The relative evolutionary roles of adaptive and non-adaptive processes remain a central question in population genetics. Resolution of this debate has been difficult as an appropriate null model incorporating...
Keywords: background selection, demographic inference, distribution of fitness effects, approximate Bayesian computation
Abstract
The question of the relative evolutionary roles of adaptive and nonadaptive processes has been a central debate in population genetics for nearly a century. While advances have been made in the theoretical development of the underlying models, and statistical methods for estimating their parameters from large-scale genomic data, a framework for an appropriate null model remains elusive. A model incorporating evolutionary processes known to be in constant operation, genetic drift (as modulated by the demographic history of the population) and purifying selection, is lacking. Without such a null model, the role of adaptive processes in shaping within- and between-population variation may not be accurately assessed. Here, we investigate how population size changes and the strength of purifying selection affect patterns of variation at “neutral” sites near functional genomic components. We propose a novel statistical framework for jointly inferring the contribution of the relevant selective and demographic parameters. By means of extensive performance analyses, we quantify the utility of the approach, identify the most important statistics for parameter estimation, and compare the results with existing methods. Finally, we reanalyze genome-wide population-level data from a Zambian population of Drosophila melanogaster, and find that it has experienced a much slower rate of population growth than was inferred when the effects of purifying selection were neglected. Our approach represents an appropriate null model, against which the effects of positive selection can be assessed.
At the founding of population genetics in the early 20th century, R.A. Fisher, J.B.S. Haldane, and S. Wright developed much of the mathematical and conceptual framework underlying the study of population-level processes controlling variation observed within and between species. However, as shown by decades of published interactions between them, they held differing views regarding the relative importance of adaptive vs. nonadaptive processes in driving evolution (Provine 2001). As pointed out by Crow (2008), these issues were not really resolved, but “rather they were abandoned in favor of more tractable studies.” With the advent of the Neutral Theory (Kimura 1968, 1983; King and Jukes 1969; Ohta 1973), the evolutionary importance of stochastic effects due to finite population size, as earlier advocated by Wright, received renewed attention.
In the following decades, further theoretical developments as well as the availability of large-scale sequencing data have validated the important role of genetic drift (Kimura 1983; Walsh and Lynch 2018). However, subsequent research on the indirect effects of selection on patterns of variability at “linked neutral” sites has reignited previous debates (Kern and Hahn 2018; Jensen et al. 2019). In particular, it remains unclear whether the large class of deleterious variants envisaged in the Neutral Theory, and their effects on linked neutral sites [background selection (BGS)], are sufficient to explain genome-wide patterns of variation and evolution, or whether a substantial contribution from the effects of beneficial variants on linked neutral sites (i.e., selective sweeps) is also required [see review by Stephan (2010)].
The primary difficulty in answering this question stems from our lack of an appropriate neutral null model, that is, a model incorporating genetic drift as modulated by the demographic history of the population, as well as a realistic distribution of fitness effects (DFE) summarizing the pervasive effects of both direct and indirect purifying selection. Without a model incorporating these evolutionary processes, which are certain to be occurring constantly in natural populations, it is not feasible to quantify the frequency with which adaptive processes may also be acting to shape patterns of polymorphism and divergence.
However, it can be difficult to distinguish the individual contributions of positive and purifying selection from demographic factors such as changes in population size, as all of these evolutionary processes may affect allele frequency distributions and patterns of linkage disequilibrium in similar ways. For example, both purifying selection and population growth can distort gene genealogies at linked neutral sites in a similar fashion (Charlesworth et al. 1993, 1995; Kaiser and Charlesworth 2009; O’Fallon et al. 2010; Charlesworth 2013; Nicolaisen and Desai 2013), and result in a skewing of the site frequency spectrum (SFS) toward rare variants. In fact, demographic inference is often performed using either synonymous or intronic sites, which are close to sites in coding regions, but the contributions of the effects of selection at linked sites are generally ignored. Patterns of variation in these regions may be skewed by the effects of either negative selection (Zeng 2013; Ewing and Jensen 2016) or positive selection (Messer and Petrov 2013), and this could strongly affect the accuracy of the inferred demographic model (Ewing and Jensen 2016; Schrider et al. 2016). In other words, selection may cause demographic parameters to be misestimated in such a way that population size changes are over or underestimated.
In addition, the extent of BGS can vary considerably across the genome. Although it is necessarily a function of the number and selective effects of directly selected sites, as well as the rate of recombination (Hudson and Kaplan 1995; Charlesworth 1996, 2013; Nordborg et al. 1996), the interaction between these parameters and the underlying demographic history of the population remains poorly understood, even for simple models. Furthermore, existing analytical work (Zeng and Charlesworth 2010b; Nicolaisen and Desai 2013; Zeng 2013) has often been done under the assumption of demographic equilibrium, and is mostly restricted to describing strongly selected mutations with a fixed selection coefficient. However, population genomic data suggest the existence of a wide DFE of deleterious mutations, with a significant proportion of weakly selected mutations with 2Nes < 10 [reviewed by Bank et al. (2014b)], where Ne is the effective population size and s is the reduction in fitness for mutant homozygotes. In regions of low crossing over, interference among such mutations may result in large distortions of the underlying genealogies (Kaiser and Charlesworth 2009; O’Fallon et al. 2010; Good et al. 2014), so that the consequences of a wide DFE are not well described by the analytical results.
We first investigate the joint effects of demography, the shape of the DFE, and the number of selected sites in shaping linked neutral variation. Next, we utilize the decay of BGS effects away from the targets of selection, by examining regions spanning coding/noncoding boundaries, to jointly infer the DFE of the coding region and the demographic history of the population. By performing extensive performance analyses, and quantifying both power and error associated with our approximate Bayesian computation (ABC) approach (Beaumont et al. 2002), the method is shown to perform well across arbitrary demographic histories and DFE shapes. Importantly, by utilizing patterns of variation and divergence across coding and noncoding boundaries, this approach avoids the assumption of synonymous site neutrality inherent to approaches based on comparisons of nonsynonymous and synonymous site variability and divergence, an assumption that has been shown to be violated in many organisms of interest (Chamary and Hurst 2005; Lynch 2007; Zeng and Charlesworth 2010a; Lawrie et al. 2013; Choi and Aquadro 2016; Jackson et al. 2017), and which can result in serious misinference (Matsumoto et al. 2016). In applying this approach to genome-wide data from a Zambian population of Drosophila melanogaster, our results show that this population has experienced a very mild 1.2-fold growth in size, considerably less than previous estimates that did not correct for the BGS-induced skew of the SFS (e.g., Ragsdale and Gutenkunst 2017; Kapopoulou et al. 2018). In addition, we estimate that ∼25% of all mutations in exons are effectively neutral in this population, and find little evidence for widespread selection on synonymous sites.
Methods
Simulations
The discrete generation simulation package SLiM 3.1 (Haller and Messer 2019) was used to simulate a functional element of length L, which is flanked by neutral nonfunctional regions. The functional region experiencing purifying selection is described by a DFE that is modeled as a discrete distribution with four bins (Figure 1A) representing effectively neutral (γ < 1), weakly deleterious (1 ≤ γ < 10), moderately deleterious (10 ≤ γ < 100), and strongly deleterious (100 ≤ γ < 10,000) classes of mutations, where γ = 2Nes. Semidominance is assumed, so that the fitness of mutant heterozygotes is exactly intermediate between the values for the two homozygotes (a dominance coefficient, h, of 0.5). Fitness effects are assumed to follow a uniform distribution within each of the four bins. To infer the extent of purifying selection, we estimated the fraction of mutations in each bin, referred to as f0, f1, f2, and f3, respectively (Figure 1A), such that 0 ≤ fi ≤ 1, and Σi fi = 1, for i = 0 to 3. In addition, to limit the computational complexity, we restricted values of fi to multiples of 0.05 (i.e., fi {0.0, 0.05, 0.10, …, 0.95, 1.0} i). These constraints allowed us to sample 1771 different DFE realizations and to work independently of arbitrary assumptions regarding DFE shape.
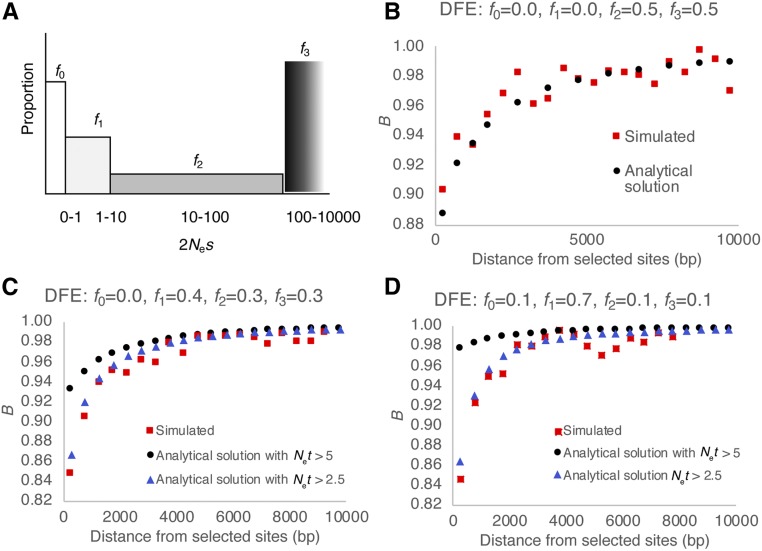
Figure 1.
(A) An example of a discrete DFE with four classes of mutations. The proportion of each class of mutation, fi, lies between 0 and 1. (B) Nucleotide-site diversity relative to the neutral expectation (B = π/π0) as a function of the distance from the directly selected sites (length 1 kb), as predicted by the analytical solution (black points) and as observed in simulations (red points). (C and D) Analytical predictions and simulated values for a DFE with larger contributions from the weakly deleterious class of mutations. Note that, for the analytical solutions, the two classes of results represent cases where mutations with 2Net < 5 (black circles) and 2Net < 2.5 (blue triangles) were ignored. DFE, distribution of fitness effects.
Simulations under demographic equilibrium
Simulations were performed for four different values of L – 0.5, 1, 5, and 10 kb. The intergenic regions were assumed to be 10 kb in length, and simulations were restricted to the intergenic region on one side of the functional region. For the purpose of power analyses and testing, we used population genetic parameter values that approximately resemble those for Drosophila populations. The population size in nature was assumed to be 106, and the recombination rate (1 × 10−8 per site per generation) and mutation rate (1 × 10−8 per site per generation) were constant across the simulated region. Although we have not included gene conversion in this study, it will be an important addition in future studies. The simulations were performed with Nsim = 5000 diploid individuals, and the recombination and mutation rates were scaled proportionally to maintain realistic values of their products with Ne. Such scaling is important for reducing computation time and has been found to be largely accurate, with some exceptions that are not relevant here (Comeron and Kreitman 2002; Hoggart et al. 2007; Kim and Wiehe 2009; Uricchio and Hernandez 2014).
We used a burn-in period of 80,000 generations, and an additional 20,000 (= 4Nsim) generations were allowed for further evolution. For every set of parameter combination (i.e., f0, f1, f2, and f3) we performed 1000 replicate simulations.
Simulations under nonequilibrium demography
Simulations with demographic changes were performed specifically to match the details of the D. melanogaster genome. A set of 94 exons belonging to the D. melanogaster genome were chosen according to certain criteria (see Results and Discussion). Simulations were performed using the length of each exon, together with 4 kb of flanking intergenic sequence. The mutation rate was assumed to be 3.0 × 10−9 per site per generation, which is consistent with both pedigree (Keightley et al. 2014) and mutation accumulation (Keightley et al. 2009) studies, although somewhat higher mutation rates have been estimated in other studies (Schrider et al. 2013; Assaf et al. 2017). Ancestral and current population sizes were sampled from a uniform prior between 105 and 107, and with values of fi chosen as described above. The nucleotide-site diversity at fourfold degenerate sites was 0.019 for the Zambian population of D. melanogaster, giving an estimate of Ne of 1.6 × 106. A scaling factor of 320 corresponding to Ne/Nsim (= 1.6 × 106/5000) was used to perform all simulations with demographic changes. With population size changes, the time of change was assumed to be fixed at Nsim generations, as inferred in previous studies (Terhorst et al. 2017; Kapopoulou et al. 2018). A total of 10 replicates were performed for each exon, resulting in 940 replicates for every parameter combination. These simulations were conducted using the computational resources of Open Science Grid (Pordes et al. 2007; Sfiligoi et al. 2009).
Calculation of summary statistics
First, we fitted a logarithmic function to the recovery of nucleotide-site diversity (π) around the functional region such that π = slope*ln(x) + intercept, where x is the distance of the site from the functional region in base pairs. We used the slope and intercept of the fit to define the number of bases required for a 50, 75, and 90% recovery of nucleotide diversity, with 50% and below being defined as the linked neutral region and 50% and above as the neutral region. This analysis provides for three nonoverlapping regions: (1) functional (experiencing direct selection), (2) linked neutral (experiencing observable levels of BGS), and (3) neutral (experiencing low/unobservable levels of BGS). The following statistics were calculated for each of these three types of regions: nucleotide-site diversity (π), Watterson’s θ, Tajima’s D, Fay and Wu’s H (both absolute and normalized), number of singletons, haplotype diversity, linkage disequilibrium-based statistics (r2, D, and D′), and divergence (i.e., number of fixed mutations per site per generation after the burn-in period). Simulations for any particular set of parameters were run with 1000 replicates, and the means and variances of the above statistics across replicates were used as summary statistics for ABC. In addition to these variables, the six statistics summarizing the characteristics of the recovery of π in linked neutral regions were included. Together, these amount to 72 initial summary statistics. All statistics were calculated using the Python package pylibseq (Thornton 2003). The sample size was kept constant at 100 genomes (i.e., 50 diploid individuals). It should be noted that some statistics are strongly dependent on the number of sites used in the calculations, and the sizes of linked and neutral regions varied for every set of parameter combination, although this effect is captured in the individual prior distributions.
ABC:
We used an ABC approach, using the R package “abc” (Csilléry et al. 2012), to coestimate the DFE characterizing a functional region, as well as the population history. The relationship between the parameters and summary statistics were modeled with a linear regression method (ridge regression) and a nonlinear correction regression method (a neural net), using default parameters provided by the package: https://cran.r-project.org/web/packages/abc/abc.pdf. The neural net method in the abc package by default fits a single-hidden-layer neural network with five units in the hidden layer. To infer posterior estimates, a tolerance of 0.05 was applied (i.e., 5% of the total number of simulations were accepted by ABC to estimate the posterior probability of each parameter). Cross-validation was performed by leaving out one randomly chosen simulation from which the summary statistics from that simulation were used to infer the parameters. A 100-fold cross-validation procedure was used to assess performance as well as to choose the tolerance value determining acceptance. The weighted medians of the posterior estimates for each parameter were used as point estimates.
Ranking of summary statistics:
Ranking of summary statistics was performed separately for both demographic equilibrium and nonequilibrium cases, using two different methods. The first approach consisted of performing Box–Cox transformations on all 72 summary statistics to correct for nonlinear relations between statistics and parameters, using the function “boxcox” in R. Specifically, we used the code provided in Figure 9 of the ABCtoolbox manual (Wegmann et al. 2009) to find partial least squares components using R. The squared correlation coefficient, r2, between the transformed statistics and parameters was then used to rank each statistic for every parameter separately, and a statistic was considered to be significantly correlated with the parameter if the P-value was < 0.05 (Bonferroni corrected for multiple testing with a significance cutoff of 0.05/72). The second approach involved a modified version of the algorithm proposed by Joyce and Marjoram (2008) for ranking statistics. With this algorithm, we started with the entire set of 72 statistics. Each statistic was removed from the set and cross-validation using 20 randomly sampled simulations was used to identify the statistic that corresponded to the least error (i.e., the removal of which causes the least reduction in accuracy). The same algorithm was performed iteratively until only two statistics remained. This method was performed for each parameter separately, was replicated 10 times, and the average ranking across these replicates was used to obtain the final ranking. The second approach was extremely time-consuming and was thus only used to rank the statistics for inferring the DFE under demographic equilibrium.
Comparisons with DFE-α:
Simulations were performed under demographic-nonequilibrium models, with 100 replicates of 94 exons each, and ancestral population sizes of 10,000 for all. Functional regions were simulated with 30% of sites being neutral, which were used to calculate the neutral SFS required by DFE-α. Est_dfe (Schneider et al. 2011) was used on the unfolded SFS to perform demographic inference and to infer the deleterious DFE. The proportion of adaptive mutations was fixed at 0.0. Final estimates of the DFE were obtained as Nws, where Nw is the weighted population size inferred by est_dfe.
Drosophila data application:
Release 5 of the D. melanogaster genome assembly (Hoskins et al. 2007) and annotation version 5.57 were used, downloaded from ftp://ftp.flybase.net/genomes/Drosophila_melanogaster/dmel_r5.57_FB2014_03/gff/. Crossing over rates estimated by Comeron et al. (2012) for every exon and flanking intergenic region were obtained from the D. melanogaster Recombination Rate Calculator (https://petrov.stanford.edu/cgi-bin/recombination-rates_updateR5.pl) (Fiston-Lavier et al. 2010), and explicitly utilized for each specific region considered. These rates were halved to obtain sex-averaged rates of recombination (Campos et al. 2017) as all regions were restricted to autosomes. We excluded all genes that have a crossing over rate fivefold larger or smaller than the average (i.e., we used only genes with a crossing over rate of between 0.44 and 11 cM/Mb). Consensus sequences of all Zambian lines were downloaded from http://www.johnpool.net/genomes.html (Lack et al. 2015). Identity by descent (IBD) tracks and admixture tracks were masked using scripts provided by the same site. Individuals with known inversions were entirely excluded from the analysis (Kapopoulou et al. 2018).
The final set consisted of 76 haploid genomes. PhastCons scores calculated with respect to 15 insect taxa were downloaded from the University of California, Santa Cruz (UCSC) genome browser (https://genome.ucsc.edu/). For each of the 94 exons, summary statistics were calculated using pylibseq (Thornton 2003) for the coding region and for 2-kb intergenic regions flanking both sides. To exclude sites in intergenic regions that might be under direct selection, a phastCons cutoff score of 0.8 was used to calculate all statistics. That is, sites that had a ≥ 80% probability of being a conserved noncoding element identified by phastCons were excluded when calculating statistics.
For the purpose of inferring derived alleles and for calculating branch-specific rates of substitution, we used the ancestral sequence to the D. melanogaster genome provided to us by the authors of Kolaczkowski et al. (2011). The ancestral sequence reconstruction had been performed by maximum likelihood over 15 insect genomes available in the UCSC genome browser (Karolchik et al. 2004). Sites with missing ancestral sequence were excluded from analysis. Branch-specific rates of substitution (also referred to as divergence in this study) were calculated by identifying derived alleles that were fixed in the D. melanogaster Zambian population (i.e., polymorphic sites were removed). After excluding sites with missing ancestral information, with IBD and admixture tracks, and which were likely to belong to a noncoding conserved element, we had on average 1062 sites per exon, 556 sites per linked region, and 666 sites per neutral regions.
It should be noted that for the purpose of performing inference using ABC, substitution rates in simulations were calculated per base pair for 25,000 generations. We thus normalized all rates obtained from simulations by the expected neutral substitution rate (i.e., µsimτsim = 320 × 3 × 10−9 × 25,000 = 0.024, where μsim is the scaled mutation rate and τsim is the number of generations used in the simulations for calculating divergence). Divergence estimates from D. melanogaster were normalized by an expected neutral substitution rate of μτ = 3 × 10−9 × 21333333 (the estimated divergence time) = 0.064 (Li et al. 1999; Halligan and Keightley 2006), where μ is the mutation rate in D. melanogaster and τ is the time to the ancestor of D. melanogaster and D. simulans in number of generations. In addition, inference was performed using divergence estimates only in the exonic regions. ABC inference for Drosophila was performed using the abc package in R, with linear regression aided by neural net with default parameters. Each inference was performed 50 times, and the mean of point estimates obtained were reported as the final parameter estimates.
Data availability
The following data are publicly available on https://github.com/paruljohri/BGS_Demography_DFE: (1) aligned sequences of the single-exon genes and their corresponding intergenic regions used in this study, including derived alleles and fixed substitutions; (2) scripts to calculate statistics from simulations and from empirical data, as well as the code used to perform simulations; (3) values of all calculated statistics obtained for all parameter combinations; and (4) a Mathematica notebook as well as a Python script for analytically calculating the reduction in linked neutral diversity caused by BGS caused by a functional element. Supplemental tables and figures have been uploaded to figshare. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11879025.
Results and Discussion
Recovery of nucleotide diversity away from a functional region as predicted under equilibrium
The nucleotide-site diversity at neutral sites linked to sites experiencing direct purifying selection, relative to its value in the absence of selection (B), can be obtained by modifying equation 6 of Nordborg et al. (1996), which is of the form:
where π0 is the nucleotide-site diversity in the absence of selection and π is its value in the presence of BGS. The term E in the exponent is a function of the heterozygous selection coefficient (t = hs) against a deleterious mutation at a selected site, and the physical distance (z) between the neutral and selected sites. Here, s is the reduction in fitness of mutant homozygotes and h is the dominance coefficient; φ(t) is the probability density function for t.
For the purpose of the current study, equation S1a of the Supplemental Information of Campos and Charlesworth (2019) was modified to model a neutral site outside a gene, located y base pairs from the end of the functional region. If a selected site is x bp from the end of the gene (in the opposite direction), the distance between the two sites is z = x + y. The integral of E (t, z) over z is equal to:
| (1) |
where U is the total mutation rate to deleterious alleles over the entire gene, l is the length of the gene in base pairs, g is the rate of gene conversion, and rc is the rate of crossing over per base pair. The crossover map is assumed to be linear, so that the net rate of recombination between the two sites is g + rcz, and z is assumed to be sufficiently large that the effect of gene conversion is independent of z.
By evaluating the integral in Equation 1, we have:
| (2) |
Note that this equation implies that, if t is small compared with y, BGS effects outside the coding region will be minimal.
We can integrate E(t) over the distribution of selection coefficients, as described in the Appendix. The expectation of E(t) for a given bin of t values is then given by the sum of the following two terms:
| (3a) |
| (3b) |
where a = g + rcy and b = g + rc(y + l), and the ti’s correspond to the boundaries of the discrete bins. For the case when b << 1, the sum of the two components is approximately equal to:
| (3c) |
Figure 1B shows the theoretical and simulation results for rc = 10−6, l = 1000, U = lμ, μ = 10−6, g = 0, t0 = 0, t1 = 0.00005, t2 = 0.0005, t3 = 0.005, and t4 = 0.5. It should be noted that these derivations assume that Net >>1, which is violated by the presence of the weakly deleterious DFE class (frequency f1). Most studies deal with this assumption by ignoring the contribution of mutations with Net < 5 or 10 (Charlesworth 2013; Elyashiv et al. 2016; Torres et al. 2019).
As expected from these considerations, when all classes of mutation were included, we found a significant discordance between the simulated and theoretically predicted values for the slope of the recovery of diversity as f1 increases (Figure 2, C and D and Supplemental Material, Table S1). On including only mutations with Net > 2.5 (i.e., γ = 2Nes > 5), the diversity patterns are mostly well explained, even when the DFE is highly skewed toward the weakly deleterious class. In fact, it is interesting to note that a combination of high values of f1 and f2 can result in BGS effects that extend up to 4 kb, even for very short exons, although the maximum reduction in diversity is ∼10–15%, which is consistent with the findings of Charlesworth (2012) and Campos and Charlesworth (2019).
Figure 2.
Effects of BGS under demographic equilibrium. (A) The slope of the recovery of nucleotide diversity in 10-kb linked neutral regions flanking functional regions, such that π = slope*ln (distance from functional region) + intercept, (B) nucleotide diversity in 500-bp linked neutral regions flanking functional regions relative to neutral expectation (B), and (C) Tajima’s D for 500-bp linked neutral region flanking functional regions. All of the above are shown for various sizes of functional elements (0.5–10 kb) and DFE shapes. The four DFE shapes considered are fi ≥ 0.8 for i = 0,1,2,3, with > 80% of mutations residing in DFE class fi, such that ∑fj ≤ 0.2, where j≠i. The DFE category “all” represents an average over all possible DFE shapes. The error bars are 2 × SD. Red points show the analytical predictions for B with: (1) f0 = 0.85, f1 = 0.05, f2 = 0.05, and f3 = 0.05; (2) f0 = 0.05, f1 = 0.85, f2 = 0.05, and f3 = 0.05; (3) f0 = 0.05, f1 = 0.05, f2 = 0.85, and f3 = 0.05; and (4) f0 = 0.05, f1 = 0.05, f2 = 0.05, and f3 = 0.85. BGS, background selection; DFE, distribution of fitness effects.
Joint effects of demography, the DFE, and the number of selected sites on linked neutral variation:
While the above results show that the effect of BGS on linked neutral regions can be determined analytically, there are several reasons for investigating BGS effects using simulations. First, the analytical expressions neglect the contribution of very weakly deleterious mutations (Net < 2.5) and do not predict the SFS. In addition, they assume demographic equilibrium, which is probably not true of natural populations.
Effects of the shape of the DFE and number of selected sites:
We first simulated 10-kb neutral regions linked to functional regions of varying sizes—0.5, 1, 5, and 10 kb—assuming demographic equilibrium, as shown in Figure 2. By varying the contributions from each bin of selective effects, with frequencies f0, f1, f2, and f3, it was possible to sample all possible DFE shapes, as described in the Methods section. As expected from Equation 3c, the reduction in diversity is nonlinearly proportional to the number of selected sites for a given recombination rate. A larger number of selected sites increases both the total reduction in diversity and the slope of the recovery of diversity away from functional regions (Figure S1). The maximum reduction in diversity in the linked neutral regions (immediately adjacent to the functional region), averaged across all DFE realizations, is ∼8, 12, 24, and 29% for 0.5, 1, 5, and 10-kb selected sites, respectively. Furthermore, for the chosen recombination rate, the median numbers of base pairs necessary to achieve a 50% recovery in diversity are 955, 1035, 1350, and 1650 bp, respectively (Figure 2A).
The reduction of nucleotide diversity at closely linked neutral regions was maximized when the proportions of weakly (f1) and moderately deleterious mutations (f2) were largest (Figure 2B and Table S2). The effect is greatest when purifying selection is weak, allowing mutations to segregate in the population prior to being purged (Campos et al. 2017). Although weakly deleterious mutations (f1) only reduce variation slightly, they generate significant distortions in the SFS (Figure 2C), consistent with previous studies (Charlesworth et al. 1995; Charlesworth 2012; Nicolaisen and Desai 2013). Moderately deleterious mutations cause the largest reduction in π, the highest rate of recovery of π around functional regions, and the largest skew in the SFS toward rare variants. As expected, the proportion of strongly deleterious mutations (f3) does not greatly affect levels of linked neutral variation, and these mutations skew the SFS only slightly. Furthermore, increasing the number of selected sites results in larger BGS effects for all DFE types, as is to be expected. It should be noted that these generalizations about BGS effects depend on the distance between the neutral and selected sites; in particular, the size of the region affected by deleterious mutations is expected to be an increasing function of the size of their fitness effects. As we were interested in understanding BGS effects caused by all classes of mutations, we focus our further discussion on sites close to the functional boundary, where all classes of mutation are likely to have an impact.
To summarize, at demographic equilibrium, neutral regions linked to functional regions under selection undergo a reduction in diversity and a skew in the SFS, both of which depend on the underlying shape of the DFE and the number of directly selected sites (Charlesworth et al. 1993, 1995; Charlesworth 2013; Campos and Charlesworth 2019). However, importantly for the sake of statistical inference, the three classes of deleterious mutation behave differently, suggesting the possibility of distinguishing their relative contributions (as discussed in the next section). Furthermore, these results again demonstrate the potentially important role of BGS in shaping patterns of neutral variation, highlighting the danger posed by ignoring these effects when performing demographic inference [see Ewing and Jensen (2016)]. Additionally, the dramatic differences in the extent of BGS effects as a function of the number of directly selected sites emphasize the necessity of directly modeling exon sizes in empirical applications.
Effects of demography and the shape of the DFE on BGS:
We investigated the effects of BGS after recent changes in population size. Populations with the same ancestral population size (Nanc) either experienced 10-fold exponential growth or contraction in the last 4Nanc generations; BGS effects were compared to populations that remained in equilibrium throughout, for all possible DFE shapes. Both expansion and contraction result in reduced BGS effects (i.e., there is an increase in B compared to equilibrium), irrespective of the shape of the DFE (Figure 3, A and B). This observation suggests that the extent of BGS caused by functional elements is not only determined by the strength of selection, but also by the demographic history of the population. Thus, demographic effects may in principle explain variable inferences among studies of the importance of purifying selection in shaping genome-wide patterns of variation (Cutter and Payseur 2013).
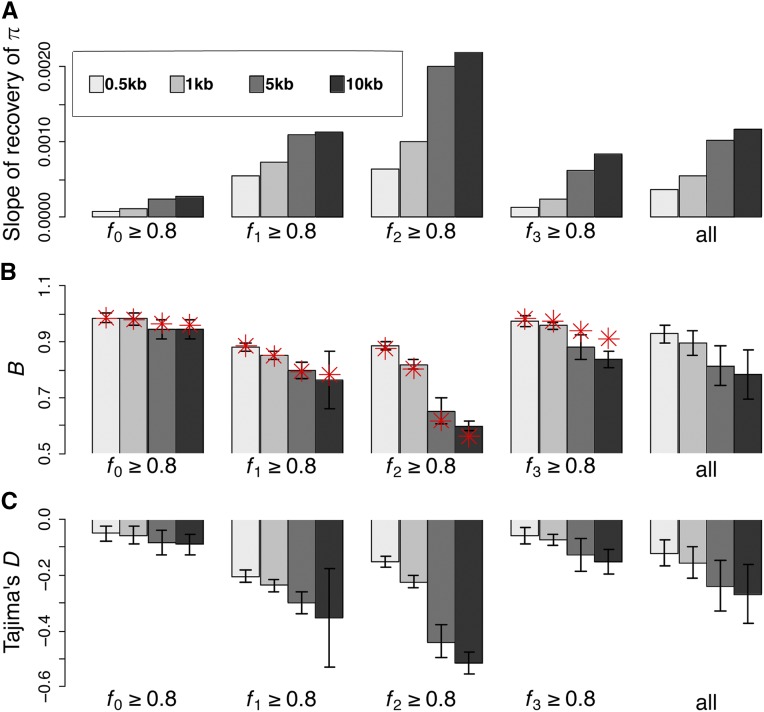
Figure 3.
Effects of BGS under nonequilibrium demography. (A) The slope of recovery of nucleotide diversity in linked neutral regions for different DFE shapes under equilibrium demography (black), population expansion (blue), and contraction (red). (B) Nucleotide-site diversity relative to neutral expectation (B), over 500 bp of linked neutral regions flanking functional regions, for varying DFE shapes and three different demographic models: equilibrium (black), 10-fold exponential expansion (blue), and 10-fold exponential decline (red). (C) Tajima’s D for the 500-bp linked neutral region flanking the functional region under equilibrium, (D) after a 10-fold expansion, and (E) after a 10-fold population size reduction. The four DFE shapes considered in all panels are fi ≥ 80% for i = 0–3, where > 80% of mutations reside in DFE class fi. The DFE category “all” represents an average over all possible DFE shapes. For nonequilibrium demography, γ = 2Nancs, where Nanc is the ancestral population size. BGS, background selection; DFE, distribution of fitness effects.
However, interestingly, there is still a significant skew in the SFS at linked neutral sites caused by BGS after a population size change (Figure 3, C–E). Thus, in more compact genomes, where BGS is pervasive, this suggests that methods that use the SFS to fit demographic models may overestimate growth, and either underestimate population contraction or misclassify contraction as expansion. It is also interesting to note that BGS effects are largest under demographic equilibrium, such that constant population size is likely to be inferred as population growth.
Inference of the DFE under demographic equilibrium:
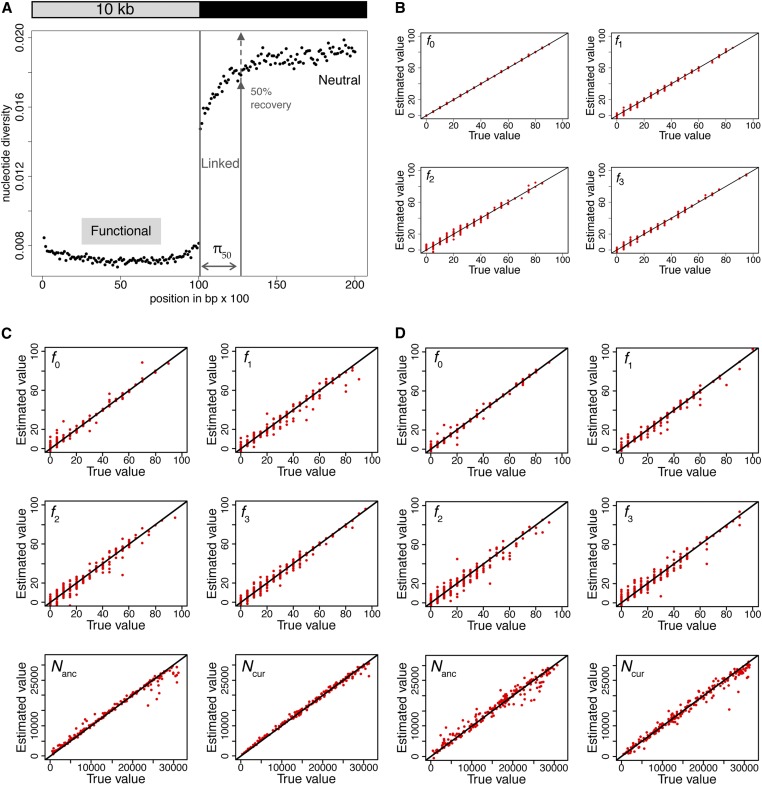
The next question we investigated was whether the parameters of the DFE can be estimated using the set of summary statistics described in the Methods section. We first determined whether it is possible to distinguish the four different classes of the DFE under demographic equilibrium, using population genomic data and divergence from the closest outgroup species. The simulations involved functional regions of lengths L = 0.5, 1, 5, and 10 kb, with linked neutral regions of 10 kb and a discrete DFE, as described previously. The ABC approach described in the Methods section was used to quantify our ability to infer the four DFE parameters. The recovery of nucleotide diversity over linked neutral regions was used to calculate the distance in base pairs (π50) required for diversity to reach 50% of its maximum value (see Methods). The neutral region within π50 base pairs from the functional region was defined as linked, and the remainder was defined as neutral (Figure 4A). Statistics were calculated for three regions (functional, linked, and neutral) separately, and the means and variances across simulation replicates of each statistic were used to infer the four parameters. The simulation replicates correspond to independently evolving loci within a genome. In the following subsections, we describe the performance of the method and its robustness to various model violations.
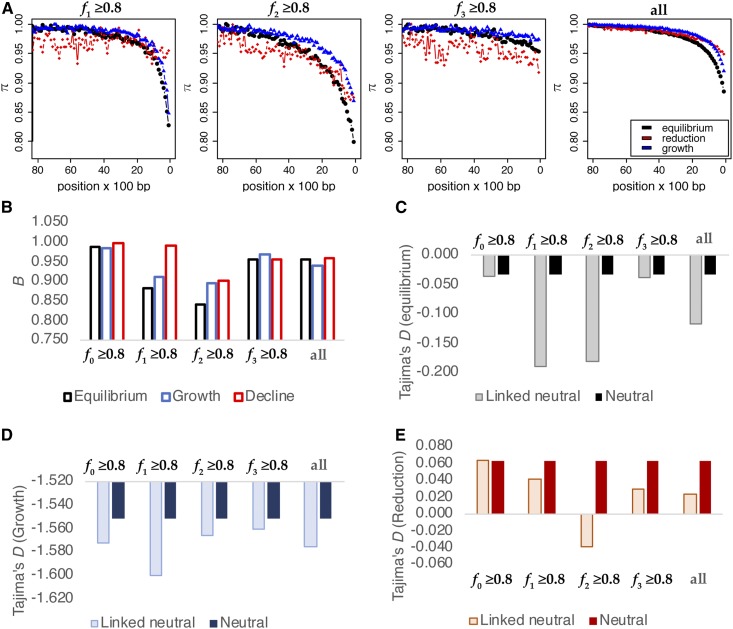
Figure 4.
(A) Values of diversity statistics across functional, linked, and neutral regions. (B) Accuracy of estimation (cross-validation) of the four classes of the DFE using statistics for functional regions only (size 1 kb), under equilibrium demography. (C) Joint estimation of population size changes and the DFE using all statistics. (D) Joint estimation of population size changes and the DFE using statistics for functional regions only. The true proportions of mutations in each DFE class and Nanc and Ncur are given on the x-axes, while the estimated values are given on the y-axes. Parameters are indicated in the upper left corners for each plot. Each dot represents 1 out of 200 different parameter combinations, sampled randomly from the entire set of simulations. DFE, distribution of fitness effects.
Accuracy of inference:
All four DFE classes were estimated fairly accurately when using all statistics (Figure S2a). However, under demographic equilibrium, the DFE is inferred much more accurately using statistics from the functional regions alone, thus sidestepping the need for the identification of linked neutral regions (Figure 4B and Figure S2b). In both cases, the accuracy of inference is highest for the neutral class and lowest for moderately deleterious mutations (class 2), and significantly improves when the size of the functional region increases (Figure S2). When using only functional regions to perform inference, the absolute difference between the true value and the estimated value of the neutral class is ∼0.034, 0.030, 0.017, and 0.010 for functional sizes of 0.5, 1, 5, and 10 kb. Therefore, for 1-kb regions, the method cannot determine whether the neutral class of mutations comprise 30 or 33% of the DFE. For the moderately deleterious class, this error is larger: 0.077, 0.060, 0.028, and 0.019, respectively. These absolute error values are not surprising, as the fi in our simulations are multiples of 0.05 out of computational necessity. The accuracy of the estimates can thus be increased by sampling the parameter space more densely.
The accuracy of estimation can also be evaluated using r2 between the true and estimated values. For instance, for 1-kb functional regions, the r2 values for f0, f1, f2, and f3 are 0.93, 0.91, 0.89, and 0.87, respectively. It is interesting to note that it is possible to infer the proportion of DFE classes when using statistics from the linked region alone (Figure S3). Although there is an increase in the absolute errors for 10-kb regions to 0.103, 0.122, 0.056, and 0.044 in f0, f1, f2, and f3, respectively, this analysis suggests that, if the population size were known to be at equilibrium, statistics for the linked neutral regions alone could distinguish between the contributions of the four DFE classes.
It should furthermore be noted that this approach does not distinguish between nonsynonymous and synonymous mutations. Indeed, no assumption is made regarding which specific bases are neutral, nearly neutral, or deleterious in the coding region. Thus, this method can be used to estimate the DFE for any type of functional region, as well as to assess the nonneutrality of synonymous sites by comparing their frequency in a given coding region with the value of f0.
Effect of misspecification of exon size and recombination rate:
In view of these results, it is important to consider whether accurate estimates depend on correctly specifying precise exon size, or if it would be sufficient to generate priors assuming, for example, a mean exon length for a given genome. To quantify this effect, simulated data sampled from the priors were based on 1-kb exons, while the test data were obtained from simulations based on alternative exon sizes. The error in inference of the DFE increases as the differences between exon sizes of the priors and those of the true sizes are increased (Figure S4), with the highest error for the moderately deleterious class 2, although when exon sizes are sufficiently large, misspecification of exon size does not strongly affect performance. A similar approach was used to determine if the presence of another functional region (also 1 kb in size), separated by an intron or intergenic region, would skew inference. As expected, smaller intron sizes result in stronger misinference than larger ones, and intronic/intergenic sizes > 4 kb performed essentially as well as those with no nearby functional exon (Figure S5). Moreover, a twofold difference between assumed and actual recombination rates inflated the errors dramatically (Figures S6 and S7). Informatively, the direction of bias generated differs by DFE class (Figure S7). For example, when true recombination rates are half of those assumed, the inferred weakly deleterious class is greatly inflated. As this class of mutations most strongly skews the linked neutral SFS, this misinference presumably arises from an attempt to fit stronger linked effects by inferring a higher proportion of mutations in this class, whereas in reality the increased BGS effects are being generated by fewer recombination events than are assumed.
These results highlight the importance of taking into account the specific exonic–intronic–intergenic structure of a particular genomic region of interest, nearby functional regions, and the specific recombination rate. Although any configuration of these details may be directly simulated, an alternative approach is simply to group exons of like size across a genome, and further reduce these to a group that is devoid of neighboring functional regions.
Joint inference of purifying selection and demography, under nonequilibrium conditions:
Based on the above results demonstrating that details of exon sizes and recombination rates are essential for accurate inference, we explicitly modeled both exon sizes and recombination rates when examining our ability to jointly infer demographic changes together with the DFE. As our example involved an African population of D. melanogaster, we chose for our simulations single-exon genes that had > 4-kb noncoding regions flanking both sides, and whose exon sizes were between 500 and 2000 bp. For this specific set of 94 exons, we simulated functional regions with specified exon sizes linked to 4-kb neutral regions and utilized the previously inferred local crossing over rate for each exonic region in question. For every parameter combination, we performed 10 replicates of each of the 94 exon sizes (resulting in a total of 940 replicates per parameter combination), with their respective recombination rates and exon sizes, and summarized the resulting means and variances of the summary statistics.
Models of exponential population size expansion and contraction assumed various ancestral population size (Nanc) and current population size (Ncur), which were both sampled uniformly between 105 and 107, following previous studies (Duchen et al. 2013; Arguello et al. 2019). As earlier work has inferred the duration of the expansion in Zambian populations to be of the order of Ne generations, this was scaled down and fixed at Nsim = 5000 generations, to attempt to infer both historical and current population sizes. Thus, for this framework, we evaluated the estimates of six parameters: f0, f1, f2, f3, Nanc, and Ncur.
Accuracy of joint inference:
Encouragingly, the results demonstrated an ability to successfully coestimate the DFE, and both ancestral and current population sizes, using the set of coding and linked noncoding summary statistics described above (Figure 4C). Under nonequilibrium demography, the estimation error for the strongly deleterious class of mutations is larger. The absolute differences between true and estimated values were 0.019, 0.027, 0.033, and 0.034 for the four DFE classes, respectively; the errors in ancestral and current sizes were 10.1 and 7.3%, respectively. The r2 values between the true and estimated values of f0, f1, f2, f3, Nanc, and Ncur were 0.97, 0.97, 0.95, 0.95, 0.99, and 0.99, respectively.
The performance of the full six-parameter estimation procedure is good, without relying on the usual stepwise approach of first utilizing putatively neutral sites to fit a demographic model, and then using this model to estimate DFE parameters. Interestingly, joint estimation is almost as accurate when using statistics from functional regions alone (Figure 4D), although it inflates the errors in the estimates of f2 and f3. The absolute differences between the true and estimated values of f0, f1, f2, and f3 were 0.015, 0.025, 0.054, and 0.049, respectively, while the errors in estimates of population sizes increase to 23 and 8% for Nanc and Ncur, respectively. Thus, the error in ancestral population size is quite large if only functional regions are used to coestimate all six parameters. Further, unlike the case of demographic equilibrium, statistics in linked regions alone can no longer be used to accurately infer parameters of the DFE (Figure S8).
Effect of misspecification of mutation rate:
We evaluated the effect of having incorrect estimates of mutation rate by inferring all six parameters under a scenario in which the assumed mutation rate is one-half of or twice the true value. Under all demographic scenarios, if the assumed mutation rate was one-half of the true rate, our method correctly estimated f0, f1, and Ncur (Figure S9). However, the strongly deleterious class 3 is consistently underestimated, the moderately deleterious class 2 is overestimated, and Nanc is biased toward a strong population size decline. A comparable magnitude of misinference is observed when the assumed mutation rate is twice the true value (Figure S9). Thus, the ABC method described in this study is sensitive to large mismatches between the true and assumed mutation rates, and is thus best suited to organisms in which pedigree- and/or mutation accumulation-based estimates are available.
Statistics important for distinguishing different classes of the DFE and demography:
As it is important to understand which statistics are needed to distinguish between the effects of demography and the different classes of the DFE, two different approaches were used to rank statistics by their importance. First, statistics were simply ranked by their regression coefficient with respect to each parameter separately. Nonlinear relationships were taken into account by using Box–Cox transformation, as suggested by Wegmann et al. (2009). With stationary population size, most of the top predictors of the fraction of neutral (f0) and strongly deleterious (f3) sites are statistics summarizing the functional region (Table S3). The top four statistics for each parameter are displayed in Figure S10. In addition, a modification of the method of Joyce and Marjoram (2008) was also employed to rank statistics (Table S4) for equilibrium demography.
As expected, the statistics that correlate most strongly with the fraction of neutral mutations are the levels of divergence and the fraction of high-frequency derived alleles, as summarized by θH (Fu 1995; Fay and Wu 2000) in functional regions. As the weakly deleterious class of mutations generate BGS effects at closely linked sites, statistics for the functional and linked region are most strongly correlated with f1. This also correlates most with H′ in functional regions, a statistic that contrasts the proportion of high-frequency derived variants with those of derived variants segregating at intermediate frequency (Fay and Wu 2000). Although this statistic was designed to identify selective sweeps, which tend to increase the proportion of high-frequency derived alleles, it is highly predictive of the fraction of the weakly deleterious class of mutations in the absence of positive selection. As shown above, larger f1 generates a stronger skew in the linked neutral SFS toward rare variants and is thus also reflected in the values of Tajima’s D in the linked neutral region. Measures of linkage disequilibrium in the functional and linked neutral regions are also correlated with the frequency of the weakly deleterious class of mutations.
Because the moderately deleterious class of mutations generates BGS effects that extend for larger distances than the more-weakly selected class, the strongest correlates of this class are generally statistics from the neutral region furthest from the directly selected sites. All the different summaries of the SFS—θW, π, and θH—correlate with this parameter, as well as the total reduction in linked neutral diversity (given by the intercept of the regression fit of π = slope*ln (distance) + intercept, where π is the diversity in linked neutral regions). The strongly deleterious class of mutations is correlated with the number of singletons and θw.
A similar analysis was performed on simulations with demographic nonequilibrium. Here, the DFE parameters are significantly correlated only with the statistics for functional regions (Tables S5 and S6). As expected intuitively, the statistics most highly correlated with the two demographic parameters are for the neutral linked regions. Ancestral population sizes correlate best with statistics that capture high-frequency derived alleles in linked neutral as well as functional regions, as these represent older mutations; current population sizes correlate most with statistics that summarize linkage disequilibrium. The same is true when ranked statistics are obtained only from functional regions. Because the class of moderately deleterious mutations and ancestral population sizes are correlated with overlapping sets of statistics, the estimates of these two parameters are partially confounded. As such, linkage disequilibrium-based statistics are essential for distinguishing between demography and purifying selection, and in estimating ancestral and current population sizes. In addition, although the variances and means of the statistics are highly correlated, the variances play a more important role in estimating current population sizes.
Comparison with DFE-α:
Because there are no other programs that simultaneously coestimate both demographic and selection parameters, we compared the performance of our method to the stepwise approach of DFE-α (Keightley and Eyre-Walker 2007; Eyre-Walker and Keightley 2009; Schneider et al. 2011), a program used widely for the inference of the DFE. DFE-α assumes that synonymous sites are neutral and uses their SFS to infer changes in population size. Conditional on the inferred demography and under the assumption that the deleterious selection coefficients follow a given distribution (generally γ), the program infers the shape and scale parameter of the assumed distribution.
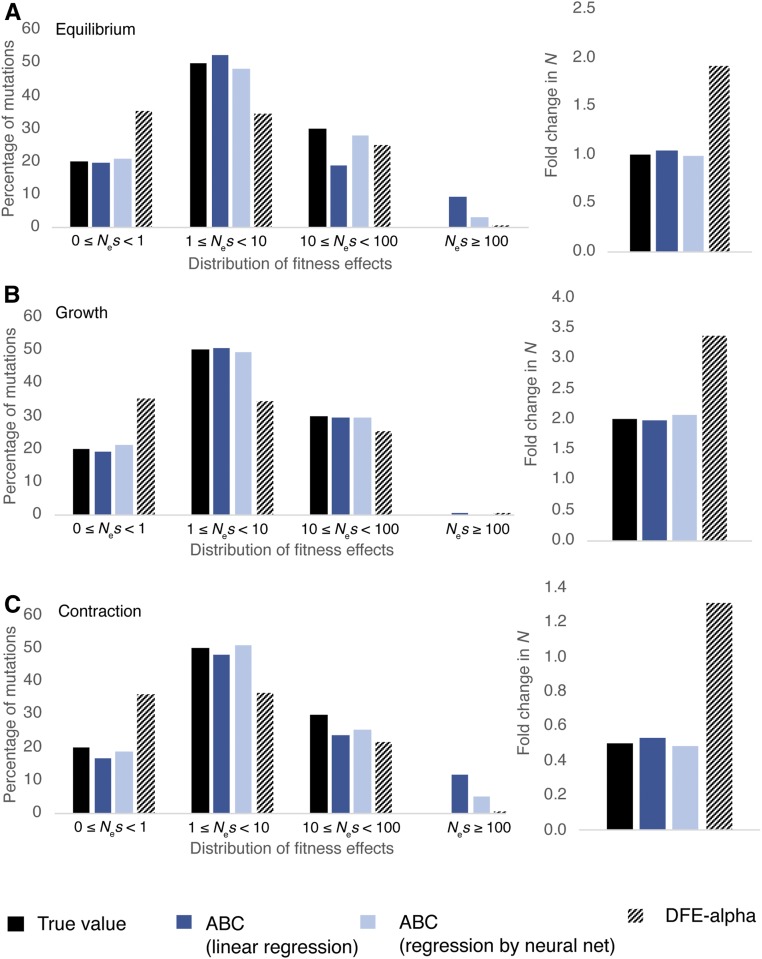
We simulated demographic equilibrium, twofold population growth, and twofold population contraction, and inferred the change in population size and the DFE using both ABC and DFE-α. Because DFE-α uses neutral sites to infer demography, in all cases we simulated a DFE consisting of 30% neutral mutations, which are a proxy for synonymous sites. These simulations were performed exactly as described previously for nonequilibrium conditions. Exon sizes between 500 and 2000 bp with flanking 4-kb linked neutral regions were simulated with recombination rates specific to the selected 94 exons (and a total of 940 replicates for every parameter combination). DFE-α performs slightly better than ABC if the true DFE is indeed γ distributed (Figure S11), although our method is able to infer the DFE with very similar accuracy.
For a discrete DFE that is skewed toward highly deleterious mutations, DFE-α and ABC perform with similar accuracy. However, our method performs better if the DFE is skewed toward slightly deleterious mutations as shown in Figure S12. It is important to note that, for the purpose of this comparison, simulations were run with numbers of directly selected sites between 500 and 2000 bp, with 30% of mutations being neutral, because neutral mutations are required to estimate demographic parameters with DFE-α. Under these conditions, BGS results in a relatively small skew in the neutral SFS (Campos and Charlesworth 2019).
As noted previously, a potential advantage of the methodology proposed here is that, by simultaneously estimating selection and demography, one is not required to make any assumptions about the neutrality of synonymous sites. We evaluated this feature by simulating a scenario where 33% of the assumed neutral sites were actually experiencing weak direct selection. As weak purifying selection generates a larger fraction of rare variants than stronger selection, programs based on neutrality would be likely to falsely infer growth. As expected, DFE-α inferred twofold growth under demographic equilibrium, and in fact inferred slight growth even for a twofold contraction (Figure 5). The resulting DFE overestimated the fraction of neutral mutations and underestimated the fraction of weakly deleterious mutations. As noted previously, such misinference will increase with the density of selected sites. However, our ABC approach accurately estimated the proportion of neutral mutations present in the selected region (Figure 5), illustrating the importance of joint inference.
Figure 5.
Comparison of the performance of the proposed ABC approach in the current study with DFE-α (when there is selection on synonymous sites), under (A) demographic equilibrium, (B) exponential growth, and (C) exponential decline. In all cases, 30% of sites were assumed to be synonymous, out of which 33% were weakly selected. Solid black bars are the true simulated values, dark blue bars give the ABC performance using ridge regression, and light blue bars give the ABC performance using linear regression aided by neural nets. Patterned bars show the performance of DFE-α. A total of 998,300 sites were analyzed in the functional region for each parameter combination, with ∼332,767 representing synonymous and 665,533 representing nonsynonymous sites. ABC, approximate Bayesian computation; DFE, distribution of fitness effects.
Application to D. melanogaster:
When simulating genomic regions, the presence of nearby coding regions that are not included in the models can generate additional BGS effects and thus bias inference. We thus restricted our analyses to protein-coding exons in the D. melanogaster genome between 500 and 2000 bp in length that are single-exon genes, and are flanked on both sides by intergenic regions > 4 kb, so that effects of linkage with other nearby functional elements are avoided. It should be noted that any genic structure could readily be chosen for inference by directly simulating the associated details when constructing the priors; we have simply chosen this realization to provide an illustrative application.
The recombination rates of both the 5′ and 3′ flanking intergenic regions are highly correlated (Figure S13), and span a considerable magnitude (Figure S14) with a mean rate of 2.78 cM/Mb (i.e., the average recombination rate for these chosen single-exon genes is very near the autosomal genome-wide average of 2.32 cM/Mb). We also verified that this set of genes was not unusual with respect to genome-wide coding sequence divergence (Figure S15). Furthermore, because sites in intergenic regions in D. melanogaster may also experience direct selection (Halligan and Keightley 2006; Casillas et al. 2007), we used phastCons scores to exclude intergenic sites that might potentially be functionally important. All sites with a phastCons score larger than 0.8 were excluded (Siepel et al. 2005).
Table 1 provides the observed summary statistics for each region class, where intergenic sites that had an ≥ 80% probability of belonging to a conserved element (i.e., with phastCons score ≥ 0.8) were excluded. It should be noted that the absence of a large difference between divergence (i.e., number of fixed substitutions specific to D. melanogaster) in exonic vs. intergenic regions is consistent with previous studies [table 1 in Andolfatto (2005)]. In addition, because we have restricted our analyses to sites where the ancestor of D. melanogaster could be predicted with high confidence, our analyses may be skewed toward more conserved sites, potentially resulting in lower divergence in intergenic regions. Previous estimates of divergence along the D. melanogaster lineage at fourfold degenerate sites were ∼0.05–0.06 (Halligan and Keightley 2006; Langley et al. 2012; Charlesworth et al. 2018), while that in coding regions was 0.023 (Langley et al. 2012). Although our estimates are lower than previous estimates, this discrepancy is explained by the larger number of individuals used to subtract polymorphic sites in this study (Table S7). With a sample size of 1 allele (corresponding to pairwise divergence), our estimates of divergence at fourfold degenerate and in coding regions are 0.05 and 0.023, respectively, consistent with previous studies. In addition, a very similar relation between pairwise divergence and polymorphism-adjusted divergence is found with simulated data (Table S8).
Table 1. Statistics calculated for the 94 single-exon genes including their 3′ flanking intergenic sequences, for 76 haploid genomes (devoid of any inversion) from the Zambian population of D. melanogaster.
| Mean | SD | |||||
|---|---|---|---|---|---|---|
| Functional | Linked | Neutral | Functional | Linked | Neutral | |
| π | 0.0083 | 0.0106 | 0.0107 | 0.0039 | 0.0042 | 0.0038 |
| θW | 0.0120 | 0.0166 | 0.0162 | 0.0045 | 0.0053 | 0.0049 |
| θH | 0.0088 | 0.0098 | 0.0097 | 0.0054 | 0.0053 | 0.0056 |
| H′ | −0.0633 | 0.0871 | 0.1169 | 0.5371 | 0.4118 | 0.3829 |
| Tajima’s D | −1.0537 | −1.1469 | −1.1103 | 0.5338 | 0.4874 | 0.4694 |
| Singleton density | 0.0215 | 0.0303 | 0.0307 | 0.0086 | 0.0116 | 0.0117 |
| Haplotype diversity | 0.9711 | 0.9680 | 0.9762 | 0.0452 | 0.0458 | 0.0444 |
| r2 | 0.0328 | 0.0364 | 0.0363 | 0.0109 | 0.0136 | 0.0128 |
| D | 0.0005 | 0.0005 | 0.0006 | 0.0009 | 0.0010 | 0.0012 |
| Branch-specific divergence | 0.01378 | 0.0156 | 0.0159 | 0.0075 | 0.0077 | 0.0071 |
Sites with phastCons scores > 0.8 were excluded. Functional refers to exons, linked refers to intergenic region (∼1 kb) adjacent to exons, and neutral refers to intergenic regions further away from exons that are adjacent to linked regions (Figure 4A). Derived alleles were identified by polarizing alleles with respect to the ancestral sequence of D. melanogaster obtained from ancestral reconstruction over 15 insect species.
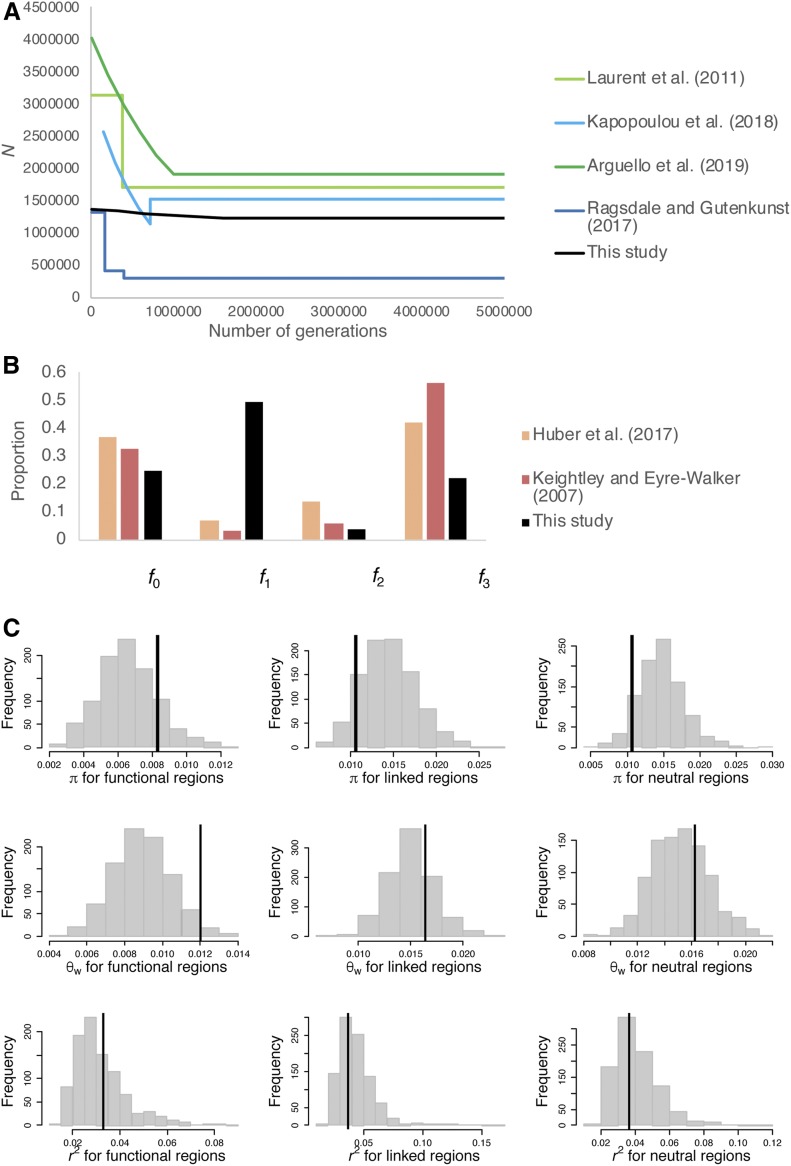
Interestingly, although previous studies have inferred ∼two- to fourfold growth in the Zambian population of D. melanogaster (Ragsdale and Gutenkunst 2017; Kapopoulou et al. 2018), we infer only 1.2-fold growth, with an ancestral Ne of 1,225,393 and current Ne of 1,357,760. Our estimates of ancestral Ne are comparable to those inferred by previous studies of African populations of D. melanogaster (Li and Stephan 2006; Laurent et al. 2011; Duchen et al. 2013; Arguello et al. 2019; Figure 6). As shown in Figure 6, we infer a much larger proportion of mildly deleterious mutations and a smaller proportion of highly deleterious mutations than in previous studies (Keightley and Eyre-Walker 2007; Huber et al. 2017), with f0 = 24.7%, f1 = 49.4%, f2 = 3.9%, and f3 = 21.9%, but this reflects the fact that our procedure includes synonymous sites among the total. Because we have inferred the DFE for a select class of single-exon genes, which have slightly higher than average divergence (Figure S15), it is possible that these exons are experiencing weaker purifying selection than the genome-wide mean. Furthermore, because we have obtained the DFE of both coding sequences and UTR regions, fourfold degenerate and UTR sites comprise 12 and 29% of the total, respectively. Previous studies have estimated 6–10% of all mutations at nonsynonymous sites to be effectively neutral. Thus, assuming that all fourfold degenerate sites are neutral, ∼40% of UTR regions are neutral (Andolfatto 2005; Campos et al. 2017), and ∼6–10% of nonsynonymous mutations are neutral, we expect f0 to be ∼27–30%. Encouragingly, we infer f0 = 25%. This observation implies that the majority of synonymous sites are not experiencing direct selection, consistent with previous results for D. melanogaster (Jackson et al. 2017). Further, although we infer a larger proportion of weakly deleterious mutations than previous studies from the distribution of γ = 2Nes, the distribution of s is quite comparable (Table S9) to that inferred by Huber et al. (2017).
Figure 6.
Joint inference of demography and purifying selection in the Zambian population of D. melanogaster. (A) Demographic model inferred in previous studies of the Zambian population (blue lines), the Zimbabwe population (green lines), and the current study (black lines). (B) The DFE for deleterious mutations in coding regions (including synonymous and nonsynonymous sites) as inferred by previous studies of other populations (colored bars) and at exonic sites of single-exon genes as inferred in the current study (black bars). The x-axis is for f0: 0 ≤ 2Nes < 1, f1: 1 ≤ 2Nes < 10, f2: 10 ≤ 2Nes < 100, and f3: 100 ≤ 2Nes < 10,000. For the previous studies, the DFE shown in this figure includes the fraction of synonymous sites in the neutral f0 class. (C) Distribution of key summary statistics (π, θW, and r2) for functional, linked, and neutral regions when simulating 100 replicates of 94 exons each using the inferred parameters. The vertical lines represent values of the statistics obtained from 76 individuals of D. melanogaster from Zambia, after excluding noncoding sites with phastCons score ≥ 0.8. DFE, distribution of fitness effects.
To test whether our inferred parameters explain the observed D. melanogaster data, we simulated 10 replicates of each of the 94 exons using the parameter estimates, and evaluated whether the means of the observed D. melanogaster values are in the 5% tails of the simulated distribution of statistics. Our parameter estimates result in a very good fit to empirical D. melanogaster population data (Figure 6 and Figure S16) for all three categories—functional (i.e., exonic), linked (i.e., noncoding region adjacent to exons), and neutral (i.e., noncoding region adjacent to the linked region)—except for Tajima’s D (linked region P = 0.011 and neutral region P = 0.010) and divergence (linked region P = 0.029 and neutral region P = 0.0) in intergenic regions, although these statistics are well fitted in functional regions.
As both positive selection in exons and purifying selection in noncoding regions could partially drive these patterns, we investigated both of these model violations. Noncoding regions flanking 2 kb of the selected exons (which were used to perform inference) were found to have 777 sites that had phastCons scores > 0.8, with a mean and median length of 25 and 15 bp, respectively. We therefore simulated conserved elements in noncoding regions that were 20 bp in length, uniformly distributed, and which made up 40% of the flanking neutral sites (i.e., 800 sites in total). Conserved elements were simulated with either weak (f1 = 1), moderate (f2 = 1), or strong (f3 = 1) purifying selection. Upon masking these sites, as was done in our Drosophila data analysis, there was no observed difference in the distribution of all statistics (Figure S17), suggesting that BGS caused by small conserved elements does not significantly affect our inference, and in fact does not alter the fit of our inferred model to the data. Interestingly, without masking sites—that is, by allowing sites that experience direct weak purifying selection to remain in the flanking sequences—our model is much better able to explain the lower Tajima’s D and divergence in intergenic regions (Figure S18). Thus, it appears likely that weak purifying selection on sequences in intergenic regions could contribute to the discrepancies between observed and expected.
Next, we simulated positive selection with selection coefficient sb for beneficial mutations under four different scenarios, representing rare and strong (1% of all mutations in exonic regions are beneficial with 2Nesb = 1000), common and strong (5% of mutations in exonic regions are beneficial with 2Nesb = 1000), common and weak (5% of mutations in exonic regions are beneficial with 2Nesb = 10), and rare and weak (1% of mutations in exonic regions are beneficial with 2Nesb = 10) selection. We find that, although strong positive selection, whether common or rare, better explains the lower Tajima’s D values in intergenic regions, it also drastically alters the distribution of most other statistics, resulting in an overall much poorer fit (Figures S19 and S20). For instance, common and strong positive selection reduces θH by an order of magnitude relative to our fitted model, and drastically increases the variance while decreasing the mean of haplotype diversity. In contrast to strong positive selection, weakly positively selected mutations do not alter the distribution of Tajima’s D in intergenic regions, but slightly increase θH in functional regions, which improves the fit to the observed data (Figures S21 and S22). In addition, all cases of positive selection significantly increase divergence in functional regions. For comparison, we also simulated the two scenarios of positive selection used by Lange and Pool (2018): 0.2% of all mutations are beneficial with 2Nesb = 60, and 0.00013% of all mutations are beneficial with 2Nesb = 10,000. As the frequency of positively selected alleles is lower in these scenarios, there was no observed difference between the distribution of statistics resulting from including or excluding positive selection (Figures S23 and S24). Thus, if the frequency of strongly positively selected mutations is much lower than 1%, as was proposed by Campos et al. (2017), our estimates of both demography and DFE shape should be unbiased, and beneficial fixations would be virtually undetectable. Future studies will investigate the ability of our approach to quantify the properties of beneficial mutations.
Conclusion
Independent of specific views on the roles of adaptive vs. nonadaptive explanations for observed levels and patterns of DNA sequence variation and divergence, it has been widely accepted that natural populations are not at demographic equilibrium, but are often characterized by fluctuating population sizes and other demographic perturbations. Additionally, a rich empirical and experimental literature has demonstrated the pervasive importance of purifying selection in eliminating the constant input of deleterious variants. It has also been found that ignoring direct effects of purifying selection and its impact on linked sites can strongly bias demographic inference (Ewing and Jensen 2016), and that ignoring demographic effects biases estimates of parameters of selection (Jensen et al. 2005; Thornton and Jensen 2007; Crisci et al. 2012, 2013). Yet, despite agreement that these processes are certain to be occurring constantly in populations and shaping patterns of variation and evolution, the construction of a statistical approach capable of simultaneously estimating parameters of the concerned processes has been difficult. Here, we provide one such approach, for which we demonstrate an ability to coestimate the parameters of a generalized DFE along with those underlying the population history.
By fitting a four-parameter DFE model that includes weak, intermediate, and strong purifying selection, as well as neutrally evolving sites, this approach avoids two common, and potentially perilous, assumptions: (1) synonymous sites are not assumed to be neutral, consistent with a growing body of literature (Chamary and Hurst 2005; Lynch 2007; Zeng and Charlesworth 2010a; Lawrie et al. 2013; Choi and Aquadro 2016; Jackson et al. 2017), and (2) the DFE is not assumed to follow a specific parameterized distribution, such as the widely-used γ distribution.
Our results demonstrate that it is possible to jointly infer the deleterious DFE and past demographic changes using an ABC framework, by including various summary statistics capturing aspects of the SFS, linkage disequilibrium, and divergence, compared between coding and flanking noncoding sequences. Ancestral population sizes and the frequency of the most deleterious classes of the DFE are estimated with relatively low accuracy, whereas the current population sizes and the neutral mutation class are estimated with high accuracy. In addition, we demonstrated that, if synonymous sites are indeed experiencing substantial purifying selection, existing programs such as DFE-α will overestimate recent growth and underestimate the proportion of mildly deleterious mutations. Importantly, the approach proposed here performs equally well regardless of whether synonymous sites are neutral or selected. However, our approach continues to assume the neutrality of flanking noncoding regions, though putatively conserved sites were masked; the impact of this masking on inference was thoroughly assessed via simulation.
Because we make no assumptions about which sites in the functional region of interest are neutral, it is in principle possible to estimate the DFE for any functional element using this methodology. The results further suggest that the accurate coestimation of these parameters is possible using only functional regions. Such an approach may be extremely useful in genomes for which it is difficult to characterize putatively neutral sites, as well as for compact genomes in which noncoding regions may be limited. However, we have only tested relatively simple demographic models, and future studies evaluating our ability to jointly estimate more complex population histories would be of value.
This approach can, in principle, be applied to any organism and functional class of interest, although power analyses suggest the utility of prior knowledge of the boundaries of functional regions as well as mutation and recombination rates. Here, we have provided an illustrative example with D. melanogaster. The results suggest that the Zambian population of this species has been largely stable in size, and that exonic regions have a large proportion of mildly deleterious mutations. Although this result might seem surprising, the DFE inferred by the current method provides the distribution of selective effects over all sites, including synonymous sites and sites in UTRs. Hence, in comparing the DFE estimated in the current study with previous estimates of the neutral class of mutations, it appears unnecessary to invoke widespread selection on synonymous sites in D. melanogaster. This result is consistent with most previous studies (Akashi 1995; Jackson et al. 2017), and our estimate of the strength of purifying selection acting on synonymous sites in the Zambian population is in line with earlier estimates for African populations (Zeng and Charlesworth 2010a; Jackson et al. 2017).
In addition to the proposed inference framework, we have derived an analytical expression for the reduction in variation caused by BGS at neutral sites outside functional regions for the case of a discrete DFE, making it feasible to obtain analytical predictions for any chosen DFE. Not only does a discrete DFE provide flexibility in inference, it may also be a more realistic representation of the true DFE (Kousathanas and Keightley 2013; Bank et al. 2014b). Although gamma distributions represent a reasonably good fit to the DFE inferred from genome-wide studies (Eyre-Walker and Keightley 2007), the DFE will be misinferred if the true distribution is multimodal (Kousathanas and Keightley 2013), as has been widely observed [e.g., in yeast (Bank et al. 2014a), viruses (Sanjuán 2010), and Escherichia coli (Jacquier et al. 2013)]. In addition, the best-fitting parameterized continuous distribution appears to be extremely specific to the particular data set being tested, and most alternative distributions fit the data nearly as well as the best-fitting distribution (Huber et al. 2017; Kim et al. 2017). The discrete DFE proposed here thus reduces the number of necessary assumptions, and has been shown to perform well in the plausible scenario in which common assumptions are indeed violated (e.g., if the true DFE is not gamma-distributed). Both the analytical results under demographic equilibrium and simulations under demographic nonequilibrium show that the number of selected sites, and the specific shape of the DFE (for instance, the frequency of mildly and moderately deleterious mutations), both decrease linked neutral variation around functional regions more than previously appreciated, and skew the SFS even when there is no reduction in diversity. Such variation in exon lengths and DFE shapes across a genome can increase the variance of statistics in linked neutral regions, which could contribute to false positives when detecting positive selection using outlier approaches.
There are at least three important caveats, which will be the subject of future study. The first concerns the estimates of ancestral and current effective population sizes. As the effective population size varies across the genome in a fashion correlated with local recombination rates (Becher et al. 2020), the estimates provided here ought to be viewed as a mean across the loci in question. While we have improved upon the common assumption of a singular genome-wide value by directly modeling each locus-specific recombination rate when performing inferences, the general importance of this effect in demographic modeling remains in need of further study. The second caveat concerns biases in inference using ABC-based methods under model violations, such as a misspecification of the mutation or recombination rate. As the method is sensitive to such violations, it will be best applied to organisms in which these parameters have been experimentally measured. Moreover, we have not included other types of mutations such as insertions/deletions, gene duplications, and transposable element insertions, all of which will increase the deleterious mutation rate and thus the effects of BGS (Comeron 2014).
The third caveat concerns inferences about selection. This study represents a proof-of-concept in demonstrating that the simultaneous inference of demography and the DFE is feasible, thereby avoiding common assumptions underlying a step-wise inference approach. While this interplay of genetic drift and purifying selection is in fact alone sufficient to fit all features of the data (consistent with previous claims: Comeron 2014, 2017; Harris et al. 2018; Jensen et al. 2019), this is not the same as claiming that positive selection is not also occurring. As our simulation results demonstrate, the presence of rare, weakly beneficial mutations is consistent with the data, though the inclusion of these parameters does not result in an improved fit. The question is less about presence/absence, than it is about statistical identifiability. Conversely, the addition of a strongly beneficial mutational class was found to be inconsistent with observed data. To investigate this further, future work will evaluate the ability to coestimate a beneficial class of fitness effects within this framework. It should also be noted that the example chosen to highlight our approach focuses on only a subset of genes in the D. melanogaster genome, and the observed DFE in this class is not necessarily universal across all coding regions in the population under consideration. In fact, the means of the scaled selection coefficients of deleterious mutations have been shown to be negatively correlated with divergence at nonsynonymous sites (Campos et al. 2017). However, importantly, a general inference approach that incorporates these two dominant processes will be a valuable tool in future genomic scans, and our appropriate null is anticipated to greatly reduce the notoriously high false-positive rates associated with the identification of positively selected loci. The ability of our approach to reject the hypothesis of frequent hard selective sweeps involving strongly selected beneficial mutations is encouraging, as hypothesis rejection is often scientifically more robust than model fitting.
Acknowledgments
We thank Rebecca Harris for discussions related to this project. This research was conducted using resources provided by Research Computing at Arizona State University (http://www.researchcomputing.asu.edu) and the Open Science Grid, which is supported by the National Science Foundation and the U.S. Department of Energy’s Office of Science. We especially thank Lauren Michael and Christina Koch from the Open Science Grid for their efforts to provide technical assistance. This work was funded by National Institutes of Health grant R01 GM-135899 to J.D.J. The authors declare no conflicts of interests.
Appendix
Derivation of the Analytical Expression for the Reduction in Diversity due to BGS
We assume a discrete DFE with four bins, such that t is uniformly distributed within each bin. The expectation of E(t) within each bin is proportional to its integral with respect to t. The overall expectation of E(t) is given by:
| (A1) |
where the ti correspond to the boundaries of the discrete bins. These are such that 0 ≤ 2Nes ≤ 1, 1 ≤ 2Nes ≤ 10, 10 ≤ 2Nes ≤ 100, and 100 ≤ 2Nes ≤ 10,000, respectively. In our case, these correspond to t0 = 0, t1 = 0.00005, t2 = 0.0005, t3 = 0.005, and t4 = 0.5. While this mirrors the DFE considered here, a similar procedure can be used for any set of bins for a given DFE.
To determine E(t) from the first line of Equation 2 of the main text, we write a = g + rcy and b = g + rc (y + l), and note that:
| (A2) |
The second integral on the right-hand side of this equation can be evaluated by substituting u = a + t (1 – a) for t, so that dt = du/(1 – a). This gives:
| (A3) |
With this change in variable, the normalizing factor for the probability density function is now (u1 – u0)−1= (1 – a)−1 (t1 – t0)−1. The contribution of this component to the expectation of E(t) yields Equation 3a of the main text.
A similar expression can be written for the integral of – t/[(1 – t)[b + t (1– b)] in the first line of Equation 2. When adding this to the integral of t/[(1 – t)[a + t (1– a)], the integrals involving 1/(1 – t) cancel out, so that this term simply contributes Equation 3b to the expectation of E(t).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11879025.
Communicating editor: R. Nielsen
Literature Cited
- Akashi H., 1995. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics 139: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437: 1149–1152. 10.1038/nature04107 [DOI] [PubMed] [Google Scholar]
- Arguello J. R., Laurent S., and Clark A. G., 2019. Demographic history of the human commensal Drosophila melanogaster. Genome Biol. Evol. 11: 844–854. 10.1093/gbe/evz022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf Z. J., Tilk S., Park J., Siegal M. L., and Petrov D. A., 2017. Deep sequencing of natural and experimental populations of Drosophila melanogaster reveals biases in the spectrum of new mutations. Genome Res. 27: 1988–2000. 10.1101/gr.219956.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank C., Hietpas R. T., Wong A., Bolon D. N., and Jensen J. D., 2014a A Bayesian MCMC approach to assess the complete distribution of fitness effects of new mutations: uncovering the potential for adaptive walks in challenging environments. Genetics 196: 841–852. 10.1534/genetics.113.156190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank C., Ewing G. B., Ferrer-Admettla A., Foll M., and Jensen J. D., 2014b Thinking too positive? Revisiting current methods of population genetic selection inference. Trends Genet. 30: 540–546. 10.1016/j.tig.2014.09.010 [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., Zhang W., and Balding D. J., 2002. Approximate Bayesian computation in population genetics. Genetics 162: 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher H., Jackson B. C., and Charlesworth B., 2020. Patterns of genetic variability in genomic regions with low rates of recombination. Curr. Biol. 30: 94–100.e3. 10.1016/j.cub.2019.10.047 [DOI] [PubMed] [Google Scholar]
- Campos J. L., and Charlesworth B., 2019. The effects on neutral variability of recurrent selective sweeps and background selection. Genetics 212: 287–303. 10.1534/genetics.119.301951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J. L., Zhao L., and Charlesworth B., 2017. Estimating the parameters of background selection and selective sweeps in Drosophila in the presence of gene conversion. Proc. Natl. Acad. Sci. USA 114: E4762–E4771. 10.1073/pnas.1619434114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casillas S., Barbadilla A., and Bergman C. M., 2007. Purifying selection maintains highly conserved noncoding sequences in Drosophila. Mol. Biol. Evol. 24: 2222–2234. 10.1093/molbev/msm150 [DOI] [PubMed] [Google Scholar]
- Chamary J., and Hurst L. D., 2005. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 6: R75 10.1186/gb-2005-6-9-r75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1996. Background selection and patterns of genetic diversity in Drosophila melanogaster. Genet. Res. 68: 131–149. 10.1017/S0016672300034029 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 2012. The effects of deleterious mutations on evolution at linked sites. Genetics 190: 5–22. 10.1534/genetics.111.134288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 2013. Background selection 20 years on. The Wilhelmine E. Key 2012 invitational lecture. J. Hered. 104: 161–171. 10.1093/jhered/ess136 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., and Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Campos J. L., and Jackson B. C., 2018. Faster-X evolution: theory and evidence from Drosophila. Mol. Ecol. 27: 3753–3771. 10.1111/mec.14534 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., and Morgan M. T., 1995. The pattern of neutral molecular variation under the background selection model. Genetics 141: 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. Y., and Aquadro C. F., 2016. Recent and long term selection across synonymous sites in Drosophila ananassae. J. Mol. Evol. 83: 50–60. 10.1007/s00239-016-9753-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., 2014. Background selection as baseline for nucleotide variation across the Drosophila genome. PLoS Genet. 10: e1004434 10.1371/journal.pgen.1004434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., 2017. Background selection as null hypothesis in population genomics: insights and challenges from Drosophila studies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20160471 10.1098/rstb.2016.0471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., and Kreitman M., 2002. Population, evolutionary and genomic consequences of interference selection. Genetics 161: 389–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., and Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905 10.1371/journal.pgen.1002905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisci J. L., Poh Y.-P., Bean A., Simkin A., and Jensen J. D., 2012. Recent progress in polymorphism-based population genetic inference. J. Hered. 103: 287–296. 10.1093/jhered/esr128 [DOI] [PubMed] [Google Scholar]
- Crisci J. L., Poh Y.-P., Mahajan S., and Jensen J. D., 2013. The impact of equilibrium assumptions on tests of selection. Front. Genet. 4: 235 10.3389/fgene.2013.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., 2008. Mid-century controversies in population genetics. Annu. Rev. Genet. 42: 1–16. 10.1146/annurev.genet.42.110807.091612 [DOI] [PubMed] [Google Scholar]
- Csilléry K., François O., and Blum M. G. B., 2012. abc: an R package for approximate Bayesian computation (ABC). Methods Ecol. Evol. 3: 475–479. 10.1111/j.2041-210X.2011.00179.x [DOI] [PubMed] [Google Scholar]
- Cutter A. D., and Payseur B. A., 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat. Rev. Genet. 14: 262–274. 10.1038/nrg3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen P., Živković D., Hutter S., Stephan W., and Laurent S., 2013. Demographic inference reveals African and European admixture in the North American Drosophila melanogaster population. Genetics 193: 291–301. 10.1534/genetics.112.145912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyashiv E., Sattath S., Hu T. T., Strutsovsky A., McVicker G. et al. , 2016. A genomic map of the effects of linked selection in Drosophila. PLoS Genet. 12: e1006130 10.1371/journal.pgen.1006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing G. B., and Jensen J. D., 2016. The consequences of not accounting for background selection in demographic inference. Mol. Ecol. 25: 135–141. 10.1111/mec.13390 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., and Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. 10.1038/nrg2146 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., and Keightley P. D., 2009. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol. Biol. Evol. 26: 2097–2108. 10.1093/molbev/msp119 [DOI] [PubMed] [Google Scholar]
- Fay J. C., and Wu C. I., 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiston-Lavier A.-S., Singh N. D., Lipatov M., and Petrov D. A., 2010. Drosophila melanogaster recombination rate calculator. Gene 463: 18–20. 10.1016/j.gene.2010.04.015 [DOI] [PubMed] [Google Scholar]
- Fu Y.-X., 1995. Statistical properties of segregating sites. Theor. Popul. Biol. 48: 172–197. 10.1006/tpbi.1995.1025 [DOI] [PubMed] [Google Scholar]
- Good B. H., Walczak A. M., Neher R. A., and Desai M. M., 2014. Genetic diversity in the interference selection limit. PLoS Genet. 10: e1004222 10.1371/journal.pgen.1004222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller B. C., and Messer P. W., 2019. SLiM 3: forward genetic simulations beyond the Wright–Fisher model. Mol. Biol. Evol. 36: 632–637. 10.1093/molbev/msy228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan D. L., and Keightley P. D., 2006. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 16: 875–884. 10.1101/gr.5022906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. B., Sackman A., and Jensen J. D., 2018. On the unfounded enthusiasm for soft selective sweeps II: examining recent evidence from humans, flies, and viruses. PLoS Genet. 14: e1007859 10.1371/journal.pgen.1007859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart C. J., Chadeau-Hyam M., Clark T. G., Lampariello R., Whittaker J. C. et al. , 2007. Sequence-level population simulations over large genomic regions. Genetics 177: 1725–1731. 10.1534/genetics.106.069088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Carlson J. W., Kennedy C., Acevedo D., Evans-Holm M. et al. , 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316: 1625–1628. 10.1126/science.1139816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber C. D., Kim B. Y., Marsden C. D., and Lohmueller K. E., 2017. Determining the factors driving selective effects of new nonsynonymous mutations. Proc. Natl. Acad. Sci. USA 114: 4465–4470. 10.1073/pnas.1619508114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., and Kaplan N. L., 1995. Deleterious background selection with recombination. Genetics 141: 1605–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson B. C., Campos J. L., Haddrill P. R., Charlesworth B., and Zeng K., 2017. Variation in the intensity of selection on codon bias over time causes contrasting patterns of base composition evolution in Drosophila. Genome Biol. Evol. 9: 102–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier H., Birgy A., Nagard H. L., Mechulam Y., Schmitt E. et al. , 2013. Capturing the mutational landscape of the beta-lactamase TEM-1. Proc. Natl. Acad. Sci. USA 110: 13067–13072. 10.1073/pnas.1215206110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J. D., Kim Y., DuMont V. B., Aquadro C. F., and Bustamante C. D., 2005. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics 170: 1401–1410. 10.1534/genetics.104.038224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J. D., Payseur B. A., Stephan W., Aquadro C. F., Lynch M. et al. , 2019. The importance of the neutral theory in 1968 and 50 years on: a response to Kern and Hahn 2018. Evolution 73: 111–114. 10.1111/evo.13650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce P., and Marjoram P., 2008. Approximately sufficient statistics and bayesian computation. Stat. Appl. Genet. Mol. Biol. 7: Article26 10.2202/1544-6115.1389 [DOI] [PubMed] [Google Scholar]
- Kaiser V. B., and Charlesworth B., 2009. The effects of deleterious mutations on evolution in non-recombining genomes. Trends Genet. 25: 9–12. 10.1016/j.tig.2008.10.009 [DOI] [PubMed] [Google Scholar]
- Kapopoulou A., Pfeifer S. P., Jensen J. D., and Laurent S., 2018. The demographic history of African Drosophila melanogaster. Genome Biol. Evol. 10: 2338–2342. 10.1093/gbe/evy185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Hinrichs A. S., Furey T. S., Roskin K. M., Sugnet C. W. et al. , 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32: D493–D496. 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., and Eyre-Walker A., 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177: 2251–2261. 10.1534/genetics.107.080663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S. et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201. 10.1101/gr.091231.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Ness R. W., Halligan D. L., and Haddrill P. R., 2014. Estimation of the spontaneous mutation rate per nucleotide site in a Drosophila melanogaster full-sib family. Genetics 196: 313–320. 10.1534/genetics.113.158758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern A. D., and Hahn M. W., 2018. The neutral theory in light of natural selection. Mol. Biol. Evol. 35: 1366–1371. 10.1093/molbev/msy092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. Y., Huber C. D., and Lohmueller K. E., 2017. Inference of the distribution of selection coefficients for new nonsynonymous mutations using large samples. Genetics 206: 345–361. 10.1534/genetics.116.197145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., and Wiehe T., 2009. Simulation of DNA sequence evolution under models of recent directional selection. Brief. Bioinform. 10: 84–96. 10.1093/bib/bbn048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1968. Evolutionary rate at the molecular level. Nature 217: 624–626. 10.1038/217624a0 [DOI] [PubMed] [Google Scholar]
- Kimura M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge: 10.1017/CBO9780511623486 [DOI] [Google Scholar]
- King J. L., and Jukes T. H., 1969. Non-Darwinian evolution. Science 164: 788–798. 10.1126/science.164.3881.788 [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., and Begun D. J., 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260. 10.1534/genetics.110.123059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A., and Keightley P. D., 2013. A comparison of models to infer the distribution of fitness effects of new mutations. Genetics 193: 1197–1208. 10.1534/genetics.112.148023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack J. B., Cardeno C. M., Crepeau M. W., Taylor W., Corbett-Detig R. B. et al. , 2015. The Drosophila Genome Nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics 199: 1229–1241. 10.1534/genetics.115.174664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J. D., and Pool J. E., 2018. Impacts of recurrent hitchhiking on divergence and demographic inference in Drosophila. Genome Biol. Evol. 10: 1882–1891. 10.1093/gbe/evy142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., Stevens K., Cardeno C., Lee Y. C. G., Schrider D. R. et al. , 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192: 533–598. 10.1534/genetics.112.142018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent S. J. Y., Werzner A., Excoffier L., and Stephan W., 2011. Approximate bayesian analysis of Drosophila melanogaster polymorphism data reveals a recent colonization of Southeast Asia. Mol. Biol. Evol. 28: 2041–2051. 10.1093/molbev/msr031 [DOI] [PubMed] [Google Scholar]
- Lawrie D. S., Messer P. W., Hershberg R., and Petrov D. A., 2013. Strong purifying selection at synonymous sites in D. melanogaster. PLoS Genet. 9: e1003527 10.1371/journal.pgen.1003527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Stephan W., 2006. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2: e166 10.1371/journal.pgen.0020166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. J., Satta Y., and Takahata N., 1999. Paleo-demography of the Drosophila melanogaster subgroup: application of the maximum likelihood method. Genes Genet. Syst. 74: 117–127. 10.1266/ggs.74.117 [DOI] [PubMed] [Google Scholar]
- Lynch M., 2007. The Origins of Genome Architecture. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Matsumoto T., John A., Baeza-Centurion P., Li B., and Akashi H., 2016. Codon usage selection can bias estimation of the fraction of adaptive amino acid fixations. Mol. Biol. Evol. 33: 1580–1589. 10.1093/molbev/msw027 [DOI] [PubMed] [Google Scholar]
- Messer P. W., and Petrov D. A., 2013. Frequent adaptation and the McDonald–Kreitman test. Proc. Natl. Acad. Sci. USA 110: 8615–8620. 10.1073/pnas.1220835110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaisen L. E., and Desai M. M., 2013. Distortions in genealogies due to purifying selection and recombination. Genetics 195: 221–230. 10.1534/genetics.113.152983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Charlesworth B., and Charlesworth D., 1996. The effect of recombination on background selection. Genet. Res. 67: 159–174. 10.1017/S0016672300033619 [DOI] [PubMed] [Google Scholar]
- O’Fallon B. D., Seger J., and Adler F. R., 2010. A continuous-state coalescent and the impact of weak selection on the structure of gene genealogies. Mol. Biol. Evol. 27: 1162–1172. 10.1093/molbev/msq006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., 1973. Slightly deleterious mutant substitutions in evolution. Nature 246: 96–98. 10.1038/246096a0 [DOI] [PubMed] [Google Scholar]
- Pordes R., Petravick D., Kramer B., Olson D., Livny M. et al. , 2007. The open science grid. J. Phys. Conf. Ser. 78: 012057 10.1088/1742-6596/78/1/012057 [DOI] [Google Scholar]
- Provine W. B., 2001. The Origins of Theoretical Population Genetics: With a New Afterword. University of Chicago Press, Chicago, IL. [Google Scholar]
- Ragsdale A. P., and Gutenkunst R. N., 2017. Inferring demographic history using two-locus statistics. Genetics 206: 1037–1048. 10.1534/genetics.117.201251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R., 2010. Mutational fitness effects in RNA and single-stranded DNA viruses: common patterns revealed by site-directed mutagenesis studies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1975–1982. 10.1098/rstb.2010.0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A., Charlesworth B., Eyre-Walker A., and Keightley P. D., 2011. A method for inferring the rate of occurrence and fitness effects of advantageous mutations. Genetics 189: 1427–1437. 10.1534/genetics.111.131730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Houle D., Lynch M., and Hahn M. W., 2013. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics 194: 937–954. 10.1534/genetics.113.151670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Shanku A. G., and Kern A. D., 2016. Effects of linked selective sweeps on demographic inference and model selection. Genetics 204: 1207–1223. 10.1534/genetics.116.190223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfiligoi, I., D. C. Bradley, B. Holzman, P. Mhashilkar, S. Padhi et al., 2009 The pilot way to grid resources using glideinWMS, pp. 428–432 in Proceedings of the 2009 WRI World Congress on Computer Science and Information Engineering, Los Angeles, CA. Institute of Electrical and Electronic Engineers, Piscataway, NJ 10.1109/CSIE.2009.950 10.1109/CSIE.2009.950 [DOI] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M. et al. , 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15: 1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., 2010. Genetic hitchhiking vs. background selection: the controversy and its implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1245–1253. 10.1098/rstb.2009.0278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhorst J., Kamm J. A., and Song Y. S., 2017. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 49: 303–309. 10.1038/ng.3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K., 2003. Libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics 19: 2325–2327. 10.1093/bioinformatics/btg316 [DOI] [PubMed] [Google Scholar]
- Thornton K. R., and Jensen J. D., 2007. Controlling the false-positive rate in multilocus genome scans for selection. Genetics 175: 737–750. 10.1534/genetics.106.064642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R., Stetter M. G., Hernandez R., and Ross-Ibarra J., 2020. The temporal dynamics of background selection in non-equilibrium populations. Genetics. 10.1534/genetics.119.302892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uricchio L. H., and Hernandez R. D., 2014. Robust forward simulations of recurrent hitchhiking. Genetics 197: 221–236. 10.1534/genetics.113.156935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh B., and Lynch M., 2018. Evolution and Selection of Quantitative Traits. Oxford University Press, Oxford: 10.1093/oso/9780198830870.001.0001 [DOI] [Google Scholar]
- Wegmann D., Leuenberger C., and Excoffier L., 2009. Efficient approximate Bayesian computation coupled with Markov chain Monte Carlo without likelihood. Genetics 182: 1207–1218. 10.1534/genetics.109.102509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., 2013. A coalescent model of background selection with recombination, demography and variation in selection coefficients. Heredity 110: 363–371. 10.1038/hdy.2012.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., and Charlesworth B., 2010a Studying patterns of recent evolution at synonymous sites and intronic sites in Drosophila melanogaster. J. Mol. Evol. 70: 116–128. 10.1007/s00239-009-9314-6 [DOI] [PubMed] [Google Scholar]
- Zeng K., and Charlesworth B., 2010b The effects of demography and linkage on the estimation of selection and mutation parameters. Genetics 186: 1411–1424. 10.1534/genetics.110.122150 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following data are publicly available on https://github.com/paruljohri/BGS_Demography_DFE: (1) aligned sequences of the single-exon genes and their corresponding intergenic regions used in this study, including derived alleles and fixed substitutions; (2) scripts to calculate statistics from simulations and from empirical data, as well as the code used to perform simulations; (3) values of all calculated statistics obtained for all parameter combinations; and (4) a Mathematica notebook as well as a Python script for analytically calculating the reduction in linked neutral diversity caused by BGS caused by a functional element. Supplemental tables and figures have been uploaded to figshare. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11879025.