Disease arises from the interaction of two genomes, host and pathogen genomes. To highlight these genomic interactions, Soltis et al. performed genome-wide association (GWA) based on genetic variation in the pathogen...
Keywords: Host–pathogen interaction, pathosystem, Arabidopsis, Botrytis cinerea, RNA-sequencing, genome-wide association, dual transcriptome
Abstract
In plant–pathogen relations, disease symptoms arise from the interaction of the host and pathogen genomes. Host–pathogen functional gene interactions are well described, whereas little is known about how the pathogen genetic variation modulates both organisms’ transcriptomes. To model and generate hypotheses on a generalist pathogen control of gene expression regulation, we used the Arabidopsis thaliana–Botrytis cinerea pathosystem and the genetic diversity of a collection of 96 B. cinerea isolates. We performed expression-based genome-wide association (eGWA) for each of 23,947 measurable transcripts in Arabidopsis (host), and 9267 measurable transcripts in B. cinerea (pathogen). Unlike other eGWA studies, we detected a relative absence of locally acting expression quantitative trait loci (cis-eQTL), partly caused by structural variants and allelic heterogeneity hindering their identification. This study identified several distantly acting trans-eQTL linked to eQTL hotspots dispersed across Botrytis genome that altered only Botrytis transcripts, only Arabidopsis transcripts, or transcripts from both species. Gene membership in the trans-eQTL hotspots suggests links between gene expression regulation and both known and novel virulence mechanisms in this pathosystem. Genes annotated to these hotspots provide potential targets for blocking manipulation of the host response by this ubiquitous generalist necrotrophic pathogen.
Introduction
Infectious disease results from an interaction between host and pathogen driven by the genetics of both organisms. The mechanisms of plant–pathogen interactions are often divided into qualitative, in which a few genetic variants of large effect shape binary disease outcomes, or quantitative, in which a spectrum of outcomes arise from the interaction of polygenic variation in the host and pathogen. The past decades have witnessed the unraveling of the molecular basis of large-effect loci on both the host and the pathogen sides that control qualitative interactions (Giraldo and Valent 2013; Marone et al. 2013; Meng and Zhang 2013; Cui et al. 2015; Lo Presti et al. 2015). In the qualitative model, alternative alleles at these genes create sweeping differences in the transcriptome and phenotypic responses to infection in the host and pathogen via differential recognition events surrounding their proteins. However, plant–pathogen interactions cover a full range of genetic architectures, from few genes of large effect to many genes of small effect (Poland et al. 2009; Kou and Wang 2010; Lannou 2012). In contrast to qualitative systems, quantitative plant–pathogen interactions exhibit a lack of virulence/resistance genes that explain large proportions of the variance in the disease outcome in the population (Poland et al. 2009; Kou and Wang 2010; St. Clair 2010; Roux et al. 2014). These interactions are highly polygenic, with genetic variation influencing diverse molecular mechanisms, extending beyond host–pathogen perception and large-effect arms-race loci (Glazebrook 2005; Nomura et al. 2005; Goss and Bergelson 2006; Rowe and Kliebenstein 2008; Barrett et al. 2009; Corwin et al. 2016a; Bartoli and Roux 2017; Wu et al. 2017; Atwell et al. 2018 preprint; Fordyce et al. 2018; Soltis et al. 2019). It is, however, unclear how these polygenic molecular interactions alter higher-order phenotypes such as virulence or the transcriptome of both species. There is conflicting evidence on the balance of such pathosystem. Some studies and traits indicate that genetic variation in the pathogen dominates the interaction (Bartha et al. 2017; Wang et al. 2018a). Other studies find a balanced contribution of plant and pathogen genetics (Corwin et al. 2016a; Soltis et al. 2019). Thus, there is a need to develop genomic approaches to understand how polygenic variations affect the genomic response of both organisms.
Polygenic variation in the pathogen should influence numerous genes that shift the pathogen’s transcriptome and cause differential expression of various virulence mechanisms. This virulence mechanism variation will affect the host and alter the host’s resistance-associated transcriptome. Thus, by measuring the transcriptome in both the pathogen and the host, it should be possible to map how genetic variation in the pathogen is conveyed through the pathogen’s transcriptome, and concurrently, how the host’s transcriptome responds. Recent work has shown that it is possible to measure the pathogen’s transcriptome in planta in the Arabidopsis thaliana–Pseudomonas syringae pathosystem, leading to new hypotheses about virulence (Nobori et al. 2018). In the A. thaliana–Botrytis cinerea pathosystem, the genetic interactions are dominated by complex small-effect loci that display a high degree of interaction between the host and pathogen (Denby et al. 2004; Finkers et al. 2007; Finkers et al. 2008; Rowe and Kliebenstein 2008; Anuradha et al. 2011; Fu et al. 2017; Fordyce et al. 2018). In this pathosystem, a cotranscriptome study with simultaneous analysis of the host and pathogen’s transcripts was recently done through single-sample RNA-sequencing (Zhang et al. 2017; Zhang et al. 2019). This cotranscriptome approach allowed the description of key virulence networks in the pathogen and resistance responses within the host (Zhang et al. 2017; Zhang et al. 2019). Further, this study revealed a single network of transcripts from pathogen and host species. However, these studies did not assess the genetic architecture behind these cotranscriptome interactions.
Genome-wide association mapping (GWA) that identifies expression quantitative trait loci (eQTL; SNPs correlated with variations in transcript expression) can reveal the genetic architecture behind these cotranscriptome interactions. Previous eQTL studies revealed that the SNPs that cause differential transcript accumulation can be parsed into cis or trans effects. Locally acting (cis) eQTL indicate regulatory variation within or near the expressed gene itself. trans-eQTL reveal SNPs that are acting at a distance to affect regulatory processes influencing the expression of the transcript. A trans-eQTL that affects many transcripts is classified as a hotspot. Such trans-acting hotspot SNPs may influence regulatory processes that, in turn, influence numerous transcripts.
eQTL analysis has been utilized to study host–pathogen interactions, albeit with a focus either on host or pathogen. Frequently, these studies focus on the host’s response, such as mapping how host loci control host gene expression over time, using either traditional QTL mapping or GWA analysis (Chen et al. 2010; Hsu and Smith 2012; Zou et al. 2012; Allen et al. 2016; Christie et al. 2017). Additional studies have begun to invert this scheme by looking at how genetic variation in the pathogen influences the host transcriptome to identify pathogen loci modulating host expression levels, and thus to identify candidate loci for interspecific signals (Saeij et al. 2007; Wu et al. 2015; Guo et al. 2017). These studies attest to the potential to identify pathogen loci that influence host gene expression. However, previous studies have thus far addressed pathogen populations with limited genetic variation, and thus identify the few polymorphic loci between strains with strongest effects on transcriptomic variation (Wu et al. 2015; Guo et al. 2017).
The genomes of both the host and the pathogen harbor extensive genetic diversity that has been successfully used for genetic mapping to identify loci controlling virulence in combination with transcriptomics and genomics (Denby et al. 2004; Rowe and Kliebenstein 2008; Dalmais et al. 2011; Schumacher et al. 2012; Zhang et al. 2017; Atwell et al. 2018 preprint; Soltis et al. 2019). Expanding the scope of these studies, we performed a cotranscriptome analysis using a single wild-type A. thaliana Columbia-0 (Col-0) host accession, and B. cinerea pathogen transcriptomes were measured using a diverse B. cinerea population (Zhang et al. 2017; Zhang et al. 2019). We conducted a GWA analysis of both host and pathogen transcriptomes to identify loci in the B. cinerea genome that may affect the transcriptomes of either or both organisms (Zhang et al. 2017; Zhang et al. 2019). The loci tagged by these SNPs have an explicit directionality of effect, as genetic causality must arise within the pathogen and then extend to the host. Our analysis found mostly small-effect polymorphisms dispersed throughout the B. cinerea genome, with several trans-eQTL hotspots. These hotspot loci are associated with specific host or pathogen transcript coexpression modules and variation in lesion size. There was no identifiable overlap in the hotspots that influenced the host’s or the pathogen’s transcriptome, suggesting a surprisingly independent basis of transcriptional regulation of host and pathogen by the B. cinerea genome. Most of the hotspot-loci-tagged genes have no previous association to plant–pathogen virulence interactions. This generates a set of B. cinerea loci that have regulatory potential in controlling the A. thaliana and B. cinerea interaction via modulation of gene expression to influence the lesions outcome.
Materials and Methods
Previously published transcriptomics
For this study, we utilized the exact previously published transcriptome data involving 1164 RNA-sequencing libraries available under the NCBI accession number SRP149815 (Zhang et al. 2017; Zhang et al. 2019). A brief recitation of the materials and methods used to generate the transcriptome data are provided below. This large number of libraries measure the interaction of three A. thaliana genotypes (Col-0, coi1-1, and npr1-1) with a collection of 96 B. cinerea genotypes that were isolated as single spores from natural infections of fruit and vegetable tissues collected in California and internationally (Zhang et al. 2017; Atwell et al. 2018 preprint; Zhang et al. 2019; Caseys et al. 2020 preprint). We focused the analysis for this eGWA study on the A. thaliana wild-type accession Col-0. The transcriptome data were generated using fourfold replication of the full-randomized complete block experimental design across two independent experiments for all interactions (i.e., two fully independent randomized biological replicates in each of two experiments). Leaves from Col-0 were harvested 5 weeks after sowing, and individually inoculated in a detached leaf assay with spores of each of 96 B. cinerea isolates (Zhang et al. 2017; Zhang et al. 2019). Whole leaves were sampled at 16 hr postinoculation, before visible lesion formation, and flash-frozen for RNA isolation while lesion area was measured at 72 hr postinoculation. RNA-sequencing libraries were prepared as described in Kumar et al. 2012; Zhang et al. 2017; and Zhang et al. 2019. Reads were aligned to both the A. thaliana TAIR10.25 and the B. cinerea B05.10 ASM83294v1 complementary DNA reference genomes, and gene counts were pooled, summed across gene models, and normalized (Langmead et al. 2009; Li et al. 2009; Van Kan et al. 2017).
GWA of gene expression profiles
As phenotype for the GWA, we used the z-scaled model-adjusted least square means of normalized gene counts obtained from a negative binomial generalized linear model for both the A. thaliana and B. cinerea transcriptomes (Zhang et al. 2017; Zhang et al. 2019). GWA was implemented using a univariate linear mixed model in a genome-wide efficient mixed model association (GEMMA; Zhou and Stephens 2012). GWA was performed using PLINK binary ped format files with a standardized relatedness matrix calculated in GEMMA. The relatedness matrix accounts for population structure among B. cinerea isolates. Individual SNP significances were extracted as P-values from the score test. We used haploid binary SNP calls with <10% missing values and the default 1% minor allele frequency (MAF) was increased to 20%. The use of MAF > 0.20 should help to limit the false positive error given the population size (Tabangin et al. 2009). The mapping was based on 96 isolates with a total of 237,878 SNPs mapped to the B. cinerea B05.10 genome (Atwell et al. 2018 preprint). We ran GEMMA once per phenotype, across 9267 B. cinerea gene expression profiles and 23,947 A. thaliana gene expression profiles.
Significance threshold by GWA of permuted transcripts
Given the large number of traits used for GWA (33,214 traits), full permutation tests assessing the significance of all associations is unfeasible. To query for potential patterns of association that may exist randomly and fix global thresholds of likely nonrandom association of SNPs with transcript variation, we performed a comparative GWA analysis of randomized assignment of each transcriptional profile across the 96-isolate collection. This analysis included five independent permutations of 9267 randomized B. cinerea transcripts and 23,947 randomized A. thaliana transcripts. Using these permutations, we compiled the top 5% of random P-values from each gene across all the permutations and utilized the median of these values as conservative genome-wide permutation thresholds, 1.96e−5 for the B. cinerea transcriptome and 2.9e−5 for the A. thaliana transcriptome. In B. cinerea, this threshold identified an average of 4756 SNPs associated with the expression of 461 transcripts (range 3843–5584 SNPs and 410–499 transcripts) across the five random permutations. In A. thaliana, this threshold identified an average of 16,446 SNPs associated with the expression of 1201 transcripts (range 13,040–22,359 SNPs and 1129–1260 transcripts) across the five random permutations. Permutation approaches are often more effective than a priori P-value thresholds to determine significance thresholds of GWA studies with large number of phenotypes (Evans and Cardon 2006).
eQTL hotspot identification, significance, and annotation
For the hotspot analysis we utilized the top single SNP per transcript using either the B. cinerea or A. thaliana transcripts alone to query for hotspots. This provides 9267 SNP associations for the 9267 B. cinerea transcripts and 23,947 SNP associations for the 23,947 A. thaliana transcripts. An eQTL hotspot was defined as a SNP having the most significant effect (top SNP) on the abundance of multiple transcripts. For an initial approach to deciding upon significant hotpots in each data set, we randomized the position of these positions across the full 237,878 SNPs and repeated this 1000 times. This analysis gave a maximum hotspot across all permutations as three transcripts for B. cinerea and six transcripts for 23,947 A. thaliana. This permutation approach, however, does not account for potential effects of coexpression on hotspot detection. Thus, we also estimated thresholds by using the five random permutations GWA analysis to query and measure the number of transcripts randomly associated with each SNP. From each permutation, we identified the position of the top SNP for each B. cinerea or A. thaliana transcript and found the top hotspot in each permutation. These maximum eQTL hotspot sizes across all the permutations were 11 B. cinerea and 80 A. thaliana transcripts. Thus, we conservatively defined significant hotspots as SNPs associated with >20 transcripts in B. cinerea and 150 transcripts in A. thaliana.
To document the putative functionality of eQTL hotspots, we annotated SNPs to genes using the identity of the nearest gene within a 2-kb window. This window was chosen because the average linkage disequilibrium decay in the B. cinerea genome is <1 kb (Atwell et al. 2018 preprint). This loss of allelic association over short distances allows tagging SNPs to particular genes with some confidence. While the majority of annotated genes had single SNP hotspots, three genes had two independent SNP hotspots. Two genes on chromosome 12 denoting A. thaliana gene expression hotspots appear closely linked, but are separated by ∼80 kb on the B. cinerea genome.
Annotation of gene ontology and modules
We annotated functions to B. cinerea genes using the BotPortal resource (http://dx.doi.org/10.15454/IHYJCX) and looked for patterns indicating signal peptides for secretion using the SignalP-5.0 Server (http://www.cbs.dtu.dk/services/SignalP-5.0/). We looked for functional overrepresentation among the genes targeted by each A. thaliana eQTL hotspot using the PANTHER overrepresentation test implemented by plant gene ontology (GO) term enrichment from TAIR (Lamesch et al. 2011; Cadic et al. 2013). Overenrichment of genes found in the previous B. cinerea and A. thaliana transcriptome modules and the eQTL hotspots were tested by the hypergeometric test (Subramanian et al. 2005; Zhang et al. 2017; Zhang et al. 2019).
Biosynthetic-pathway-focused cis-eQTL analysis
All of the SNPs within the three clustered biosynthetic pathways were described previously (Zhang et al. 2019). Within each pathway, the SNPs were used to cluster the isolates by hierarchical clustering using the R package pvclust. Distances were estimated using the unweighted pair group method with arithmetic mean distance, with correlation distance and 1000 bootstrap replications (Suzuki and Shimodaira 2015). The 95% bootstrap probability values are show in green, while the approximately unbiased P-values are reported in red. Clustering is drawn according to those edges with strong support under both estimations.
Data availability statement
Strains are available upon request. Supplemental Material, File S1 contains information on functional annotation of the B. cinerea genes targeted by the B. cinerea trans-eQTL hotspots. File S2 contains information on functional annotation of the A. thaliana genes targeted by the B. cinerea trans-eQTL hotspots. File S3 contains GO summary analysis of the A. thaliana genes targeted within each B. cinerea trans-eQTL hotspot. File S4 contains summary information on the top SNP hits from GWA of each B. cinerea expression trait (transcript). SNP data and R scripts are available in Dryad (https://doi.org/10.25338/B83P56). Transcriptome data involving 1164 RNA-sequencing libraries are available under the NCBI accession number SRP149815 (Zhang et al. 2017; Zhang et al. 2019). Gene annotations for B. cinerea genes are available on the BotPortal online platform (http://dx.doi.org/10.15454/IHYJCX). Supplemental material available at figshare: https://doi.org/10.25386/genetics.8069480.
Results
eQTL indicate polygenic transcriptome modulation
To understand how natural genetic variation in the pathogen influences both the host and pathogen transcriptomes, we performed expression GWA across all genes expressed in each species within the A. thaliana–B. cinerea pathosystem. This analysis incorporated the z-scaled expression profiles of 9267 B. cinerea genes and 23,947 Col-0 A. thaliana genes, mapped as individual traits across 96 diverse B. cinerea isolates. For each trait, we used a GEMMA univariate linear mixed model that estimates the significance of effects of each SNP on the focal trait as a P-value after accounting for potential effects of population structure within the B. cinerea isolates (Zhou and Stephens 2012). For GEMMA, we used a previously described B. cinerea genome-wide SNP data set of 237,878 SNPs with a conservative minimum MAF of 0.20 (Atwell et al. 2018 preprint; Soltis et al. 2019).
Using permutation estimated thresholds (see Materials and Methods), we identified 461,723 B. cinerea SNPs associated with transcriptional variation in 1616 B. cinerea genes and 978,693 SNPs associated with 5213 A. thaliana genes (Figure 1). In comparison to the random permutations, this suggests a false discovery rate for SNPs of ∼1% using the 1.96e-5 threshold for B. cinerea (4756 permutation/461,723 empirical) and 1.6% using the 2.9e−5 threshold for A. thaliana (16,446 permutation/978,693 empirical).
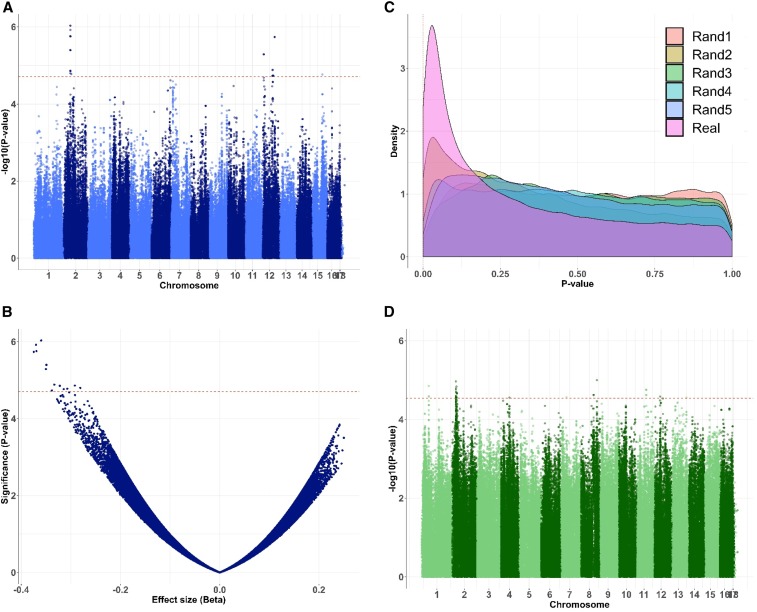
Figure 1.
Examples of expression-based GWA mapping in B. cinerea genome. The two transcripts plotted correspond to the mean of significant SNPs with 50 and 41 significant SNPs, respectively. (A–C) GWA results for the mapping of the expression of Bcin01g09490, a methyltransferase. Red dashed lines indicate significance at P = 1.96e−05. (A) Manhattan plot of log-scaled P-values of SNPs associations across the 18 chromosomes of B. cinerea. (B) Relation between significance and effect size as estimated by GEMMA. (C) P-value density in the real expression data vs. the five randomizations. (D) A cross-species eQTL Manhattan plot for the host’s expression of AT1G28370 (an ethylene response factor) across the 18 chromosomes of B. cinerea. The red dashed line indicates significance at P = 2.9e−5.
The range of significant SNPs per transcript is wide with 0–16,814 SNPs (median = 0, mean = 49.8) for B. cinerea and 0–24,622 SNPs (median = 0, mean = 40.8) for A. thaliana. The observation of transcript variation associated with a large number of significant SNPs has been seen in other eQTL analyses (Figure S1) (West et al. 2007; Wang et al. 2018b). Given the wide range of significant SNPs per transcript and how it could shape the analysis, we restricted the analysis of the top SNP per transcript (SNP with the lowest P-value) to survey genomic patterns. We further tested how conservative our threshold was by comparing the permuted top to the empirical top P-value across all SNPs per transcript. This showed that the P-value of empirical top SNP per trait for 69% of the B. cinerea transcripts, and for 58% of the A. thaliana transcripts, is lower than permuted transcripts top SNP across all five permutations. By comparing the pattern of trans-hotspots across the genome using all the top SNP per transcripts and only top significant SNP per transcripts showed a similar image of the trans-hotspots, with SNP below the threshold reinforcing the trans-hotspots (Figure 2, A and B).
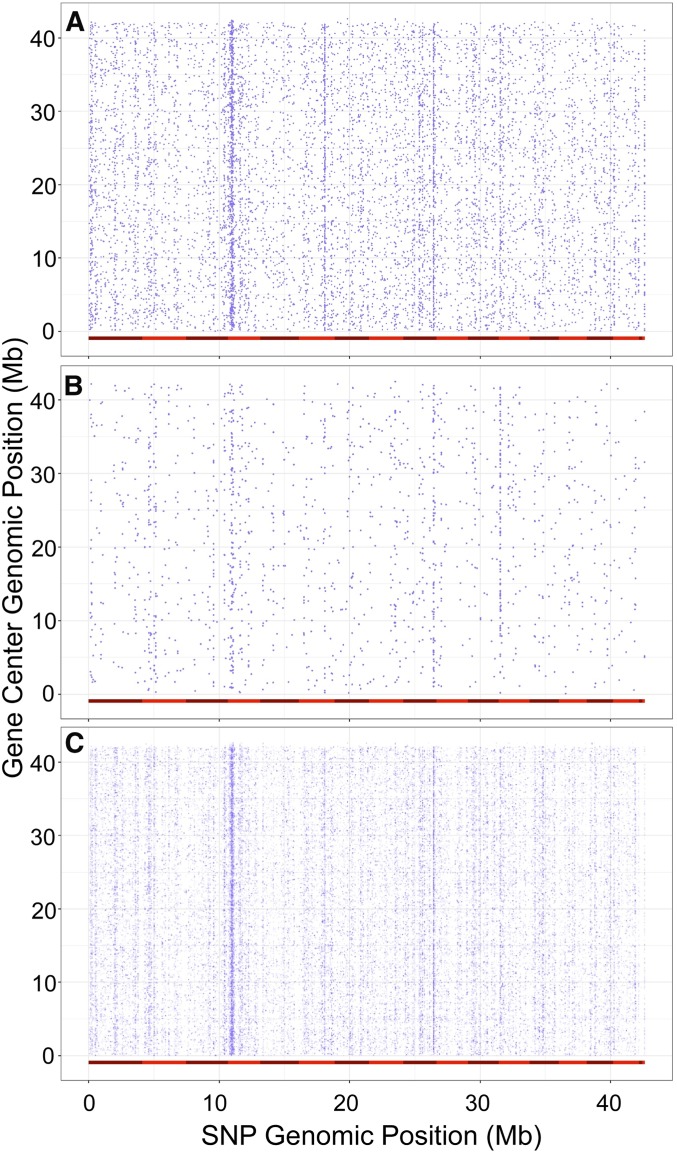
Figure 2.
Comparing B. cinerea gene center to position of top associated SNP. For all 9267 transcripts mapped to the B. cinerea genome, we retained only the SNPs with highest probability (lowest P-value) of significant effect on expression for each transcript. (A) The single top SNP per all transcripts. (B) The single top SNP for only those transcripts that have at least one SNP crossing the 1.96e−5 significance threshold. (C) The top 10 SNPs per all transcripts. The 18 chromosomes of B. cinerea are delimited by differentially shaded red bars along the x-axis, while positions indicate individual SNPs. The y-axis depicts the same chromosome alignment, but positions are the center of each mapped transcript. Vertical striping of SNP positions indicates genomic locations of putative trans-eQTL hotspots.
Therefore, we focused on all the top significant SNP per transcript for the remaining analysis. This restrictive analysis on SNPs that are most likely to be associated with transcript variation accounts for the fact that nearly every transcript has a highly significant heritability ascribed to the B. cinerea genome (Zhang et al. 2017; Zhang et al. 2019). To account for the potentially low resolution in genomic signal encompassed by the single top SNP, we also assessed any general pattern using the top 10 SNPs.
Absence of observed transcriptome cis-effect
A hallmark of within-species eQTL mapping studies using either GWA or structured mapping populations in a wide range of species is the occurrence of cis-diagonals (Brem et al. 2002; Schadt et al. 2003; Monks et al. 2004; Keurentjes et al. 2007; West et al. 2007; Zou et al. 2012). This occurs because polymorphisms proximal to the gene, cis-eQTL or cis-SNPs, frequently have large effects on the transcripts accumulation measured. To test if our analysis identifies a similar cis-diagonal in the B. cinerea transcriptome, we plotted the position of the transcript’s genomic position against the top SNP for all the B. cinerea transcripts. As cis-diagonals are generally enriched in SNPs with the largest effects on transcript abundance, the top GWA SNP should identify this diagonal, assuming there are no intervening technical or biological issues. However, there was no evidence supporting a cis-diagonal (Figure 2). This pattern held whether we examined the top SNP per transcript (Figure 2A) or the top 10 SNPs per transcript (Figure 2C). In contrast, there is evidence for trans-eQTL hotspots: distant loci that modulate expression variation across many of the pathogen genes (Figure 2).
Because any significance threshold may complicate the identification of a cis-signal, we converted all the P-values assigned to SNPs per transcript into their percentile rank. This conversion was performed by sorting all P-values in ascending order, assigning ranks values to each P-value and converting these values to percentage of the total number of SNPs. We then identified all the SNPs within each B. cinerea transcript or within 1 kb of the start or end of the transcript, and extracted the percentile ranks for these SNPs. The number of local SNPs ranged from 1 to 352 SNPs, with an average of 16 local SNPs per transcripts. The distribution of ranks across all the local SNPs was flat and similar to a random sampling of P-values, further indicating no general cis-enrichment within this data set (Figure S2). We next limited this percentile rank analysis to the local top SNP for each transcript because each cis-region may have only one or a few true associations masked by noncausal SNPs. As a random comparison, we simulated a data set of percentile rank by random draw of 16 SNPs out of all SNPs within defined cis-defined boundaries and repeated this 9267 times (the number of B. cinerea transcripts). For each simulated transcript, we extracted the top SNP and repeated the whole simulation five times. This showed that that cis-SNPs are not enriched for higher significance than the genome, and might even be less frequent than expected by random chance (Figure S2). As such, we do not detect evidence for overrepresentation of cis-effect loci.
Allelic heterogeneity and structural variation potentially masking cis-effects
Haplotype heterogeneity or allele frequency may complicate the accurate identification of cis-polymorphisms and hide a cis-diagonal (Chan et al. 2010; Rivas et al. 2011; Visscher et al. 2017). To assess these possibilities, we used three clustered biosynthetic pathways: the botcinic acid biosynthetic pathway (13 genes, 55.8 kb, chromosome 1), botrydial biosynthetic pathway (7 genes, 26 kb, chromosome 12), and a putative cyclic peptide pathway (10 genes, 46.5 kb, chromosome 1) (Deighton et al. 2001; Colmenares et al. 2002; Porquier et al. 2016; Zhang et al. 2019). These pathways have known presence/absence polymorphisms segregating in the studied B. cinerea population and were expected to hold cis-patterns, yet none were detected by our GWA analysis (Siewers et al. 2005; Pinedo et al. 2008). Additionally, genes of the botcinic acid and botrydial biosynthetic pathways are variable across the Botrytis genus (Valero-Jiménez et al. 2019). Critically, the transcription profiles of the genes within these pathways are highly correlated across isolates, suggesting pathway-specific control of expression variation (Zhang et al. 2019). This known presence/absence variation suggested that the absence of cis-SNPs in these loci may represent false negatives. To test if these pathways have undetected cis-SNPs, we extracted all the SNPs within each biosynthetic cluster and investigated haplotype diversity across the 96 B. cinerea isolates.
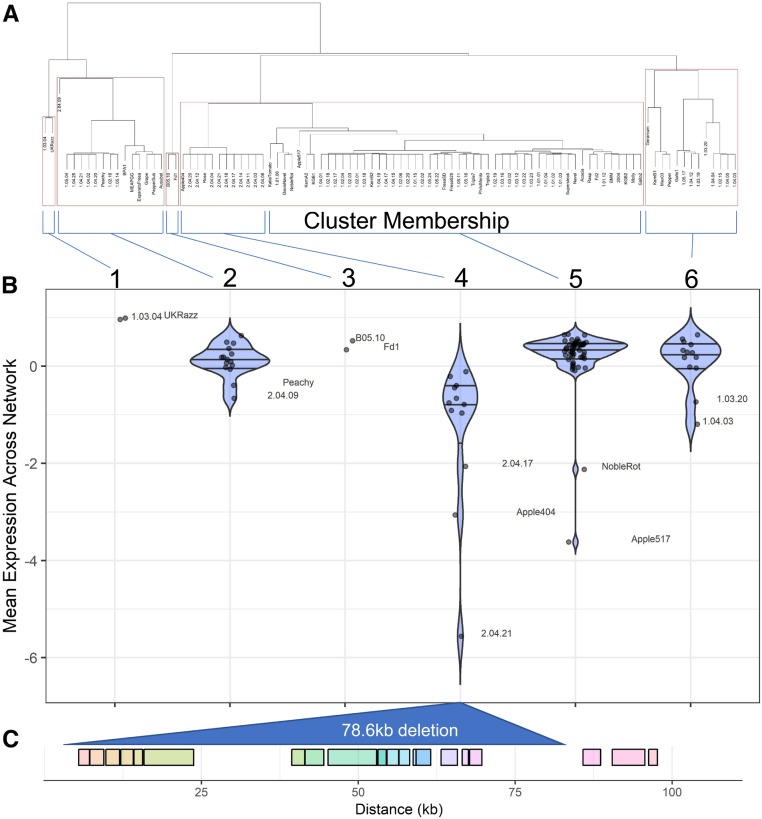
We first investigated the botcinic acid cluster that contains multiple distinct haplotypes (Figure 3A). To test for haplotype-specific effects on transcript expression within a pathway, we z-scaled each individual transcript and averaged across all the genes in the pathway to get a pathway expression value (Kliebenstein et al. 2006). The pathway expression identified a group of 12 isolates (cluster 4 in Figure 3B) with distinctly lower expression level than the other groups. Investigating the short-read sequences and SNPs showed that these 12 isolates share a 78.6-kb deletion that removes the entire botcinic acid biosynthetic cluster, from Bcboa1 to Bcboa17 (Figure 3). After accounting for this major deletion, we found no remaining significant effect of group membership in the remaining groups on the expression profile (F(1,74) = 0.36; P = 0.55). However, within each of these groups, independent isolates have low pathway expression, suggesting loss-of-expression polymorphisms (e.g., isolates Noble Rot and Apple517) (Figure 3B). While each of these isolates contain independent smaller deletions, the origins of the loss of expression of the genes of botcinic acid pathway remain unknown. Functional analysis of the botcinic acid biosynthetic cluster has thus far identified one transcription factor (Bcboa13) that controls the expression of the cluster (Porquier et al. 2019). However, none of the SNPs within or near Bcboa13 were significantly associated with variation in expression of the botcinic acid biosynthesis genes. These patterns may suggest undetected cis-effect polymorphisms in addition to the large common deletion and independent additional events.
Figure 3.
Analysis of the botcinic acid gene cluster on B. cinerea chromosome 1. (A) An SNP-based hierarchical clustering of B. cinerea isolates. The hierarchical clustering was based on unweighted pair group method with arithmetic mean distance, with 1000 bootstrap replications. Red boxes indicate clustering with >95% confidence. (B) Violin plots of botcinic acid network expression within B. cinerea groups. The mean network expression was obtained by converting expression of each gene across the isolates into its corresponding z-score and then averaging across the z-scores. Below is the gene model of the gene cluster, with each rainbow-colored box delimiting single genes (including Bcboa1 to Bcboa13 and 5 additional genes). The large 78.6-kb deletion in the isolate cluster 4 major deletion is indicated as a triangle.
We then investigated the botrydial and putative cyclic peptide biosynthetic pathways for additional evidence of undetected cis-acting genetic variation. These two pathways exhibit a lack of cis-effect SNP patterns similar to the botcinic acid pathway. Hierarchical clustering within each of these pathways by SNP variation divided the isolate population into two groups that were not associated with mean pathway expression (Figure S3 and Figure S4, both bipartite divisions were supported by >95% of bootstraps). While there was no obvious structural variation in the botrydial pathway, the cyclic peptide pathway contained small deletions within the intergenic regions that correlate with low expression. Furthermore, two isolates with partial deletions within the genes early in the pathway exhibited very low pathway expression (1.05.16 and 1.05.22) (Figure S4).
Overall, we identified cis-acting deletions in two of three of the clustered biosynthetic pathways. This highlights the potential of structural variants that often fall below the minor allele cutoffs and compromise the detection of cis-effects by GWA within B. cinerea. Testing whether insertion and deletion events account for the majority of localized control of expression variation would require both long-read sequencing to accurately identify these structural variants and computational approaches that can blend SNP and insertion/deletion information.
Detection of trans-eQTL hotspots
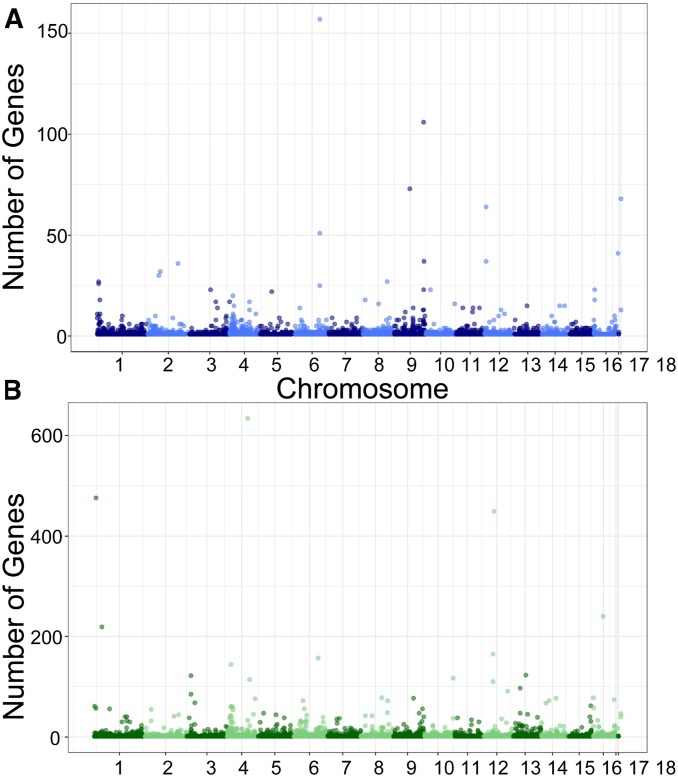
Using the top SNP per transcript, the GWA analysis identified a strong signature of trans-eQTL hotspots (Figure 2): polymorphisms that may influence the regulation of numerous genes in trans. These hotspots were detected using both the B. cinerea and A. thaliana transcriptome with solely the top SNP per transcript (Figure 4 and Figure 5). Using a permutation approach, we fixed conservative thresholds for eQTL hotspots to 20 transcripts for B. cinerea and 150 transcripts for A. thaliana. This identified 25 trans-eQTL hotspots dispersed across the B. cinerea genome that modulate either the host (12 SNPs) or pathogen (13 SNPs) transcriptomes (Figure 5, Figure 6, and Table 1). Hotspot SNPs had an average MAF of 27% (1% SE) with no correlation between the MAF and the number of transcripts associated with a particular SNP within either the Arabidopsis or Botrytis transcripts. Further, the SNPs at different hotspot were not cosegregating across the isolates.
Figure 4.
Frequency and positions of trans-eQTL hotspots in the B. cinerea genome. (A) Number of Botrytis transcripts associated with trans-eQTL SNPs aligned along the 18 chromosomes of B. cinerea (x-axis). (B) Number of Arabidopsis transcripts associated with cross-species eQTL SNPs aligned along the 18 chromosomes of B. cinerea (x-axis).
Figure 5.
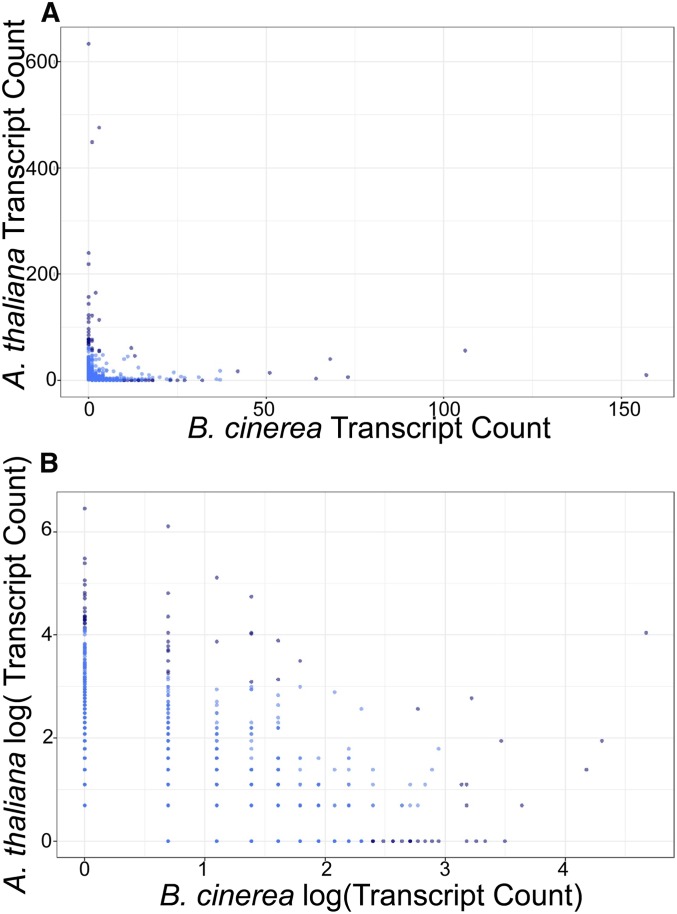
Cross-species hotspot comparison in the B. cinerea genome. For each SNP that is a top hit for one or more transcripts, the number of associated transcripts is counted, across both the B. cinerea transcriptome and the A. thaliana transcriptome.
Figure 6.
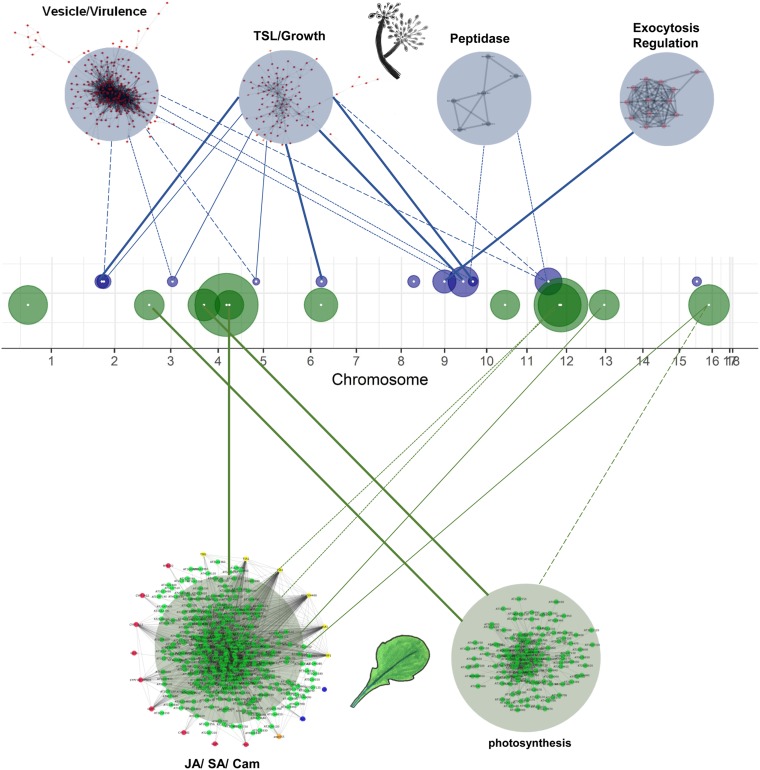
Genes associated with eQTL hotspots are in virulence and defense coexpression networks. Circles along the B. cinerea 18 chromosomes are eQTL hotspots, centered at the gene containing the eQTL, and with radius proportional to the number of transcripts associated with this hotspot. The gene center is marked with a white dot. Hotspots for B. cinerea transcripts are drawn in blue, hotspots for A. thaliana transcripts are drawn in green. The network names are based on biological functions from gene ontology analysis of network members, from Figure 4 of Zhang et al. (2019) and Figure 6 of Zhang et al. (2017). The A. thaliana networks depicted are the most inclusive of the host-dependent networks, from npr1-1. Links between hotspots and coexpression networks are drawn according to the number of genes shared between them. Variable line weight represents the percent of hotspot target genes shared with the coexpression network; 1–25% is dashed, 25–50% is dotted, 50–75% is solid, 75–100% is bold.
Table 1. Annotation of the genes identified from B. cinerea and A. thaliana eQTL hotspots.
| Hotspot gene | Hotspot SNP | Botrytis transcripts | Arabidopsis transcripts | Name | Gene function | Virulence correlation | Virulence GWA | Arabidopsis GO overrepresentation |
|---|---|---|---|---|---|---|---|---|
| Bcin01g01610 | 629157 | 0 | 219 | Glucose/ribitol dehydrogenase | NA | Amylopectin, glycogen, chlorophyll, chloroplast, photosynthesis | ||
| Bcin02g02480 | 910858 | 31 | 6 | P = 0.008 | NA | |||
| Bcin02g02850 | 1032242 | 32 | 0 | Fructosamine-3-kinase | NA | NA | ||
| Bcin03g00960 | 336467 | 1 | 122 | GTP cyclohydrolase I | NA | Carotenoid biosynthesis, chloroplast organization | ||
| Bcin03g05020 | 1695103 | 24 | 15 | NA | NA | |||
| Bcin04g00830 | 310509 | 0 | 144 | NACHT nucleoside triphosphatase | NA | Nucleic acid metabolism | ||
| Bcin04g04700 | 1633072 | 0 | 634 | Heterokaryon incompatibility | NA | Photosynthesis, light, translation | ||
| Bcin04g05160 | 1791561 | 3 | 114 | NA | Chitin response | |||
| Bcin05g02780 | 1015184 | 22 | 2 | NA | NA | |||
| Bcin06g05680 | 1952544 | 0 | 157 | WLM | P < 0.001 | NA | ||
| Bcin06g05790 | 1988179 | 25 | 0 | NA | NA | |||
| Bcin08g05340 | 2014228 | 27 | 0 | F-box domain | NA | NA | ||
| Bcin09g03390 | 1235841 | 73 | 6 | 6-Phosphogluconate dehydrogenase | NA | NA | ||
| Bcin09g06590 | 2330312 | 106 | 56 | SNF2-related; Helicase, superfamily 1/2 | NA | Metabolism | ||
| Bcin09g06590 | 2334368 | 23 | 0 | SNF2-related; Helicase, superfamily 1/2 | NA | NA | ||
| Bcin10g00940 | 383007 | 23 | 1 | P < 0.001 | NA | |||
| Bcin10g05900 | 2268522 | 0 | 117 | Winged helix-turn-helix transcription factor | NA | Water stress | ||
| Bcin12g00330 | 115491 | 37 | 1 | Bcpat1 | Topoisomerase-II-associated protein PAT1 | P < 0.001 | NA | |
| Bcin12g00330 | 115511 | 64 | 3 | Bcpat1 | Topoisomerase-II-associated protein PAT1 | P < 0.001 | NA | |
| Bcin12g02130 | 758420 | 0 | 110 | NA | Response to stimulus | |||
| Bcin12g02130 | 760499 | 2 | 265 | NA | Jasmonic acid, fungal response, microbe defense, biotic stress | |||
| Bcin12g02340 | 842369 | 1 | 449 | Bccds1 | Phosphatidate cytidylyltransferase | P < 0.001 | Primary metabolism, amino acid biosynthesis, salt stress, biotic response | |
| Bcin13g02930 | 1026752 | 0 | 123 | SET domain | NA | Transport | ||
| Bcin16g00010 | 76596 | 23 | 2 | SsuA/THI5-like | NA | Yes | NA | |
| Bcin16g01950 | 787512 | 0 | 240 | Bccwh41 | Glycoside hydrolase, family 63 | NA | Photosynthesis |
Each row identifies a significant eQTL hotspot SNP associated with transcript variations in B. cinerea or in A. thaliana. Gene functions are from BotPortal, Arabidopsis GO overrepresentation are from PANTHER. The table lists if the gene’s transcript is correlated with virulence or if the SNP was associated with virulence via GWA (Atwell et al. 2018 preprint; Zhang et al. 2019).
The use of a cotranscriptome approach should theoretically identify trans-eQTL hotspot affecting B. cinerea transcripts that create an eQTL hotspot affecting the host’s transcriptome. However, the GWA analysis detected no cross-species-connected eQTL hotspots across the two transcriptomes (Figure 5). This result was consistent using either the single top SNP or the top 10 SNPs per transcript (Figure S5). The absence of cross-species overlap in eQTL hotspots suggests that the pathogen’s influence on the host’s transcriptome is not limited to major interactions between trans-eQTL hotspots, but can involve molecularly constrained changes in the pathogen that are magnified in the host’s response. One possible scenario is a SNP that alters the expression of a single effector gene or mechanism in the pathogen that does not affect the pathogen but instead affects the host. For example, altering expression of the botcinic acid biosynthetic cluster would alter the accumulation of that phytotoxic metabolite and cause large responses in the host. Similarly, mutations that do not affect transcript abundance, like missense polymorphisms that alter protein function, could equally lead to a lack of overlap in hotspots. Finally, it is possible that the high level of genetic variation in B. cinerea may decrease our power to detect the cross-species trans-eQTL hotspots. Deeper analysis into the transcriptome and downstream responses could elucidate how restricted responses in the pathogen transcriptome translate to sweeping responses in the host. Future studies using these eQTL hotspots as a priori candidates for control of transcript variation in both host and pathogen may need to increase power to detect more modulation overlap across the two transcriptomes.
eQTL hotspot modules
To better understand the trans-eQTL hotspots, we examined the genes influenced by each hotspot. We first collected the GO annotations within each species to test if the hotspot transcripts were enriched for some specific functionality. The B. cinerea GO annotations showed a preponderance of enzyme, signal peptides for secretion, and transcription factor annotations, but no specific molecular insights arose, largely because the majority of genes had no annotation (Table 1, Table S1, File S1, and File S4). In contrast, GO analysis of the A. thaliana transcripts showed that three of the hotspots have an overrepresentation of photosynthesis-related functions within their targeted genes (Table 1, File S2, and File S3). Downregulation of photosynthesis transcripts is a hallmark of plant immune processes (Bilgin et al. 2010; Jiang et al. 2017). Two other A. thaliana hotspots primarily affect genes associated with abiotic stress responses. Only two of the A. thaliana hotspots influence expected plant defense loci, including chitin response and microbe defenses. This suggests that the B. cinerea genes underlying these hotspots specifically influence defined networks within the host.
In previous work, we had defined key transcript modules within both the host and pathogen transcriptomes that connected to virulence (Zhang et al. 2017; Zhang et al. 2019). Thus we tested for overlap between the trans-eQTL hotspot-defined modules and previously defined transcript modules using a hypergeometric enrichment test (Zhang et al. 2017; Atwell et al. 2018 preprint; Zhang et al. 2019). Nine of the 11 B. cinerea eQTL hotspots were enriched for transcripts present in one or more of four major B. cinerea coexpression modules identified when grown on A. thaliana (Figure 6). An additional six B. cinerea modules did not share any transcript membership with detected eQTL hotspots. Similarly, nine of the A. thaliana eQTL hotspots were enriched for transcripts from two of the major A. thaliana modules when infected with B. cinerea (Figure 6). These two A. thaliana modules contain genes that function in jasmonate/salicylic acid signaling processes and camalexin biosynthesis (network I), or photosynthesis (network IV). Interestingly, these links are not limited to a single hotspot, but have strong connections across several different eQTL hotspots, suggesting that these A. thaliana modules are influenced by the pathogens polygenic architecture (Figure 6).
eQTL hotspot candidate genes
To generate working hypotheses on the possible causal basis of the eQTL hotspots, we investigated the candidate genes underlying the associated SNPs. The 12 B. cinerea hotspots that influence A. thaliana transcripts are located within 11 genes, including four enzymes and two genes associated with isolate compatibility (Table 1). The 13 B. cinerea hotspots that influence B. cinerea expression profiles were associated with 11 genes, including four enzymes (Table 1). However, only one of these 22 genes was previously linked to virulence functions in B. cinerea or other fungi. Bccwh41 (Bcin16g01950), a glycoside hydrolase whose homolog shows increased expression in virulent strains of Ustilago maydis on A. thaliana (Martínez-Soto et al. 2013). To test if any of these 22 eQTL hotspot candidate genes may influence virulence in B. cinerea, we used the existing coexpression and virulence data to compare the level of expression of these 22 genes to existing virulence measurements (Zhang et al. 2017; Atwell et al. 2018 preprint; Zhang et al. 2019). Transcript accumulation for three B. cinerea hotspot genes and two of the A. thaliana hotspot genes are strongly positively correlated to lesion size variation, while none are negatively correlated with lesion size (Table 1) (Zhang et al. 2019). Further, we utilized a previous GWA analysis of virulence of these same isolates on A. thaliana to test for any overlap. This showed that one of the B. cinerea hotspot genes (Bcin16g00010, SsuA/THI5-like) is a top GWA hit, controlling lesion size across host genotypes and association methods (Table 1) (Atwell et al. 2018 preprint). Together, this suggests that these genes are likely candidates for controlling both the host and pathogen transcriptomes.
Discussion
Dispersed interactions across host and pathogen genomes
Using cotranscriptome GWA, we identified 25 trans-eQTL hotspots dispersed across the B. cinerea genome that modulate either the host or pathogen transcriptomes. This contrasts with previous cross-species eQTL studies, which identified one or only a few cross-species eQTL hotspots (Wu et al. 2015; Guo et al. 2017). Further, most of the B. cinerea genetic variation detected in our study is distant from the affected B. cinerea transcripts, i.e., located in trans. These trans-eQTL hotspots influence expression variation for five major B. cinerea modules containing genes dispersed across the genome (Zhang et al. 2019). In particular, the eQTL hotspots influenced the expression of specific B. cinerea coexpression networks (vesicle/virulence, translation/growth, exocytosis regulation, and peptidase). These candidate polymorphisms are spread throughout the genome and the detected eQTL hotspots are not in regions of the genome with significantly elevated genetic variation. Further, the genetic targets of these eQTL are dispersed across the plant and pathogen genomes (Zhang et al. 2019). As such, B. cinerea does not fit the model of what might be expected in filamentous fungi showing multiple-speed genome evolution due to varying selective pressures influencing the genome. In such specialist fungi with closer coevolution to their host species than B. cinerea, diverse fungal virulence effectors are clustered in regions of the genome containing enhanced rates of mutation and polymorphism, while the rest of the genome shows slower evolutionary rates (Dong et al. 2015). If this scenario were true for the Botrytis genome, it would predict clustering of the GWA hits to a few locations rather than distribution across the genome, as was found. This is consistent with previous findings of high genome-wide diversity in B. cinerea, and virulence and host specificity mapping to large swaths of the pathogen genome, including 16 of 18 chromosomes (Atwell et al. 2018 preprint; Caseys et al. 2020 preprint). Similar eQTL analyses in the multispeed genome filamentous fungi are required to test whether eQTL in pathogens with a multispeed genome truly cluster within the highly polymorphic regions. Together, these findings provide evidence for polygenic trans-regulation of gene expression in B. cinerea interactions that then coalesces around specific transcriptional modules to influence virulence.
Polygenic modules and pleiotropy in cross-species eQTL
Previous pathogen eQTL studies identified qualitative patterns whereby each host expression profile was explained by a single major-effect pathogen locus (Guo et al. 2017) or each pathogen eQTL connected to a specific host network (Wu et al. 2015). In contrast, cotranscriptome GWA with B. cinerea identified a more complex picture, with numerous trans-eQTL hotspots altering multiple transcriptome modules in either the host or the pathogen. This suggests that the polygenic architecture of the pathogen may partially function by influencing these defined modules rather than working as thousands of individual genes, each independently targeting the host. It remains to be ascertained if this system functions to create robustness in these connections in the face of changes to the pathogen or host genetics, or if this is an indication of existence of a discrete set of interaction mechanisms between the host and the pathogen.
Candidate causal loci encode diverse mechanisms
Querying the putative function of the candidate loci underlying the different trans-eQTL hotspots identified an array of potential molecular mechanisms. While one might assume that transcription factors are the most likely genes for containing genetic variation that would lead to trans-eQTL hotspots, we instead found enrichment for enzyme-encoding genes among these loci. Eight enzyme-encoding genes contained the SNPs for four of the B. cinerea trans-eQTL and four of the A. thaliana trans-eQTL hotspots. Interestingly, these enzymes have potential activities in sugar release from the plant cell wall, or reactions involving sugar phosphates (Table 1). In addition to enzymatic activity, four of the Botrytis trans-eQTL candidate genes encoded transcriptional regulators. Bcin10g05900, a putative winged helix transcription factor (TF), is predicted to have pathway-specific effects, while the other three are putative general transcriptional regulators, including Bcin12g00330, a putative topoisomerase-II-associated protein PAT1, and Bcin09g06590, a putative helicase (Table 1). Interestingly, the putative winged helix TF trans-eQTL hotspot alters the expression of A. thaliana genes associated with water deprivation responses. This suggests that this putative winged helix TF may influence a specific virulence factor that influences this A. thaliana network. Interestingly, while the candidate genes connect to processes that likely influence virulence, none of them have been explicitly shown to influence virulence in B. cinerea. Future work is necessary to test these loci and discover the mechanisms by which they influence virulence and the host/pathogen cotranscriptome.
Complications in detection of cis-acting loci
The vast majority of eQTL studies identify a strong signature of cis-acting loci. However, in B. cinerea, the main detected pattern was trans-eQTL, with few identified cis-eQTL. A deeper investigation suggested that this may be due to genetic factors that complicate the ability to identify cis-acting SNPs. B. cinerea has high haplotype diversity, and in three gene clusters investigated, potential rare cis-acting variants fell below the MAF cutoff for GWA, potentially leading to false-negative detection errors (Tabangin et al. 2009). Further, a number of deletions were found in this locus, which complicates the detection of a cis-eQTL signature by introducing non-SNP variation unused by current GWA algorithms. Additional cis-acting variants may also be hidden by undetected transposon variation (Porquier et al. 2019).
A full understanding of the pattern of potential cis-acting loci in B. cinerea would require a detailed investigation of structural variation by incorporating long-read sequencing in a larger population size. Additionally, the GWA algorithms would need to be written to allow for simultaneous use of both SNP and presence/absence polymorphism data; one option is to code deletions as an additional state for each genotyped variant (Wang et al. 2018a). It should be noted that these same difficulties would also create undetected trans-eQTL. This does suggest that there is likely a significant fraction of undetected cis-eQTLs within B. cinerea, caused by the high polymorphism rate within this species.
To summarize, previous work in the A. thaliana–B. cinerea pathosystem established connections between host polymorphisms and lesion growth, between gene expression and lesion size, and between transcriptomes of the host and pathogen (Corwin et al. 2016b; Zhang et al. 2017; Zhang et al. 2019). This study establishes a foundation to study how genetic variation in the pathogen can manipulate the host transcriptome to influence disease progression. In the Arabidopsis–Botrytis pathosystem, this connection had a preponderance of trans-acting polymorphisms with mainly moderate to small effects, suggesting that a polygenic architecture underlies the transcriptome variation, similar to the polygenic architecture observed for virulence. Using previously defined transcriptome modules showed that there may be a modular structure to these effects, with specific pathogen SNPs linking to specific modules in either the host or the pathogen. However, future validation work will be required to test the directionality and mechanism of this cross-talk. Similar work in other systems will help to build our functional knowledge of cross-kingdom communication between host and pathogen.
Acknowledgments
Funding for this work was provided by the National Science Foundation award IOS-1339125 to D.J.K., the United States Department of Agriculture National Institute of Food and Agriculture Hatch project number CA-D-PLS-7033-H to D.J.K., and by the Danish National Research Foundation grant DNRF99 to D.J.K.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.8069480.
Communicating editor: T. Juenger
Literature Cited
- Allen M., Carrasquillo M. M., Funk C., Heavner B. D., Zou F. et al. , 2016. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci. Data 3: 160089 10.1038/sdata.2016.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anuradha C., Gaur P. M., Pande S., Gali K. K., Ganesh M. et al. , 2011. Mapping QTL for resistance to botrytis grey mould in chickpea. Euphytica 182: 1–9. 10.1007/s10681-011-0394-1 [DOI] [Google Scholar]
- Atwell S., Corwin J., Soltis N., Zhang W., Copeland D. et al. , 2018. Resequencing and association mapping of the generalist pathogen Botrytis cinerea. bioRxiv. doi: (Preprint posted December 7, 2018). 10.1101/489799 [DOI] [Google Scholar]
- Barrett L. G., Kniskern J. M., Bodenhausen N., Zhang W., and Bergelson J., 2009. Continua of specificity and virulence in plant host–pathogen interactions: causes and consequences. New Phytol. 183: 513–529. 10.1111/j.1469-8137.2009.02927.x [DOI] [PubMed] [Google Scholar]
- Bartha I., McLaren P. J., Brumme C., Harrigan R., Telenti A. et al. , 2017. Estimating the respective contributions of human and viral genetic variation to HIV control. PLOS Comput. Biol. 13: e1005339 10.1371/journal.pcbi.1005339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli C., and Roux F., 2017. Genome-wide association studies in plant pathosystems: toward an ecological genomics approach. Front. Plant Sci. 8: 763 10.3389/fpls.2017.00763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin D. D., Zavala J. A., Zhu J., Clough S. J., Ort D. R. et al. , 2010. Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 33: 1597–1613. 10.1111/j.1365-3040.2010.02167.x [DOI] [PubMed] [Google Scholar]
- Brem R. B., Yvert G., Clinton R., and Kruglyak L., 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. 10.1126/science.1069516 [DOI] [PubMed] [Google Scholar]
- Cadic E., Coque M., Vear F., Grezes-Besset B., Pauquet J. et al. , 2013. Combined linkage and association mapping of flowering time in sunflower (Helianthus annuus L.). Theor Appl Genet. 126: 1337–1356. 10.1007/s00122-013-2056-2 [DOI] [PubMed] [Google Scholar]
- Caseys C., Shi G., Soltis N., Gwinner R., Corwin J. et al. , 2020. Quantitative interactions drive Botrytis cinerea disease outcome across the plant kingdom. bioRxiv. doi: (Preprint posted February 6, 2020). 10.1101/507491 [DOI] [Google Scholar]
- Chan E. K., Rowe H. C., Hansen B. G., and Kliebenstein D. J., 2010. The complex genetic architecture of the metabolome. PLoS Genet. 6: e1001198 10.1371/journal.pgen.1001198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hackett C. A., Niks R. E., Hedley P. E., Booth C. et al. , 2010. An eQTL analysis of partial resistance to Puccinia hordei in barley. PLoS One 5: e8598 10.1371/journal.pone.0008598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie N., Myburg A. A., Joubert F., Murray S. L., Carstens M. et al. , 2017. Systems genetics reveals a transcriptional network associated with susceptibility in the maize–grey leaf spot pathosystem. Plant J. 89: 746–763. 10.1111/tpj.13419 [DOI] [PubMed] [Google Scholar]
- Colmenares A. J., Aleu J., Duran-Patron R., Collado I. G., and Hernandez-Galan R., 2002. The putative role of botrydial and related metabolites in the infection mechanism of Botrytis cinerea. J. Chem. Ecol. 28: 997–1005. 10.1023/A:1015209817830 [DOI] [PubMed] [Google Scholar]
- Corwin J. A., Copeland D., Feusier J., Subedy A., Eshbaugh R. et al. , 2016a The quantitative basis of the Arabidopsis innate immune system to endemic pathogens depends on pathogen genetics. PLoS Genet. 12: e1005789 10.1371/journal.pgen.1005789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin J. A., Subedy A., Eshbaugh R., and Kliebenstein D. J., 2016b Expansive phenotypic landscape of Botrytis cinerea shows differential contribution of genetic diversity and plasticity. Mol. Plant Microbe Interact. 29: 287–298. 10.1094/MPMI-09-15-0196-R [DOI] [PubMed] [Google Scholar]
- Cui H., Tsuda K., and Parker J. E., 2015. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66: 487–511. 10.1146/annurev-arplant-050213-040012 [DOI] [PubMed] [Google Scholar]
- Dalmais B., Schumacher J., Moraga J., Le Pecheur P., Tudzynski B. et al. , 2011. The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Mol. Plant Pathol. 12: 564–579. 10.1111/j.1364-3703.2010.00692.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton N., Muckenschnabel I., Colmenares A. J., Collado I. G., and Williamson B., 2001. Botrydial is produced in plant tissues infected by Botrytis cinerea. Phytochemistry 57: 689–692. 10.1016/S0031-9422(01)00088-7 [DOI] [PubMed] [Google Scholar]
- Denby K. J., Kumar P., and Kliebenstein D. J., 2004. Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J. 38: 473–486. 10.1111/j.0960-7412.2004.02059.x [DOI] [PubMed] [Google Scholar]
- Dong S., Raffaele S., and Kamoun S., 2015. The two-speed genomes of filamentous pathogens: waltz with plants. Curr. Opin. Genet. Dev. 35: 57–65. 10.1016/j.gde.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Evans D. M., and Cardon L. R., 2006. Genome-wide association: a promising start to a long race. Trends Genet. 22: 350–354. 10.1016/j.tig.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Finkers R., van den Berg P., van Berloo R., ten Have A., van Heusden A. W. et al. , 2007. Three QTLs for Botrytis cinerea resistance in tomato. Theor. Appl. Genet. 114: 585–593. 10.1007/s00122-006-0458-0 [DOI] [PubMed] [Google Scholar]
- Finkers R., Bai Y. L., van den Berg P., van Berloo R., Meijer-Dekens F. et al. , 2008. Quantitative resistance to Botrytis cinerea from Solanum neorickii. Euphytica 159: 83–92. 10.1007/s10681-007-9460-0 [DOI] [Google Scholar]
- Fordyce R. F., Soltis N. E., Caseys C., Gwinner R., Corwin J. A. et al. , 2018. Digital imaging combined with genome-wide association mapping links loci to plant-pathogen interaction traits. Plant Physiol. 178: 1406–1422. 10.1104/pp.18.00851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. Q., van Silfhout A., Shahin A., Egberts R., Beers M. et al. , 2017. Genetic mapping and QTL analysis of Botrytis resistance in Gerbera hybrida. Mol. Breed. 37: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo M. C., and Valent B., 2013. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11: 800–814. 10.1038/nrmicro3119 [DOI] [PubMed] [Google Scholar]
- Glazebrook J., 2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227. 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Goss E. M., and Bergelson J., 2006. Variation in resistance and virulence in the interaction between Arabidopsis thaliana and a bacterial pathogen. Evolution 60: 1562–1573. 10.1111/j.0014-3820.2006.tb00501.x [DOI] [PubMed] [Google Scholar]
- Guo Y., Fudali S., Gimeno J., DiGennaro P., Chang S. et al. , 2017. Networks underpinning symbiosis revealed through cross-species eQTL mapping. Genetics 206: 2175–2184. 10.1534/genetics.117.202531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J., and Smith J. D., 2012. Genome wide studies of gene expression relevant to coronary artery disease. Curr. Opin. Cardiol. 27: 210–213. 10.1097/HCO.0b013e3283522198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., He F., and Zhang Z., 2017. Large-scale transcriptome analysis reveals arabidopsis metabolic pathways are frequently influenced by different pathogens. Plant Mol. Biol. 94: 453–467. 10.1007/s11103-017-0617-5 [DOI] [PubMed] [Google Scholar]
- Keurentjes J. J., Fu J., Terpstra I. R., Garcia J. M., van den Ackerveken G. et al. , 2007. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc. Natl. Acad. Sci. USA 104: 1708–1713. 10.1073/pnas.0610429104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D. J., West M. A., van Leeuwen H., Loudet O., Doerge R. W. et al. , 2006. Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics 7: 308 10.1186/1471-2105-7-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou Y., and Wang S., 2010. Broad-spectrum and durability: understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 13: 181–185. 10.1016/j.pbi.2009.12.010 [DOI] [PubMed] [Google Scholar]
- Kumar R., Ichihashi Y., Kimura S., Chitwood D. H., Headland L. R. et al. , 2012. A high-throughput method for Illumina RNA-seq library preparation. Front. Plant Sci. 3: 202 10.3389/fpls.2012.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., Berardini T. Z., Li D., Swarbreck D., Wilks C. et al. , 2011. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannou C., 2012. Variation and selection of quantitative traits in plant pathogens. Annu. Rev. Phytopathol. 50: 319–338. 10.1146/annurev-phyto-081211-173031 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L., Lanver D., Schweizer G., Tanaka S., Liang L. et al. , 2015. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66: 513–545. 10.1146/annurev-arplant-043014-114623 [DOI] [PubMed] [Google Scholar]
- Marone D., Russo M., Laidò G., De Leonardis A., and Mastrangelo A., 2013. Plant nucleotide binding site–leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int. J. Mol. Sci. 14: 7302–7326. 10.3390/ijms14047302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Soto D., Robledo-Briones A. M., Estrada-Luna A. A., and Ruiz-Herrera J., 2013. Transcriptomic analysis of Ustilago maydis infecting Arabidopsis reveals important aspects of the fungus pathogenic mechanisms. Plant Signal. Behav. 8: e25059. 10.4161/psb.25059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., and Zhang S., 2013. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51: 245–266. 10.1146/annurev-phyto-082712-102314 [DOI] [PubMed] [Google Scholar]
- Monks S., Leonardson A., Zhu H., Cundiff P., Pietrusiak P. et al. , 2004. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 75: 1094–1105. 10.1086/426461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobori T., Velásquez A. C., Wu J., Kvitko B. H., Kremer J. M. et al. , 2018. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc. Natl. Acad. Sci. USA 115: E3055–E3064 (erratum: Proc. Natl. Acad. Sci. USA 115: E11564). 10.1073/pnas.1800529115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Melotto M., and He S.-Y., 2005. Suppression of host defense in compatible plant–Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 8: 361–368. 10.1016/j.pbi.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Pinedo C., Wang C.-M., Pradier J.-M., Dalmais B., Choquer M. et al. , 2008. Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea. ACS Chem. Biol. 3: 791–801. 10.1021/cb800225v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J. A., Balint-Kurti P. J., Wisser R. J., Pratt R. C., and Nelson R. J., 2009. Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14: 21–29. 10.1016/j.tplants.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Porquier A., Morgant G., Moraga J., Dalmais B., Luyten I. et al. , 2016. The botrydial biosynthetic gene cluster of Botrytis cinerea displays a bipartite genomic structure and is positively regulated by the putative Zn(II)2Cys6 transcription factor BcBot6. Fungal Genet. Biol. 96: 33–46. 10.1016/j.fgb.2016.10.003 [DOI] [PubMed] [Google Scholar]
- Porquier A., Moraga J., Morgant G., Dalmais B., Simon A. et al. , 2019. Botcinic acid biosynthesis in Botrytis cinerea relies on a subtelomeric gene cluster surrounded by relics of transposons and is regulated by the Zn 2 Cys 6 transcription factor BcBoa13. Curr. Genet. 65: 965–980. 10.1007/s00294-019-00952-4 [DOI] [PubMed] [Google Scholar]
- Rivas M. A., Beaudoin M., Gardet A., Stevens C., Sharma Y. et al. , 2011. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 43: 1066–1073. 10.1038/ng.952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux F., Voisin D., Badet T., Balagué C., Barlet X. et al. , 2014. Resistance to phytopathogens e tutti quanti: placing plant quantitative disease resistance on the map. Mol. Plant Pathol. 15: 427–432. 10.1111/mpp.12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H. C., and Kliebenstein D. J., 2008. Complex genetics control natural variation in Arabidopsis thaliana resistance to Botrytis cinerea. Genetics 180: 2237–2250. 10.1534/genetics.108.091439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij J. P. J., Coller S., Boyle J. P., Jerome M. E., White M. W. et al. , 2007. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature 445: 324–327. 10.1038/nature05395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E. E., Monks S. A., Drake T. A., Lusis A. J., Che N. et al. , 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302. 10.1038/nature01434 [DOI] [PubMed] [Google Scholar]
- Schumacher J., Pradier J. M., Simon A., Traeger S., Moraga J. et al. , 2012. Natural variation in the VELVET gene bcvel1 affects virulence and light-dependent differentiation in Botrytis cinerea. PLoS One 7: e47840 10.1371/journal.pone.0047840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewers V., Viaud M., Jimenez-Teja D., Collado I. G., Gronover C. S. et al. , 2005. Functional analysis of the cytochrome P450 monooxygenase gene bcbot1 of Botrytis cinerea indicates that botrydial is a strain-specific virulence factor. Mol. Plant Microbe Interact. 18: 602–612. 10.1094/MPMI-18-0602 [DOI] [PubMed] [Google Scholar]
- Soltis N. E., Atwell S., Shi G., Fordyce R., Gwinner R. et al. , 2019. Restricted access interactions of tomato and Botrytis cinerea genetic diversity: parsing the contributions of host differentiation, domestication, and pathogen variation. Plant Cell 31: 502–519. 10.1105/tpc.18.00857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Clair D. A., 2010. Quantitative disease resistance and quantitative resistance loci in breeding. Annu. Rev. Phytopathol. 48: 247–268. 10.1146/annurev-phyto-080508-081904 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L. et al. , 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R., and Shimodaira H., 2015. pvclust: Hierarchical Clustering with P-Values via Multiscale Bootstrap Resampling. R package Version 2.0–0. Available at . Accessed: January 1, 2019.
- Tabangin M. E., Woo J. G., and Martin L. J., 2009. The effect of minor allele frequency on the likelihood of obtaining false positives. BMC Proc. 3: S41 10.1186/1753-6561-3-S7-S41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero-Jiménez C. A., Veloso J., Staats M., and van Kan J. A., 2019. Comparative genomics of plant pathogenic Botrytis species with distinct host specificity. BMC Genomics 20: 203 10.1186/s12864-019-5580-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kan J. A., Stassen J. H., Mosbach A., Van Der Lee T. A., Faino L. et al. , 2017. A gapless genome sequence of the fungus Botrytis cinerea. Mol. Plant Pathol. 18: 75–89. 10.1111/mpp.12384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Wray N. R., Zhang Q., Sklar P., McCarthy M. I. et al. , 2017. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101: 5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Roux F., Bartoli C., Huard-Chauveau C., Meyer C. et al. , 2018a Two-way mixed-effects methods for joint association analysis using both host and pathogen genomes. Proc. Natl. Acad. Sci. USA 115: E5440–E5449. 10.1073/pnas.1710980115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. F., Chen Q. Y., Wu Y. Y., Lemmon Z. H., Xu G. H. et al. , 2018b Genome-wide analysis of transcriptional variability in a large maize-teosinte population. Mol. Plant 11: 443–459. 10.1016/j.molp.2017.12.011 [DOI] [PubMed] [Google Scholar]
- West M. A. L., Kim K., Kliebenstein D. J., van Leeuwen H., Michelmore R. W. et al. , 2007. Global eQTL mapping reveals the complex genetic architecture of transcript level variation in Arabidopsis. Genetics 175: 1441–1450. 10.1534/genetics.106.064972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Cai B., Sun W., Huang R., Liu X. et al. , 2015. Genome-wide analysis of host-Plasmodium yoelii interactions reveals regulators of the type I interferon response. Cell Rep. 12: 661–672. 10.1016/j.celrep.2015.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. Q., Sakthikumar S., Dong C., Zhang P., Cuomo C. A. et al. , 2017. Comparative genomics integrated with association analysis identifies candidate effector genes corresponding to Lr20 in phenotype-paired Puccinia triticina isolates from Australia. Front. Plant Sci. 8: 148 10.3389/fpls.2017.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Corwin J. A., Copeland D., Feusier J., Eshbaugh R. et al. , 2017. Plastic transcriptomes stabilize immunity to pathogen diversity: the jasmonic acid and salicylic acid networks within the Arabidopsis/Botrytis pathosystem. Plant Cell 29: 2727–2752. 10.1105/tpc.17.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Corwin J. A., Copeland D., Feusier J., Eshbaugh R. et al. , 2019. Plant-necrotroph co-transcriptome networks illuminate a metabolic battlefield. eLife 8: e44279. 10.7554/eLife.44279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., and Stephens M., 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44: 821–824. 10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F., Chai H. S., Younkin C. S., Allen M., Crook J. et al. , 2012. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 8: e1002707 10.1371/journal.pgen.1002707 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. Supplemental Material, File S1 contains information on functional annotation of the B. cinerea genes targeted by the B. cinerea trans-eQTL hotspots. File S2 contains information on functional annotation of the A. thaliana genes targeted by the B. cinerea trans-eQTL hotspots. File S3 contains GO summary analysis of the A. thaliana genes targeted within each B. cinerea trans-eQTL hotspot. File S4 contains summary information on the top SNP hits from GWA of each B. cinerea expression trait (transcript). SNP data and R scripts are available in Dryad (https://doi.org/10.25338/B83P56). Transcriptome data involving 1164 RNA-sequencing libraries are available under the NCBI accession number SRP149815 (Zhang et al. 2017; Zhang et al. 2019). Gene annotations for B. cinerea genes are available on the BotPortal online platform (http://dx.doi.org/10.15454/IHYJCX). Supplemental material available at figshare: https://doi.org/10.25386/genetics.8069480.