Highlights
-
•
High sensitivity of Simplexa™ COVID-19 Direct assay.
-
•
100 % specificity of this new assay.
-
•
“Almost perfect” agreement with Corman’s method (κ = 0.938).
-
•
High-speed detection, all-in-one reagent mix and no separate extraction required.
-
•
Promising for laboratory diagnosis of COVID-19 and field application.
Keywords: SARS-CoV-2, Real-time RT-PCR, Rapid response, Surveillance
Abstract
Background
So far, one of the major drawbacks of the available molecular assays for the diagnosis of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) is the need for viral nucleic acid extraction from clinical specimens.
Objective
The aim of this study was to evaluate the performances of a newly designed real-time RT-PCR (Simplexa™ COVID-19 Direct assay), that is established with an all-in-one reagent mix and no separate extraction required.
Results
The lower limit of detection (LOD) for both target genes resulted the same: 3.2 (CI: 2.9–3.8) log10 cp/mL and 0.40 (CI: 0.2–1.5) TCID50/mL for S gene while 3.2 log10 (CI: 2.9–3.7) log10 cp/mL and 0.4 (CI: 0.2–1.3) TCID50/mL for ORF1ab. The LOD obtained with extracted viral RNA for both S gene or ORF1ab was 2.7 log10 cp/mL. Crossreactive analysis performed in 20 nasopharyngeal swabs confirmed a 100% of clinical specificity of the assay. Clinical performances of Simplexa™ COVID-19 Direct assay were assessed in 278 nasopharyngeal swabs tested in parallel with Corman's method. Concordance analysis showed an "almost perfect" agreement in SARS-CoV-2 RNA detection between the two assays, being κ = 0.938; SE = 0.021; 95% CI = 0.896-0.980.
Conclusions
The high sensitivity and specificity of this new assay indicate that it is promising for laboratory diagnosis, enabling highspeed detection in just over one hour, which is significantly faster than the up to five hours currently required by traditional extraction followed by amplification technologies, thus allowing prompt decision making regarding isolation of infected patients.
1. Background
The pandemic caused by the severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) has generated global concern given its rapid spread in multiple countries and fatal progression of the Coronavirus infection disease (COVID-19) [1,2]. However, asymptomatic or paucisymptomatic cases especially in the early phase of the infection have also been reported [3,4]. Since the spread of SARS-CoV-2 out of China, one of the main concerns on the diagnosis of this new virus has been the development of rapid and sensitive diagnostic assays. So far, nucleic acid amplification testing is the gold standard for the diagnosis of SARS-CoV-2 in respiratory samples [5,6]. Molecular assays as previously demonstrated for epidemics caused by other coronaviruses (SARS-CoV and MERS-CoV), are crucial for rapid application of infection control measures, case identification and contact tracing [6,7]. However, one of the major drawbacks of these assays is the need for viral nucleic acid extraction from clinical specimens. Recently, DiaSorin S.p.A. (Gerenzano, Italy) has developed a SARS-CoV-2-specific real-time RT-PCR (Simplexa™ COVID-19 Direct assay) performed directly on clinical samples.
2. Objectives
This study was performed at two regional reference laboratories, with the following aims: I) to assess the analytical performances of the newly designed Simplexa™ COVID-19 Direct assay; II) to evaluate its clinical performance as compared to WHO protocols [7,8] using different types of clinical specimens from patients with laboratory-confirmed COVID-19.
3. Study design
3.1. Viruses
Viral stock used to establish analytical sensitivity was the first Italian isolate obtained from a patient hospitalized in the Biosafety Level 3 facility at National Institute for Infectious Diseases “Lazzaro Spallanzani” (INMI) in Rome, named 2019-nCoV/Italy-INMI1 [9]. The infectious titre of the viral stock used in the study, performed by Reed and Muench method on VeroE6 cells, was 107 TCDI50/mL; for the establishment of analytical sensitivity, the viral stock was spiked into oral swab-UTM matrix and serial dilutions (from 100 to 0.001 TCDI50/mL) were tested in multiple replicates. The evaluation of the corresponding RNA copies/mL was performed based on standard curve obtained by Corman’s E-SARS-CoV-2 gene.
3.2. Clinical specimens
A total of 278 consecutive respiratory samples (nasal and nasopharyngeal swabs) collected for COVID-19 diagnosis between 20 February and 24 March 2020 were included in the study; twenty additional nasopharyngeal swabs from patients known to be positive for other Human Coronaviruses were used to establish clinical specificity of the Simplexa™ COVID-19 Direct assay. Moreover, to evaluate the performance of the test in a different clinical specimen, a total of 33 Broncho-Alveolar Lavage (BAL) collected for COVID-19 diagnosis between 20 March and 03 April 2020 were also analysed in parallel with the Simplexa™ COVID-19 Direct assay and the routine laboratory method, based on the WHO protocols [7,8], using the Abbot m2000 extraction platform.
3.3. Simplexa™ COVID-19 direct assay
For Simplexa™ COVID-19 Direct assay, one vial of Reaction Mix was thawed for each sample. Respiratory samples were collected with a flexible nasopharyngeal nylon flocked swabs (FLOQSwabs™, Copan Italia, Brescia, Italy) dipped in 2–3 mL universal transport medium (UTM™, Copan Italia). After swirling, the swab was discarded and 50 μL of sample and 50 μL of Reaction Mix were added to their specific wells on a direct amplification disk, which was loaded onto the LIAISON® MDX instrument (DiaSorin). For BAL, a pre-treatement with 1:1 SV40 Sputagest Selectavial (Mast Diagnostic GmbH, Reinfeld, Germany) was performed.
3.4. COVID-19 real-time RT-PCR assays
Total nucleic acids (DNA/RNA) were extracted from 200 μL of nasal and nasopharyngeal specimens. Clinical samples were pre-treated with 1:1 ATL lysis buffer and extracted using the QIAsymphony® instrument with QIAsymphony® DSP Virus/Pathogen Midi Kit (Complex 400 protocol) according to the manufacturer’s instructions (QIAGEN, Qiagen, Hilden, Germany). The presence of SARS-CoV-2 RNA was assessed by Corman’s protocol, targeting E and RNA-dependent RNA polymerase (RdRp) genes as reference method [7]. According to the WHO guidelines Corman’s protocol was carried out using the E assay for screening and the RdRp assay for confirmation [8]. Total nucleic acids (DNA/RNA) were extracted from 600 μL of BAL samples, pre-treated with 1:1 SV40 Sputagest Selectavial and then with 1:1 ATL lysis buffer, using the Abbott m2000 system according to the manufacturer’s instructions (Abbott molecular, Des Plaines, IL, USA).
3.5. Statistical analysis
The analytical sensitivity (SARS-Cov-2 copy number at a 95% detection rate and SARS-Cov-2 TCDI50 at a 95% detection rate) was calculated with probit analysis, using the MedCalc statistical software, on the basis of results obtained by several replicates of serial dilutions of the 2019-nCoV/Italy-INMI1 spiked into oral swab-UTM matrix. The evaluation of the concordance between results obtained with Simplexa™ COVID-19 Direct assay and with the Corman’s reference method was performed using GraphPad Prism statistical package.
4. Results
4.1. Analytical sensitivity and lower limit of detection (LOD)
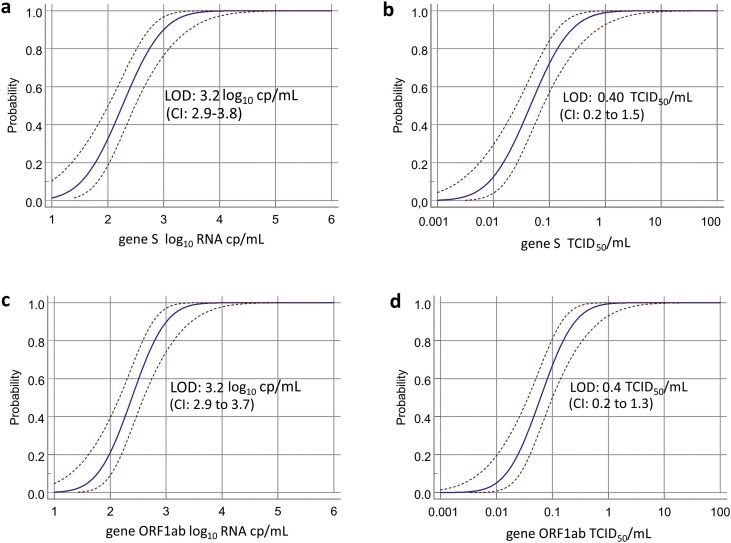
To assess the sensitivity of the prototype assay, both live SARS-CoV-2 and extracted viral RNA were tested. More in detail, for LOD calculation with live virus, SARS-CoV-2 particles from 2019nCoV/Italy-INMI1 isolate were spiked into oral swab-UTM matrix and serial dilutions from 100 to 0.001 TCDI50/mL were tested in replicates. We observed 100 % detection down to about 1 TCID50/ml, corresponding to 4 × 103 cp/mL (Table 1 ). The analytical sensitivity (i.e. the LOD, corresponding to the concentration of SARS-CoV-2-RNA detected with response probability of 95% for either S or ORF1ab) was determined by Probit regression model (Fig. 1 ). More in detail, the LOD for both target genes resulted the same: 3.2 (CI: 2.9 to 3.8) log10 cp/mL (Fig. 1a) and 0.40 (CI: 0.2 to 1.5) TCID50/mL (Fig. 1b) for S; 3.2 log10 (CI: 2.9 to 3.7) log10 cp/mL (Fig. 1c) and 0.4 (CI: 0.2 to 1.3) TCID50/mL (Fig. 1d) for ORF1ab. Consistent, although slightly better results, were obtained for LOD calculated on extracted viral RNA. This was spiked in universal transport medium (UTM) additioned with RNasin (1.6 U/μL) to avoid RNA degradation, and replicate 10-fold serial dilutions of were loaded on the MDx instrument and tested with the Simplexa™COVID-19 Direct assay. The LOD obtained with extracted viral RNA for both S gene or ORF1ab was 2.7 log10 cp/mL.
Table 1.
Limit of Detection of the Simplexa™ COVID-19 Direct assay.
| Target | Viral stock dilution (TCD50/mL) | RNA cp/mL* | Gene 1 (S gene) |
Gene2 (ORF1ab) |
Overall % Determinations (replicates) | ||||
|---|---|---|---|---|---|---|---|---|---|
| % Det (reps) | Mean Ct | SD | % Det (reps) | Mean Ct | SD | ||||
| SARS-CoV2 Viral particles 2019nCoV/ Italy-INMI1 | 10^-5 (100 TCID50/ml) | 4 × 105 | 100% (3/3) | 23.5 | 0.06 | 100% (3/3) | 24.1 | 0.12 | 100% (3/3) |
| 10^-6 (10 TCID50/ml) | 4 × 104 | 100% (3/3) | 26.8 | 0.26 | 100% (3/3) | 27.4 | 0.42 | 100% (3/3) | |
| 10^-7 (1 TCID50/ml) | 4 × 103 | 100% (40/40) | 29.8 | 0.66 | 100% (40/40) | 30.2 | 0.74 | 100% (40/40) | |
| 10^-8 (0.1 TCID50/ml) | 4 × 102 | 73.7% (14/19) | 33.4 | 0.83 | 68.4% (13/19) | 33.8 | 0.81 | 84.2% (16/19) | |
| 5*10^-9 (0.05 TCID50/ml) | 2 × 102 | 20% (1/5) | 33.5 | _ | 20% (1/5) | 33.7 | _ | 20% (1/5) | |
| 2.5*10^-9 (0.025 TCID50/ml) | 1 × 102 | 0% (0/5) | _ | _ | 0% (0/5) | _ | _ | 0% (0/5) | |
| 1.25*10^-9 (0.0125 TCID50/ml) | 50 | 20% (1/5) | 34.2 | _ | 20% (1/5) | 34.2 | _ | 20% (1/5) | |
| 10^-9 (0.01 TCID50/ml) | 40 | 50% (3/6) | 34.4 | 0.12 | 16.7% (1/6) | 34.6 | _ | 50% (3/6) | |
| 6.25*10^-10 (0.00625 TCID50/ml) | 25 | 0% (0/4) | _ | _ | 0% (0/4) | _ | _ | 0% (0/4) | |
| 3.13*10^-10 (0.00312 TCID50/ml) | 12.5 | 0% (0/5) | _ | _ | 0% (0/5) | _ | _ | 0% (0/5) | |
| 10^-10 (0.001 TCID50/ml) | 4 | 0% (0/3) | _ | _ | 0% (0/3) | _ | _ | 0% (0/3) | |
RNA copies/mL are based on standard curve for gene E SARS-CoV-2.
Fig. 1.
Probit analysis for Simplexa™ COVID-19 Direct assay.
4.2. Analytical specificity
In silico evaluation and preliminary in vitro analytical specificity study showed that the Simplexa™COVID-19 Direct only detects all COVID-19 virus strains, without cross reactivity with other viruses (Simplexa COVID-19 direct, instruction manual available at https://www.fda.gov/media/136286/download. We established the clinical specificity using clinical samples from patients known to be positive for other respiratory viruses, including Human Coronaviruses (Table 2 ). Results obtained for 20 nasopharyngeal swabs confirmed a 100 % of clinical specificity of the assay, based on the absence of non-specific signal detection due to any type of cross-reactivity event.
Table 2.
Analytical specificity of the Simplexa™ COVID-19 Direct assay.
| N# | ID # | Gene1 (S) | Gene2 (ORF1ab) | Simplexa™ COVID-19 Direct assay | Corman’s method | Differential diagnosis |
|---|---|---|---|---|---|---|
| 1 | 7760 | Not Detected | Not Detected | Negative | Negative | Rhino/ Enterovirus |
| 2 | 7775 | Not Detected | Not Detected | Negative | Negative | CoV HKU1 |
| 3 | 7957 | Not Detected | Not Detected | Negative | Negative | CoV HKU1 |
| 4 | 7686 | Not Detected | Not Detected | Negative | Negative | CoV HKU1 |
| 5 | 7972 | Not Detected | Not Detected | Negative | Negative | CoV NL63, Rhino/Enterovirus |
| 6 | 7914 | Not Detected | Not Detected | Negative | Negative | CoV HKU1 |
| 7 | 7736 | Not Detected | Not Detected | Negative | Negative | CoV 229E |
| 8 | 7708 | Not Detected | Not Detected | Negative | Negative | CoV OC43 |
| 9 | 7977 | Not Detected | Not Detected | Negative | Negative | CoV HKU1 |
| 10 | 7969 | Not Detected | Not Detected | Negative | Negative | CoV NL63, Adenovirus |
| 11 | 7555 | Not Detected | Not Detected | Negative | Negative | CoV HKU1 |
| 12 | 7571 | Not Detected | Not Detected | Negative | Negative | CoV OC43 |
| 13 | 7693 | Not Detected | Not Detected | Negative | Negative | CoV HKU1 |
| 14 | 7903 | Not Detected | Not Detected | Negative | Negative | CoV NL63 |
| 15 | 7551 | Not Detected | Not Detected | Negative | Negative | CoV HKU1 |
| 16 | 5036 | Not Detected | Not Detected | Negative | Negative | CoV NL63 |
| 17 | 7991 | Not Detected | Not Detected | Negative | Negative | CoV HKU1 |
| 18 | 5040 | Not Detected | Not Detected | Negative | Negative | CoV NL63 |
| 19 | 7771 | Not Detected | Not Detected | Negative | Negative | CoV HKU1 |
| 20 | 7901 | Not Detected | Not Detected | Negative | Negative | CoV HKU1 |
4.3. Performance evaluation on clinical specimens
A total of 278 nasopharyngeal swab were tested in parallel with the Simplexa™ COVID-19 Direct assay and Corman’s reference method in two main reference centers (i.e. the Laboratory of Virology of National Institute for Infectious Diseases “Lazzaro Spallanzani” (INMI) IRCCS, Rome, and the Molecular Virology Unit, Fondazione IRCCS Policlinico San Matteo, Pavia). Results obtained from the two laboratories were all included in the study. All the 99 SARS-CoV-2 samples tested positive with the reference test showing median cycle threshold (Ct) of 24.2 (range 16.6–36.9), also resulted positive with Simplexa™ COVID-19 Direct assay, with a clinical sensitivity of 100 %. Notably, Simplexa™ COVID-19 Direct assay was able to detect 8 additional positive samples resulted negative with Corman’s method. In these samples, a median of 30.8 Ct (range 28.2–33.3) in FAM detector and 31.2 Ct (range 28.2–33.6) in JOE detector was observed, respectively. Concordance analysis showed an “almost perfect” agreement in SARS-CoV-2 RNA detection between the two assays, being κ = 0.938; SE = 0.021; 95 % CI = 0.896-0.980 (Table 3 ). Moreover, the performance of the assay in a different clinical specimen type, i.e. BAL, that is considered of utmost importance in patients with lower respiratory tract implication, was also evaluated: a total of 33 BAL samples were analysed in parallel with the Simplexa™ COVID-19 Direct assay and Corman’s method. For these samples, the Abbot m2000 extraction platform was used, due to temporary restriction of QIAsymphony® reagents. While with BAL samples, the reference method provided elevated number of invalid results (14 out of 33), with overall 12 positives out of 19 valid results, 33 out of 33 samples achieved a valid result with the Simplexa™ COVID-19 Direct assay, providing 20 positives out of 33 valid results. The concordance among the 19 samples with valid results in both assays was 100 % (12 positive and 7 negative).
Table 3.
Concordance of results obtained with Simplexa™ COVID-19 Direct assay and Corman’s method.
| Simplexa™ COVID-19 Direct assay |
||||
|---|---|---|---|---|
| Positive | Negative | TOT | ||
| Corman's method | Positive | 99 | 0 | 99 |
| Negative | 8 | 171 | 179 | |
| Total | 107 | 171 | 278 | |
5. Discussion
Since the spread of COVID-19 in China, several SARS-CoV-2-specific RT-PCRs have been developed and commercialized in response to rising diagnostic demands. Prompt and rapid diagnosis are the main goals, particularly in the early phase of epidemics and are the first step towards limiting the propagation of the infection. In this scenario, we have evaluated the performances of the newly designed real-time RT-PCR Simplexa™ COVID-19 Direct assay. Overall, the test is fast, easy-to-use, does not need extra-equipment such as centrifuges or an extraction system, and can be handled by laboratory personal with very little extra training for the procedures and interpretation of results. Furthermore, the simplicity of design and the all-in-one coupled with easy-to-use design, makes it suitable for the field settings, and for near-to-patient diagnosis.
The LOD of the prototype assay, established with both SARS-CoV-2 viral particles and SARS-CoV2 extracted viral RNA, was less than 1000 cp/ml. All the SARS-CoV-2 samples resulted positive with the reference test (Corman’s method), also resulted positive with Simplexa™ COVID-19 Direct assay, with a diagnostic sensitivity of 100% and “almost perfect” concordance. Notably, Simplexa™ COVID-19 Direct assay showed a slightly higher sensitivity with respect of the reference test, identifying near 3% additional positive samples. An high performance of the test was also observed using different clinical specimens: among the BAL samples that were able to be analysed by both methods, a perfect agreement was observed; noteworthy, Simplexa™ COVID-19 Direct assay was able to detect the virus in BAL samples resulted invalid with the reference method. The discrepancy in the processing capability between the two systems is attributable to the highly viscous composition of BAL, that could impair its accessibility to a traditional extraction method compared to a faster and easier sample processing method such as that used in the Simplexa™ COVID-19 Direct assay. Moreover a 100% of clinical specificity was observed against swabs resulted positive to other viruses, particularly human coronaviruses, indicating no cross-reactivity with other similar viruses. Key advantages of this assay are simple operation procedures and high-speed of detection in just over an hour, which is significantly faster than the up to seven hours currently required by traditional extraction followed by amplification technologies. The lack of extraction reduces turnaround time (TAT) for the diagnosis of COVID-19, thus allowing prompt decision making regarding isolation of infected patients.
The only limitation of the assay is the small number of samples which can be tested in a run, since each instrument can support a ring of maximum eight position; this limitation is offset by the rapidity of the assay, due to the lack of extraction. Overall, the good analytical and clinical performances, coupled with user friendly architecture and short TAT indicate that it is promising for laboratory diagnosis of COVID-19 and field application.
Ethical statement
Ethical approval was not required as human samples were routinely collected and patients' data remained anonymous.
Author’s contribution
LB, AP: designed the study, analyzed data, wrote manuscript; FG, FN, MT, performed laboratory testing; AB, GM, VT: design the Simplexa™ COVID-19 Direct assay; FC, GS, SM: assisted in designing the study, performed laboratory testing. CC: discussed results. EL: assisted in designing the study, read and revised manuscript. MRC, FB: designed the study and read and revised manuscript
Declaration of Competing Interest
Angela Brisci, Giulia Minnucci, Veronica Tettamanzi are employees of the DiaSorin S.p.A. (Gerenzano, Italy). In no way their contribution to the completion of the study influenced the study design and the analysis of the results. No additional conflict of interest or other competing relationships exist.
Acknowledgement
This research was supported by funds to National Institute for Infectious Diseases ‘Lazzaro Spallanzani’ IRCCS from Ministero della Salute, Ricerca Corrente, linea1; European Commission – Horizon 2020 (EU project 101003544 – CoNVat; EU project 101003551 - EXSCALATE4CoV) and to Fondazione IRCCS Policlinico San Matteo from Ministero della Salute, Ricerca Finalizzata grant no. GR-2013-02358399. We thank Daniela Sartori for manuscript editing.
References
- 1.Li Q., Guan X., Wu P. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei M., Yuan J., Liu Y., Fu T., Yu X., Zhang Z.J. Novel coronavirus infectionin hospitalized infants under 1 year of age in China. JAMA. 2020;323(13):1313–1314. doi: 10.1001/jama.2020.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020:S0140–6736. doi: 10.1016/S0140-6736(20)30154-9. (20)30154-30159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K. Yuen KY; SARS study group. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corman V.M., Landt O., Kaiser M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf.
- 9.Capobianchi M.R., Rueca M., Messina F. Molecular characterization of SARS-CoV-2 from the first case of COVID-19 in Italy [published online ahead of print, 2020 Mar 27] Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.03.025. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]