Highlights
-
•
Explore the effect of four virus inactivation methods on the rapid detection results of COVID-19 nucleic acid.
-
•
Four treatment methods and specimens without inactivated treatment have shown good consistency.
-
•
Effectively protect laboratory staff and reduce mental stress at work.
Keywords: COVID-19, RT-PCR, Viral inactivation
Abstract
Objective
This paper aims to explore the effect of four virus inactivation methods on the rapid detection results of COVID-19 nucleic acid.
Methods
Collected samples of nasopharyngeal swabs from 2 patients diagnosed with COVID-19 at the First People's Hospital of Zhaoqing City, each of sample was divided into 5 groups (groupA∼E): A:Non-inactivated raw sample; B:75 % ethanol inactivation; C:56 ℃ incubation for 30 min inactivation; D:65 ℃ incubation for 10 min inactivation; E:Pre-inactivation using RNA virus special preservation fluid added into the sampling tube to treated the nasopharyngeal swab sample separately, using real-time fluorescent RT-PCR to detect the N gene of COVID-19 and the ORF1ab gene simultaneously. All the groups are diluted in 1:2, 1:4, 1:8 ratios. The objectives are to compare the effect of the varied inactivation method on CT(Cycle Threshold)results in PCR, conduct correlation and Bland-Altman analysis.
Results
For the N gene and ORF1ab gene, the CT values of 4 inactivated and Non-inactivated treatment were correlated (P<0.001). The results of the four treatment methods and specimens without inactivated treatment have shown good consistency.
Conclusion
The treatment of nasopharyngeal swab specimens using mentioned four inactivated methods had no significant effect on the subsequent detection of the new COVID-19 nucleic acid test. Lab test-persons can flexibly adopt pre-inactivation methods to ensure the accuracy of virus nucleic acid test results, meanwhile guarantee the safety of lab test-persons.
1. Introduction
COVID-19 has infected more than 2,000,000 people all over the world by April 22nd, including 165,310 deaths, posing a huge threat to the physical and mental health of the general public. At the meantime more than 1000 medical staff were infected in China, resulting in 5 deaths. The First People's Hospital is appointed COVID-19 patients treatment institution in Zhaoqing City. By far it has accepted 20 confirmed cases of new coronary pneumonia patients, out of which 10 cases have been cured and 1 case ended in casualty. According to the COVID-19 Diagnosis and Treatment Plan (6th Trail Edition), this virus is highly contagious and can spread quickly among population. There is no specific diagnosis and treatment program against the pneumonia caused by COVID-19 (Zhu et al., 2020).
Currently, RT-PCR is the most widely adopted clinical screening method for COVID-19 due to its speed, low cost, and convenience in clinical use (Lancet, 2020; National Health Commission (NHC) of the PRC and National Administration of Traditional Chinese Medicine of the PRC, 2020).Since patient virus samples sent for lab testing are extremely infectious, the experimenter should only carry out the experimental operations when their biosecurity protection measurements are fully equipped, following relevant restrictions and safety guidance. According to documents such as the in-use guidance for the diagnosis and treatment of COVID-19, this virus can be inactivated either at 75 % alcohol, 56 degrees incubation for 30 min, or 65 degrees incubation for 10 min according to Chinese expert’s consensus (National Health Commission of the People’s Republic of China, 2020; Group of experts on medical treatment from Tongji Hospital affiliated of Wuhan Tongji Medical College, 2020; Chinese Society of Laboratory Medicine, 2020). In order to reduce the risk of in-lab infection during the experimental operation, reduce the mental stress of laboratory staff, the high-risk specimen will be inactivated before the lab-test undergoes. However it remains unknown whether the specimen inactivation would affect the subsequent RT-PCR test results or not. Therefore, this study intends to compare 4 different inactivation methods to explore impact of the 4 inactivation methods upon the results of viral nucleic acid PCR detection, and to lower the risk of medical staff in-lab infection while ensuring the accuracy of the test, and eventually to provide new solutions for the laboratory biosecurity environment protection.
2. Materials and methods
Specimen Source: 2 confirmed COVID-19 infected cases in Zhaoqing City.
Description: 2 patients were hospitalized due to fever, RT-PCR nucleic acid test results showed positive, and confirmed as COVID-19 infected cases by the Guangdong Provincial Center for Disease Control and Prevention.
3. Virus inactivation method
3.1. Specimens treatment
-
•
Group A: 100 μl nasopharyngeal swabs +300 μl Physiological saline;
-
•
Group B:100 μl nasopharyngeal swabs +300 μl Ethanol anhydrous, forming a treatment environment with a final concentration of 75 % ethanol, 30 min still placement at room temperature;
-
•
Group C:100 μl nasopharyngeal swabs +300 μl Physiological saline,56 °C incubation for 30 min;
-
•
Group D:100 μl nasopharyngeal swabs +300 μl Physiological saline,65 °C incubation 10 min;
-
•
Group E:100 μl nasopharyngeal swabs +300 μl RNA Virus-specific storage fluid(DAAN Gene Products Regulation Affairs Center,RNA Virus-specific retention fluid has been pre-added to the sample retention tube. Its inactivation strategy is that: by denaturing proteins, dissolving proteins and eliminating protein's secondary structures, it can lead to the degradation of cell structures and the quick separation of nuclein and nucleic acid to inhibit nuclease and eventually protect RNA from degradation.),30 min still placement at room temperature.
Then the nasopharyngeal swab elution sluice above will be diluted in a ratio of 1:2, 1:4, 1:8 for separated nucleic acid extraction, followed by RT-PCR nucleic acid testing.
3.2. Specimens RNA extraction and RT-PCR
Extraction kits are made by Chongqing Zhong Yuan Biotechnology Co., Ltd and the Nucleic acid extractor manufacturer is Hangzhou Allsheng Instruments Co.,Ltd.
RT-PCR reagents amplify viral genomes(DAAN Gene Co., Ltd. Of Sun Yat-sen University, Lot No.:2020017).
N gene:
Forward Primers (F):GGGGAACTTCTCCTGCTAGAAT
Reverse Primers(R):CAGACATTTTGCTCTCAAGCTG
Fluorescent Probe(P):5′-FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3′
ORF1ab gene:
Forward Primers (F):CCCTGTGGGTTTTACACTTAA
Reverse Primers(R):ACGATTGTGCATCAGCTGA
Fluorescent Probe(P):5′-FAM−CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1−3'
The thermal cycling conditions used were: an initial reverse transcription step of 50℃for 15 min,next to denaturation step of 95℃ for 15 min, followed by 45 cycles of 94℃ for 15 s, 55℃ for 45 s, and a final step of 40℃ for 20 s.
The Applied Biosystems® 7500 instrument is used in amplification test to determine whether it’s negative or positive. If each batch of tests has supplied positive and negative control, and the results are within controlled range, the batch test is valid, on the contrary, invalid. All tests are repeated for 3 times,and test-person will use the average cycle threshold for calculation.
4. Statistical analysis
Statistical analysis software SPSS 19.0 was used in this particular research. Correlation between different specimens CT values were analyzed using Pearson product-moment correlation coefficient analysis, and the graphics were illustrated by Graphpad 5.0. Furthermore, The Bland-Altman plots was drawn using MedCalc software, as well as the the consistency of CT values analysis. P < 0.05 is considered as statistically significant.
5. Results
-
1
Comparison of RT-PCR results of different dilution concentration samples after non-inactivated treatment and inactivated treatment
The results of the RT-PCR were found to be similar to those of the Non-inactive specimens after the detection of specimens with different concentrations, as shown in Table 1 .
-
2
Correlation Analysis: RT-PCR Results of the Non-inactivated Treatment and the Four Inactivated Treatments
Table 1.
CT value (average) of PCR for inactivated and Non-inactivated samples.
| Speciment Number |
Dilution Factor | N |
ORF1ab |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-inactive | 75 % ethanol 30 min | 56 ℃ 30 min | 65 ℃ 10 min | RNA Virus-specific retention fluid | Non-inactive | 75 % ethanol 30 min |
56 ℃ 30 min |
65 ℃ 10 min |
RNA Virus-specific retention fluid | ||
| 1 | 1 | 25.72 | 25.89 | 28.58 | 28.01 | 25.81 | 30.81 | 30.41 | 30.25 | 29.93 | 30.38 |
| 1 | 1/2 | 26.09 | 26.55 | 28.33 | 28.68 | 27.42 | 32.27 | 32.71 | 31.3 | 31.19 | 33.42 |
| 1 | 1/4 | 28.11 | 26.22 | 30.64 | 29.99 | 28.99 | 34.23 | 33.57 | 32.98 | 32.55 | 34.96 |
| 1 | 1/8 | 26.21 | 24.87 | 32.81 | 31.71 | 30.61 | 37.05 | 35.41 | 36.89 | 35.29 | 36.65 |
| 2 | 1 | 33.63 | 35.06 | 35.85 | 35.09 | 34.75 | 37.03 | 40.53 | 39.12 | 35.61 | 36.4 |
| 2 | 1/2 | 34.76 | 35.58 | 36.94 | 35.95 | 35.97 | 39.11 | 42.91 | 43.85 | 36.51 | 38.21 |
| 2 | 1/4 | 36.13 | 37.81 | 37.39 | 38.66 | 37.72 | 39.95 | 44.36 | 44.28 | 38.16 | 41.92 |
| 2 | 1/8 | 37.42 | 39.12 | 39.04 | 40.76 | 37.91 | 43.79 | 46.36 | 46.77 | 41.63 | 42.15 |
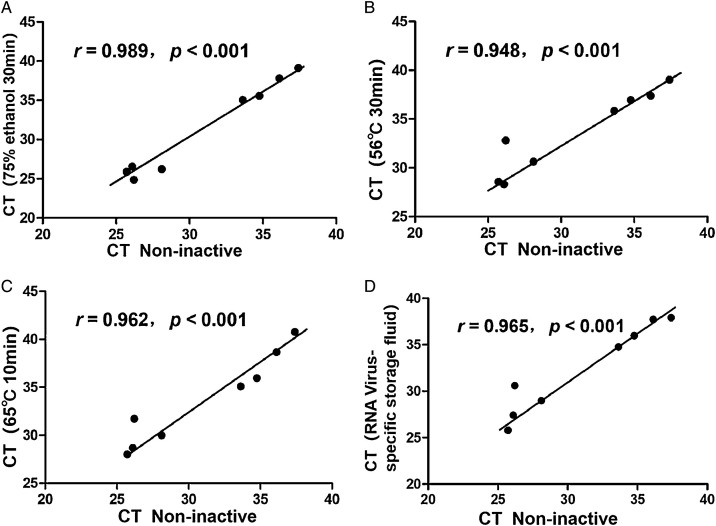
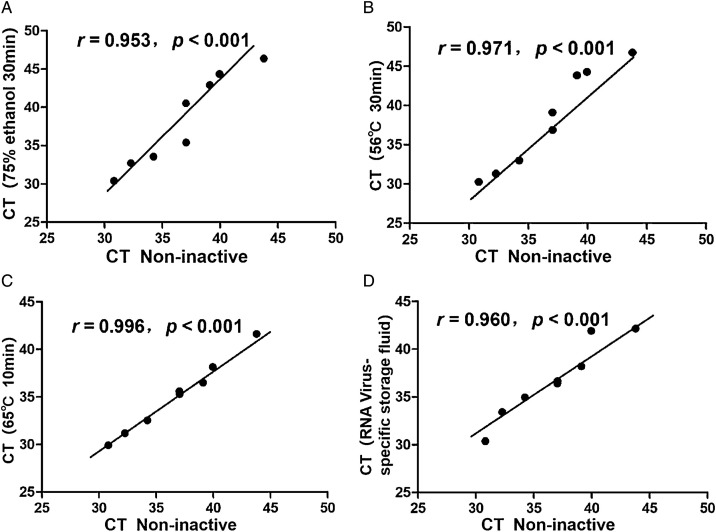
Using Pearson product-moment correlation coefficient analysis, the results showed that the CT values between the four inactivated and non-inactivated treatments were correlated with both N gene and ORF1ab gene (P<0.001)(Table 2 ), N gene(Fig. 1 ), ORF1ab gene(Fig. 2 ).
Table 2.
Analysis of correlation between non-inactivated and inactivated treatment.
| Non-inactive Variable |
N |
ORF1ab |
||||||
|---|---|---|---|---|---|---|---|---|
| 75 %ethanol(30 min) | 56 ℃ (30 min) |
65 ℃ (10 min) |
RNA Virus-specific retention fluid | 75 %ethanol (30 min) |
56 ℃ (30 min) |
65 ℃ (10 min) |
RNA Virus-specific retention fluid | |
| r | 0.989 | 0.948 | 0.962 | 0.965 | 0.953 | 0.971 | 0.996 | 0.960 |
| p | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
Fig. 1.
Analysis of correlation between non-inactivated and inactivated treatment(N gene).
Fig. 2.
Analysis of correlation between non-inactivated and inactivated treatment(ORF1ab gene).
Figure Legends:Horizontal axis represent non-inactivated treatment CT values, and vertical axis represent CT values of the four inactivated treatments.
A:Correlation analysis of CT values between non-inactivated and 75 % ethanol inactivated;
B:Correlation analysis of CT values between non-inactivated and 56 ℃ 30 min inactivated;
C:Correlation analysis of CT values between non-inactivated and 65 ℃ 10 min inactivated;
D:Correlation analysis of CT values between non-inactivated and RNA Virus-specific storage fluid inactivated.
Figure Legends:Horizontal axis represent non-inactivated treatment CT values, and vertical axis represent CT values of the four inactivated treatments.
A:Correlation analysis of CT values between non-inactivated and 75 % ethanol inactivated;
B:Correlation analysis of CT values between non-inactivated and 56 ℃ 30 min inactivated;
C:Correlation analysis of CT values between non-inactivated and 65 ℃ 10 min inactivated;
D:Correlation analysis of CT values between non-inactivated and RNA Virus-specific storage fluid inactivated.
-
3
Consistency Analysis of the RT-PCR Results of the Non-inactivated Treatment and Four Inactivated Treatments
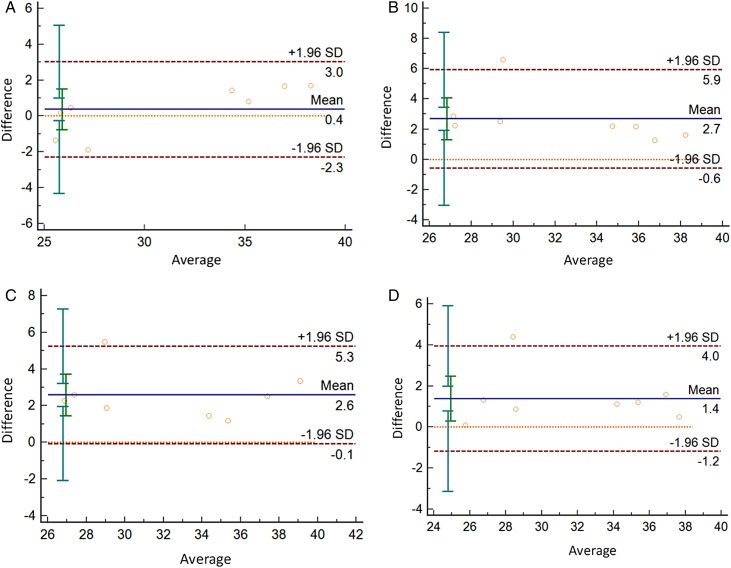
The scatter diagram will be drawn using the Bland-Altman method, for which the difference between four inactivated methods and the non-inactive treatment is the vertical axis, and the average value is the horizontal axis. As the figure shown, for the N gene, 12.5 % (1/8) of scattered points of 56℃/30 min inactivated sample, 65℃/10 min sample, RNA virus-specific preservation fluid sample were distributed outside the 95 % trust interval. For the 75 % ethanol sample, the mean of the CT value difference of inactivated and non-inactivated is among is 0.4. 95 %CI(-2.3,3.0),Fig. 3 .
Fig. 3.
Consistency analysis between non-inactivated and inactivated treatment(N gene).
A:Consistency analysis of CT values between non-inactivated and 75 % ethanol inactivated;
B:Consistency analysis of CT values between non-inactivated and 56 ℃ 30 min inactivated;
C:Consistency analysis of CT values between non-inactivated and 65 ℃ 10 min inactivated;
D:Consistency analysis of CT values between non-inactivated and RNA Virus-specific storage fluid inactivated.
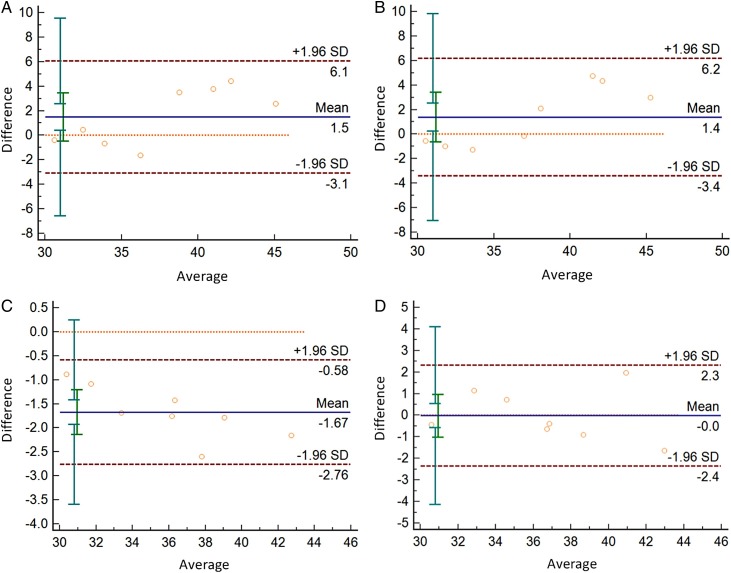
For the ORF1ab gene, the mean of the CT value difference of 75 % 30 min ethanol inactivated and non-inactivated is among is 1.5, 95 %CI(-3.1,6.1); 56℃ 30 min inactivated difference among means of CT value is 1.4, 95 %CI(-3.4,6.2); 65℃ 10 min inactivated difference among means of CT value is -1.67, 95 %CI(-2.76,-0.58);RNA Virus-specific storage fluid in inactivated difference among means of CT value is 0, 95 %CI(-2.4,2.3), All points are distributed within the 95 % trust interval (Fig. 4 ).
Fig. 4.
Consistency analysis between non-inactivated and inactivated treatment(ORF1ab gene).
A:Consistency analysis of CT values between non-inactivated and 75 % ethanol inactivated;
B:Consistency analysis of CT values between non-inactivated and 56 ℃ 30 min inactivated;
C:Consistency analysis of CT values between non-inactivated and 65 ℃ 10 min inactivated;
D:Consistency analysis of CT values between non-inactivated and RNA Virus-specific storage fluid inactivated.
6. Discuss
Recent studies have found that the COVID-19 is mainly spread through droplets, body contact, respiratory aerosols and daily contacts in an infected environment. Fever, Fatigue, dry cough, etc. are the first symptoms to some patients, while others start with diarrhea as the first symptoms, in some cases, patients are symptom-free, regarded as asymptomatic infection. A small number of patients may suffer acute respiratory distress syndrome, heavy metabolic acidosis and blood clotting dysfunction, even death (Huang et al., 2020).
According to the relevant documents, the COVID-19 is sensitive to heat, therefore solvents such as 56℃ 30 min, ether.75 % ethanol, Chlorine-containing disinfectants, Peroxyacetic acid and chloromethane can effectively eliminate the virus. Nucleic acid testing is by far the most effective method to confirm the infection of COVID-19.
In order to reduce the possibility in-lab spread of COVID-19, while ensure the accuracy of the experimental results, in this particular study, the Pearson product-moment correlation coefficient and Bland-Altman analysis method were applied to testify the PCR correlation and co-efficiency between non-inactivated samples and samples inactivated using the four methods respectively. As the data analysis results show, for the N gene and ORF1ab gene, the CT values of 4 inactivated and non-inactivated treatment were correlated (P<0.001). Besides, as the figure shown, for the N gene, 12.5 % (1/8) of scattered points of 56℃/30 min inactivated sample, 65℃/10 min sample, RNA virus-specific preservation fluid sample were distributed outside the 95 % trust interval. For the ORF1ab gene, All points are distributed within the 95 % trust interval.This indicates that the results of the four treatment methods and non-inactivation treatment have shown good consistency.
This suggests that, before conducting viral nucleic acid testing, inactivate nasopharyngeal swab specimens using 75 % ethanol, 56 ℃ 30 min, 65 ℃ 10 min, or RNA virus-specific preservation fluid will impose no significant effect on subsequent COVID-19 nucleic acid testing, which can also effectively protect laboratory staff and reduce mental stress at work. Most laboratories tend to use three other inactivation methods as the RNA virus-specific preservation fluid tubes need to be purchased separately. There are also some limitations in this research because only two specimens were analyzed. Furthermore, dilution of the sample may lead to the decreases in detection efficiency. In this case for some specimens with low CT value, the sample size needs to be expanded for further analysis. Besides in this particular study, used only one test kit box is used for analysis, and it is suggested that future researchers should expanded the scope of study, in order to provide up-to-date strategies for virus inactivation.
In addition, according to the longitudinal comparison of RNA test results from 20 COVID-19 confirmed cases in the First People's Hospital of Zhaoqing City for nearly 30 days, It is suggested that nasopharyngeal swab nucleic acid test results starts to turn and remain negative two or three week after hospitalized, while cloacal swab results show continuous positive, which signifies that, viruses may have starts metastasis to alimentary canals.
CRediT authorship contribution statement
Yueying Wang: Investigation, Methodology, Resources, Visualization, Writing - original draft. Wei Song: Investigation, Methodology, Resources, Writing - original draft. Zuguo Zhao: Methodology, Visualization, Writing - review & editing. Ping Chen: Validation. Jian Liu: Validation. Chende Li: Supervision, Conceptualization.
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- Chinese Society of Laboratory Medicine The experts’ consensus on nucleic acid detection of 2019-nCoV. Zhonghua Yi Xue Za Zhi. 2020;100(00):E003. doi: 10.3760/cma.j.issn.0376-2491.2020.0003. [Chinese] [DOI] [Google Scholar]
- Group of experts on medical treatment from Tongji Hospital affiliated of Wuhan Tongji Medical College Guidelines for diagnosis and treatment of Corona Virus Disease 2019 (Third Edition),[J] Herald of Medicine. 2020:1–9. doi: 10.3870/j.issn.1004-0781.2020.03.001[Chinese]. [DOI] [Google Scholar]
- Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China[J] Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancet T. Emerging understandings of 2019-nCoV[J] Lancet. 2020;395(10221):311. doi: 10.1016/S0140-6736(20)30186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Health Commission (NHC) of the PRC and National Administration of Traditional Chinese Medicine of the PRC . seventh edition. 2020. Guidance for Corona Virus Disease 2019: Prevention, Control, Diagnosis and Management.http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml [Chinese] [Google Scholar]
- National Health Commission of the People’s Republic of China . 2020. Handbook of Prevention and Treatment of the Pneumonia Caused by the 2019-nCoV.http://en.nhc.gov.cn/2020-02/06/c_76295.htm [Chinese] [Google Scholar]
- Zhu N., Zhang D., Wang W. A Novel Coronavirus from patients with pneumonia in China, 2019[J] N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]