Abstract
Course-based undergraduate research experiences (CUREs) are an effective way to introduce students to contemporary scientific research. Research experiences have been shown to promote critical thinking, improve understanding and proper use of the scientific method, and help students learn practical skills including writing and oral communication. We aimed to improve scientific training by engaging students enrolled in an upper division elective course in a human microbiome CURE. The “Fiber Force” course is aimed at studying the effect of a wholesome high-fiber diet (40 to 50 g/day for two weeks) on the students’ gut microbiomes. Enrolled students participated in a noninvasive diet intervention, designed health surveys, tested hypotheses on the effect of a diet intervention on the gut microbiome, and analyzed their own samples (as anonymized aggregates). The course involved learning laboratory techniques (e.g., DNA extraction, PCR, and 16S sequencing) and the incorporation of computational techniques to analyze microbiome data with QIIME2 and within the R software environment. In addition, the learning objectives focused on effective student performance in writing, data analysis, and oral communication. Enrolled students showed high performance grades on writing, data analysis and oral communication assignments. Pre- and post-course surveys indicate that the students found the experience favorable, increased their interest in science, and heightened awareness of their diet habits. Fiber Force constitutes a validated case of a research experience on microbiology with the capacity to improve research training and promote healthy dietary habits.
INTRODUCTION
Incorporating research experiences into the undergraduate curriculum is a major emphasis of national educational reform (1, 2) and the American Society for Microbiology (ASM) (3). The educational field proposes integration of research experiences into traditional lab courses in the form of course-based undergraduate research experiences (CUREs) (4). CUREs enable students to experience research first-hand, providing a more accurate understanding of how scientific research is conducted. Authentic research experiences lead to student-reported gains in general skills (e.g., oral, visual, and written communication) and more specific research-associated skills (e.g., research design, hypothesis formation, data analysis) (5–8). In addition, students in CURE courses develop scientific reasoning skills, begin to identify themselves as scientists, are more inclined to pursue graduate education or careers in science (9–11), and have increased graduation rates (12).
To maximize student engagement in research and in the classroom, CURE curricula should focus on relevant research topics and questions. One of the fastest growing research areas in the last 10 to 15 years has been the relationship between human health and the human microbiome, or the consortia of commensal microorganisms living in and on our bodies. The gut microbiome alone encompasses more than 1,000 resident microorganisms, including bacteria, viruses, fungi, and protozoa (13). The majority of these microorganisms inhabit the colon, where they contribute to human health through the biosynthesis of vitamins and essential amino acids and the generation of metabolic byproducts through fermentation of low- or non-digestible dietary carbohydrates (hereafter called fiber) (14–19). The type and proportion of fiber that reaches the colon is an important factor that can drive alterations of gut microbial composition (20–22). Diets low in fiber reduce total bacterial abundance in the gut microbiome (23), while diets high in fiber increase microbiota richness (24, 25). Interest in the role of dietary fiber in regulating the gut microbiome has led to diet interventions, which suggest that a high-fiber diet can alter metabolic parameters (21, 26) and ameliorate clinically relevant colon cancer biomarkers (25).
Given the importance of fiber for a healthy gut, further dietary studies are still needed. Especially in healthy individuals, there is a need to understand not only the stability and resilience of the “normal” microbiota, but also the amount and type of fiber that can result in positive microbial changes. Therefore, we developed a research-based learning laboratory course at University of California Irvine (UCI) to study the effect of a high-fiber diet on the gut microbiome of students. The Microbiome laboratory course at UCI, or Fiber Force, aimed to engage students in research activities by actively participating in a safe diet intervention. Students manipulated their own gut microbiomes through a safe high-fiber diet intervention and actively participated in the sample processing and data analysis. Students analyzed their own samples, gained molecular experience in the laboratory, and were instructed on computational tools to analyze the data. Similar to other microbiology modules described recently (27–36, 38), a goal of Fiber Force was for students to develop critical reasoning and problem-solving skills related to microbiology and microbiome research.
Intended audience
The microbiome activity described here uses molecular biology and next-generation sequencing and computational skills. The course is intended as a microbiology or molecular biology laboratory. The course is most effective as a small enrollment course as it requires consenting students to undergo a diet intervention. The exercise was piloted in an upper-division elective course called Advanced Molecular Biology techniques (M130L) at the UCI (spring 2018) with enrollment of 18 students (all seniors). The class comprised 10 female and 8 male students and included 8 first-generation college students. The students’ performance of the Microbiome course was compared with a previous version of the course (M130L, 2017) in which students were exposed to an inquiry module aimed at studying promoters using pClone described by Campbell et al. (37). The 2017 course had an enrollment of 20 students (all seniors): 8 female and 12 male, with 8 first-generation college students. The CURE study was approved by UC Irvine IRB # 2018-4297 (Microbiome intervention) # 2018-4211 (educational effectiveness), and # 2016-3168 (critical thinking and science literacy).
Learning time
This activity was conducted within a 10-week laboratory course meeting eight hours/week (two four-hour meetings). One four-hour meeting was dedicated to active lectures and problem-based learning and was followed by a short laboratory session. The second four-hour meeting was used for laboratory exercises and discussions. The 10-week schedule included the diet intervention (for consenting students), sample processing, DNA extraction and sequencing, QIIME2 and R workshops for data analysis, and poster presentations. After the two-week diet intervention, DNA sequencing delayed laboratory activities. During this time, we implemented R and QIIME2 workshops (Appendices 1 and 2), worked on grant proposal writing activities (outlined in Appendix 3), and introduced basic microbiology techniques (e.g., isolation of bacteria from soil [34, 38]). Schedule flexibility meant activities could be implemented within the course’s existing schedule. We provide an activity overview (Appendix 4) and syllabus (Appendix 5).
Prerequisite student knowledge
Students were expected to have completed all core biology courses (including Chemistry, Biology, Molecular Biology, and Biochemistry), a common practice for upper-division laboratories at UCI. Students had a basic knowledge of microbial cell biology, the central dogma of molecular biology, as well as metabolic and phylogenetic diversity. Many students also had experience in pipetting, PCR, DNA extraction, and DNA gel electrophoresis. However, this experience was not assumed, and we covered basic laboratory principles and practices in the course. Advanced analyses, such as phylogenetics, were covered with active lectures and discussions (Appendix 6).
Learning objectives
Upon completion of this activity, students will be able to:
Design and execute a dietary intervention plan to study the gut microbiome, with proper sample collection and storage.
Become proficient in microbiome laboratory and computational techniques and skills.
Apply basic microbiome principles and concepts to solve experimental problems.
Apply the scientific method in different ways, including writing hypotheses, analyzing results, reporting data, and proposing further experiments.
Report hypotheses, proposed research, results, and conclusions both orally and in writing to an audience of scientists and peers
Interpret and evaluate results from lab experiments and primary research articles.
Critically analyze scientific publications, research proposals, and results
PROCEDURE
Materials
Students collected daily dietary information before and after the intervention using MyFitnessPal. During the two-week intervention, we provided students with 10 fiber-rich meals each week prepared by Thistle (www.Thistle.com), along with dietary information to increase fiber intake (Appendix 14). We collected six fecal microbial samples in total (from consenting students), three samples in the week prior to the intervention, and three in the second week of their high-fiber diet (> 40 g of fiber a day). Students were instructed on stool collection and storage procedures (Appendix 7) using Eppendorf tubes (future projects should consider larger 50 mL tubes). Students dropped off samples to the lab, and these were stored at –70oC. During the laboratory component of the class, students weighed two of their fecal samples (one pre-intervention and one post-intervention) with a weighing dish/paper and metal spoon/spatula. Once all samples were collected, the students extracted DNA using the ZymoBIOMICS DNA Miniprep Kit. We provided 70% ethanol, bead beater for improved DNA lysing, a microcentrifuge, and plastic laboratory materials including tubes and tips (Appendix 9). For safety purposes, sample processing took place in a biosafety level 2 (BSL2) laboratory with a biological cabinet. After DNA extraction, the rest of the protocol can be performed in BSL1. Students performed some PCR and gel electrophoresis (see Appendix 8); however, processing of all fecal samples, including the use of barcoded primers suitable for high throughput sequencing (i.e., Illumina MiSeq), was performed by the UCI Microbiome Initiative (details in Appendix 8). For a broader application, aliquoted samples can be sent to any suitable internal or external service.
R workshop description
Due to the inherently large datasets produced by microbiome studies, students were instructed in how to import, view, and analyze various data types in R (40). Students participated in a hands-on R workshop to familiarize themselves with various data types, functions, and analyses. To start, students explored small, simple datasets to learn how to manipulate and process data, visualize data on graphs, and conduct statistical analyses (Appendix 1). In particular, the instructor walked through various lines of code on his own device with students interactively participating. For the second workshop, students were shown how to import sample microbiome datasets for downstream microbiome analyses.
QIIME2 workshop and pipeline description
Students spent two class periods becoming familiar with the command-line interface for Quantitative Insights into Microbial Ecology v2 (QIIME2) (41). QIIME2 is a computational pipeline designed to analyze microbiome sequence data, especially 16S rRNA gene data. Briefly, the program is designed to 1) import data, 2) demultiplex data, 3) quality filter data, 4) denoise data, and 5) calculate diversity and other statistical metrics. The first day of the workshop consisted of students bringing their laptop with either a MacOS or Linux operating system (Windows users were instructed to enable Windows Subsystem for Linux, a standard feature on Windows 10). Students followed installation instructions for QIIME2 (https://docs.qiime2.org/2019.4/install/) through a standard Miniconda installation. During installation, students discussed the QIIME2 pipeline and the vocabulary that comes with microbial sequence data (see ideas for discussion topics in Appendix 2). Students learned Linux-based commands, such as navigating, creating, and removing directories. After installation, students completed an abridged version of the “Moving Pictures” tutorial (https://docs.qiime2.org/2019.7/tutorials/moving-pictures/), which skips time-intensive steps such as denoising. The workshop emphasized the interpretation of data, such as alpha and beta diversity plots.
The R and QIIME2 workshops described above can be replaced by computational and data analysis training. For resources and training on microbiome analysis we recommend checking the Human Microbiome project webpage (https://www.hmpdacc.org/outreach/workshops.php) (42), Bioinformatics Inquiry through Sequencing (35, 43), Galaxy resources (https://usegalaxy.org/) (44), and the Program for Unifying Microbiome Analysis (45).
Student instructions
Instructions and protocols were provided to students via electronic files on our learning management system. The protocol for DNA extraction was used directly from the manufacturer’s manual (Zymo, https://www.zymoresearch.com/collections/zymobiomics-dna-kits/products/zymobiomics-dna-miniprep-kit). Each student carried out the protocols individually; however, students were allowed to work in groups in the lab and to discuss and analyze data. The outline for the grant proposal exercise is provided in Appendix 3. All protocols are listed in Appendix 8, and the guidelines for lab notebook keeping, lab meetings, and poster presentations are provided in Appendices 10 to 12.
Faculty instructions
The Fiber Force microbiome lab course can be run in approximately 10 weeks, including a two-week diet intervention (Appendix 4). During the diet intervention weeks and sequencing weeks (no lab activity), we suggest introducing students to computational techniques. We implemented R and QIIME2 workshops (Appendices 1 and 2) during interventions and while students waited for sample processing and sequencing. Alternatively, other laboratory modules/techniques can be introduced during this time. This course is designed for smaller laboratory courses (15 to 30 students). To accommodate larger class sizes, teaching assistants with a strong computational background can be allocated to sections of 15 to 20 students. Instructors and/or teaching assistants are in charge of facilitating discussions, teaching active lessons, supervising group work, answering questions, and demonstrating techniques (when needed). The weekly discussion (active lecture) handouts are provided in Appendix 6. To ensure students properly practice research and oral communication skills, we recommend adding “laboratory or group meetings” to the schedule to discuss research questions, hypotheses, research plans, preliminary results, and troubleshooting. At the end of the course, students share their findings as a poster presentation, to which instructors, teaching assistants, students, and research laboratories are invited. The protocols (Appendix 8) and the respective required laboratory preparation, including materials and kits (Appendix 9) are described in the appendices.
A significant prerequisite to implementing the Fiber Force course is getting Institutional Review Board (IRB) approval in advance, a process that can take up to six months. UCI IRB reviewed the educational (educational research) and diet intervention (clinical study) components separately. To ensure timely approval of IRB requests, we recommend 1) providing a detailed description of how the data will be anonymized for privacy protection, 2) writing a detailed consent form for students that includes a protocol for opting out of the study without affecting course performance (see the consent form in Appendix 15), and 3) using a third-party (other than the instructor) as holder of the key that connects student’s names with sample codes. The IRB background should cite previous studies performed in the context of classrooms (see, for example, 46–48) or the present manuscript (UCI IRB # 2018-4297) as precedent.
Suggestions for determining student learning
An essential component of research involves the ability to write a research proposal, conduct experiments in a rigorous and reproducible manner, and communicate scientific findings in oral and written forms. Accordingly, the assessment task for Fiber Force involved 1) a step-by-step proposal writing activity (Appendix 3) to evaluate ability to design and propose new research; 2) weekly discussion handouts and lab notebook evaluations to assess laboratory performance; 3) data meeting reports and oral poster presentations to evaluate science communication. The assessments are described in full in the course syllabus (Appendix 5). The data meeting reports and poster format followed structural conventions of a scientific publication. The activities required students to outline the background of the field, provide the aims and hypotheses of the study, present results in both text and graphical forms with descriptive legends, and discuss the validity and significance of their findings. The integration of this assessment task with learning activities in this project directly align with our learning objectives (Table 1). The marking rubric for the proposal writing spanned numerous criteria including quality of aims, literature support, effective introduction of project aims, validity of research questions and aims, and validity of proposed research. These rubrics are part of a manuscript that is currently under preparation (contact the corresponding author). The rubrics used to evaluate lab notebooks, lab meeting presentations and poster presentations are included in Appendix 13.
TABLE 1.
Alignment between learning goals and course assessments.
| Learning Outcomes | Assessment |
|---|---|
| Design and execute a dietary intervention plan to study the gut microbiome, with proper sample collection and storage | Assessed through data meeting reports, lab notebook points, and poster presentation |
| Become proficient in microbiome laboratory and computational techniques and skills | Assessed through data meeting reports, lab notebook points, and poster presentation |
| Apply basic microbiome principles and concepts to solve experimental problems | Weekly quizzes |
| Apply the scientific method in different ways, including writing hypotheses, analyzing results, reporting data, and proposing further experiments | Assessed by poster presentation, lab meeting reports and grant proposal writing |
| Report hypotheses, proposed research, results, and conclusions both orally and in writing to an audience of scientists and peers | Discussions of research articles, lab reports, and grant proposal writing |
| Interpret and evaluate results from lab experiments and primary research articles | Lab reports, lab meetings, poster presentations |
| Critically analyze scientific publications, research proposals and results | Lab discussions and problems and analysis of research articles, grant proposal writing, data meetings, poster presentations |
Sample data
Students were required to read the literature on the effects of diet on the gut microbiome. This led to predictions of and/or hypotheses on the impact of fiber before and after the diet intervention.
After the samples were sequenced, students were expected to interpret OTU tables, use QIIME2/R to visualize data, and compare samples to validate (or not) their predictions. Students then presented their results during a poster symposium (samples in Appendix 19). One example predicted that the intervention would lead to an increase in fiber-degrading bacteria (for student-led literature review see Appendix 19) as these bacteria should possess a specialized ability to degrade complex carbohydrates. After analysis, the students found that the > 40 g fiber intervention caused no significant change in diversity or abundance of microbes at the genus or phylum levels, but registered increases in some relevant bacterial genera. A second example hypothesized that a 40- to 50-g fiber intervention would exhibit increases in short-chain-fatty-acid-producing bacteria known to be responsible for gastrointestinal fermentation of fiber. However, the students concluded that there was no significant difference in the relative abundance of short-chain-fatty-acid-producing bacteria before versus after the diet intervention. This shows that the same dataset can be used by different groups of students to study different predictions or hypotheses.
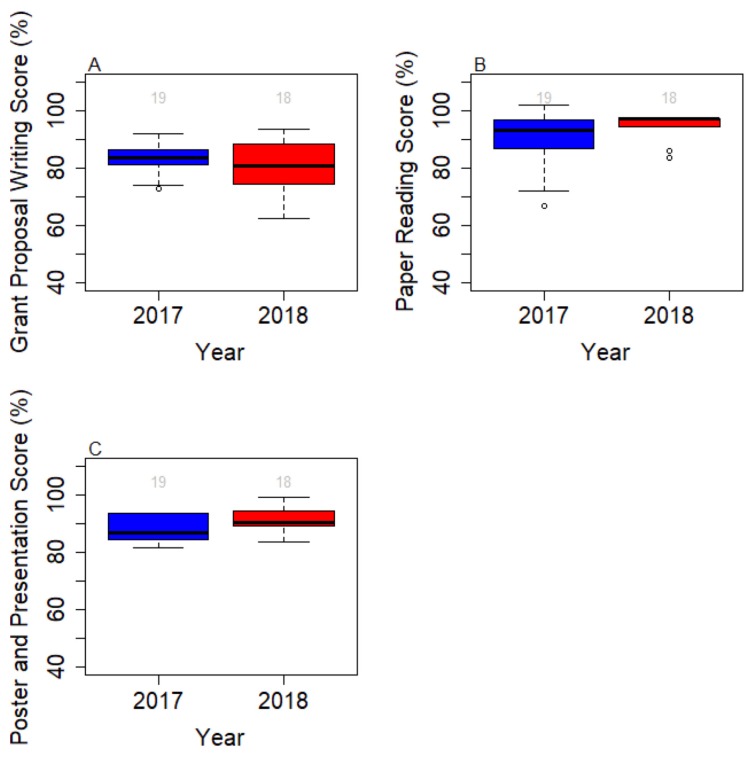
In congruency with the quality of posters shown in Appendix 19, students of the Fiber Force course registered high scores on writing, data analysis, and presentation, with median scores of 81%, 83% and 90%, respectively (Fig. 1).
FIGURE 1.
Course performance between 2017 and 2018 courses were compared using the following grade scores A) grade on a step-by-step proposal-writing activity (outlined in Appendix 3) to evaluate ability to design and propose new research; B) weekly discussion handouts to assess data analysis performance; C) data meeting reports and oral poster presentation to evaluate science communication.
Safety issues
The Fiber Force course involves collection of fecal samples from healthy individuals (according to answers from the health survey, Appendix 16), but potentially pathogenic bacteria can be present in fecal samples. As part of the course, instructors and teaching assistants receive BSL2 safety training to instruct students on the necessary personal protective equipment (PPE), the safe handling of samples, and the proper disposal of biological waste in accordance with BSL2 regulations. Aliquoting of fecal samples, as well as bead-beating (the first step in DNA extraction) should be handled inside a biosafety cabinet, and DNA extraction should be conducted in a BSL2 laboratory. The subsequent steps (PCR, electrophoresis) can be performed in a BSL1 laboratory. Students with health conditions such as pregnancy, allergies, or immune-compromised status should not directly handle or come into contact with fecal samples. Laboratory bench surfaces and notebooks were decontaminated with 70% ethanol before and after each session and all students washed their hands with antibacterial detergent. Electronic devices were used (if needed) inside sealed plastic bags that were wiped before and after each use. All waste was disposed of in accordance with BSL2 regulations and decontaminated by autoclaving. The biosafety rules and policies for the laboratory are included in the syllabus (Appendix 5).
DISCUSSION
Field testing
Fiber Force was tested in an Advanced Molecular Biology course at the University of California, Irvine, with an enrollment of 18 students. The project protocols and interventions were cleared in accordance with the UCI IRB, and participants provided consent with regard to their de-identified microbiome data and responses to health and course surveys. Teaching and lab assistants increased participants to 22 (Fig. 2). To ensure participants consumed a diet rich in wholesome fiber for two weeks, each participant was provided with 10 meals per week from a food delivery company called Thistle. These lunch and dinner meals had between 15 and 30 grams of fiber each (https://www.thistle.co/menu/). For breakfast and snacks, as well as on weekends, students supplied their own meals. In our experience, supplying ready-made meals ensured that participants ate a varied and wholesome diet. However, this was not a necessary part of the course, as instructors can provide resources for students to design their own diets (see Appendix 14). For instance, legumes such as split peas, lentils, lima beans, black beans, and chickpeas are particularly rich in fiber (12 to 15 g of fiber per cup) and are economical for students’ budgets. Students frequently discussed their diet choices during class, sharing ideas and recipes with each other, and the diet intervention also raised awareness about healthy food habits. Participation in the study as a subject was not required to take the course, nor was it needed to receive meal compensation. In addition, course grades were not attached to participation as a study subject (i.e., samples were de-identified).
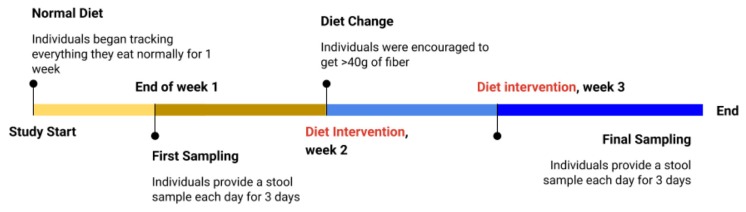
FIGURE 2.
Experimental design for the high-fiber study intervention and collection of fecal samples.
The first week of class consisted of discussing the intervention and designing health surveys with the students to evaluate their diet intervention. Students were asked to find examples of surveys in the literature and design (as a class) a 10- to 25-question survey about health status, usual (pre-intervention) food habits, and other relevant questions for the study (e.g., body mass index [BMI]). Students answered their designed survey (Appendix 16), indicating that all students were generally healthy. After the intervention, we asked students to complete an open-ended survey that included questions about their experience in the Fiber Force course (see questions in Appendix 17 and answers in Appendix 18).
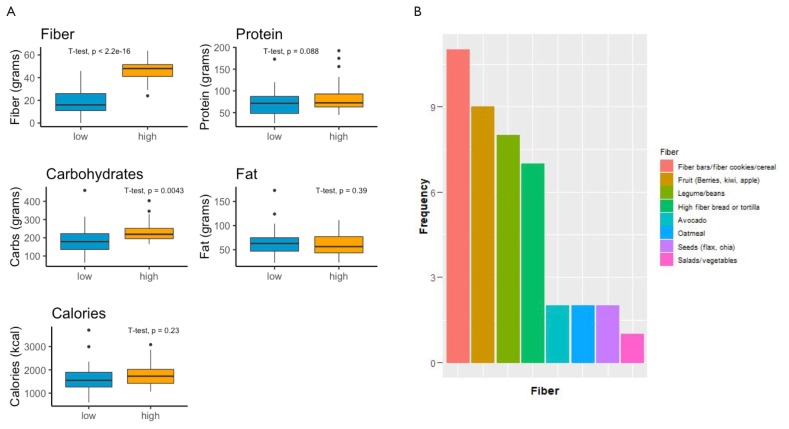
Participants in the intervention increased their fiber ingestion from an average of approximately 15 g (lowest pre-intervention individual averaged 1.8 g) to an average of approximately 40 g of fiber per day (highest intervention individual averaged 54 g) with a concomitant increase in carbohydrate consumption (Fig. 3B). The intervention minimally altered fat and protein consumption with similar caloric intake (Fig. 3B). To reach their daily fiber goals, students preferred legumes and fruits (e.g., berries) due to their high content of fiber per serving (Fig. 3A). As a consequence of this dramatic increase in dietary fiber, 42% of students reported an increase in flatulence and bloating, and 31% reported appetite loss (Appendix 18). These are expected secondary effects due to the increase of bulk and fermentation. Students did not report increased time in meal preparation or cooking during the intervention, suggesting that the intervention does not drastically increase time commitment outside the classroom.
FIGURE 3.
Fiber type and quantity and dietary changes by participants in the fiber intervention study. A) Quantities (in grams) of fiber, protein, carbohydrates, fat, and calories pre- and post-intervention (N=24). B) Participants in the diet intervention were asked to answer the following: “What were your ‘go to’ or staple high-fiber foods? Name your top 3 and provide a description.” Answers were tallied and plotted by frequency (n=18).
Evidence of student learning
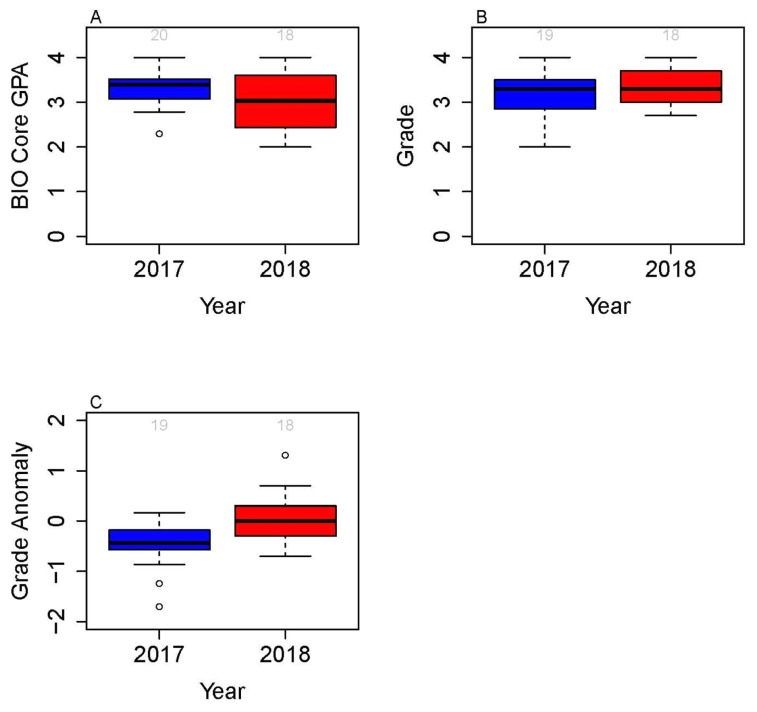
To determine whether students in the Fiber Force course mastered important research-based skills, we measured course grade on important skill-based assignments. We also compared performance in the Fiber Force course (2018) with a previous version of the course (2017) that used a research module (37) using synthetic biology to study bacterial promoters (Fig. 4). In the 2017 course, students designed primers to study their own promoters and made predictions, but they did not participate in the experimental design nor were they active participants in the study. The types of course assignments, rubrics and grade weight in 2017 and 2018 (Table 1) were the same, except that the topic of proposal writing and discussions in 2017 was synthetic biology. The 2018 cohort exhibited a lower GPA in prior lower-division biology core courses than the 2017 cohort (Fig. 4A). The 2018 cohort also had less laboratory experience, with 62% of the students indicating that they had taken zero or one laboratory before Fiber Force, compared with 42% in 2017. This means that the 2018 cohort had less preparation for this upper-division research course. However, both groups showed similar average final course grades at the end of the quarter (Fig. 4B). The 2018 cohort also performed better in the Fiber Force course than in other classes taken simultaneously that quarter, compared with the 2017 cohort (Fig. 4C). Students from both cohorts (2017 and 2018) scored similarly in writing, data analysis, and communication assessments (Fig. 1), with median scores between 82.67 and 90.29, indicating high levels of mastery.
FIGURE 4.
Performance of students in the Advanced Molecular Biology course M130L in Fiber Force (2018) compared with control (inquire module, studying promoters using pClone by 37) 2017 course. In 2017, each group picked a guided inquiry project module and collected data. Students studied their own promoters but did not participate in experimental design. In the 2018 course, the class as a whole volunteered as participants in interventions, designed surveys, and discussed data. They also worked in small groups for presentation and discussion. Panel A compares the students’ GPA in biology-related core courses (Biochemistry, Molecular Biology) followed before the 2017 and 2018 courses, as a way to compare incoming level. Panel B compares the average final course grade in the 2017 and 2018 courses. Panel C compares the grade average in the 2017/2018 courses with grades in other courses that students followed simultaneously that quarter. Grade anomaly is the average GPA on other courses that quarter minus M130L course grade.
To gather feedback, we used a published CURE survey (https://www.grinnell.edu/academics/resources/ctla/assessment/cure-survey, 49), for self-reported estimates on the course experience. The (post-CURE) survey responses on the course experience were very positive, and students estimated the gains in research-based aspects of the experience (e.g., on small group work, writing proposal, data collection, and analysis, etc.) as “large” or “very large” gains (49, not shown).
In our post-course open-ended survey, we asked students to comment on particular aspects of their experience in the Fiber Force course (see questions asked in Appendix 17 and answers in Appendix 18n = 16). The qualitative responses were thematically classified and tallied. About a third (35%) of the students listed research exposure, 25% being part of an intervention, and 25% teamwork as their favorite aspect of the course. When asked about what skills showed highest gains in the course, 37% of students listed writing, 31% research skills, 20% oral presentation, 20% teamwork, and 20% computer/data analysis skills. These answers suggest that after following the Fiber Force course, students perceive themselves as possessing increased competence in these skills, and their achieved gains align with our intended learning outcomes.
All in all, we show the Fiber Force course promoted student development of fundamental research skills, meeting our learning outcomes. The positive impact on student development of research skills is consistent with results published for other CURE courses (9, 10, 32, 36, 47). It has been shown that integrating personal microbiome studies in the classroom improves student engagement and interest in science courses (47). Our manuscript presents an option for a research experience on fiber microbiome intervention suitable for undergraduate classes in which the students can participate as study subjects. Additional research would be needed to validate this CURE course on educational effectiveness, motivation, and engagement.
In addition to our learning outcomes, we highlight the benefit the diet intervention had on student awareness on healthy food choices. When asked if the intervention changed any of their food habits, 63% of students indicated that they now check food labels for fiber content, while 20% indicated that they are trying to eat more fiber or chose food based on fiber content. In addition, students were more likely to communicate the health benefits of a high-fiber diet to others in their lives, which inspired the name of our course: Fiber Force, as a force for awareness.
SUPPLEMENTARY MATERIALS
ACKNOWLEDGMENTS
We thank the UCI Microbiome Initiative for making the Fiber Force course possible and Thistle for sponsoring the diet intervention. AO was supported by a NIH T32 training grant (1T32AI14134601A1) from UC Irvine’s Training Program in Microbiology and Infectious Diseases. Special thanks to all the students in the 2018 M130L course at UC Irvine who made this study possible. The authors have no conflicts of interest to declare.
Footnotes
Supplemental materials available at http://asmscience.org/jmbe
REFERENCES
- 1.American Association for the Advancement of Science. Vision and change in undergraduate biology education: a call to action: a summary of recommendations made at a national conference organized by the American Association for the Advancement of Science; July 15–17, 2009; Washington, DC. 2011. [Google Scholar]
- 2.National Research Council. BIO2010: transforming undergraduate education for future research biologists. The National Academies Press; Washington, DC: 2003. [PubMed] [Google Scholar]
- 3.Merkel S. The development of curricular guidelines for introductory microbiology that focus on understanding. J Microbiol Biol Educ. 2012;13:32–38. doi: 10.1128/jmbe.v13i1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handelsman J, Ebert-May D, Beichner R, Bruns P, Chang A, DeHaan R, Gentile J, Lauffer S, Stewart J, Tilghman SM, Wood SB. Scientific teaching. Science. 2004;304:521–522. doi: 10.1126/science.1096022. [DOI] [PubMed] [Google Scholar]
- 5.Seymour E, Hunter AB, Laursen SL, DeAntoni T. Establishing the benefits of undergraduate research for undergraduates in the sciences: first findings from a three-year study. Sci Educ. 2004;88:493–534. doi: 10.1002/sce.10131. [DOI] [Google Scholar]
- 6.Lopatto D. Undergraduate research as a catalyst for liberal learning. Peer Rev. 2006;8:22–25. [Google Scholar]
- 7.Lopatto D. Undergraduate research experiences support science career decisions and active learning. CBE Life Sci Educ. 2007;6:297–306. doi: 10.1187/cbe.07-06-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laursen S, Hunter A, Seymour E, Thiry H, Melton G. Undergraduate research in the sciences: engaging students in real science. Jossey-Bass; San Francisco, CA: 2010. [Google Scholar]
- 9.Lopatto D, Alvarez C, Barnard D, Chandrasekaran C, Chung HM, Du C, Eckdahl T, Goodman AL, Hauser C, Jones CJ, Kopp OR, Kuleck GA, McNeil G, Morris R, Myka JL, Nagengast A, Overvoorde PJ, Poet JL, Reed K, Regisford G, Revie D, Rosenwald A, Saville K, Shaw M, Skuse GR, Smith C, Smith M, Spratt M, Stamm J, Thompson JS, Wilson BA, Witkowski C, Youngblom J, Leung W, Shaffer CD, Buhler J, Mardis E, Elgin SC. Undergraduate research. Genomics education partnership. Science. 2008;322:684–685. doi: 10.1126/science.1165351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiry H, Laursen S. The role of student-advisor interactions in apprenticing undergraduate researchers into a scientific community of practice. J Sci Educ Technol. 2011;20(6):771–784. doi: 10.1007/s10956-010-9271-2. [DOI] [Google Scholar]
- 11.Brownell SE, Hekmat-Scafe DS, Singla V, Chandler Seawell P, Conklin Imam JF, Eddy SL, Stearns T, Cyert MS. A high-enrollment course-based undergraduate research experience improves student conceptions of scientific thinking and ability to interpret data. CBE Life Sci Educ. 2015;14(2):ar21. doi: 10.1187/cbe.14-05-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodenbusch SE, Hernandez PR, Simmons SL, Dolan EL. Early engagement in course-based research increases graduation rates and completion of science, engineering, and mathematics degrees. CBE Life Sci Educ. 2016;15:ar20. doi: 10.1187/cbe.16-03-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bird AR, Brown IL, Topping DL. Starches, resistant starches, the gut microflora and human health. Curr Issues Intest Microbiol. 2000;1:25–37. [PubMed] [Google Scholar]
- 15.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 16.Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 17.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014;20:779–786. doi: 10.1016/j.cmet.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Vrese M, Schrezenmeir J. Probiotics, prebiotics, and synbiotics. Adv Biochem Eng Biotechnol. 2008;111:1–66. doi: 10.1007/10_2008_097. [DOI] [PubMed] [Google Scholar]
- 20.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haro C, Garcia-Carpintero S, Alcala-Diaz JF, Gomez-Delgado F, Delgado-Lista J, Perez-Martinez P, Rangel Zuñiga OA, Quintana-Navarro GM, Landa BB, Clemente JC, Lopez-Miranda J, Camargo A, Perez-Jimenez F. The gut microbial community in metabolic syndrome patients is modified by diet. J Nutr Biochem. 2015;27:27–31. doi: 10.1016/j.jnutbio.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Costabile A, Deaville ER, Morales AM, Gibson GR. Prebiotic potential of a maize-based soluble fibre and impact of dose on the human gut microbiota. PLOS One. 2016;11(1):e0144457. doi: 10.1371/journal.pone.0144457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halmos EP, Christophersen CT, Bird AR, Shepherd SJ, Gibson PR, Muir JG. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut. 2015;64:93–100. doi: 10.1136/gutjnl-2014-307264. [DOI] [PubMed] [Google Scholar]
- 24.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, Gougis S, Rizkalla S, Batto JM, Renault P, Doré J, Zucker JD, Clément K, Ehrlich SD ANR MicroObes consortium. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 25.O’Keefe SJD, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, Vipperla K, Naidoo V, Mtshali L, Tims S, Puylaert PGB, DeLany J, Krasinskas A, Benefiel AC, Kaseb HO, Newton K, Nicholson JK, de Vos WM, Gaskins HR, Zoetendal EG. Fat, fibre and cancer risk in African Americans and rural Africans. Nature Communications. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, Mao Y, Zhang X, Pang X, Wei C, Zhao G, Chen Y, Zhao L. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–41. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- 27.Boomer SM, Lodge DP, Dutton BE. Bacterial diversity studies using the 16S rRNA gene provide a powerful research-based curriculum for molecular biology laboratory. Microbiol Educ. 2002;3:18–25. doi: 10.1128/154288102X14285807655107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drew JC, Triplett EW. Whole genome sequencing in the undergraduate classroom: outcomes and lessons from a pilot course. J Microbiol Biol Educ. 2008;9:311. doi: 10.1128/jmbe.v9.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campo D, Garcia-Vazquez E. Inquiry-based learning of molecular phylogenetics. J Biol Educ. 2008;43:15–20. doi: 10.1080/00219266.2008.9656144. [DOI] [Google Scholar]
- 30.Gasper B, Gardner S. Engaging students in authentic microbiology research in an introductory biology laboratory course is correlated with gains in student understanding of the nature of authentic research and critical thinking. J Microbiol Biol Educ. 2013;14(1):25–34. doi: 10.1128/jmbe.v14i1.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SC, Findley AM, Gissendanner CR, Golebiewska UP, Guild N, Hartzog GA, Grillo WH, Hollowell GP, Hughes LE, Johnson A, King RA, Lewis LO, Li W, Rosenzweig F, Rubin MR, Saha MS, Sandoz J, Shaffer CD, Taylor B, Temple L, Vazquez E, Ware VC, Barker LP, Bradley KW, Jacobs-Sera D, Pope WH, Russell DA, Cresawn SG, Lopatto D, Bailey CP, Hatfull GF. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio. 2014;5(1):e01051–13. doi: 10.1128/mBio.01051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang JT, Daly JN, Willner DL, Patil J, Hall RA, Schembri MA, Tyson G, Hugenholtz P. Do you kiss your mother with that mouth? An authentic large-scale undergraduate research experience in mapping the human oral microbiome. J Microbiol Biol Educ. 2015;16(1):50–60. doi: 10.1128/jmbe.v16i1.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farnham KR, Dube DH. A semester-long project-oriented biochemistry laboratory based on Helicobacter pylori urease. Biochem Mol Biol Educ. 2015;43:333–340. doi: 10.1002/bmb.20884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caruso JP, Israel N, Rowland K, Lovelace MJ, Saunders MJ. Citizen science: the small world initiative improved lecture grades and California critical thinking skills test scores of nonscience major students at Florida Atlantic University. J Microbiol Biol Educ. 2016;17(1):156–162. doi: 10.1128/jmbe.v17i1.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartman MR, Harrington KT, Etson CM, Fierman MB, Slonim DK, Walt DR. Personal microbiomes and next-generation sequencing for laboratory-based education. FEMS Microbiol Lett Dec. 2016;363(23):fnw266. doi: 10.1093/femsle/fnw266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakshi A, Webber A, Patrick L, Wischusen W, Thrash C. The CURE for cultivating fastidious microbes. J Microbiol Biol Educ. 2019;20(1) doi: 10.1128/jmbe.v20i1.1635. pii:20.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell AM, Eckdahl T, Cronk B, Andresen C, Frederick P, Huckuntod S, Shinneman C, Wacker A, Yuan J. pClone: synthetic biology tool makes promoter research accessible to beginning biology students. CBE Life Sci Educ. 2014;13(2):285–296. doi: 10.1187/cbe.13-09-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis E, Sloan T, Aurelius K, Barbour A, Bodey E, Clark B, Dennis C, Drown R, Fleming M, Humbert A, Glasgo E, Kerns T, Lingro K, McMillin M, 1, Meyer A, Pope B, Stalevicz A, Steffen B, Steindl A, Williams C, Wimberley C, Zenas R, Butela K, Wildschutte H. Antibiotic discovery throughout the small world initiative: a molecular strategy to identify biosynthetic gene clusters involved in antagonistic activity. Microbiologyopen. 2017;6(3) doi: 10.1002/mbo3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez-Baeza Y, González A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R The Earth Microbiome Project Consortium; The Earth Microbiome Project Consortium. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2018. https://www.R-project.org/ [Google Scholar]
- 41.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet C, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ. 2018. https://peerJ.com/preprints/27295/ [DOI] [PMC free article] [PubMed]
- 42.Rosenwald AG, Arora GS, Madupu R, Roecklein-Canfield J, Russell JS. The human microbiome project: an opportunity to engage undergraduates in research. Proc Comp Sci. 2012;9:540–549. doi: 10.1016/j.procs.2012.04.058. [DOI] [Google Scholar]
- 43.BioSeq. Bioinformatics Inquiry through Sequencing. http://ase.tufts.edu/chemistry/walt/sepa/index.html.
- 44.Batut B, Gravouil K, Defois C, Hiltemann S, Brugère JF, Peyretaillade E, Peyret P. ASaiM: a Galaxy-based framework to analyze microbiota data. GigaScience. 2018;7(6):giy057. doi: 10.1093/gigascience/giy057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell K, Dao C, Freise A, Mangul S, Parker JM. PUMA: a tool for processing 16S rRNA taxonomy data for analysis and visualization. doi: 10.1101/482380. bioRxiv 482380. [DOI] [Google Scholar]
- 46.Venkataraman A, Sieber JR, Schmidt AW, Waldron C, Theis KR, Schmidt TM. Variable responses of human microbiomes to dietary supplementation with resistant starch. Microbiome. 2016;4:33. doi: 10.1186/s40168-016-0178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber KS, Bridgewater LC, Jensen JL, Breakwell DP, Nielsen BL, Johnson SM. Personal microbiome analysis improves student engagement and interest in immunology, molecular biology, and genomics undergraduate courses. PLOS One. 2018;13(4):e0193696. doi: 10.1371/journal.pone.0193696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, Leff JW, Vázquez-Baeza Y, Gonzalez A, Knight R, Dunn RR, Fierer N. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15:531. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denofrio LA, Russell B, Lopatto D, Lu Y. Linking student interests to science curricula. Science. 2007;318:1872–1873. doi: 10.1126/science.1150788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.