Abstract
Translational geroscience is an interdisciplinary field descended from basic gerontology that seeks to identify, validate, and clinically apply interventions to maximize healthy, disease-free lifespan. In this review, we describe a research pipeline for the identification and validation of lifespan extending interventions. Beginning in invertebrate model systems, interventions are discovered and then characterized using other invertebrate model systems (evolutionary translation), models of genetic diversity, and disease models. Vertebrate model systems, particularly mice, can then be utilized to validate interventions in mammalian systems. Collaborative, multi-site efforts, like the Interventions Testing Program (ITP), provide a key resource to assess intervention robustness in genetically diverse mice. Mouse disease models provide a tool to understand the broader utility of longevity interventions. Beyond mouse models, we advocate for studies in companion pets. The Dog Aging Project is an exciting example of translating research in dogs, both to develop a model system and to extend their healthy lifespan as a goal in itself. Finally, we discuss proposed and ongoing intervention studies in humans, unmet needs for validating interventions in humans, and speculate on how differences in survival among human populations may influence intervention efficacy.
Keywords: Translational geroscience, healthspan, longevity, aging, rapamycin, metformin, mTOR, companion animals
Introduction
What is Translational geroscience?
The hope of dramatically extending our lifespan has captivated humanity for millennia. Over the last two decades, the biology of aging has matured as a field of study and led to greater engagement and investment in aging as a biological problem that can be understood at the molecular level [1–3]. In addition to the functional decline and loss of vigor associated with age, a generalized increase in disease susceptibility is now regarded as a consequence of biological aging [4, 5]. Numerous chronic diseases manifest during aging. In fact, of the ten leading causes of mortality in high income countries (as of 2016, the last year with available data), eight have advanced age as their greatest predisposing factor [6]. Developing interventions that target the molecular mechanisms of aging (or “hallmarks of aging”, as they’re commonly referred) should not only add vigorous years to our lives, but also reduce overall human disease burden. Translational geroscience is an emerging, interdisciplinary field descended from basic gerontology that seeks to identify, validate, and clinically apply interventions to maximize healthy, disease-free lifespan [7–9].
Why drug aging?
In some ways, the secrets of healthy aging are not enigmatic. Proper diet with care to include necessary micronutrients, exercise, adequate sleep, and effective management of stress are all well-known and intuitive ways to add to our healthy years. An important question to address in light of this is why we should focus on developing interventions if lifestyle management alone is sufficient to extend healthy lifespan? One answer is that many people do not have the resources (either in terms of money, time, or both) to proactively invest in maintaining their health. For some, making sure there is enough to eat is a priority over eating healthy. Instead of exercise after a long day of work, many instead prioritize family time and relaxation. This situation is common, even in economically-thriving countries. Those people, whose limited resources keep them focused on day-to-day survival, are as equally deserving of living long, disease-free, lives as those with resources to invest in their long-term health. All of this is not to say that lifestyle choices should not be pursued, or even prioritized, as healthy aging strategies, only that pharmacological interventions that increase healthy lifespan are an option to address an important inequity that exists in human health.
Another reason to consider pharmacological intervention to maintain our health as we age comes from early successes in preclinical geroscience to identify compounds that can impressively extend lifespan in model systems. Several compounds are now known to extend lifespan across broad evolutionary distances [10–14]. For example, rapamycin, among the most promising current interventions, can extend lifespan in yeast, worms, flies, and multiple mouse models [15–20]. While the magnitude of effect varies between organisms and genetic backgrounds, experiments in genetically heterogeneous mice show average lifespan extension between 10–25% [19]. Applied to human populations, a 15% increase in average life expectancy at birth in the US would change from 78.8 (as of 2015, the last year with available data) to 90.6 years [21]. This would be a dramatic improvement in human survival. In addition to improving lifespan in wild type model systems, rapamycin also promotes extended lifespan in multiple mouse disease models [22, 23], including heart disease [24] and cancer models [25–28]. This bolsters the hypothesis that there are broad clinical applications for rapamycin and other mTOR inhibitors. In a first of its kind test, short-term rapamycin administration improved measures of cardiac function in pet dogs [29]. While no lifespan data are available for humans, the short-term treatment with mTOR inhibitors appear to broadly improve immune function in the elderly [30, 31].
In addition to rapamycin, multiple other compounds show promise as healthspan and lifespan extending interventions. Metformin, typically used to treat type II diabetes, extends lifespan in worms and mice [32, 33]. In humans, numerous meta analyses suggest an association between metformin use and lowered cancer incidence [34–42]. Further tests in non-diabetic individuals, like the proposed Targeting Aging with Metformin (TAME) study, will better establish metformin’s potential to broadly reduce cancer and other age-related disease incidence in humans [10]. Nicotinamide riboside (NR) and nicotinamide mononucleotide (NMN) are NAD(+) precursors that improve several features of aging, including muscle and cognitive function and vascular aging, and treatment with NR beginning at midlife is reported to increase lifespan in mice [43–45]. Senolytics, compounds that specifically target senescent cells for destruction, are important therapies that could increase lifespan and reduce age-related disease burden, particularly cancer and diseases driven by chronic inflammation [46–48]. Other compounds commonly used by humans, including caffeine, aspirin, and ibuprofen have successfully extended lifespan in model systems [12, 14, 49–51]. This raises the question: what other commonly consumed and FDA-regulated compounds alter physiology in such a way as to promote longevity?
How do we identify and validate lifespan promoting compounds?
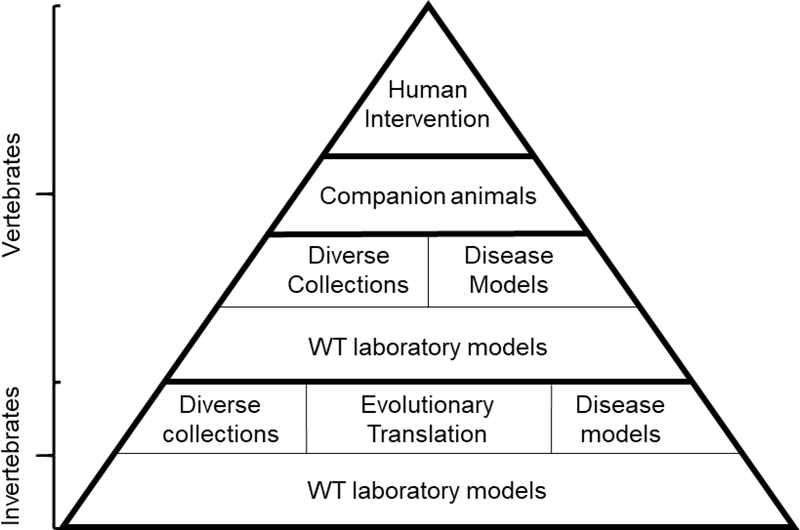
What other molecules extend healthspan and lifespan? What combinations of molecules can be designed such that they target multiple pathways associated with aging and increased disease risk? Most importantly, how does individual genetic variation influence success of these interventions? We are only beginning to explore this “intervention space” and understand what is possible with regards to healthspan and lifespan intervention. In addition to broader options with regard to mTOR inhibition, compounds that target other pathways known to regulate aging, or stated more precisely, other nodes in the “aging network”, are critical to develop and validate. Breakthroughs in interventions that extend healthy longevity can lead the way to a precision medicine like approach to maximize individual health by utilizing combinatorial treatment strategies coupled with individualized dosing based on genotype. In what follows, we describe a translational geroscientific workflow to identify and validate lifespan-extending interventions. We envision this as a translational pyramid with identification of compounds in invertebrate systems, like yeast, forming the base of our translational research pipeline and leading to experiments in other invertebrate systems, in models of genetic diversity, into wild type and genetically diverse vertebrate systems, then finally, in companion pets and humans (Figure 1).
Figure 1. A translational pyramid for longevity intervention from invertebrates to companion animals and humans.
Interventions are first screened using common laboratory models (either invertebrate or vertebrate) in wild type (WT) laboratory genetic backgrounds. Successful interventions can then be studied for their efficacy among genetically-diverse collections. Disease models and other short-lived backgrounds can also be utilized to identify particular translational opportunities for interventions. Studies in genetically-diverse companion animals can validate interventions that are then tested in humans.
Invertebrate systems
For basic biology, single-celled yeast and invertebrate systems (referred to hereafter simply as “invertebrates”) boast unrivaled benefits. These organisms are inexpensive to culture in large populations and have a variety of phenotypes and disease models that can be analyzed using multiple techniques. There are also well-developed genetic tools, models of genetic diversity, and cost-effective genome sequencing methods that are broadly utilized. Taken together, invertebrate systems are optimal for discovery-driven research. For biology of aging in particular, most invertebrates are short-lived, which allows experiments to be conducted in a reasonable time frame. Using multiple invertebrate models to identify and validate interventions provides assessment of evolutionary translatability that is important to consider when developing mammalian interventions [52]. Beyond these general characteristics, each of the three major invertebrate genetic model systems (yeast, worms, and flies) have unique strengths and weaknesses.
1. Wild type (WT) lab models
The term wild type (WT) originally described the collective traits of organisms isolated from the wild, namely flies [53]. Today, the term is more commonly used to denote the genetic background of laboratory organisms used in a given experimental system. There is growing appreciation that these organisms really no longer represent the ancestral state but are instead adapted to conditions in the lab [54, 55]. Laboratory conditions often alter developmental and reproductive life history traits and not always the same way depending on the model system [54, 56, 57]. These adaptations may be particularly important in studies of longevity, where factors like environmental nutrient status and reproductive timing strongly influence aging. It remains an open and important question whether genetic and environmental interventions that increase longevity in lab-evolved backgrounds have similar effects in genotypes that are not domesticated to the lab (discussed further below).
Within each model organism the WT strain used in one study may differ substantially from the WT strain used in a different study, even among studies from the same laboratory. Such strain background differences have the potential to confound interpretation if not carefully considered and addressed by the researchers performing the studies. For example, understanding the relationship between Sir2 and caloric restriction (CR) in budding yeast was complicated by the fact that Sir2 overexpression was shown to increase lifespan in one yeast WT strain (W303–1A)[58], while CR extended lifespan in a second yeast WT strain (PSY316) [59, 60]. Although a model was initially proposed that Sir2 and CR act within the same genetic pathway, it was not appreciated for several years that neither intervention extended lifespan in the other strain background [61, 62]. In a third WT strain background (BY4742), however, both interventions significantly extend lifespan allowing for more formal analysis of their genetic relationship [62].
Historically, there have been numerous yeast and fly genetic backgrounds commonly used across different labs. In recent years, the yeast aging field has mostly adopted the strain background used for the yeast ORF deletion collection [63, 64]: BY4743 for diploids and the corresponding haploid derivatives BY4741 (MATa) and BY4742 (MATα). In C. elegans, the vast majority of research is performed in a single genetic background, Bristol N2.
Yeast
The budding yeast Saccharomyces cerevisiae is an important workhorse in geroscience [65]. Some of the most well-established longevity regulating pathways were first identified in yeast, including sirtuins and mTOR signaling [15, 58, 66, 67]. Two major aging paradigms exist in yeast: chronological and replicative [68]. In the chronological model, populations of yeast are grown to quiescence in liquid culture and changes in viability are assessed over time by outgrowth challenge in rich media [69–71]. At the cellular level, yeast chronological aging can be thought of as a model of post-mitotic cell aging [72]. These cells, like neurons in humans, are non-dividing and must maintain cellular function while bearing whatever burden is imposed upon them by the environment and their continued cellular metabolism. This is particularly important during yeast chronological aging, where media acidification resulting from carbon metabolism is a strong driver of lost cellular viability [73].
Yeast replicative aging, instead of monitoring population viability over time, measures the replicative potential of individual yeast cells [74, 75]. To do this, individual “mother” cells are arrayed on plates using a compound microscope equipped with an optical fiber dissection needle [76]. Cycles of incubation, microdissection of daughter cells, and quantification are then performed until all mother cells cease dividing. This model of aging is akin to stem cell aging, where numbers of divisions that cells undergo before replicative senescence importantly influences tissue-level health and function. Newer methods of quantifying replicative lifespan and age-related changes in cell biology using microfluidics hold promise to increase throughput by decreasing assay time (from three weeks using microdissection to three days using microfluidics) and allow for longitudinal studies of cellular aging to be performed at the single cell level [77, 78].
Worms
The nematode Caenorhabditis elegans is one of the simplest animal model systems with tissue-level differentiation, which includes a nervous system, muscle, intestine, and a pharynx whose neuromuscular function resembles that of a mammalian heart. The first breakthrough in molecular genetics of aging that established the role of insulin-like signaling as a key longevity pathway was made using worms, underscoring the importance of this model system [79–82]. As opposed to yeast, measuring worm lifespan is conceptually more straightforward [83]. Synchronized populations of animals are plated on media containing a bacterial lawn (typically E.coli) as a food source and monitored until all animals cease moving [84]. The lifespan of a typical wild type animal is around three weeks. Lifespan in worms, however, is highly sensitive to changes in environment, including temperature and food source [85, 86]. As with yeast replicative lifespan, new technologies are utilizing platforms like microfluidics and so-called “lifespan machines”, incubation and culturing systems combined with image capturing to facilitate automated lifespan analysis [87, 88]. Utilizing these standardized technologies to perform lifespan analysis reduces technical hurdles to performing assays while increasing output, allowing for higher-throughput survival analysis.
Flies
The fruit fly Drosophila melanogaster is the most physiologically complex of the three major invertebrate aging models [89]. Along with greater complexity comes more opportunities to study tissue level aging, sex-specific lifespan differences, and more complex health measures like age-related behavioral changes and impaired learning [90–93]. This allowed for some of the first targeted studies on the evolution of lifespan characteristics where researchers evolved short- or long-lived fly populations by controlling reproductive age of parents [94, 95]. Early validation, and better understanding, of the role that insulin signaling plays in regulating aging was performed using flies [96–98]. Other processes likely to play a role in mammalian health and aging, like sleep and circadian rhythms, are also effectively studied using fruit flies [99]. An interesting difference from other invertebrate models is that fruit flies do not have a dominant wild type background in which the majority of research is performed. Multiple genetic backgrounds are utilized, including Oregon R, Canton S, w118, and yw. Importantly, these backgrounds have different survival characteristics [100, 101]. The typical fruit fly lifespan is 2–3 months but, just as with nematodes, this is highly dependent on culture conditions [102, 103]. Again, like nematodes, cohorts of age-matched animals are separately cultured [104, 105]. Fruit flies are typically reared in vials containing a yeast-based food source and transferred to new vials periodically.
Once a lifespan extending intervention is identified using a WT invertebrate system, a next step is to validate its translatability, identify natural genetic variants that influence intervention efficacy, and evaluate how the intervention impacts age-associated and other disease models. There are three common ways these goals are achieved using other invertebrate models and genetic collections.
2. Evolutionary translation between invertebrate model systems
Utilizing short-lived invertebrate models is ideal for rapidly identifying lifespan-extending molecules and extracts. Once an intervention is identified, how do we proceed? We can divide lifespan extending interventions by whether they impact private or public mechanisms of aging [106]. Private mechanism interventions target cellular processes that uniquely contribute to aging in certain model systems. For instance, yeast accumulate a type of genome instability with replicative age marked by proliferation of repeating ribosomal DNA (rDNA) in the form of extrachromosomal circular rDNA [107]. Inhibiting rDNA instability and accumulation of extrachromosomal rDNA circles leads to extended lifespan in yeast [108]. While the role of rDNA instability in some disease states is becoming better appreciated [109], rDNA circle accumulation is likely a private mechanism of yeast aging that is not shared, at least by mammals.
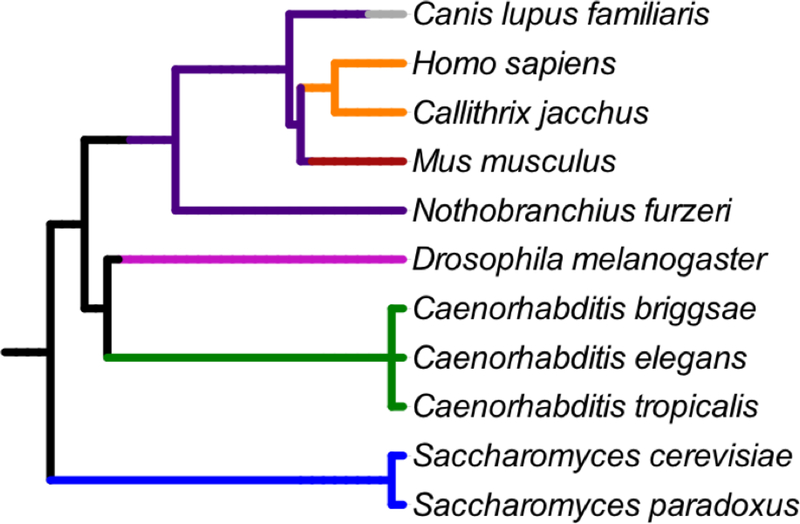
Interventions that target public mechanisms impact cellular mechanisms that contribute to aging in many different organisms. These shared mechanisms comprise the “Hallmarks of Aging” and include genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication [3]. Interventions that target fundamental cellular processes that fail as organisms age, or those that elicit evolutionarily conserved cellular stress responses are likely to be public. These interventions have the greatest potential to extend lifespan in multiple organisms, including mammals. One way to validate whether an intervention impacts public or private mechanisms of aging is to test for conserved longevity and/or healthspan-related measures in multiple model systems. Utilizing invertebrate systems to validate interventions harnesses the research power of each system while also testing whether the intervention effect is conserved across hundreds of millions of years of evolutionary divergence (Figure 2) [110, 111]. This strategy is effectively used to validate newly identified and commonly used compounds [12, 14, 33]. Evolutionary conservation of this sort provides an important foundation for choosing interventions to move forward in translational geroscience and compliments traditional genetic and mechanistic approaches.
Figure 2. Phylogenetic relationships of key organisms used in the translational geroscience research pyramid.
Fungi = blue, nematodes = green, Arthropods = magenta, vertebrates = dark purple, rodents = dark red, primates = orange, and canines = gray. Phylogenetic tree contructed using NCBI phyloT and Interactive Tree of Life (iTOL) v3 [111].
3. Genetically-diverse collections
Besides testing interventions across invertebrate model organisms, other diverse invertebrate collections and genetic libraries are useful for evaluating lifespan extending interventions. Some of these collections and libraries represent engineered genetic variation, while others capture natural genetic diversity. There are genetically diverse panels and libraries for all three major invertebrate systems that can be utilized to develop a more sophisticated understanding of lifespan and healthspan interventions.
Yeast
The most utilized collection of engineered genetic mutants is the yeast single gene deletion collection [63, 112]. This library of nearly 6000 mutants is constructed by systematic replacement of every non-essential yeast gene with a selectable marker. Using this collection, comprehensive genome wide analyses of genes that modulate both replicative and chronological lifespan have been performed [15, 113–115]. These studies have revealed numerous pathways that regulate cellular aging. In a screen of deletion mutants, the particular benefits of CR in suppressing diminished lifespan in mitochondrial mutant was identified [116]. Insights here led to using the mTOR inhibitor rapamycin as a novel treatment for mitochondrial disease in mice (discussed in detail below). For intervention studies, this collection is particularly valuable as a means of performing epistasis analysis that allows a better understanding of which cellular pathways the intervention interacts. Testing interventions in deletion mutants is also a useful means of identifying new interventions that target known longevity pathways [117].
For natural genetic diversity, the Saccharomyces Genome Resequencing Project (SGRP) collects a global assortment of two different Saccharomyces species (S. cerevisiae and S. paradoxus) representing wild and domesticated yeast from multiple ecotypes [118]. Initial characterization of strains from this collection reveal substantial natural variation in replicative lifespan [57]. Given the large evolutionary distance between yeast isolates [119], this collection represents another resource for evolutionary translation approaches. Perhaps more interesting for translational geroscience, this collection captures a substantial amount of natural genetic variation that can be probed in the context of intervention evaluation. Across these strains, there are over 235,000 SNPs [118]. Understanding drug efficacy across a range of genotypes provides insights into safety of the drug when given to genetically diverse populations and how well individual genotypes respond. Such an understanding will be critical for precision medicine approaches to translational geroscience.
Worms
Worms, like yeast, boast a large genetic library that allows for whole genome analysis at the single gene level. In contrast to yeast, however, this library is not composed of genetic mutants, but is instead made up of E. coli housing unique vectors, each of which targets a worm gene for post-transcriptional knockdown using feeding-based RNA interference (RNAi) [120–124]. Several large-scale screens have identified lifespan extending mutants using RNAi in WT and other mutant backgrounds [125–129]. The important roles of the mitochondria and mRNA translation in determining longevity were conclusively established in these early screens. While there are challenges when comparing RNAi results across labs, primarily due to methodological and RNAi induction variability, these screens are a particularly useful means of generating hypotheses that can be independently validated.
A new program to investigate lifespan extending interventions in nematodes utilizes natural genetic diversity not just within C. elegans, but also in closely related species. The Caenorhabditis Interventions Testing Program (CITP) is a multi-site effort meant to validate longevity enhancing treatments using worms [130, 131]. This project is modeled after another NIA-supported Interventions Testing Program (ITP) which evaluates well-studied interventions for increased lifespan and healthspan in mice (see below). In addition to their efforts to develop standardized protocols for worm lifespan studies, the CITP is notable for its use of multiple independent strains of genetically diverse Caenorhabditis species [131–133]. In addition to eight different C. elegans isolates, the CITP utilizes eight isolates of C. briggsae and six isolates of C. tropicalis. This is particularly remarkable given historical difficulties culturing Caenorhabditis species from the wild [134, 135]. Whole genome sequences of the CITP strains will be an important resource for investigating intervention efficacy among this set of animals.
Flies
Cost-effective genome sequencing methods have allowed expansion of genetic model systems from a few wild type lab organisms to large panels of organisms representing genetically diverse natural populations. An example of this is the Drosophila Genetic Reference Panel (DGRP) [136]. The DGRP is a collection of ~200 inbred Drosophila melanogaster lines descended from a population of flies isolated at a farmer’s market in Raleigh NC, USA. Even though the parental lines were all isolated from the same location, the DGRP represents a wealth of genetic diversity [137]. Amongst these 205 lines, over 4.8 million SNPs and 1.2 million larger polymorphisms are captured. Disease phenotypes are screened in the DGRP to identify naturally segregating genetic variants that modify human disease states [138, 139]. The population structure of the DGRP makes this collection well-suited for genome-wide association studies (GWAS) [140]. While there is substantial variation in survival amongst the DGRP, a GWAS for longevity failed to statistically validate variants associated with longevity within this collection [141]. This likely rules out alleles of large effect segregating in this population. Epistatic interactions between multiple alleles, however, may account for a larger amount of variation in aging than currently appreciated [142]. These researchers suggested expanding the number of lines within the collection to increase power to resolve longevity associated effects of segregating SNPs. Another GWAS using 800 lines from a different genetically diverse population, the Drosophila Synthetic Population Resource (DSPR), identified multiple quantitative trait loci (QTLs) that explain up to 22% of lifespan variation [143–145]. Capturing broader genetic variation in sexually isolated populations may reveal novel longevity associated alleles of greater effect [146, 147]. Studies addressing the efficacy of lifespan extending compounds on survival and health have not been completed with the DGRP, but this collection holds promise as a model to investigate how natural genetic variation influences intervention outcomes.
4. Age associated disease models
The promise of targeting aging is not only that we can extend the quantity of our lifespan, but that we can enhance the quality. This is not possible without decreasing the burden of age-associated disease. The core hypothesis of translational geroscience is that there is a fundamental connection between aging and age-associated disease such that, delaying aging will consequently delay the onset of disease. In particular, a major emphasis of translational geroscience is compression of morbidity, meaning that the functional decline and chronic diseases associated with age are pushed back as far as possible toward the end of life [148]. A key test of this hypothesis is showing that lifespan extending interventions broadly reduce age-associated disease. There are numerous invertebrate models that attempt to recapitulate aspects of human age-associated diseases. Testing compounds of interest in these disease models is useful for understanding the full utility of lifespan interventions. Interestingly, in some cases, lifespan extending compounds have proven to be effective in disease models not obviously linked to normative aging, such as the ability of rapamycin to suppress severe mitochondrial disease in mice. This lends support for additional studies that screen lifespan extending compounds widely across disease models.
Yeast
Yeast provide a window into fundamental features of normal and disease cell biology that translate to humans in many cases [149]. In terms of age-related diseases, many models of protein aggregation diseases have been developed using yeast [150–152]. These have helped elucidate basic mechanisms of the disease state. Yeast are very well-suited for measuring cellular phenotypes associated with a given disease, like mitochondrial dysfunction or protein aggregation [153]. Fundamental drivers of cancer, like genome instability and mutagenesis, are effectively modeled using mutator yeast that are defective for mismatch repair and polymerase proofreading [154–158]. With few complex behaviors that can be tracked with disease progression, however, yeast is limited beyond understanding cell-intrinsic aspects of basic molecular and cellular biology of disease.
Worms
Multicellularity and tissue differentiation in C. elegans provide a more relevant context to study the effects of lifespan extending interventions on age-associated disease. Like yeast, worms are also a popular model system to study protein aggregation diseases [159, 160]. With a simple neuromuscular system, age-related paralysis and other behavioral changes provide healthspan-related phenotypes that can be measured in the context of these disease models [161]. Without a circulatory system, worms are less amenable to studying vascular aging. However, the pharynx, an appendage in the head that grinds bacteria as they are ingested, has been described as similar to the heart in its pumping activity and regulation [162]. Age-associated changes in pharyngeal pumping are another commonly used functional measure of organismal health [163, 164]. Mitochondrial disease and mtDNA genome instability are modeled using Pol gamma proofreading or polymerase dead mutator worms, multiple mutants defective for components of the electron transport chain, and mutants for other critical mitochondrial functions [165–167]. Worms also provide useful models of conditions involved in dysregulated lipid metabolism, like obesity and related metabolic disorders [168, 169]. While they do not possess a kidney, worms have even proven useful in understanding kidney disease, particularly as its pathogenesis relates to defective ciliary function [170, 171].
Flies
Combining genetic disease models with pharmacological interventions is well established in flies [172]. Flies are well-suited for studying similar neurodegenerative and protein aggregation diseases as those of yeast and worms [173–175]. Fundamental connections between mitochondrial fission and fusion cycles and Parkinson’s disease onset were identified in flies [176, 177]. Additionally, thanks to their more advanced nervous system and complex behavior, flies allow other neurological and psychiatric diseases to be studied [178, 179]. Mitochondrial disease models that impact the electron transport chain (ETC) and mitochondrial energetics are also well characterized [180]. Interestingly, rapamycin improves survival in a fly model of Leigh syndrome, a disease driven by defects in ETC function [17]. Flies also provide useful models of cardiac aging, cancer, and even as a model for immunoaging [181–183].
Vertebrates
Validating interventions in vertebrate systems
Invertebrate systems are a discovery engine for longevity interventions. The goal, however, is to translate these interventions into vertebrate systems. Short-lived vertebrate systems are essential for validating interventions in a timely manner, but few options are available. An emerging model for aging research is the African turquoise killifish (Nothobranchius furzeri) [184]. With a median lifespan between 4–6 months, the African turquoise killifish is the shortest-lived vertebrate that can currently be bred in the lab [185, 186]. In addition to survival, many age-associated phenotypes, including enhanced cognitive decline, can be studied using the killifish [187]. While few longitudinal studies have been reported, resveratrol and low temperature both extend lifespan in this model [188, 189]. Interestingly, transfer of gut microbiome from young to old fish extends lifespan, showing the importance of gut flora in aging [190]. With more genome editing and analysis resources now available [191, 192], we can hope to see the killifish develop as a vertebrate discovery and validation engine for lifespan interventions.
While non-mammalian fish models and rats are occasionally used, mouse models are the major vertebrate workhorse in translational geroscience [193, 194]. Wild type mouse models are genetically well-established and age-associated pathologies are deeply understood. Ever increasing disease models provide complimentary approaches to test interventions. The NIA-sponsored Interventions Testing Program (ITP) is a multi-site effort utilizing genetically heterogeneous mice to validate longevity interventions. These resources establish mice as the major mammalian system for intervention validation.
5. WT vertebrate models
If all roads in the ancient world led to Rome, then the mouse, Mus musculus, is truly the Rome of the translational geroscientific research pipeline. Mice are ubiquitous in aging research and translational research in general. All of the most promising longevity interventions are being actively researched using mouse models [195–198]. Mice are among the shortest-lived mammals and have a typical lifespan in the lab of 2–3 years [199]. There are multiple common genetic backgrounds that are used for general research [200]. Mice are anatomically and physiologically very well characterized. This allows for detailed pathological analyses to be conducted, even though reporting of this data is sometimes sparse [201, 202]. Efforts to standardize pathological grading of aged tissues will allow detailed assessment of organismal aging that can be applied to testing intervention efficacy [203]. Assessing cause of death in mice is difficult given care and maintenance requirements, but the most common pathologies that contribute to mortality are cancer, kidney disease, and inflammation [204]. Animal care and maintenance is rigorous [205, 206]. This is necessary to maintain healthy and humane conditions and it provides a measure of standardization across research sites that is underappreciated. Compared to humans, it is estimated that 9 days is approximately equivalent to a human year for a mouse [200]. This is interesting to consider in the context of drug studies when thinking about dosing windows and comparing to humans.
As described above for invertebrate models, it is important to carefully consider genetic background when designing and interpreting geroscience studies in mice. Several different WT mouse strains are utilized for such studies, with the most common being inbred C57BL/6J and genetically heterogeneous UMHET3. A comparison of longevity across 31 different mouse strains at The Jackson Laboratory found substantial differences in morbidity and mortality [207, 208]. Also, as mentioned above, multiple generations of breeding under laboratory conditions is likely to have selected for genotypes that develop rapidly and produce large litter sizes among inbred mouse strains.
6. Genetically-diverse collections
The relative complexity of mice slows the development of genetic tools compared to invertebrate systems. Efforts are underway to systematically construct the knockout mouse collection, a genome wide set of null mutants for every non-essential mouse gene [209]. This will unlock similar genetic tools for intervention epistasis studies currently used in yeast. Many recombinant lines exist that are produced typically by mating lab strains and establishing diverse lines from progeny. The ILSXISS are a set of recombinant inbred lines generated by mating mutants isolated within the lab [210]. These lines provided key insights into the importance of genetic background in modifying the effect of CR on lifespan [211]. The largest effort to create diverse genetic models using mice is the Collaborative Cross, a genetic reference panel of recombinant inbred mice descended from an eight-way cross and mating scheme utilizing five lab mouse models and three mouse models representing natural genetic variation [212, 213]. By mating lines from the Collaborative Cross, researchers are now constructing the Diversity Outbred (DO) mouse populations [214, 215]. This set of lines is different than the other genetically diverse organism collections discussed because these lines are kept as heterozygotes instead of being inbred to homozygosity [216]. This makes the DO population the closest model to human genetic variation available to biomedical researchers. This will be an important population for testing intervention robustness in genetically diverse mammals.
A key resource for translational geroscience is the NIH-sponsored Interventions Testing Program (ITP). This is an NIA-sponsored multi-site effort bringing together labs from University of Michigan, The Jackson Laboratory, and University of Texas-San Antonio to test proposed pro-longevity interventions [217, 218]. The ITP uses a heterogeneous genetic mouse model constructed from crosses of multiple unique mouse strains (in this case, a four-way cross between common lab strains) [219–221]. Interventions are selected for ITP testing based on proposal and then evaluated by committee. To date, 42 compounds have been tested, nine of these at different doses, in combination with other compounds, or tested in different aged mice (Table 1) [19, 20, 51, 199, 222–225]. Of these, two (rapamycin and acarbose) extend lifespan in both sexes and another four (aspirin, 17αEstradiol, nordihydroguaiaretic acid, and protandim®) extend only in males [226].
Table 1. Summary of lifespan data from the NIA Interventions Testing Progam by Cohort.
Interventions and concentrations tested, age treament started, median lifespan with 95% confidence intervals (CIs), number of animals enrolled (n), and percent change (compared to cohort controls) separated by sex, year enrolled (Cohort), and reference for compounds screened and in progress by the Interventions Testing Program, 2004–2018.
| Median lifespan and 95% CIs (days) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Intervention | Concentration | Age started | Females | n | % change | Males | n | % change | Cohort | Reference |
| Control (combined) | NA | NA | 886 (875–894) | 2269 | NA | 803 (792–812) | 2599 | NA | Compiled | NA |
| Control | NA | NA | 892 (860–916) | 288 | NA | 790 (765–819) | 379 | NA | 2004 | Strong et al., 2008 |
| 4-OH-PBN | 315 ppm | 4 months | 850 (827–905) | 144 | −4.7 | 813 (773–866) | 180 | 2.9 | 2004 | Strong et al., 2008 |
| Aspirin | 20 ppm | 4 months | 851 (835–878) | 144 | −4.6 | 852 (825–889) | 180 | 7.8* | 2004 | Strong et al., 2008 |
| Nordihydroguaiaretic acid (NDGA) | 2,500 ppm | 9 months | 895 (862–923) | 149 | 0.3 | 883 (847–911) | 180 | 11.8* | 2004 | Strong et al., 2008 |
| Nitrofluribprofen (NFP) | 200 ppm | 4 months | 884 (845–915) | 144 | −0.9 | 815 (774–855) | 181 | 3.2 | 2004 | Strong et al., 2008 |
| Control | NA | NA | 887 (862–905) | 296 | NA | 797 (769–835) | 378 | NA | 2005 | Harrrison et al., 2009 |
| caffeic acid phenethyl ester (CAPE) | 30 ppm | 4 months | 884 (855–920) | 144 | −0.3 | 817 (774–875) | 180 | 2.5 | 2005 | Harrrison et al., 2009 |
| caffeic acid phenethyl ester (CAPE) | 300 ppm | 4 months | 926 (886–965) | 148 | 4.4 | 815 (748–852) | 183 | 2.3 | 2005 | Harrrison et al., 2009 |
| Enalapril Maleate | 120 ppm | 4 months | 868 (839–934) | 144 | −2.1 | 849 (807–875) | 180 | 6.5 | 2005 | Harrrison et al., 2009 |
| Rapamycin | 14 ppm | 20 months | 1005 (988–1058) | 144 | 13.3* | 952 (902–1004) | 177 | 19.4* | 2005 | Harrrison et al., 2009 |
| Control | NA | NA | 891 (873–911) | 272 | NA | 834 (800–869) | 342 | NA | 2006 | Miller et al., 2011 |
| Rapamycin | 14 ppm | 9 months | 1052 (1001–1112) | 144 | 18.1* | 902 (854–966) | 171 | 8.2* | 2006 | Miller et al., 2011 |
| Resveratrol | 300 ppm | 12 months | 906 (870–939) | 144 | 1.7 | 830 (794–879) | 165 | −0.5 | 2006 | Miller et al., 2011 |
| Resveratrol | 1200 ppm | 12 months | 905 (867–944) | 144 | 1.6 | 884 (845–908) | 168 | 6.0 | 2006 | Miller et al., 2011 |
| Simvastatin | 12 ppm | 10 months | 916 (883–952) | 143 | 2.8 | 858 (809–888) | 167 | 2.9 | 2006 | Miller et al., 2011 |
| Simvastatin | 120 ppm | 10 months | 898 (869–933) | 144 | 0.8 | 793 (753–853) | 168 | −4.9 | 2006 | Miller et al., 2011 |
| Control | NA | NA | 866 (835–892) | 281 | NA | 786 (749–827) | 294 | NA | 2007 | Strong et al., 2013 |
| Curcumin | 2000 ppm | 4 months | 905 (845–936) | 132 | 4.5 | 808 (758–840) | 158 | 2.8 | 2007 | Strong et al., 2013 |
| Green tea extract | 2000 ppm | 4 months | 923 (898–944) | 130 | 6.6 | 822 (778–881) | 153 | 4.6 | 2007 | Strong et al., 2013 |
| Medium Chain Triglyceride Oil | 60000 ppm | 4 months | 882 (852–931) | 136 | 1.8 | 777 (734–853) | 153 | −1.1 | 2007 | Strong et al., 2013 |
| Oxaloacetic acid | 2200 ppm | 4 months | 892 (855–940) | 134 | 3.0 | 819 (760–858) | 153 | 4.2 | 2007 | Strong et al., 2013 |
| Resveratrol | 300 ppm | 4 months | 907 (877–939) | 131 | 4.7 | 813 (767–869) | 153 | 3.4 | 2007 | Strong et al., 2013 |
| Control | NA | NA | 891 (869–915) | 280 | NA | 807 (782–841) | 300 | NA | 2009 | Harrison et al., 2014 |
| 17α-Estradiol | 4.8 ppm | 10 months | 902 (867–930) | 136 | 1.2 | 900 (852–935) | 156 | 11.5* | 2009 | Harrison et al., 2014 |
| Acarbose | 1000 ppm | 4 months | 939 (883–985) | 136 | 5.4* | 984 (952–1027) | 156 | 21.9* | 2009 | Harrison et al., 2014 |
| Methylene Blue | 28 ppm | 4 months | 903 (866–945) | 136 | 1.3 | 790 (738–848) | 156 | −2.1 | 2009 | Harrison et al., 2014 |
| Rapamycin | 42 ppm | 9 months | 1131 (1115–1174) | 136 | 26.9* | 992 (911–1037) | 156 | 22.9* | 2009 | Miller et al., 2014 |
| Rapamycin | 4.7 ppm | 9 months | 1037 (977–1079) | 136 | 16.4* | 834 (767–897) | 156 | 3.3 | 2009 | Miller et al., 2014 |
| Rapamycin | 14 ppm | 9 months | 1084 (1031–1149) | 136 | 21.7* | 909 (820–974) | 156 | 12.6* | 2009 | Miller et al., 2014 |
| Control | NA | NA | 902 (873–940) | 280 | NA | 784 (739–819) | 300 | NA | 2010 | Strong et al., 2016 |
| Fish Oil | 15000 ppm | 9 months | 863 (834–920) | 136 | −4.3 | 812 (771–876) | 156 | 3.6 | 2010 | Strong et al., 2016 |
| Fish Oil | 50000 ppm | 9 months | 923 (883–947) | 136 | 2.3 | 734 (696–808) | 156 | −6.4 | 2010 | Strong et al., 2016 |
| Nordihydroguaiaretic acid (NDGA) | 5000 ppm | 6 months | 880 (849–934) | 136 | −2.4 | 852 (818–892) | 156 | 8.7* | 2010 | Strong et al., 2016 |
| Nordihydroguaiaretic acid (NDGA) | 800 ppm | 6 months | NA | NA | NA | 855 (820–931) | 156 | 9.1* | 2010 | Strong et al., 2016 |
| Nordihydroguaiaretic acid (NDGA) | 2500 ppm | 6 months | NA | NA | NA | 875 (828–917) | 156 | 11.6* | 2010 | Strong et al., 2016 |
| Control | NA | NA | 874 (862–896) | 288 | NA | 785 (749–810) | 306 | NA | 2011 | Strong et al., 2016 |
| 17α-Estradiol | 14.4 ppm | 10 months | 893 (858–931) | 136 | 2.2 | 931 (867–966) | 156 | 18.6* | 2011 | Strong et al., 2016 |
| Bile Acids (ursodeoxycholic acid) | 5000 ppm | 5 months | 884 (848–913) | 136 | 1.1 | 839 (775–875) | 156 | 6.9 | 2011 | Strong et al., 2016 |
| Metformin | 1000 ppm | 9 months | 875 (839–922) | 144 | 0.1 | 847 (800–887) | 162 | 7.9 | 2011 | Strong et al., 2016 |
| Protandim® | 600 ppm | 10 months | 887 (868–930) | 136 | 1.5 | 853 (791–891) | 156 | 8.7* | 2011 | Strong et al., 2016 |
| Rapamycin + Metformin | 14 ppm + 1000 ppm | 9 months | 1079 (1037–1123) | 144 | 23.5* | 955 (906–999) | 162 | 21.7* | 2011 | Strong et al., 2016 |
| Control | NA | NA | 877 (852–914) | 284 | NA | 821 (789–849) | 300 | NA | 2012 | Strong et al., 2016 |
| Acarbose | 1000 ppm | 16 months | 903 (873–950) | 136 | 3.0 | 880 (814–916) | 156 | 7.2* | 2012 | Strong et al., 2016 |
| 2-(2-hydroxyphenyl)-benzoxazole | 1 ppm | 15 months | 856 (832–900) | 136 | −2.4 | 822 (790–853) | 156 | 0.1 | 2012 | NA |
| INT-767 FXR/TG5R agonist | 180 ppm | 10 months | 868 (840–895) | 136 | −1.0 | 793 (762–836) | 156 | −3.4 | 2012 | NA |
| Control | NA | NA | NA | NA | NA | NA | NA | NA | 2013 | In progress |
| Acarbose | 2500 ppm | 8 months | NA | NA | NA | NA | NA | NA | 2013 | In progress |
| Acarbose | 1000 ppm | 8 months | NA | NA | NA | NA | NA | NA | 2013 | In progress |
| Acarbose | 400 ppm | 8 months | NA | NA | NA | NA | NA | NA | 2013 | In progress |
| Ursolic Acid | 2000 ppm | 10 months | NA | NA | NA | NA | NA | NA | 2013 | In progress |
| Control | NA | NA | NA | NA | NA | NA | NA | NA | 2014 | In progress |
| Aspirin | 60 ppm | 11 months | NA | NA | NA | NA | NA | NA | 2014 | In progress |
| Aspirin | 200 ppm | 11 months | NA | NA | NA | NA | NA | NA | 2014 | In progress |
| Inulin | 600 ppm | 11 months | NA | NA | NA | NA | NA | NA | 2014 | In progress |
| Glycine | 80,000 ppm | 9 months | NA | NA | NA | NA | NA | NA | 2014 | In progress |
| TM5441 | 60 ppm | 11 months | NA | NA | NA | NA | NA | NA | 2014 | In progress |
| Control | NA | NA | NA | NA | NA | NA | NA | NA | 2015 | In progress |
| 17-DMAG | 30 ppm | 6 months | NA | NA | NA | NA | NA | NA | 2015 | In progress |
| Minocycline | 300 ppm | 6 months | NA | NA | NA | NA | NA | NA | 2015 | In progress |
| MitoQ | 100 ppm | 7 months | NA | NA | NA | NA | NA | NA | 2015 | In progress |
| Rapamycin – intermittent | 42 ppm | 20 months | NA | NA | NA | NA | NA | NA | 2015 | In progress |
| β-guanadinopropionic acid | 3300 ppm | 6 months | NA | NA | NA | NA | NA | NA | 2015 | In progress |
| Control | NA | NA | NA | NA | NA | NA | NA | NA | 2016 | In progress |
| 17α-Estradiol | 14.4 ppm | 16 + 20 months | NA | NA | NA | NA | NA | NA | 2016 | In progress |
| Canagliflozin | 180 ppm | 7 months | NA | NA | NA | NA | NA | NA | 2016 | In progress |
| Candesartan Cilexetil | 30 ppm | 8 months | NA | NA | NA | NA | NA | NA | 2016 | In progress |
| Geranylgeranyl acetone | 600 ppm | 9 months | NA | NA | NA | NA | NA | NA | 2016 | In progress |
| MIF098 | 240 ppm | 8 months | NA | NA | NA | NA | NA | NA | 2016 | In progress |
| Nicotinamide riboside | 1000 ppm | 8 months | NA | NA | NA | NA | NA | NA | 2016 | In progress |
| Control | NA | NA | NA | NA | NA | NA | NA | NA | 2017 | In progress |
| 1,3-butanediol | 100,000 ppm | 6 months | NA | NA | NA | NA | NA | NA | 2017 | In progress |
| Captopril | 180 ppm | 5 months | NA | NA | NA | NA | NA | NA | 2017 | In progress |
| L-Leucine | 40,000 ppm | 5 months | NA | NA | NA | NA | NA | NA | 2017 | In progress |
| PB125 | 100 ppm | 5 months | NA | NA | NA | NA | NA | NA | 2017 | In progress |
| Rapamycin + Acarbose | 14.7 ppm + 1000 ppm | 9 + 16 months | NA | NA | NA | NA | NA | NA | 2017 | In progress |
| Sulindac | 5 ppm | 5 months | NA | NA | NA | NA | NA | NA | 2017 | In progress |
| Syringaresinol | 300 ppm | 5 months | NA | NA | NA | NA | NA | NA | 2017 | In progress |
= significantly long-lived compared to cohort matched control.
7. Disease models
A particular strength of mouse research is the variety of disease models that can be utilized. Multiple reviews are devoted to documenting and comparing the wealth of models for different pathologies available, including age-associated diseases [227–229]. Of particular interest in biology of aging research are mouse progeria models, like the Zmpste−/− and LmnaL530P/L530P mice (Hutchinson Guilford Syndrome and laminopathy models), Ercc1−/− mice (Xeroderma pigmentosum model), and Bubr1−/− mice [230–235]. Mouse models of elevated oxidative stress, like the Cu/Zn superoxide dismutase knockout mouse, are an emerging progeroid model that shows accelerated frailty and molecular damage, especially in the liver, lungs, and kidneys [236, 237].
One notable example for the use of lifespan extending interventions beyond impacting age-related survival comes from research into mitochondrial disease that began using yeast. In a screen for yeast deletion mutants that differentially respond to low glucose CR, multiple mitochondrial mutants responded best to the intervention, with some seeing their lifespans more than double [116]. This led to the hypothesis that CR, or a CR mimetic, may have clinical application in the context of mitochondrial disease. To test this, researchers utilized the Ndufs4 knockout mouse model that recapitulates many aspects of Leigh syndrome, a human childhood mitochondrial disease [238]. Due to the frail nature of the Ndufs4 knockout mice and the early onset of disease, rapamycin was administered beginning at postnatal day 10 instead of CR. Impressively, rapamycin treatment nearly doubled median lifespan in this disease model [23]. This groundbreaking study opened up new interest in utilizing mTOR inhibitors as treatments for mitochondrial disease [22, 239]. It is exciting to consider that this study began in yeast and ended with a successful mammalian disease intervention in little more than a few years. Interestingly, more recent work has shown that hypoxia increases lifespan of Ndufs4 knockout mice even more robustly than rapamycin [240, 241]. Like rapamycin, hypoxia increases lifespan in C. elegans [242, 243] and may explain longevity among people living at high altitude [244]. These observations suggest that this mouse model of severe mitochondrial disease can be used as a short-lived discovery platform for interventions likely to extend lifespan in WT mice and perhaps people.
Reaching the goal
Applying interventions in companion animals and humans
Using the translational geroscience pipeline, we can go from identifying interventions in diverse invertebrate model systems to validating interventions using mice. From here, where do we go? In studies of CR, the first well-studied longevity extending intervention, substantial efforts were made to validate findings in a non-human primate, the rhesus monkey [245]. Parallel studies lasting more than 30 years each at the University of Wisconsin-Madison and NIA ultimately converged on CR having positive effects on primate health and lifespan, despite differences in study design and early controversies about CR having inconsistent effects between sites [245–247]. Given the exceptionally long lifespan and correspondingly high costs of rhesus monkeys, however, it seems unlikely that additional longevity studies will be performed in these animals. More recently, efforts have been made to establish the common marmoset (Callithrix jacchus) as a shorter-lived non-human primate model in geroscience [248–250]. This small primate has an average lifespan in the lab of 4–6 years and a maximum lifespan upwards of 16 years [251]. Efforts are underway now to establish whether rapamycin extends lifespan in this primate [252, 253]
While primate studies are the intuitive next step for testing putative geroscience interventions from an evolutionary point of view (Figure 2), they lack an important component for translational validation: the human environment. In this context an exciting approach is to utilize companion animals for understanding aging biology and intervention testing.
8. Companion animals
Companion animals as a non-laboratory model system for translational geroscience represent both a unique scientific model and potentially a realization of the goals of geroscience itself. Companion animals share a number of similarities with humans that are not recapitulated in lab organisms, including their environment and microbiota [254]. These non-genetic factors are critically important to model and understand in translational geroscience as it is estimated they account for approximately 80% of human lifespan variation [255–259]. In terms of pathology, companion animals share many age-associated diseases with humans, and we have a sophisticated understanding of disease progression for many species through veterinary records [260–262]. It is even suggested that studies in companion animals be built into the FDA drug approval process to span the “valley of death” between basic biomedical research and successful human disease treatment [260, 263]. Beyond their utility as it relates to developing interventions for ourselves, delaying disease and extending lifespan in companion animals is an important end in itself. Considering the important role of pets in our lives and our efforts to care for them to our fullest abilities, it is clear that we generally regard these animals with the same deference as we do other people. Investing in the healthy life of companion animals, then, is an important way that we fulfill an obligation to them that is similar, if not identical, to the obligation we have in biomedical research to help each other.
The Dog Aging Project is a research program designed to develop companion dogs as a model system for aging and test the efficacy of rapamycin as a healthy longevity intervention in pets [9]. This national study is enrolling dogs for longitudinal and intervention studies. In addition to survival, an important measure of intervention success in the rapamycin trials will be delayed disease onset. Veterinary records provide a good resource to establish breed specific age associated disease risk and life expectancy [261, 262, 264–266]. This allows a comparison of types of morbidities and comorbidities and when they occur between rapamycin treated and historical populations. Middle aged mid and large breed dogs (≥ 40 pounds) were selected for the first rapamycin experiments. Pilot studies with these dogs identified higher rates of asymptomatic heart valve degeneration than previously understood and provided evidence that short-term rapamycin treatment may improve age-associated cardiac decline [29, 267].
9. Humans
The last translational step to humans is the most difficult. The ideal intervention, if started between 40–55 years old, would take decades to be clearly assessed in terms of delayed morbidity and mortality. This adds to the already substantial challenges associated with longitudinal research in humans [268]. Despite its difficulty, clinical research that firmly establishes intervention efficacy in humans is critical to obtain. Proposed and ongoing human studies with metformin and rapamycin analogs (rapalogs) use more proximal measures of health and resilience in older individuals to assess efficacy. A further challenge is that no validated biomarker of biological age currently exists that can be used to validate and measure intervention success in human populations.
Initial clinical studies
The two best studied pharmacological longevity interventions to date are metformin and rapamycin. Metformin is a biguanide that was initially synthesized in the early 1920’s for its potential antidiabetic properties. Interest in biguanides as a diabetes medication originates from early use of French lilac extract as a diabetes treatment [269]. This extract is rich in guanidine, an otherwise toxic compound with long-known antidiabetic properties [270]. Metformin is FDA-approved for type-II diabetes treatment [271, 272]. There is increasing evidence that metformin also broadly reduces cancer risk [42, 273–278]. Epidemiological studies and meta-analysis also suggest that diabetics taking metformin have greater survival than age-matched non-diabetic controls [279, 280]. In the near future, Targeting Aging with Metformin (TAME), a study led by researchers at Albert Einstein College of Medicine, will test metformin as a healthy aging intervention in the elderly [10]. TAME intends to recruit thousands of non-diabetic individuals 65–79 years of age across the US for a randomized, placebo-controlled trial with metformin. Survival, as well as morbidity and comorbidities, will be monitored during and after treatment.
Rapamycin was originally identified as an antifungal compound secreted by soil Streptomyces species on Easter Island (Rapa Nui) [281]. These effects were known long before the mechanistic target of rapamycin (mTOR) was identified as a kinase complex that acts as a signaling hub in nutrient sensing [282–285]. In the clinic, rapamycin is used in high doses as an immunosuppressive, particularly during organ transplant [286, 287]. Like metformin, rapamycin and related analogs (rapalogs) are being explored for their anticancer potential [288, 289]. Current efforts to establish the benefits of rapamycin during human aging are focused on delaying age-associated immunosenescence [30]. Over 200 elderly (≥65 years old) individuals participated in an initial study that tested whether administration of the rapalog everolimus improved immune response to an annual flu vaccination. Those treated with everolimus showed greater antibody response to flu vaccine than placebo treated controls with only minor side effects (mouth ulceration or headache). In another study by this group, combined treatment of everolimus with a catalytic mTOR inhibitor (BEZ235) also improved immune function and largely avoided these side effects [31]. Interestingly, a short-term (6 weeks) treatment with BEZ235 alone resulted in reduced annual rate of infection in the elderly compared to control over at least one year. This suggests that immune changes associated with mTOR inhibition persist long after treatment is stopped.
Unmet needs
A standing challenge for human studies of healthspan intervention is measuring successful intervention. Unlike a traditional clinical trial, geroscience interventions are not aimed at treating or curing a specific disease after diagnosis. Instead, they should keep people healthy and alive longer, more akin to a preventative therapy. Due to the long lifespans of humans, this means that the length of time needed to assess efficacy is much longer than a typical clinical trial. As seen in the rapalog tests in elderly patients more proximal measures, like immune function, can be used to assess intervention success. While this can be an effective path to FDA approval, it does not really demonstrate that an intervention is impacting the biological aging process.
Perhaps the greatest unmet need for translational geroscience are clinically validated biomarkers of aging that can be measured to assess changes in biological age (which is in contrast to chronological age, typically measured as the time since birth). The American Federation for Aging Research propose that ideal biomarkers would be non-invasive, accurately measure cellular health as it relates to age-related physiological decline, and be translatable to other systems in such a way that basic research can be performed utilizing the marker [290]. Identifying such biomarkers has proven difficult; however, the advent of relatively inexpensive -omics technologies combined with powerful computational approaches such as machine learning have given new hope to finding predictive biomarkers [291]. Recent efforts have focused on developing panels of multiple biomarkers that can be assayed together to meet these proposed guidelines [292, 293]. Overall, however, there remains no consensus on what marker(s) reliably predict age at this time.
An emerging biomarker of aging is the epigenetic clock. The epigenetic clock is composed of a set of methylated genomic CpG residues that, taken together, are highly predictive of biological age [294]. DNA methylation (DNAm) patterns are predictive of age in multiple tissues and respond to healthy lifestyle and diet [295–298]. Epigenetic clocks have been characterized for mice (under normal and caloric restricted conditions) and domesticated dogs [299, 300]. Understanding methylation age, particularly in mice, provides new means of validating age-associated changes of other interventions that can be related to humans. The epigenetic clock is accelerated in progeroid conditions like Werner syndrome [301]. Interestingly, the epigenetic clock is found to be accelerated in conditions not typically thought of as accelerating aging, such as HIV infection and Down syndrome [302, 303]. Lifestyle factors, like stress, are also being recognized as driving accelerated epigenetic aging [304–306].
What to expect
Increased disease-free lifespan is without doubt a great benefit to individuals, but what are the broader societal impacts of extending our healthy lifespan? One answer to this is found in the concept of the longevity dividend [307, 308]. The longevity dividend refers to economic and other societal benefits that result from wide improvements in healthy aging [309]. Healthcare expenditure, for instance, will be fundamentally changed by breakthroughs in extended healthspan. In 2016, healthcare in the US cost $3.3 trillion dollars and 37% of that was paid through Medicare or Medicaid [310]. A great deal of these costs (37.9% as of 2013) are generated by treating chronic disease in elderly individuals, like heart disease, hypertension, periodontal disease, and diabetes [311–313]. Breakthroughs in healthspan interventions that delay or prevent these conditions will likely save considerable money for individuals as well as local, state, and federal governments. Due to the similarities in healthcare between humans and companion animals [314], a key test of the longevity dividend concept as it relates to healthcare economics may be provided by successful geroscience intervention in pet dogs.
Beyond the question of whether interventions will extend human healthspan and lifespan, a related question that is less frequently asked is whether interventions will work the same for everyone. As seen in mice, many interventions preferentially extend lifespan in males [20, 51, 226]. In invertebrate CR studies using genetically diverse models, shorter lived strains tend to reap the largest benefits from the intervention [116, 315]. Additionally, in the studies that revealed genotype-dependent responses to CR using the ILSXISS mice lines, the best responders to CR were also among the shorter lived [316]. An explanation proposed for this is that, the closer a population is to its maximum lifespan, the harder it will be to improve upon its lifespan [317]. While this is certainly true, it is unclear if organisms studied in the lab are close to their physiological maximum lifespan. In yeast, for instance, even the longest lived-strain tested (sgf73Δ median RLS = 42, WT median RLS = 25) showed a small increase in lifespan under CR [116]. It seems, instead, that interventions like CR are inherently equitable, meaning that those who get the largest benefit from the intervention are those that need it the most.
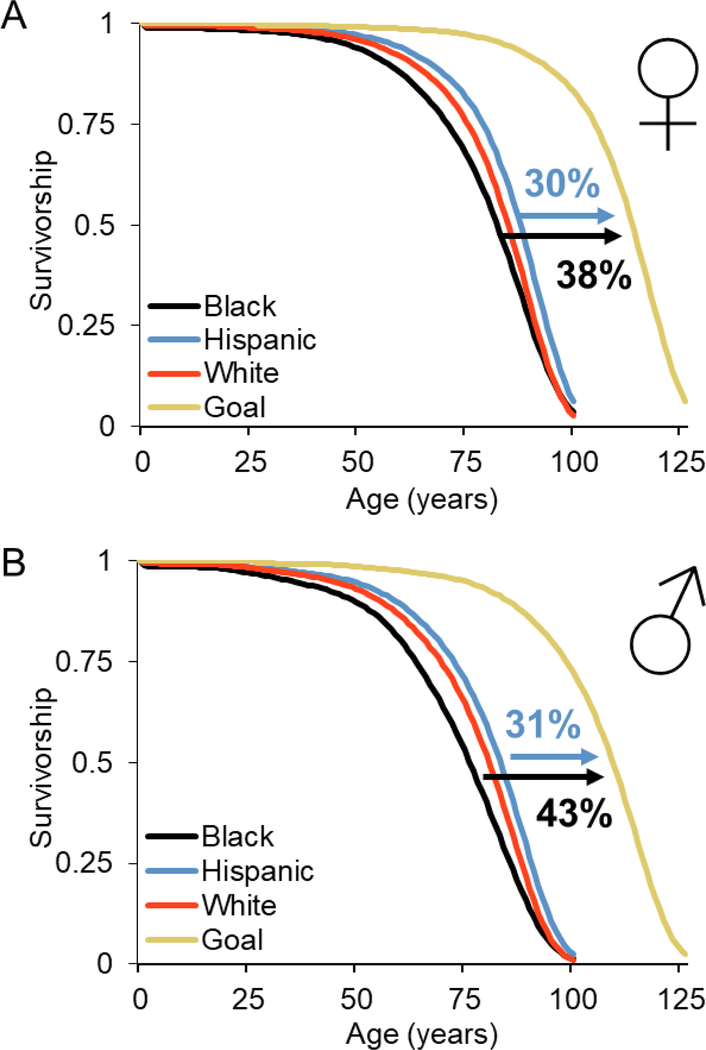
Another important longevity dividend produced by successful healthy aging intervention may be decreased health disparities between different human populations. In the US, human survival is stratified based on sex, race, and ethnicity (Figure 3) [318]. As of 2014, there is a 12-year difference in median survival between the longest-lived group, Hispanic women (median lifespan = 88) and the shortest lived, black men (median lifespan = 76). Black men and women, along with white men, are the three shortest lived groups. We can model idealized human survival by assuming that the maximum human lifespan is close to what we’ve already witnessed [319], ~125 years, and that age-associated survival dynamics will be similar to the longest-lived subgroup (Hispanic males and females) (Figure 3a and 3b). To achieve this ideal, an intervention (or combinations of interventions) would need to extend median lifespan by ~30% in the longest-lived populations and by closer to 40% in the shortest-lived populations (black males and females). In terms of age-associated morbidities, comparing black and white populations (data for other groups is less reliable), the biggest difference in causes of death between these groups is due to diabetes [320]. If healthspan interventions are similarly equitable in human populations as they are in model systems, we may see the largest benefits in health among those with the shortest lifespans, particularly black populations. Decreased health disparities that result from healthspan intervention can be thought of as another important longevity dividend.
Figure 3. Human survival curves separated by sex, race, and Hispanic origin.
A) females B) males. Gold curves represent idealized survival based on maximum human lifespan. Data from US life tables, 2014 [318].
Conclusions
Identifying and utilizing lifespan and healthspan extending interventions holds particular promise in extending lifespan and reducing human and companion animal disease burden. Common invertebrate systems provide models for discovery of lifespan extending compounds. Using diverse invertebrate systems, as well as models of genetic diversity and disease models, treatment efficacy can be evaluated in genetically diverse and evolutionarily distant organisms. Vertebrate model systems, particularly mice, provide a mammalian system to validate interventions and understand efficacy as it relates to genetic diversity and disease models. Translating successful interventions to companion pets, particularly dogs, provides a large, genetically diverse mammalian model to better evaluate interventions while potentially extending healthy lifespan of these animals, which is desirable and a biomedical breakthrough in itself. Finally, human healthspan interventions, while promising for all, may yield particular benefits for those already predisposed to shorter lifespan.
Acknowledgements
We thank Rich Miller and Francesca Macchiarini for datasets and helpful information regarding the Interventions Testing Program. We would like to thank Daniel Promislow, Alessandro Bitto, Benjamin Blue, and Michael Kiflezghi for careful review of the manuscript. MBL was supported by the Howard Hughes Medical Institute (HHMI) Gilliam Fellowship for Advanced Study, the National Institutes of Health (NIH) Cellular and Molecular Biology training grant (NIH T32GM007270), and the University of Washington Graduate Opportunities and Minority Achievement Program (UW GO-MAP) Bank of America Fellowship. MK is co-director of the University of Washington Nathan Shock Center of Excellence in the Basic Biology of Aging (NIH P30AG013280).
References:
- 1.Vijg J and Suh Y, Genetics of longevity and aging, in Annual Review of Medicine. 2005, Annual Reviews: Palo Alto. p. 193–212. [DOI] [PubMed] [Google Scholar]
- 2.Pitt JN and Kaeberlein M, Why is aging conserved and what can we do about it? PLoS Biol, 2015. 13(4): p. e1002131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Otin C, et al. , The hallmarks of aging. Cell, 2013. 153(6): p. 1194–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sierra F, The Emergence of Geroscience as an Interdisciplinary Approach to the Enhancement of Health Span and Life Span. Cold Spring Harbor Perspectives in Medicine, 2016. 6(4): p. a025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaeberlein M, Longevity and aging. F1000Prime Rep, 2013. 5: p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. Global Health Estimates 2016: Deaths by Cause, Age, Sex, by Country and by Region, 2000–2016. 2018. [cited 2018 05/3½018]; Available from: http://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death.
- 7.Seals DR and Melov S, Translational Geroscience: Emphasizing function to achieve optimal longevity. Aging-Us, 2014. 6(9): p. 718–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy BK, et al. , Geroscience: linking aging to chronic disease. Cell, 2014. 159(4): p. 709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaeberlein M, Creevy KE, and Promislow DEL, The dog aging project: translational geroscience in companion animals. Mammalian Genome, 2016. 27(7–8): p. 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barzilai N, et al. , Metformin as a Tool to Target Aging. Cell Metabolism, 2016. 23(6): p. 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy BK and Lamming DW, The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging. Cell Metabolism, 2016. 23(6): p. 990–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He C, et al. , Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import. Plos Genetics, 2014. 10(12): p. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips T and Leeuwenburgh C, Lifelong aspirin supplementation as a means to extending life span. Rejuvenation Research, 2004. 7(4): p. 243–251. [DOI] [PubMed] [Google Scholar]
- 14.Wan QL, et al. , Aspirin extends the lifespan of Caenorhabditis elegans via AMPK and DAF-16/FOXO in dietary restriction pathway. Experimental Gerontology, 2013. 48(5): p. 499–506. [DOI] [PubMed] [Google Scholar]
- 15.Powers RW, et al. , Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes & Development, 2006. 20(2): p. 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robida-Stubbs S, et al. , TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab, 2012. 15(5): p. 713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang A, et al. , Rapamycin enhances survival in a Drosophila model of mitochondrial disease. Oncotarget, 2016. 7(49): p. 80131–80139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjedov I, et al. , Mechanisms of Life Span Extension by Rapamycin in the Fruit Fly Drosophila melanogaster. Cell Metabolism, 2010. 11(1): p. 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller RA, et al. , Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell, 2014. 13(3): p. 468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison DE, et al. , Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature, 2009. 460(7253): p. 392–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy SL, et al. , Deaths: Final data for 2015, N.C.f.H. Statistics, Editor. 2017, National Vital Statistics Reports: Hyattsville, MD. [PubMed] [Google Scholar]
- 22.Siegmund SE, et al. , Low-dose rapamycin extends lifespan in a mouse model of mtDNA depletion syndrome. Human Molecular Genetics, 2017. 26(23): p. 4588–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson SC, et al. , mTOR Inhibition Alleviates Mitochondrial Disease in a Mouse Model of Leigh Syndrome. Science, 2013. 342(6165): p. 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos FJ, et al. , Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med, 2012. 4(144): p. 144ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popovich IG, et al. , Lifespan extension and cancer prevention in HER-2/neu transgenic mice treated with low intermittent doses of rapamycin. Cancer Biology & Therapy, 2014. 15(5): p. 586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livi CB, et al. , Rapamycin extends life span of Rb1(+/−) mice by inhibiting neuroendocrine tumors. Aging-Us, 2013. 5(2): p. 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anisimov VN, et al. , Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle, 2011. 10(24): p. 4230–6. [DOI] [PubMed] [Google Scholar]
- 28.Huber S, et al. , Inhibition of the mammalian target of rapamycin impedes lymphangiogenesis. Kidney International, 2007. 71(8): p. 771–777. [DOI] [PubMed] [Google Scholar]
- 29.Urfer SR, et al. , A randomized controlled trial to establish effects of short-term rapamycin treatment in 24 middle-aged companion dogs. Geroscience, 2017. 39(2): p. 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mannick JB, et al. , mTOR inhibition improves immune function in the elderly. Science Translational Medicine, 2014. 6(268): p. 7. [DOI] [PubMed] [Google Scholar]
- 31.Mannick JB, et al. , TORC1 inhibition enhances immune function and reduces infections in the elderly. Science Translational Medicine, 2018. 10(449). [DOI] [PubMed] [Google Scholar]
- 32.Martin-Montalvo A, et al. , Metformin improves healthspan and lifespan in mice. Nature Communications, 2013. 4: p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabreiro F, et al. , Metformin Retards Aging in C. elegans by Altering Microbial Folate and Methionine Metabolism. Cell, 2013. 153(1): p. 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans JMM, et al. , Metformin and reduced risk of cancer in diabetic patients. British Medical Journal, 2005. 330(7503): p. 1304–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, et al. , Metformin and reduced risk of hepatocellular carcinoma in diabetic patients: a meta-analysis. Scandinavian Journal of Gastroenterology, 2013. 48(1): p. 78–87. [DOI] [PubMed] [Google Scholar]
- 36.Preston MA, et al. , Metformin Use and Prostate Cancer Risk. European Urology, 2014. 66(6): p. 1012–1020. [DOI] [PubMed] [Google Scholar]
- 37.Zhang ZJ, et al. , Reduced Risk of Lung Cancer With Metformin Therapy in Diabetic Patients: A Systematic Review and Meta-Analysis. American Journal of Epidemiology, 2014. 180(1): p. 11–14. [DOI] [PubMed] [Google Scholar]
- 38.Provinciali N, et al. , Metformin: risk-benefit profile with a focus on cancer. Expert Opinion on Drug Safety, 2015. 14(10): p. 1573–1585. [DOI] [PubMed] [Google Scholar]
- 39.Li LF, et al. , The effects of metformin on ovarian cancer: an updated systematic review and meta-analysis. International Journal of Clinical and Experimental Medicine, 2016. 9(9): p. 17559–17568. [Google Scholar]
- 40.Nie ZH, Zhu HL, and Gu MJ, Reduced colorectal cancer incidence in type 2 diabetic patients treated with metformin: a meta-analysis. Pharmaceutical Biology, 2016. 54(11): p. 2636–2642. [DOI] [PubMed] [Google Scholar]
- 41.Seebacher V, et al. , The prognostic role of metformin in patients with endometrial cancer: a retrospective study. European Journal of Obstetrics & Gynecology and Reproductive Biology, 2016. 203: p. 291–296. [DOI] [PubMed] [Google Scholar]
- 42.Noto H, et al. , Cancer Risk in Diabetic Patients Treated with Metformin: A Systematic Review and Meta-analysis. Plos One, 2012. 7(3): p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryu D, et al. , NAD+ repletion improves muscle function in muscular dystrophy and counters global PARylation. Sci Transl Med, 2016. 8(361): p. 361ra139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, et al. , NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science, 2016. 352(6292): p. 1436–43. [DOI] [PubMed] [Google Scholar]
- 45.Das A, et al. , Impairment of an Endothelial NAD(+)-H2S Signaling Network Is a Reversible Cause of Vascular Aging. Cell, 2018. 173(1): p. 74–89 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tchkonia T, et al. , Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. Journal of Clinical Investigation, 2013. 123(3): p. 966–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker DJ, et al. , Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature, 2016. 530(7589): p. 184–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker DJ, et al. , Clearance of p16(Ink4a)-positive senescent cells delays ageing-associated disorders. Nature, 2011. 479(7372): p. 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutphin GL, et al. , Caffeine extends life span, improves healthspan, and delays age-associated pathology in Caenorhabditis elegans. Longev Healthspan, 2012. 1: p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wanke V, et al. , Caffeine extends yeast lifespan by targeting TORC1. Mol Microbiol, 2008. 69(1): p. 277–85. [DOI] [PubMed] [Google Scholar]
- 51.Strong R, et al. , Nordihydroguaiaretic acid and aspirin increase lifespan of genetically heterogeneous male mice. Aging Cell, 2008. 7(5): p. 641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bitto A, et al. , Biochemical Genetic Pathways that Modulate Aging in Multiple Species. Cold Spring Harb Perspect Med, 2015. 5(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgan TH, Sex limited inheritence in Drosophila. Science, 1910. 32(812): p. 120–2. [DOI] [PubMed] [Google Scholar]
- 54.Sterken MG, et al. , The laboratory domestication of Caenorhabditis elegans. Trends in genetics : TIG, 2015. 31(5): p. 224–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mortimer RK and Johnston JR, Genealogy of principal strains of the yeast genetic stock center. Genetics, 1986. 113(1): p. 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sgro CM and Partridge L, Evolutionary responses of the life history of wild-caught Drosophila melanogaster to two standard methods of laboratory culture. American Naturalist, 2000. 156(4): p. 341–353. [Google Scholar]
- 57.Kaya A, et al. , Defining Molecular Basis for Longevity Traits in Natural Yeast Isolates. NPJ Aging Mech Dis, 2015. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaeberlein M, McVey M, and Guarente L, The SIR2/¾ complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Development, 1999. 13(19): p. 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin SJ, et al. , Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature, 2002. 418(6895): p. 344–8. [DOI] [PubMed] [Google Scholar]
- 60.Lin SJ, Defossez PA, and Guarente L, Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science, 2000. 289(5487): p. 2126–8. [DOI] [PubMed] [Google Scholar]
- 61.Kaeberlein M, et al. , Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev, 2005. 126(4): p. 491–504. [DOI] [PubMed] [Google Scholar]
- 62.Kaeberlein M, et al. , Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol, 2004. 2(9): p. E296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winzeler EA, et al. , Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science, 1999. 285(5429): p. 901–906. [DOI] [PubMed] [Google Scholar]
- 64.Giaever G, et al. , Functional profiling of the Saccharomyces cerevisiae genome. Nature, 2002. 418(6896): p. 387–91. [DOI] [PubMed] [Google Scholar]
- 65.Longo VD, et al. , Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab, 2012. 16(1): p. 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaeberlein M, et al. , Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science, 2005. 310(5751): p. 1193–6. [DOI] [PubMed] [Google Scholar]
- 67.Fabrizio P, et al. , Regulation of longevity and stress resistance by Sch9 in yeast. Science, 2001. 292(5515): p. 288–90. [DOI] [PubMed] [Google Scholar]
- 68.Mirisola MG, Braun RJ, and Petranovic D, Approaches to study yeast cell aging and death. Fems Yeast Research, 2014. 14(1): p. 109–118. [DOI] [PubMed] [Google Scholar]
- 69.Murakami C and Kaeberlein M, Quantifying Yeast Chronological Life Span by Outgrowth of Aged Cells. Journal of Visualized Experiments : JoVE, 2009(27): p. 1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu J, et al. , Assessing chronological Aging in Saccharomyces cerevisiae. Methods in molecular biology (Clifton, N.J.), 2013. 965: p. 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fabrizio P and Longo VD, The chronological life span of Saccharomyces cerevisiae. Methods Mol Biol, 2007. 371: p. 89–95. [DOI] [PubMed] [Google Scholar]
- 72.MacLean M, Harris N, and Piper PW, Chronological lifespan of stationary phase yeast cells; a model for investigating the factors that might influence the ageing of postmitotic tissues in higher organisms. Yeast, 2001. 18(6): p. 499–509. [DOI] [PubMed] [Google Scholar]
- 73.Burtner CR, et al. , A molecular mechanism of chronological aging in yeast. Cell Cycle, 2009. 8(8): p. 1256–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mortimer RK and Johnston JR, Life Span of Individual Yeast Cells. Nature, 1959. 183: p. 1751–1752. [DOI] [PubMed] [Google Scholar]
- 75.Wasko BM and Kaeberlein M, Yeast replicative aging: a paradigm for defining conserved longevity interventions. Fems Yeast Research, 2014. 14(1): p. 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Steffen KK, Kennedy BK, and Kaeberlein M, Measuring replicative life span in the budding yeast. J Vis Exp, 2009(28). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen KL, Crane MM, and Kaeberlein M, Microfluidic technologies for yeast replicative lifespan studies. Mechanisms of Ageing and Development, 2017. 161: p. 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee SS, et al. , Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform. Proceedings of the National Academy of Sciences of the United States of America, 2012. 109(13): p. 4916–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tissenbaum HA, Using C. elegans for aging research. Invertebrate Reproduction & Development, 2015. 59: p. 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Friedman DB and Johnson TE, A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics, 1988. 118(1): p. 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kenyon C, et al. , A C. elegans mutant that lives twice as long as wild-type. Nature, 1993. 366(6454): p. 461–464. [DOI] [PubMed] [Google Scholar]
- 82.Kimura KD, et al. , daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science, 1997. 277(5328): p. 942–946. [DOI] [PubMed] [Google Scholar]
- 83.Klass MR, Aging in nematode Caenorhabditis elegans - Major biological and environmental factors influencing life span. Mechanisms of Ageing and Development, 1977. 6(6): p. 413–429. [DOI] [PubMed] [Google Scholar]
- 84.Sutphin GL and Kaeberlein M, Measuring Caenorhabditis elegans life span on solid media. J Vis Exp, 2009(27). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller H, et al. , Genetic interaction with temperature is an important determinant of nematode longevity. Aging Cell, 2017. 16(6): p. 1425–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han B, et al. , Microbial Genetic Composition Tunes Host Longevity. Cell, 2017. 169(7): p. 1249–1262.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stroustrup N, et al. , The Caenorhabditis elegans Lifespan Machine. Nature Methods, 2013. 10(7): p. 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xian B, et al. , WormFarm: a quantitative control and measurement device toward automated Caenorhabditis elegans aging analysis. Aging Cell, 2013. 12(3): p. 398–409. [DOI] [PubMed] [Google Scholar]
- 89.He Y and Jasper H, Studying aging in Drosophila. Methods, 2014. 68(1): p. 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cannon L, et al. , Expression patterns of cardiac aging in Drosophila. Aging Cell, 2017. 16(1): p. 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Toivonen JM and Partridge L, Endocrine regulation of aging and reproduction in Drosophila. Molecular and Cellular Endocrinology, 2009. 299(1): p. 39–50. [DOI] [PubMed] [Google Scholar]
- 92.Jones MA and Grotewiel M, Drosophila as a model for age-related impairment in locomotor and other behaviors. Experimental Gerontology, 2011. 46(5): p. 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grotewiel MS, et al. , Functional senescence in Drosophila melanogaster. Ageing Research Reviews, 2005. 4(3): p. 372–397. [DOI] [PubMed] [Google Scholar]
- 94.Rose MR, Laboratory evolution of postponed senescence in Drosophila melanogaster. Evolution, 1984. 38(5): p. 1004–1010. [DOI] [PubMed] [Google Scholar]
- 95.Mueller LD, Evolution of accelerated senescence in laboratory populations of Drosophila. Proceedings of the National Academy of Sciences of the United States of America, 1987. 84(7): p. 1974–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Clancy DJ, et al. , Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science, 2001. 292(5514): p. 104–6. [DOI] [PubMed] [Google Scholar]
- 97.Tu MP, Epstein D, and Tatar M, The demography of slow aging in male and female Drosophila mutant for the insulin-receptor substrate homologue chico. Aging Cell, 2002. 1(1): p. 75–80. [DOI] [PubMed] [Google Scholar]
- 98.Tatar M, et al. , A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science, 2001. 292(5514): p. 107–10. [DOI] [PubMed] [Google Scholar]
- 99.Bushey D, et al. , Sleep, aging, and lifespan in Drosophila. Bmc Neuroscience, 2010. 11: p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Iliadi KG, Iliadi NN, and Boulianne GL, Regulation of Drosophila life-span: Effect of genetic background, sex, mating and social status. Experimental Gerontology, 2009. 44(8): p. 546–553. [DOI] [PubMed] [Google Scholar]
- 101.Grandison RC, et al. , Effect of a Standardised Dietary Restriction Protocol on Multiple Laboratory Strains of Drosophila melanogaster. Plos One, 2009. 4(1): p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hollingsworth MJ, Temperature and length of life in Drosophila. Exp Gerontol, 1969. 4(1): p. 49–55. [DOI] [PubMed] [Google Scholar]
- 103.Chapman T and Partridge L, Female fitness in Drosophila melanogaster: An interaction between the effect of nutrition and of encounter rate with males. Proceedings of the Royal Society B-Biological Sciences, 1996. 263(1371): p. 755–759. [DOI] [PubMed] [Google Scholar]
- 104.Linford NJ, et al. , Measurement of Lifespan in Drosophila melanogaster. Jove-Journal of Visualized Experiments, 2013(71): p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Piper MDW and Partridge L, Protocols to study ageing in Drosophila. Methods in molecular biology (Clifton, N.J.), 2016. 1478: p. 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Partridge L and Gems D, Mechanisms of aging: public or private? Nature Reviews Genetics, 2002. 3: p. 165–175. [DOI] [PubMed] [Google Scholar]
- 107.Sinclair DA and Guarente L, Extrachromosomal rDNA circles - A cause of aging in yeast. Cell, 1997. 91(7): p. 1033–1042. [DOI] [PubMed] [Google Scholar]
- 108.Defossez PA, et al. , Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Molecular Cell, 1999. 3(4): p. 447–455. [DOI] [PubMed] [Google Scholar]
- 109.Hallgren J, et al. , Neurodegeneration-associated instability of ribosomal DNA. Biochimica Et Biophysica Acta-Molecular Basis of Disease, 2014. 1842(6): p. 860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.dos Reis M, et al. , Uncertainty in the Timing of Origin of Animals and the Limits of Precision in Molecular Timescales. Current Biology, 2015. 25(22): p. 2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Letunic I and Bork P, Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res, 2016. 44(W1): p. W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Giaever G and Nislow C, The Yeast Deletion Collection: A Decade of Functional Genomics. Genetics, 2014. 197(2): p. 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McCormick MA, et al. , A Comprehensive Analysis of Replicative Lifespan in 4,698 Single-Gene Deletion Strains Uncovers Conserved Mechanisms of Aging. Cell Metabolism, 2015. 22(5): p. 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Burtner CR, et al. , A genomic analysis of chronological longevity factors in budding yeast. Cell Cycle, 2011. 10(9): p. 1385–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fabrizio P, et al. , Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet, 2010. 6(7): p. e1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schleit J, et al. , Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging Cell, 2013. 12(6): p. 1050–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee MB, et al. , A system to identify inhibitors of mTOR signaling using high-resolution growth analysis in Saccharomyces cerevisiae. Geroscience, 2017. 39(4): p. 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liti G, et al. , Population genomics of domestic and wild yeasts. Nature, 2009. 458(7236): p. 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yue JX, et al. , Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat Genet, 2017. 49(6): p. 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kamath RS and Ahringer J, Genome-wide RNAi screening in Caenorhabditis elegans. Methods, 2003. 30(4): p. 313–321. [DOI] [PubMed] [Google Scholar]
- 121.Kamath RS, et al. , Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature, 2003. 421(6920): p. 231–7. [DOI] [PubMed] [Google Scholar]
- 122.Fire A, et al. , Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 1998. 391: p. 806. [DOI] [PubMed] [Google Scholar]
- 123.Rual JF, et al. , Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res, 2004. 14(10B): p. 2162–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kamath RS, et al. , Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol, 2001. 2(1): p. Research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hamilton B, et al. , A systematic RNAi screen for longevity genes in C. elegans. Genes Dev, 2005. 19(13): p. 1544–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yanos ME, Bennett CF, and Kaeberlein M, Genome-Wide RNAi Longevity Screens in Caenorhabditis elegans. Current Genomics, 2012. 13(7): p. 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dillin A, et al. , Rates of behavior and aging specified by mitochondrial function during development. Science, 2002. 298(5602): p. 2398–401. [DOI] [PubMed] [Google Scholar]
- 128.Lee SS, et al. , A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet, 2003. 33(1): p. 40–8. [DOI] [PubMed] [Google Scholar]
- 129.Hansen M, et al. , New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet, 2005. 1(1): p. 119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Plummer WT, et al. , Standardized Protocols from the Caenorhabditis Intervention Testing Program 2013–2016: Conditions and Assays used for Quantifying the Development, Fertility and Lifespan of Hermaphroditic Caenorhabditis Strains. Protocol Exchange, 2017. [Google Scholar]
- 131.Lucanic M, et al. , Impact of genetic background and experimental reproducibility on identifying chemical compounds with robust longevity effects. Nature Communications, 2017. 8: p. 14256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lithgow GJ, Driscoll M, and Phillips P, A long journey to reproducible results. Nature, 2017. 548(7668): p. 387–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kiontke KC, et al. , A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. Bmc Evolutionary Biology, 2011. 11: p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schulenburg H and Felix MA, The Natural Biotic Environment of Caenorhabditis elegans. Genetics, 2017. 206(1): p. 55–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Barriere A and Felix MA, High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr Biol, 2005. 15(13): p. 1176–84. [DOI] [PubMed] [Google Scholar]
- 136.Mackay TFC, et al. , The Drosophila melanogaster Genetic Reference Panel. Nature, 2012. 482: p. 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang W, et al. , Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Research, 2014. 24(7): p. 1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chow CY and Reiter LT, Etiology of Human Genetic Disease on the Fly. Trends in Genetics, 2017. 33(6): p. 391–398. [DOI] [PubMed] [Google Scholar]
- 139.Lavoy S, et al. , Genetic Modifiers of Neurodegeneration in a <em>Drosophila</em> Model of Parkinson's Disease. Genetics, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mackay TFC and Huang W, Charting the genotype-phenotype map: lessons from the Drosophila melanogaster Genetic Reference Panel. Wiley Interdiscip Rev Dev Biol, 2018. 7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ivanov DK, et al. , Longevity GWAS Using the Drosophila Genetic Reference Panel. Journals of Gerontology Series a-Biological Sciences and Medical Sciences, 2015. 70(12): p. 1470–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huang W, et al. , Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci U S A, 2012. 109(39): p. 15553–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Highfill CA, Reeves GA, and Macdonald SJ, Genetic analysis of variation in lifespan using a multiparental advanced intercross Drosophila mapping population. Bmc Genetics, 2016. 17: p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.King EG, Macdonald SJ, and Long AD, Properties and Power of the Drosophila Synthetic Population Resource for the Routine Dissection of Complex Traits. Genetics, 2012. 191(3): p. 935-U522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.King EG, et al. , Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Research, 2012. 22(8): p. 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kao JY, et al. , Postmating reproductive barriers contribute to the incipient sexual isolation of the United States and Caribbean Drosophila melanogaster. Ecol Evol, 2015. 5(15): p. 3171–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yukilevich R and True JR, Incipient sexual isolation among cosmopolitan Drosophila melanogaster populations. Evolution, 2008. 62(8): p. 2112–21. [DOI] [PubMed] [Google Scholar]
- 148.Fries JF, Aging, natural death, and the compression of morbidity. The New England journal of medicine, 1980. 303(3): p. 130–5. [DOI] [PubMed] [Google Scholar]
- 149.Bassett DE, Boguski MS, and Hieter P, Yeast genes and human disease. Nature, 1996. 379(6566): p. 589–590. [DOI] [PubMed] [Google Scholar]
- 150.Gitler AD, et al. , The Parkinson’s disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proceedings of the National Academy of Sciences of the United States of America, 2008. 105(1): p. 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tenreiro S and Outeiro TF, Simple is good: yeast models of neurodegeneration. Fems Yeast Research, 2010. 10(8): p. 970–979. [DOI] [PubMed] [Google Scholar]
- 152.Outeiro TF and Lindquist S, Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science, 2003. 302(5651): p. 1772–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen X and Petranovic D, Amyloid-beta peptide-induced cytotoxicity and mitochondrial dysfunction in yeast. Fems Yeast Research, 2015. 15(6): p. 10. [DOI] [PubMed] [Google Scholar]
- 154.Herr AJ, et al. , Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet, 2011. 7(10): p. e1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Herr AJ, et al. , DNA replication error-induced extinction of diploid yeast. Genetics, 2014. 196(3): p. 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hanahan D and Weinberg RA, Hallmarks of cancer: the next generation. Cell, 2011. 144(5): p. 646–74. [DOI] [PubMed] [Google Scholar]
- 157.Rayner E, et al. , A panoply of errors: polymerase proofreading domain mutations in cancer. Nature Reviews Cancer, 2016. 16(2): p. 71–81. [DOI] [PubMed] [Google Scholar]
- 158.Tomasetti C, Li L, and Vogelstein B, Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science, 2017. 355(6331): p. 1330–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kumar DKV, et al. , Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease. Science Translational Medicine, 2016. 8(340): p. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.McColl G, et al. , Utility of an improved model of amyloid-beta (Aβ1–42) toxicity in Caenorhabditis elegansfor drug screening for Alzheimer’s disease. Molecular Neurodegeneration, 2012. 7(1): p. 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Keowkase R, Aboukhatwa M, and Luo YA, Fluoxetine protects against amyloid-beta toxicity, in part via daf-16 mediated cell signaling pathway, in Caenorhabditis elegans. Neuropharmacology, 2010. 59(4–5): p. 358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Haun C, et al. , Rescue of Caenorhabditis elegans pharyngeal development by a vertebrate heart specification gene. Proceedings of the National Academy of Sciences of the United States of America, 1998. 95(9): p. 5072–5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Huang C, Xiong CJ, and Kornfeld K, Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America, 2004. 101(21): p. 8084–8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ayyadevara S, et al. , Aspirin Inhibits Oxidant Stress, Reduces Age-Associated Functional Declines, and Extends Lifespan of Caenorhabditis elegans. Antioxidants & Redox Signaling, 2013. 18(5): p. 481–490. [DOI] [PubMed] [Google Scholar]
- 165.Haroon S, et al. , Multiple Molecular Mechanisms Rescue mtDNA Disease in C-elegans. Cell Reports, 2018. 22(12): p. 3115–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bratic I, Hench J, and Trifunovic A, Caenorhabditis elegans as a model system for mtDNA replication defects. Methods, 2010. 51(4): p. 437–443. [DOI] [PubMed] [Google Scholar]
- 167.Maglioni S and Ventura N, C. elegans as a model organism for human mitochondrial associated disorders. Mitochondrion, 2016. 30: p. 117–125. [DOI] [PubMed] [Google Scholar]
- 168.Jones KT and Ashrafi K, Caenorhabditis elegans as an emerging model for studying the basic biology of obesity. Disease Models & Mechanisms, 2009. 2(5–6): p. 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.McKay RM, et al. , C elegans: a model for exploring the genetics of fat storage. Dev Cell, 2003. 4(1): p. 131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ganner A and Neumann-Haefelin E, Genetic kidney diseases: Caenorhabditis elegans as model system. Cell and Tissue Research, 2017. 369(1): p. 105–118. [DOI] [PubMed] [Google Scholar]
- 171.Barr MM, Caenorhabditis elegans as a model to study renal development and disease: Sexy cilia. Journal of the American Society of Nephrology, 2005. 16(2): p. 305–312. [DOI] [PubMed] [Google Scholar]
- 172.Pandey UB and Nichols CD, Human Disease Models in Drosophila melanogaster and the Role of the Fly in Therapeutic Drug Discovery. Pharmacological Reviews, 2011. 63(2): p. 411–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xu ZH, et al. , Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proceedings of the National Academy of Sciences of the United States of America, 2013. 110(19): p. 7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lee EB, Lee VMY, and Trojanowski JQ, Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nature Reviews Neuroscience, 2012. 13(1): p. 38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dawson TM, Ko HS, and Dawson VL, Genetic Animal Models of Parkinson’s Disease. Neuron, 2010. 66(5): p. 646–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Poole AC, et al. , The PINK1/Parkin pathway regulates mitochondrial morphology. Proceedings of the National Academy of Sciences of the United States of America, 2008. 105(5): p. 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Matsuda N, et al. , PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. Journal of Cell Biology, 2010. 189(2): p. 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.van Alphen B and van Swinderen B, Drosophila strategies to study psychiatric disorders. Brain Research Bulletin, 2013. 92: p. 1–11. [DOI] [PubMed] [Google Scholar]
- 179.Guo F, et al. , Circadian neuron feedback controls the Drosophila sleep-activity profile. Nature, 2016. 536(7616): p. 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Burman JL, et al. , A Drosophila model of mitochondrial disease caused by a complex I mutation that uncouples proton pumping from electron transfer. Disease Models & Mechanisms, 2014. 7(10): p. 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sonoshita M and Cagan RL, Chapter Nine - Modeling Human Cancers in Drosophila, in Current Topics in Developmental Biology, Pick L, Editor. 2017, Academic Press; p. 287–309. [DOI] [PubMed] [Google Scholar]
- 182.Nishimura M, et al. , Drosophila as a model to study cardiac aging. Experimental Gerontology, 2011. 46(5): p. 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zerofsky M, et al. , Aging of the innate immune response in Drosophila melanogaster. Aging Cell, 2005. 4(2): p. 103–108. [DOI] [PubMed] [Google Scholar]
- 184.Hu CK and Brunet A, The African turquoise killifish: A research organism to study vertebrate aging and diapause. Aging Cell, 2018. 17(3): p. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Valdesalici S and Cellerino A, Extremely short lifespan in the annual fish Nothobranchius furzeri. Proc Biol Sci, 2003. 270 Suppl 2: p. S189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dodzian J, et al. , A Protocol for Laboratory Housing of Turquoise Killifish (Nothobranchius furzeri). J Vis Exp, 2018(134). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kim Y, Nam HG, and Valenzano DR, The short-lived African turquoise killifish: an emerging experimental model for ageing. Disease Models & Mechanisms, 2016. 9(2): p. 115–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Valenzano DR, et al. , Temperature affects longevity and age-related locomotor and cognitive decay in the short-lived fish Nothobranchius furzeri. Aging Cell, 2006. 5(3): p. 275–8. [DOI] [PubMed] [Google Scholar]
- 189.Valenzano DR, et al. , Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr Biol, 2006. 16(3): p. 296–300. [DOI] [PubMed] [Google Scholar]
- 190.Smith P, et al. , Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Harel I, Valenzano DR, and Brunet A, Efficient genome engineering approaches for the short-lived African turquoise killifish. Nat Protoc, 2016. 11(10): p. 2010–2028. [DOI] [PubMed] [Google Scholar]
- 192.Valenzano Dario R., et al. , The African Turquoise Killifish Genome Provides Insights into Evolution and Genetic Architecture of Lifespan. Cell, 2015. 163(6): p. 1539–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Speakman JR and Mitchell SE, Caloric restriction. Molecular Aspects of Medicine, 2011. 32(3): p. 159–221. [DOI] [PubMed] [Google Scholar]
- 194.Genade T, et al. , Annual fishes of the genus Nothobranchius as a model system for aging research. Aging Cell, 2005. 4(5): p. 223–233. [DOI] [PubMed] [Google Scholar]
- 195.Bielas J, et al. , Long term rapamycin treatment improves mitochondrial DNA quality in aging mice. Experimental Gerontology, 2018. 106: p. 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Karnewar S, et al. , Metformin regulates mitochondrial biogenesis and senescence through AMPK mediated H3K79 methylation: Relevance in age-associated vascular dysfunction. Biochimica Et Biophysica Acta-Molecular Basis of Disease, 2018. 1864(4): p. 1115–1128. [DOI] [PubMed] [Google Scholar]
- 197.Hou YJ, et al. , NAD(+) supplementation normalizes key Alzheimer’s features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proceedings of the National Academy of Sciences of the United States of America, 2018. 115(8): p. E1876–E1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Luan YA, et al. , Chronic Caffeine Treatment Protects Against alpha-Synucleinopathy by Reestablishing Autophagy Activity in the Mouse Striatum. Frontiers in Neuroscience, 2018. 12: p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wilkinson JE, et al. , Rapamycin slows aging in mice. Aging Cell, 2012. 11(4): p. 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dutta S and Sengupta P, Men and mice: Relating their ages. Life Sciences, 2016. 152: p. 244–248. [DOI] [PubMed] [Google Scholar]
- 201.Pettan-Brewer C and Treuting PM, Practical pathology of aging mice. Pathobiology of Aging & Age Related Diseases, 2011. 1: p. 10.3402/pba.v1i0.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Snyder JM, Ward JM, and Treuting PM, Cause-of-Death Analysis in Rodent Aging Studies. Vet Pathol, 2016. 53(2): p. 233–43. [DOI] [PubMed] [Google Scholar]
- 203.Ladiges W, et al. , A New Preclinical Paradigm for Testing Anti-Aging Therapeutics. Journals of Gerontology Series a-Biological Sciences and Medical Sciences, 2017. 72(6): p. 760–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Brayton CF, Treuting PM, and Ward JM, Pathobiology of Aging Mice and GEM: Background Strains and Experimental Design. Veterinary Pathology, 2012. 49(1): p. 85–105. [DOI] [PubMed] [Google Scholar]
- 205.Leach MC, Thornton PD, and Main DCJ, Identification of appropriate measures for the assessment of laboratory mouse welfare. Animal Welfare, 2008. 17(2): p. 161–170. [Google Scholar]
- 206.Conour LA, Murray KA, and Brown MJ, Preparation of animals for research - Issues to consider for rodents and rabbits. Ilar Journal, 2006. 47(4): p. 283–293. [DOI] [PubMed] [Google Scholar]
- 207.Bogue MA, et al. , Accessing Data Resources in the Mouse Phenome Database for Genetic Analysis of Murine Life Span and Health Span. J Gerontol A Biol Sci Med Sci, 2016. 71(2): p. 170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yuan R, et al. , Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell, 2009. 8(3): p. 277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Austin CP, et al. , The knockout mouse project. Nature Genetics, 2004. 36(9): p. 921–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Williams RW, et al. , Genetic structure of the LXS panel of recombinant inbred mouse strains: a powerful resource for complex trait analysis. Mammalian Genome, 2004. 15(8): p. 637–647. [DOI] [PubMed] [Google Scholar]
- 211.Liao CY, et al. , Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging cell, 2010. 9(1): p. 92–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chesler EJ, et al. , The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mammalian Genome, 2008. 19(6): p. 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Churchill G, et al. , The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nature Genetics, 2004. 36(11): p. 1133–1137. [DOI] [PubMed] [Google Scholar]
- 214.Churchill GA, et al. , The Diversity Outbred mouse population. Mamm Genome, 2012. 23(9–10): p. 713–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Logan RW, et al. , High-precision genetic mapping of behavioral traits in the diversity outbred mouse population. Genes Brain Behav, 2013. 12(4): p. 424–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Svenson KL, et al. , High-Resolution Genetic Mapping Using the Mouse Diversity Outbred Population. Genetics, 2012. 190(2): p. 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Warner HR, et al. , Program for testing biological interventions to promote healthy aging. Mechanisms of Ageing and Development, 2000. 115(3): p. 199–208. [DOI] [PubMed] [Google Scholar]
- 218.Nadon NL, et al. , Design of aging intervention studies: the NIA interventions testing program. Age, 2008. 30(4): p. 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jackson AU, et al. , Multiple-trait quantitative trait loci analysis using a large mouse sibship. Genetics, 1999. 151(2): p. 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Miller RA, et al. , Exotic mice as models for aging research: polemic and prospectus. Neurobiology of aging, 1999. 20(2): p. 217–31. [DOI] [PubMed] [Google Scholar]
- 221.Miller RA, et al. , An Aging Interventions Testing Program: study design and interim report. Aging Cell, 2007. 6(4): p. 565–75. [DOI] [PubMed] [Google Scholar]
- 222.Miller RA, et al. , Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci, 2011. 66(2): p. 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Strong R, et al. , Evaluation of Resveratrol, Green Tea Extract, Curcumin, Oxaloacetic Acid, and Medium-Chain Triglyceride Oil on Life Span of Genetically Heterogeneous Mice. Journals of Gerontology Series a-Biological Sciences and Medical Sciences, 2013. 68(1): p. 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Harrison DE, et al. , Acarbose, 17-alpha-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell, 2014. 13(2): p. 273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Strong R, et al. , Longer lifespan in male mice treated with a weakly estrogenic agonist, an antioxidant, an alpha-glucosidase inhibitor or a Nrf2-inducer. Aging Cell, 2016. 15(5): p. 872–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Nadon NL, et al. , NIA Interventions Testing Program: Investigating Putative Aging Intervention Agents in a Genetically Heterogeneous Mouse Model. EBioMedicine, 2017. 21: p. 3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Day C-P, Merlino G, and Van Dyke T, Preclinical Mouse Cancer Models: A Maze of Opportunities and Challenges. Cell, 2015. 163(1): p. 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Janus C and Welzl H, Mouse models of neurodegenerative diseases: criteria and general methodology. Methods Mol Biol, 2010. 602: p. 323–45. [DOI] [PubMed] [Google Scholar]
- 229.Bellantuono I and Potter PK, Modelling ageing and age-related disease. Drug Discovery Today: Disease Models, 2016. 20: p. 27–32. [Google Scholar]
- 230.Varela I, et al. , Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature, 2005. 437: p. 564. [DOI] [PubMed] [Google Scholar]
- 231.Mounkes LC, et al. , A progeroid syndrome in mice is caused by defects in A-type lamins. Nature, 2003. 423(6937): p. 298–301. [DOI] [PubMed] [Google Scholar]
- 232.Gurkar AU and Niedernhofer LJ, Comparison of mice with accelerated aging caused by distinct mechanisms. Experimental Gerontology, 2015. 68: p. 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Osorio FG, et al. , Accelerated ageing: from mechanism to therapy through animal models. Transgenic Research, 2009. 18(1): p. 7–15. [DOI] [PubMed] [Google Scholar]
- 234.Baker DJ, et al. , BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet, 2004. 36(7): p. 744–9. [DOI] [PubMed] [Google Scholar]
- 235.Weeda G, et al. , Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol, 1997. 7(6): p. 427–39. [DOI] [PubMed] [Google Scholar]
- 236.Deepa SS, et al. , A new mouse model of frailty: the Cu/Zn superoxide dismutase knockout mouse. Geroscience, 2017. 39(2): p. 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Snider TA, et al. , The Geropathology Grading Platform demonstrates that mice null for Cu/Zn-superoxide dismutase show accelerated biological aging. Geroscience, 2018. 40(2): p. 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kruse SE, et al. , Mice with Mitochondrial Complex I Deficiency Develop a Fatal Encephalomyopathy. Cell Metabolism, 2008. 7(4): p. 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zheng X, et al. , Alleviation of neuronal energy deficiency by mTOR inhibition as a treatment for mitochondria-related neurodegeneration. eLife, 2016. 5: p. e13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ferrari M, et al. , Hypoxia treatment reverses neurodegenerative disease in a mouse model of Leigh syndrome. Proc Natl Acad Sci U S A, 2017. 114(21): p. E4241–E4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Jain IH, et al. , Hypoxia as a therapy for mitochondrial disease. Science, 2016. 352(6281): p. 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Leiser SF, et al. , Life-span extension from hypoxia in Caenorhabditis elegans requires both HIF-1 and DAF-16 and is antagonized by SKN-1. J Gerontol A Biol Sci Med Sci, 2013. 68(10): p. 1135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mehta R, et al. , Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science, 2009. 324(5931): p. 1196–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Li Y, et al. , Hypoxia potentially promotes Tibetan longevity. Cell Res, 2017. 27(2): p. 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mattison JA, et al. , Caloric restriction improves health and survival of rhesus monkeys. Nature Communications, 2017. 8: p. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Mattison JA, et al. , Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature, 2012. 489(7415): p. 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Colman RJ, et al. , Caloric restriction delays disease onset and mortality in rhesus monkeys. Science, 2009. 325(5937): p. 201–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Salmon AB, Moving toward ‘common’ use of the marmoset as a non-human primate aging model. Pathobiol Aging Age Relat Dis, 2016. 6: p. 32758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lelegren M, et al. , Pharmaceutical inhibition of mTOR in the common marmoset: effect of rapamycin on regulators of proteostasis in a non-human primate. Pathobiol Aging Age Relat Dis, 2016. 6: p. 31793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tardif SD, et al. , The marmoset as a model of aging and age-related diseases. ILAR J, 2011. 52(1): p. 54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ross CN, et al. , Aging Phenotypes of Common Marmosets (Callithrix jacchus). Journal of Aging Research, 2012. 2012: p. 567143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ross C, et al. , Metabolic consequences of long-term rapamycin exposure on common marmoset monkeys (Callithrix jacchus). Aging (Albany NY), 2015. 7(11): p. 964–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tardif S, et al. , Testing efficacy of administration of the antiaging drug rapamycin in a nonhuman primate, the common marmoset. J Gerontol A Biol Sci Med Sci, 2015. 70(5): p. 577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Song SJ, et al. , Cohabiting family members share microbiota with one another and with their dogs. Elife, 2013. 2: p. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hjelmborg J, et al. , Genetic influence on human lifespan and longevity. Hum Genet, 2006. 119(3): p. 312–21. [DOI] [PubMed] [Google Scholar]
- 256.Passarino G, De Rango F, and Montesanto A, Human longevity: Genetics or Lifestyle? It takes two to tango. Immunity & Ageing : I & A, 2016. 13: p. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Dato S, et al. , The genetics of human longevity: an intricacy of genes, environment, culture and microbiome. Mech Ageing Dev, 2017. 165(Pt B): p. 147–155. [DOI] [PubMed] [Google Scholar]
- 258.Herskind AM, et al. , The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet, 1996. 97(3): p. 319–23. [DOI] [PubMed] [Google Scholar]
- 259.Skytthe A, et al. , Longevity studies in GenomEUtwin. Twin Res, 2003. 6(5): p. 448–54. [DOI] [PubMed] [Google Scholar]
- 260.Kol A, et al. , Companion animals: Translational scientist’s new best friends. Science translational medicine, 2015. 7(308): p. 308ps21-308ps21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Hoffman JM, et al. , The companion dog as a model for human aging and mortality. Aging Cell, 2018. 17(3): p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Jin K, et al. , Multiple morbidities in companion dogs: a novel model for investigating age-related disease. Pathobiology of Aging and Age-related Diseases, 2016. 6: p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Gamo NJ, et al. , Valley of death: A proposal to build a “translational bridge” for the next generation. Neuroscience research, 2017. 115: p. 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Bonnett BN, et al. , Mortality in over 350,000 insured Swedish dogs from 1995–2000: I. Breed-, gender-, age- and cause-specific rates. Acta Veterinaria Scandinavica, 2005. 46(3): p. 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Egenvall A, et al. , Mortality in over 350,000 insured Swedish dogs from 1995–2000: II. Breed-specific age and survival patterns and relative risk for causes of death. Acta Veterinaria Scandinavica, 2005. 46(3): p. 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Mattin MJ, et al. , Prevalence of and Risk Factors for Degenerative Mitral Valve Disease in Dogs Attending Primary-care Veterinary Practices in England. Journal of Veterinary Internal Medicine, 2015. 29(3): p. 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Urfer SR, et al. , Asymptomatic heart valve dysfunction in healthy middle-aged companion dogs and its implications for cardiac aging. Geroscience, 2017. 39(1): p. 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Caruana EJ, et al. , Longitudinal studies. Journal of Thoracic Disease, 2015. 7(11): p. E537–E540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Bailey CJ and Day C, Traditional plant medicines as treatments for diabetes. Diabetes Care, 1989. 12(8): p. 553–564. [DOI] [PubMed] [Google Scholar]
- 270.Watanabe C, Studies in the metabolic changes induced by administration of guanidine bases. J. biol. Chem, 1918. 33: p. 253–265. [Google Scholar]
- 271.Adak T, et al. , A reappraisal on metformin. Regulatory Toxicology and Pharmacology, 2018. 92: p. 324–332. [DOI] [PubMed] [Google Scholar]
- 272.Thomas I and Gregg B, Metformin; a review of its history and future: from lilac to longevity. Pediatric Diabetes, 2017. 18(1): p. 10–16. [DOI] [PubMed] [Google Scholar]
- 273.Soranna D, et al. , Cancer Risk Associated with Use of Metformin and Sulfonylurea in Type 2 Diabetes: A Meta-Analysis. Oncologist, 2012. 17(6): p. 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lee MS, et al. , Type 2 diabetes increases and metformin reduces total, colorectal, liver and pancreatic cancer incidences in Taiwanese: a representative population prospective cohort study of 800,000 individuals. Bmc Cancer, 2011. 11: p. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Decensi A, et al. , Metformin and cancer risk in diabetic patients: a systematic review and meta-analysis. Cancer Prev Res (Phila), 2010. 3(11): p. 1451–61. [DOI] [PubMed] [Google Scholar]
- 276.Zhang ZJ, et al. , Reduced risk of colorectal cancer with metformin therapy in patients with type 2 diabetes: a meta-analysis. Diabetes Care, 2011. 34(10): p. 2323–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Libby G, et al. , New Users of Metformin Are at Low Risk of Incident Cancer A cohort study among people with type 2 diabetes. Diabetes Care, 2009. 32(9): p. 1620–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Bodmer M, et al. , Long-Term Metformin Use Is Associated With Decreased Risk of Breast Cancer. Diabetes Care, 2010. 33(6): p. 1304–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Bannister CA, et al. , Can people with type 2 diabetes live longer than those without? A comparison of mortality in people initiated with metformin or sulphonylurea monotherapy and matched, non-diabetic controls. Diabetes Obesity & Metabolism, 2014. 16(11): p. 1165–1173. [DOI] [PubMed] [Google Scholar]
- 280.Campbell JM, et al. , Metformin reduces all-cause mortality and diseases of ageing independent of its effect on diabetes control: A systematic review and meta-analysis. Ageing Research Reviews, 2017. 40: p. 31–44. [DOI] [PubMed] [Google Scholar]
- 281.Vezina C, Kudelski A, and Sehgal SN, Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo), 1975. 28(10): p. 721–6. [DOI] [PubMed] [Google Scholar]
- 282.Sabatini DM, et al. , RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell, 1994. 78(1): p. 35–43. [DOI] [PubMed] [Google Scholar]
- 283.Sabers CJ, et al. , Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem, 1995. 270(2): p. 815–22. [DOI] [PubMed] [Google Scholar]
- 284.Lorenz MC and Heitman J, TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem, 1995. 270(46): p. 27531–7. [DOI] [PubMed] [Google Scholar]
- 285.Stan R, et al. , Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J Biol Chem, 1994. 269(51): p. 32027–30. [PubMed] [Google Scholar]
- 286.Saunders RN, Metcalfe MS, and Nicholson ML, Rapamycin in transplantation: A review of the evidence. Kidney International, 2001. 59(1): p. 3–16. [DOI] [PubMed] [Google Scholar]
- 287.Eisen HJ, et al. , Everolimus for the prevention of allograft rejection and vasculopathy in cardiac-transplant recipients. New England Journal of Medicine, 2003. 349(9): p. 847–858. [DOI] [PubMed] [Google Scholar]
- 288.Cloughesy TF, et al. , Antitumor activity of rapamycin in a phase I trial for patients with recurrent PTEN-Deficient glioblastoma. Plos Medicine, 2008. 5(1): p. 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Meric-Bernstam F and Gonzalez-Angulo AM, Targeting the mTOR Signaling Network for Cancer Therapy. Journal of Clinical Oncology, 2009. 27(13): p. 2278–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.AFAR, A.F.f.A.R., Biomarkers of Aging: An introduction to aging science brought to you by the American Federation for Aging Research. 2016, American Federation for Aging Research: New York, NY. [Google Scholar]
- 291.Fabris F, Magalhães J.P.d., and Freitas AA, A review of supervised machine learning applied to ageing research. Biogerontology, 2017. 18(2): p. 171–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Burkle A, et al. , MARK-AGE biomarkers of ageing. Mechanisms of Ageing and Development, 2015. 151: p. 2–12. [DOI] [PubMed] [Google Scholar]
- 293.Lara J, et al. , A proposed panel of biomarkers of healthy ageing. BMC Med, 2015. 13: p. 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Horvath S and Raj K, DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nature Reviews Genetics, 2018. 19(6): p. 371–384. [DOI] [PubMed] [Google Scholar]
- 295.Horvath S, DNA methylation age of human tissues and cell types. Genome Biology, 2013. 14(10): p. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Quach A, et al. , Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging-Us, 2017. 9(2): p. 419–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Bocklandt S, et al. , Epigenetic Predictor of Age. Plos One, 2011. 6(6): p. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Hannum G, et al. , Genome-wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Molecular Cell, 2013. 49(2): p. 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Stubbs TM, et al. , Multi-tissue DNA methylation age predictor in mouse. Genome Biology, 2017. 18: p. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Thompson MJ, et al. , An epigenetic aging clock for dogs and wolves. Aging-Us, 2017. 9(3): p. 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Maierhofer A, et al. , Accelerated epigenetic aging in Werner syndrome. Aging (Albany NY), 2017. 9(4): p. 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Horvath S and Levine AJ, HIV-1 Infection Accelerates Age According to the Epigenetic Clock. Journal of Infectious Diseases, 2015. 212(10): p. 1563–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Horvath S, et al. , Accelerated epigenetic aging in Down syndrome. Aging Cell, 2015. 14(3): p. 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Zannas AS, et al. , Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol, 2015. 16: p. 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Palma-Gudiel H, et al. , Maternal psychosocial stress during pregnancy alters the epigenetic signature of the glucocorticoid receptor gene promoter in their offspring: a meta-analysis. Epigenetics, 2015. 10(10): p. 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Gassen NC, et al. , Life stress, glucocorticoid signaling, and the aging epigenome: Implications for aging-related diseases. Neurosci Biobehav Rev, 2017. 74(Pt B): p. 356–365. [DOI] [PubMed] [Google Scholar]
- 307.Olshansky SJ, et al. , In pursuit of the longevity dividend. Scientist, 2006. 20(3): p. 28–36. [Google Scholar]
- 308.Olshansky SJ, Articulating the Case for the Longevity Dividend. Cold Spring Harbor Perspectives in Medicine, 2016. 6(2): p. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Olshansky SJ, The Future of Health. Journal of the American Geriatrics Society, 2018. 66(1): p. 195–197. [DOI] [PubMed] [Google Scholar]
- 310.Hartman M, et al. , National Health Care Spending In 2016: Spending And Enrollment Growth Slow After Initial Coverage Expansions. Health Affairs, 2017. 37(1): p. 150–160. [DOI] [PubMed] [Google Scholar]
- 311.Lassman D, et al. , US Health Spending Trends By Age And Gender: Selected Years 2002–10. Health Affairs, 2014. 33(5): p. 815–822. [DOI] [PubMed] [Google Scholar]
- 312.Dieleman JL, et al. , US Spending on Personal Health Care and Public Health, 1996–2013. Jama-Journal of the American Medical Association, 2016. 316(24): p. 2627–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.An JY, et al. , Rapamycin treatment attenuates age-associated periodontitis in mice. Geroscience, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Creevy KE, et al. , The Companion Dog as a Model for the Longevity Dividend. Cold Spring Harbor Perspectives in Medicine, 2016. 6(1): p. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Wilson KA, et al. , Genome-wide analysis reveals distinct genetic mechanisms of diet-dependent lifespan and healthspan in D. melanogaster. bioRxiv, 2018. [Google Scholar]
- 316.Rikke BA, et al. , Genetic dissection of dietary restriction in mice supports the metabolic efficiency model of life extension. Experimental gerontology, 2010. 45(9): p. 691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Olshansky SJ, Has the Rate of Human Aging Already Been Modified? Cold Spring Harbor Perspectives in Medicine, 2015. 5(12): p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Arias E, Heron M, and Xu J, United States Life Tables, 2014. Natl Vital Stat Rep, 2017. 66(4): p. 1–64. [PubMed] [Google Scholar]
- 319.Dong X, Milholland B, and Vijg J, Evidence for a limit to human lifespan. Nature, 2016. 538: p. 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Heron M, Deaths: Leading Causes for 2014. Natl Vital Stat Rep, 2016. 65(5): p. 1–96. [PubMed] [Google Scholar]