Abstract
Glioblastoma (GBM) is the most common and malignant type of primary brain tumor, showing rapid development and resistance to therapies. On average, patients survive 14.6 months after diagnosis and less than 5% survive five years or more. Several pieces of evidence have suggested that the DNA damage signaling and repair activities are directly correlated with GBM phenotype and exhibit opposite functions in cancer establishment and progression. The functions of these pathways appear to present a dual role in tumorigenesis and cancer progression. Activation and/or overexpression of ATRX, ATM and RAD51 genes were extensively characterized as barriers for GBM initiation, but paradoxically the exacerbated activity of these genes was further associated with cancer progression to more aggressive stages. Excessive amounts of other DNA repair proteins, namely HJURP, EXO1, NEIL3, BRCA2, and BRIP, have also been connected to proliferative competence, resistance and poor prognosis. This scenario suggests that these networks help tumor cells to manage replicative stress and treatment-induced damage, diminishing genome instability and conferring therapy resistance. Finally, in this review we address promising new drugs and therapeutic approaches with potential to improve patient survival. However, despite all technological advances, the prognosis is still dismal and further research is needed to dissect such complex mechanisms.
Keywords: Glioblastoma, DNA repair, biomarkers, tumor progression, therapy resistance
Introduction
Gliomas are brain cancers that present glial differentiation and represent a group of highly heterogeneous tumors with diverse histological, immunohistochemical and molecular characteristics (Louis et al., 2016; Wirsching and Weller, 2016). They correspond to only 2% of the overall cases of cancer. However, despite the low incidence, these cancers represent an important cause of death due to the elevated mortality associated, especially regarding the most malignant and common form, glioblastoma (GBM). Patients diagnosed with a GBM present an average survival of 14.6 months and only 5% survives for more than 5 years (McNeill, 2016). GBM is characterized by prominent dedifferentiation, diffuse infiltration, exacerbated proliferation, presence of necrosis and angiogenesis, resistance to apoptosis and conspicuous genomic instability. These tumors are seriously aggressive and resistant to available treatments, which involve surgical resection, radiotherapy, and chemotherapy with temozolomide (TMZ) (Louis et al., 2016). In this review, we focused on alterations of DNA damage response (DDR) and DNA repair genes encountered in astrocytomas from different grades, aiming to draw an integrated view of how dysfunctions in these pieces of machinery are orchestrated to allow tumorigenesis, cancer progression, resistance to therapy, as well as its potential involvement in controlling the marked genomic instability of GBM cells. Also, we suggest some future perspectives of promising approaches that could possibly improve GBM treatment.
Glioma classification and frequent genetic alterations
The first complete and robust classification of the tumors from the central nervous system (CNS) proposed by the World Health Organization (WHO) was published in 2007 (Louis et al., 2007). This publication redesigned the previous grading system (Kernohan and Mabon, 1949), as follows: grade I was defined as benign tumors with low proliferative potential that can be cured after surgical resection; grade II referred to lesions with moderate mitotic activity, infiltrative capability and tendency to progress to higher grades of malignancy; grade III correspond to tumors with histological evidence of malignancy, as nuclear atypia and high mitotic activity; and grade IV that present higher levels of atypia, exacerbated mitotic activity, besides angiogenesis and necrosis, which are associated with rapid tumor growth and fatal outcome for patients. In summary, the assortment was largely based on differentiation levels and histopathological features (Louis et al., 2007).
More recently, in 2016, the newest edition of the WHO classification was published (Louis et al., 2016), integrating genetic features and novel molecular biomarkers with the traditional histology examination. The current update changed grouping criteria, redefined diffuse gliomas, included new entities and discouraged the diagnosis of tumors difficult to be defined, such as oligoastrocytomas. Three major types of gliomas were distinguished, diffuse astrocytic and oligodendroglial, other astrocytic, and ependymal tumors. Grade I to IV assignment as a malignancy ruler was kept and mutations in IDH1 and histone H3 (H3K27M) were included, as well as the 1p/19q-codeletion (Table 1) (Louis et al., 2007, 2016). Moreover, grade II tumors were considered low-grade glioma (LGG) due to their less aggressive behavior, and grades III and IV as high grade (HGG), as they present worse prognoses (Louis et al., 2016). Considering that approximately 76% of all gliomas exhibit astrocytic origin (Louis et al., 2016; McNeill, 2016), hereafter we use the malignancy scale and LGG and HGG to refer only to astrocytoma.
Table 1. WHO 2016 types and grade of glioma.
| Diffuse astrocytic and oligodentroglial tumors | Grade |
|---|---|
| Diffuse astrocytoma, IDH-mutant | II |
| anaplastic astrocytoma, IDH-mutant | III |
| glioblastoma, IDH-wildtype | IV |
| glioblastoma, IDH-mutant | IV |
| diffuse midline glioma, H3K27M-mutant | IV |
| oligodendroglioma, IDH mutant and 1p/19q-codeleted | II |
| Anaplastic oligodendroglioma, IDH-mutant and 1p/19q-codeleted | III |
| Other astrocitic tumors | |
| Pilocytic astrocytoma | I |
| Subependymal giant cell astrocytoma | I |
| Pleomorphic xanthoastrocytoma | II |
| anaplastic pleomorphic xanthoastrocytoma | III |
| Ependymal tumors | |
| Subependymoma | I |
| myxopapillary ependymoma | I |
| ependymoma | II |
| ependymoma, RELA fusion-positive | II or III |
| Anaplastic ependymoma | III |
LGG usually present an indolent behavior, but about 70% of cases undergo progression to grades III and IV within 5 to 10 years after diagnosis. Occurring mainly in childhood, LGG represents more than 30% of central nervous system neoplasms in this population (Louis et al., 2016). Despite the typical heterogeneity, LGG harbor alterations in the BRAF gene that commonly lead to the loss of its regulatory N-terminal region. Other genetic abnormalities are also described, but in all cases, the defects frequently lead to constitutive activation of the MAP (mitogen-activated protein) kinase pathway (Jones et al., 2012; Zhang et al., 2013). Besides BRAF mutations, translocations involving tyrosine kinase receptors have been likewise documented. For example, neurotrophic tyrosine kinase receptors (NTRK) 2 and 3 were found fused by its N-terminus with other genes, acquiring the ability to interact with actin or topoisomerase I. Interestingly, NTRK fusions have also been noticed in pediatric HGG (Wu et al., 2014), suggesting a potential general role for these types of fusions in glioma development. Furthermore, mutations in fibroblast growth factor receptor 1 (FGFR1) are the second most common point mutation in LGG, after BRAF V600E (Jones et al., 2013).
Glioblastoma (astrocytoma grade IV) is the most common and aggressive HGG, accounting for 16% of brain tumors and 60-75% of astrocytomas (Thakkar et al., 2014). GBM occurs mainly in elderly individuals among 45-75 years of age, usually leading the patient to death in 12-15 months after diagnosis. Even under rigorous therapy, the majority of cases relapses in 1-2 years after surgery and less than 5% of patients survive for 5 years or more (McNeill, 2016). GBM is further classified as primary or secondary according to their clinical history. Primary GBM occurs in a de novo manner without evidence of previous lesion and accounts for 90% of cases; secondary GBM is a result of LGG progression into HGG and represents 10% of cases (Ohgaki and Kleihues, 2013; Louis et al., 2014). Primary and secondary GBMs present marked genetic differences and distinct transcriptional activity that identify unique entities, predict prognosis and delineate a progression pattern (Maher et al., 2006; Ohgaki and Kleihues, 2013).
The Cancer Genome Atlas (TCGA) Research Network performed detailed genome-wide analyses and disclosed the intricate genetic profile of GBMs, and grade II and III gliomas, by characterizing more than 1000 human samples. The majority of cases harbor alterations in the following genes: MGMT, IDH1, TP53, RB1, RTK, RAS, EGFR, cyclin D1/3, MDM2, PTEN, CDK4, PDGFRA, PIK3CA, NF1, PIK3R1, LZTR1, BRAF, FGFR1, FGFR2, FGFR3, ATRX, TERT, NOTCH1, FUBP1, CIC (Cancer Genome Atlas Research Network, 2008, 2015). Considering the landscape of alterations characterized, three core signaling pathways underlying GBM pathogenesis were identified: tyrosine kinase receptors, p53, and retinoblastoma. Additionally, global transcriptional profiling allowed a more refined classification of GBMs into four molecularly distinct subgroups: proneural, neural, classical and mesenchymal that are also characterized by a particular set of high frequent mutations (Table 2) (Verhaak et al., 2010; Brennan et al., 2013). It was also characterized a subtype of proneural GBMs that presents a hypermethylated phenotype of CpG islands (G-CIMP), which is associated with improved survival and is more prevalent in LGG (Noushmehr et al., 2010; Brennan et al., 2013). The response to the different therapy protocols currently applied varies considerably among these transcriptional subgroups. Classical and mesenchymal subtypes obtain benefit from more intensive treatment, while patients with the neural profile apparently get only a small increase in survival and the proneural show no increment (Verhaak et al., 2010). However, even for patients who benefit from intensive therapy, the survival gain corresponds to a few months only, and to the best of our knowledge, there is no literature evidence of the clinical use of the subclassification.
Table 2. GBM subgroups and their main genetic changes.
| Classical | Mesenchymal | Neural | Proneural |
|---|---|---|---|
| EGFR mutation/overexpression | NF1 loss/mutation | EGFR overexpression | PDGFRA amplification |
| PTEN loss/mutation | TP53 loss/mutation | neuron markers expression | IDH1 mutation |
| CDKN2A loss | PTEN loss/mutation | PIK3A/PIK3R1 mutations | |
| NES overexpression | MET, CHI3L1, CD44, MERTK overexpression | TP53, CDKN2A, PTEN loss/mutation | |
| Notch and Shh pathways activation | TNF family and NFkB pathways activation | proneural markers expression |
Despite the diversity of genetic alterations underlying GBM pathogenesis, all subtypes present remarkable proliferation rate, diffuse infiltration, enhanced survival capacity and robust angiogenesis, which provide high resistance to the available therapies and unavoidable recurrence. All of these characteristics added to prominent intra-tumor heterogeneity and genomic instability, make GBM one of the most complex types of cancer frequently associated with dismal prognosis (Noushmehr et al., 2010; Verhaak et al., 2010; Brennan et al., 2013). The robust characterization of gliomas now available, encompassing large-scale genetic and epigenetic profiling, high throughput transcriptomic and proteomic analysis, revealed novel important GBM features, as new biomarkers and unique signatures capable of providing better diagnosis, predict prognosis and/or treatment response (Cancer Genome Atlas Research Network, 2008; Noushmehr et al., 2010). The list of significant biomarkers is growing in an astonishing manner and includes a wide range of molecules such as lncRNAs and microRNAs (e.g. HOTAIR and miR-141), and several DNA repair genes (Boccard et al., 2015; Bian et al., 2016; Reon et al., 2016), which are importantly correlated with genome stability and will be explored in more detail in the next sections. Table 3 summarizes the main DNA repair biomarkers discussed in this review.
Table 3. DNA repair genes considered biomarkers* of GBM susceptility and/or progression.
| Gene | Alteration | Impact on disease progression | References |
|---|---|---|---|
| MGMT | promoter methylation | response to TMZ treatment | Hegi et al., 2005; Hegi et al., 2008; Stupp et al., 2009; Binabaj et al., 2018 |
| APNG | overexpression | controversial | Agnihotri et al., 2012; Fosmark et al., 2017 |
| HJURP | overexpression | poor outcome, worse overall survival | de Tayrac et al., 2011; Valente et al., 2013 |
| DDB2 | reduction | reduced survival | de Sousa et al., 2017 |
| BRCA2 | overexpression | reduced survival | de Sousa et al., 2017 |
| BRIP1 | overexpression | reduced survival | de Sousa et al., 2017 |
| XRCC3 | polymorphism | increased GBM susceptibility | Custodio et al., 2012 |
| EXO1 | polymorphism | increased GBM susceptibility | Chang et al., 2008 |
| EXO1 | overexpression | reduced survival | de Sousa et al., 2017 |
| NEIL3 | overexpression | reduced survival | de Sousa et al., 2017 |
| MSH6 | mutations | controversial | Hunter et al., 2006; Maxwell et al., 2008; Yip et al., 2009; Sun et al., 2018 |
*This table shows only the biomarkers discussed throughout the review. Please refer to the text for further information.
DNA repair genes as biomarkers of astrocytoma aggressiveness
The methylation status of MGMT (O-6-Methylguanine-DNA-Methyltransferase) promoter was the first biomarker to be used for patient stratification in clinical trials as a predictor of GBM response to treatment with alkylating agents (Hegi et al., 2005, 2008). The MGMT gene encodes a DNA repair protein responsible for the removal of alkylation at guanines O6 position, a site that is commonly altered by TMZ, the gold standard chemotherapeutic for GBM treatment. Methylation of the MGMT promoter reduces protein expression, thus impairing the repair capacity of TMZ-induced damage, boosting the response to treatment (Hegi et al., 2008).
In a randomized phase III clinical trial with a set of 206 GBM patients, Stupp and colleagues observed that 45% of patients presented methylations in the MGMT promoter. This feature was associated with a better overall survival, 21.7 months after chemotherapy associated with radiotherapy, in comparison to 15.3 months for patients carrying non-methylated genotype (Stupp et al., 2009). More recently, a meta-analysis of 10 eligible studies, including the MGMT methylation status of more than four thousand subjects, confirmed that patients bearing this genotype present longer overall survival (Binabaj et al., 2018), emphasizing MGMT status as an independent indicator of a favorable prognosis. MGMT methylation could also be found in patient serum and strongly correlated with its presence in the tumor tissues (Fiano et al., 2014), suggesting that detection in blood samples could represent a reliable tool to predict response to TMZ treatment. MGMT methylation also disclosed its relevance as biomarker for other types of malignancies, including breast (Neto et al., 2012), colorectal (Lee et al., 2009), prostate (Cortese et al., 2012), cervical (Sun et al., 2015), gastric (Jin et al., 2014) and lung (Ostrow et al., 2010) cancers.
Therefore, MGMT promoter methylation, i.e., reduced MGMT protein expression, is a frequent epigenetic alteration in GBM patients related to better outcome, survival and response to treatment. On the other hand, when MGMT promoter is not methylated, i.e. normal MGMT expression, this protein is considered a good therapeutic target, once it is available and susceptible to pharmacological inhibition. Hence, several groups are concentrating efforts to develop strategies to reduce MGMT activity and/or expression, enhancing TMZ sensitivity.
Additionally, among patients carrying the MGMT methylated phenotype, those with high levels of the alkyl purine-DNA-N-glycosylase (APNG) enzyme present better overall survival and this result was supported by data from TCGA database (Fosmark et al., 2017), making the APNG expression levels an important factor to be associated to MGMT methylation status. APNG is a DNA repair enzyme involved in the base excision repair (BER) pathway, which is responsible for removing methyl of adducts, induced by alkylating agents, creating apurinic or apyrimidinic sites (Evans et al., 2000). Curiously, APNG overexpression was also associated with poor survival (Agnihotri et al., 2012), suggesting a controversial role for this enzyme as a prognostic biomarker. Taken together, these pieces of evidence highlight the importance of complementary studies toward the development of different therapy approaches and novel drugs that do not rely exclusively on MGMT methylation phenotype.
Expression levels of the Holiday Junction Recognizing Protein (HJURP) were also correlated with prognosis of astrocytoma patients. HJURP was reported as highly overexpressed in tumors from different grades and showed an independent capacity of survival prediction (Valente et al., 2009, 2013). HJURP was also shown to be involved in DNA double-strand breaks (DSB) restoration (Kato et al., 2007) by mechanisms not yet characterized. It has also important roles in the deposition of Centromere Protein A (CENP-A) at the centromeric chromatin (Foltz et al., 2009), presenting capacity to allow centromeric chromatin expansion (Perpelescu et al., 2015) and assembly of ectopic kinetochores (Barnhart et al., 2011). Other studies have described the association between HJURP overexpression, combined with additional alterations, and a higher risk of death for GBM patients (de Tayrac et al., 2011; Valente et al., 2013). Moreover, it was also reported that HJURP overexpression has an independent prognostic value for breast (Hu et al., 2010), lung (Kato et al., 2007), liver (Hu et al., 2017) and ovarian (Li et al., 2018) cancer patients, reinforcing the involvement of this protein with cancer aggressiveness and poor outcome.
Furthermore, several expression signatures of DNA repair genes were strongly associated with poor prognosis of astrocytoma patients. Among the alterations included in these signatures, reduction of DDB2 and overexpression of EXO1, NEIL3, BRCA2 and BRIP1, were independently correlated with worse prognoses, revealing single-gene signatures that represent new feasible biomarkers. EXO1 and NEIL3 exhibited remarkable overexpression and showed to be involved in DSB restoration kinetics and radiation resistance of GBM cell lines, respectively (de Sousa et al., 2017). Additional studies are necessary to better describe their roles in GBM biology and potential enrollment as biomarkers. In agreement, a study including 539 GBM cases identified and validated a gene expression signature including 15 key DNA repair genes significantly correlated to prognosis, and five of them (CDK7, DDB2, RNH1, RFC2 and FAH) were highly predictive of recurrence and disease-free survival (Kun et al., 2017).
In a recent study, an expression profiling of selected 154 genes involved in DNA damage signaling/repair and cell cycle was accomplished in cohorts containing paired samples of primary and recurrent GBM. Gobin et al. (2019) identified and validated a 27-gene signature that was able to stratify patients in two well-defined groups (G1 and G3) showing co-regulation and inverse expression patterns. A third subset containing samples with a more neutral profile formed a separate group named G2. Although no correlation with prognosis was found when only primary or paired GBM cohorts were considered, when analyzing only the cases of recurrence, the progression-free and overall survival were significantly worse in patients whose tumors progressed from G3 to G1 profile. Additionally, the use of inhibitors targeting RAD51 and mitotic kinases in tumor-derived cell cultures promoted a decrease in the viability of G3 cells. These data suggested that specific targets, selected on the basis of prognosis-correlated signatures, might represent vulnerabilities of a subset of tumors and can provide guidelines for personalized therapies (Gobin et al., 2019).
DNA repair functions in tumorigenesis and progression
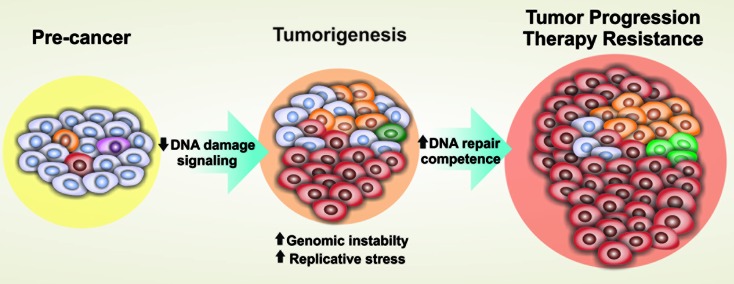
The DNA damage response (DDR) and the downstream recruited DNA repair machinery cross-communicate to form an intricate network of genome surveillance that identifies and repairs DNA injuries, protecting cells from intrinsic and microenvironmental genotoxic stress. Increasing pieces of evidence have been showing that this system presents antagonistic roles in tumorigenesis and tumor progression. Several studies have demonstrated that DDR and DNA repair genes may be inactivated in early tumorigenesis, enabling genomic instability and tumor development (Golmard et al., 2013; Nissar et al., 2014), whereas secondary mutations grant a selective advantage to the tumor (Gorgoulis et al., 2005). Once the tumor is established, the repair activity undergoes reactivation, which avoids cell collapsing and allows tumor progression (Kauffmann et al., 2008; Turner et al., 2015), and is also associated with resistance to treatments (Eich et al., 2013; Atkins et al., 2015) (Figure 1).
Figura 1. DNA damage signaling and repair pathways show opposite regulation in tumorigenesis and tumor progression. Early in tumorigenesis, oncogene activation leads to replication stress and DNA damage, usually triggering the DDR machinery and leading to checkpoint-imposed senescence or cell death. When this barrier is overcome, by loss-of-function mutations in DDR and/or DNA repair genes, tumor establishment ensues. Progressively advanced tumors experience increasing levels of replication stress and genetic instability and often adapt to this environment, developing exacerbated DNA repair competences that avoid cell death and favor tumor progression.
Several studies have demonstrated that activation of DDR-DNA repair system works as an oncogene-induced barrier against tumor establishment (Bartek et al., 2007; Squatrito and Holland, 2011), and mutations or alterations that lead to loss of function or downregulation could represent a trigger for gliomagenesis (Cha and Yim, 2013). In a study using a mouse model for glioma development, it was demonstrated that the induced expression of RAD51, a central protein for repair by homologous recombination (HR), decreases both the incidence of oncogene-induced glioma and the genomic instability, impairing carcinogenesis (Westermark et al., 2011). Using a similar model, Squatrito and colleagues showed that components of the DDR pathway are frequently altered in gliomas and loss of ATM or its downstream targets accelerates tumor formation (Squatrito et al., 2010). Loss of ATRX, a protein required for non-homologous end joining (NHEJ), was also reported to promote GBM growth in an animal model (Koschmann et al., 2016).
A study conducted within the Brazilian population showed that patients carrying the Thr241Met polymorphism in the XRCC3 gene presented an increased risk of tumor development, suggesting that its malfunction contributes to astrocytoma and glioblastoma susceptibility. XRCC3 encodes a protein involved in HR repair, and this polymorphism can potentially affect the enzyme function as well as its interaction with other repair proteins (Custodio et al., 2012). Additionally, a nonsynonymous single nucleotide polymorphism (SNP) in the EXO1 gene was found to be potentially associated with GBM susceptibility. EXO1 is an important exonuclease of HR and the evaluated SNP promotes a drastic amino acid change that could affect the protein internal structure, as well as its protein-protein binding interface, impairing its normal function (Chang et al., 2008). Other polymorphisms of this nature have been also associated with GBM risk (Franceschi et al., 2016; Qi et al., 2016).
Paradoxically, DDR and the repair machinery act as a double-edged sword during tumorigenesis and cancer progression. Once the tumor has been established, replicative stress can promote the aberrant constitutive activation of DDR and repair execution (Bartkova et al., 2010; Carruthers et al., 2018), allowing tumor progression and driving treatment resistance. In turn, the exacerbated activity of these pathways defend malignantly transformed cells from replicative stress, high mutation rates, and the rampant genome instability (Kauffmann et al., 2008; Turner et al., 2015). Bartkova and colleagues observed that DDR is constitutively active in LGG and GBM samples, but not in normal brain tissues nor in regions adjacent to the tumor. Interestingly, in GBMs, which present the highest proliferation rates, the amounts of DNA damage detected were diminished in comparison to LGG (Bartkova et al., 2010). This data suggest that the DDR machinery is more effective in GBM than in LGG, helping highly malignant cells to manage their unstable genome and avoid collapse and death. Moreover, an increase in NHEJ and HR activities, mediated by the RTK/RAS pathway, was observed along with glioma progression (Turner et al., 2015).
More recently, EXO1 and NEIL3, DNA repair genes extremely overexpressed in different grade astrocytomas, showed a strong correlation with patient survival and GBM cells viability (de Sousa et al., 2017). EXO1 is a 5’ to 3’ exonuclease that resects the blunt ends of DSBs generating the single-strand tail necessary to invade the double-strand DNA used as a template in HR repair (Kim and Wilson, 2012). Silencing of EXO1 in T98G cells led to faster restoration of DNA injury induced by ionizing radiation (IR), suggesting that the absence of EXO1 possibly directs the DSB repair to the faster and error-prone NHEJ pathway (de Sousa et al., 2017). These results indicate a potential role of EXO1 to facilitate DNA repair during astrocytoma progression. Those data are in agreement with the increase in NHEJ and HR activities during glioma progression observed by Turner et al. (2015).
Additionally, NEIL3 knockdown was associated with a higher percentage of DNA damage and cell death after IR (de Sousa et al., 2017). NEIL3 is a DNA glycosidase that participates of BER by removing oxidized bases, which can be induced secondarily by IR, giving rise to apurinic/apyrimidinic sites that are recognized and converted to single-strand breaks (SSB) by the endonuclease APEX2 (Takao et al., 2009). These observations suggested a potential role for NEIL3 in the management of oxidative stress, supporting tumor progression. Taken together, these data imply the progressive requirement of DDR signaling and enhanced DNA repair competence accompanying tumor progression.
Along with progression, malignant cells usually acquire genetic alterations that enable metastasis and several models have been proposed to clarify how this intricate process occurs (Hunter, 2015). It is well known that GBM cells are highly infiltrative and relapse mainly locally, but they can also easily migrate and spread along nerves, meninges, and local blood vessels, inducing CNS metastasis (Scherer, 1938; Awan et al., 2015). The interactions between endothelial and GBM cells in microenvironmental niches seem to be important for progression and dispersion (Gilbertson and Rich, 2007). However, less than 2% of GBM cases metastasize outside the CNS (Beauchesne, 2011) and the roles of DNA repair genes during metastasis onset are controversial. In melanoma, for example, the upregulation of DNA repair genes is related to metastasis, while in many other tumors the opposite has been reported (Broustas and Lieberman, 2014).
DNA repair and GBM resistance to treatment
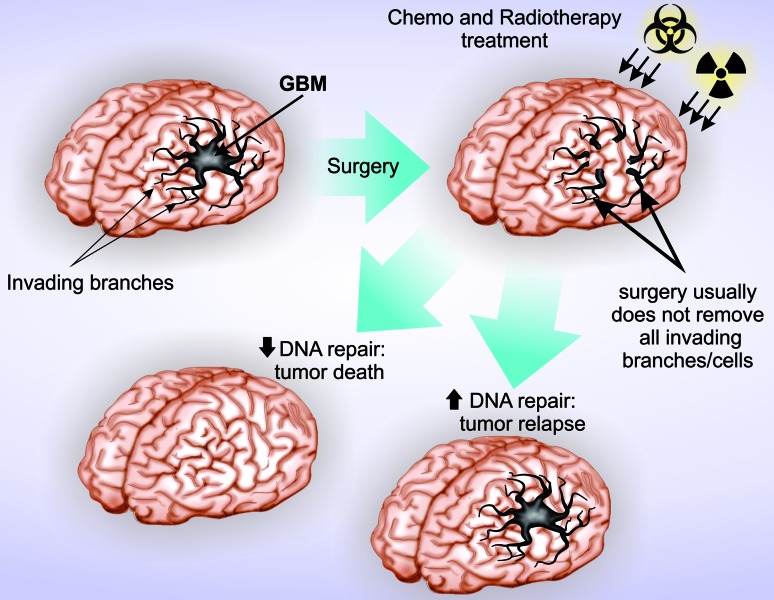
The therapy employed for GBM patients is usually multimodal and involves surgical resection, as much as considered safe, followed by adjuvant chemo and radiotherapy (CRT). However, the specificities of the therapeutic protocols are established according to each case’s necessity. The best available protocol for GBM treatment is the one reported by Stupp and colleagues, which indicates surgery followed by 6 weeks of CRT in tumor bed plus 6 additional cycles of TMZ-only (Stupp et al., 2005). This treatment reduces death risk by 37% but survival prolongation is still minimal due to the high resistance of GBM cells and frequent recurrence (Stupp et al., 2005, 2009) (Figure 2), emphasizing the urgency of better disease control and improvement of patient’s survival and life quality. In this section, we exploit literature that suggest the enrollment of the exacerbated activity of DNA repair pathways in treatment resistance, as well as their potential roles in subpopulations of cells that drive tumor relapse.
Figura 2. DNA repair competences predispose GBM to treatment resistance. The current standard treatment for GBM consists of surgery followed by chemoradiotherapy. Surgery frequently presents low efficiency due to the invasive nature of GBM cells, making difficult the complete resection. Tumors cells with higher DNA repair capabilities can efficiently manage chemoradiotherapy-induced DNA damage and support subsequent tumor relapse. Successful treatment is attainable when surgery can safely remove at least 75% of tumor tissue and the remaining cells present low DNA repair activity.
MGMT is the most studied DNA repair protein regarding associations with treatment response. The methylation status of the MGMT promoter affects protein expression and directly modulates TMZ response (Hegi et al., 2005, 2008). However, some patients did not present any clinical improvement after TMZ administration, even when MGMT levels are reduced, indicating that methylated MGMT promoter uniquely does not mean a successful treatment (Hegi et al., 2005). Additionally, it is also known that TMZ triggers the activity of different repair pathways, such as, BER (Yoshimoto et al., 2012), MMR, NER, NHEJ and HR (Nagel et al., 2017), which may also be considered to predict resistance.
Moreover, N-methylated bases are not recognized by MGMT and trigger BER pathway activation, suggesting that BER machinery could be targeted to enhance TMZ effectiveness. In fact, the inhibition of APNG, promoted a decrease in TMZ resistance, even in MGMT competent cells. It was also shown that the disruption of Ape1 (Bobola et al., 2001) or DNA polymerase β (Trivedi et al., 2008) activity sensitizes cells to TMZ. Moreover, increased NER activity was detected after treatment with TMZ (Nagel et al., 2017), while downregulation of ERCC1 promoted higher sensitivity to TMZ treatment, similarly to MGMT inhibition (Boccard et al., 2015).
MMR machinery defects have also been associated with TMZ resistance. The MMR pathway acts primarily during replication, recognizing and correcting insertions, deletions and base misincorporations, such as the mismatch between a TMZ-induced O6-methylguanine (O6-MeG) and thymidine. However, only daughter strands can undergo this type of repair and, consequently, the misincorporated thymidine is removed but the O6-MeG remains on the template strand. This mechanism occurs repeatedly and produces a futile cycle of repair, generates DSBs, activates ATR and Chk1, and ultimately can cause cell cycle arrest and cell death (Zhang et al., 2012). Thus, cells defective in MMR do not recognize DNA lesions and evade cell cycle arrest. Reduction of MSH6, a protein involved in mismatch recognition and MMR initiation, increased TMZ resistance of A172TR3 cells, which primarily exhibited a resistant phenotype (Yip et al., 2009). Additionally, clinical data showed the presence of MSH6 mutations in GBM specimens after TMZ therapy but not in pre-treatment samples, suggesting that these changes results from CT and may contribute to recurrence (Hunter et al., 2006; Yip et al., 2009). However, other studies suggest that MMR deficiency does not mediate clinical resistance to temozolomide (Maxwell et al., 2008) and that the upregulation of MSH6 could be associated with resistance, opposing the relationship between MMR and TMZ resistance (Sun et al., 2018). Thus, further studies are necessary to better elucidate the complex network involved in DNA repair and its relationship TMZ resistance.
Two major pathways, NHEJ and HR, are responsible for the DSBs repair. In contrast to NHEJ, which works at any moment of the cell cycle, HR pathway acts preferentially during S and G2 phases, when sister chromatids are available to be used as a repair template. HR restores DSBs generated by IR, as well as those produced by replication fork collapse (Helleday, 2010). Although TMZ does not cause DSB directly, O6-MeG can lead to futile repair cycles, stalling replication forks and inducing DSBs subsequently (Roos and Kaina, 2013). Once TMZ can produce DSB during S phase, HR is more relevant than NHEJ to repair these lesions. It has been demonstrated that mutation or knockdown of ATM or ATR kinases, which are sensors of DSB occurrence, sensitizes cells to TMZ (Eich et al., 2013). Moreover, the pharmacological inhibition or knockdown of downstream proteins in the HR pathway, such as BRCA2, RAD51, and CHK2, also increased sensitivity to treatment (Quiros et al., 2011; Eich et al., 2013). Furthermore, GBM cells treated with other alkylating agents were more sensitive after inhibition of MRE11, a component of MRN complex that recognizes DNA lesions and recruits ATM kinase (Berte et al., 2016). In contrast, some reports suggest that ATM/ATR activation is required to induce cell death after the MMR futile cycle is established (Caporali et al., 2004) and that MRE11 knockdown decreases sensitivity to TMZ (Mirzoeva et al., 2006). Altogether, these data highlight the complexity of responses to treatment and imply that the whole genetic background must be considered to better predict therapy efficiency.
IR causes several types of DNA damage and is frequently employed as a complementary therapy to GBM patients. Several studies have suggested that RAD51 (HR) and DNA-PKcs (NHEJ) activities may be related to GBM resistance to IR (Short et al., 2011; King et al., 2017). Russell and colleagues showed that GBM cells pretreated with Gleevec, an indirect inhibitor of RAD51, presented a reduction in RAD51 foci formation and higher sensitivity to IR (Russell et al., 2003). Likewise, GBM cells with DNA-PKcs deficiency or inhibition showed suppression of IR-induced migration, invasion, and microvascular formation and presented higher levels cell death (Zhuang et al., 2011; Gustafsson et al., 2014; Liu et al., 2015). Taken together, these data suggest that NHEJ and HR inhibitory agents present a great therapeutic potential.
Poly ADP-ribose polymerase 1 (PARP1) is a nuclear enzyme crucial for the overall repair process. PARP1 is rapidly activated by strand breaks and signals the presence of DNA lesions by attaching ADP-ribose units to chromatin proteins, leading to the recruitment of the downstream targets (Kim et al., 2005; Luo and Kraus, 2012). In DSB repair, PARP1 has been shown to be responsible for the recruitment of MRE11 and NBS1 (Haince et al., 2008), which are essential for both HR and NHEJ pathways (Roos and Krumm, 2016). Therefore, pharmacological inhibition of PARP1 may represent an excellent adjuvant therapy, since its inhibition could sensitize GBM cells to TMZ and IR, even when MGMT is normally expressed or MMR is deficient (Tentori et al., 2002; Barazzuol et al., 2013). Indeed, using MGMT non-methylated GBM models, a recent study showed that the association of veliparib, a PARP1 inhibitor, with IR inhibited colony formation, reduced the levels of MRE11 (HR pathway) and increased apoptosis. Furthermore, the oral administration of veliparib plus concomitant RT induced apoptosis and diminished cell proliferation in mice (Jue et al., 2017).
Additionally, inhibition of PARP1 intensifies SSBs occurrence, which are converted to DSBs during replication. This effect is greater in BRCA1/2 defective cells because missed DSBs restoration is secondly impaired and cell death is activated (Helleday, 2011). A great variety of studies and clinical trials have been conducted to evaluate the safety and efficacy of several PARP1 inhibitors (PARPi) exploiting its synthetic lethality in BRCA-defective tumors, mostly in breast and ovarian cancer (Yap et al., 2019). Although mutations in BRCA genes are rare in GBM, PTEN mutations similarly impair HR and are found in one-third of cases, enabling the exploration of PARPi synthetic lethality also for GBM (McEllin et al., 2010). However, the literature is still not conclusive about the benefits of the use of PARPi for GBM patients (Gupta et al., 2018). There are in vivo/in vitro conflicting data (Gupta et al., 2014) and PARPi have limited permeability and an efflux liability through the blood-brain barrier, showing heterogeneous response (Sarkaria et al., 2018). So far, the inhibition of constitutively active DDR and repair proteins, a common alteration in GBM, shows great potential to improve treatment effectiveness.
The high resistance of GBM to therapies has also been associated with the presence of cancer stem cells, which differ from other GBM cells by being capable of unlimited self-renewal and presenting low proliferative ratios. These characteristics certainly contribute to resistance once usual treatments aim to eliminate highly proliferative cells, making this population very relevant to clinical treatment. GBM cancer stem cells (CSC) subpopulations can be identified by the expression of CD133, SOX-2 and Nestin (Colleoni and Torrente, 2008; Ma et al., 2008) that were all negatively correlated with patient survival (Zeppernick et al., 2008), highlighting the importance to uncover mechanisms underlying CSCs competences.
Recently, Carruthers and coworkers showed that GBM CSCs present constitutively high levels of replicative stress, both in vitro and in vivo, and provided evidence that this phenotype underlies the activation of DDR and consequent radiation resistance (Carruthers et al., 2018). In agreement, recent studies have demonstrated that GBM CSCs exhibit greater efficiency in the activation of DNA damage sensors, as ATM, 53BP1 and H2AX, and are more resistant to CT. These cells can also become dormant in drug presence and usually restart proliferation after drug withdrawal (Annovazzi et al., 2015). In addition, inhibition of RAD51 in glioma stem cells reduced IR-induced foci formation and DSB repair, diminishing CSC population (Short et al., 2011; King et al., 2017). The CSC population was also affected by the combination of talazoparib, a PARP1 inhibitor, with IR. The combined treatment induced prolonged G2/M arrest and reduced proliferation rates of GBM CSCs (Lesueur et al., 2018). Altogether, these data suggest that the impairment of repair machinery can sensitize CSCs to IR and could improve therapy efficiency.
Altogether, these observations show the requirement of a more comprehensive analysis of all repair pathways, which can potentially reveal novel opportunities for therapeutic strategies, with either the sensitization of GBM cells to available treatments or the identification of new targets for drug development. It is important to emphasize that resistance is a competence derived from many factors and, besides DNA repair enhancement, drug efflux pumps and alterations in other signaling pathways, as those involved in the control of cell survival, proliferation and apoptosis, surely represent complementary mechanisms of resistance.
New therapies and future prospective
Researchers have been putting a lot of effort in the development of new therapies and/or approaches that potentially sensitizes GBM to TMZ or IR, as well as in the identification of novel promising drugs. In this ambit, genome-wide and synthetic lethality studies are gaining attention. To enhance efficacy of TMZ, Johannessen and coworkers screened for synthetic lethality using RNAi technology (Johannessen et al., 2019). They used an shRNA library targeting 5,046 human genes to seek for essential genes during TMZ treatment. The knockdown of a 292-gene cluster reduced cell growth of U87 cells when combined with a sublethal dose of TMZ. They also showed that the antipsychotic drug thioridazine mimics the gene cluster silencing, improving TMZ sensitivity in vitro and reducing tumor growth in vivo. Thioridazine was mechanistically shown to interfere in the autophagy process, impairing the fusion between autophagosomes and lysosomes, and preventing the metabolic adaptive changes related to TMZ resistance in GBM cells (Johannessen et al., 2019).
Other FDA-approved compounds have shown in vitro anti-GBM activity, both isolated and in various combinations. Jiang and coworkers observed that 22 compounds were active against GBM cells. Remarkably, the combination of pitavastatin, used in dyslipidemia control, with low doses of irinotecan, a topoisomerase I inhibitor, showed the greatest potential. Pitavastatin reduced the irinotecan IC50 by 40 to 70-fold (Jiang et al., 2014). Additionally, the repurpose of antihypertensive, beta-blockers (Rundle-Thiele et al., 2016), antidiabetics (Wurth et al., 2013) and anti-alcoholism (Triscott et al., 2012) drugs for GBM adjuvant treatment also demonstrated good potential. As these drugs already have known properties and are approved by drug administration agencies, the clinical application could be facilitated and benefit patients earlier than developing a new drug.
Although studies of new drugs take more time and are more laborious, they are also extremely important and largely conducted worldwide. New drugs for GBM treatment, such as bevazicumab, erlotinib (Raizer et al., 2016), 1,3-bis (2-chloroethyl)-1 nitrosourea (BCNU) (Vinjamuri et al., 2009), and gliadel (Xing et al., 2015), are being massively exploited with good results and some of them have already reached phase II clinical trials. In phase III trials, the implantation of gliadel and BCNU wafers in the after surgery tumor bed was assessed and improved overall survival average in 2 months (Westphal et al., 2003). Sorafenib, a kinase inhibitor that targets multiple RTK receptors, was demonstrated to have in vitro and in vivo antitumor properties in GBM cell lines (Siegelin et al., 2010). This drug was also able to selectively inhibit GBM CSC proliferation by affecting MAPK and PI3K/Akt pathways (Yang et al., 2010). Sorafenib is under clinical trials to evaluate its safety and efficacy, both as monotherapy and in combination with TMZ, bevazicumab or RT (Wurth et al., 2014).
In addition to the repurpose of FDA-approved drugs and the study of new ones, the development of immunotherapies has also been drawing attention as an important intervention strategy in the last few years. Although pieces of evidence suggest that resistance to radiotherapy can be overcome by immunotherapy in many types of cancer (Gameiro et al., 2014; Maxwell et al., 2017), most clinical trials show no improvement in progression free and/or overall survival in GBM patients (Filley et al., 2017). It is widely known that GBM presents a highly immune-suppressive microenvironment and induces T-cell apoptosis or qualitative defects. Additionally, patients usually present lymphopenia as GBM induces sequestration of T-cells in the bone marrow, reducing their amount at the tumor site and in lymphoid organs; and the blood-brain barrier blocks and/or effluxes antibodies and other large molecules (Sevenich, 2019). Altogether, these features ultimately lead to the poor or no GBM response to immunotherapies observed in clinical trials, e.g., CheckMate 143 (Filley et al., 2017).
However, some strategies have been assessed in order to improve GBM’s response to immunotherapy. For example, Mathios and coworkers showed that local chemotherapy improved the response to anti-programmed cell death protein 1 (anti-PD1) - a checkpoint blockade immunotherapy - in mice by restoring T-cell function and its antitumor activity, while systemic chemotherapy is immunosuppressive (Mathios et al., 2016). Moreover, a recently identified hypermutated GBM subtype, harboring mutations in the exonuclease domain of the polymerase epsilon gene (POLE) and/or biallelic MMR deficiency (bMMRD) (Erson-Omay et al., 2015), respond better to immunotherapies such as immune checkpoint inhibition and neoantigen loads (Bouffet et al., 2016). Hypermutated GBM cells present defective proofreading during DNA replication, which increases mutation rates, stimulating the arising of neoantigens that could activate T-cells and consequently augment the chances of immunotherapy effectiveness (Bouffet et al., 2016). Furthermore, the hypermutated phenotype could also cause the arising of key mutations that provide new tumor competencies, opening the possibility to exploit their synthetic lethality potential and improve treatment response through a combination of radio-chemo-immunotherapy. These evidences highlight the great potential of a combined multi-therapy strategy to overcome GBM’s resistance to the currently available therapies, yet studies in further detail would be necessary to determine the best strategies and protocols.
Moreover, non-classical approaches have been prospected and demonstrated great potential to improve GBM treatment. Tumor treating fields (TTF) is an interesting approach that has been employed in clinics since 2004. TTF modality employs alternating electric fields at an intermediate frequency, from 100 to 300 kHz, to inhibit tumor cell proliferation. It was observed that TTF could suppress invasion and migration of U87 and U373 GBM cell lines, and angiogenesis in endothelial cell lines by downregulation of PI3K/AKT/NF-kB pathway. Impairing of epithelial to mesenchymal transition (EMT) was also observed after TTF, which promoted both increased E-cadherin and diminished vimentin expression (Kim et al., 2016). Additionally, patients treated with TTF presented enhanced overall survival when compared to those who received conventional therapy only (Rulseh et al., 2012).
Another alternative approach of great potential for GBM treatment is the photodynamic therapy (PDT). PDT is based on the administration of a photosensitizing agent (PS), topic or systemically, to the patient with subsequent local exposure to a light source of a specific wavelength, leading to the formation of highly cytotoxic reactive oxygen species. Among its several advantages, worthy of note are the minimum systemic adverse effects due to its double selectivity (de Freitas et al., 2017). In the past decade, the application of PDT to treat GBM has been investigated and proven to be a promising approach, both in vitro and in vivo (Chakrabarti et al., 2013; Akimoto, 2016). In fact, the Japanese health insurance coverage has introduced, since September 2013, the PDT as a new intraoperative therapy with an indication for malignant brain tumors (Akimoto, 2016).
When applied during surgery, PDT has a double action. Firstly, it helps the surgeon to locate the whole tumor through the fluorescence emission of the photosensitizers (Fluorescence Image Guided Surgical Resection), providing higher rates of complete resection (Eljamel, 2015). Secondly, PDT increases the success of tumor ablation by eliminating its roots while normal brain tissue is spared due to a preferable accumulation of PSs in tumor cells, improving progression-free and overall survival of patients (Akimoto, 2016). However, several variables were found among the studies, including light dose and delivery method, photosensitizer utilized, and sensitivity of different tumor types, evidencing the requirement of standardized clinical trials to effectively evaluate PDT as a treatment option for GBM patients (Quirk et al., 2015). Other modern attempts under study include the use of gamma knife surgery (Skeie et al., 2013), gene therapy (Wilson et al., 2014), modulation of the immune system (Garris and Pittet, 2013), molecular imaging (Jarzabek et al., 2013) and nanoparticles (Tivnan et al., 2017).
Concluding remarks
Despite all progress made over the last few years concerning both molecular knowledge and novel therapeutic methods, GBM is still poorly responsive to current treatments and extremely lethal. To improve positive impact on patients’ outcome and overall survival, we believe that drug resistance mechanisms and potential novel therapeutic targets should be deeply assessed. Advances in the characterization of CSC are also mandatory for comprehension of resistance and recurrence. To achieve this aim, the understanding of molecular features within the cellular and tissue contexts is imperative. Certainly, each tumor is a unique entity that relies on complex signaling networks and specific competencies working cooperatively to sustain its aggressive phenotype. The research community is only entering a new era of exploring the huge amount of genetic information now available. Hopefully, we expect that in the upcoming years there will be an integrated and meaningful knowledge about the fast progression and high resistance of GBM, allowing the identification of weaknesses of this devastating disease. In this scenario, the appropriate exploration of good molecular targets and adjuvant compounds, allied to refined diagnoses approaches, should permit the development of personalized therapies and establishment of adequate health care for GBM patients.
Acknowledgments
VV was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo - FAPESP (grant #2013/13465-1; grant #2018/05018-9) and by Center for Cell-Based Therapy (CEPID/FAPESP; grant #2013/08135-2). JFS and RBS received fellowships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001. LMF received fellowship from FAPESP (grant # 2014/24581-5). CRF was supported by FAPESP (grant # 2016/05345-4). The authors acknowledge Douglas Elias Santos for sharing his abilities and developing the illustrations for this review.
Footnotes
Associate Editor: Nicholas Hoch
Conflict of Interest
The authors declare no potential conflicts of interest.
Author contributions
JFdS conceived and designed the review, wrote the manuscript and put together the others authors’ contributions, RBS contributed on the “DNA repair and resistance to treatment” section, LMdF contributed on the “New therapies and future prospective” section, CRF and VV made valuable suggestions and corrected the manuscript, all authors read and approved the final version.
References
- Agnihotri S, Gajadhar AS, Ternamian C, Gorlia T, Diefes KL, Mischel PS, Kelly J, McGown G, Thorncroft M, Carlson BL, et al. Alkylpurine-DNA-N-glycosylase confers resistance to temozolomide in xenograft models of glioblastoma multiforme and is associated with poor survival in patients. J Clin Invest. 2012;122:253–266. doi: 10.1172/JCI59334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto J. Photodynamic therapy for malignant brain tumors. Neurol Med Chir. 2016;56:151–157. doi: 10.2176/nmc.ra.2015-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annovazzi L, Caldera V, Mellai M, Riganti C, Battaglia L, Chirio D, Melcarne A, Schiffer D. The DNA damage/repair cascade in glioblastoma cell lines after chemotherapeutic agent treatment. Int J Oncol. 2015;46:2299–2308. doi: 10.3892/ijo.2015.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins RJ, Ng W, Stylli SS, Hovens CM, Kaye AH. Repair mechanisms help glioblastoma resist treatment. J Clin Neurosci. 2015;22:14–20. doi: 10.1016/j.jocn.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Awan M, Liu S, Sahgal A, Das S, Chao ST, Chang EL, Knisely JP, Redmond K, Sohn JW, Machtay, et al. Extra-CNS metastasis from glioblastoma: a rare clinical entity. Expert Rev Anticancer Ther. 2015;15:545–552. doi: 10.1586/14737140.2015.1028374. [DOI] [PubMed] [Google Scholar]
- Barazzuol L, Jena R, Burnet NG, Meira LB, Jeynes JC, Kirkby KJ, Kirkby NF. Evaluation of poly (ADP-ribose) polymerase inhibitor ABT-888 combined with radiotherapy and temozolomide in glioblastoma. Radiat Oncol. 2013;8:65. doi: 10.1186/1748-717X-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Hamerlik P, Stockhausen MT, Ehrmann J, Hlobilkova A, Laursen H, Kalita O, Kolar Z, Poulsen HS, Broholm H, et al. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene. 2010;29:5095–5102. doi: 10.1038/onc.2010.249. [DOI] [PubMed] [Google Scholar]
- Beauchesne P. Extra-neural metastases of malignant gliomas: myth or reality? Cancers (Basel) 2011;3:461–477. doi: 10.3390/cancers3010461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berte N, Piee-Staffa A, Piecha N, Wang M, Borgmann K, Kaina B, Nikolova T. Targeting homologous recombination by pharmacological inhibitors enhances the killing response of glioblastoma cells treated with alkylating drugs. Mol Cancer Ther. 2016;15:2665–2678. doi: 10.1158/1535-7163.MCT-16-0176. [DOI] [PubMed] [Google Scholar]
- Bian EB, Ma CC, He XJ, Wang C, Zong G, Wang HL, Zhao B. Epigenetic modification of miR-141 regulates SKA2 by an endogenous ‘sponge’ HOTAIR in glioma. Oncotarget. 2016;7:15. doi: 10.18632/oncotarget.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binabaj MM, Bahrami A, ShahidSales S, Joodi M, Joudi Mashhad M, Hassanian SM, Anvari K, Avan A. The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials. J Cell Physiol. 2018;233:378–386. doi: 10.1002/jcp.25896. [DOI] [PubMed] [Google Scholar]
- Bobola MS, Blank A, Berger MS, Stevens BA, Silber JR. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin Cancer Res. 2001;7:3510–3518. [PubMed] [Google Scholar]
- Boccard SG, Marand SV, Geraci S, Pycroft L, Berger FR, Pelletier LA. Inhibition of DNA-repair genes Ercc1 and Mgmt enhances temozolomide efficacy in gliomas treatment: a pre-clinical study. Oncotarget. 2015;6:12. doi: 10.18632/oncotarget.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffet E, Larouche V, Campbell BB, Merico D, de Borja R, Aronson M, Durno C, Krueger J, Cabric V, Ramaswamy V, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34:2206–2211. doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broustas CG, Lieberman HB. DNA damage response genes and the development of cancer metastasis. Radiat Res. 2014;181:111–130. doi: 10.1667/RR13515.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporali S, Falcinelli S, Starace G, Russo MT, Bonmassar E, Jiricny J, D’Atri S. DNA damage induced by temozolomide signals to both ATM and ATR: role of the mismatch repair system. Mol Pharmacol. 2004;66:478–491. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- Carruthers RD, Ahmed SU, Ramachandran S, Strathdee K, Kurian KM, Hedley A, Gomez-Roman N, Kalna G, Neilson M, Gilmour L, et al. Replication stress drives constitutive activation of the DNA damage response and radioresistance in glioblastoma stem-like cells. Cancer Res. 2018;78:5060–5071. doi: 10.1158/0008-5472.CAN-18-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha HJ, Yim H. The accumulation of DNA repair defects is the molecular origin of carcinogenesis. Tumour Biol. 2013;34:3293–3302. doi: 10.1007/s13277-013-1038-y. [DOI] [PubMed] [Google Scholar]
- Chakrabarti M, Banik NL, Ray SK. Photofrin based photodynamic therapy and miR-99a transfection inhibited FGFR3 and PI3K/Akt signaling mechanisms to control growth of human glioblastoma In vitro and in vivo. PLoS One. 2013;8:e55652. doi: 10.1371/journal.pone.0055652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JS, Yeh RF, Wiencke JK, Wiemels JL, Smirnov I, Pico AR, Tihan T, Patoka J, Miike R, Sison JD, et al. Pathway analysis of single-nucleotide polymorphisms potentially associated with glioblastoma multiforme susceptibility using random forests. Cancer Epidemiol Biomarkers Prev. 2008;17:1368–1373. doi: 10.1158/1055-9965.EPI-07-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni F, Torrente Y. The new challenge of stem cell: brain tumour therapy. Cancer Lett. 2008;272:1–11. doi: 10.1016/j.canlet.2008.05.046. [DOI] [PubMed] [Google Scholar]
- Cortese R, Kwan A, Lalonde E, Bryzgunova O, Bondar A, Wu Y, Gordevicius J, Park M, Oh G, Kaminsky Z, et al. Epigenetic markers of prostate cancer in plasma circulating DNA. Hum Mol Genet. 2012;21:3619–3631. doi: 10.1093/hmg/dds192. [DOI] [PubMed] [Google Scholar]
- Custodio AC, Almeida LO, Pinto GR, Santos MJ, Almeida JR, Clara CA, Rey JA, Casartelli C. Variation in DNA repair gene XRCC3 affects susceptibility to astrocytomas and glioblastomas. Genet Mol Res. 2012;11:332–339. doi: 10.4238/2012.February.10.4. [DOI] [PubMed] [Google Scholar]
- de Freitas LM, Serafim RB, de Sousa JF, Moreira TF, Dos Santos CT, Baviera AM, Valente V, Soares CP, Fontana CR. Photodynamic therapy combined to cisplatin potentiates cell death responses of cervical cancer cells. BMC Cancer. 2017;17:123. doi: 10.1186/s12885-017-3075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa JF, Torrieri R, Serafim RB, Di Cristofaro LF, Escanfella FD, Ribeiro R, Zanette DL, Paco-Larson ML, da Silva WA, Jr., Tirapelli DP, et al. Expression signatures of DNA repair genes correlate with survival prognosis of astrocytoma patients. Tumour Biol. 2017;39:1010428317694552. doi: 10.1177/1010428317694552. [DOI] [PubMed] [Google Scholar]
- de Tayrac M, Aubry M, Saikali S, Etcheverry A, Surbled C, Guenot F, Galibert MD, Hamlat A, Lesimple T, Quillien V, et al. A 4-gene signature associated with clinical outcome in high-grade gliomas. Clin Cancer Res. 2011;17:317–327. doi: 10.1158/1078-0432.CCR-10-1126. [DOI] [PubMed] [Google Scholar]
- Eich M, Roos WP, Nikolova T, Kaina B. Contribution of ATM and ATR to the resistance of glioblastoma and malignant melanoma cells to the methylating anticancer drug temozolomide. Mol Cancer Ther. 2013;12:2529–2540. doi: 10.1158/1535-7163.MCT-13-0136. [DOI] [PubMed] [Google Scholar]
- Eljamel S. 5-ALA fluorescence image guided resection of glioblastoma multiforme: A meta-analysis of the literature. Int J Mol Sci. 2015;16:10443–10456. doi: 10.3390/ijms160510443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erson-Omay EZ, Caglayan AO, Schultz N, Weinhold N, Omay SB, Ozduman K, Koksal Y, Li J, Serin Harmanci A, Clark V, et al. Somatic POLE mutations cause an ultramutated giant cell high-grade glioma subtype with better prognosis. Neuro Oncol. 2015;17:1356–1364. doi: 10.1093/neuonc/nov027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- Fiano V, Trevisan M, Trevisan E, Senetta R, Castiglione A, Sacerdote C, Gillio-Tos A, De Marco L, Grasso C, Magistrello M, et al. MGMT promoter methylation in plasma of glioma patients receiving temozolomide. J Neurooncol. 2014;117:347–357. doi: 10.1007/s11060-014-1395-4. [DOI] [PubMed] [Google Scholar]
- Filley AC, Henriquez M, Dey M. Recurrent glioma clinical trial, CheckMate-143: the game is not over yet. Oncotarget. 2017;8:91779–91794. doi: 10.18632/oncotarget.21586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosmark S, Hellwege S, Dahlrot RH, Jensen KL, Derand H, Lohse J, Sorensen MD, Hansen S, Kristensen BW. APNG as a prognostic marker in patients with glioblastoma. PLoS One. 2017;12:e0178693. doi: 10.1371/journal.pone.0178693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi S, Tomei S, Mazzanti CM, Lessi F, Aretini P, La Ferla M, De Gregorio V, Pasqualetti F, Zavaglia K, Bevilacqua G, et al. Association between RAD 51 rs1801320 and susceptibility to glioblastoma. J Neurooncol. 2016;126:265–270. doi: 10.1007/s11060-015-1974-z. [DOI] [PubMed] [Google Scholar]
- Gameiro SR, Jammeh ML, Wattenberg MM, Tsang KY, Ferrone S, Hodge JW. Radiation-induced immunogenic modulation of tumor enhances antigen processing and calreticulin exposure, resulting in enhanced T-cell killing. Oncotarget. 2014;5:403–416. doi: 10.18632/oncotarget.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris C, Pittet MJ. Therapeutically reeducating macrophages to treat GBM. Nat Med. 2013;19:1207–1208. doi: 10.1038/nm.3355. [DOI] [PubMed] [Google Scholar]
- Gilbertson RJ, Rich JN. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7:733–736. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- Gobin M, Nazarov PV, Warta R, Timmer M, Reifenberger G, Felsberg J, Vallar L, Chalmers AJ, Herold-Mende CC, Goldbrunner R, et al. A DNA repair and cell cycle gene expression signature in primary and recurrent glioblastoma: prognostic value and clinical implications. Cancer Res. 2019;79:1226–1238. doi: 10.1158/0008-5472.CAN-18-2076. [DOI] [PubMed] [Google Scholar]
- Golmard L, Caux-Moncoutier V, Davy G, Al Ageeli E, Poirot B, Tirapo C, Michaux D, Barbaroux C, d’Enghien CD, Nicolas A, et al. Germline mutation in the RAD51B gene confers predisposition to breast cancer. BMC Cancer. 2013;13:484. doi: 10.1186/1471-2407-13-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr., Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Mladek AC, Carlson BL, Boakye-Agyeman F, Bakken KK, Kizilbash SH, Schroeder MA, Reid J, Sarkaria JN. Discordant in vitro and in vivo chemopotentiating effects of the PARP inhibitor veliparib in temozolomide-sensitive versus -resistant glioblastoma multiforme xenografts. Clin Cancer Res. 2014;20:3730–3741. doi: 10.1158/1078-0432.CCR-13-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SK, Smith EJ, Mladek AC, Tian S, Decker PA, Kizilbash SH, Kitange GJ, Sarkaria JN. PARP inhibitors for sensitization of alkylation chemotherapy in glioblastoma: Impact of blood-brain barrier and molecular heterogeneity. Front Oncol. 2018;8:670. doi: 10.3389/fonc.2018.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson AS, Abramenkovs A, Stenerlow B. Suppression of DNA-dependent protein kinase sensitize cells to radiation without affecting DSB repair. Mutat Res. 2014;769:1–10. doi: 10.1016/j.mrfmmm.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, De Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:8. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis. 2010;31:955–960. doi: 10.1093/carcin/bgq064. [DOI] [PubMed] [Google Scholar]
- Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol. 2011;5:387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wang Q, Wang Y, Chen J, Li P, Han M. Holliday junction-recognizing protein promotes cell proliferation and correlates with unfavorable clinical outcome of hepatocellular carcinoma. Onco Targets Ther. 2017;10:2601–2607. doi: 10.2147/OTT.S127738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Huang G, Sadanandam A, Gu S, Lenburg ME, Pai M, Bayani N, Blakely EA, Gray JW, Mao JH. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010;12:R18. doi: 10.1186/bcr2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, Greenman C, Edkins S, Bignell G, Davies H, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter K. The role of individual inheritance in tumor progression and metastasis. J Mol Med. 2015;93:719–725. doi: 10.1007/s00109-015-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarzabek MA, Sweeney KJ, Evans RL, Jacobs AH, Stupp R, O’Brien D, Berger MS, Prehn JH, Byrne AT. Molecular imaging in the development of a novel treatment paradigm for glioblastoma (GBM): an integrated multidisciplinary commentary. Drug Discov Today. 2013;18:1052–1066. doi: 10.1016/j.drudis.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Jiang P, Mukthavaram R, Chao Y, Bharati IS, Fogal V, Pastorino S, Cong X, Nomura N, Gallagher M, Abbasi T, et al. Novel anti-glioblastoma agents and therapeutic combinations identified from a collection of FDA approved drugs. J Transl Med. 2014;12:13. doi: 10.1186/1479-5876-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Xie L, Xie CH, Zhou YF. Aberrant DNA methylation of MGMT and hMLH1 genes in prediction of gastric cancer. Genet Mol Res. 2014;13:4140–4145. doi: 10.4238/2014.May.30.9. [DOI] [PubMed] [Google Scholar]
- Johannessen TC, Hasan-Olive MM, Zhu H, Denisova O, Grudic A, Latif MA, Saed H, Varughese JK, Rosland GV, Yang N, et al. Thioridazine inhibits autophagy and sensitizes glioblastoma cells to temozolomide. Int J Cancer. 2019;144:1735–1745. doi: 10.1002/ijc.31912. [DOI] [PubMed] [Google Scholar]
- Jones DT, Gronych J, Lichter P, Witt O, Pfister SM. MAPK pathway activation in pilocytic astrocytoma. Cell Mol Life Sci. 2012;69:12. doi: 10.1007/s00018-011-0898-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Hutter B, Jäger N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DA, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:6. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jue TR, Nozue K, Lester AJ, Joshi S, Schroder LB, Whittaker SP, Nixdorf S, Rapkins RW, Khasraw M, McDonald KL. Veliparib in combination with radiotherapy for the treatment of MGMT unmethylated glioblastoma. J Transl Med. 2017;15:61. doi: 10.1186/s12967-017-1164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Rosselli F, Lazar V, Winnepenninckx V, Mansuet-Lupo A, Dessen P, van den Oord JJ, Spatz A, Sarasin A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27:565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- Kernohan JW, Mabon RF. A simplified classification of the gliomas. Proc Staff Meet Mayo Clin. 1949;24:71–75. [PubMed] [Google Scholar]
- Kim EH, Song HS, Yoo SH, Yoon M. Tumor treating fields inhibit glioblastoma cell migration, invasion and angiogenesis. Oncotarget. 2016;7:65125–65136. doi: 10.18632/oncotarget.11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Wilson DM., 3rd Overview of base excision repair biochemistry. Curr Mol Pharmacol. 2012;5:3–13. doi: 10.2174/1874467211205010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HO, Brend T, Payne HL, Wright A, Ward TA, Patel K, Egnuni T, Stead LF, Patel A, Wurdak, et al. RAD51 is a selective DNA repair target to radiosensitize glioma stem cells. Stem Cell Rep. 2017;8:125–139. doi: 10.1016/j.stemcr.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschmann C, Calinescu AA, Nunez FJ, Mackay A, Fazal-Salom J, Thomas D, Mendez F, Kamran N, Dzaman M, Mulpuri L, et al. ATRX loss promotes tumor growth and impairs nonhomologous end joining DNA repair in glioma. Sci Transl Med. 2016;8:328ra328. doi: 10.1126/scitranslmed.aac8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kun S, Duan Q, Liu G, Lu JM. Prognostic value of DNA repair genes based on stratification of glioblastomas. Oncotarget. 2017;8:58222–58230. doi: 10.18632/oncotarget.17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, Park J, Kim DH. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res. 2009;15:6185–6191. doi: 10.1158/1078-0432.CCR-09-0111. [DOI] [PubMed] [Google Scholar]
- Lesueur P, Chevalier F, El-Habr EA, Junier MP, Chneiweiss H, Castera L, Muller E, Stefan D, Saintigny Y. Radiosensitization effect of Talazoparib, a Parp inhibitor, on glioblastoma stem cells exposed to low and high linear energy transfer radiation. Sci Rep. 2018;8:3664. doi: 10.1038/s41598-018-22022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li X, Meng Q, Khan AQ, Chen X. Increased expression of Holliday junction-recognizing protein (HJURP) as an independent prognostic biomarker in advanced-stage serous ovarian carcinoma. Med Sci Monit. 2018;24:3050–3055. doi: 10.12659/MSM.906647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang L, Liu Y, Sun C, Zhang H, Miao G, Di CX, Zhou X, Zhou R, Wang Z. DNA-PKcs deficiency inhibits glioblastoma cell-derived angiogenesis after ionizing radiation. J Cell Physiol. 2015;230:1094–1103. doi: 10.1002/jcp.24841. [DOI] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:12. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, Von Deimling A, Aldape K, Brat D, Collins VP, Eberhart C. International Society Of Neuropathology-Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014;24:6. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YH, Mentlein R, Knerlich F, Kruse ML, Mehdorn HM, Held-Feindt J. Expression of stem cell markers in human astrocytomas of different WHO grades. J Neurooncol. 2008;86:31–45. doi: 10.1007/s11060-007-9439-7. [DOI] [PubMed] [Google Scholar]
- Maher EA, Brennan C, Wen PY, Durso L, Ligon KL, Richardson A, Khatry D, Feng B, Sinha R, Louis DN, et al. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res. 2006;66:11502–11513. doi: 10.1158/0008-5472.CAN-06-2072. [DOI] [PubMed] [Google Scholar]
- Mathios D, Kim JE, Mangraviti A, Phallen J, Park CK, Jackson CM, Garzon-Muvdi T, Kim E, Theodros D, Polanczyk M, et al. Anti-PD-1 antitumor immunity is enhanced by local and abrogated by systemic chemotherapy in GBM. Sci Transl Med. 2016;8:370ra180. doi: 10.1126/scitranslmed.aag2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell JA, Johnson SP, McLendon RE, Lister DW, Horne KS, Rasheed A, Quinn JA, Ali-Osman F, Friedman AH, Modrich PL, et al. Mismatch repair deficiency does not mediate clinical resistance to temozolomide in malignant glioma. Clin Cancer Res. 2008;14:4859–4868. doi: 10.1158/1078-0432.CCR-07-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell R, Jackson CM, Lim M. Clinical trials investigating immune checkpoint blockade in glioblastoma. Curr Treat Options Oncol. 2017;18:51. doi: 10.1007/s11864-017-0492-y. [DOI] [PubMed] [Google Scholar]
- McEllin B, Camacho CV, Mukherjee B, Hahm B, Tomimatsu N, Bachoo RM, Burma S. PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res. 2010;70:5457–5464. doi: 10.1158/0008-5472.CAN-09-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill KA. Epidemiology of brain tumors. Neurol Clin. 2016;34:981–998. doi: 10.1016/j.ncl.2016.06.014. [DOI] [PubMed] [Google Scholar]
- Mirzoeva OK, Kawaguchi T, Pieper RO. The Mre11/Rad50/Nbs1 complex interacts with the mismatch repair system and contributes to temozolomide-induced G2 arrest and cytotoxicity. Mol Cancer Ther. 2006;5:2757–2766. doi: 10.1158/1535-7163.MCT-06-0183. [DOI] [PubMed] [Google Scholar]
- Nagel ZD, Kitange GJ, Gupta SK, Joughin BA, Chaim IA, Mazzucato P, Lauffenburger DA, Sarkaria JN, Samson LD. DNA repair capacity in multiple pathways predicts chemoresistance in glioblastoma multiforme. Cancer Res. 2017;77:198–206. doi: 10.1158/0008-5472.CAN-16-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JC, Neto, Ikoma MM, Carvalho KC, Vassallo J, De Brot M, Gobbi H, Soares FA, Rocha RM. MGMT and PTEN as potential prognostic markers in breast cancer. Exp Mol Pathol. 2012;92:6. doi: 10.1016/j.yexmp.2011.09.019. [DOI] [PubMed] [Google Scholar]
- Nissar S, Sameer AS, Rasool R, Rashid F. DNA repair gene—XRCC1 in relation to genome instability and role in colorectal carcinogenesis. Oncol Res Treat. 2014;37:418–422. doi: 10.1159/000364898. [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:12. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19:764–772. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- Ostrow KL, Hoque MO, Loyo M, Brait M, Greenberg A, Siegfried JM, Grandis JR, Gaither Davis A, Bigbee WL, Rom W, et al. Molecular analysis of plasma DNA for the early detection of lung cancer by quantitative methylation-specific PCR. Clin Cancer Res. 2010;16:3463–3472. doi: 10.1158/1078-0432.CCR-09-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M, Hori T, Toyoda A, Misu S, Monma N, Ikeo K, Obuse C, Fujiyama A, Fukagawa T. HJURP is involved in the expansion of centromeric chromatin. Mol Biol Cell. 2015;26:2742–2754. doi: 10.1091/mbc.E15-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Yu HQ, Zhang Y, Ding LJ, Zhao DH, Lv P, Wang WY, Xu Y. A comprehensive meta-analysis of genetic associations between key polymorphic loci in DNA repair genes and gliomar risk. Mol Neurobiol. 2016;54:1314–1325. doi: 10.1007/s12035-016-9725-5. [DOI] [PubMed] [Google Scholar]
- Quirk BJ, Brandal G, Donlon S, Vera JC, Mang TS, Foy AB, Lew SM, Girotti AW, Jogal S, LaViolette PS, et al. Photodynamic therapy (PDT) for malignant brain tumors—where do we stand? Photodiagnosis Photodyn Ther. 2015;12:530–544. doi: 10.1016/j.pdpdt.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Quiros S, Roos WP, Kaina B. Rad51 and BRCA2—New molecular targets for sensitizing glioma cells to alkylating anticancer drugs. PLoS One. 2011;6:e27183. doi: 10.1371/journal.pone.0027183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizer JJ, Giglio P, Hu J, Groves M, Merrell R, Conrad C, Phuphanich S, Puduvalli VK, Loghin M, Paleologos N, et al. A phase II study of bevacizumab and erlotinib after radiation and temozolomide in MGMT unmethylated GBM patients. J Neurooncol. 2016;126:185–192. doi: 10.1007/s11060-015-1958-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reon BJ, Anaya J, Zhang Y, Mandell J, Purow B, Abounader R, Dutta A. Expression of lncRNAs in low-grade gliomas and glioblastoma multiforme: An in silico analysis. PLoS Med. 2016;13:e1002192. doi: 10.1371/journal.pmed.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–248. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Roos WP, Krumm A. The multifaceted influence of histone deacetylases on DNA damage signalling and DNA repair. Nucleic Acids Res. 2016;44:10017–10030. doi: 10.1093/nar/gkw922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulseh AM, Keller J, Klener J, Sroubek J, Dbaly V, Syrucek M, Tovarys F, Vymazal J. Long-term survival of patients suffering from glioblastoma multiforme treated with tumor-treating fields. World J Surg Oncol. 2012;10:220. doi: 10.1186/1477-7819-10-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle-Thiele D, Head R, Cosgrove L, Martin JH. Repurposing some older drugs that cross the blood-brain barrier and have potential anticancer activity to provide new treatment options for glioblastoma. Br J Clin Pharmacol. 2016;81:199–209. doi: 10.1111/bcp.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JS, Brady K, Burgan WE, Cerra MA, Oswald KA, Camphausen K, Tofilon PJ. Gleevec-mediated inhibition of Rad51 expression and enhancement of tumor cell radiosensitivity. Cancer Res. 2003;63:7377–7383. [PubMed] [Google Scholar]
- Sarkaria JN, Hu LS, Parney IF, Pafundi DH, Brinkmann DH, Laack NN, Giannini C, Burns TC, Kizilbash SH, Laramy JK, et al. Is the blood-brain barrier really disrupted in all glioblastomas? A critical assessment of existing clinical data. Neuro Oncol. 2018;20:184–191. doi: 10.1093/neuonc/nox175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer H. Structural development in gliomas. Am J Cancer. 1938;34:18. [Google Scholar]
- Sevenich L. Turning “cold” into “hot” tumors - opportunities and challenges for radio-immunotherapy against primary and metastatic brain cancers. Front Oncol. 2019;9:163. doi: 10.3389/fonc.2019.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SC, Giampieri S, Worku M, Alcaide-German M, Sioftanos G, Bourne S, Lio KI, Shaked-Rabi M, Martindale C. Rad51 inhibition is an effective means of targeting DNA repair in glioma models and CD133+ tumor-derived cells. Neuro Oncol. 2011;13:487–499. doi: 10.1093/neuonc/nor010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegelin MD, Raskett CM, Gilbert CA, Ross AH, Altieri DC. Sorafenib exerts anti-glioma activity in vitro and in vivo. Neurosci Lett. 2010;478:165–170. doi: 10.1016/j.neulet.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeie BS, Wang J, Dodoo E, Heggdal JI, Gronli J, Sleire L, Bragstad S, Ganz JC, Chekenya M, Mork S, et al. Gamma knife surgery as monotherapy with clinically relevant doses prolongs survival in a human GBM xenograft model. Biomed Res Int. 2013;2013:139674. doi: 10.1155/2013/139674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Brennan CW, Helmy K, Huse JT, Petrini JH, Holland EC. Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell. 2010;18:619–629. doi: 10.1016/j.ccr.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Holland EC. DNA damage response and growth factor signaling pathways in gliomagenesis and therapeutic resistance. Cancer Res. 2011;71:5945–5949. doi: 10.1158/0008-5472.CAN-11-1245. [DOI] [PubMed] [Google Scholar]
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- Stupp R, Mason WP, Van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:9. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Sun Q, Pei C, Li Q, Dong T, Dong Y, Xing W, Zhou P, Gong Y, Zhen Z, Gao Y, et al. Up-regulation of MSH6 is associated with temozolomide resistance in human glioblastoma. Biochem Biophys Res Commun. 2018;496:1040–1046. doi: 10.1016/j.bbrc.2018.01.093. [DOI] [PubMed] [Google Scholar]
- Sun Y, Li S, Shen K, Ye S, Cao D and Yang J. DAPK1, MGMT and RARB promoter methylation as biomarkers for high-grade cervical lesions. Int J Clin Exp Pathol. 2015;8:14939–14945. [PMC free article] [PubMed] [Google Scholar]
- Takao M, Oohata Y, Kitadokoro K, Kobayashi K, Iwai S, Yasui A, Yonei S, Zhang QM. Human Nei-like protein NEIL3 has AP lyase activity specific for single-stranded DNA and confers oxidative stress resistance in Escherichia coli mutant. Genes Cells. 2009;14:261–270. doi: 10.1111/j.1365-2443.2008.01271.x. [DOI] [PubMed] [Google Scholar]
- Tentori L, Portarena I, Torino F, Scerrati M, Navarra P, Graziani G. Poly(ADP-ribose) polymerase inhibitor increases growth inhibition and reduces G(2)/M cell accumulation induced by temozolomide in malignant glioma cells. Glia. 2002;40:44–54. doi: 10.1002/glia.10113. [DOI] [PubMed] [Google Scholar]
- Thakkar JP, Dolecek TA, Horbinski C, Ostrom QT, Lightner DD, Barnholtz-Sloan JS, Villano JL. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomarkers Prev. 2014;23:1985–1996. doi: 10.1158/1055-9965.EPI-14-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivnan A, Heilinger T, Ramsey J, O’Connor G, Pokorny J, Sarkaria J, Stringer B, Day B, Boyd A, Kim E, et al. Anti-GD2-ch14.18/CHO coated nanoparticles mediate glioblastoma (GBM)-specific delivery of the aromatase inhibitor, Letrozole, reducing proliferation, migration and chemoresistance in patient-derived GBM tumor cells. Oncotarget. 2017;8:16605–16620. doi: 10.18632/oncotarget.15073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triscott J, Lee C, Hu K, Fotovati A, Berns R, Pambid M, Luk M, Kast RE, Kong E, Toyota E, et al. Disulfiram, a drug widely used to control alcoholism, suppresses the self-renewal of glioblastoma and over-rides resistance to temozolomide. Oncotarget. 2012;3:1112–1123. doi: 10.18632/oncotarget.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi RN, Wang XH, Jelezcova E, Goellner EM, Tang JB, Sobol RW. Human methyl purine DNA glycosylase and DNA polymerase beta expression collectively predict sensitivity to temozolomide. Mol Pharmacol. 2008;74:505–516. doi: 10.1124/mol.108.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KM, Sun Y, Ji P, Granberg KJ, Bernard B, Hu L, Cogdell DE, Zhou X, Yli-Harja O, Nykter M, et al. Genomically amplified Akt3 activates DNA repair pathway and promotes glioma progression. Proc Natl Acad Sci U S A. 2015;112:3421–3426. doi: 10.1073/pnas.1414573112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente V, Serafim RB, de Oliveira LC, Adorni FS, Torrieri R, Tirapelli DP, Espreafico EM, Oba-Shinjo SM, Marie SK, Paco-Larson ML, et al. Modulation of HJURP (Holliday Junction-Recognizing Protein) levels is correlated with glioblastoma cells survival. PLoS One. 2013;8:e62200. doi: 10.1371/journal.pone.0062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente V, Teixeira SA, Neder L, Okamoto OK, Oba-Shinjo SM, Marie SK, Scrideli CA, Paco-Larson ML, Carlotti CG., Jr Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. BMC Mol Biol. 2009;10:17. doi: 10.1186/1471-2199-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinjamuri M, Adumala RR, Altaha R, Hobbs GR, Crowell EB., Jr Comparative analysis of temozolomide (TMZ) versus 1,3-bis (2-chloroethyl)-1 nitrosourea (BCNU) in newly diagnosed glioblastoma multiforme (GBM) patients. J Neurooncol. 2009;91:221–225. doi: 10.1007/s11060-008-9702-6. [DOI] [PubMed] [Google Scholar]
- Westermark UK, Lindberg N, Roswall P, Brasater D, Helgadottir HR, Hede SM, Zetterberg A, Jasin M, Nister M, Uhrbom L. RAD51 can inhibit PDGF-B-induced gliomagenesis and genomic instability. Neuro Oncol. 2011;13:1277–1287. doi: 10.1093/neuonc/nor131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal M, Hilt DC, Bortey E, Delavault P, Olivares R, Warnke PC, Whittle IR, Jaaskelainen J, Ram Z. A phase 3 trial of local chemotherapy with biodegradable carmustine (BCNU) wafers (Gliadel wafers) in patients with primary malignant glioma. Neuro Oncol. 2003;5:79–88. doi: 10.1215/S1522-8517-02-00023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TA, Karajannis MA, Harter DH. Glioblastoma multiforme: State of the art and future therapeutics. Surg Neurol Int. 2014;5:64. doi: 10.4103/2152-7806.132138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsching HG, Weller M. The role of molecular diagnostics in the management of patients with gliomas. Curr Treat Options Oncol. 2016;17:51. doi: 10.1007/s11864-016-0430-4. [DOI] [PubMed] [Google Scholar]
- Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46:444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurth R, Pattarozzi A, Gatti M, Bajetto A, Corsaro A, Parodi A, Sirito R, Massollo M, Marini C, Zona G, et al. Metformin selectively affects human glioblastoma tumor-initiating cell viability: A role for metformin-induced inhibition of Akt. Cell Cycle. 2013;12:145–156. doi: 10.4161/cc.23050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurth R, Barbieri F, Florio T. New molecules and old drugs as emerging approaches to selectively target human glioblastoma cancer stem cells. Biomed Res Int. 2014;2014:126586. doi: 10.1155/2014/126586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing WK, Shao C, Qi ZY, Yang C, Wang Z. The role of Gliadel wafers in the treatment of newly diagnosed GBM: a meta-analysis. Drug Des Devel Ther. 2015;9:3341–3348. doi: 10.2147/DDDT.S85943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Brown C, Buettner R, Hedvat M, Starr R, Scuto A, Schroeder A, Jensen M, Jove R. Sorafenib induces growth arrest and apoptosis of human glioblastoma cells through the dephosphorylation of signal transducers and activators of transcription 3. Mol Cancer Ther. 2010;9:953–962. doi: 10.1158/1535-7163.MCT-09-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap TA, Plummer R, Azad NS, Helleday T. The DNA damaging revolution: PARP inhibitors and beyond. Am Soc Clin Oncol Educ Book. 2019;39:185–195. doi: 10.1200/EDBK_238473. [DOI] [PubMed] [Google Scholar]
- Yip S, Miao J, Cahill DP, Iafrate AJ, Aldape K, Nutt CL, Louis DN. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15:4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Mizoguchi M, Hata N, Murata H, Hatae R, Amano T, Nakamizo A, Sasaki T. Complex DNA repair pathways as possible therapeutic targets to overcome temozolomide resistance in glioblastoma. Front Oncol. 2012;2:186. doi: 10.3389/fonc.2012.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeppernick F, Ahmadi R, Campos B, Dictus C, Helmke BM, Becker N, Lichter P, Unterberg A, Radlwimmer B, Herold-Mende CC. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res. 2008;14:123–129. doi: 10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- Zhang J, Stevens MF, Bradshaw TD. Temozolomide: Mechanisms of action, repair and resistance. Curr Mol Pharmacol. 2012;5:102–114. doi: 10.2174/1874467211205010102. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:10. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang W, Li B, Long L, Chen L, Huang Q, Liang ZQ. Knockdown of the DNA-dependent protein kinase catalytic subunit radiosensitizes glioma-initiating cells by inducing autophagy. Brain Res. 2011;1371:7–15. doi: 10.1016/j.brainres.2010.11.044. [DOI] [PubMed] [Google Scholar]