Abstract
Prostate cancer (PCa) is one of the leading causes of death among men. Genes such as PCA3, PSA, and Fra-1 are suggested to serve as potential tools for the detection of PCa, as they are deregulated during this pathology. A similar event occurs with small non-coding RNAs, called miRNAs, specifically miR-195-5p, miR-133a-3p, and miR-148b-3p, which were analyzed in a Chinese population and suggested to be possible candidates for PCa diagnosis. We evaluated the expression levels of three miRNAs and three genes in tissue samples of PCa and benign prostate disease, such as benign prostatic hyperplasia, or prostatitis, in order to determine their potential as candidates for PCa detection. Our results showed a statistically significant overexpression of 279-fold increase in PSA levels and a 1,012-fold increase in PCA3 levels in PCa patients compared to benign prostate disease patients (p = 0.001 and p = 0.002, respectively). We observed a positive correlation between the expression of miR-148b-3p and the expression of PSA and PCA3 genes, two established biomarkers in PCa. The expression of miR-148b-3p was not related to clinical characteristics, such as age and weight, as observed for the other miRNAs analyzed, suggesting its potential as a biomarker for detection of this pathology.
Keywords: Gene expression, biomarker, miRNAs, correlation, prostate cancer
Introduction
Prostate cancer (PCa) is the fourth most common cancer worldwide and the second most common type of cancer diagnosed in men. It is also recognized as the most frequent malignant tumor in men older than 50 years (GLOBOCAN, 2012). Several genes involved in the progression of PCa, such as the Prostate Cancer Associated 3 (PCA3), the Fos-related antigen 1 (Fra-1) and the Prostate Specific Antigen (PSA), present different expression levels in individuals with and without cancer. In this context, PCA3 is one of the most reported genes (Bussemakers et al., 1999; Floriano-Sánchez et al., 2009). This gene is exclusively expressed in prostatic tissue and is found highly overexpressed in patients with PCa. For this reason, the PCA3 gene is suggested as an interesting biomarker for this pathology (Xue et al., 2014).
Recent studies have shown that the Fra-1 gene is also deregulated in patients with PCa. Fra-1 is a proto-oncogene that encodes a transcription factor with a central role in the regulation of several biological processes, including cell proliferation, differentiation, transformation, and inflammation. Thus, Fra-1 is also in the spotlight for its potential as a biomarker in PCa (Adiseshaiah et al., 2005; Zhu et al., 2014).
Current biomarkers for the detection of PCa have low specificity and sensitivity, as is the case of PSA, a protein that is not specific for this malignancy, and that may be elevated by different pathologies (Daniyal et al., 2014). It is important to mention that most studies on PSA are directed to quantifying serum PSA levels, but only a few have focused on measuring gene expression. These studies suggested that PSA gene expression may be used in combination with other biomarkers to offer a better diagnosis (ACS, 2012; Coelho et al., 2015).
Small non-coding RNAs (known as microRNAs or miRNAs) in charge of regulating gene expression have been reported deregulated in several pathologies (McDonald et al., 2017). In PCa studies, an amounting evidence has demonstrated the importance of miRNAs for this pathology. This is especially true for miR-195-5p, miR-133a-3p, and miR-148b-3p as their deregulation causes the inhibition of proliferation, migration, and invasion of PCa cells (Xu et al., 2009; Li et al., 2011; Tao et al., 2012; Kojima et al., 2012; Dai and Grant, 2015; Wu et al., 2015). MiR-195-5p plays a regulatory role in migration, invasion, proliferation, epithelial-mesenchymal transition, angiogenesis, and metastasis of tumor cells by targeting the 3’UTR sequence of RPS6KB1, FGF2, Fra-1, and BCOX1 genes (Cai et al., 2015; Liu et al., 2015; Wu et al., 2015; Guo et al., 2015). On the other hand, miR-133a-3p deregulation is suggested to be a key step in oncogenesis and progression of PCa due to regulation of the PNP gene (Kojima et al., 2012). Lastly, miR-148b-3p is associated with various carcinogenic genes, and its expression level is found deregulated in several types of cancer. In general, downregulation of this miRNA relates to high-grade tumors. More importantly, miR-148b-3p is suggested as an indicator to distinguish malignant from benign prostate disease (Watahiki et al., 2011; Walter et al., 2013).
Clinical evidence demonstrates that early diagnosis of PCa is determinant to improve the treatment outcome. Therefore, the aim of this study was to analyze the expression and correlation of different miRNAs (miR-195-5p, 133a-3p and 148b-3p), genes (Fra-1, PSA, and PCA3), and clinicopathological characteristics (age, body weight, and serum PSA) to be used independently or in combination as biomarkers for early diagnosis of PCa.
Subjects and Methods
Patient recruitment
Our study group consisted of 19 patients: 13 diagnosed with PCa and 6 with benign prostate disease (BPD). The BPD group included men with benign prostatic hyperplasia or prostatitis. All PCa and BPD patients met the inclusion criteria: Mexican men older than 18 years with a diagnosis for PCa or BPD confirmed by histopathology, neither receiving chemotherapy nor radiotherapy, and not presenting any other type of cancer. All patients were recruited either from the Mexican Social Security Institute (MSSI) or from the Alvarez and Arrazola Radiologists Clinic, both located in Sinaloa, Mexico, from August 2016 to September 2017. Clinicopathological data (age, weight, height, serum PSA, and other diseases) were collected through direct questionnaire and hospital or clinic database and the Gleason score was provided by a pathologist. All patients granted approval by signing an informed consent that was previously reviewed and approved by the Ethics and Research Committee of the MSSI and Alvarez and Arrazola Radiologists Clinic.
Tissue samples and RNA extraction
Tissue samples were obtained through transrectal biopsy and were used for RNA extraction. Total RNA, including miRNAs, was isolated using the miRNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The concentration of isolated RNA was measured with the assistance of the GENESYS 10S UV-Vis Spectrophotometer (Thermo Scientific TM).
Relative expression of miRNAs
Reverse transcription (RT) was performed from 10 ng of total RNA with the TaqMan Advanced miRNA cDNA Synthesis kit (Applied Biosystems,). Quantitative estimations of miR-195-5p, miR-133a-3p, and miR-148b-3p were performed by real-time polymerase chain reaction (RT-PCR) method using TaqMan® MicroRNA Assays (Applied Biosystems). We normalized the expression of our miRNAs of interest using miR-16-5p as the reference gene. Briefly, we followed the procedure described in Andersen et al. (2004) to identify the most appropriate gene based on expression stability. The NormFinder Software allowed us to identify miR-16-5p as the best candidate (stability value = 0.013). The results are presented as the ratio of the number of copies of a given gene to that of the reference gene. To obtain data about the relative expression of the miRNAs, we used the ΔΔCq method (2-DDCq algorithm) (Livak and Schmittgen, 2001).
Relative expression of Fra-1, PSA, and PCA3
RT was performed from 1 μg of total RNA with ImProm-IITM Reverse Transcriptase (Promega Corporation, Madison, WI, USA) and random primers. Quantitative estimation of Fra-1, PSA, and PCA3 transcripts were performed by real-time PCR method with TaqMan® Assays (Applied Biosystems) on StepOnePlusTM Real-Time PCR system (Applied Biosystems). We used beta actin (Act-β) as reference gene. The reaction conditions consisted of enzyme activation at 95 °C for 20 s, followed by 40 cycles of 95 °C for 1 s and 60 °C for 20 s. All samples were assessed in technical duplicates. If Cq (quantification cycle) values obtained from technical duplicates were in discrepancy, the sample was reassessed. To obtain data about relative gene expression we used the algorithm of Livak and Schmittgen (2001).
Statistical and correlational analysis
The Student’s t-test and Mann-Whitney U-test (when appropriate) were used to compare differences between continuous variables. Due to small sample size, the Fisher’s exact test was used to compare differences between dichotomous variables. Pearson and Spearman tests (when appropriate) were used to calculate the correlation coefficient between the following variables: expression of miRNAs, expression of Fra-1, PSA, and PCA3, serum PSA, age, and body weight. All these variables were contrasted among each other to observe relationships.
The Statistical Package for the Social Sciences (SPSS, Inc., Chicago, IL,) version 20 software was used for all statistical calculations. Results with a p-value < 0.05 were considered statistically significant.
Results
Clinicopathological characteristics
Our study population had an average age of 67.11 ± 7.04 years. In PCa patients the average age was 67.54 ± 7.95 years and in BPD patients, 66.67 ± 5.09 (p = 0.810). Table 1 shows the results of the clinicopathological characteristics analysis. Evaluation of the body mass index (BMI) yielded an average of 25.54 ± 1.56 in our total population (Table 1). The results showed no statistical significance (p = 0.359) between groups.
Table 1. Clinical characterization of prostate cancer (PCa) and benign prostate disease (BPD) groups.
| Variable | PCa (n=13) | BPD (n=6) | p |
|---|---|---|---|
| Mean age (years) | 67.54 ± 7.95 | 66.67 ± 5.09 | 0.81 |
| Mean BMI | 25.74 ± 1.69 | 24.78 ± 0.79 | 0.359 |
| PCa family history (yes) % | 23.1 | 66.7 | 0.046 * |
| Mean PSA (μg/L) | 102.78 ± 214.90 | 17.15 ± 1.62 | 0.513 |
Statistically significant value. BMI: body mass index; PSA, Prostate specific antigen.
An exhaustive medical examination of our groups revealed that 10.5% of the subjects presented prostatitis, 15.8% exhibited dyslipidemias and other neoplasms, and 36.8% suffered from diabetes and hypertension (Table 1). However, we found no statistical difference (pe = 0.233) between groups.
Concerning family history of cancer, we found that 15.4% of our patients with PCa had first-degree relatives who suffered from the same disease, and 7.7% mentioned relatives with breast cancer, for a total of 23.1% of patients with at least one family member affected by some type of cancer. Regarding our BPD group, 33.3% had no family history with malignant neoplasms, and 66.7% had first-degree relatives with PCa (Table 1).
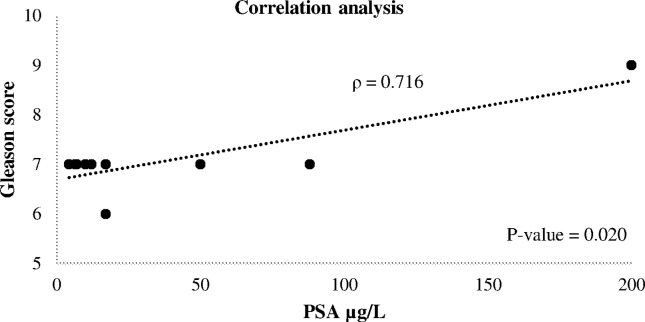
Classifying our patients using the Gleason score, we observed that in the PCa group, 9.1% were classified as 6 (3+3), 36.36% as 7 (3+4), 36.36% as 7 (4+3), and 18.18% as 9 (5+4), corresponding to low, low-intermediate, intermediate-high, and high aggressiveness, respectively. Patients with a Gleason score of 6 (3+3), 7 (3+4), 7 (4+3), and 9 (5+4) exhibited mean levels of PSA of 16, 8.83, 39.82, and 200 μg/L, respectively. Performing a Spearman correlation test, we observed that there was a relationship between the level of serum PSA and the aggressiveness of the tumor (p = 0.020, ρ = 0.716) (Figure 1).
Figure 1. Correlation between levels of serum PSA and Gleason score. The correlation coefficient of 0.716 with p-value < 0.05 indicates that serum PSA was moderately correlated with Gleason score.
Relative gene expression levels
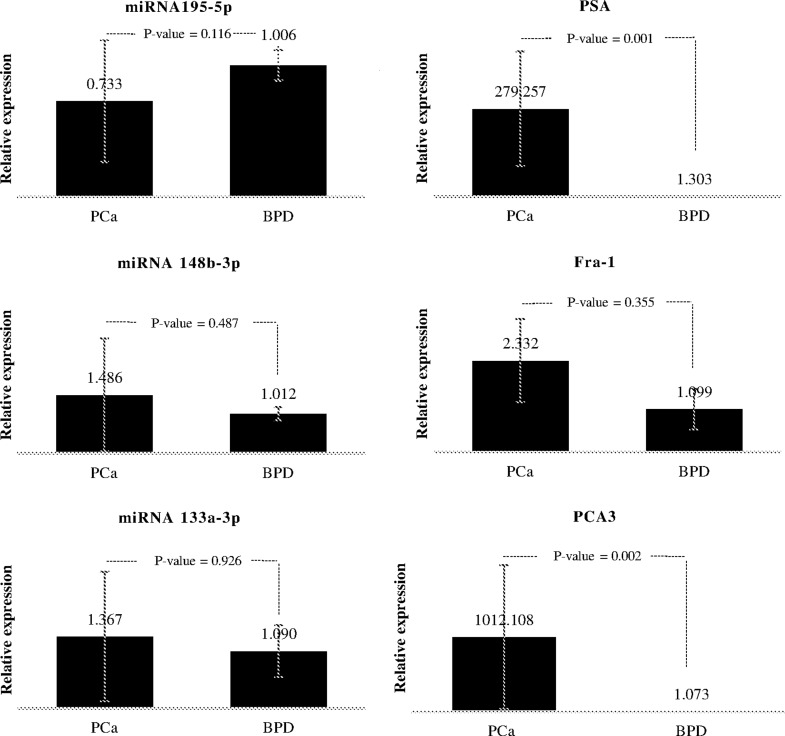
For miR-195-5p, 148-3p, 133a-3p, and the Fra-1 gene, we found no statistically significant difference in expression levels in prostatic tissue between men with PCa and BPD (p = 0.116, p = 0.487, p = 0.926, and p = 0.355, respectively) as shown in Figure 2. However, our results showed a statistically significant overexpression of 279-fold increase in the PSA levels and a 1,012-fold increase in the PCA3 levels of PCa patients compared to BPD (p = 0.001 and p = 0.002, respectively) (Figure 2).
Figure 2. Expression levels of miRNAs, PSA, PCA3, and Fra-1 genes. Real-time PCR exhibited similar expression levels of the three miRNAs (left column) and Fra-1 gene (mid-right) in PCa and BPD tissue samples. Expression levels of PCA3 (bottom-right) and PSA (top-right) genes were significantly higher in PCa than in BPD samples. Error bars indicate ± SD.
Correlation analysis
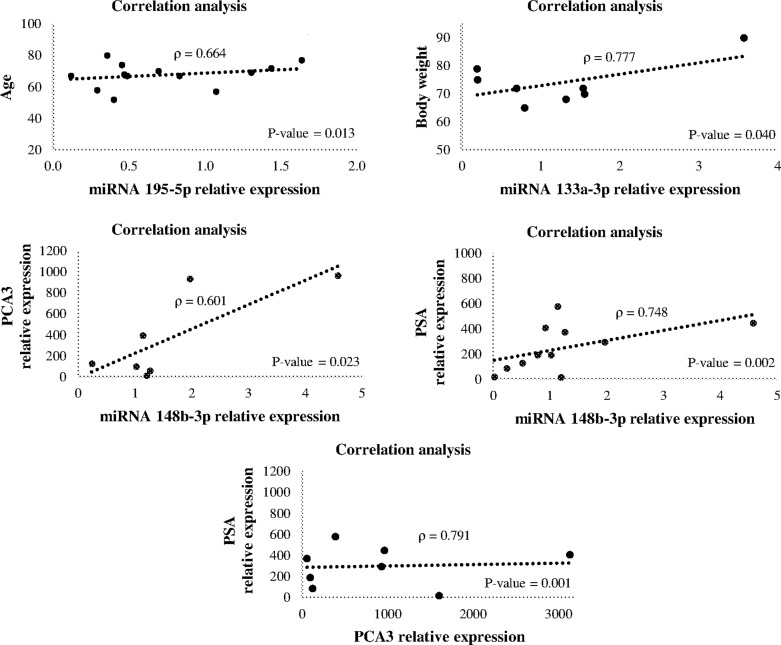
In the correlation analysis, we observed a relationship between miR-195-5p expression and the age of patients with PCa (p = 0.013) with a correlation coefficient of 0.664. In addition, the expression of miR-133a-3p correlated with an increase in body weight with a correlation coefficient of 0.777 (p = 0.040). Regarding the expression of miR-148b-3p, a positive relationship was observed when compared with the expression levels of PCA3 and PSA genes, with correlation coefficients of 0.601 and 0.748, respectively (p = 0.023 and p = 0.002). Lastly, PSA and PCA3 expression levels showed a strong correlation with a coefficient of 0.791 (p = 0.001) (Figure 3).
Figure 3. Correlation analysis between expression levels of miRNAs, clinicopathological characteristics, PSA, and PCA3 genes. The correlation coefficient of 0.664 with p-value < 0.05 indicates that miR-195-5p expression was moderately correlated with age in PCa patients. Also, there was a strong correlation between body weight and miR-133a-3p expression with a correlation coefficient of 0.777 (p < 0.05). There was also a moderate correlation between miR-148b-3p and PSA or PCA3 expressions with a correlation coefficient of 0.601 and 0.748, respectively. Correlation coefficient of 0.791 (p < 0.05) indicated a strong correlation of PCA3 with PSA expression.
Discussion
Deregulation in gene expression, that is, an increase or decrease in the production of mRNA, is a frequent event in PCa. Thus, the analysis of genes that are associated with the development and progression of PCa offers a new strategy for specific and early detection, addressing the most important problem in PCa and avoiding unnecessary biopsies.
We analyzed the expression of PCA3 and PSA, two genes commonly reported in this pathology due to their clinical value as biomarkers. Our results showed an overexpression of PCA3 in tissues from PCa patients, similar to previous reports where its expression level was evaluated in different body fluids and tissues (Bussemakers et al., 1999; Li et al., 2018). Nonetheless, only a handful of analyses has been conducted in the Mexican population (Floriano-Sánchez et al., 2009), highlighting the necessity to investigate the expression of PCA3 and PSA in these individuals. Moreover, as previously reported by de la Taille et al. (2011), we observed a correlation between the expression of PCA3 and the Gleason score; a high level of PCA3 was related to values ??above 7 on the Gleason score, suggesting that the expression level of this gene could serve as an indicator of aggressiveness in Mexicans as well (de la Taille et al., 2011).
PSA is a widely used protein for PCa detection, due to its ability to identify abnormalities in the prostate. However, it is not specific for PCa. In this regard, analyses of the PSA gene in prostatic tissue could provide more accurate information about the presence of this disease. Thus, we analyzed the relationship between the expression of the PSA gene and the expression of five other genes (Fra-1, PCA3, miR-195-5p, miR-133a-3p, and miR-148b-3p) in prostatic tissue, as well as the protein levels in the serum. Interestingly, we observed no relationship between the expression of the PSA gene and the concentration of serum PSA. This can be due to different factors. In 1995, it was discovered that PSA is expressed not only in the prostate but also in various tissues and structures, such as the periurethral glands, perianal glands, and apocrine sweat glands (Graves, 1995), showing that serum PSA is not prostate-specific. This makes it necessary to search for alternative biomarkers for the detection of PCa.
In this study we investigated different genes that are confirmed to be of importance in PCa, such as Fra-1. Fra-1 is a pro-oncogenic and pro-angiogenic gene known for activating the IL-6/JAK/Stat3 signaling pathway, and for promoting the release of MMP-9, MMP-2, VEGF, and TGF-β in breast and lung cancer (Adiseshaiah et al., 2005; Luo et al., 2010). In addition, Fra-1 has been shown to be involved in regulating growth, migration, and invasion in two different prostate cancer cell lines, DU145 and PC3 (Wu et al., 2015). However, these findings have yet to be confirmed in PCa patients, as in vitro results not always translate to humans. We found this to be true in Mexican PCa patients, in which Fra-1 was observed at similar expression levels as the control group, suggesting that in Mexican men, this gene may not serve as a positive indicator of PCa.
To explore the potential of different molecules as biomarkers, we also evaluated the expression levels of three miRNAs suggested to be involved in this pathology. We decided to investigate miR-195-5p, miR-133a-3p, and miR-148b-3p. With a little less than a half a decade of study, these three miRNAs became rapidly associated with cancer. Underexpression of miR-195-5p and 133a-3p is related to proliferation, invasion, and migration in PCa cells and, in the case of miR-195-5p, also metastasis. miR-195-5p mediates cellular processes through targeting the fibroblast growth factor 2 (FGF2) and BCOX1 genes (Guo et al., 2015; Liu et al., 2015). On the other hand, miR-133a-3p regulates genes such as EGFR, CASP9, and IGF1R (Tao et al., 2012; Pashaei et al., 2017). These genes have already been reported to be implicated in different processes of PCa, such as apoptosis, metastasis, and androgen-independence (Wu and Yu, 2014; Day et al., 2017; Yilmaz et al., 2017). miR-148b-3p ha, so far,s not been studied in depth in PCa. However, it is known to play a role in gastric, lung, and bladder cancer through the regulation of the Wnt, MAPK, and Jak-STAT signaling pathways, which are involved in PCa (Luo et al., 2015; Huang, 2016). The Wnt family plays an important role in cell proliferation and differentiation (Angers and Moon, 2009, Whyte et al., 2012). It has been observed that Wnt signaling is related to the development of prostate cancer (Murillo-Garzón and Kypta, 2017). The MAPK pathway has been reported to be involved in the growth and metastasis of PCa (Bradham and McClay, 2006; Wagner and Nebreda, 2009) and the JAK/STAT pathway has been observed upregulated in PCa. Furthermore, by suppressing JAK/STAT3 signaling, cell growth is suppressed and apoptosis is promoted (Aalinkeel et al., 2010; Aghaee-Bakhtiari et al., 2015).
Our results showed, that when compared to BPD patients, PCa patients expressed 27.1% lower miR-195-5p levels, 25.4% higher miR-133a-3p levels, and 46.8% higher miR-148b-3p levels; however, none of these results were statistically significant. These results differ from previous reports conducted on the same set of miRNAs carried out in Chinese populations (Guo et al., 2015; Liu et al., 2015; Wu et al., 2015), whereas ours was conducted in Mexicans. When analyzing the ancestry of Mexican men, Martínez-Cortés et al. (2012) showed that in the Sinaloa, region, where our patients were from, European ancestry is of 63%, whereas and the Asian is as low as 1% This could explain the discrepancy between the previous and our study.
It is important to identify whether clinicopathological characteristics are capable of modifying the expression levels of the miRNAs involved in our study. Hence, correlation analyses were performed to determine elationships between the variables. We observed an association between the age of patients and expression of miR-195-5p, suggesting that levels of this transcript increase over time. There is limited information about miR-148b-3p in PCa and, although we did not observe a significant difference in expression between men with PCa and BPD, correlations were identified between miR-148b-3p and the expression levels of PCA3 and PSA, two genes highly related to PCa. The observed relationship demonstrates the importance of studying alternative molecules involved in the control of gene expression in PCa to understand the control mechanisms underlying this pathology, improve detection methods, and propose new therapeutic approaches.
Here, we present evidence that PCA3 and PSA expression levels in tissue are suitable for differentiating between men with benign or malignant PCa disease. We observed a correlation between the expression of miR-148b-3p in tissue with the overexpression of PSA and PCA3 genes, which are established biomarkers in PCa. The expression of this miRNA was not related to variables such as age and weight, as observed in the other miRNAs analyzed, suggesting its potential as a biomarker for this pathology.
Acknowledgments
This work was supported by Universidad Politécnica de Sinaloa. We thank MSc Martín Irigoyen Arredondo, MSc Alejandra Paola Martínez Camberos, and MSc Saul Armando Beltrán Ontiveros for their assistance during this work.
Footnotes
Associate Editor: Houtan Noushmer
Conflict of Interest
The authors declare that they have no competing interests.
Author Contributions
EAM, FBH, MAA and NGM designed the study. FBH and MAA performed the data collection. FBH and NGM performed the experiments. FBH, JRQ, ERM and NGM performed statistical analysis and data interpretation. EAM, FBH and NGM wrote the manuscript. ELL, ERM, VPC and FLO revised the manuscript critically for important intellectual content.
References
- Aalinkeel R, Hu Z, Nair BB, Sykes DE, Reynolds JL, Mahajan SD, Schwartz SA. Genomic analysis highlights the role of the JAK-STAT signaling in the antiproliferative effects of dietary - flavonoid “ashwagandha” in prostate cancer cells. Evid Based Complement Alternat Med. 2010;7:177–187. doi: 10.1093/ecam/nem184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACS Prostate Cancer Overview. 2012. http://www.cancer.org/cancer/prostatecancer/index.
- Adiseshaiah P, Peddakama S, Zhang Q, Kalvakolanu DV, Reddy SP. Mitogen regulated induction of FRA-1 proto-oncogene is controlled by the transcription factors binding to both serum and TPA response elements. Oncogene. 2005;24:4193–4205. doi: 10.1038/sj.onc.1208583. [DOI] [PubMed] [Google Scholar]
- Aghaee-Bakhtiari SH, Arefian E, Naderi M, Noorbakhsh F, Nodouzi V, Asgari M, Fard-Esfahani P, Mahdian R, Soleimani M. MAPK and JAK/STAT pathways targeted by miR-23a and miR-23b in prostate cancer: Computational and in vitro approaches. Tumour Biol. 2015;36:4203–4212. doi: 10.1007/s13277-015-3057-3. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Ledet-Jensen J, Ørntoft T. Normalization of real-time quantitative RT-PCR data: A model-based variance estimation approach to identify genes suited for normalization - applied to bladder- and colon-cancer data-sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Angers S, Moon RT. Proximal events in Wnt signal transduction. Rev Mol Cell Biol. 2009;10:468–77. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- Bradham C, McClay DR. P38 MAPK in development and cancer. Cell Cycle. 2006;5:824–828. doi: 10.4161/cc.5.8.2685. [DOI] [PubMed] [Google Scholar]
- Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP. DD3: A new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- Cai C, Chen QB, Han ZD, Zhang YQ, He HC, Chen JH, Chen YR, Yang HB, Wu YD, Zeng YR, et al. miR-195 inhibits tumor progression by targeting RPS6KB1 in human prostate cancer. Clin Cancer Res. 2015;21:4922–4934. doi: 10.1158/1078-0432.CCR-15-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Grant S. BCL2L11/Bim as a dual-agent regulating autophagy and apoptosis in drug resistance. Autophagy. 2015;11:416–418. doi: 10.1080/15548627.2014.998892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniyal M, Ali S, Zamir A, Muhammad A, Sultana HM, Khan A. Epidemiology, etiology, diagnosis and treatment of prostate cancer. Asian Pac J Cancer Prev. 2014;15:9575–9578. doi: 10.7314/apjcp.2014.15.22.9575. [DOI] [PubMed] [Google Scholar]
- de la Taille A, Irani J, Graefen M, Chun F, de Reijke T, Kil P, Gontero P, Mottaz A, Haese A. Clinical evaluation of the PCA3 assay in guiding initial biopsy decisions. J Urol. 2011;185:2119–2125. doi: 10.1016/j.juro.2011.01.075. [DOI] [PubMed] [Google Scholar]
- Day KC, Lorenzatti Hiles G, Kozminsky M, Dawsey SJ, Paul A, Broses LJ, Shah R, Kunja LP, Hall C, Palanisamy N, et al. HER2 and EGFR overexpression support metastatic progression of prostate cancer to bone. Cancer Res. 2017;77:74–85. doi: 10.1158/0008-5472.CAN-16-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floriano-Sánchez E, Cárdenas-Rodríguez N, Castro-Marín M, Álvarez-Grave P, Lara-Padilla E. DD3: PCA3 gene expression in cancer and prostatic hyperplasia. Clin Invest Med. 2009;32:258–264. [PubMed] [Google Scholar]
- Coelho FF, Loli Guimarães F, Cabral WLR, Salles PGO, Mateo EC, Nogueira e Nogueira LM, Fonseca CEC, Gomes KB. Expression of PCA3 and PSA genes as a biomarker for differential diagnosis of nodular hyperplasia and prostate cancer. Genet Mol Res. 2015;14:13519–13531. doi: 10.4238/2015.October.28.13. [DOI] [PubMed] [Google Scholar]
- Graves HC. Nonprostatic sources of prostate-specific antigen: A steroid hormone-dependent phenomenon? Clin Chem. 1995;41:7–9. [PubMed] [Google Scholar]
- Guo J, Wang M, Liu X. MicroRNA-195 suppresses tumor cell proliferation and metastasis by directly targeting BCOX1 in prostate carcinoma. J Exp Clin Cancer Res. 2015;34:91. doi: 10.1186/s13046-015-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MX. Down-expression of circulating micro ribonucleic acid (miRNA)-148/152 family in plasma samples of non-small cell lung cancer patients. J Cancer Res Ther. 2016;12:671–675. doi: 10.4103/0973-1482.150420. [DOI] [PubMed] [Google Scholar]
- Kojima S, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M, et al. Tumor suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer. 2012;106:405–413. doi: 10.1038/bjc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y, Zou C, Zhang X, Liu S, Wang X, et al. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin Cancer Res. 2011;17:1722–1730. doi: 10.1158/1078-0432.CCR-10-1800. [DOI] [PubMed] [Google Scholar]
- Li M, Zhou D, Zhang W, Gao S, Zhou X. Urine PCA3 mRNA level in diagnostic of prostate cancer. J Cancer Res Ther. 2018;14:864–866. doi: 10.4103/jcrt.JCRT_734_17. [DOI] [PubMed] [Google Scholar]
- Liu C, Guan H, Wang Y, Chen M, Xu B, Zhang L, Lu K, Tao T, Zhang X, Huang Y. miR-195 Inhibits EMT by targeting FGF2 in prostate cancer cells. PLoS One. 2015;10:1–13. doi: 10.1371/journal.pone.0144073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo YP, Zhou H, Krueger J, Kaplan C, Liao D, Markowitz D, Liu C, Chen T, Chuang TH, Xiang R, et al. The Role of proto-oncogene Fra-1 in remodeling the tumor microenvironment in support of breast tumor cell invasion and progression. Oncogene. 2010;29:662–673. doi: 10.1038/onc.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Zhang C, Tang F, Zhao J, Shen C, Wang C, Yu P, Wang M, Li Y, Di JI, et al. Bioinformatics identification of potentially involved microRNAs in Tibetan with gastric cancer based on microRNA profiling. Cancer Cell Int. 2015;15:115. doi: 10.1186/s12935-015-0266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cortés G, Salazar-Flores J, Fernández-Rodríguez LG, Rubi-Castellanos R, Rodríguez-Loya C, Velarde-Félix JS, Muñoz-Valle JF, Parra-Rojas I, Rangel-Villalobos H. Admixture and population structure in Mexican-Mestizos based on paternal lineages. J Hum Genet. 2012;57:568–574. doi: 10.1038/jhg.2012.67. [DOI] [PubMed] [Google Scholar]
- McDonald AC, Vira M, Shen J, Sanda M, Raman JD, Liao J, Patil D, Taioli E. Circulating microRNAs in plasma as potential biomarkers for the early detection of prostate cancer. Prostate. 2017;78:411–418. doi: 10.1002/pros.23485. [DOI] [PubMed] [Google Scholar]
- Murillo-Garzón V, Kypta R. WNT signalling in prostate cancer. Nat Rev Urol. 2017;11:683–696. doi: 10.1038/nrurol.2017.144. [DOI] [PubMed] [Google Scholar]
- Pashaei E, Pashaei E, Ahmady M, Ozen M, Aydin N. Meta-analysis of miRNA expression profiles for prostate cancer recurrence following radical prostatectomy. PLoS One. 2017;12:e0179543. doi: 10.1371/journal.pone.0179543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Wu D, Xu B, Qian W, Li P, Lu Q, Yin C, Zhang W. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol Rep. 2012;27:1967–1975. doi: 10.3892/or.2012.1711. [DOI] [PubMed] [Google Scholar]
- Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- Walter BA, Valera Vladimir A, Pinto Peter A, Merino MJ. Comprehensive microRNA profiling of prostate cancer. J Cancer. 2013;4:350–357. doi: 10.7150/jca.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watahiki A, Wang Y, Morris J, Dennis K, O’Dwyer M, Gleave M, Gout P, Wang Y. MicroRNAs associated with metastatic prostate cancer. PLoS One. 2011;6:e24950. doi: 10.1371/journal.pone.0024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte JL, Smith AA, Helms JA. Wnt signaling and injury repair. Cold Spring Harb Perspect Biol. 2012;4:a008078. doi: 10.1101/cshperspect.a008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Yu E. Insulin-like growth factor receptor-1 (IGF-IR) as a target for prostate cancer therapy. Cancer Metastasis Rev. 2014;33:607–617. doi: 10.1007/s10555-013-9482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Ji A, Wang X, Zhu Y, Yu Y, Lin Y, Liu Y, Li S, Liang Z, Xu X, et al. MicroRNA-195-5p, a new regulator of Fra-1, suppresses the migration and invasion of prostate cancer cells. J Transl Med. 2015;13:289. doi: 10.1186/s12967-015-0650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–121. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- Xue W, Ying X, Jiang J, Xu YH. Prostate cancer antigen 3 as a biomarker in the urine for prostate cancer diagnosis: a meta-analysis. J Cancer Res Ther. 2014;10:218–220. doi: 10.4103/0973-1482.145881. [DOI] [PubMed] [Google Scholar]
- Yilmaz SG, Yencilek F, Yildirim A, Yencilek E, Isbir T. Effects of Caspase 9 gene polymorphism in patients with prostate cancer. In Vivo. 2017;31:205–208. doi: 10.21873/invivo.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Li J, Su J, Li J, Li J, Deng B, Shi Q, Zhou Y, Chen X. FOS-like antigen 1 is highly expressed in human psoriasis tissues and promotes the growth of HaCaT cells in vitro . Mol Med Rep. 2014;10:2489–2494. doi: 10.3892/mmr.2014.2509. [DOI] [PubMed] [Google Scholar]
Internet Resources
- GLOBOCAN - Global Cancer Observatory web-based platform [accessed September 2018]. http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx.