Abstract
MicroRNAs (miRNAs) play an essential role in gene expression and affect the development of tumours, including breast cancer (BC). Polymorphisms in miRNA genes can affect the interaction of miRNAs with their target messenger RNA by interfering, creating or disrupting target sites. The single nucleotide polymorphism (SNP) rs2910164, located in the seed region of miR146a, was shown to be associated with BC among different populations. In the present study, we investigated whether rs2910164 is associated with BC in 326 patients and 411 controls from a Brazilian population of predominantly European ancestry. The presence of the allele rs2910164*C was associated with an increased risk of BC (OR=1.4, 95% CI=1.03-1.85, p = 0.03). We also analysed publicly available RNA-seq data to evaluate if miR146a is differentially expressed in different subtypes of BC. Genotyping was performed by polymerase chain reaction with sequence-specific primers (PCR-SSP). By leveraging public data from TCGA database, we analysed 461 patients and found that miR146a is significantly more expressed in BC than in non-tumor tissue (1.47 fold, p = 0.02) and is expressed to a greater degree in aggressive BC subtypes.
Keywords: miRNA polymorphism, miR146a, rs2910164, Breast cancer, Case-control study
Introduction
Breast cancer (BC) is the second most frequent type of cancer in Brazilian women (INCA, 2018) and is considered the leading cause of death by malignant disease among women worldwide (Bray et al., 2018). Identifying genetic factors associated with BC can aid in predicting disease risk at the individual and population levels, and provide information about the underlying disease mechanism. Some of the genetic variants implicated in BC are SNPs (single nucleotide polymorphisms) located in microRNA (miRNA) genes (Alshatwi et al., 2012).
miRNAs are a highly conserved class of small (~22 nucleotide) endogenous non-coding RNAs (Bartel, 2004). They play an essential role in cancer development by modulating the expression of oncogenes and tumour suppressor genes (Croce, 2009). Small alterations in the miRNA sequence can lead to substantial phenotypic consequences. Their function can be modified in three main ways: 1) altered transcription levels of the primary transcript; 2) altered processing and transport of the pri-miRNA and pre-miRNA; 3) altered interaction of the miRNA with their target mRNAs (Ryan et al., 2010).
Genome-wide association studies (GWAS) previously identified 102 loci associated with BC in Europeans (Michailidou et al., 2017). GWAS showed that 90% of the SNPs associated with complex diseases were located in non-protein coding regions, including the ones transcribed to miRNAs (Hrdlickova et al., 2014; Almeida et al., 2018). Notwithstanding, SNPs have been found in only 10% of the known pre-miRNAs. Within the seed region, Variation occurs in only less than 1% of the known seed regions, which consists of the nucleotides 2-8 from the mature miRNA sequence. The seed region specifies to which mRNAs the particular miRNA will be able to bind and the lack of variation suggests a purifying selection (Saunders et al., 2007).
The mir146a gene is located at the chromosomal region 5q33.3 and encodes the miR146a-3p and the miR146a-5p. The SNP rs2910164 is a G > C alteration at position +60 from the first nucleotide of the pre-miR146a (Jazdzewski et al., 2008), on the seed region of miR146a-3p. The result is a G:U to C:U change leading to a weaker pairing within the pre-miRNA hairpin structure (Hu et al., 2008), which alters the processing efficiency of pre-miR146a. It has been shown that the amount of mature miR146a is two times lower with the C allele. This difference is due to a less efficient pri-miRNA processing and a reduced transport of pre-miR146a from the nucleus to the cytoplasm (Jazdzewski et al., 2008).
There are few case-controls studies focusing the polymorphism rs2910164 in association with BC. Five studies were described in the European population (Catucci et al., 2010; Pastrello et al., 2010; Garcia et al., 2011; Cardeñosa et al., 2012; McVeigh et al., 2017) and one in an Australian Caucasian population (Uphadhyaya et al., 2015). The majority of these studies have focused on familial BC (Catucci et al., 2010; Pastrello et al., 2010; Garcia et al., 2011; Cardeñosa et al., 2012). The results are conflicting showing positive association in only two studies, with C allele (GC+CC) in Pastrello et al. (2010) and with G allele in sporadic BC in Uphadhyaya et al. (2015).
Comparing patients with triple-negative breast cancer (TNBC) and no TNBC, we recently demonstrated that miR146a-5p presents a higher level of expression in TNBC (Sugita et al., 2016). This result motivated the investigation of whether rs2910164 is associated with BC in a Southern Brazilian population and to evaluate the expression of miR146a-5p among subtypes of BC.
Subjects and Methods
Study cohort
The study population consisted of 326 Brazilian women diagnosed with sporadic BC, from the Nossa Senhora das Graças Hospital, Curitiba, Brazil. The control group comprised 411 healthy Brazilian women from the bone marrow donor biobank of the Laboratory of Immunogenetics and Histocompatibility, Genetics Department, Federal University of Paraná, Curitiba. Both patients and controls were from the same region in South Brazil and belong to the same ethnic background, which is of predominantly European ancestry (Probst et al., 2000; Sugita et al., 2016). This study was approved by the Brazilian Commission of Ethics in Research (CONEP) under the number CAAE 67400917.3.0000.55 following Brazilian Federal laws. All participants signed informed written consent following the principles of the Declaration of Helsinki. The mean ages of the case and the control groups were 56.23 ± 15 and 47.66 ± 4.69, respectively. Histopathological parameters, immunohistochemical features, and status of regional lymph node invasion from cancer samples are summarized in Table 1. The immunohistochemical classification was based on Goldhirsch et al. (2013).
Table 1. Histopathological and immunohistochemical parameters of breast cancer patients.
| Breast cancer patients n=326 | ||||||
|---|---|---|---|---|---|---|
| Histology of carcinoma | n | % | Tumour Grade | n | % | |
| Invasive ductal | 233 | 75% | Grade I | 37 | 16% | |
| Invasive lobular | 28 | 9% | Grade II | 125 | 53% | |
| Ductal in situ | 12 | 4% | Grade III | 75 | 32% | |
| Invasive mucinous | 9 | 3% | Without Information | 89 | - | |
| Others | 28 | 9% | ||||
| Without information | 16 | - | ||||
| Immunohistochemical classification | n | % | Lymph node invasion | n | % | |
| Luminal A | 84 | 40% | Present | 116 | 47% | |
| Luminal B | 78 | 38% | Absent | 133 | 53% | |
| HER2+ | 15 | 7% | Without information | 77 | - | |
| Triple-Negative | 31 | 15% | ||||
| Without information | 118 | - | ||||
Genotyping
The SNP rs2910164 was genotyped using polymerase chain reaction with specific sequence primers (PCR-SSP). The primer sequences used were: forward 5’-GGTTGTGTCAGTGTCAGACCTC-3’, forward 5’-GGTTGTGTCAGTGTCAGACCTG-3’, reverse 5’-GAGCCTGAGACTCTGCCTTCT-3’ and the product size was 196 bp. PCR amplifications were carried out in a 20 μL reaction containing: 40 ng DNA, 0.2 μM each primer, 0.2mM dNTPs, KCl 50 mM, Tris-HCl 10 mM and 1.25 U Taq polymerase (Invitrogen, Carlsbad, CA, USA). PCR conditions were: 95 °C for 1 min, followed by 35 cycles of 95 °C for 30 s, 59 °C for 30 s and 72 °C for 30 s, followed by 5 min at 72 °C. After the reaction, the products were analysed by gel electrophoresis in 2% agarose gel stained with GelRed® (Biotium, Fremont, CA, USA). As an internal amplification control, we amplified the galactosylceramidase gene (GALC) using the following primers: forward 5’- TTACCCAGAGCCCTATCGTTCT -3’ and reverse 5’- GTCTGCCCATCACCACCTATT -3’. The GALC amplicon (352 bp) was expected in all reactions. We validated the specificity and accuracy of our PCR-SSP method by sequencing five random individuals with Sanger methods and verifying their genotypes.
Expression analysis from publicly available RNA-Seq data
The Cancer Genome Atlas Program (TCGA) is a public database that contains data from thousands of matched cancer and non-cancer samples (TCGA Network, 2012). We analysed RNA-Seq data from 837 BC patients and 105 adjacent tissues to search for possible differential expression levels of miR146a in BC compared to non-tumour tissue and among intrinsic subtypes. Pre-processing and normalization of RNA-seq data were performed by TCGA biolinks package from R/Bioconductor (Colaprico et al., 2016). For expression analysis from molecular subtypes, we analysed 461 patients, being 211 Luminal A, 112 Luminal B, 85 Basal-like, and 53 HER2-Enriched.
Statistical analysis
Association analysis was performed using chi-square test of independence and using odds ratios (OR) and 95% confidence intervals (Mantel and Haenszel, 1959). We used R package Hardy-Weinberg (Graffelman, 2015) to test if genotype distributions were under Hardy Weinberg equilibrium. A chi-square test of independence was used to compare the distribution of rs2910164 genotypes among the several parameters (histopathological, subgroups defined by immunohistochemistry, tumour grade, and lymph node invasion). Bartlett’s test was applied to compare the age of cancer diagnosis in different genotypes (Bartlett, 1937). Analysis of the expression data from TCGA was performed using Kruskal-Wallis test followed by Dunn test (Kruskal and Wallis, 1952; Dunn, 1964). Spearman correlation was used to check the correlation of miR146a and the genes BRCA1, TRAF6, IRAK1, TRAF3IP1.
Alpha (α) was set at 0.05 for all tests and all analyses were performed with R Software (R core team, 2017) with the packages Nortest and readxl (Gross and Ligges, 2015; Wickham and Bryan, 2017).
Results
The presence of the allele rs2910164C is associated with breast cancer risk
The genotype GC was more frequent in patients compared to controls (Table 2). Considering that the difference in the distribution of the three genotypes was not significant (p = 0.10), we determined the magnitude of the effect on BC susceptibility testing three models by odds ratio tests: G vs. C for additive, CC + GC vs. GG for dominant and CC vs. GC + GG for recessive inheritance. The dominant model (presence of allele C) was associated with an increased risk of BC (OR = 1.4, p = 0.03). The C allele was observed 4% more frequently in the case than in the control population. The distribution of genotypes followed Hardy-Weinberg equilibrium in the control population (p = 0.70).
Table 2. Genotypic and allelic frequencies of the SNP rs2910164 in breast cancer patients and controls.
| Patients (n = 326) | Controls (n = 411) | ||||
|---|---|---|---|---|---|
| Allele | f | f | OR | p | 95%CI |
| G | 0.730 | 0.769 | ns | ||
| C | 0.270 | 0.231 | ns | ||
| Genotype | % (n) | % (n) | |||
| G+ | 95.1 (310) | 94.9 (390) | ns | ||
| C+ | 49.1 (160) | 41.1 (169) | 1.4 | 0.03 | 1.03 - 1.85 |
| GG | 50.9 (166) | 58.9 (242) | ns | ||
| GC | 44.2 (144) | 36 (148) | 1.4 | 0.03 | 1.04 - 1.89 |
| CC | 4.9 (16) | 5.1 (21) | ns |
Legend: G+ = carriers of allele G (GG+GC), C+ = carriers of allele C (CC+GC), f =allelic frequency, OR = odds ratio; p = p-value; 95% CI = 95% confidence interval; ns = not significant.
We observed no differences in distribution of lymph node invasion (p = 0.25), tumour grade (p = 0.30) and immunohistochemical subgroups (p = 0.90) comparing risk group GC+CC vs. GG group from BC patients. Applying Bartlett test for homogeneity, no difference was observed in the variance analysis for assessing the risk of developing BC in younger ages for the risk group GC+CC vs. the GG group.
miR146a is differentially expressed in breast cancer
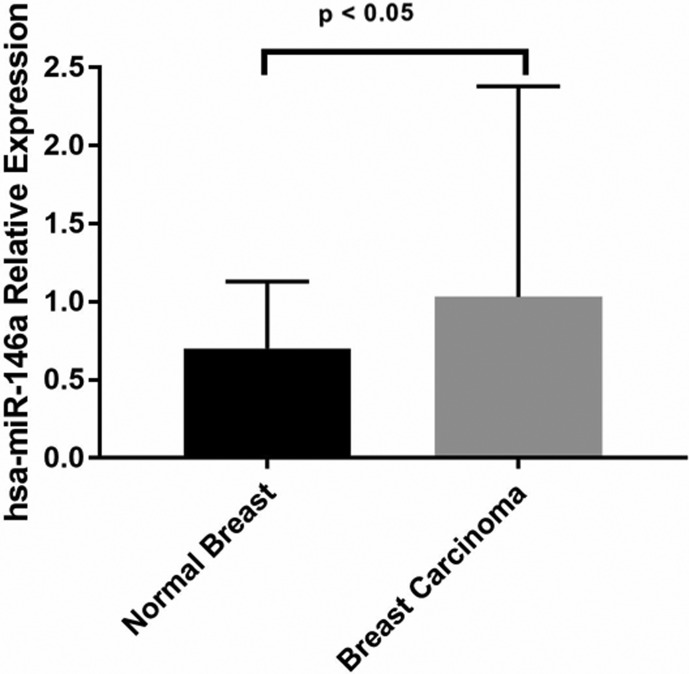
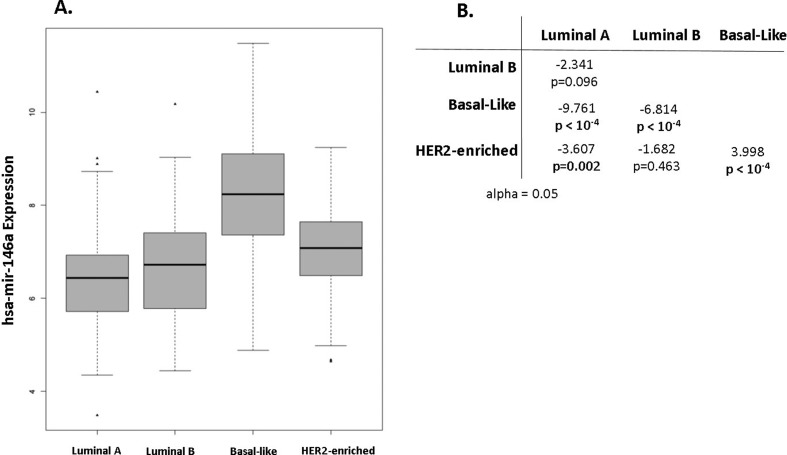
Accessing publicly available RNA-Seq data from TCGA database, we found miR146a significantly more expressed in tumour tissue compared to non-tumour adjacent tissue (Figure 1). Kruskal-Wallis analysis and Dunn’s test indicated significant differences in the expression of miR146a between each molecular subtype (Figure 2).
Figure 1. Higher expression levels of hsa-miR-146a in breast cancer analysis from TCGA RNA-seq data.
Figure 2. A. Comparison of hsa-mir-146a expression in breast cancer intrinsic subtypes; B. Dunn’s test results summarized.
Spearman correlation analysis was performed to analyse the expression of miR146a-5p and predicted target genes, BRCA1, IRAK1, TRAF3IP1, and TRAF6 using TCGA RNA-seq data. No correlation with BRCA1 was found (rho= -0.02, p=0.58) However, we observed a negative correlation between miR146a-5p expression levels with TRAF6 (rho= -0.13, p= 0.0002) and TRAF3IP1 (rho=-0.27, p= 8.5e-14), and a positive correlation of miR146a-5p with IRAK1 expression (rho= 0.27, p= 3.5x10-14). For BRCA1 we tested the correlation with miR146a-5p within each molecular subtype but did not find correlation in any of them. Also, rs2910164 was not found in linkage disequilibrium with any other variant described at ENSEMBL database and 1000genomes database in any population (The 1000 Genomes Project Consortium, 2015; Zerbino et al., 2018).
Discussion
The allelic frequencies of rs2910164 vary between populations; as a result, some alleles that may confer risk in a particular population may be the same that are protective in others (Upadhyaya et al., 2016). The allelic frequencies of rs2910164 observed in the control group of the present study were similar to the Europeans from Hapmap (CEU G-76% and C-24%) (International HapMap Consortium, 2003), as well as the Europeans from NHLBI Exome Sequencing Project (NHLBI E_A G-77% and C-23%) (The 1000 Genomes Project Consortium, 2015). This result is following the previously reported predominantly European ancestry of the population from Southern Brazil (Probst et al., 2000; Sugita et al., 2016).
The association of rs2910164 and BC was studied in several studies from different populations (Xu et al., 2013; Dai et al., 2015; Uphadhyaya et al., 2015; Barjui et al., 2017; Zhang et al., 2017). Dai et al. (2015) showed that the effect of rs2910164 on BC susceptibility is different across populations. The authors suggest that the allele C is associated with BC risk in Europeans (2777 cases and 2898 controls) but not in Asians (1537 cases and 1587 controls). This discrepancy could be explained by the interaction of rs2910164 with different genetic backgrounds, by different exposure to carcinogens, or due to linkage disequilibrium with a different causal variant. However, we have not found significant LD between rs2910164 with any other polymorphism described in either ENSEMBL or the 1000genomes database. Our results, therefore, suggest that LD is not the primary factor influencing the different associations observed for rs2910164 with BC. Also evidencing the differences of the effect of rs2910164 on the risk of BC in diverse populations, the allele rs2910164G was associated with increased risk of sporadic BC in Australians (OR =1.77, p < 10-4; Uphadhyaya et al., 2015) and Iranians (OR = 1.91, p = 0.03; Barjui et al., 2017). Other studies, on the other hand, reported no association (Zhang et al., 2016; Mu et al., 2017).
We observed a higher frequency of patients carrying the allele rs2910164C in comparison to controls. Although the rates of the genotype CC were similar in both groups, this genotype was observed in relatively low frequency (5%). It is plausible to expect that studying larger cohorts could reveal differences between rates of the genotype CC in cases and controls. In fact, increased risk for CC genotype was observed by different studies that analysed Europeans (Lian et al., 2012; Dai et al., 2015).
Younger age at the time of diagnosis is an important prognostic marker for BC, also associated with hereditary cases (Armaou et al., 2009). In a study involving 40 American women with hereditary BC, the presence of the allele rs2910164C was increased in women diagnosed at younger ages, with a median age of 45 versus 56 (p=0.029; Shen et al., 2008). A study of 348 Italian women with hereditary BC showed similar results with a median age at diagnosis of 35 versus 52; (p=0.028; Pastrello et al., 2010). On the other hand, a third study reported a lack of association with younger age (Catucci et al., 2010). Studying sporadic BC cases, we did not find any association of rs2910164 with the age of diagnosis. These findings were similar to those observed in other studies with sporadic BC, and this possible difference in the association between sporadic and hereditary BC with rs2910164 is still not understood (Cardeñosa et al., 2012; Qi et al., 2015; Barjui et al., 2017; Mcveigh et al., 2017).
We also did not find any association between genotypes of rs2910164 and regional lymph node invasion, immunohistochemical subgroups or tumour grade, which suggests a lack of association of rs2910164 with tumour progression. This result is similar to studies in the Irish population (McVeigh et al., 2017) and the Chinese (Qi et al., 2015).
We found that miR146a was significantly more expressed in BC than in healthy tissue. The highest expression was observed in basal-like tumours, which have partial overlap with TNBC in immunohistochemical classification. Among the BC subgroups defined by immunohistochemistry, triple-negative is recognized to inactivate BRCA1 tumour suppressor gene (Turner et al., 2007). A reduced expression level of BRCA1 is observed in a significant proportion of sporadic BC cases, despite the absence of somatic mutations in BRCA1 (Mueller and Roskelley, 2003). Interestingly, miR146a-5p was predicted to target BRCA1 and thereby reduce its expression (Shen et al., 2008; Garcia et al., 2011). Also, the expression levels of miR146a-5p were observed three times higher in triple-negative tumours compared to other subgroups of mammary tumours (M’Hamed et al., 2015). The hypothesis of regulation of BRCA1 expression by miR146a-5p was not corroborated in our analysis using RNA-Seq data. This analysis did not show a correlation between expression levels of miR146a and BRCA1 in BC in general, neither in basal-like patients.
Studies suggest that TRAF6, IRAK1 (Bhaumik et al., 2008) and EGFR, a protein associated with tumour progression and metastasis (Kumaraswamy et al., 2016), are targets for mir146a-5p. Our analysis showed a negative correlation between the expression of mir146a-5p with TRAF6 as well as with TRAF3IP1. However, we found a positive correlation of miR146a-5p with IRAK1. It has been shown that reduced TRAF6 and IRAK1 levels lessen the activity of NF-kB, a potential inducer of proliferation, survival, angiogenesis, and metastasis (Bhaumik et al., 2008). In summary, the studies on miR146a and their targets suggest that rs2910164 can exert their effects on cancer mainly by changing the binding to the target rather than by modulating the expression levels of miR146a-5p. However, further studies are needed to validate these targets and to provide direct evidence of the regulation by miR146a.
In conclusion, our study provides insights into the role of miR146a in a Brazilian population of predominantly European ancestry. We show the presence of rs2910164C increasing the susceptibility to sporadic BC among women despite its apparent lack of influence in disease progression. We observed significantly higher expression levels of miR146a-5p associated with the severity of BC, the highest expression observed being in patients with basal-like tumours. Our results point to the importance of miRNAs in the susceptibility to cancer and suggest that functional studies will further elucidate the impact of miR146a in breast cancer.
Acknowledgments
We thank all participants for their collaboration with this study. Special thanks to Kirsten Anderson for kindly revising this manuscript and also to the Immunogenetics and Histocompatibility Laboratory (LIGH) of the Federal University of Paraná for sharing their biobank and their physical structure. This research was supported by Coordenação de Aperfeicoamento de Pessoal de Nivel Superior (CAPES), Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq) and Fundação Araucaria de Apoio ao Desenvolvimento Cientifico e Tecnologico do Parana (PRONEX 2016).
Footnotes
Associate Editor: Houtan Noushmehr
Conflicts of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicial to the impartiality of the reported research.
Authors Contribution
HMB, RAC, DGA, DFG, JCO and EMSFR conceived the study, HMB, CM, DGA, DFG and JCO conducted and supervised the experiments; HMB, CM, DGA, RCA, IJC, EMSFR analysed the data, HMB, RAC, EMSFR wrote the draft manuscript, RSL, FK and CAU collected the samples and revised all clinical data; all authors read and approved the final version.
References
- Almeida RC, Chagas VS, Castro MAA, Petzl-Erler ML. Integrative Analysis Identifies Genetic Variants Associated With Autoimmune Diseases Affecting Putative MicroRNA Binding Sites. Front. Genet. 2018;9:1–13. doi: 10.3389/fgene.2018.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armaou S, Pertesi M, Fostira F, Thodi G, Athanasopoulos PS, Kamakari S, Athanasiou A, Gogas H, Yannoukakos D, Fountzilas G, et al. Contribution of BRCA1 germ-line mutations to breast cancer in Greece: a hospital-based study of 987 unselected breast cancer cases. Br J Cancer. 2009;101:32–37. doi: 10.1038/sj.bjc.6605115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barjui SP, Reiisi S, Ebrahimi SO, Shekari B. Study of correlation between genetic variants in three microRNA genes (hsa-miR-146a, hsa-miR-502 binding site, hsa-miR-27a) and breast cancer risk. Curr Res Transl Med. 2017;5:1–7. doi: 10.1016/j.retram.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartlett MS. Properties of sufficiency and statistical tests. Proc R Soc Lond Ser A. 1937;160:268–282. [Google Scholar]
- Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-κB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27:5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Cardeñosa EE, Jiménez IJ, Suela SP, González IC, Huerta ÁS, Beltran AS, El Busto MC, González EB, Lluch OF, Edo JB, et al. Low penetrance alleles as risk modifiers in familial and sporadic breast cancer. Fam Cancer. 2012;11:629–636. doi: 10.1007/s10689-012-9563-1. [DOI] [PubMed] [Google Scholar]
- Catucci I, Yang R, Verderio P, Pizzamiglio S, Heesen L, Hemminki K, Sutter C, Wappenschmidt B, Dick M, Arnold N, et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum Mutat. 2010;31:E1052–E1057. doi: 10.1002/humu.21141. [DOI] [PubMed] [Google Scholar]
- Colaprico A, Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z-J, Shao Y-P, Wang X-J, Xu D, Kang H-F, Ren H-T, Min W-L, Lin S, Wang M, Song Z-J. Five common functional polymorphisms in microRNAs (rs2910164, rs2292832, rs11614913, rs3746444, rs895819) and the susceptibility to breast cancer: evidence from 8361 cancer cases and 8504 controls. Curr Pharm Des. 2015;21:455–63. doi: 10.2174/1381612821666141208143533. [DOI] [PubMed] [Google Scholar]
- Dunn OJ. Multiple comparisons using rank sums. Technometrics. 1964;6:241–252. [Google Scholar]
- Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, Lidereau R, Mikaélian I, Mazoyer S. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med. 2011;3:279–290. doi: 10.1002/emmm.201100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, Senn HJ, Albain KS, André F, Bergh J, et al. Personalizing the treatment of women with early breast cancer: Highlights of the St. Gallen International Expert Consensus on the primary therapy of early breast Cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graffelman J. Exploring Diallelic Genetic Markers: The HardyWeinberg Package. J Stat Softw. 2015;64:1–23. [Google Scholar]
- Hrdlickova B, Almeida RC, Borek Z, Withoff S. Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta. 2014;1842:1910–1922. doi: 10.1016/j.bbadis.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, et al. Genetic variants of miRNA sequences and non – small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Instituto Nacional de Câncer (INCA) Estimativas 2018: Incidências de Câncer no Brasil. 2018. https://www.inca.gov.br/sites/ufu.sti.inca.local/files//media/document//estimativa-incidencia-de-cancer-no-brasil-2018.pdf.
- International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J Am Stat Assoc. 1952;47:583–621. [Google Scholar]
- Kumaraswamy E, Wendt KL, Augustine LA, Stecklein SR, Sibala EC, Li D, Gunewardena S, Jensen RA. BRCA1 regulation of epidermal growth factor receptor (EGFR) expression in human breast cancer cells involves microRNA-146a and is critical for its tumor suppressor function. Oncogene. 2016;34:4333–4346. doi: 10.1038/onc.2014.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H, Wang L, Zhang J. Increased Risk of Breast Cancer Associated with CC Genotype of Has-miR-146a Rs2910164 Polymorphism in Europeans. PLoS One. 2012;7:e31615. doi: 10.1371/journal.pone.0031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M’Hamed IF, Privat M, Ponelle F, Penault-Llorca F, Kenani A, Bignon YJ. Identification of miR-10b, miR-26a, miR-146a and miR-153 as potential triple-negative breast cancer biomarkers. Cell Oncol. 2015;38:433–442. doi: 10.1007/s13402-015-0239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- Mcveigh TP, Mulligan RJ, McVeigh UM, Owens PW, Miller N, Bell M, Sebag F, Guerin C, Quill DS, Weidhaas JB, et al. Investigating the association of rs2910164 with cancer predisposition in an Irish cohort. Endocr Connect. 2017;6:614–624. doi: 10.1530/EC-17-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S, Lemaçon A, Soucy P, Glubb D, Rostamianfar A, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu K, Wu Z, Yu J, Guo W, Wu N, Wei L, Zhang H. Meta-analysis of the association between three microRNA polymorphisms and breast cancer susceptibility. Oncotarget. 2017;8:68809–68824. doi: 10.18632/oncotarget.18516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CR, Roskelley CD. Regulation of BRCA1 expression and its relationship to sporadic breast cancer. Breast Cancer Res. 2003;5:45–52. doi: 10.1186/bcr557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrello C, Polesel J, Della Puppa L, Viel A, Maestro R. Association between hsa-mir-146a genotype and tumor age-of-onset in BRCA1 / BRCA2 -negative familial breast and ovarian cancer patients. Carcinogenesis. 2010;31:2124–2126. doi: 10.1093/carcin/bgq184. [DOI] [PubMed] [Google Scholar]
- Probst CM, Bompeixe EP, Pereira NF, O Dalalio MM, Visentainer JE, Tsuneto LT, Petzl-Erler ML. HLA polymorphism and evaluation of European, African, and Amerindian contribution to the white and mulatto populations from Paraná, Brazil. Hum Biol. 2000;72:597–617. [PubMed] [Google Scholar]
- Qi P, Wang L, Zhou B, Yao WJ, Xu S, Zhou Y, Xie ZB. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet Mol Res. 2015;14:6289–6296. doi: 10.4238/2015.June.11.2. [DOI] [PubMed] [Google Scholar]
- Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl Acad Sci U S A. 2007;104:3300–5. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Ambrosone CB, Dicioccio RA, Odunsi K, Lele SB, Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- Sugita B, Gill M, Mahajan A, Duttargi A, Kirolikar S, Almeida R, Regis K, Oluwasanmi OL, Marchi F, Marian C, et al. Differentially expressed miRNAs in triple negative breast cancer between African-American and non-Hispanic white women. Oncotarget. 2016;7:79274–79291. doi: 10.18632/oncotarget.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, Savage K, Gillett CE, Schmitt FC, Ashworth A, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- Upadhyaya A, Smith RA, Chacon-Cortes D, Revêchon G, Bellis C, Lea RA, Haupt LM, Chambers SK, Youl PH, Griffiths LR. Association of the microRNA-Single Nucleotide Polymorphism rs2910164 in miR146a with sporadic breast cancer susceptibility: A case control study. Gene. 2016;576:256–260. doi: 10.1016/j.gene.2015.10.019. [DOI] [PubMed] [Google Scholar]
- Xu Y, Gu L, Pan Y, Li R, Gao T, Song G, Nie Z, Chen L, Wang S, He B. Different Effects of Three Polymorphisms in MicroRNAs on Cancer Risk in Asian Population: Evidence from Published Literatures. PLoS One. 2013;8:1–16. doi: 10.1371/journal.pone.0065123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang Y, Yan W, Wang W, Zhao X, Ma X, Gao X, Zhang S. Association between three functional microRNA polymorphisms (miR-499 rs3746444, miR-196a rs11614913 and miR-146a rs2910164) and breast cancer risk: a meta-analysis. Oncotarget. 2016;8:393–407. doi: 10.18632/oncotarget.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- Gross J, Ligges U. Nortest: Tests for Normality. 2015. https://CRAN.R-project.org/package=nortest.
- R Development Core Team R: A language and environment for statistical computing, reference index version 2.12.1. 2010. https://www.R-project.org/
- Wickham H, Bryan J. Readxl: Read Excel Files. 2017. https://CRAN.R-project.org/package=readxl.