenberg M. The exonuclease activity of DNA polymerase γ is required for ligation during mitochondrial DNA replication. Nat Commun. 2015;6:7303. doi: 10.1038/ncomms8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic S, Jiang M, Nicholls TJ, Uhler JP, Dirksen-Schwanenland C, Polosa PL, Simard ML, Li X, Atanassov I, Rackham O, et al. Mice lacking the mitochondrial exonuclease MGME1 accumulate mtDNA deletions without developing progeria. Nat Commun. 2018;9:1202. doi: 10.1038/s41467-018-03552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr JA, Haack TB, Freisinger P, Karall D, Makowski C, Koch J, Feichtinger RG, Zimmermann FA, Rolinski B, Ahting U, et al. Spectrum of combined respiratory chain defects. J Inherit Metab Dis. 2015;38:629–640. doi: 10.1007/s10545-015-9831-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EA, Oliveira MT. Replicating animal mitochondrial DNA. Genet Mol Biol. 2013;36:308–315. doi: 10.1590/S1415-47572013000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza O, Bourdoncle A, Boulé JB, Brosh RM, Mergny JL. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44:1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, Rizzuto R, Moraes CT, Shanske S, Arnaudo E, Fabrizi GM, Koga Y, DiMauro S, Schon EA. Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA. Nucleic Acids Res. 1990;18:561–567. doi: 10.1093/nar/18.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel-Sosa JF, Herrero MD, Munoz ML, Aguirre-Campa LE, Pérez-Ramírez G, García-Ramírez R, Ruiz-Pesini E, Montoya J. Phylogenetic analysis of mitochondrial DNA in a patient with Kearns-Sayre syndrome containing a novel 7629-bp deletion. Mitochondrial DNA. 2013;24:420–431. doi: 10.3109/19401736.2012.760550. [DOI] [PubMed] [Google Scholar]
- Moraes CT, Shanske S, Tritschler HJ, Aprille JR, Andreetta F, Bonilla E, Schon EA, DiMauro S. mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am J Hum Genet. 1991;48:492–501. [PMC free article] [PubMed] [Google Scholar]
- Moraes CT, Sciacco M, Ricci E, Tengan CH, Hao H, Bonilla E, Schon EA, DiMauro S. Phenotype-genotype correlations in skeletal muscle of patients with mtDNA deletions. Muscle Nerve Suppl. 1995;3:S150–S153. doi: 10.1002/mus.880181429. [DOI] [PubMed] [Google Scholar]
- Moretton A, Morel F, Macao B, Lachaume P, Ishak L, Lefebvre M, Garreau-Balandier I, Vernet P, Falkenberg M, Farge G. Selective mitochondrial DNA degradation following double-strand breaks. PLoS One. 2017;12:e0176795. doi: 10.1371/journal.pone.0176795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem MM, Maheshan R, Costford SR, Kaufman BA, Sondheimer N. Targeted reduction of pathogenic heteroplasmy through binding of G-quadruplex DNA. bioRxiv:10.1101/358101 2018 [Google Scholar]
- Nelson I, Degoul F, Obermaier-Kusser B, Romero N, Borrone C, Marsac C, Vayssiere JL, Gerbitz K, Fardeau M, Ponsot G, et al. Mapping of heteroplasmic mitochondrial DNA deletions in Kearns-Sayre syndrome. Nucleic Acids Res. 1989;17:8117–8124. doi: 10.1093/nar/17.20.8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng YS, Turnbull DM. Mitochondrial disease: Genetics and management. J Neurol. 2016;263:179–191. doi: 10.1007/s00415-015-7884-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KV, Sharief FS, Chan SS, Copeland WC, Naviaux RK. Molecular diagnosis of Alpers syndrome. J Hepatol. 2006;45:108–116. doi: 10.1016/j.jhep.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Nissanka N, Bacman SR, Plastini MJ, Moraes CT. The mitochondrial DNA polymerase gamma degrades linear DNA fragments precluding the formation of deletions. Nat Commun. 2018;9:2491. doi: 10.1038/s41467-018-04895-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurminen A, Farnum GA, Kaguni LS. Pathogenicity in POLG syndromes: DNA polymerase gamma pathogenicity prediction server and database. BBA Clin. 2017;7:147–156. doi: 10.1016/j.bbacli.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira MT, Kaguni LS. Reduced stimulation of recombinant DNA polymerase γ and mitochondrial DNA (mtDNA) helicase by variants of mitochondrial single-stranded DNA-binding protein (mtSSB) correlates with defects in mtDNA replication in animal cells. J Biol Chem. 2011;286:40649–40658. doi: 10.1074/jbc.M111.289983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira MT, Haukka J, Kaguni LS. Evolution of the metazoan mitochondrial replicase. Genome Biol Evol. 2015;7:943–959. doi: 10.1093/gbe/evv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira PH, da Silva CL, Cabral JM. An appraisal of human mitochondrial DNA instability: new insights into the role of non-canonical DNA structures and sequence motifs. PLoS One. 2013;8:e59907. doi: 10.1371/journal.pone.0059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten ABC, Sallevelt SCEH, Carling PJ, Dreesen JCFM, Drüsedau M, Spierts S, Paulussen ADC, de Die-Smulders CEM, Herbert M, Chinnery PF, et al. Mutation-specific effects in germline transmission of pathogenic mtDNA variants. Hum Reprod. 2018;33:1331–1341. doi: 10.1093/humrep/dey114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallotti F, Chen X, Bonilla E, Schon EA. Evidence that specific mtDNA point mutations may not accumulate in skeletal muscle during normal human aging. Am J Hum Genet. 1996;59:591–602. [PMC free article] [PubMed] [Google Scholar]
- Peeva V, Blei D, Trombly G, Corsi S, Szukszto MJ, Rebelo-Guiomar P, Gammage PA, Kudin AP, Becker C, Altmüller J, et al. Linear mitochondrial DNA is rapidly degraded by components of the replication machinery. Nat Commun. 2018;9:1727. doi: 10.1038/s41467-018-04131-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F, Soares P, Carneiro J, Pereira L, Richards MB, Samuels DC, Amorim A. Evidence for variable selective pressures at a large secondary structure of the human mitochondrial DNA control region. Mol Biol Evol. 2008;25:2759–2770. doi: 10.1093/molbev/msn225. [DOI] [PubMed] [Google Scholar]
- Pereira GC, Branco AF, Matos JA, Pereira SL, Parke D, Perkins EL, Serafim TL, Sardão VA, Santos MS, Moreno AJ, et al. Mitochondrially targeted effects of berberine [Natural Yellow 18, 5,6-dihydro-9,10-dimethoxybenzo(g)-1,3-benzodioxolo(5,6-a) quinolizinium] on K1735-M2 mouse melanoma cells: comparison with direct effects on isolated mitochondrial fractions. J Pharmacol Exp Ther. 2007;323:636–649. doi: 10.1124/jpet.107.128017. [DOI] [PubMed] [Google Scholar]
- Persson Ö, Muthukumar Y, Basu S, Jenninger L, Uhler JP, Berglund AK, McFarland R, Taylor RW, Gustafsson CM, Larsson E, Falkenberg M. Copy-choice recombination during mitochondrial L-strand synthesis causes DNA deletions. Nat Commun. 2019;10:759. doi: 10.1038/s41467-019-08673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AF, Millet AR, Tigano M, Dubois SM, Crimmins H, Babin L, Charpentier M, Piganeau M, Brunet E, Sfeir A. Single-molecule analysis of mtDNA replication uncovers the basis of the common deletion. Mol Cell. 2017;65:527–538.e6. doi: 10.1016/j.molcel.2016.12.014. [DOI] [PubMed] [Google Scholar]
- Picard M, Vincent AE, Turnbull DM. Expanding our understanding of mtDNA deletions. Cell Metab. 2016;24:3–4. doi: 10.1016/j.cmet.2016.06.024. [DOI] [PubMed] [Google Scholar]
- Pitceathly RD, Rahman S, Hanna MG. Single deletions in mitochondrial DNA-molecular mechanisms and disease phenotypes in clinical practice. Neuromuscul Disord. 2012;22:577–586. doi: 10.1016/j.nmd.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Pohjoismäki JL, Goffart S, Tyynismaa H, Willcox S, Ide T, Kang D, Suomalainen A, Karhunen PJ, Griffith JD, Holt IJ, et al. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J Biol Chem. 2009;284:21446–21457. doi: 10.1074/jbc.M109.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjoismäki JL, Goffart S, Spelbrink JN. Replication stalling by catalytically impaired Twinkle induces mitochondrial DNA rearrangements in cultured cells. Mitochondrion. 2011;11:630–634. doi: 10.1016/j.mito.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Qian Y, Ziehr JL, Johnson KA. Alpers disease mutations in human DNA polymerase gamma cause catalytic defects in mitochondrial DNA replication by distinct mechanisms. Front Genet. 2015;6:135. doi: 10.3389/fgene.2015.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Copeland WC. POLG-related disorders and their neurological manifestations. Nat Rev Neurol. 2019;15:40–52. doi: 10.1038/s41582-018-0101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakulendran S, Pitceathly RD, Taanman JW, Costello H, Sweeney MG, Woodward CE, Jaunmuktane Z, Holton JL, Jacques TS, Harding BN, et al. A clinical, neuropathological and genetic study of homozygous A467T POLG-related mitochondrial disease. PLoS One. 2016;11:e0145500. doi: 10.1371/journal.pone.0145500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala N, Gerhold JM, Martinsson P, Klymov A, Spelbrink JN. Replication factors transiently associate with mtDNA at the mitochondrial inner membrane to facilitate replication. Nucleic Acids Res. 2014;42:952–967. doi: 10.1093/nar/gkt988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve AK, Krishnan KJ, Elson JL, Morris CM, Bender A, Lightowlers RN, Turnbull DM. Nature of mitochondrial DNA deletions in substantia nigra neurons. Am J Hum Genet. 2008;82:228–235. doi: 10.1016/j.ajhg.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Yasukawa T, Cluett TJ, Holt IJ. Analysis of mitochondrial DNA by two-dimensional agarose gel electrophoresis. Methods Mol Biol. 2009;554:15–35. doi: 10.1007/978-1-59745-521-3_2. [DOI] [PubMed] [Google Scholar]
- Reyes A, Yasukawa T, Holt IJ. Analysis of replicating mitochondrial DNA by two-dimensional agarose gel electrophoresis. Methods Mol Biol. 2007;372:219–232. doi: 10.1007/978-1-59745-365-3_16. [DOI] [PubMed] [Google Scholar]
- Ribeyre C, Lopes J, Boulé JB, Piazza A, Guédin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha MC, Rosa HS, Grady JP, Blakely EL, He L, Romain N, Haller RG, Newman J, McFarland R, Ng YS, et al. Pathological mechanisms underlying single large-scale mitochondrial DNA deletions. Ann Neurol. 2018;83:115–130. doi: 10.1002/ana.25127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos S, Macao B, Fusté JM, Lindberg C, Jemt E, Holme E, Moslemi AR, Oldfors A, Falkenberg M. Subnormal levels of POLγA cause inefficient initiation of light-strand DNA synthesis and lead to mitochondrial DNA deletions and progressive external ophthalmoplegia [corrected] Hum Mol Genet. 2013;22:2411–2422. doi: 10.1093/hmg/ddt094. [DOI] [PubMed] [Google Scholar]
- Russell OM, Fruh I, Rai PK, Marcellin D, Doll T, Reeve A, Germain M, Bastien J, Rygiel KA, Cerino R, et al. Preferential amplification of a human mitochondrial DNA deletion in vitro and in vivo. Sci Rep. 2018;8:1799. doi: 10.1038/s41598-018-20064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage JM, Gildemeister OS, Knight KL. Discovery of a novel function for human Rad51: Maintenance of the mitochondrial genome. J Biol Chem. 2010;285:18984–18990. doi: 10.1074/jbc.M109.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldaña-Martínez A, Muñoz ML, Pérez-Ramírez G, Montiel-Sosa JF, Montoya J, Emperador S, Ruiz-Pesini E, Cuevas-Covarrubias S, López-Valdez J, Ramírez RG. Whole sequence of the mitochondrial DNA genome of Kearns Sayre Syndrome patients: Identification of deletions and variants. Gene. 2019;688:171–181. doi: 10.1016/j.gene.2018.11.085. [DOI] [PubMed] [Google Scholar]
- Sallevelt SC, de Die-Smulders CE, Hendrickx AT, Hellebrekers DM, de Coo IF, Alston CL, Knowles C, Taylor RW, McFarland R, Smeets HJ. De novo mtDNA point mutations are common and have a low recurrence risk. J Med Genet. 2017;54:73–83. doi: 10.1136/jmedgenet-2016-103876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DC, Schon EA, Chinnery PF. Two direct repeats cause most human mtDNA deletions. Trends Genet. 2004;20:393–398. doi: 10.1016/j.tig.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Schaefer AM, McFarland R, Blakely EL, He L, Whittaker RG, Taylor RW, Chinnery PF, Turnbull DM. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- Schon EA, Rizzuto R, Moraes CT, Nakase H, Zeviani M, DiMauro S. A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science. 1989;244:346–349. doi: 10.1126/science.2711184. [DOI] [PubMed] [Google Scholar]
- Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: Roles of inherited and somatic mutations. Nat Rev Genet. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D, Patel G, Patel SS. Homologous DNA strand exchange activity of the human mitochondrial DNA helicase TWINKLE. Nucleic Acids Res. 2016;44:4200–4210. doi: 10.1093/nar/gkw098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoffner JM, Lott MT, Voljavec AS, Soueidan SA, Costigan DA, Wallace DC. Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: a slip-replication model and metabolic therapy. Proc Natl Acad Sci U S A. 1989;86:7952–7956. doi: 10.1073/pnas.86.20.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Cao Y, Samuels DC. Replication pauses of the wild-type and mutant mitochondrial DNA polymerase gamma: a simulation study. PLoS Comput Biol. 2011;7:e1002287. doi: 10.1371/journal.pcbi.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink JN, Toivonen JM, Hakkaart GA, Kurkela JM, Cooper HM, Lehtinen SK, Lecrenier N, Back JW, Speijer D, Foury F, et al. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J Biol Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Moraes CT. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum Mol Genet. 2005;14:893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiban J, Farnum GA, Hovde SL, Kaguni LS. The N-terminal domain of the Drosophila mitochondrial replicative DNA helicase contains an iron-sulfur cluster and binds DNA. J Biol Chem. 2014;289:24032–24042. doi: 10.1074/jbc.M114.587774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojkovic G, Makarova AV, Wanrooij PH, Forslund J, Burgers PM, Wanrooij S. Oxidative DNA damage stalls the human mitochondrial replisome. Sci Rep. 2016;6:28942. doi: 10.1038/srep28942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A, Isohanni P. Mitochondrial DNA depletion syndromes - many genes, common mechanisms. Neuromuscul Disord. 2010;20:429–437. doi: 10.1016/j.nmd.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Suomalainen A, Majander A, Wallin M, Setälä K, Kontula K, Leinonen H, Salmi T, Paetau A, Haltia M, Valanne L, et al. Autosomal dominant progressive external ophthalmoplegia with multiple deletions of mtDNA: clinical, biochemical, and molecular genetic features of the 10q-linked disease. Neurology. 1997;48:1244–1253. doi: 10.1212/wnl.48.5.1244. [DOI] [PubMed] [Google Scholar]
- Szczepanowska K, Foury F. A cluster of pathogenic mutations in the 3’-5’ exonuclease domain of DNA polymerase gamma defines a novel module coupling DNA synthesis and degradation. Hum Mol Genet. 2010;19:3516–3529. doi: 10.1093/hmg/ddq267. [DOI] [PubMed] [Google Scholar]
- Tadi SK, Sebastian R, Dahal S, Babu RK, Choudhary B, Raghavan SC. Microhomology-mediated end joining is the principal mediator of double-strand break repair during mitochondrial DNA lesions. Mol Biol Cell. 2016;27:223–235. doi: 10.1091/mbc.E15-05-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torregrosa-Muñumer R, Forslund JME, Goffart S, Pfeiffer A, Stojkovic G, Carvalho G, Al-Furoukh N, Blanco L, Wanrooij S, Pohjoismäki JLO. PrimPol is required for replication reinitiation after mtDNA damage. Proc Natl Acad Sci U S A. 2017;114:11398–11403. doi: 10.1073/pnas.1705367114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunov S, Pyle A, Valentino ML, Liguori R, Yu-Wai-Man P, Burté F, Duff J, Kleinle S, Diebold I, Rugolo M, et al. Clonal expansion of mtDNA deletions: different disease models assessed by digital droplet PCR in single muscle cells. Sci Rep. 2018;8:11682. doi: 10.1038/s41598-018-30143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT, Larsson NG. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci U S A. 2005;102:17993–17998. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2010;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, Jalanko A, Spelbrink JN, Paetau A, Suomalainen A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci U S A. 2005;102:17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoulis C, Engelsen BA, Telstad W, Aasly J, Zeviani M, Winterthun S, Ferrari G, Aarseth JH, Bindoff LA. The spectrum of clinical disease caused by the A467T and W748S POLG mutations: a study of 26 cases. Brain. 2006;129:1685–1692. doi: 10.1093/brain/awl097. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Dermaut B, Löfgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- Vanderstraeten S, Van den Brûle S, Hu J, Foury F. The role of 3’-5’ exonucleolytic proofreading and mismatch repair in yeast mitochondrial DNA error avoidance. J Biol Chem. 1998;273:23690–23697. doi: 10.1074/jbc.273.37.23690. [DOI] [PubMed] [Google Scholar]
- Viguera E, Canceill D, Ehrlich SD. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20:2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008;40:1484–1488. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- Wanrooij S, Goffart S, Pohjoismäki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanrooij S, Luoma P, van Goethem G, van Broeckhoven C, Suomalainen A, Spelbrink JN. Twinkle and POLG defects enhance age-dependent accumulation of mutations in the control region of mtDNA. Nucleic Acids Res. 2004;32:3053–3064. doi: 10.1093/nar/gkh634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IJ, Carling PJ, Alston CL, Floros VI, Pyle A, Hudson G, Sallevelt SC, Lamperti C, Carelli V, Bindoff LA, et al. Mitochondrial DNA sequence characteristics modulate the size of the genetic bottleneck. Hum Mol Genet. 2016;25:1031–1041. doi: 10.1093/hmg/ddv626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LJ. Diagnostic challenges of mitochondrial DNA disorders. Mitochondrion. 2007;7:45–52. doi: 10.1016/j.mito.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Xu W, Zhao W, Morehouse N, Tree MO, Zhao L. Divalent cations alter the rate-limiting step of PrimPol-catalyzed DNA elongation. J Mol Biol. 2019;431:673–686. doi: 10.1016/j.jmb.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylikallio E, Tyynismaa H, Tsutsui H, Ide T, Suomalainen A. High mitochondrial DNA copy number has detrimental effects in mice. Hum Mol Genet. 2010;19:2695–2705. doi: 10.1093/hmg/ddq163. [DOI] [PubMed] [Google Scholar]
- Young MJ, Longley MJ, Li FY, Kasiviswanathan R, Wong LJ, Copeland WC. Biochemical analysis of human POLG2 variants associated with mitochondrial disease. Hum Mol Genet. 2011;20:3052–3066. doi: 10.1093/hmg/ddr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeviani M, Di Donato S. Mitochondrial disorders. Brain. 2004;127:2153–2172. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
- Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]